Abstract
In Escherichia coli K-12, the major glucose transporter with a central role in carbon catabolite repression and in inducer exclusion is the phosphoenolpyruvate-dependent glucose:phosphotransferase system (PTS). Its membrane-bound subunit, IICBGlc, is encoded by the gene ptsG; its soluble domain, IIAGlc, is encoded by crr, which is a member of the pts operon. The system is inducible by d-glucose and, to a lesser degree, by l-sorbose. The regulation of ptsG transcription was analyzed by testing the induction of IICBGlc transporter activity and of a single-copy Φ(ptsGop-lacZ) fusion. Among mutations found to affect directly ptsG expression were those altering the activity of adenylate cyclase (cyaA), the repressor DgsA (dgsA; also called Mlc), the general PTS proteins enzyme I (ptsI) and histidine carrier protein HPr (ptsH), and the IIAGlc and IIBGlc domains, as well as several authentic and newly isolated UmgC mutations. The latter, originally thought to map in the repressor gene umgC outside the ptsG locus, were found to represent ptsG alleles. These affected invariably the substrate specificity of the IICBGlc domain, thus allowing efficient transport and phosphorylation of substrates normally transported very poorly or not at all by this PTS. Simultaneously, all of these substrates became inducers for ptsG. From the analysis of the mutants, from cis-trans dominance tests, and from the identification of the amino acid residues mutated in the UmgC mutants, a new regulatory mechanism involved in ptsG induction is postulated. According to this model, the phosphorylation state of IIBGlc modulates IICGlc which, directly or indirectly, controls the repressor DgsA and hence ptsG expression. By the same mechanism, glucose uptake and phosphorylation also control the expression of the pts operon and probably of all operons controlled by the repressor DgsA.
In Escherichia coli K-12, d-glucose (Glc) is taken up and concomitantly phosphorylated either by the glucose-specific enzyme II (EII) transporter (IIGlc) or the mannose-specific EII transporter (IIMan) (genes manXYZ) of the phosphoenolpyruvate-dependent carbohydrate phosphotransferase system (PTS) (for reviews, see references 10 and 21). As for most other PTS carbohydrates, the phosphoryl groups are sequentially transferred from PEP through two common intermediates, enzyme I (EI; gene: ptsI) and the phosphohistidine carrier protein (HPr; gene: ptsH), to sugar-specific EII (IICBGlc; see below) and to glucose (for a review see reference 41).
IIGlc consists of two subunits, IIAGlc (crr [catabolite repression resistance]) and membrane-bound IICBGlc (ptsG) (8). The crr gene is part of the ptsHI crr operon (46) separated from the ptsG gene, which maps at 25.0 min (4). IIAGlc is a small hydrophilic protein which has, in addition to its transport function, a central regulatory role in carbon catabolite repression and inducer exclusion (for a review, see reference 22). The IICBGlc subunit is composed of an amino-terminal, hydrophobic IICGlc domain, which largely determines substrate specificity, and a carboxy-terminal, hydrophilic IIBGlc domain, which is phosphorylated at the Cys421 residue (32). The system normally recognizes glucose as well as methyl-α-d-glucoside (αMG), 5-thio-d-glucoside, l-sorbose and, with a low affinity, 2-deoxyglucose (2DG) (for a review, see reference 40).
Only little attention has been paid to the regulation of ptsG expression, although IICBGlc has an outstanding regulatory function in establishing glucose as a favored carbon source. Moreover, for both E. coli (45) and Salmonella enterica serovar Typhimurium (55), it was demonstrated that the activity of IICBGlc is the rate-limiting step in glucose utilization. Both ptsG expression (19, 37, 43) and manXYZ expression (36) are positively regulated by the cyclic AMP (cAMP)-cAMP receptor protein (CrpA) complex and negatively controlled by the DgsA (Mlc) protein. The dgsA locus (deoxyglucose sensitive) at 35.9 min on the E. coli chromosome was discovered as a suppressor mutation that enables ptsG-negative mutants to grow anaerobically on glucose via a constitutively expressed IIMan system and enhanced sensitivity to 2DG, a major substrate of this transport system (44). The DgsA protein was rediscovered recently and renamed Mlc (making large colonies) (16). Plumbridge (36) demonstrated that dgsA and mlc are the same gene; for priority reasons and according to Berlyn (4), we call this gene dgsA. The DgsA protein represses its own synthesis as well as the expression of the ptsHI crr operon (18, 38) and the mal regulon (7). It may represent a novel global repressor and may counteract the global regulator cAMP-CrpA to ensure the expression of those genes, which are linked to glucose metabolism (19, 38, 39, 44). The inducer for DgsA, however, has not been identified.
A different type of ptsG regulatory mutation, called umgC (uptake of αMG control; umg is the former name of ptsG), was described by Jones-Mortimer and Kornberg (17). This mutation, which was claimed to map close to but not in ptsG, enabled E. coli cells with inactive IIMan to grow on mannose (Man) and glucosamine (GlcN or Glm). The authors concluded that mannose and glucosamine are not inducers of the glucose PTS, that the umgC mutation causes constitutive ptsG expression, and that umgC encodes a repressor, UmgC. In this paper, we describe the isolation and characterization of UmgC-like mutants which were selected as described by Jones-Mortimer and Kornberg (17). Moreover, we reinvestigated one of their UmgC mutants and showed that umgC mutations map within the ptsG allele and alter characteristically the ptsG induction pattern and IICBGlc transporter activity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The E. coli K-12 strains and plasmids used in this study are listed in Table 1. LJ110 (Fnr+) was obtained from a cross of K-12(P1) with W3110 and by selection for growth on minimal agar plates supplemented with 0.2% glycerol and 10 mM KNO3 under anaerobic conditions. Cells were routinely grown either in standard phosphate minimal medium (54) supplemented with 0.2% various carbon sources, in Lennox broth without glucose and calcium ions, or in 2xTY medium as described by Ausubel et al. (2). The utilization of various carbohydrates was screened on MacConkey agar plates (Difco) containing 1% of the indicated carbon source. Antibiotics were used at the following concentrations: tetracycline, 10 mg/liter; ampicillin, 50 mg/liter; chloramphenicol, 25 mg/liter; kanamycin, 25 mg/liter; and spectinomycin, 1,500 mg/liter for the multicopy plasmid system or 100 mg/liter in rich medium and 500 mg/liter in minimal medium for the single-copy system. Transductions were carried out with P1 vir essentially as described by Arber (1).
TABLE 1.
E. coli K-12 strains and plasmids used in this studya
| Strain or plasmid | Relevant genotype or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| CAG12078 | F−zce-726::Tn10 | 50 |
| ZSC112LΔG | F− Glk− Δ(ptsG::cat) | 6 |
| ZSC112LΔLPM | F− Glk− Δ(manXYZ::cat) | 12 |
| JWL359 | F− ΔlacU169 zah-735::Tn10 | J. W. Lengeler |
| JM1100 | Hfr man-8 | 13 |
| PS8 | F− ΔlacU169 recA56 | 56 |
| TP2811 | F− Δ(ptsHI crr::kan) | 25 |
| W3110 | F− Fnr− | 15 |
| LJ110 | W3110 Fnr+ | This study |
| LJ120 | Δ(ptsG::cat) | This study |
| LJ121 | Δ(ptsG::cat) man-8 zea-225::Tn10 | This study |
| LJ130 | LJ110 Δ(manXYZ::cat) | This study |
| LJ132 | LJ130 zce-726::Tn10 | This study |
| LJ132-3 | LJ132 ptsG3 | This study |
| LJ333 | LJ132-3 ptsG33 | This study |
| LZ1 | LJ130 ptsG1 | This study |
| LZ22 | LZ1 ptsG22 | This study |
| LZ23 | LZ1 ptsG23 | This study |
| LZ100 | LZ1 ptsG1 ΔlacU169 zah-735::Tn10 | This study |
| LZ110 | LJ110 ptsG+ ΔlacU169 zah-735::Tn10 | This study |
| LZ120 | LZ110 ΔcyaA854 | This study |
| LZ140 | LZ110 Δ(ptsHI crr::kan) | This study |
| LZ150 | LZ110 Δ(ptsG::cat) | This study |
| LZ160 | LZ110 Δ(ptsG::cat) Δ(ptsHI crr::kan) | This study |
| HK727 | F−ptsG727ptsI19(Ts) crr(Ts) manXYZ727 | 35 |
| LZ727 | LJ130 ptsG727zce-726::Tn10 | This study |
| JWL184-1 | F−ptsG184manXYZ184 | 23 |
| JWL184-20 | JWL184-1 Δ(ptsG::cat) | This study |
| JWL184-30 | JWL184-1 Δ(manXYZ::cat) | This study |
| KM563 | F−dgsA::Tn10kan | 7 |
| LJ138 | LJ130 dgsA::Tn10kan | This study |
| LZ138 | LJ138 ptsG+ ΔlacU169 zah-735::Tn10 | This study |
| LZ139 | LZ138 Δ(ptsG::cat) | This study |
| LZ170 | LZ1 dgsA::Tn10kan | This study |
| Plasmids | ||
| pSU19 | Cmr | 27 |
| pPSO110 | tnpA+ Cmr | 54 |
| pJBH | Apr | 6 |
| pJCH | Apr | 6 |
| pTM30 | Apr | 29 |
| pTM110 | AprptsG+ | This study |
| pTM1 | AprptsG1 | This study |
| pTM32-8 | AprptsG3 | This study |
| pTM727 | AprptsG727 | This study |
| pTM184-1 | AprptsG184 | This study |
| pSTJ30Δ320 | Apr ΔptsG320 | This study |
| pSTJ30Δ328 | Apr ΔptsG328 | This study |
| pSTJ30Δ396 | Apr ΔptsG396 | This study |
| pSTJ30Δ436 | Apr ΔptsG436 | This study |
| pSTJ30Δ459 | Apr ΔptsG459 | This study |
| pTIM101 | Apr Scr | This study |
| pTIM103 | Apr Φ(ptsGop-lacZ) | This study |
| pTIM104 | Apr Φ(ptsHop-lacZ) | This study |
| F′8 (gal+) | gal+ | 11 |
| F′8::Tn Φ(ptsGop-lacZ) | Scr | This study |
| F′8::Tn Φ(ptsHop-lacZ) | Scr | This study |
The nomenclature for the pts genes is that of Postma et al. (40); the nomenclature for other genes is that of Berlyn (4). For clarity, ptsG or manXYZ mutations in the manA locus from different strains were named according to their source (subscript numbers). The dgsA (2DG sensitivity) gene encodes the repressor DgsA or Mlc (making large colonies).
Construction of single-copy Φ(ptsGop-lacZ) and Φ(ptsHp-lacZ) fusions.
For the construction of a single-copy Φ(ptsGop-lacZ) fusion, the promoter-operator region was amplified from genomic DNA of LJ110 by PCR using the oligonucleotides ptsG4 (5′-AATCAACCTGCGATGGTTCC-3′; hybridizing to bp −337 to −318 upstream of the ptsG start codon) and ptsG3 (5′-AATACCTGCGATAGGCAGTACGGATACCGG-3′, hybridizing to codons 19 to 28 for IICBGlc). The product was treated with Klenow DNA polymerase to produce blunt ends and was inserted into the EcoRV restriction site of pTIM101. Vector pTIM101 essentially is a derivative of plasmid pIC-19H (26) in which the multiple cloning site was deleted. It carries a truncated Tn1721 transposon (52) which consists of the inverted repeat regions, the multiple cloning site of pBluescript II SK(+) (HaeII box) (2), and the so-called Ω element of plasmid pHP45Ω (42). The Ω element provides transcriptional and translational stop signals to prevent read-through from any potential upstream promoter and an spc gene for spectinomycin and streptomycin resistance. The orientation of the PCR insert was controlled by DNA sequencing. Downstream of the ptsG promoter region, the promoterless lacZ gene from plasmid pRU869 (53) was inserted into the HindIII/SalI restriction sites to produce plasmid pTIM103.
For the construction of a single-copy Φ(ptsHp-lacZ) fusion, the promoter-operator region was amplified from genomic DNA of LJ110 by PCR using the oligonucleotides pts1 (5′-GATCTCTTCACTGAGAAAGAATTGC-3′, hybridizing to codons 313 to 321 of CysK) and pts2 (5′-ACATTGTATTTCCCCAACTTATAGG-3′, hybridizing to 21 bp upstream of ptsH and codon 1 of HPr). The 420-bp fragment was treated with Klenow DNA polymerase to produce blunt ends and was inserted into the EcoRV restriction site of pTIM101. The orientation of the PCR insert was controlled by DNA sequencing. Downstream of the ptsH promoter region, the promoterless lacZ gene from plasmid pRU869 was inserted into the HindIII/SalI restriction sites to produce plasmid pTIM104.
PS8/F′8 (gal+) (11) was transformed with plasmids pTIM103 and pTIM104. To allow transposition onto the F′8 plasmid, cells were transformed with plasmid pPSO110 (54), which carries the gene for the Tn1721 transposase. F′ plasmids containing the Φ(ptsGop-lacZ) or Φ(ptsHp-lacZ) transcriptional fusions on the truncated, artificial transposon were transferred into appropriate test strains. Loss of mobilized pTIM103 or pTIM104 was controlled by simultaneous loss of Apr.
Isolation of plasmid DNA, restriction analysis, and cloning procedures.
All manipulations with recombinant DNA were carried out using standard procedures as described previously (2). Plasmid DNA was prepared either by using standard phenol extraction protocols as described previously (48) or by using the JETstar DNA purification system (Genomed, Bad Oeynhausen, Germany). Restriction enzymes were purchased from New England Biolabs (Schwalbach, Germany). They were used according to the recommendations of the supplier. Oligonucleotides for sequencing or PCR were purchased from Interactiva (Ulm, Germany).
Mutation analysis.
DNA amplification of the ptsG alleles was done as described by Saiki et al. (47) using Taq DNA polymerase from Roche Diagnostics, Mannheim, Germany, or Goldstar polymerase from Eurogentec, Seraing, Belgium. The forward PCR primer ptsG+ (5′-AACTGCAGGTGTTTAAGAATGCATTTGCT-3′) for the amplification of ptsG contained an engineered PstI restriction site (underlined) upstream of an artificial GTG start codon (boldface; the original ATG was changed to GTG to lower the levels of expression of ptsG after subcloning into pSU19 [see below]). The reverse PCR primer ptsG− (5′-CTTAAAGCTTAGTGGTTACGGATGTA-3′) introduced an engineered HindIII restriction site (underlined) immediately downstream of the TAA stop codon (Fig. 1). The reaction profile consisted of 32 cycles of denaturing at 94°C for 1 s, annealing at 50°C for 1 s, and extension at 72°C for 45 s in an Air Thermo-Cycler 1605 from Idaho Technology Inc., Idaho Falls, Idaho. PCR products were directly purified using a Wizard PCR Preps DNA purification system (Promega Corp., Mannheim, Germany). All DNA sequencing reactions were performed by the dideoxy chain termination method (2) using an ALFexpress AutoRead or dATP labeling mix sequencing kit from Amersham-Pharmacia Biotech, Freiburg, Germany. The nucleotide sequences of both strands were determined after subcloning into vector pSU19 (27) using 5′ cyanine fluorescent dye (Cy-5)-labeled universal and reverse primers or unlabeled internal ptsG sequencing oligonucleotides priming about every 250 bp within the gene. Computer analysis was done with DNASIS sequencing analysis software (Hitachi) and by using the BLAST programs and database services provided by the National Center for Biotechnology Information, Bethesda, Md.
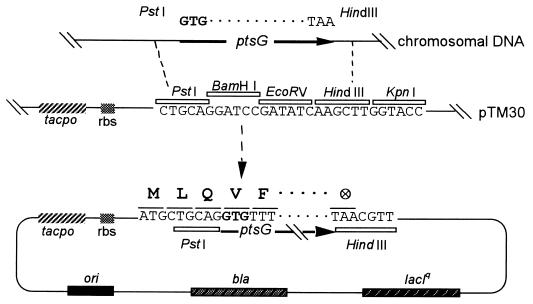
FIG. 1.
Construction of ptsG expression plasmids. The ptsG alleles from various strains were amplified by PCR as described in Materials and Methods, introducing artificial PstI (upstream) and HindIII (downstream) restriction sites. The original ATG start codon of ptsG was changed to GTG (boldface). Fragments were subcloned into pTM30 cut with PstI and HindIII in frame with an artificial ATG start codon provided by the expression vector. All constructs provided three additional, amino-terminal amino acid residues. The expression vector also provided a strictly regulated tac promoter-operator (tacpo), a lacIq gene, and a ribosome binding site (rbs) in an optimal position with respect to the start codon.
Construction of defined carboxy-terminal deletions in IICBGlc.
Plasmid pSTJ30Δ320 is a derivative of pTM110 which was digested with restriction enzymes StuI (bp 957 to 963 in ptsG) and HindIII (in the polylinker region downstream of ptsG) (Fig. 1). DNA was treated with Klenow DNA polymerase to generate blunt ends and was religated. The open reading frame encoded a protein which consisted of up to amino acid 320 of IICBGlc and six additional, plasmid-encoded amino acids (AWYLTN). All other pSTJ30 deletion plasmids were generated using pTM110 as a template, the ptsG+ primer as an upstream primer, and an appropriate downstream primer in a PCR. The following reverse PCR primers were used for the different deletion plasmids: for pSTJ30Δ396, ptsG21 (5′-CCTGAAGCTTTTGCATCTTCAGTCG-3′); for pSTJ30Δ436, ptsG22 (5′-TGATAAGCTTTAGACACATCAGCAA-3′); and for pSTJ30Δ459, ptsG23 (5′-GAAAAGCTTCTGAACACCAGAACC-3′). All primers created an artificial HindIII restriction site (underlined). The last number in each plasmid name indicates the last original PtsG amino acid encoded by the particular construct. PCR fragments were treated with HindIII and PstI and cloned into pTM30. All plasmid constructs were confirmed by DNA sequencing. The addition of at least 50 μM isopropyl-β-d-thiogalactopyranoside (IPTG) to cells carrying these deletion plasmids led to complete growth inhibition, indicating that truncated proteins were expressed from these plasmids and, like wild-type PtsG, were lethal for the cells (data not shown).
Transport and enzyme assays.
Transport of α-d-[methyl-14C]glucopyranoside (final concentration, 25 μM) was measured in exponentially grown cells as described previously (49). Samples were taken after 10, 20, and 30 s. Transport activities were calculated from the initial uptake rates. The β-galactosidase assay was performed as described by Pardee and Prestige (34). All enzyme activities are in nanomoles per milligram of protein per minute.
RESULTS
Isolation and characterization of UmgC mutants of E. coli.
Our isogenic E. coli derivatives which lack the IIMan transporter due to a mutation in the manA locus (genes manXYZ) (9) are unable to utilize d-glucosamine (GlcN or Glm) or d-mannose (Man) as a sole carbon source (Table 2). Cells of the ManA− mutants LJ130 and LJ132 had to be incubated for 3 days at 37°C on minimal glucosamine plates before small colonies appeared. One hundred of these were isolated, purified, and tested for their ability to grow on various carbohydrates. The majority (80%) of the isolated colonies had a pleiotropic phenotype, i.e., they became Glm+ Man+ and at the same time became sensitive to d-arabinitol (Atls) and ribitol (Rtls). Each of these four carbohydrates has been discussed to be a minor gratuitous substrate for IICBGlc, and transport occurs only when the transporter is expressed constitutively (reviewed in references 39 and 40). No degradation pathways for Atl-5-phosphate and Rtl-5-phosphate are present in E. coli K-12, thus explaining the sensitivity (14). Two mutants, LZ1 and LJ132-3, derived from LJ130 and LJ132, respectively, were further characterized, compared to an authentic UmgC mutant (HK727), and used to map the new mutations (Table 2).
TABLE 2.
Phenotypes of various ptsG mutants from E. coli K-12 in the presence of different carbon sourcesa
| Strain | Relevant mutation(s) | Phenotype in the presence of the following carbon source:
|
||||
|---|---|---|---|---|---|---|
| Glc | GlcN | Man | Rtl | Atl | ||
| LJ110 | ptsG+ manXYZ+ | + (70) | + (126) | + (120) | r | r |
| LJ130 or LJ132 | ptsG+ Δ(manXYZ::cat) | + (70) | − | − | r | r |
| LJ120 | Δ(ptsG::cat) manXYZ+ | + (125) | + (126) | + (120) | r | r |
| LZ1 | ptsG1 Δ(manXYZ::cat) | + (72) | + (280) | + (144) | s | s |
| LJ132-3 | ptsG3 Δ(manXYZ::cat) | + (72) | + (210) | + (125) | s | s |
| LJ138 | ptsG+ Δ(manXYZ::cat) dgsA::Tn10kan | + (72) | − | − | r | r |
| HK727 (at 30°C) | ptsG727manXYZ727ptsI19(Ts) crr727(Ts) | + | + | + | s | s |
| HK727 (at 42°C) | − | − | − | r | r | |
| LZ727 | ptsG727 Δ(manXYZ::cat) pts+ crr+ | + | + | + | s | s |
| JWL184-1 | ptsG184manXYZ184 | + | − | + | r | r |
| JWL184-20 | Δ(ptsG::cat) | − | − | − | r | r |
| JWL184-30 | Δ(manXYZ::cat) | + | − | + | r | r |
Growth was tested on MacConkey indicator plates with 1% glucose (Glc), ribitol (Rtl), and d-arabinitol (Atl) or 0.5% mannose (Man) and on minimal agar plates with 0.2% glucosamine (GlcN). Reactions were classified as growth (+) and no growth (−) on minimal agar plates, as red (+, good fermentation) and colorless (−, no fermentation) colonies on MacConkey-glucose or MacConkey-mannose plates, and as sensitive (s) and resistant (r) colonies on MacConkey-ribitol or MacConkey-arabinitol plates. Generation times (in parentheses) determined in minimal medium with 0.2% carbohydrate are given in minutes. All cells were grown at 37°C except for HK727 cells, which were grown at 30 or 42°C as indicated.
Mapping by P1 transduction for both LZ1 and LJ132-3 placed the new mutations close to zce-726::Tn10 (95% coupling to ptsG) and also close to or in ptsG (data not shown). Strain HK727 carries an authentic UmgC mutation, a ManA− allele, the ptsI19(Ts) allele, and a crr(Ts) mutation, which prevent growth on PTS carbohydrates at temperatures above 30°C (30, 35). As expected for a strain lacking IIMan but carrying the UmgC mutation, HK727 is able to grow on GlcN plates; like LZ1 and LJ132-3, it is also Man+ Atls Rtls. When incubation is done at 42°C, this phenotype changes to Glm− Man− Atlr Rtlr, indicating that cells are sensitive to the two pentitols only in the presence of a functional PTS. To differentiate between PTS- and UmgC-dependent effects, the ptsG727 allele from HK727 was transduced into LJ130 after the zce-727::Tn10 cassette from CAG12078 was transferred into HK727. The majority (76%) of Tetr transductants of LJ130 (e.g., LZ727 in Table 2) exhibited a temperature-resistant Pts+ phenotype and were Glm+ Man+ Atls Rtls, i.e., the UmgC phenotype. According to these mapping data, mutants LZ1 and LJ132-3 thus seem to correspond to authentic UmgC mutants. Any further attempts to uncouple the ptsG and umgC markers in LJ132-3, LZ1-2, or LZ727 failed (data not shown). Thus, a mutation in ptsG or a mutation very close to it seems to be responsible for GlcN, Man, Rtl, and Atl uptake in the UmgC mutants.
Exact growth rates were determined for the various wild-type and mutant strains on different carbon sources (Table 2). Strains included a derivative of LJ130 carrying a defined dgsA::Tn10kan mutation from KM563. This mutant, LJ138, did not show a UmgC phenotype, although it expressed ptsG in a constitutive way (see below). Thus, a change in the substrate specificity of IICBGlc rather than constitutive expression of ptsG was responsible for the characteristic change in UmgC mutants. The generation times for LJ110 and isogenic derivatives on glycerol (90 to 96 min) and on glucose (72 to 75 min) were almost equal, except for the DgsA− mutant, which had a generation time of 115 min on glycerol. This result seems to corroborate the hypothesis that DgsA is a global rather than a glucose-specific regulator (7, 38, 39). While growth on mannose was relatively similar for a ManA+ strain and a UmgC mutant, growth of the latter on glucosamine was retarded (210 and 280 min compared to 126 min). Finally, the addition of 1% ribitol or arabinitol to cells of LZ1 or LJ132-3 growing exponentially on glycerol caused growth inhibition after three cell division cycles, while mutant but not wild-type cells preinduced with glucose stopped growing almost immediately (data not shown). The induction of ptsG by the two pentitols in the mutants but not in the wild type was responsible for this effect (see below).
Induction of ptsG expression in various isogenic mutants.
To test the ptsG expression levels, various uninduced and induced strains were tested for αMG uptake (Table 3). IICBGlc activity in the wild type (LJ130) was induced about threefold by glucose, while LJ138 exhibited the expected constitutive transport activity. LZ1 and LJ132-3 showed increased and decreased uninduced transport activities, respectively. Both strains could be induced by the addition of glucose but, interestingly and in contrast to LJ130, also by the addition of glucosamine, mannose, or ribitol. The induced UmgC mutant LZ1 always exhibited higher transport activity than LJ130, thus resembling the DgsA− mutant LJ138. Introduction of the dgsA::Tn10kan mutation into LZ1 (producing strain LZ170) led to fully constitutive uptake activity which resembled the fully constitutive ptsG expression of LZ727 and that of other authentic UmgC strains, e.g., JM1110 (17). Interestingly, the induction levels in the wild type and the mutants LZ1 and LJ132-3 were always lower than those in the fully constitutive mutants LZ170 and LZ727.
TABLE 3.
Induction by the uptake of αMG in isogenic strainsa
| Strain | Relevant mutation(s) | αMG uptake (nmol mg−1 min−1) in the presence of the following inducer:
|
||||
|---|---|---|---|---|---|---|
| None | Glc | GlcN | Man | Rtl | ||
| LJ130 | Δ(manXYZ::cat) | 2.3 | 6.4 | 2.1 | 2.1 | 2.1 |
| LJ138 | dgsA::Tn10kan | 9.8 | 5.0 | 9.3 | 7.7 | 8.4 |
| LZ1 | ptsG1 | 6.5 | 12.1 | 16.6 | 11.4 | 12.9 |
| LZ170 | ptsG1dgsA::Tn10kan | 15.8 | 15.1 | 19.2 | 15.8 | 20.3 |
| LJ132-3 | ptsG3 | 1.1 | 6.4 | 6.6 | 6.2 | 5.2 |
| LZ727 | ptsG727 | 22.6 | 20.1 | 22.5 | 21.4 | 18.5 |
Cells were grown in liquid minimal medium with 0.2% glycerol plus a second carbohydrate (0.2%) as indicated and harvested during exponential growth. The uptake of αMG was tested at 25 μM (final concentration). The mean values of at least three measurements are given.
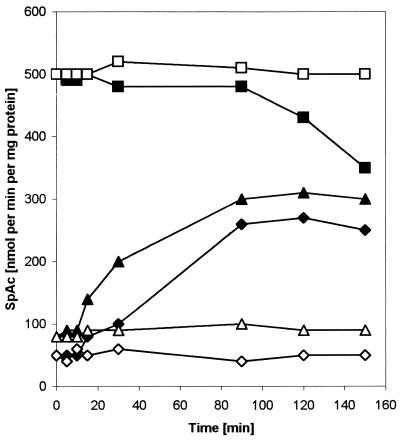
The inducibility of ptsG expression in LZ1 was confirmed both by measuring the mRNA levels in cells growing either on glycerol or glucose (data not shown) and by monitoring the ptsG induction of both wild-type LZ110 and mutants LZ100 and LZ138 by use of the single-copy Φ(ptsGop-lacZ) fusion. At 15 min after the addition of glucose, in both wild-type LZ110 and ptsG1 mutant LZ100, β-galactosidase activities started to increase, reaching a maximum at about 120 min after induction (Fig. 2). β-Galactosidase activities slowly decreased at the end of the exponential growth phase (data not shown). Basal LacZ activity was slightly higher in LZ100 but could be induced threefold, as in the wild-type strain. In the DgsA− mutant LZ138, maximum enzyme activity was high compared to those in LZ110 and LZ100; the addition of glucose caused a decrease in β-galactosidase activity probably as a consequence of a reduction in the intracellular cAMP level. The Φ(ptsGop-lacZ) fusion was also used to test for induction by substrates other than glucose (Table 4). Induction in the wild type occurred only after the addition of glucose, whereas ptsG expression in LZ100 was also induced by glucose, glucosamine, mannose, ribitol, d-fructose (Fru), and d-mannitol (Mtl) but not by the non-PTS carbohydrate l-arabinose (Ara).
FIG. 2.
Kinetics of induction of Φ(ptsGop-lacZ) in strains LZ110 (wild type), LZ100 (ptsG1), and LZ138 (dgsA::kan). Cells harboring a single copy of the Φ(ptsGop-lacZ) translational fusion on F′8 were pregrown overnight in minimal medium with 0.2% glycerol and used to inoculate fresh medium with 0.2% glycerol. Glucose (0.2%) was added in the early exponential growth phase at 0 min. Samples were harvested and analyzed for β-galactosidase activity (SpAc). Symbols: ◊, uninduced LZ110; ⧫, induced LZ110; ▵, uninduced LZ100; ▴, induced LZ100; □, uninduced LZ138; ■, induced LZ138.
TABLE 4.
Induction tested by Φ(ptsGop-lacZ) expression in various isogenic mutantsa
| Strain | Relevant chromosomal mutation(s) | β-Galactosidase activity (nmol mg−1 min−1) in the presence of the following inducer:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| None | Glc | GlcN | Man | Rtl | Mtl | Fru | Ara | ||
| LZ110 | ptsG+ | 52 | 265 | 49 | 51 | 53 | 48 | 61 | 56 |
| LZ100 | ptsG1 | 103 | 298 | 305 | 271 | 310 | 261 | 266 | 60 |
| LZ120 | ΔcyaA854 | 55 | 51 | 49 | 56 | 60 | ND | ND | ND |
| LZ150 | Δ(ptsG::cat) | 48 | 52 | 47 | 50 | 50 | ND | ND | ND |
| LZ140 | Δ(ptsHI crr::kan) | 295 | 280 | 278 | 302 | 290 | ND | ND | ND |
| LZ160 | Δ(ptsG::cat) Δ(ptsHI crr::kan) | 30 | 29 | 27 | 32 | 25 | ND | ND | ND |
| LZ138 | Δ(dgsA::kan) | 549 | 420 | 532 | 511 | 530 | ND | ND | ND |
| LZ139 | Δ(ptsG::cat) Δ(dgsA::kan) | 450 | 450 | ND | ND | ND | ND | ND | ND |
Various mutants derived from LJ110 (ptsG+ manXYZ+) and all carrying Φ(ptsGop-lacZ) on F′8 were tested for ptsG expression as transcribed from the wild-type ptsG promoter-operator. Cells were grown in minimal medium with 0.2% glycerol plus a second carbohydrate (0.2%) as indicated and harvested during exponential growth. The mean values of at least three measurements of β-galactosidase activities are given. ND, not determined.
The fusions were also used to test in our isogenic mutants, all derived from E. coli W3110, the influence of defined mutations known to affect ptsG expression (19, 37). The ΔcyaA854 mutant LZ120 and the Δ(ptsG::cat) mutant LZ150 had low uninduced levels of β-galactosidase activities that could not be induced by any of the tested carbohydrates. This result implies that the induction of ptsG depends on the presence of cAMP-CrpA and of IICBGlc, even though glucose can be transported by the IIMan system in a Δ(ptsG::cat) mutant, as shown by the ability to grow on glucose (Table 2). Introduction of a defined Δ(ptsHI crr::kan) mutation led to complete derepression of ptsG expression in LZ140, whereas the double mutant LZ160 [Δ(ptsHI crr::kan) Δ(ptsG::cat)] exhibited β-galactosidase activity that was below even the basal expression level in LZ150. Thus, intracellular glucose is not sufficient and the presence of nonphosphorylated IICBGlc is required for ptsG induction. Strains carrying the Δ(dgsA::kan) mutation exhibited constitutive β-galactosidase activity independently of the presence (LZ138) or absence (LZ139) of ptsG. These results confirm and extend data obtained by Plumbridge (38), who used strain JM101 and derivatives thereof for similar experiments (see Discussion).
Subcloning and sequencing of ptsG alleles from various IICBGlc mutants.
All carbohydrates (Table 4) that induce ptsG expression in LZ1 have been postulated to be substrates of either wild type (40, 41) or mutant (3) IICBGlc. We speculated that a mutation in this strain and in the other mutants changing the induction and substrate specificity of IICBGlc and not overexpression of the transporter per se was responsible for the UmgC phenotype. This hypothesis implied that there may be no distinct gene for a UmgC repressor protein, thus explaining our failure to detect a umgC gene in the vicinity of ptsG. To test this hypothesis, the ptsG alleles from all the mutants were amplified by a PCR. Additionally, strain JWL184-1, which was described as being semiconstitutive for ptsG expression (23), was included in this approach. It exhibited a strong Glc+ phenotype (55-min generation time) and weak Man+ and Glm− Atlr Rtlr phenotypes. The introduction of defined ptsG or manXYZ mutations revealed that JWL184-1 carries a thus-far-unidentified manXYZ mutation and perhaps a umgC-like mutation in ptsG (Table 2).
Sequencing analysis showed that each mutant carried a single-base-pair substitution which caused an amino acid exchange (Table 5). Interestingly, the same Ser169 is replaced either by a Phe residue in LZ1 or by a Pro residue in LJ132-3. The mutation Glu387Gly in the HK727 ptsG allele is located in the putative linker region between the IIC and IIB domains of the glucose permease (Fig. 3), whereas the mutation Phe195Leu in JWL184-1 appears to be located in putative transmembrane helix 6 of the IIC domain (5).
TABLE 5.
Influence of various ptsG alleles on ptsG induction as measured in LZ150 by the F′8::Tn Φ(ptsGop-lacZ) test systema
| Plasmid | Allele | Mutation | Amino acid exchange | β-Galactosidase activity (nmol mg−1 min−1) in the presence of the following inducer:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| None | Glc | GlcN | Man | Rtl | Mtl | Fru | Ara | ||||
| pTM30 | Control | 70 | 70 | 77 | 80 | 88 | 89 | 99 | 90 | ||
| pTM110 | ptsG+ | 75 | 202 | 62 | 125 | 83 | 74 | 99 | 98 | ||
| pTM1 | ptsG1 | TCC→TTC | S169F | 72 | 212 | 298 | 297 | 175 | 199 | 166 | 101 |
| pTM32-3 | ptsG3 | TCC→CCC | S169P | 75 | 206 | 277 | 258 | 194 | 165 | 175 | 111 |
| pTM727 | ptsG727 | GAA→GGA | E387G | 492 | 226 | 528 | 398 | 537 | 317 | 414 | 406 |
| pTM184 | ptsG184 | TTT→CTT | F195L | 67 | 262 | 92 | 177 | 58 | 104 | 110 | 80 |
LZ150 Δ(ptsG::cat) man+/F′8::Tn Φ(ptsGop-lacZ) harboring various plasmids was grown in liquid minimal medium with 0.2% glycerol, 0.1% Casamino Acids, and 50 mg of ampicillin per liter. For induction, a second carbohydrate (0.2%) was added, and cells were harvested during exponential growth. The mean values of at least three measurements of β-galactosidase activities are given.
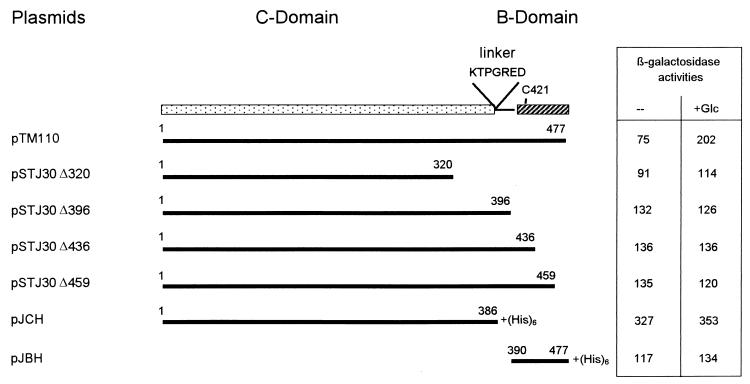
FIG. 3.
Construction of carboxy-terminal deletions of IICBGlc and testing of trans-activation of a Φ(ptsGop-lacZ) fusion. The functional domains of IICBGlc are indicated as follows: the amino-terminal, hydrophobic IICGlc domain is shown as a stippled box; the carboxy-terminal, hydrophilic IIBGlc domain with the phosphorylated Cys421 residue is shown as a hatched box; and the linker with the conserved 382-KTPGRED-388 motif is shown as a thin line. Deletion plasmids are shown as thick lines, and the associated numbers refer to the first and the last original amino acids of the truncated IICBGlc protein. Plasmids pJCH and pJBH (kindly provided by B. Erni) encode six additional His residues. LZ150/F′8::Tn Φ(ptsGop-lacZ) harboring various plasmids was grown in liquid minimal medium with 0.2% glycerol, 0.1% Casamino Acids, and 10 mg of ampicillin per liter. Glucose (0.2%) was added after one cell division cycle where indicated (+Glc). Cells were harvested during exponential growth. β-Galactosidase activities are given as nanomoles of protein per milligram per minute. The mean values of at least two measurements are given.
For a complementation analysis of the various ptsG alleles, the genes were cloned and expressed as PstI/HindIII fragments in pTM30 (Fig. 1). The IICBGlc quantities expressed from these plasmid constructs without induction were as high as those in induced wild-type LJ110, as indicated by Western blot analysis with a polyclonal antiserum against the IIBGlc domain (kindly provided by B. Erni). The addition of at least 50 μM IPTG led to complete growth inhibition (data not shown).
The various ptsG alleles were able to complement the ptsG manXYZ double mutant LJ121 to a positive phenotype on MacConkey-Glc plates. However, only the mutated IICBGlc transporters expressed from pTM1, pTM32-3, and pTM727 produced the typical UmgC phenotype shown by their parent strains LZ1, LJ132-3, and LZ727 (Table 2) and were transdominant over ptsG+, e.g., in LJ130 (data not shown). LJ121 harboring pTM184 (from JWL184-1) became Glc+ and Man+ but remained Glm− Atlr Rtlr. These results were further confirmed by measuring the pattern of induction of Φ(ptsGop-lacZ) expression in LZ150/F′8::Tn Φ(ptsGop-lacZ) carrying the various ptsG alleles on pTM30. In the presence of the wild-type allele on pTM110, only glucose (2.5-fold) and perhaps mannose (1.7-fold) produced induction (Table 5). The mutated transporters expressed from pTM1 and pTM32-3 also caused inducibility by glucosamine, mannose, ribitol, mannitol, and fructose but not by the non-PTS carbohydrate arabinose, whereas cells harboring pTM184 were significantly induced by glucose (4-fold) and mannose (2.6-fold). Moreover, by expression of the original UmgC mutant allele from pTM727, constitutive β-galactosidase activity was observed.
These results corroborate the hypothesis that altered IICBGlc from LZ1, LJ132-3, and HK727 alone is responsible for the typical UmgC phenotype and that no additional mutation is involved. JWL184-1 carries a different type of ptsG mutation. The substrate specificity of this mutated IICBGlc is less relaxed than that of the transporters from the other mutants in that it results in only enhanced mannose transport. It is important to note that the two newly isolated mutations from LZ1 and LJ132-3 and the one from JWL184-1 differ from the HK727 ptsG allele in that they do not cause transdominant constitutive ptsG expression.
Rtlr derivatives of LZ1 and LJ132-3 were isolated on MacConkey agar plates with 1% ribitol. The majority (93%) of these either had ptsG completely deleted or inactivated by IS10 insertions or carried other mutations in ptsG (Glc− Man− Glm− phenotype), as revealed by Southern hybridization and DNA sequencing analysis (data not shown), confirming that IICBGlc is the only transport system for these carbohydrates in our isogenic strains. Others (4%) had a pleiotropic negative phenotype for PTS carbohydrates, resembling PtsI− and PtsH− mutants (data not shown). Inactivation of ptsHI crr probably occurred less frequently than inactivation of ptsG because of the reduced viability of mutants with a lesion in general PTS proteins.
Three Rtlr derivatives, LZ22 and LZ23 from LZ1 and LJ333 from LJ132-3, with a Glc+ Man− Glm− phenotype, were further characterized. DNA sequencing analysis of both ptsG promoter-operator regions and reading frames revealed that in each case, the original mutation was still present. However, LZ22 showed one base-pair substitution, a T-to-G transversion (boldface), in the previously identified −10 region (TTTACTCT to TTGACTCT) (19, 37); this change most likely leads to a strong reduction in ptsG expression. The ptsG allele of LZ23 carried an additional Thr246Ser amino acid exchange, whereas LJ333 had additional Ala82Pro and Val83Ile exchanges in IICBGlc. These additional mutations apparently reduce transport activity.
Effects of plasmid-encoded and truncated IICBGlc on the level of expression of a Φ(ptsGop-lacZ) fusion in a Δ(ptsG::cat) strain.
Deletion of ptsG completely prevented the induction of a Φ(ptsGop-lacZ) fusion by the addition of glucose, even in a Man+ strain, or any other tested carbohydrate (Table 4). We previously reported (T. Zeppenfeld, C. Larisch, J. W. Lengeler, and K. Jahreis, Abstr. Int. Meet. Fachgr. Biochem. GDCh and SF431, p. 68, 1999) that IICGlc expression in the presence of pJCH alone (6), which encodes the IICGlc domain and parts of the linker region (amino acids 1 to 386 plus six additional His residues), resulted in strong constitutive β-galactosidase activity (Fig. 3) in LZ150/F′8::Tn Φ(ptsGop-lacZ). The truncated protein was not capable of transporting glucose; i.e., there was no complementation for glucose uptake in the ptsG manXYZ double mutant LJ121 (data not shown). Plumbridge (38) suggested that the IIBGlc domain of PtsG may be responsible for generating the inducing signal for ptsG expression. Therefore, we also tested plasmid pJBH (6), which encodes the IIBGlc domain (amino acids 390 to 477 plus six additional His residues), for transactivation in this assay. Cells harboring pJBH exhibited only slightly, if any, increased β-galactosidase activity from the Φ(ptsGop-lacZ) fusion compared to the wild-type control. These results correspond to the results obtained by Plumbridge (38), who also observed only a very weak positive regulatory effect of overproduction of the IIBGlc domain.
To further investigate what part of the IICBGlc domain might be responsible for the interaction with DgsA, we constructed different plasmids that encode proteins with defined carboxy-terminal deletions within the IIBGlc domain (Fig. 3). The addition of 50 μM IPTG to cells harboring these plasmid constructs led to immediate growth arrest, providing some evidence for the production of the truncated proteins. However, attempts to detect these truncated proteins by Western blot analysis with a polyclonal antiserum against the hydrophilic IIBGlc domain failed. None of these deletion plasmids could complement for glucose uptake, e.g., in LJ121; however, like pJBH, all pSTJ30 derivatives slightly increased β-galactosidase activity from a Φ(ptsGop-lacZ) fusion in LZ150, compared to the uninduced wild type or the pTM30 control (Fig. 3 and Table 5). This increased β-galactosidase activity was independent of the addition of glucose.
Effects of mutated IICBGlc from LZ1 on the level of expression of a Φ(ptsHp-lacZ) fusion.
Besides regulating ptsG expression, DgsA is also involved in the regulation of the pts operon (18, 38). To test whether the ptsG1 allele has an effect on the pts operon, a single-copy Φ(ptsHp-lacZ) operon fusion was constructed. In wild-type ptsG+ strain LZ110, ptsH expression was induced about 3.3-fold by the addition of glucose (Table 6), slightly by mannose and mannitol, but not at all by ribitol. In the ptsG1 mutant LZ100, the induction pattern changed as described for the Φ(ptsGop-lacZ) fusion; i.e., the basal expression level and the level of expression with glucose were increased, and the mutant was clearly inducible by the addition of mannose, ribitol, and mannitol (three- to fivefold). These changes, caused by alteration of the IICBGlc transporter, are consistent with the hypothesis that the substrate binding and/or transport activities of IICBGlc directly modulate DgsA binding activity for all DgsA-regulated operons (see Discussion).
TABLE 6.
Induction of Φ(ptsHp-lacZ) expression in LZ110 (ptsG+) and LZ100 (ptsG1) by different carbon sourcesa
| Strain | β-Galactosidase activity (nmol mg−1 min−1) in the presence of the following inducer:
|
||||
|---|---|---|---|---|---|
| None | Glc | Man | Rtl | Mtl | |
| LZ110 Φ(ptsHp-lacZ) | 100 | 330 | 140 | 100 | 140 |
| LZ100 Φ(ptsHp-lacZ) | 150 | 400 | 510 | 310 | 280 |
LZ110 (ptsG+) and LZ100 (ptsG1) harboring F′8::Tn Φ(ptsHp-lacZ) were grown in minimal medium with 0.2% glycerol plus a second carbohydrate (0.2%) as indicated and harvested during exponential growth. The mean values of at least two measurements of β-galactosidase activities are given.
DISCUSSION
In this study, we were able to show the mechanism of previously described umuC mutations in E. coli K-12 (17). We provide evidence for the nonexistence of a postulated distinct umgC regulatory gene for ptsG expression and show that mutations causing the UmgC phenotype map within ptsG. From the analysis of existing and newly isolated UmgC mutants emerges a novel regulatory model for ptsG expression. It contains, besides classical regulatory proteins, new elements of a glucose sensory system which correspond to parts of the glucose PTS. Essential components of this new model have been described by Kimata et al. (19) and by Plumbridge (38).
Central regulatory elements for the ptsG promoter-operator are the repressor DgsA (also known as Mlc), the PTS-dependent phosphorylation state of the IICBGlc complex, and the global cAMP-CrpA activator. As has been shown by direct in vitro binding assays, ptsG transcription is repressed by DgsA binding to the operator and activated by cAMP-CrpA (19, 37). In accordance with the new model, the expression of IICBGlc cannot be induced above the basal level in a ΔcyaA background, whereas it is constitutively high in a ΔdgsA cyaA+ background (Table 4). No molecular inducer for DgsA could be found in gel retardation assays which included glucose and glucose-6-phosphate (18, 19, 37). The new model postulates that IICBGlc is the glucose sensor responsible for the induction of ptsG and that IICGlc occurs in two conformations: one (IIC) with a low affinity and the other (IIC*) with a high affinity for DgsA. IIC* prevents DgsA from binding to the operator, either by competition for a limited amount of DgsA or by allosterically reducing the affinity of DgsA for the operator. In the absence of IICBGlc (in a ΔptsG strain), DgsA remains permanently bound to its operator, resulting in complete repression of transcription. In the absence of the DgsA repressor (in a ΔdgsA strain), transcription becomes constitutive, independent of the presence of IICBGlc but still dependent on activation by cAMP-CrpA. We speculate that phosphorylated IIBGlc either shifts the equilibrium toward the low-affinity IICGlc conformation or that phosphorylated IIBGlc binds to the IICGlc domain and thereby excludes DgsA from binding. As predicted by the model, mutants with defects in EI (ptsI), HPr (ptsH), or IIAGlc (crr) all show constitutive expression of ptsG, provided an intact or a mutated IICBGlc complex is also present (Table 4). Thus, ΔptsG Δpts mutants are noninducible. Final support for the model comes from the observation that mutants expressing DgsA and the IICGlc domain in the absence of the IIBGlc and/or IIAGlc domain, i.e., the putative IIC* state, show constitutive ptsG expression (Table 4 and Fig. 3). The failure of the C-terminal deletion proteins of IICBGlc encoded by the pSTJ30 deletion derivatives (Fig. 3) to generate (like pJCH) enhanced constitutive ptsG expression might be caused by a permanent “locked-off” conformation of the IICGlc domain or by reduced protein stability.
In contrast to the bgl and other operons controlled through a PTS-dependent antitermination system (reviewed in reference 51), no antiterminator protein is involved in ptsG control in E. coli, and there is no evidence thus far for PTS-dependent phosphorylation or dephosphorylation of either the repressor DgsA (18) or the global activator CrpA. This observation again corroborates the new model in which the PTS-dependent phosphorylation of IICBGlc acts through alteration of the conformation of the inducer domain IICGlc rather than through phosphotransfer to a cognate regulator. Based on the new model, the UmgC and other ptsG mutations can now be understood in the following way.
(i) The fully constitutive allele ptsG727 from the original HK727 mutant is a “locked-in” mutation which blocks IIC permanently in the IIC* state (Table 5). Moreover, all mutated ptsG alleles exhibit trans-dominance over the wild-type ptsG allele. The corresponding E387G mutation affects a linker motif postulated before as being essential for the communication between IIBGlc and IICGlc (5, 6, 24). In E. coli, there is no efficient ATP-dependent glucosamine kinase which could generate the first obligatory intermediate, glucosamine-6-phosphate. Furthermore, glucosamine, normally transported and phosphorylated through IIMan, is a noninducing and poor substrate for IICBGlc. Therefore, selecting for Glm+ derivatives in a ManA− mutant selects for mutations allowing a high level of expression of IICBGlc and efficient phosphorylation of glucosamine by the glucose PTS. Thus, the constitutive DgsA mutant (LJ138 in Table 2) remains unable to take up and phosphorylate this substrate efficiently, while all UmgC-like mutants accept this substrate fairly well. In contrast, other mutants are able to grow on d-ribose and d-xylose via the IICBGlc transporter but in the absence of concomitant substrate phosphorylation (33). Such mutations always arise in combination with a dgsA mutation, indicating that substrates transported through IICBGlc by facilitated diffusion do not induce ptsG due to the lack of dephosphorylation of IIBGlc during the process.
(ii) Besides the fully constitutive type of UmgC mutations represented by ptsG727, there are semiconstitutive and inducible UmgC mutations; i.e., they require the presence of a substrate which is taken up and phosphorylated by the IICBGlc transporter for full induction. They range from efficiently accepting all four substrates and having a clearly increased (≥2-fold) basal induction level (S169F) to allowing only glucosamine (P238L; data not shown) or mannose (F195L) and not arabinitol and ribitol transport and phosphorylation. Similar UmgC mutations, but in different amino acid residues (G176D, A288V, G320S, and P384R), have been isolated by Notley-McRobb and Ferenci (31). It is tempting to speculate that all of these mutations lead to a relaxed substrate specificity of the IICBGlc transporter. In accordance with this hypothesis, this group isolated another class of UmgC-like mutants (V12G, V12F, and G13C) with increased growth on glucosamine and enhanced growth on glucose and mannose (31).
It is more difficult to explain how mutations (boldface) in the hydrophilic linker motif (382-KTPGRED-388) cause changes in substrate specificity relative to mutations located in the membrane-bound IICGlc domain, which is supposed to carry central parts of the substrate binding and catalytic site. The function of this interdomain linker in transport was systematically investigated by alanine-scanning mutagenesis (20). Amino acid substitutions T383A and G385A caused a strong reduction in phosphotransfer activity. The linker perhaps plays an important role in the conformational changes of the protein, precisely coupling the interaction between the IIBGlc and the IICGlc domains in the process of phosphotransfer to the substrate bound in IICGlc (5, 6, 24). Substantial changes in the conformation of a PTS transporter during the process of substrate binding and phosphorylation have been reported for IICBAMtl (28). Using isothermal titration calorimetry, these authors found that approximately 50 to 60 residues are involved in the binding and phosphorylation of the substrate mannitol and the interaction between IICMtl and IIBMtl. This interaction seems to be necessary for phosphotransfer from IIBMtl to the IICMtl-bound substrate and the release of the phosphorylated substrate into the cytoplasm. The fact that the E387G mutant exhibits altered substrate specificity, enhanced transport activity, and a locked-in conformation for the interaction with DgsA seems to indicate that substrate binding, dephosphorylation of IIBGlc, and a change into the inducing conformation (IIC*) are related processes. The observation that the S169F mutation caused an increased level of basal expression also fits this hypothesis.
The unexpected finding that our laboratory strain JWL184-1 carries a mutation in ptsG indicates that other laboratory strains of E. coli K-12 also may carry unidentified ptsG mutations. Plumbridge (38), for example, reported that in her JM101 copy, a Φ(ptsGop-lacZ) fusion was inducible by glucose, N-acetylglucosamine, mannitol, trehalose and, to a lesser extent, glucosamine and mannose. This finding is in contrast to our finding that the E. coli K-12 reference strain W3110 and its isogenic derivatives can be induced only by glucose. Characteristically, older transport studies had indicated d-glucose, αMG, and 5-thio-d-glucoside as the only substrates for the IICBGlc transporter (39).
An interesting question is how DgsA could be titrated by IICBGlc if the dgsA gene itself is autoregulated. DgsA binding sites cloned on low-copy-number plasmids indeed do not titrate DgsA and therefore do not lead to constitutive expression of DgsA-controlled genes (unpublished results). Cloning of the same DgsA binding sites on a multicopy-number vector, however, led to titration of DgsA and to constitutive Φ(ptsGop-lacZ) expression (38). This result indicates that dgsA expression is rather limiting and that titration of DgsA by IICBGlc present at about 2,000 molecules per cell might be possible.
The results presented here extend and contribute to understanding of the function of IICBGlc not only in glucose transport but also in glucose sensing and response. In this process, the transport of glucose triggers a dual-signal transduction pathway. One branch consists of the glucose-dependent modulation of the level of phosphorylation of IIAGlc. Unphosphorylated IIAGlc binds to and reversibly inhibits non-PTS transporters, e.g., for lactose, maltose, or glycerol (inducer exclusion); in its phosphorylated form, IIAGlc activates adenylate cyclase to synthesize cAMP (reviewed in reference 22). In the second branch, the level of phosphorylation of IICBGlc directly modulates the activity of the anticatabolite repressor DgsA. DgsA, in interaction with cAMP-CrpA, can be used by E. coli to precisely regulate carbon catabolite gene expression.
ACKNOWLEDGMENTS
We thank H. L. Kornberg, B. Erni, W. Boos, and K. Schmid for helpful discussions and generous gifts of strains, plasmids, and antiserum against IICBGlc; T. Ferenci and J. Plumbridge for helpful discussions of unpublished information; M. Berlyn (E. coli Genetic Stock Center, New Haven, Conn.) and P. Henderson for donating strains; and S. Tebbe for expert technical help.
We thank the Deutsche Forschungsgemeinschaft for financial support through SFB171 TP C4 and SFB431 TP K2.
REFERENCES
- 1.Arber W. Transduction of chromosomal genes and episomes in E. coli. Virology. 1958;11:250–272. doi: 10.1016/0042-6822(60)90066-0. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidmann J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley-Interscience; 1990. [Google Scholar]
- 3.Begley G S, Warner K A, Arents J C, Postma P W, Jacobson G R. Isolation and characterization of a mutation that alters the substrate specificity of the Escherichia coli glucose permease. J Bacteriol. 1996;178:940–942. doi: 10.1128/jb.178.3.940-942.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berlyn M K B. Linkage map of Escherichia coli K-12, edition 10: the traditional map. Microbiol Mol Biol Rev. 1998;62:814–984. doi: 10.1128/mmbr.62.3.814-984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buhr A, Erni B. Membrane topology of the glucose transporter of Escherichia coli. J Biol Chem. 1993;268:11599–11603. [PubMed] [Google Scholar]
- 6.Buhr A, Flukiger K, Erni B. The glucose transporter of Escherichia coli. Overexpression, purification, and characterization of functional domains. J Biol Chem. 1994;269:23437–23443. [PubMed] [Google Scholar]
- 7.Decker K, Plumbridge J, Boos W. Negative transcriptional regulation of a positive regulator: the expression of malT, encoding the transcriptional activator of the maltose regulon of Escherichia coli, is negatively controlled by Mlc. Mol Microbiol. 1998;27:381–390. doi: 10.1046/j.1365-2958.1998.00694.x. [DOI] [PubMed] [Google Scholar]
- 8.Erni B, Zanolari B. Glucose-permease of the bacterial phosphotransferase system. Gene cloning, overproduction, and amino acid sequence of enzyme IIGlc. J Biol Chem. 1986;261:16398–16403. [PubMed] [Google Scholar]
- 9.Erni B, Zanolari B, Kocher H P. The mannose permease of Escherichia coli consists of three different proteins. Amino acid sequence and function in sugar transport, sugar phosphorylation, and penetration of phage lambda DNA. J Biol Chem. 1987;262:5238–5247. [PubMed] [Google Scholar]
- 10.Ferenci T. Adaptation to life at micromolar nutrient levels: the regulation of Escherichia coli glucose transport by endoinduction and cAMP. FEMS Microbiol Rev. 1996;18:301–317. doi: 10.1111/j.1574-6976.1996.tb00246.x. [DOI] [PubMed] [Google Scholar]
- 11.Fiethen L, Starlinger P. Mutations in the galactose operator. Mol Gen Genet. 1970;108:322–330. doi: 10.1007/BF00267769. [DOI] [PubMed] [Google Scholar]
- 12.Gutknecht R, Lanz R, Erni B. Mutational analysis of invariant arginines in the IIABMan subunit of the Escherichia coli phosphotransferase system. J Biol Chem. 1998;273:12234–12238. doi: 10.1074/jbc.273.20.12234. [DOI] [PubMed] [Google Scholar]
- 13.Henderson P J, Giddens R A, Jones-Mortimer M C. Transport of galactose, glucose and their molecular analogues by Escherichia coli K-12. Biochem J. 1977;162:309–320. doi: 10.1042/bj1620309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heuel H, Shakeri-Garakani A, Turgut S, Lengeler J W. Genes for d-arabinitol and ribitol catabolism from Klebsiella pneumoniae. Microbiology. 1998;144:1631–1639. doi: 10.1099/00221287-144-6-1631. [DOI] [PubMed] [Google Scholar]
- 15.Hill C W, Harnish B W. Inversions between ribosomal RNA genes of Escherichia coli. Proc Natl Acad Sci USA. 1981;78:7069–7072. doi: 10.1073/pnas.78.11.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosono K, Kakuda H, Ichihara S. Decreasing accumulation of acetate in a rich medium by Escherichia coli on introduction of genes on a multicopy plasmid. Biosci Biotechnol Biochem. 1995;59:256–261. doi: 10.1271/bbb.59.256. [DOI] [PubMed] [Google Scholar]
- 17.Jones-Mortimer M C, Kornberg H L. Amino-sugar transport systems of Escherichia coli K12. J Gen Microbiol. 1980;117:369–376. doi: 10.1099/00221287-117-2-369. [DOI] [PubMed] [Google Scholar]
- 18.Kim S-Y, Nam T-W, Shin D, Koo B-M, Seok Y-J, Ryu S. Purification of Mlc and analysis of its effects on pts expression in Escherichia coli. J Biol Chem. 1999;274:25398–25402. doi: 10.1074/jbc.274.36.25398. [DOI] [PubMed] [Google Scholar]
- 19.Kimata K, Inada T, Tagami H, Aiba H. A global repressor (Mlc) is involved in glucose induction of the ptsG gene encoding major glucose transporter in Escherichia coli. Mol Microbiol. 1998;29:1509–1519. doi: 10.1046/j.1365-2958.1998.01035.x. [DOI] [PubMed] [Google Scholar]
- 20.Lanz R, Erni B. The glucose transporter of the Escherichia coli phosphotransferase system. Mutant analysis of the invariant arginines, histidines, and domain linker. J Biol Chem. 1998;273:12239–12243. doi: 10.1074/jbc.273.20.12239. [DOI] [PubMed] [Google Scholar]
- 21.Lengeler J W. Carbohydrate transport in bacteria under environmental conditions: a black box? Antonie Leeuwenhoek. 1993;63:275–288. doi: 10.1007/BF00871223. [DOI] [PubMed] [Google Scholar]
- 22.Lengeler J W. The phosphoenolpyruvate-dependent carbohydrate:phosphotransferase system and control of carbon source utilization. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes; 1995. pp. 231–254. [Google Scholar]
- 23.Lengeler J W, Auburger A M, Mayer R, Pecher A. The phosphoenolpyruvate-dependent carbohydrate:phosphotransferase system enzymes II as chemoreceptors in chemotaxis of Escherichia coli K-12. Mol Gen Genet. 1981;183:163–170. doi: 10.1007/BF00270156. [DOI] [PubMed] [Google Scholar]
- 24.Lengeler J W, Titgemeyer F, Vogler A P, Wöhrl B M. Structure and homologies of carbohydrate:phosphotransferase system (PTS) proteins. Philos Trans R Soc London Ser B. 1990;326:489–504. doi: 10.1098/rstb.1990.0027. [DOI] [PubMed] [Google Scholar]
- 25.Levy S, Zeng G-Q, Danchin A. Cyclic AMP synthesis in Escherichia coli strains bearing known deletions in the pts phosphotransferase operon. Gene. 1990;86:27–33. doi: 10.1016/0378-1119(90)90110-d. [DOI] [PubMed] [Google Scholar]
- 26.Marsh J L, Erfle M, Wykes E J. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene. 1984;32:481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- 27.Martinez E, Bartolome B, de la Cruz F. pACYC184-derived cloning vectors containing the multiple cloning site and lacZ, a reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988;68:159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 28.Meijberg W, Schuurman-Wolters G K, Robilliard G T. Thermodynamic evidence for conformational coupling between the B and C domains of the mannitol transporter of Escherichia coli, enzyme IIMtl. J Biol Chem. 1998;273:7949–7956. doi: 10.1074/jbc.273.14.7949. [DOI] [PubMed] [Google Scholar]
- 29.Morrison T B, Parkinson J S. A fragment liberated from the Escherichia coli CheA kinase that blocks stimulatory, but not inhibitory, chemoreceptor signaling. J Bacteriol. 1997;179:5543–5550. doi: 10.1128/jb.179.17.5543-5550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson S O, Lengeler J W, Postma P W. Role of IIIGlc of the phosphoenolpyruvate-glucose phosphotransferase system in inducer exclusion in Escherichia coli. J Bacteriol. 1984;160:360–364. doi: 10.1128/jb.160.1.360-364.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Notley-McRobb L, Ferenci T. Substrate specificity and signal transduction pathways in the glucose-specific enzyme II (EIIGlc) component of the Escherichia coli phosphotransferase system. J Bacteriol. 2000;182:4437–4442. doi: 10.1128/jb.182.16.4437-4442.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nuoffer C, Zanolari B, Erni B. Glucose permease of Escherichia coli. The effect of cysteine to serine mutations on the function, stability, and regulation of transport and phosphorylation. J Biol Chem. 1988;263:6647–6655. [PubMed] [Google Scholar]
- 33.Oh H, Park Y, Park C. A mutated PtsG, the glucose transporter, allows uptake of d-ribose. J Biol Chem. 1999;274:14006–14011. doi: 10.1074/jbc.274.20.14006. [DOI] [PubMed] [Google Scholar]
- 34.Pardee A B, Prestige L S. The initial kinetics of enzyme induction. Biochim Biophys Acta. 1961;49:77–88. doi: 10.1016/0006-3002(61)90871-x. [DOI] [PubMed] [Google Scholar]
- 35.Parra F, Mortimer M C, Kornberg H L. Phosphotransferase-mediated regulation of carbohydrate utilization in Escherichia coli K12: the nature of the iex (crr) and gsr (tgs) mutations. J Gen Microbiol. 1983;129:337–348. doi: 10.1099/00221287-129-2-337. [DOI] [PubMed] [Google Scholar]
- 36.Plumbridge J. Control of the expression of the manXYZ operon in Escherichia coli: Mlc is a negative regulator of the mannose PTS. Mol Microbiol. 1998;27:369–380. doi: 10.1046/j.1365-2958.1998.00685.x. [DOI] [PubMed] [Google Scholar]
- 37.Plumbridge J. Expression of ptsG, the gene for the major glucose PTS transporter in Escherichia coli, is repressed by Mlc and induced by growth on glucose. Mol Microbiol. 1998;29:1053–1063. doi: 10.1046/j.1365-2958.1998.00991.x. [DOI] [PubMed] [Google Scholar]
- 38.Plumbridge J. Expression of the phosphotransferase system both mediates and is mediated by Mlc regulation in Escherichia coli. Mol Microbiol. 1999;32:260–273. doi: 10.1046/j.1365-2958.1999.01462.x. [DOI] [PubMed] [Google Scholar]
- 39.Postma P W, Lengeler J W. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1985;49:232–269. doi: 10.1128/mr.49.3.232-269.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1149–1174. [Google Scholar]
- 42.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 43.Rephaeli A W, Saier M., Jr Regulation of genes coding for enzyme constituents of the bacterial phosphotransferase system. J Bacteriol. 1980;141:658–663. doi: 10.1128/jb.141.2.658-663.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roehl R A, Vinopal R T. Genetic locus, distant from ptsM, affecting enzyme IIA/IIB function in Escherichia coli K-12. J Bacteriol. 1980;142:120–130. doi: 10.1128/jb.142.1.120-130.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruyter G J G, Postma P W, van Dam K. Control of glucose metabolism by enzyme IIGlc of the phosphoenolpyruvate-dependent phosphotransferase system in Escherichia coli. J Bacteriol. 1991;173:6184–6191. doi: 10.1128/jb.173.19.6184-6191.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saffen D W, Presper K A, Doering T L, Roseman S. Sugar transport by the bacterial phosphotransferase system. Molecular cloning and structural analysis of the Escherichia coli ptsH, ptsI, and crr genes. J Biol Chem. 1987;262:16241–16253. [PubMed] [Google Scholar]
- 47.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 49.Schmid K, Schupfner M, Schmitt R. Plasmid-mediated uptake and metabolism of sucrose by Escherichia coli K-12. J Bacteriol. 1982;151:68–72. doi: 10.1128/jb.151.1.68-76.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. Collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stülke J, Arnaud M, Rapoport G, Martin-Verstraete I. PRD—a protein domain involved in PTS-dependent induction and catabolite repression of catabolic operons in bacteria. Mol Microbiol. 1998;28:865–874. doi: 10.1046/j.1365-2958.1998.00839.x. [DOI] [PubMed] [Google Scholar]
- 52.Ubben D, Schmitt R. Tn1721 derivatives for transposon mutagenesis, restriction mapping and nucleotide sequence analysis. Gene. 1986;41:145–152. doi: 10.1016/0378-1119(86)90093-4. [DOI] [PubMed] [Google Scholar]
- 53.Ubben D, Schmitt R. A transposable promoter and transposable promoter probes derived from Tn1721. Gene. 1987;53:127–134. doi: 10.1016/0378-1119(87)90100-4. [DOI] [PubMed] [Google Scholar]
- 54.Ulmke C, Kreth J, Lengeler J W, Welte W, Schmid K. Site-directed mutagenesis of loop L3 of sucrose porin ScrY leads to changes in substrate selectivity. J Bacteriol. 1999;181:1920–1923. doi: 10.1128/jb.181.6.1920-1923.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van der Vlag J, Van't Hof R, Van Dam K, Postma P W. Control of glucose metabolism by the enzymes of the glucose phosphotransferase system in Salmonella typhimurium. Eur J Biochem. 1995;230:170–182. doi: 10.1111/j.1432-1033.1995.0170i.x. [DOI] [PubMed] [Google Scholar]
- 56.Wöhrl B M, Wehmeier U F, Lengeler J W. Positive and negative regulation of the expression of the l-sorbose (sor) operon by SorC in Klebsiella pneumoniae. Mol Gen Genet. 1990;224:193–200. doi: 10.1007/BF00271552. [DOI] [PubMed] [Google Scholar]