Abstract
Many bacterial promoters possess multiple sites for binding of transcriptional activator proteins. The uhpT promoter, which controls expression of the sugar phosphate transport system in Escherichia coli, possesses multiple sites for its specific activator protein, UhpA, and a single site for binding of the global regulator, the catabolite gene activator protein (CAP). The binding of UhpA to the uhpT promoter was determined by DNase protection assays; UhpA displayed different affinities for the target sites. The upstream or strong sites, between positions −80 and −50, exhibited a higher affinity for UhpA than did the downstream or weak sites, between positions −50 and −32, adjoining the RNA polymerase-binding site. Phosphorylation of UhpA strongly increased its affinity for both sites. To examine the possible roles of the two sets of UhpA-binding sites, a series of insertion and deletion mutations were introduced at the boundary between them, as suggested from the positions that were protected by UhpA against hydroxyl radical cleavage. Deletions extended in the direction of the weak sites. The insertion or deletion of one helical turn of DNA resulted in the loss of promoter activity and of occupancy by UhpA of the remaining weak-site sequences but was accompanied by normal occupancy of the strong site and no change in the gel retardation behavior of the promoter fragments. However, the deletion of two helical turns of DNA, i.e., 20, 21, or 22 bp, resulted in the novel appearance of UhpA-independent expression and in an additional level of expression that was dependent on UhpA but independent of an inducing signal. The UhpA-independent promoter activity was shown to result from activation by CAP at its more proximal position. UhpA-dependent activity under noninducing conditions appears to result from the binding of unphosphorylated UhpA to the strong sites, which are now in the position normally occupied by the weak sites. Thus, regulated phosphorylation of the response regulator UhpA enhances its occupancy of the weak sites where favorable contacts can allow the binding of RNA polymerase to the promoter.
The proper placement of transcription-regulatory proteins within their target promoter is likely to be important for their function. Many promoters contain multiple sites for binding of one or more transcription-regulatory proteins (4). Expression of the Escherichia coli uhpT gene, encoding the sugar phosphate transport system (20), is controlled by two transcription activators which bind to sites within the region 120 bp upstream of the transcription start. The effect of changes in the locations of their binding sites may provide information about their role in assembly of the transcription initiation complex. Expression of the uhpT promoter is induced by extracellular glucose-6-phosphate (Glu6P) acting through an unusual two-component regulatory system (17) in which the membrane-bound UhpBC sensor kinase complex regulates the phosphorylation and activation of the response regulator UhpA. Perhaps owing to the absence of a −35 element, the uhpT promoter is absolutely dependent on phosphorylated UhpA (P-UhpA), but unphosphorylated UhpA can activate transcription when it is overexpressed or altered by certain mutations. UhpA-dependent transcription is further stimulated 10- to 15-fold by the catabolite gene activator protein (CAP) in complex with cyclic AMP.
The mechanism of CAP stimulation at promoters in which CAP binds to sites located at positions −40 to −80 has been extensively studied, especially the role of specific contacts with the C-terminal domain of the RpoA subunit (α-CTD) of RNA polymerase (RNAP) (reviewed in references 2 and 6). Merkel et al. (22) showed that the insertion of an integral number of DNA-helical turns between the uhpT promoter and the CAP-binding site centered at position −105.5 (all coordinates are relative to the in vitro transcription start site) led to a substantial and progressive decline in stimulation by CAP. Insertion of a nonintegral number of helical turns resulted in the loss of stimulation by CAP. Thus, CAP action depends on both its proper helical phasing and its proximity to the remainder of the uhpT promoter. The binding of CAP confers a slight increase of UhpA binding to its upstream sites, and the stimulation of the uhpT promoter by CAP appears to require the functioning of α-CTD (24) but not the activating surfaces used at other CAP-dependent promoters (21).
UhpA belongs to the NarL family of response regulators (11, 29). The sequence of its C-terminal DNA-binding and activation domain is related to the helix-turn-helix motif in NarL and is conserved among otherwise unrelated transcription activators (1). UhpA binds to multiple sites in the uhpT promoter between positions −80 and −32 (5, 23). DNase I and hydroxyl radical footprinting suggested that lower concentrations of UhpA were required for occupancy of the strong binding sites between positions −80 and −50 than for occupancy of the weak sites between −50 and −32, but the relative affinities and the effect of UhpA phosphorylation were not quantified.
The existence of multiple binding sites with differing affinities is found for other response regulators, including OmpR (14, 19, 25, 26), NarL (18), and BvgA (37), as well as members of the LysR and AraC families of activators (reviewed in references 8 and 30). These distinct protein-binding sites with different affinities probably have specific roles in transcription activation. One model for UhpA action proposes that its occupancy of the upstream, strong binding sites does not directly result in transcription activation but facilitates occupancy of the downstream, weak sites adjoining the RNAP-binding region. Occupancy of the weak sites may be critical for transcription activation and may occur by oligomerization of UhpA molecules along the DNA in response to its phosphorylation. Another possibility is that occupancy of both the strong and weak sites contributes independently to transcription activation. To explore these models, we describe here the effect of the phosphorylation of UhpA on its occupancy of sites in the uhpT promoter, as measured by DNase footprinting. Based on these results, we tested whether UhpA activation was affected by the helical phasing between the strong and weak sites and whether deletion of the weak sites to bring the strong sites into proximity to the RNAP-binding region would allow UhpA to activate uhpT transcription without the need for phosphorylation. In parallel with studies of the consequences of deletion or insertion of sequences at the boundary between the strong and weak sites, we examined transcription by a uhpT variant promoter containing a canonical −35 element.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli K-12 strains RK1280 and RK1271 are derived from strain MC4100 [Δ(argF-lac)U169 araD139 flhD5301 ptsF25 relA1 rpsL150 rbsR22 deoC1] (31) by phage P1-mediated cotransduction of uhp+ and ΔuhpA(A15–A189), respectively, through linkage to pyrE+ zib631::Tn10. The ΔuhpA(A15–A189) allele contains an in-frame deletion of the coding sequences for the region between amino acids 15 and 189 of UhpA (15). Both strains were made recA by cotransduction with an srl::Tn10 marker. The host strain for plasmid constructions was JM109. Growth media were Luria broth for rich medium and minimal salts medium A supplemented with casein hydrolysate (0.5%) and glycerol (1%). Strains were grown in the presence of ampicillin (100 μg/ml).
Recombinant DNA techniques.
Plasmid DNA was isolated using the QIAprep spin miniprep kit (Qiagen, Valencia, Calif.). Oligonucleotides used in this study were synthesized by Gibco-BRL (Rockville, Md.). DNA fragments were purified from agarose gel slices by using the QIAquick gel extraction kit (Qiagen). PCRs were carried out using VENT polymerase (New England Biolabs, Inc., Beverly, Mass.). PCR products were purified using the QIAquick PCR purification kit (Qiagen). The nucleotide sequence of PCR-mutagenized DNA was performed at the Biomolecular Research Center, University of Virginia School of Medicine, using an ABI Prism 377 DNA sequencer.
Mutagenesis.
Two oligonucleotides, oQC1 (−154 5′-GCA GGA ATT CTT TTT GAA CGC −134) and oQC2 (+51 5′ TAC AGG ATC CAA AGC CAG CAT GG +29), were used as primers in PCR with plasmid pRJK10 as the DNA template. These primers amplify the promoter fragment flanked by upstream EcoRI and downstream BamHI sites (underlined sequences). The 185-bp EcoRI-BamHI PCR product, extending from positions −144 to +41, was cloned into pGEM3Z(f) for sequence determination and then into plasmid pRS415 to generate a uhpT-lacZ transcriptional fusion (32). Deletion and insertion mutations were introduced into this promoter region by PCR-based overlap extension mutagenesis using appropriate primers (12). The CAP-binding site in the promoter variants was inactivated by site-directed mutagenesis with an oligonucleotide that introduces seven base substitutions to change the sequence CGTGATGCATCTCACC to CCTAGTGCATCCTAGG. The sequences of primers used to generate all mutations are available upon request. All introduced mutational changes were verified by DNA sequence determination.
Genetic techniques.
Each uhpT promoter derivative cloned as a lacZ fusion in plasmid pRS415 was transferred by homologous recombination to bacteriophage λRZ5, as previously described (22, 27, 32). The resulting uhpT-lacZ-bearing phages were used to isolate single lysogens in the indicated strains by integration in the attλ site.
β-Galactosidase assay.
The activity of β-galactosidase expressed from uhpT-lacZ fusions was measured as previously described (23). All assays were repeated at least three times in duplicate; the standard error was ±10% of the mean value.
DNase footprinting.
DNA was isolated as EcoRI-BamHI fragments released from the respective plasmids and labeled at the 3′ end of the bottom strand by incubation with [α-32P]dATP (1 μCi/ml; 3,000 Ci/mmol; ICN) and Klenow fragment (40 U/ml; Boehringer-Mannheim). Nucleotide precursors were removed by gel filtration through G-50 QuickSpin columns (Boehringer-Mannheim). DNase I footprinting reactions were carried out as previously described (7, 23). The radioactive label in DNA fragments was quantified by using a PhosphorImager and ImageQuant program (Molecular Dynamics, Sunnyvale, Calif.). Band intensities were normalized by reference to nearby bands outside the UhpA-binding regions. Fractional occupancy was calculated as {1 − [(band intensity in the presence of UhpA − background from empty lane)/(band intensity of nearby band − background)]/[(band intensity in the absence of UhpA − background)/(band intensity of nearby band − background)]}.
Proteins and chemicals.
UhpA protein was purified to >95% homogeneity as previously described (5, 23). CAP was purified by cyclic AMP affinity chromatography (36). RNAP was purchased from Amersham-Pharmacia Biotech, Inc. (Piscataway, N.J.). Protein concentrations were determined by the Bradford dye-binding assay (Bio-Rad, Hercules, Calif.) with bovine serum albumin as the standard.
Purified UhpA was phosphorylated as described previously (5) by incubation of 12.5 μM UhpA at 37°C for 1 h with 10 mM acetyl phosphate in buffer D (50 mM Tris-HCl, 6 mM MgCl2, 1 mM dithiothreitol [pH 7.5]). Under these conditions, >80% of UhpA molecules are phosphorylated. Unphosphorylated UhpA was incubated in the same manner but without acetyl phosphate. The samples were used immediately for a footprinting assay.
RESULTS
Effect of UhpA phosphorylation on site occupancy.
To examine the effect of the phosphorylation of UhpA on its binding to DNA sites in the uhpT promoter between positions −80 and −32, DNase I-protection experiments were performed on a uhpT promoter fragment extending from −144 to +41. There are few DNase I-susceptible sites in the A+T-rich region between −80 and −50 (5, 23). UhpA did protect against cleavage at positions −69, −70, −75, and −76, and occupancy of these positions was measured as representative of strong-site binding. Protection from DNase cleavage of the cluster of positions between −47 and −31 was determined as weak-site binding. Comparable results were obtained whether occupancy was measured from the intensity of individual bands or of the ensemble. Fractional occupancy was measured in the presence of increasing concentrations of UhpA or of P-UhpA, prepared by incubation with acetyl phosphate.
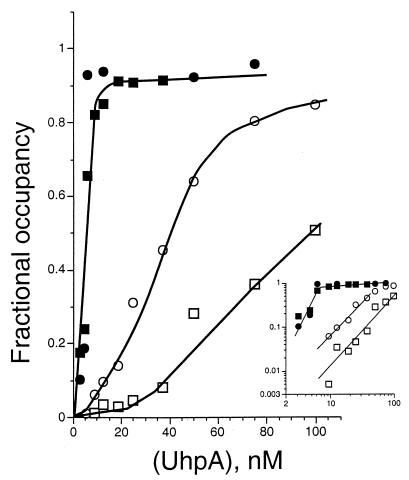
Unphosphorylated UhpA (Fig. 1) showed a substantial difference in affinity for the strong site and the weak sites. Binding activity, especially to the weak sites, exhibited sigmoidal concentration dependence. Half-maximal protection of the strong and weak sites occurred at around 40 and 100 nM UhpA, respectively. Appreciable occupancy of the weak sites was not seen until the strong site was at least half occupied. Phosphorylation of UhpA (Fig. 1) markedly increased its binding to the uhpT promoter, with half-maximal protection of all sites at 5 to 8 nM P-UhpA. However, comparison of the affinities of UhpA and P-UhpA was complicated by the decreased solubility or stability of P-UhpA relative to UhpA. Phosphorylation of UhpA greatly increased site occupancy and affinity, especially of the weak sites, but was not necessary for binding.
FIG. 1.
Effect of the phosphorylation of UhpA on binding to sites in the uhpT promoter. Fractional occupancy represents the intensity of DNase cleavage of bands in the strong (positions −76 to −69, circles) and weak (positions −47 to −31, squares) sites in the uhpT promoter, corrected for background and normalized to the intensity of nearby bands. The UhpA protein was added at the indicated concentrations in its native state (open symbols) or after phosphorylation by incubation with acetyl phosphate (solid symbols). The inset shows the binding data plotted logarithmically.
Promoter variants with altered spacing.
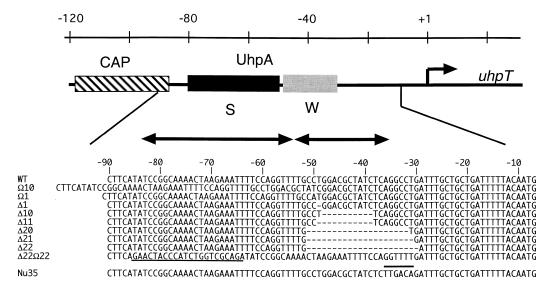
If occupancy of the weak binding sites is necessary for uhpT transcription and if it occurs by oligomerization of UhpA molecules along the DNA from the strong sites, changes in the spacing or helical phasing of the strong and weak sites should affect uhpT promoter function. PCR-based mutagenesis was used to change the spacing between the strong and weak sites. A series of promoter variants carrying insertions or deletions into the weak sites from around position −50 (Fig. 2) were cloned into plasmid pRS415 to create uhpT-lacZ transcriptional fusions, which were then integrated into the chromosome of strain RK1280 as single-copy λRZ5 lysogens. The −50 position was chosen as the boundary for insertions or deletions because it lies between regions that UhpA protects from hydroxyl radical cleavage (5). The β-galactosidase expression of cells carrying the multicopy plasmids or the lysogens was determined in the absence or presence of the inducer Glu6P (Table 1). Expression in the absence of a promoter insert was very low, as expected for this transcription-isolated reporter (32). Uninduced expression from the wild-type single-copy uhpT-lacZ fusion was as low as that from the empty vector and showed substantial induction by Glu6P. The multicopy wild-type fusion exhibited a 4.6-fold increase over the single-copy level, which is less than expected from the increase in gene copy number and probably reflects the limiting amount of chromosome-encoded UhpA activator for its multicopy targets (34).
FIG. 2.
Promoter mutations. The schematic representation of the uhpT promoter shows the location and extent of protein-binding regions along the top. CAP, the CAP-binding site; S, the higher-affinity UhpA-binding region; W, the weak or lower-affinity UhpA-binding region. The sequence changes in the promoter variants used in this study are depicted along the bottom. Deleted residues are indicated by dashes. The 22-base insertion in Δ22 Ω22 is underlined, and the sequence changes to introduce a consensus −35 sequence in the Nu35 variant are indicated by an overbar.
TABLE 1.
Expression of uhpT promoter variants and the effect of presence of UhpA and the CAP-binding site
| uhpTplacZd | β-Galactosidase activity in indicated host strain when uhpT-lacZ fusion is carried one:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Plasmida
|
Lysogenb
|
|||||||||
| RK1280 uhp+
|
RK1271 ΔuhpA
|
RK1280 uhp+
|
RK1271 ΔuhpA
|
RK1280 ΔCAP sitec
|
||||||
| − | + | − | + | − | + | − | + | − | + | |
| None | 2 | 1 | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 1 |
| WTg | 42 | 1,790 | 3 | 3 | 2 | 387 | 1 | 1 | 1 | 31 |
| Ω10 | 5 | 3 | 5 | 4 | 1 | 1 | 1 | 1 | 1 | 1 |
| Ω1 | 2 | 385 | 2 | 7 | 1 | 45 | NDf | ND | 3 | 5 |
| Δ1 | 17 | 145 | 6 | 6 | 1 | 75 | ND | ND | 1 | 1 |
| Δ10 | 5 | 13 | 5 | 4 | 1 | 3 | 1 | 1 | 1 | 1 |
| Δ11 | 21 | 65 | 20 | 22 | 2 | 4 | ND | ND | 1 | 1 |
| Δ20 | 123 | 545 | 96 | 96 | 5 | 37 | 13 | 13 | 2 | 8 |
| Δ21 | 207 | 1,650 | 117 | 121 | 126 | 139 | 19 | 22 | 1 | 3 |
| Δ22 | 232 | 885 | 194 | 200 | 232 | 313 | 85 | 93 | 1 | 1 |
| Δ22 Ω22 | 52 | 70 | 53 | 54 | 22 | 26 | 4 | 4 | ND | ND |
| Nu35 | 4,360 | 4,480 | 4,780 | 5,010 | 1,390 | 1,550 | 1,765 | 1,870 | ND | ND |
The indicated promoter variants were inserted at the EcoRI-BamHI fragment upstream of the promoterless lacZ gene in plasmid pRS415. Plasmids were introduced by transformation into strains RK1280 (uhp+) and RK1271 (ΔuhpA), as indicated.
The uhpT promoter variants were transferred from plasmid pRS415 derivative to λRZ5 by homologous recombination, and phage lysates were used to isolate ampicillin-resistant lysogens in the indicated host strains. Single lysogens were used.
The series of uhpT promoter variants were subjected to a second round of PCR-based mutagenesis in which the CAP recognition sequence was altered by seven base changes.
The sequence changes in the promoter variants are indicated in Fig. 2.
The β-galactosidase activity was measured in triplicate in at least three independent experiments. Cells were grown in the absence (+) or presence (−) of 0.25 mM Glu6P as indicated and assayed in a microplate reader (Molecular Devices, Inc.).
ND, not determined.
WT, wild type.
The promoter variants with altered spacing showed various regulatory responses (Table 1). The Ω10 variant carries a tandem duplication of the 10-bp sequence from −49 to −40, while the Δ10 variant carries a deletion of that same 10-bp segment extending into the weak sites (Fig. 2). Both mutations inactivated promoter function to <1% of the wild-type level. Removal of an 11-bp segment in mutant Δ11 also resulted in an almost complete loss of activity. Insertion or deletion of a single base pair at position −50 in mutants Ω1 and Δ1 resulted in an 80 to 90% decrease in expression. In all these mutants, the expression that occurred required induction by Glu6P. These results suggest that the proper helical phasing of protein-binding sites is not sufficient for promoter function and that spacing may also play a role.
In contrast to the drastic reduction of promoter function upon the insertion or deletion of one DNA-helical turn, considerable uhpT-lacZ expression in the absence or presence of Glu6P was seen upon the deletion of two helical turns in the Δ20, Δ21, or Δ22 variants. These deletions removed the weak UhpA-binding sites and brought the strong sites closer to the RNAP-binding site. The different levels of basal and induced activity in the Δ20, Δ21, and Δ22 variants suggest that the helical phasing between the binding sites for UhpA and RNAP may be important for activation. Examination of the sequence of the deletion variants (Fig. 2) did not reveal the creation of a new −35 element. Primer extension analysis of RNA from Glu6P-induced cells showed that all active promoters used the same transcription site as the wild-type promoter (data not shown).
Somewhat different responses were seen when the promoter variants were present on single- or multigene copy reporters. The uninduced activities of the Δ21 and Δ22 variants present in a single copy were 33 and 60%, respectively, of the induced level from the wild-type promoter and were only modestly increased by Glu6P induction to 36 and 81%. When these promoter variants were carried on the multicopy plasmid pRS415, their basal level expression was three to five times higher than in the wild-type promoter and there was a further 3.8- to 8-fold induction by Glu6P, unlike their single-copy behavior. The relative activities of the mutant promoters showed a different rank order on plasmids than when present in a single copy. Despite these quantitative differences in response to gene dosage, it is clear that the presence of the weak UhpA-binding sites is not required for promoter function when they are replaced by the strong sites.
UhpA-independent transcription.
The marked elevation in basal expression upon removal of the weak sites could result from the proximity of RNAP to unphosphorylated UhpA bound at the strong sites. To test whether expression of the Δ20, Δ21, and Δ22 variants depended on UhpA function, all uhpT-lacZ promoter fusions were transferred to strain RK1271 carrying an in-frame deletion in uhpA. As expected (16), expression from the wild-type uhpT promoter required UhpA (Table 1). The low-level expression in the Ω1 and Δ1 promoter variants was also dependent on UhpA, while the Ω10 and Δ10 promoters remained silent.
In contrast, the Δ20, Δ21, and Δ22 variants exhibited substantial expression but no further induction by Glu6P in the ΔuhpA strain. The level of UhpA-independent expression was lower than the uninduced levels from the same promoters in the UhpA+ strain but was much higher than that conferred by the wild-type promoter in the absence of Glu6P or UhpA (Table 1). These results showed that deletion of the 20- to 22-bp sequence containing the weak sites resulted in a marked increase in both UhpA-dependent and UhpA-independent expression under noninducing conditions. These results are consistent with the premise that occupancy and activation from the strong sites are less dependent on UhpA phosphorylation than is activation from the weak site.
CAP dependence.
To test whether the increased UhpA-independent expression occurred because the deletions brought the CAP-binding site close enough to the promoter to allow direct activation by CAP, the CAP-binding site in each variant promoter was inactivated by changes in seven key residues. Expression was assayed in a single gene copy in the uhp+ strain RK1280 (Table 1). Disruption of the CAP-binding site from the wild-type promoter decreased activity to 8% of the wild-type level, as previously seen in crp mutants or upon deletion of the CAP-binding sequences (21). Expression from all of the variant promoters was very low, indicating that their UhpA-dependent and UhpA-independent activities were highly CAP dependent.
To test whether the greater proximity of the CAP-binding site at position −83.5 in the Δ22 promoter allowed increased UhpA-independent activity, the double mutant Δ22 Ω22 was constructed. This promoter combined the Δ22 deletion with the insertion of a random 22-bp sequence at position −85 between the UhpA site and the CAP site, to return the CAP-binding site to the same position as in the wild-type promoter. The uhpT-lacZ expression in this mutant decreased more than 10-fold relative to the Δ22 variant, and it was further reduced in the absence of UhpA (Table 1). Thus, deletion of the weak UhpA-binding sequences allows the operation of two new modes of transcription activation. First, UhpA-independent activation occurred because the CAP-binding site was close enough to allow CAP to directly activate RNAP. Second, activation by unphosphorylated UhpA occurred because its strong sites are adjacent to the RNAP-binding site; this activation required the proximity of CAP and UhpA on the DNA.
UhpA binding to variant promoters.
The binding of UhpA to the variant promoters was examined by gel electrophoretic mobility shift and DNase footprinting assays. In gel shift assays, all of the variant promoters showed the same behavior as the wild-type fragment with respect to the degree of retardation and the dependence on the amount of UhpA added (data not shown). This result showed that changing the spacing or removal of the weak sites did not interfere with UhpA binding to the strong sites remaining in the uhpT promoter region.
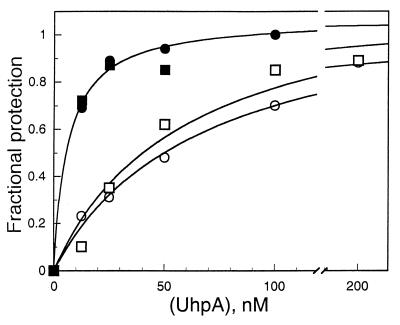
In DNase footprinting experiments, occupancy of the strong-site sequences in the wild-type and Δ22 promoter fragments showed similar dependence on the concentration of UhpA and the same increase in occupancy in response to phosphorylation of UhpA (Fig. 3). Protection of the strong site in all promoter variants showed dependence on P-UhpA concentration comparable to that in the wild-type promoter (data not shown). These results showed that the presence of the weak sites did not affect the binding of UhpA to the strong sites. Protection of the weak sites in the Ω1 and Δ1 variants required higher concentrations of UhpA, i.e., >100 nM, than were needed for the wild-type promoter. There was no detectable protection by 100 nM UhpA of the region downstream of position −50 in any of the promoter variants in which the weak site was deleted, i.e., Δ10, Δ11, Δ20, Δ21, or Δ22. In the Ω10 variant, P-UhpA protects about 20 bp of the weak-site sequences, as well as the strong site. These results showed that binding of UhpA to the weak sites depends on both the proper nucleotide sequence and the orientation relative to the strong sites.
FIG. 3.
DNase protection assay of the binding of UhpA (open symbols) and P-UhpA (closed symbols) to the strong site of the wild-type and Δ22 uhpT promoters. Occupancy of the strong-site residues that are cleaved by DNase I in the wild-type (circles) and Δ22 variant (squares) promoter fragments is shown. The DNase protection assay is carried out as described for Fig. 1.
A −35 element eliminates the need for UhpA.
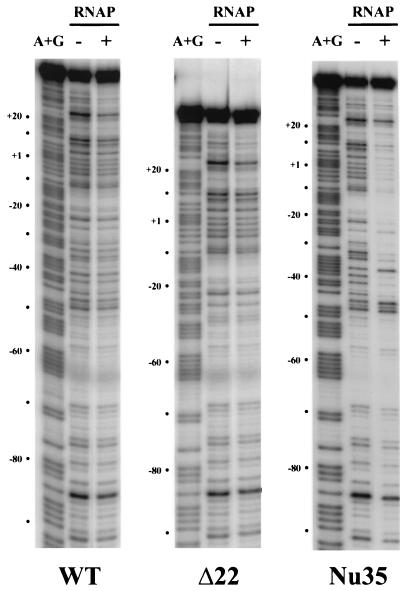
The requirement for UhpA for transcription may result from the absence of an effective −35 element. The Nu35 variant promoter was constructed to introduce a canonical −35 element, TTGACA, 17 bp upstream of the −10 element. Expression of lacZ from this promoter was 2.5 to 4 times higher than that from the Glu6P-induced wild-type promoter and was little affected by the presence of Glu6P (Table 1). The absence of UhpA led to a slight increase in expression. The high activity of the Nu35 promoter variant confirms that the inactivity of the wild-type promoter in the absence of UhpA results from the absence of a −35 element and that UhpA does not interfere with RNAP binding to the Nu35 promoter. RNAP binding to some uhpT promoters was investigated by DNase footprinting experiments (Fig. 4). There was no obvious binding of RNAP to the wild-type or Δ22 promoter, but RNAP protected the Nu35 promoter between positions −45 and +20.
FIG. 4.
DNase footprinting assay of RNAP binding to uhpT promoter variants. Promoter fragments carrying the wild-type (WT), Δ22, and Nu35 sequences in the indicated absence or presence of 50 nM RNAP at 37°C for 30 min, followed by digestion with DNase I for 30 s at 25°C. The lanes marked A+G present the purine cleavage products from Maxam-Gilbert sequencing of the same fragment. The coordinates on the left of each panel are relative to the transcription start site of the wild-type promoter.
DISCUSSION
The classical concept in which a single operator site controls regulated promoters has been revised with the recognition of the presence in many promoters of multiple regulatory protein-binding sites. Multiple binding sites allow some repressors, such as LacI or GalR, to form DNA loops which decrease transcription by restricting the conformational flexibility needed for DNA to melt or wrap around RNAP (3, 28). Many activators have several binding sites whose occupancy contributes to promoter expression, although usually one site is of major importance. Well-studied examples are found among members of the AraC, LysR, and LuxR families, as well as in the OmpR and NarL families of phosphorylation-dependent response regulator proteins. For example, the OmpR protein, which regulates porin gene expression in E. coli, binds with different affinities to three or four sites in the ompC and ompF promoters, respectively (13, 14, 25). Phosphorylation of OmpR results in a marked increase in affinity for all sites (10). Strong and weak binding sites for BvgA, which regulates the expression of several virulence factors in Bordetella pertussis, are present in its target promoters, and occupancy of the weak sites is strongly increased by phosphorylation of BvgA (37). The promoters regulated by OmpR and BvgA are activated in a graded manner in response to the level of the phosphorylated protein. The strong sites are usually upstream of the weak sites and the promoter. Occupancy of the UhpA-binding sites in the uhpT promoter shows behavior similar to that of OmpR and BvgA, namely, different affinities for strong and weak sites, apparently cooperative occupancy of the weak sites, and a marked increase in affinity for all sites upon phosphorylation of UhpA.
This current study was designed to explore how occupancy of the strong and weak sites contributes to transcription activation and whether occupancy of both types of sites is coupled. Previous studies showed that some linker substitutions in either the strong or weak sites strongly decreased promoter activity (22), but it was not known whether these mutations affected the binding of UhpA or RNAP. Other studies suggested that UhpA at the −35 region interacts with the ς70 subunit of the RNAP holoenzyme to enhance RNAP binding and to allow low-level transcription (24). Strong stimulation of transcription requires the interaction of the α-CTD with UhpA and CAP in the −80 region.
The coupling of occupancy of the strong and weak sites was indicated by observations that single base changes in the UhpA-binding region do not strongly affect promoter function and that 4-bp changes on one end of the strong site can affect binding to the other end (T. J. Merkel and I. N. Olekhnovich, unpublished data). Also, the insertion or deletion of a single base pair between the sites resulted in a considerable reduction in promoter activity and an apparent decrease in the affinity of UhpA for the weak sites. Further work to characterize the binding to isolated strong and weak sites is necessary to ensure that these changes did not disrupt an important recognition element for UhpA binding. The insertion or deletion of one DNA-helical turn of the weak site resulted in the almost complete loss of promoter function and of occupancy of the remaining weak-site sequences but had no obvious effect on occupancy of the strong site. In the Ω10 insertion, the binding of UhpA to two helical turns in the weak site occurred normally, but the additional helical turn introduced by the insertion appeared not to be occupied, leaving an empty span between the binding sites for UhpA and for RNAP. These results are interpreted to mean that the weak sites are occupied in units of two DNA-helical turns, suggesting that UhpA binds as a dimer to each pair of sites. The dimeric nature of UhpA binding is indicated by the pattern of DNA cleavage by UhpA molecules carrying a hydroxyl radical-generating moiety near its DNA-binding domain (I. N. Olekhnovich and R. J. Kadner, unpublished data).
The deletion of two helical turns in the Δ20, Δ21, and Δ22 variants replaced the weak site at residues −51 to −30 with strong-site sequences. These deletion variants exhibited a substantial increase in UhpA-independent and constitutive activity. The UhpA-independent expression could be explained by the ability of CAP to activate natural and constructed promoters when it is bound near position −80 (9, 33). This UhpA-independent activity was fully dependent on CAP action and was lost when the CAP-binding site was inactivated or moved back to its normal position. Thus, the UhpA-independent expression resulted from the greater proximity of the CAP site to the promoter.
The two-turn deletion variants also exhibited expression that required UhpA but not induction by Glu6P. This behavior fits the initial premise that the strong sites are at least partially occupied by UhpA in the absence of its phosphorylation, as was suggested from the binding process in vitro. Transcription activation by the phosphorylation-independent binding of UhpA occurs only when the strong sites are near the RNAP-binding region. The level of constitutive expression in some of the deletion variants approached that of the induced wild-type promoter, indicating that UhpA makes contacts with RNAP similar to those of the wild-type promoter. The addition of Glu6P resulted in a further increase in expression when the promoter variants were present in multiple copies but not when they were present in a single copy. This copy number effect can be explained by the competition of the plasmid-borne promoters for the limiting amounts of the chromosome-encoded UhpA protein, as manifested by the limitation in maximal expression by the multicopy wild-type promoter. When present in a single copy, the deletion promoters can be fully occupied by UhpA, so that phosphorylation of UhpA does not increase binding and gene expression noticeably. When the number of promoter copies is increased by plasmid carriage, the promoters on some plasmids are occupied and others are not, but now phosphorylation of UhpA allows the binding of all available UhpA molecules, leading to the further inducibility.
The UhpA-dependent expression in the Δ22 variant was more strongly dependent on CAP than was that in the wild-type promoter, as shown by the almost complete loss of expression upon inactivation of the CAP-binding site. Moreover, the CAP site must be near the UhpA-binding region to activate transcription. Insertion of a random 22-bp sequence between the UhpA-binding region and the CAP-binding region in the Δ22 Ω22 variant reduced promoter activity considerably. In this variant the CAP site is separated from the UhpA sites by three DNA-helical turns rather than by the one turn in the wild-type promoter. We conclude that CAP and UhpA must bind in proximity for either to activate transcription. The α-CTD, which is necessary for substantial activation of uhpT transcription (24), may contact both CAP and UhpA at the −80 region, as suggested by the formation of a DNase-hypersensitive site there when in the presence of all three proteins. Formation of the hypersensitive site at −80 does not occur on the Δ22 Ω22 variant promoter fragment.
RNAP does not form an obvious DNase protection footprint at the wild-type uhpT promoter, although it cooperates with UhpA and CAP to form the DNase-hypersensitive site at position −80. The promoter carrying a canonical −35 element showed high promoter activity, which was unaffected by UhpA. RNAP exhibited a typical footprint at this promoter, and the DNase-hypersensitive site at −80 was apparent in the presence of RNAP + UhpA or RNAP + UhpA + CAP, even though UhpA and CAP had no apparent effect on transcription. The effect of CAP on expression of the Nu35 promoter was not tested, since CAP activation has not been previously described for strong promoters. As shown by its high activity in the presence of an effective −35 region, the inactivity of the uhpT promoter in the absence of UhpA is the result of its weak −35 element and not the consequence of silencing activity, as proposed for the blocking by NarL of the silencing of the nir promoter by FIS (factor for inversion stimulation) and another protein (35). Taken together, these results support the theory that activation of the uhpT promoter occurs by two independent interactions of domains of RNAP with UhpA molecules at opposite ends of the UhpA-binding region.
ACKNOWLEDGMENTS
This work was supported by research grant GM38681 from the National Institute of General Medical Sciences and funds from the University of Virginia.
We are indebted to Igor Olekhnovich for helpful discussions, reagents, and advice.
REFERENCES
- 1.Baikalov I, Schröder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus R P, Dickerson R E. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- 2.Busby S, Ebright R H. Transcription activation at class II CAP-dependent promoters. Mol Microbiol. 1997;23:853–859. doi: 10.1046/j.1365-2958.1997.2771641.x. [DOI] [PubMed] [Google Scholar]
- 3.Choy H, Adhya S. Negative control. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. I. Washington, D.C.: ASM Press; 1996. pp. 1287–1299. [Google Scholar]
- 4.Collado-Vides J, Magasanik B, Gralla J D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahl J L, Wei B Y, Kadner R J. Protein phosphorylation affects binding of the Escherichia coli transcription activator UhpA to the uhpT promoter. J Biol Chem. 1997;272:1910–1919. doi: 10.1074/jbc.272.3.1910. [DOI] [PubMed] [Google Scholar]
- 6.Ebright R H. Transcription activation at class I CAP-dependent promoters. Mol Microbiol. 1993;8:797–802. doi: 10.1111/j.1365-2958.1993.tb01626.x. [DOI] [PubMed] [Google Scholar]
- 7.Galas D, Schmitz A. DNase footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallegos M-T, Schleif R, Bairoch A, Hofmann K, Ramos J L. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaston K, Bell A, Kolb A, Buc H, Busby S. Stringent spacing requirements for transcription activation by CRP. Cell. 1990;62:733–743. doi: 10.1016/0092-8674(90)90118-x. [DOI] [PubMed] [Google Scholar]
- 10.Head C G, Tardy A, Kenney L J. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J Mol Biol. 1998;281:857–870. doi: 10.1006/jmbi.1998.1985. [DOI] [PubMed] [Google Scholar]
- 11.Henikoff S, Wallace J C, Brown J P. Finding protein similarities with nucleotide sequence databases. Methods Enzymol. 1990;183:111–132. doi: 10.1016/0076-6879(90)83009-x. [DOI] [PubMed] [Google Scholar]
- 12.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 13.Huang K-J, Igo M M. Identification of the bases in the regulatory region, which interact with the transcription factor OmpR. J Mol Biol. 1996;262:615–628. doi: 10.1006/jmbi.1996.0540. [DOI] [PubMed] [Google Scholar]
- 14.Huang K-J, Lan C-Y, Igo M M. Phosphorylation stimulates the cooperative DNA-binding properties of the transcription factor OmpR. Proc Natl Acad Sci USA. 1997;94:2828–2832. doi: 10.1073/pnas.94.7.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Island M D, Kadner R J. Interplay between the membrane-associated UhpB and UhpC regulatory proteins. J Bacteriol. 1993;175:5028–5034. doi: 10.1128/jb.175.16.5028-5034.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Island M D, Wei B-Y, Kadner R J. Structure and function of the uhp genes for the sugar phosphate transport system in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1992;174:2754–2762. doi: 10.1128/jb.174.9.2754-2762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadner R J. Expression of the Uhp sugar-phosphate transport system of Escherichia coli. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: ASM Press; 1995. pp. 263–274. [Google Scholar]
- 18.Li J, Kustu S, Stewart V. In vitro interaction of nitrate-responsive regulatory protein NarL with DNA target sequences in the fdnG, narG, narK, and frdA operon control regions of Escherichia coli K-12. J Mol Biol. 1994;241:150–165. doi: 10.1006/jmbi.1994.1485. [DOI] [PubMed] [Google Scholar]
- 19.Maeda S, Mizuno T. Evidence for multiple OmpR-binding sites in the upstream activation sequence of the ompC promoter in Escherichia coli: a single OmpR-binding site is capable of activating the promoter. J Bacteriol. 1990;172:501–503. doi: 10.1128/jb.172.1.501-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maloney P C, Ambudkar S V, Anantharam V, Sonna L A, Varadhachary A. Anion-exchange mechanisms in bacteria. Microbiol Rev. 1990;54:1–17. doi: 10.1128/mr.54.1.1-17.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merkel T J, Dahl J L, Ebright R H, Kadner R J. Transcription activation at the Escherichia coli uhpT promoter by the catabolite gene activator protein. J Bacteriol. 1995;177:1712–1718. doi: 10.1128/jb.177.7.1712-1718.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merkel T J, Nelson D M, Brauer C L, Kadner R J. Promoter elements required for positive control of transcription of the Escherichia coli uhpT gene. J Bacteriol. 1992;174:2763–2770. doi: 10.1128/jb.174.9.2763-2770.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olekhnovich I N, Dahl J L, Kadner R J. Separate contributions of UhpA and CAP to activation of transcription of the uhpT promoter of Escherichia coli. J Mol Biol. 1999;292:973–986. doi: 10.1006/jmbi.1999.3127. [DOI] [PubMed] [Google Scholar]
- 24.Olekhnovich I N, Kadner R J. RNA polymerase α and ς70 subunits participate in transcription of the Escherichia coli uhpT promoter. J Bacteriol. 1999;181:7266–7273. doi: 10.1128/jb.181.23.7266-7273.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rampersaud A, Harlocker S L, Inouye M. The OmpR protein of Escherichia coli binds to sites in the ompF promoter region in a hierarchical manner determined by its degree of phosphorylation. J Biol Chem. 1994;269:12559–12566. [PubMed] [Google Scholar]
- 26.Rampersaud A, Norioka S, Inouye M. Characterization of OmpR binding sequences in the upstream region of the ompF promoter essential for transcriptional activation. J Biol Chem. 1989;264:18693–18700. [PubMed] [Google Scholar]
- 27.Roland K L, Liu C, Turnbough C L. Role of the ribosome in suppressing transcriptional termination at the pyrB1 attenuator of Escherichia coli K12. Proc Natl Acad Sci USA. 1988;85:7149–7153. doi: 10.1073/pnas.85.19.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy S, Garges S, Adhya S. Activation and repression of transcription by differential contact: two sides of a coin. J Biol Chem. 1998;273:14059–14062. doi: 10.1074/jbc.273.23.14059. [DOI] [PubMed] [Google Scholar]
- 29.Saier M H J. Bacterial sensor kinase/response regulator systems. Res Microbiol. 1994;145:349–355. doi: 10.1016/0923-2508(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 30.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 31.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 32.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 33.Ushida C, Aiba H. Helical phase-dependent action of CRP: effect of the distance between the CRP site and the −35 region on promoter activity. Nucleic Acids Res. 1990;18:6325–6330. doi: 10.1093/nar/18.21.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weston L A, Kadner R J. Role of uhp genes in expression of the Escherichia coli sugar-phosphate transport system. J Bacteriol. 1988;170:3375–3383. doi: 10.1128/jb.170.8.3375-3383.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu H, Tyson K L, Cole J A, Busby S J. Regulation of transcription initiation at the Escherichia coli nir operon promoter: a new mechanism to account for codependence on two transcription factors. Mol Microbiol. 1998;27:493–505. doi: 10.1046/j.1365-2958.1998.00699.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, Gunasekera A, Ebright Y, Ebright R H. Derivatives of CAP having no solvent-accessible cysteine residues, or having a unique solvent-accessible cysteine residue at amino acid 2 of the helix-turn-helix motif. J Biomol Struct Dyn. 1991;9:463–473. doi: 10.1080/07391102.1991.10507929. [DOI] [PubMed] [Google Scholar]
- 37.Zu T, Manetti R, Rappuoli R, Scarlato V. Differential binding of BvgA to two classes of virulence genes of Bordetella pertussis directs promoter selectivity by RNA polymerase. Mol Microbiol. 1996;21:557–565. doi: 10.1111/j.1365-2958.1996.tb02564.x. [DOI] [PubMed] [Google Scholar]