Abstract
To assess the diagnostic accuracy of the rapid antigen test (RAT) compared with RT-PCR (reference standard) for SARS-CoV-2, we searched MEDLINE/PubMed and Web of Science for relevant records. The QUADAS-2 tool was used to assess study quality, and quantitative synthesis was conducted using a bivariate random-effects model. The meta-analysis included 135 studies (166,943 samples). The pooled sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, and diagnostic odds ratio were 0.76 (95%CI: 0.73–0.79), 1.00 (95%CI: 1.00–1.00), 276.1 (95% CI, 184.1–414.1), 0.24 (95% CI, 0.21–0.27), and 1171 (95% CI, 782–1755), respectively. Compared to other sample types, nasal samples had the best RAT sensitivity [0.79 (95%CI: 0.71–0.85)]. The sensitivities of the different RAT kits ranged from 0.41 (95%CI: 0.23–0.61) to 0.90 (95%CI: 0.70–0.97). Sensitivity was markedly better in samples with lower Ct, and RAT achieved excellent pooled sensitivity at 1.00 (95%CI: 0.70–1.00) among samples with Ct < 20. Testing within 10 days of symptom onset resulted in a high sensitivity. For ≤ 3, ≤ 7, and ≤ 10 days, the sensitivities were 0.91 (95%CI: 0.83–0.96), 0.89 (95%CI: 0.84–0.93), and 0.88 (95%CI: 0.83–0.92), respectively. RAT kits show high sensitivity and specificity in early infection, especially when the viral load is high. Moreover, using nasal samples for antigen testing, which are moderately sensitive and patient-friendly, is a reliable alternative to nasopharyngeal sampling. RAT might be effective for fighting the COVID-19 pandemic; however, it must be complemented by the careful handling of negative test results.
Keywords: Rapid antigen test, SARS-CoV-2, COVID-19, RT-PCR, Screening
1. Introduction
Coronavirus disease COVID-19 (COVID-19), an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), became a global pandemic within a short period (Lai et al., 2020; “WHO Director-General’s opening remarks at the media briefing on COVID-19 - 11 March 2020,” 2021). The rapid and precise diagnosis of COVID-19 is essential to enable prompt and accurate public health surveillance, prevention, and control (Jin et al., 2020).
Two major types of diagnostic tests for COVID-19 are currently available: a direct method to examine clinical specimens for the presence of viral particles, viral antigens, or viral nucleic acids, and a serological test to detect anti-SARS-CoV-2 antibodies (Borges et al., 2021). Reverse transcription polymerase chain reaction (RT-PCR) is currently the gold standard for detecting SARS-CoV-2 because of its ability to directly measure the genomic portion of the virus (Yüce et al., 2021). However, RT-PCR may be unsuitable in emergency settings because it may take several hours to obtain results. It also requires expensive technology and competent operators, and these may be unavailable in remote health clinics, especially in underdeveloped countries (Khandker et al., 2021). To improve this situation, the rapid antigen test (RAT) for COVID-19, which does not require specific and costly machinery, emerged as an essential alternative tool to aid the clinical diagnosis of COVID-19 (Yamayoshi et al., 2020, p. 1). It is low-cost and straightforward, with a shorter turnaround time. Thus, it can be used as a point-of-care test, allowing for the immediate isolation of infected individuals and permitting the early implementation of appropriate infection control measures, which is critical in a pandemic (Torres et al., 2021).
As numerous COVID-19 antigen tests are rapidly evolving, a growing number of independent validations have been conducted. Studies on the diagnostic accuracy of RAT vary widely in terms of quality, methodology, and results, generally showing excellent specificity but variable sensitivity. The different results may be due to differences in study design, manufacturers of RAT kits, patient selection, type of specimen, and stage of disease at the time of sample collection (Khandker et al., 2021). Therefore, the efficacy of RAT still needs to be thoroughly investigated. This systematic review and meta-analysis aimed to assess the diagnostic accuracy of RAT compared to RT-PCR methods as a reference standard.
2. Materials and methods
This systematic review and meta-analysis were conducted following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines, and the protocol of this study was registered in the PROSPERO database (CRD42022339683).
2.1. Literature search
The MEDLINE/PubMed and Web of Science databases were searched for relevant studies published up to 10 May 2022. We used a combination of free text and MeSH terms to identify relevant studies. The main search terms were: “SARS-CoV-2″, “COVID-19″, “antigen test”, “Specificity”, and “Sensitivity”. The detailed search strategies are presented in Table S1. Two researchers independently conducted a literature search to minimize potential biases.
2.2. Inclusion and exclusion criteria
Any study that satisfied the following requirements was considered eligible for inclusion in our meta-analysis: (i) use of RAT as an index test, (ii) measurement of the performance of RAT against RT-PCR as a reference standard, and (iii) availability of the sensitivity and specificity of RAT. The following were the exclusion standards: (i) duplicate original investigation, reviews, editorials, letters, comments, and meta-analysis articles, and (ii) unavailability of data (by article review or calculation) necessary for a meta-analysis.
2.3. Data extraction and quality assessment
We extracted the following data from the full texts and supplemental materials of all qualified articles: the first author's last name, the publication year, the country of residence of the study participants, and true positive (TP), false positive (FP), false negative (FN), and true negative (TN) values. The diagnostic parameters were calculated using the sensitivity and specificity values if they were unavailable. The risk of bias of each included publication was assessed using The Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) tool. (Whiting et al., 2011). Two researchers carried out the assessment process independently. A third researcher was invited to reach a settlement in case of disagreement.
2.4. Statistical analysis
The extracted data were recorded for further analysis using STATA software (Stata Corporation, College Station, TX, USA). We used a bivariate random-effects model to perform the quantitative synthesis. We calculated each parameter of individual studies by the following formulas to derive the pooled sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, and diagnostic odds ratio:
Sensitivity=TP/(TP+FN).
Specificity=TN/(TN+FP).
Positive likelihood ratio=Sensitivity/(1–Specificity).
Negative likelihood ratio= (1–Sensitivity)/Specificity.
Diagnostic odds ratio= Positive likelihood ratio/Negative likelihood ratio.
The forest plots were employed to show the overall effects. The area under the curve (AUC) was calculated using an optimal cutoff value by a summary receiver operating characteristic (SROC) curve. To access interstudy heterogeneity, bivariate boxplots, qualitative Q tests, and quantitative I 2 tests were utilized. Publication bias was evaluated by Deeks’ funnel plot. The Fagan nomogram and the likelihood ratio scattergram were used to access the diagnostic value and clinical application value, respectively. All tests were two-sided. A P value < 0.05 was considered statistically significant.
3. Results
3.1. Features of eligible studies
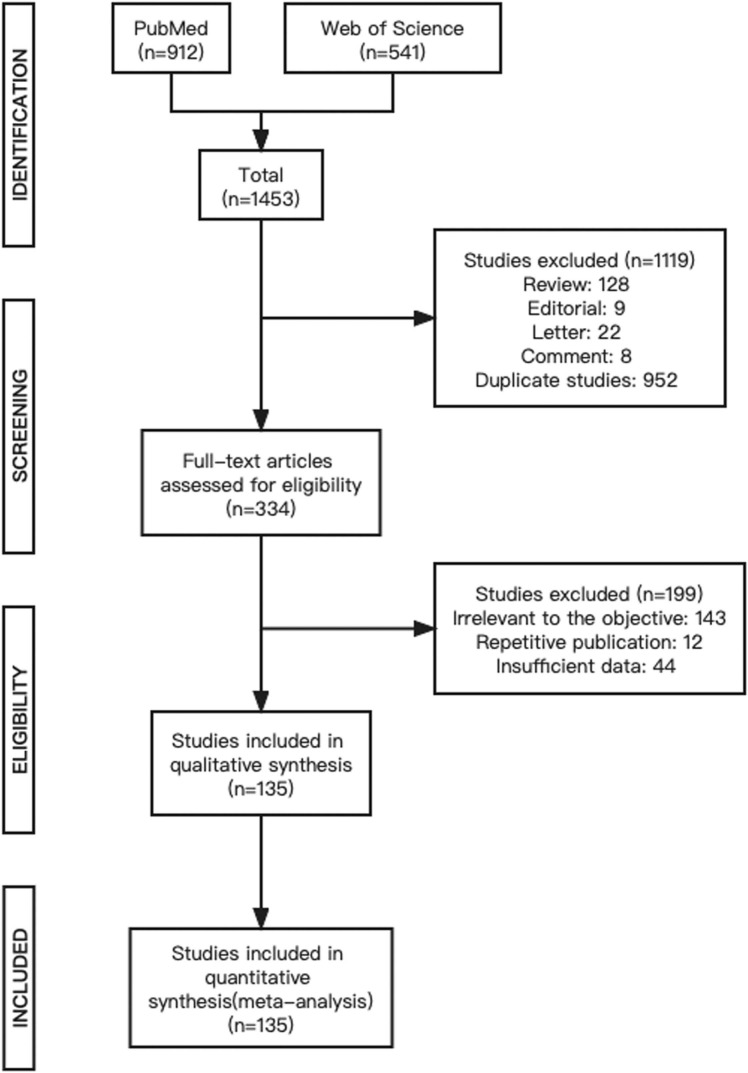
According to our search criteria, 1453 publications were initially selected. A total of 1119 articles were excluded during the initial screening process, including reviews (n = 128), editorials (n = 9), letters (n = 22), commentaries (n = 8), and duplicates (n = 952). A total of 334 articles were selected for full-text review to determine if they qualified for the meta-analysis. Of these, 199 articles including articles irrelevant to the objective of the meta-analysis (n = 143), repetitive studies (n = 12), and articles with insufficient data (n = 44) were excluded. The remaining 135 studies met the eligibility criteria and were included in the meta-analysis. The inclusion and exclusion processes are shown in Fig. 1.
Fig. 1.
PRISMA flow diagram.
The 135 studies selected for this meta-analysis included 166,943 samples. All included studies were published between 2020 and 2022 and involved 37 countries; the top three countries in terms of the number of studies were Italy (n = 18), the USA (n = 15), and Spain (n = 15). All articles were not pre-prints. Except for three manufacturer-dependent studies, the remaining were all manufacturer-independent studies. Forty different RAT kits were investigated. Different types of specimens (nasal, nasopharyngeal, oropharyngeal, throat, and saliva swabs) were collected from suspected symptomatic or asymptomatic participants ( Table 1). Ninety-three studies evaluated the diagnostic efficacy with nasopharyngeal swabs, and 26 studies assessed the performance with nasal swabs. Cycle threshold (Ct) values for positive RT-PCR were provided in 27 studies. Thirteen studies reported the time of symptom onset in RT-PCR-positive patients.
Table 1.
Characterization of included studies.
| DOI | Author | Year | Country | Rapid Antigen Test Kit | Specimen Types | TP | TN | FP | FN | SS |
|---|---|---|---|---|---|---|---|---|---|---|
| 10.3390/pathogens10060658 | Fiedler | 2021 | Germany | LIAISON® SARS-CoV-2 Ag assay (Diasorin, Saluggia, Italy), | nasopharyngeal swabs | 77 | 72 | 0 | 33 | 182 |
| 10.1016/j.jviromet.2020.114024 | Krüttgen | 2021 | Germany | Roche SARS-CoV-2 antigen assay | nasopharyngeal swabs | 53 | 72 | 3 | 22 | 150 |
| 10.1016/j.jcv.2021.104991 | Abdelhanin | 2021 | Belgium | Elecsys® SARS-CoV-2 Antigen assay | nasopharyngeal swabs | 81 | 102 | 0 | 42 | 225 |
| 10.1016/j.ijid.2021.09.069 | Sberna | 2021 | Italy | Lumipulse® G SARS-CoV-2 Ag assay | nasopharyngeal swabs | 231 | 212 | 26 | 44 | 513 |
| 10.1007/s40121–021–00510-x | Nörz | 2021 | Germany | Elecsys® SARS-CoV-2 Antigen assay | oro-nasopharyngeal swabs | 236 | 2743 | 4 | 156 | 3139 |
| 10.1002/jmv.27459 | García‐Salguero | 2021 | Spain | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 30 | 69 | 0 | 8 | 107 |
| 10.1016/j.jcv.2021.104909 | Moeren | 2021 | Netherlands | LIAISON® SARS-CoV-2 Ag assay (Diasorin, Saluggia, Italy), | oro-nasopharyngeal swabs | 54 | 174 | 0 | 20 | 248 |
| 10.1016/j.jiac.2020.11.021 | Aoki | 2021 | Japan | Lumipulse® G SARS-CoV-2 Ag assay | nasopharyngeal swabs | 22 | 516 | 8 | 2 | 548 |
| 10.1016/j.jviromet.2021.114409 | Randriamahazo | 2022 | Madagascar | STANDARD Q COVID-19 Ag Test | nasopharyngeal swabs | 94 | 106 | 0 | 56 | 256 |
| 10.1016/j.ijmmb.2021.07.003 | Kanaujia | 2021 | India | Coris bioconcept COVID-19 ag respi-strip test | nasopharyngeal swabs | 136 | 293 | 2 | 53 | 484 |
| 10.1016/j.ijid.2021.05.082 | Mayanskiy | 2021 | Russia | CoviNAg ELISA kit | nasopharyngeal swabs | 164 | 72 | 23 | 18 | 277 |
| 10.1128/JCM.00896–21 | Korenkov | 2021 | Germany | STANDARD Q COVID-19 Ag Test | oro-nasopharyngeal swabs | 90 | 1816 | 2 | 120 | 2028 |
| 10.3390/vaccines10020198 | Lau | 2022 | Singapore | Elecsys® SARS-CoV-2 Antigen assay | nasopharyngeal swabs | 26 | 288 | 1 | 35 | 350 |
| 10.1002/jmv.26830 | Ciotti | 2021 | Italy | Coris bioconcept COVID-19 ag respi-strip test | nasopharyngeal swabs | 12 | 11 | 0 | 27 | 50 |
| 10.1016/j.jcv.2021.104838 | Bianco | 2021 | Italy | LumiraDx SARS-CoV-2 Ag Test | nasal swabs | 269 | 561 | 48 | 29 | 907 |
| 10.1002/jcla.23745 | Peña‐Rodríguez | 2021 | Mexico | STANDARD Q COVID-19 Ag Test | nasopharyngeal swabs | 79 | 265 | 0 | 25 | 369 |
| 10.1016/j.jcv.2020.104713 | Toptan | 2021 | Germany | R-Biopharm | oro-nasopharyngeal swabs | 45 | 9 | 0 | 13 | 67 |
| 10.1002/jcla.23906 | Mueller | 2021 | Italy | Elecsys® SARS-CoV-2 Antigen assay | nasopharyngeal swabs | 12 | 356 | 0 | 35 | 403 |
| 10.3390/v14030468 | Salcedo | 2022 | USA | Rapid antigen tests (E25Bio, Inc., Cambridge, MA, USA) | nasal swabs | 51 | 113 | 1 | 8 | 173 |
| 10.1186/s12985–020–01452–5 | Chaimayo | 2020 | Thailand | STANDARD Q COVID-19 Ag Test | nasopharyngeal and throat swabs | 59 | 389 | 5 | 1 | 454 |
| 10.1186/s12879–021–06716–1 | Mitchell | 2021 | USA | Sofia SARS rapid antigen | nasal swabs | 36 | 107 | 0 | 5 | 148 |
| 10.3390/diagnostics12030650 | Polvere | 2022 | Italy | FAST COVID-19 SARS-CoV-2 Antigen Rapid Test kit | nasal swabs | 113 | 383 | 3 | 2 | 501 |
| 10.1016/j.ijid.2021.02.005 | Hirotsu | 2021 | Japan | Lumipulse® G SARS-CoV-2 Ag assay | nasopharyngeal swabs | 37 | 989 | 0 | 3 | 1029 |
| 10.3390/jcm10102099 | Osmanodja | 2021 | Germany | Dräger Antigen Test SARS-CoV-2 | nasal swabs | 62 | 308 | 1 | 8 | 379 |
| 10.1016/j.jcv.2021.105048 | Fourati | 2022 | France | COVID‐VIRO® analysis | nasopharyngeal swabs | 215 | 1614 | 0 | 77 | 1906 |
| 10.1080/1354750X.2021.1876769 | Möckel | 2021 | Germany | Roche SARS-CoV-2 antigen assay | oro-nasopharyngeal swabs | 85 | 358 | 1 | 29 | 473 |
| 10.1016/j.jiac.2021.02.029 | Takeuchi | 2021 | Japan | QuickNavi™-COVID19 Ag | nasopharyngeal swabs | 91 | 1081 | 0 | 14 | 1186 |
| 10.1007/s00430–021–00706–5 | Häuser | 2021 | Germany | LIAISON® SARS-CoV-2 Ag assay (Diasorin, Saluggia, Italy), | nasopharyngeal swabs | 68 | 1632 | 0 | 101 | 1801 |
| 10.3389/fped.2021.647274 | Jung | 2021 | France | BIOSYNEX Ag-RDT | nasopharyngeal swabs | 29 | 271 | 4 | 4 | 308 |
| 10.3390/v14010017 | Klajmon | 2022 | Poland | Humasis COVID-19 Ag Test kit | nasopharyngeal swabs | 43 | 140 | 2 | 4 | 189 |
| 10.1093/ajcp/aqab173 | Drain | 2022 | USA | LumiraDx SARS-CoV-2 Ag Test | nasal swabs | 23 | 194 | 0 | 5 | 222 |
| 10.1016/j.jcv.2021.104789 | Landaas | 2021 | Norway | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal and throat swabs | 186 | 3738 | 3 | 64 | 3991 |
| 10.1016/j.ijid.2021.10.027 | Thirion-Romero | 2021 | Mexico | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 256 | 579 | 9 | 216 | 1060 |
| 10.1515/cclm-2021–0569 | Hartard | 2021 | France | LIAISON® SARS-CoV-2 Ag assay (Diasorin, Saluggia, Italy), | nasopharyngeal swabs | 39 | 330 | 2 | 7 | 378 |
| 10.1128/JCM.01742–21 | Almendares | 2022 | USA | BinaxNOW COVID-19 Ag Card test kit | nasal swabs | 157 | 3116 | 4 | 142 | 3419 |
| 10.1515/cclm-2021–0182 | Menchinelli | 2021 | Italy | Lumipulse® G SARS-CoV-2 Ag assay | nasopharyngeal swabs | 155 | 397 | 3 | 39 | 594 |
| 10.1016/j.heliyon.2021.e08455 | Rahman | 2021 | Bangladesh | STANDARD Q COVID-19 Ag Test | nasopharyngeal swabs | 261 | 593 | 0 | 46 | 900 |
| 10.1016/j.jcv.2021.104941 | Escrivá | 2021 | Spain | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 99 | 331 | 0 | 18 | 448 |
| 10.1016/j.jcv.2021.104961 | Merino-Amador | 2021 | Spain | Clinitest Rapid COVID-19 Antigen Test (ClinitestRT) (Siemens, Healthineers, Erlangen, Germany) | nasopharyngeal swabs | 179 | 256 | 2 | 13 | 450 |
| 10.1016/j.diagmicrobio.2021.115591 | Onsongo | 2022 | Kenya | NowCheck SARS-CoV-2 Ag test | oro-nasopharyngeal swabs | 129 | 845 | 0 | 23 | 997 |
| 10.1016/j.jcv.2020.104659 | Linares | 2020 | Spain | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 44 | 195 | 0 | 16 | 255 |
| 10.1016/j.ijid.2020.10.073 | Nalumansi | 2021 | Uganda | STANDARD Q COVID-19 Ag Test | nasopharyngeal swabs | 63 | 159 | 13 | 27 | 262 |
| 10.3390/ijerph19073826 | Cattelan | 2022 | Italy | LumiraDx SARS-CoV-2 Ag Test | nasal swabs | 174 | 51 | 3 | 54 | 282 |
| 10.1016/j.cmi.2020.11.004 | Albert | 2021 | Spain | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 43 | 358 | 0 | 11 | 412 |
| 10.1097/INF.0000000000003101 | González-Donapetry | 2021 | Spain | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 14 | 422 | 0 | 4 | 440 |
| 10.3201/eid2705.204688 | Igloi | 2021 | Netherlands | Roche SARS-CoV-2 antigen assay | nasopharyngeal swabs | 158 | 780 | 4 | 28 | 970 |
| 10.1002/jmv.26896 | Courtellemont | 2021 | France | COVID‐VIRO® analysis | nasopharyngeal swabs | 117 | 127 | 0 | 4 | 248 |
| 10.1371/journal.pone.0247918 | Krüger | 2021 | Germany | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 92 | 1001 | 1 | 14 | 1108 |
| 10.1016/j.jiac.2021.03.021 | Asai | 2021 | Japan | Lumipulse® G SARS-CoV-2 Ag assay | saliva | 49 | 238 | 4 | 14 | 305 |
| 10.3390/v13050818 | Cento | 2021 | Italy | LumiraDx SARS-CoV-2 Ag Test | nasopharyngeal swabs | 297 | 596 | 17 | 50 | 960 |
| 10.3389/fpubh.2021.728969 | Alqahtani | 2021 | Bahrain | Panbio COVID-19 Ag Rapid Test Device | nasal swabs | 602 | 3420 | 30 | 131 | 4183 |
| 10.3346/jkms.2021.36.e101 | Oh | 2021 | Korea | STANDARD Q COVID-19 Ag Test | nasopharyngeal swabs | 7 | 78 | 0 | 33 | 118 |
| 10.1371/journal.pone.0259527 | Thell | 2021 | Austria | Roche SARS-CoV-2 antigen assay | nasopharyngeal swabs | 171 | 325 | 3 | 42 | 541 |
| 10.1017/ice.2021.281 | Smith | 2021 | Maryland | Sofia SARS rapid antigen | nasopharyngeal swabs | 180 | 2645 | 7 | 55 | 2887 |
| 10.4269/ajtmh.21–0809 | Mungomklang | 2021 | Thailand | STANDARD Q COVID-19 Ag Test | nasopharyngeal swabs | 35 | 1024 | 3 | 38 | 1100 |
| 10.1017/ice.2021.20 | James | 2021 | USA | BinaxNOW COVID-19 Ag Card test kit | nasal swabs | 86 | 2184 | 3 | 66 | 2339 |
| 10.1016/j.cmi.2020.09.057 | Diao | 2021 | China | FIC assay | nasopharyngeal swabs | 152 | 50 | 0 | 49 | 251 |
| 10.1016/j.jiac.2021.07.005 | Kiyasu | 2021 | Japan | QuickNavi™-COVID19 Ag | nasopharyngeal swabs | 151 | 1746 | 0 | 37 | 1934 |
| 10.1016/j.cmi.2021.02.001 | Merino | 2021 | Spain | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 325 | 592 | 7 | 34 | 958 |
| 10.3390/v13050796 | Kim | 2021 | Korea | GenBody™ COVID-19 Ag test | nasopharyngeal swabs | 121 | 198 | 2 | 9 | 330 |
| 10.1371/journal.pone.0258394 | Jo | 2022 | Korea | STANDARD Q COVID-19 Ag Test | nasopharyngeal swabs | 34 | 110 | 0 | 26 | 170 |
| 10.3201/eid2711.211449 | Surasi | 2021 | USA | BinaxNOW COVID-19 Ag Card test kit | nasal swabs | 55 | 642 | 0 | 72 | 769 |
| 10.3390/diagnostics11112110 | Altawalah | 2021 | Kuwait | LIAISON® SARS-CoV-2 Ag assay (Diasorin, Saluggia, Italy), | nasopharyngeal swabs | 113 | 150 | 0 | 37 | 300 |
| 10.1111/apm.13189 | Jakobsen | 2021 | Denmark | STANDARD Q COVID-19 Ag Test | nasal swabs | 32 | 7008 | 0 | 34 | 7074 |
| 10.3390/diagnostics12020447 | Yin | 2022 | Belgium | Lumipulse® G SARS-CoV-2 Ag assay | nasopharyngeal swabs | 95 | 396 | 4 | 7 | 502 |
| 10.1007/s15010–021–01723–5 | Fitoussi | 2021 | France | BIOSYNEX Ag-RDT | nasopharyngeal swabs | 121 | 816 | 3 | 27 | 967 |
| 10.1371/journal.pone.0260862 | Pollreis | 2021 | USA | BinaxNOW COVID-19 Ag Card test kit | nasal swabs | 25 | 177 | 0 | 12 | 214 |
| 10.1016/j.ijid.2021.04.048 | Caputo | 2021 | Italy | Lumipulse® G SARS-CoV-2 Ag assay | nasopharyngeal swabs | 436 | 3661 | 102 | 67 | 4266 |
| 10.3201/eid2710.210080 | Tinker | 2021 | USA | BinaxNOW COVID-19 Ag Card test kit | nasal swabs | 8 | 1500 | 0 | 32 | 1540 |
| 10.1016/j.jcv.2021.105023 | Okoye | 2022 | USA | BinaxNOW COVID-19 Ag Card test kit | nasal swabs | 45 | 3759 | 2 | 4 | 3810 |
| 10.1016/j.jviromet.2021.114299 | Paul | 2021 | India | COVID‐VIRO® analysis | nasopharyngeal swabs | 72 | 50 | 0 | 26 | 148 |
| 10.1016/j.jiac.2021.10.024 | Suzuki | 2022 | Japan | RapidTesta SARS-CoV-2 | nasopharyngeal swabs | 53 | 1045 | 8 | 21 | 1127 |
| 10.1080/23744235.2021.1914857 | Homza | 2021 | Czech Republic | ECOTEST Covid-19 Antigen Rapid Test | nasopharyngeal swabs | 125 | 321 | 9 | 39 | 494 |
| 10.1002/jmv.27220 | Carbonell‐Sahuquillo | 2021 | Spain | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 24 | 323 | 0 | 10 | 357 |
| 10.1016/j.ijid.2021.03.051 | Bouassa | 2021 | France | SIENNA™ COVID-19 Antigen Rapid Test | nasopharyngeal swabs | 90 | 50 | 0 | 10 | 150 |
| 10.3390/diagnostics11122300 | Sazed | 2021 | Bangladesh | OnSite® COVID-19 Ag Rapid Test | nasal swabs | 121 | 245 | 2 | 12 | 380 |
| 10.1016/j.ijid.2021.07.010 | Jegerlehner | 2021 | Switzerland | Roche SARS-CoV-2 antigen assay | nasopharyngeal swabs | 92 | 1319 | 2 | 49 | 1462 |
| 10.1016/j.jinf.2021.02.014 | Bulilete | 2021 | Spain | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 100 | 1220 | 2 | 40 | 1362 |
| 10.4103/ijmr.IJMR_3305_20 | Gupta | 2021 | India | STANDARD Q COVID-19 Ag Test | nasopharyngeal swabs | 63 | 252 | 1 | 14 | 330 |
| 10.1007/s41999–021–00584–3 | Paap | 2021 | Netherlands | Roche SARS-CoV-2 antigen assay | nasopharyngeal swabs | 27 | 363 | 45 | 26 | 461 |
| 10.1371/journal.pone.0250886 | Moeren | 2021 | Netherlands | BD Veritor System for Rapid Detection of SARS-CoV-2 | nasopharyngeal and throat swabs | 16 | 334 | 0 | 1 | 351 |
| 10.1136/bmj.n1637 | Fiñana | 2021 | UK | SARS-CoV-2 antigen rapid lateral flow test (LFT) | nasopharyngeal and throat swabs | 28 | 5431 | 3 | 42 | 5504 |
| 10.1128/Spectrum.00342–21 | Chiu | 2021 | USA | LFA-based INDICAID COVID-19 rapid antigen test (INDICAID rapid test) | nasal swabs | 158 | 23462 | 42 | 30 | 23692 |
| 10.1016/j.ajem.2021.10.022 | Turcato | 2022 | Italy | STANDARD Q COVID-19 Ag Test | nasopharyngeal swabs | 329 | 3470 | 32 | 68 | 3899 |
| 10.3390/diagnostics11122217 | Tonen-Wolyec | 2021 | France | BIOSYNEX Ag-RDT | nasopharyngeal swabs | 20 | 84 | 0 | 2 | 106 |
| 10.1007/s11845–021–02863–1 | Kolesova | 2021 | Italy | Elecsys® SARS-CoV-2 Antigen assay | nasopharyngeal swabs | 64 | 34 | 0 | 12 | 110 |
| 10.1007/s11845–021–02776-z | Denina | 2021 | Italy | LumiraDx SARS-CoV-2 Ag Test | nasal swabs | 16 | 160 | 14 | 1 | 191 |
| 10.1038/s41598–021–90026–8 | Takeuchi | 2021 | Japan | QuickNavi™-COVID19 Ag | nasal swabs | 37 | 811 | 0 | 14 | 862 |
| 10.1002/jcla.24203 | Begum | 2022 | Bangladesh | InTec Rapid SARS‐CoV‐2 Antigen Test | nasopharyngeal swabs | 101 | 102 | 0 | 11 | 214 |
| 10.1016/j.jcv.2021.104878 | Ferté | 2021 | France | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 33 | 636 | 0 | 19 | 688 |
| 10.1128/JCM.03077–20 | Pollock | 2021 | USA | MSD S-PLEX SARS-CoV-2 N assay | nasopharyngeal swabs | 112 | 89 | 1 | 24 | 226 |
| 10.3390/ijerph18179151 | Kyritsi | 2021 | Greece | Rapid Test Ag 2019-nCoV (PROGNOSIS, BIOTECH, Larissa, Greece) | nasopharyngeal swabs | 141 | 458 | 1 | 24 | 624 |
| 10.1155/2021/3893733 | Loconsole | 2021 | Italy | Lumipulse® G SARS-CoV-2 Ag assay | nasopharyngeal swabs | 205 | 677 | 18 | 11 | 911 |
| 10.1016/j.ijid.2021.09.008 | Leiner | 2021 | Germany | Standard F COVID-19 Ag FIA | oro-nasopharyngeal swabs | 491 | 3208 | 80 | 297 | 4076 |
| 10.1371/journal.pone.0253321 | Nsoga | 2021 | Switzerland | Panbio COVID-19 Ag Rapid Test Device | oropharyngeal swabs | 136 | 232 | 2 | 32 | 402 |
| 10.3390/diagnostics12040847 | Ahmed | 2022 | Malaysia | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 101 | 51 | 2 | 3 | 157 |
| 10.23749/mdl.v112i5.12097 | Visci | 2021 | Italy | LIAISON® SARS-CoV-2 Ag assay (Diasorin, Saluggia, Italy), | nasopharyngeal swabs | 78 | 113 | 8 | 10 | 209 |
| 10.1371/journal.pone.0263327 | Mori | 2022 | Japan | Roche SARS-CoV-2 antigen assay | nasopharyngeal swabs | 42 | 1014 | 0 | 14 | 1070 |
| 10.1371/journal.pone.0249710 | Sood | 2021 | USA | BinaxNOW COVID-19 Ag Card test kit | nasal swabs | 127 | 539 | 9 | 99 | 774 |
| 10.1093/ofid/ofab059 | Masiá | 2021 | Spain | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 118 | 709 | 0 | 77 | 904 |
| 10.1002/jmv.27249 | Cassuto | 2021 | France | COVID‐VIRO® analysis | nasal swabs | 31 | 202 | 0 | 1 | 234 |
| 10.1007/s15010–020–01542–0 | Lanser | 2020 | Austria | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 31 | 2 | 0 | 20 | 53 |
| 10.1128/JCM.00083–21 | Pollock | 2021 | USA | BinaxNOW COVID-19 Ag Card test kit | nasal swabs | 227 | 2003 | 12 | 66 | 2308 |
| 10.3390/diagnostics12030710 | Lee | 2022 | Korea | STANDARD Q COVID-19 Ag Test | nasopharyngeal swabs | 58 | 104 | 0 | 13 | 175 |
| 10.1016/j.diagmicrobio.2021.115531 | Bräunlich | 2022 | Germany | Roche SARS-CoV-2 antigen assay | nasopharyngeal swabs | 45 | 2867 | 21 | 45 | 2978 |
| 10.1128/Spectrum.01008–21 | Siddiqui | 2021 | USA | BinaxNOW COVID-19 Ag Card test kit | nasal swabs | 179 | 5826 | 13 | 43 | 6061 |
| 10.1016/j.ijid.2021.05.063 | Leixner | 2021 | Austria | AMP Rapid Test SARS-CoV-2 Ag | nasopharyngeal swabs | 65 | 297 | 1 | 29 | 392 |
| 10.1128/JCM.00991–21 | L′Huillier | 2021 | Switzerland | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 79 | 703 | 0 | 40 | 822 |
| 10.1016/j.jiph.2021.06.002 | Amer | 2021 | Egypt | STANDARD Q COVID-19 Ag Test | oro-nasopharyngeal swabs | 54 | 9 | 5 | 15 | 83 |
| 10.3390/jcm10071471 | Amendola | 2021 | Italy | Lumipulse® G SARS-CoV-2 Ag assay | saliva | 22 | 80 | 5 | 20 | 127 |
| 10.1016/j.ijid.2021.04.087 | Peña | 2021 | Chile | STANDARD Q COVID-19 Ag Test | nasopharyngeal swabs | 51 | 766 | 3 | 22 | 842 |
| 10.1016/j.jiac.2021.07.006 | Kurihara | 2021 | Japan | QuickChaser® Auto SARS-CoV-2 | nasopharyngeal swabs | 62 | 1316 | 2 | 21 | 1401 |
| 10.1016/j.jcv.2020.104455 | Scohy | 2020 | Brussels | Coris bioconcept COVID-19 ag respi-strip test | nasopharyngeal swabs | 32 | 42 | 0 | 74 | 148 |
| 10.1016/j.cmi.2020.12.022 | Torres | 2021 | Spain | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 38 | 555 | 0 | 41 | 634 |
| 10.1016/j.ijid.2020.05.098 | Porte | 2020 | Chile | Bioeasy 2019-Novel Coronavirus (2019-nCoV) Fluorescence Antigen Rapid Test Kit | oro-nasopharyngeal swabs | 77 | 45 | 0 | 5 | 127 |
| 10.1016/j.jcv.2020.104654 | Cerutti | 2020 | Italy | STANDARD Q COVID-19 Ag Test | nasopharyngeal swabs | 77 | 221 | 0 | 32 | 330 |
| 10.1002/jmv.27378 | Treggiari | 2021 | Italy | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 282 | 3741 | 4 | 140 | 4167 |
| 10.1002/jmv.27412 | Villalba | 2021 | Cuba | Elecsys® SARS-CoV-2 Antigen assay | nasopharyngeal swabs | 288 | 183 | 19 | 33 | 523 |
| 10.1016/j.ijid.2021.11.034 | Jian | 2022 | China | COVID-19 Antigen Rapid Test Kit (Eternal Materials, New Taipei City, Taiwan) | nasopharyngeal swabs | 55 | 2009 | 15 | 17 | 2096 |
| 10.1007/s15010–021–01681-y | Krüger | 2022 | Germany | LumiraDx SARS-CoV-2 Ag Test | nasal swabs | 120 | 611 | 4 | 26 | 761 |
| 10.1002/jmv.26855 | Veyrenche | 2020 | France | Coris bioconcept COVID-19 ag respi-strip test | nasopharyngeal swabs | 13 | 20 | 0 | 32 | 65 |
| 10.1007/s10096–021–04346–8 | Aranaz-Andrés | 2022 | Spain | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 25 | 510 | 1 | 5 | 541 |
| 10.1016/j.jiac.2021.08.015 | Nomoto | 2021 | Japan | Lumipulse® G SARS-CoV-2 Ag assay | nasopharyngeal swabs | 66 | 19 | 1 | 14 | 100 |
| 10.1002/jmv.27033 | Holzner | 2021 | Germany | STANDARD Q COVID-19 Ag Test | nasopharyngeal swabs | 379 | 1816 | 8 | 172 | 2375 |
| 10.1002/jmv.27149 | Eleftheriou | 2021 | Greece | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 42 | 693 | 0 | 9 | 744 |
| 10.1016/j.eclinm.2021.100954 | Fernandez-Montero | 2021 | Spain | Roche SARS-CoV-2 antigen assay | nasopharyngeal swabs | 35 | 2486 | 8 | 14 | 2543 |
| 10.1016/j.ijid.2021.07.043 | Leli | 2021 | Italy | LumiraDx SARS-CoV-2 Ag Test | nasal swabs | 114 | 596 | 30 | 52 | 792 |
| 10.1016/j.jpeds.2021.01.027 | Villaverde | 2021 | Spain | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 35 | 1540 | 3 | 42 | 1620 |
| 10.1093/jpids/piab081 | Ford | 2022 | USA | BinaxNOW COVID-19 Ag Card test kit | nasal swabs | 267 | 1774 | 2 | 67 | 2110 |
| 10.1093/labmed/lmab033 | Thakur | 2021 | India | SARS-CoV-2 antigen rapid lateral flow test (LFT) | nasopharyngeal swabs | 29 | 592 | 1 | 55 | 677 |
| 10.1016/j.jviromet.2021.114201 | Orsi | 2021 | Italy | FREND™ COVID-19 Ag assay | nasopharyngeal swabs | 56 | 50 | 0 | 4 | 110 |
| 10.3390/healthcare9070868 | Ifko | 2021 | Slovenia | NADAL COVID-19 antigen test | nasopharyngeal swabs | 20 | 90 | 12 | 3 | 125 |
| 10.1128/JCM.00374–21 | Lefever | 2021 | Belgium | LIAISON® SARS-CoV-2 Ag assay (Diasorin, Saluggia, Italy), | nasopharyngeal swabs | 134 | 210 | 0 | 70 | 414 |
| 10.1002/jmv.27505 | Roger | 2021 | France | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 86 | 4204 | 33 | 102 | 4425 |
| 10.37201/req/054.2021 | Gras-Valenti | 2021 | Spain | Panbio COVID-19 Ag Rapid Test Device | nasopharyngeal swabs | 58 | 398 | 1 | 37 | 494 |
3.2. Quality assessment
Fig. S1 and Table S2 show the quality of the studies in our meta-analysis, based on the QUADAS-2 tool. In the majority (78.5%, 106/135) of the included studies, all patients were consecutively or randomly included, and inappropriate exclusions and case-control designs were avoided. All the studies were judged to have a low risk of bias in the index test and reference standard domains. Regarding the flow and time domains, 73.3% (99/135) of the studies were considered to have a low risk of bias, as they received the same reference standard, and all selected patients were enrolled in the analysis. The patient selection, index tests, and reference standards were considered to meet the objectives of this meta-analysis.
3.3. Publication bias
Deeks’ funnel plot (Fig. S2) did not display significant asymmetry on visual inspection; the P-value of 0.78 for the slope coefficient also suggested symmetry in the data and no striking publication bias in this study.
3.4. Analysis of heterogeneity
We found that P values of the Q test for sensitivity and specificity were both < 0.001 based on heterogeneity statistics, suggesting significant interstudy heterogeneity. In addition, as the bivariate boxplot shows in Fig. S3, most studies clustered within the median distribution with 28 outliers, further indicating the presence of interstudy heterogeneity. Thus, a bivariate random-effects model was appropriate for quantitative synthesis.
3.5. Diagnostic performance
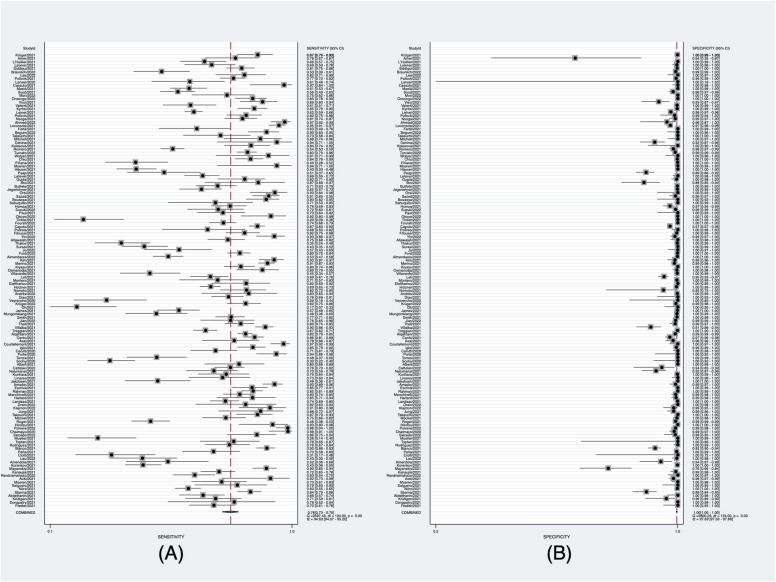
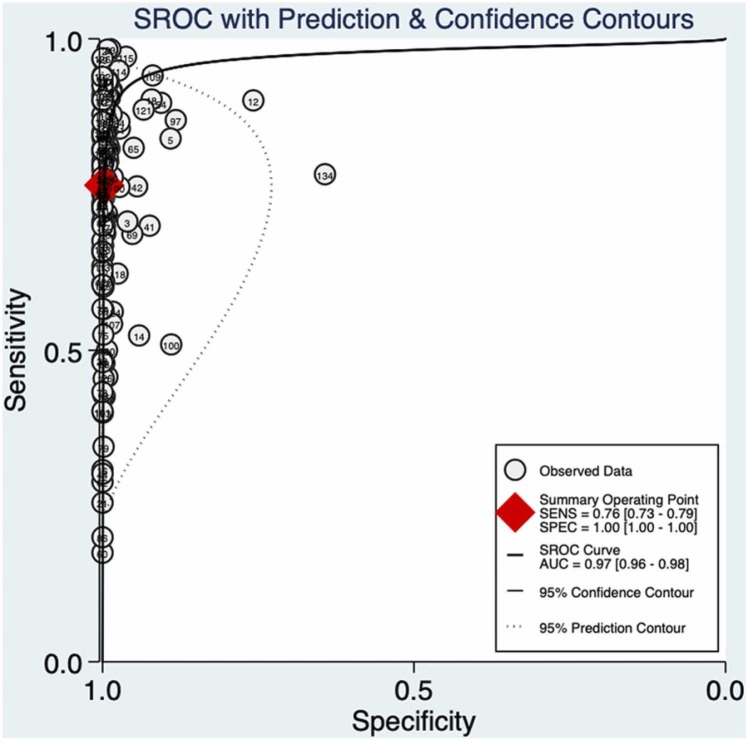
The meta-analysis demonstrated a pooled sensitivity of 0.76 (95% CI: 0.73–0.79) ( Fig. 2A) and a pooled specificity of 1.00 (95% CI: 1.00–1.00) (Fig. 2B). The pooled positive likelihood ratio, negative likelihood ratio, and diagnostic odds ratio were 276.1 (95% CI, 184.1–414.1), 0.24 (95% CI, 0.21–0.27), and 1171 (95% CI, 782–1755), respectively. Additionally, the summary AUC was 0.97 (95% CI, 0.96–0.98) ( Fig. 3), which reveals that RAT is of high diagnostic value for COVID-19. As shown in Fig. 3, there was no shoulder-arm-shaped distribution in the SROC curve, and the proportion of heterogeneity due to the threshold effect was 0.12, indicating that the heterogeneity of this meta-analysis was independent of the threshold effect.
Fig. 2.
Pooled sensitivity and pooled specificity of RAT. (A) Forest plots of pooled sensitivity. (B) Forest plots of pooled specificity.
Fig. 3.
Summary ROC curve and its area under curve.
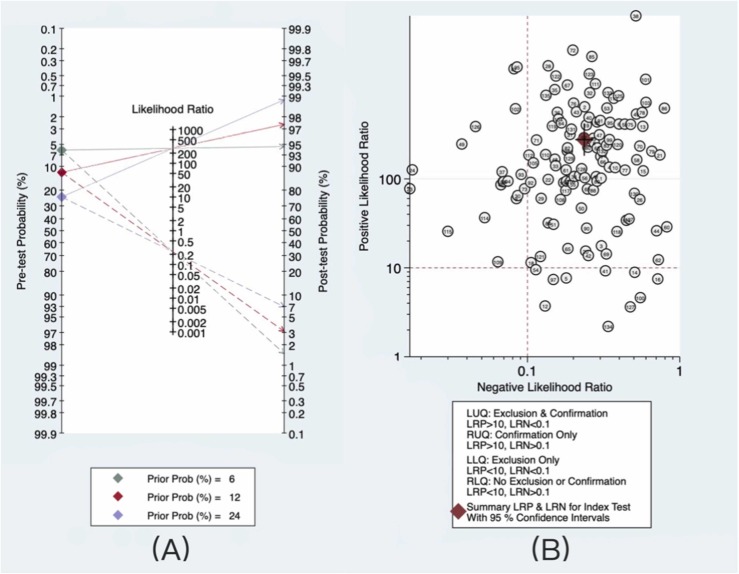
The results of the statistical analysis were used to set the pretest probability to 12%. The Fagan plot presented in Fig. 4A shows that the posttest probability increased to 97% if the antigen test was positive and was as low as 3% if the antigen test was negative. When we assumed a higher pretest probability of infection of 24% (doubling the prevalence rate), both the positive and negative posttest probabilities improved to 99% and 7%, respectively. When the prevalence rate was halved to 6%, the probability of a positive posttest dropped to 95%, and the probability of a negative posttest dropped to 1%.
Fig. 4.
Diagnostic value and clinical application value of RAT. (A) Fagan plot for evaluating diagnostic value: The solid line represents the positive post-test probability, and the dotted line represents the negative post-test probability. (B) Likelihood ratio scattergram for evaluating clinical application value.
The likelihood ratio scattergram (Fig. 4B) showed that more than three-quarters of the studies (77.8%, 105/135) along with the summary point of likelihood ratios obtained as functions of mean sensitivity and specificity were in the right upper quadrant. These findings suggest that RAT helps confirm the presence of SARS-CoV-2 when the test result is positive and not for its exclusion when negative.
3.6. Subgroup analyses
In Table 2, all the samples achieved a specificity of 1.00. When assessing studies evaluating nasopharyngeal swab as the sample type for Ag-RDT, the pooled sensitivity from 93 studies with 76,945 samples was 0.76 (95% CI: 0.72–0.79). Analysis of performance with a nasal swab (26 studies, 64,125 samples) showed a higher pooled sensitivity of 0.79 (95% CI: 0.71–0.85). For samples from other parts including combined nasopharyngeal and throat, oropharyngeal, combined oropharyngeal, and nasopharyngeal and saliva swabs (16 studies, 22,372 samples), the pooled sensitivity was 0.76 (95% CI: 0.66–0.84).
Table 2.
Pooled sensitivity and specificity among subgroups of studies.
| Subgroups | No. of study | Total Sample Size | Polled Sensitivity (95% CI) | Polled Specificity (95% CI) |
|---|---|---|---|---|
| Sample Types | ||||
| nasopharyngeal | 93 | 76,945 | 0.76(0.72–0.79) | 1.00(1.00–1.00) |
| nasal | 26 | 64,125 | 0.79(0.71–0.85) | 1.00(0.99–1.00) |
| other | 16 | 22,372 | 0.76(0.66–0.84) | 1.00(0.99–1.00) |
| RATt Kit | ||||
| COVID‐VIRO® analysis | 4 | 2536 | 0.90(0.70–0.97) | 1.00(1.00–1.00) |
| Lumipulse® G SARS-CoV-2 Ag assay | 10 | 8895 | 0.86(0.79–0.91) | 0.98(0.96–0.99) |
| BIOSYNEX Ag-RDT | 3 | 1382 | 0.85(0.77–0.90) | 0.99(0.98–1.00) |
| LumiraDx SARS-CoV-2 Ag Test | 7 | 4115 | 0.83(0.76–0.88) | 0.97(0.94–0.99) |
| QuickNavi™-COVID19 Ag | 3 | 3982 | 0.81(0.76–0.85) | 1.00(1.00–1.00) |
| Panbio COVID-19 Ag Rapid Test Device | 25 | 30,332 | 0.73(0.67–0.79) | 1.00(1.00–1.00) |
| LIAISON® SARS-CoV-2 Ag assay | 7 | 3532 | 0.72(0.60–0.82) | 1.00(0.97–1.00) |
| Roche SARS-CoV-2 antigen assay | 9 | 10,648 | 0.71(0.62–0.78) | 0.99(0.98–1.00) |
| STANDARD Q COVID-19 Ag Test | 17 | 20,765 | 0.70(0.59–0.79) | 1.00(0.99–1.00) |
| BinaxNOW COVID-19 Ag Card test kit | 10 | 23,344 | 0.65(0.50–0.77) | 1.00(1.00–1.00) |
| Elecsys® SARS-CoV-2 Antigen assay | 6 | 4750 | 0.65(0.44–0.81) | 1.00(0.98–1.00) |
| Coris bioconcept COVID-19 ag respi-strip test | 4 | 747 | 0.41(0.23–0.61) | 1.00(0.53–1.00) |
| Days after symptom onset | ||||
| ≤ 3 days | 10 | 870 | 0.91(0.83–0.96) | / |
| ≤ 7 days | 13 | 1862 | 0.89(0.84–0.93) | / |
| ≤ 10 days | 13 | 1918 | 0.88(0.83–0.92) | / |
| > 10 days | 4 | 72 | 0.36(0.21–0.55) | / |
| Ct Values | ||||
| < 20 | 15 | 368 | 1.00(0.70–1.00) | / |
| 20–25 | 11 | 342 | 0.94(0.87–0.97) | / |
| 25–30 | 23 | 579 | 0.70(0.53–0.84) | / |
| > 30 | 24 | 715 | 0.24(0.16–0.33) | / |
Among the 40 RAT kits used in this study, 28 did not provide sufficient data for the bivariate meta-analysis. Of the remaining 12 RAT kits, COVID‐VIRO® analysis showed the highest pooled sensitivity of 0.90 (95% CI: 0.70–0.97), followed by Lumipulse® G SARS-CoV-2 Ag assay with a pooled sensitivity of 0.86 (95% CI: 0.79–0.91); the combined sensitivity of BIOSYNEX Ag-RDT, LumiraDx SARS-CoV-2 Ag Test, and QuickNavi™-COVID19 Ag Test were all above 0.80. The Coris bioconcept COVID-19 ag respi-strip test had the lowest pooled sensitivity of 0.41 (95% CI: 0.23–0.61).
The pooled sensitivity decreased as Ct values increased. Samples with a Ct value < 20 achieved excellent pooled sensitivity at 1.00 (95% CI: 0.70–1.00). Ct value using the cutoff of 20–25 also showed a high sensitivity of 0.94 (95% CI: 0.87–0.97). The pooled sensitivity decreased to 0.70 (95% CI: 0.53–0.84) when the Ct value was 25–30. For Ct value > 30, the pooled sensitivity was relatively low at 0.24 (95% CI: 0.16–0.33).
We assessed sensitivity at three different cutoff points on the days after the onset of symptoms. For ≤ 3, ≤ 7, and ≤ 10 days, the summary sensitivities were 0.91 (95% CI: 0.83–0.96), 0.89 (95% CI: 0.84–0.93), and 0.88 (95% CI: 0.83–0.92), respectively. When the number of days after symptom onset exceeded 10 days, the sensitivity notably decreased to 0.36 (95% CI: 0.21–0.55).
4. Discussion
In this meta-analysis, a comprehensive literature search was conducted, and we summarized data from 135 studies, including 163,442 samples, to evaluate the diagnostic performance of RAT in COVID-19. The pooled sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, and diagnostic odds ratio were 0.76 (95%CI: 0.73–0.79), 1.00 (95%CI: 1.00–1.00), 276.1 (95% CI, 184.1–414.1), 0.24 (95% CI, 0.21–0.27), and 1171 (95% CI, 782–1755), respectively. A positive likelihood ratio > 10 confirms the diagnosis of the disease, while a negative likelihood ratio < 0.1 excludes the possibility of the disease. When the diagnostic odds ratio > 1, the larger the value, the better the ability to distinguish between healthy people and patients. Our results indicated that RAT had a high diagnostic value.
Possibly due to differences in sensitivity, specificity, and patient population between the studies, we detected a high degree of heterogeneity; however, the bivariate random-effects model we used provided a relatively robust statistical result. Performance between manufacturer-dependent studies and manufacturer-independent studies may differ hugely, but when we removed the 3 manufacturer-dependent studies, the overall effect remained unchanged, (Sensitivity: 0.76 versus 0.76; Specificity: 1.00 versus 1.00; AUC: 0.97 versus 0.97), indicating that our results were not driven by the 3 manufacturer-dependent articles. Furthermore, SROC did not detect marked heterogeneity in the pooled sensitivity and specificity. Tests for publication bias also indicated no noticeable bias. Thus, the statistical analysis of this meta-analysis was reliable to some extent.
By analyzing the data, we hypothesized that the pretest probability was 12%, resulting in a positive posttest probability of 97% and a negative posttest probability of 3%; this suggested a very high probability that a patient with SARS-CoV-2 infection would test positive in the antigen test. According to our findings, pretest probability is positively correlated with posttest probability. This suggests that RAT is more applicable to high-risk populations. Considering that the RAT provided 1.00 specificity in our study along with its rapid turnaround time, it could be used as a screening tool in particular situations, such as highly suspicious contacts, or for triage in an emergency department. A positive antigen test will confirm the infection and prevent the virus from spreading, as well as accelerate and optimize the management of infected individuals. By quickly identifying infected patients, the decision-making process of the entire emergency department is improved.
Nasopharyngeal swabs generally have the highest detection rate for the diagnostic testing of respiratory viruses including SARS-CoV-2 (Lee et al., 2021). However, they must be collected by trained healthcare professionals using protective equipment, and their collection often causes considerable discomfort to patients (Lindner et al., 2021). In comparison, nasal sample collection is notably painless, and self-collection is possible (Lee et al., 2022). Moreover, nasal sampling is associated with less coughing or sneezing during collection, leading to less droplet exposure, thus reducing the transmission risk among healthcare workers (Takeuchi et al., 2021). Recent studies have reported that the diagnostic sensitivity of RT-PCR for nasal specimens is comparable to that for nasopharyngeal specimens (Péré et al., 2020, Tu et al., 2020). Interestingly, our analysis revealed that the sensitivity for nasal swabs (0.79) was higher than that for nasopharyngeal swabs (0.76) for RAT in cases where both swabs reached a specificity of 1.00. Therefore, the results indicate that using a more superficially collected nasal swab specimen is a good alternative for detecting SARS-CoV-2.
The overall sensitivity of the different RAT kits varies widely, ranging from 0.90 (95% CI: 0.70–0.97) to 0.41 (95% CI: 0.23–0.61). Three RAT kits (LumiraDx SARS-CoV-2 Ag Test, Panbio COVID-19 Ag Rapid Test Device, and STANDARD Q COVID-19 Ag Test) in our research have been authorized by the World Health Organization (WHO) for emergency use (Coronavirus Disease (COVID-19) Pandemic — Emergency Use Listing Procedure (EUL) Open for IVDs, 2020). For suspected patients, WHO recommends that a RAT kit reach a minimum performance criterion of 0.80 sensitivity and 0.97 specificity (Antigen-Detection in the Diagnosis of SARS-CoV-2 Infection, 2021). Only the LumiraDx SARS-CoV-2 Ag Test met this criterion, with a sensitivity of 0.83 (95% CI: 0.76–0.88) and specificity of 0.97 (95% CI: 0.94–0.99). The other two kits did not reach a sensitivity of 0.80 (sensitivity of 0.73 for Panbio and 0.70 for Standard Q), although both had a specificity of 1.00. Therefore, these results suggest an urgent need to further validate the performance of RAT kits on the emergency use list.
Previous studies have shown that lower Ct values represent higher viral loads, resulting in significantly higher RAT sensitivity, antigen concentration, and Ct values that are highly correlated (Pollock et al., 2021), and these were confirmed by our study. An outstanding sensitivity of 1.00 was achieved for Ct values < 20, after which the sensitivity of the RAT gradually declined as Ct values increased. Several studies have reported that the infectivity of SARS-CoV-2 persists for only approximately 8–10 days after the onset of symptoms (Bullard et al., 2020; Hirotsu et al., 2021; Million et al., 2020; Perera et al., 2020; van Kampen et al., 2021; Wölfel et al., 2020). Based on the results of the meta-analysis, the sensitivity within 10 days after the appearance of symptoms (0.88) was relatively favorable, which was not much lower than that within 3 days (0.91). Our findings support the use of RAT as an early stage screening tool for symptomatic patients, particularly those with high viral loads.
When Ct values were > 24, Bullard et al. observed that infectious viruses could not be isolated from the diagnostic samples (Bullard et al., 2020). In our research, although the pooled sensitivity was relatively low at 0.24 for Ct value > 30, a comparatively high sensitivity of 0.70 was maintained for Ct value using the cutoff of 25–30. Thus, it can be assumed that the missed cases of RAT will not cause a large-scale transmission. Our findings suggest that RAT sensitivity was as low as 0.36 ten days after symptom onset. However, according to the Centers for Disease Control and Prevention, 10 days after the appearance of symptoms can be considered a stage of low contagiousness (CDC, 2020). Hence, patients who have had symptoms for a more extended period may have a low risk of infecting others, even if they are incorrectly classified as negative for the SARS‐CoV‐2 antigen.
Our findings support the previous studies that RAT had high sensitivity and specificity and performed better in samples with high viral load, but in contrast to the earlier studies, we have a new finding that nasal swabs have a higher sensitivity than nasopharyngeal swabs for RAT. In addition, the strength of the present study lies in the number of studies (and samples) analyzed compared with previous studies (Arshadi et al., 2022, Chen et al., 2021, Hayer et al., 2021). Although our study did not assess the impact of the SARS-CoV-2 variant, RAT may not be influenced by the variant because RAT targets the nucleocapsid antigen whereas the mutant has a variable mutation at the spike antigen (Gupta et al., 2021).
Our study has some limitations, due to the lack of detailed information in the articles, the data of ≤ 10 days included data of both ≤ 7 days and 8–10 days, resulting in some overlap between the data of ≤ 10 days and ≤ 7 days, which may account for the similar sensitivity of the two (Sensitivity: 0.89 versus 0.88), whether the sensitivity of ≤ 7 days was similar with that of 8–10 days after symptom onset need to further study. We did not evaluate all RAT kits, but only part of them because of the limited data.
5. Conclusions
RAT kits show high sensitivity and specificity in the early stages of infection, especially when the viral load is high. In addition, using nasal samples for antigen testing, which is moderately sensitive and patient-friendly, is a reliable alternative to nasopharyngeal sampling. RAT might be an effective tool for the clinical management of patients in hospital settings, especially during the initial triage, as it aids the rapid identification of positive patients to prevent transmission, thus helping disrupt the COVID-19 pandemic. RAT also seems applicable to other areas, such as regular mass screening or airport screening, because it should allow for a more convenient and time-saving experience for people who travel. However, this important epidemiological benefit must be complemented with the thoughtful and responsible handling of negative test results.
Funding
This work was supported by the National Natural Science Foundation of China [Grant numbers 82172331, 81972028, 81802089, 81672094], the Key Projects for Province Science and Technology Program of Fujian Province, China [Grant number 2020D017] and the Natural Science Foundation of Fujian Province, China [Grant number 2020J05285]. The funders played no role in the study design, data collection, or analyses, the decision to publish, or manuscript preparation.
Ethical approval statement
No ethics approval was required for this work.
CRediT authorship contribution statement
Jia-Wen Xie: Methodology, Writing – original draft. Yun He: Software, Investigation. Ya-Wen Zheng: Formal analysis, Investigation. Mao Wang: Validation, Data curation. Yong Lin: Visualization, Supervision. Li-Rong Lin: Conceptualization, Writing – review & editing.
Conflict of interest
The authors declared that they have no conflicts of interest to this work.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.micres.2022.127185.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
Data availability
Data will be made available on request.
References
- Antigen-Detection in the Diagnosis of SARS-CoV-2 Infection, 2021. [WWW Document]. URL: 〈https://www.who.int/publications-detail-redirect/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays〉. (Accessed 22 May 2022).
- Arshadi M., Fardsanei F., Deihim B., Farshadzadeh Z., Nikkhahi F., Khalili F., Sotgiu G., Shahidi Bonjar A.H., Centis R., Migliori G.B., Nasiri M.J., Mirsaeidi M. Diagnostic accuracy of rapid antigen tests for COVID-19 detection: a systematic review with meta-analysis. Front. Med. 2022;9 doi: 10.3389/fmed.2022.870738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges L.P., Martins A.F., Silva B. de M., Dias B. de P., Gonçalves R.L., Souza D.R.V. de, Oliveira M.G.B. de, Jesus P.C. de, Serafini M.R., Quintans J.S.S., Coutinho H.D.M., Martins R.L., Júnior L.J.Q. Rapid diagnosis of COVID-19 in the first year of the pandemic: a systematic review. Int. Immunopharmacol. 2021;101 doi: 10.1016/j.intimp.2021.108144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L., Boodman C., Bello A., Hedley A., Schiffman Z., Doan K., Bastien N., Li Y., Van Caeseele P.G., Poliquin G. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa638. ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC, 2020. Healthcare Workers [WWW Document]. Centers for Disease Control and Prevention. URL: 〈https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html〉. (Accessed 31 May 2022).
- Chen C.-C., Lu S.-C., Bai C.-H., Wang P.-Y., Lee K.-Y., Wang Y.-H. Diagnostic accuracy of SARS-CoV-2 antigen tests for community transmission screening: a systematic review and meta-analysis. Int. J. Environ. Res. Public Health. 2021;18:11451. doi: 10.3390/ijerph182111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronavirus Disease (COVID-19) Pandemic — Emergency Use Listing Procedure (EUL) Open for IVDs, 2020. [WWW Document]. WHO – Prequalification of Medical Products (IVDs, Medicines, Vaccines and Immunization Devices, Vector Control). URL: 〈https://extranet.who.int/pqweb/vitro-diagnostics/coronavirus-disease-covid-19-pandemic-%E2%80%94-emergency-use-listing-procedure-eul-open〉. (Accessed 20 May 2022).
- Gupta D., Sharma P., Singh M., Kumar M., Ethayathulla A.S., Kaur P. Structural and functional insights into the spike protein mutations of emerging SARS-CoV-2 variants. Cell. Mol. Life Sci. 2021;78:7967–7989. doi: 10.1007/s00018-021-04008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayer J., Kasapic D., Zemmrich C. Real-world clinical performance of commercial SARS-CoV-2 rapid antigen tests in suspected COVID-19: a systematic meta-analysis of available data as of November 20, 2020. Int. J. Infect. Dis. 2021;108:592–602. doi: 10.1016/j.ijid.2021.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y., Maejima M., Shibusawa M., Amemiya K., Nagakubo Y., Hosaka K., Sueki H., Hayakawa M., Mochizuki H., Tsutsui T., Kakizaki Y., Miyashita Y., Omata M. Prospective study of 1308 nasopharyngeal swabs from 1033 patients using the LUMIPULSE SARS-CoV-2 antigen test: Comparison with RT-qPCR. Int. J. Infect. Dis. 2021;105:7–14. doi: 10.1016/j.ijid.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Wang M., Zuo Z., Fan C., Ye F., Cai Z., Wang Y., Cui H., Pan K., Xu A. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int. J. Infect. Dis. 2020;94:49–52. doi: 10.1016/j.ijid.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandker S.S., Nik Hashim N.H.H., Deris Z.Z., Shueb R.H., Islam M.A. Diagnostic accuracy of rapid antigen test kits for detecting SARS-CoV-2: a systematic review and meta-analysis of 17,171 suspected COVID-19 patients. J. Clin. Med. 2021;10:3493. doi: 10.3390/jcm10163493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.-C., Shih T.-P., Ko W.-C., Tang H.-J., Hsueh P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.A., Herigon J.C., Benedetti A., Pollock N.R., Denkinger C.M. Performance of saliva, oropharyngeal swabs, and nasal swabs for SARS-CoV-2 molecular detection: a systematic review and meta-analysis. J. Clin. Microbiol. 2021;59 doi: 10.1128/JCM.02881-20. e02881-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Widyasari K., Yang H.-R., Jang J., Kang T., Kim S. Evaluation of the diagnostic accuracy of nasal cavity and nasopharyngeal swab specimens for SARS-CoV-2 detection via rapid antigen test according to specimen collection timing and viral load. Diagnostics. 2022;12:710. doi: 10.3390/diagnostics12030710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner A.K., Nikolai O., Kausch F., Wintel M., Hommes F., Gertler M., Krüger L.J., Gaeddert M., Tobian F., Lainati F., Köppel L., Seybold J., Corman V.M., Drosten C., Hofmann J., Sacks J.A., Mockenhaupt F.P., Denkinger C.M. Head-to-head comparison of SARS-CoV-2 antigen-detecting rapid test with self-collected nasal swab versus professional-collected nasopharyngeal swab. Eur. Respir. J. 2021;57:2003961. doi: 10.1183/13993003.03961-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Million M., Lagier J.-C., Gautret P., Colson P., Fournier P.-E., Amrane S., Hocquart M., Mailhe M., Esteves-Vieira V., Doudier B., Aubry C., Correard F., Giraud-Gatineau A., Roussel Y., Berenger C., Cassir N., Seng P., Zandotti C., Dhiver C., Ravaux I., Tomei C., Eldin C., Tissot-Dupont H., Honoré S., Stein A., Jacquier A., Deharo J.-C., Chabrière E., Levasseur A., Fenollar F., Rolain J.-M., Obadia Y., Brouqui P., Drancourt M., La Scola B., Parola P., Raoult D. Early treatment of COVID-19 patients with hydroxychloroquine and azithromycin: a retrospective analysis of 1061 cases in Marseille. Fr. Travel Med. Infect. Dis. 2020;35 doi: 10.1016/j.tmaid.2020.101738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péré H., Podglajen I., Wack M., Flamarion E., Mirault T., Goudot G., Hauw-Berlemont C., Le L., Caudron E., Carrabin S., Rodary J., Ribeyre T., Bélec L., Veyer D. Nasal swab sampling for SARS-CoV-2: a convenient alternative in times of nasopharyngeal swab shortage. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00721-20. e00721-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R.A.P.M., Tso E., Tsang O.T.Y., Tsang D.N.C., Fung K., Leung Y.W.Y., Chin A.W.H., Chu D.K.W., Cheng S.M.S., Poon L.L.M., Chuang V.W.M., Peiris M. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg. Infect. Dis. 2020;26:2701–2704. doi: 10.3201/eid2611.203219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock N.R., Savage T.J., Wardell H., Lee R.A., Mathew A., Stengelin M., Sigal G.B. Correlation of SARS-CoV-2 nucleocapsid antigen and RNA concentrations in nasopharyngeal samples from children and adults using an ultrasensitive and quantitative antigen assay. J. Clin. Microbiol. 2021;59 doi: 10.1128/JCM.03077-20. e03077-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi Y., Akashi Y., Kato D., Kuwahara M., Muramatsu S., Ueda A., Notake S., Nakamura K., Ishikawa H., Suzuki H. Diagnostic performance and characteristics of anterior nasal collection for the SARS-CoV-2 antigen test: a prospective study. Sci. Rep. 2021;11:10519. doi: 10.1038/s41598-021-90026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres I., Poujois S., Albert E., Álvarez G., Colomina J., Navarro D. Point-of-care evaluation of a rapid antigen test (CLINITESTⓇ rapid COVID-19 antigen test) for diagnosis of SARS-CoV-2 infection in symptomatic and asymptomatic individuals. J. Infect. 2021;82:e11–e12. doi: 10.1016/j.jinf.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y.-P., Jennings R., Hart B., Cangelosi G.A., Wood R.C., Wehber K., Verma P., Vojta D., Berke E.M. Swabs collected by patients or health care workers for SARS-CoV-2 testing. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2016321. NEJMc2016321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kampen J.J.A., van de Vijver D.A.M.C., Fraaij P.L.A., Haagmans B.L., Lamers M.M., Okba N., van den Akker J.P.C., Endeman H., Gommers D.A.M.P.J., Cornelissen J.J., Hoek R.A.S., van der Eerden M.M., Hesselink D.A., Metselaar H.J., Verbon A., de Steenwinkel J.E.M., Aron G.I., van Gorp E.C.M., van Boheemen S., Voermans J.C., Boucher C.A.B., Molenkamp R., Koopmans M.P.G., Geurtsvankessel C., van der Eijk A.A. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19) Nat. Commun. 2021;12:267. doi: 10.1038/s41467-020-20568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- W.H.O. Director-General’s Opening Remarks at the Media Briefing on COVID-19 – 11 March 2020, (n.d.). [WWW Document]. URL: 〈https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020〉. (Accessed 19 May 2022).
- Whiting, P.F., Rutjes, A.W.S., Westwood, M.E., Mallett, S., Deeks, J.J., Reitsma, J.B., Leeflang, M.M.G., Sterne, J.A.C., Bossuyt, P.M.M., QUADAS-2 Group, 2011. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med., 155, pp. 529–536. 〈 10.7326/0003-4819-155-8-201110180-00009〉. [DOI] [PubMed]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Yamayoshi S., Sakai-Tagawa Y., Koga M., Akasaka O., Nakachi I., Koh H., Maeda K., Adachi E., Saito M., Nagai H., Ikeuchi K., Ogura T., Baba R., Fujita K., Fukui T., Ito F., Hattori S., Yamamoto K., Nakamoto T., Furusawa Y., Yasuhara A., Ujie M., Yamada S., Ito M., Mitsuya H., Omagari N., Yotsuyanagi H., Iwatsuki-Horimoto K., Imai M., Kawaoka Y. Comparison of rapid antigen tests for COVID-19. Viruses. 2020;12:1420. doi: 10.3390/v12121420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yüce M., Filiztekin E., Özkaya K.G. COVID-19 diagnosis—a review of current methods. Biosens. Bioelectron. 2021;172 doi: 10.1016/j.bios.2020.112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material.
Supplementary material.
Supplementary material.
Data Availability Statement
Data will be made available on request.