Abstract
A search for a potential algC homologue within the Pseudomonas aeruginosa PAO1 genome database has revealed an open reading frame (ORF) of unknown function, ORF540 in contig 54 (July 1999 Pseudomonas genome release), that theoretically coded for a 445-amino-acid-residue polypeptide (I. M. Tavares, J. H. Leitão, A. M. Fialho, and I. Sá-Correia, Res. Microbiol. 150:105–116, 1999). The product of this gene is here identified as the phosphoglucosamine mutase (GlmM) which catalyzes the conversion of glucosamine-6-phosphate to glucosamine-1-phosphate, an essential step in the formation of the cell wall precursor UDP-N-acetylglucosamine. The P. aeruginosa gene has been cloned into expression vectors and shown to restore normal peptidoglycan biosynthesis and cell growth of a glmM Escherichia coli mutant strain. The GlmM enzyme from P. aeruginosa has been overproduced to high levels and purified to homogeneity in a six-histidine-tagged form. Beside its phosphoglucosamine mutase activity, the P. aeruginosa enzyme is shown to exhibit phosphomannomutase and phosphoglucomutase activities, which represent about 20 and 2% of its GlmM activity, respectively.
Pseudomonas aeruginosa is a leading cause of nosocomial infections (30) and one of the main causes of debilitating pulmonary infections in patients with cystic fibrosis (CF) (9, 30). It can express a number of cell surface and extracellular polysaccharides that are virulence factors and that contribute to its success as an opportunistic pathogen (3, 9). Specifically, this species produces two distinct types of lipopolysaccharide (LPS) (17, 26), and strains infecting CF patients often overproduce the extracellular polysaccharide alginate (9, 18). The protein encoded by the algC gene possesses both phosphomannomutase (PMM) and phosphoglucomutase (PGM) activities, which are required for the interconversion of mannose-6-phosphate (Man-6-P) or glucose-6-phosphate (Glc-6-P) to Man-1-P or Glc-1-P, respectively (2, 33). Man-1-P and Glc-1-P are required for the formation of the precursors GDP-d-mannuronic acid, GDP-d-mannose, UDP-d-glucose, and TDP-l-rhamnose, which are used for alginate, LPS, and rhamnolipid biosynthesis (2, 23, 25, 26, 33, 34). Several pieces of evidence indicate that the algC gene product is indeed common and essential to the biosynthetic pathways of both LPS and alginate in P. aeruginosa (2, 26, 33, 34). An algC homologue has been recently identified within the P. aeruginosa genome, but the enzymatic nature of the gene product has not been established (28). This open reading frame (ORF), provisionally called ORF540, theoretically coded for a protein of 445 amino acids. It was located in contig 54 (15 July 1999 Pseudomonas genome release), and its actual position on the single PAO1 contig is from position 5333450 to 5334784 (reverse complement) (15 December 1999 release) (Pseudomonas Genome Project [http://www.pseudomonas.com]). Interestingly, the corresponding amino acid sequence showed significant homology with proteins belonging to the phosphohexomutase family found in databases (11), particularly with gene products from Escherichia coli, Staphylococcus aureus, or Helicobacter pylori that were recently characterized as phosphoglucosamine mutases (5, 12, 21). These latter enzymes (GlmM) catalyze the formation of glucosamine-1-phosphate (GlcN-1-P) from GlcN-6-P, the first step in the biosynthetic pathway leading to the formation of UDP-N-acetylglucosamine, an essential common precursor to cell envelope components such as peptidoglycan and LPS (11). We here report that the P. aeruginosa glmM gene can fully complement a well-characterized glmM deficiency in E. coli and that the protein purified to homogeneity showed the expected phosphoglucosamine mutase activity. It is also shown that GlmM enzymes from P. aeruginosa and E. coli exhibit additional PMM (relatively high) and PGM (low) activities.
MATERIALS AND METHODS
Bacterial strains, plasmid vectors, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. P. aeruginosa IST27N is a nonmucoid variant spontaneously derived from a mucoid strain isolated from a Portuguese CF patient at the Sta. Maria Hospital in Lisbon, Portugal (16, 28). This strain was used to obtain the genomic DNA used as a template to amplify ORF540 by PCR. P. aeruginosa strain 8858, carrying an algC mutation, is an alginate-negative mutant (34) and was used in complementation assays. E. coli strains JM83 (32) and GPM83 (21) were used as hosts for plasmids and for the preparation of the overproduced GlmM enzyme. Strain GPM83, which carries an inactivated copy of the glmM gene on the chromosome and a wild-type copy of glmM on a plasmid whose replication is thermosensitive, was used in complementation tests. The plasmid vector pTrc99A was purchased from Pharmacia (Uppsala, Sweden), and the pTrcHis60 vector was recently described (24). Recombinant plasmid pNZ49 carries the P. aeruginosa algC gene, encoding a protein with PMM and PGM activities, into the cloning vector pMMB66(HE) (34). This plasmid and the algC mutant 8858 were a kind gift from A. M. Chakrabarty (34). The broad-host-range controlled expression vector pMMB66(HE) was described earlier (7). Unless otherwise noted, 2YT (22) was used as a rich medium to grow E. coli strains, and Lennox L broth medium (Sigma Chemical Co., St. Louis, Mo.) was used to grow P. aeruginosa strains. Solid medium used to grow P. aeruginosa strains was Pseudomonas Isolation Agar (Difco Laboratories, Detroit, Mich.). Growth was monitored at 600 nm with a spectrophotometer (UV-1601; Shimadzu, Duisburg, Germany). For strains carrying drug resistance genes, antibiotics were used at the following concentrations, in micrograms per milliliter: ampicillin, 100; kanamycin, 30; chloramphenicol, 25 (for E. coli); and carbenicillin, 300 (for P. aeruginosa).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| IST27N | Alg−, spontaneous variant of a mucoid CF isolate | 16 |
| 8858 | Alg−, algC mutant | 34 |
| E. coli | ||
| XL1-Blue | recA1 lac[F′ proAB lacIqZΔM15] | 10 |
| HB101 | recA hsdS20(rB− mB−) | 27 |
| JM83 | ara Δ(lac-proAB) rpsL thi φ80 dlacZ ΔM15 | 32 |
| GPM83 | JM83 glmM::kan(pGMM) | 21 |
| PAE831 | JM83 glmM::kan(pMLD136) | This study |
| PAE832 | JM83 glmM::kan(pMLD137) | This study |
| Plasmids | ||
| pCR2.1 | ColE1 Apr Kmr; TA PCR cloning vector | Invitrogen |
| pRK2013 | Kmr Tra+ | 6 |
| pMMB66(HE) | Apr; broad-host-range plasmid | 7 |
| pNZ49 | P. aeruginosa algC in pMMB66(HE) | 34 |
| pIT351 | P. aeruginosa glmM in pCR2.1 | This study |
| pIT352 | P. aeruginosa glmM in pMMB66(HE) | This study |
| pTrc99A | Apr; expression vector | Pharmacia |
| pTrcHis60 | Apr; derivative of pTrc99A | 24 |
| pMLD136 | P. aeruginosa glmM in pTrc99A | This study |
| pMLD137 | P. aeruginosa glmM in pTrcHis60 | This study |
General DNA techniques.
DNA restriction and modification enzymes were obtained from New England Biolabs (Beverly, Mass.), Appligene Oncor (Illkirch, France), and Gibco BRL (Gaithersburg, Md.). DNA fragments were purified with the Wizard purification system (Promega Corporation, Madison, Wis.). Total DNA was extracted from cells of P. aeruginosa IST27N grown overnight in Lennox L broth liquid medium at 30°C with orbital agitation by the method of Goldberg and Ohman (8). Small- and large-scale plasmid isolations from E. coli cells were carried out by the alkaline lysis method, and standard procedures for endonuclease digestions, ligation, and agarose electrophoresis were used (27). E. coli cells were made competent for transformation with plasmid DNA by treatment with CaCl2 (4) or by electroporation. The ability of plasmids to complement the thermosensitive glmM mutant strain GPM83 was tested as described earlier (21). The ability of plasmids to complement the P. aeruginosa algC mutant was tested by introducing plasmid pNZ49, plasmid pIT352 (prepared as described below), or the cloning vector pMMB66(HE) into P. aeruginosa strains by triparental filter mating using the helper plasmid pRK2013 (6) as previously described (15). P. aeruginosa transconjugants were selected in Pseudomonas Isolation Agar plates with carbenicillin and incubated at 37 or 30°C (14) for up to 5 days to compare mucoidy.
Construction of plasmids.
PCR primers used to amplify ORF540 from the P. aeruginosa IST27N chromosome were designed from the P. aeruginosa PAO1 genomic database sequence found to flank this ORF, according to the results of the Pseudomonas Genome Project (15 July 1999 release) (http://www .pseudomonas.com). These primer oligonucleotides were purchased from Pharmacia, and BamHI and HindIII restriction sites (shown in boldface below) were incorporated at the 5′ ends of the sense and the antisense primers, respectively. The sense primer was specific for a 81-bp region upstream from the ORF540 initiation codon (5′-GGATCCGGCCAAGGGCGCACGGATAAT-3′ [primer A]). The antisense primer used corresponds to oligonucleotides complementary to the 53-bp sequence downstream from the TGA stop codon (5′-AAGCTTGGGGCAAAGTGGGCGCAGATGTT-3′ [primer B]). Amplification reactions were carried out in a final volume of 50 μl containing 0.3 nmol of each primer, 10 nmol of each deoxynucleoside triphosphate, 125 nmol of MgCl2, 2.5 U of Taq polymerase (Appligene), and 250 ng of genomic DNA. Thermocycling reactions were performed in a PTC-100 thermal cycler (MJ Research, Inc., Watertown, Mass.) and consisted of an initial denaturation at 95°C (120 s) and 30 cycles of primer annealing at 61.8°C (30 s), extension at 72°C (60 s), and denaturation at 95°C (120 s). The 1,504-bp amplification product was recovered, purified from the gel using the Wizard PCR Preps DNA purification kit from Promega, and cloned into the vector plasmid pCR2.1 (Invitrogen, San Diego, Calif.), giving rise to plasmid pIT351. Plasmid pIT352 was constructed by ligating the HindIII 1,562-bp fragment of pIT351, encompassing the ORF540 sequence, under the control of the tac promoter into the broad-host-range controlled expression vector pMMB66(HE).
A plasmid allowing expression of the putative glmM gene from P. aeruginosa under control of the strong IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible trc promoter was constructed as follows. PCR primers were designed to incorporate a BspHI site (in boldface) 5′ at the initiation codon (underlined) of the gene (5′-ACAAATCATGAGCAGAAAATACTTCGGGACTGAC-3′ [primer C]) and a HindIII site (in boldface) 3′ to the gene after the stop codon (5′-CTCAAAAGCTTCCAGACTGGCAAGCAAAATCAAGC-3′ [primer D]). These primers were used to amplify the gene from plasmid pIT352. PCR amplification of DNA was performed in a thermocycler 60 apparatus (Bio-Med) using Taq polymerase (Appligene). The resulting material was treated with BspHI and HindIII and was ligated between the compatible sites NcoI and HindIII of vector pTrc99A. The thermosensitive glmM mutant strain GPM83 (21) was then transformed with this ligation mixture, and clones were selected for both ampicillin resistance and growth at 42°C. All transformants isolated in that way carried the expected plasmid, named pMLD136. As shown by their sensitivity to chloramphenicol, these clones had lost the thermosensitive plasmid pGMM initially present in strain GPM83, and they consequently now expressed the P. aeruginosa enzyme as the sole source of phosphoglucosamine mutase activity. One of these clones, named PAE831, was chosen for further investigations. Essentially the same procedure was used for the expression of the protein in a C-terminal His6-tagged form. In that case, the gene was amplified with primer C (see above) and primer E (5′-AGCAGGATCCAGCACATACCTCAGAAACAATTTTTGCG-3′). The resulting fragment was cut with BspHI and BamHI (boldface) and was ligated between the compatible NcoI and BglII sites of vector pTrcHis60 (24), generating plasmid pMLD137. Transformation of the glmM mutant strain GPM83 with this plasmid and selection for growth at 42°C on ampicillin plates provided strain PAE832, which expressed the His6-tagged P. aeruginosa enzyme as the sole source of phosphoglucosamine mutase activity. DNA sequencing was performed to confirm that the sequences of the chromosomal fragments that had been cloned into plasmids were correct.
Preparation of crude enzyme.
JM83, PAE831, and PAE832 cells were grown at 37°C in 2YT-ampicillin medium (0.5-liter cultures). When the optical density of the culture reached 0.1, IPTG (Eurogentec Seraing, Belgium) was added at a final concentration of 1 mM, and growth was continued for 3 h. Cells of thermosensitive mutant strain GPM83 were grown at 30°C, or first at 30°C and then for 5 h at the restrictive temperature of 42°C, as previously described (21). Cells were harvested and washed with 40 ml of cold 20 mM potassium phosphate buffer (pH 7.4) containing 0.5 mM MgCl2 and 0.1% β-mercaptoethanol (buffer A). The cell pellet was then resuspended in 5 ml of the same buffer and disrupted by sonication (VibraCell sonicator; Bioblock, Illkirch, France) for 3 min in the cold. The resulting suspension was centrifuged at 4°C for 30 min at 200,000 × g, and the supernatant was dialyzed overnight at 4°C against 100 volumes of buffer A. The resulting solution (5 ml; 10 to 12 mg of protein · ml−1), designated crude enzyme, was stored at −20°C. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of proteins was performed as previously described using 12% polyacrylamide gels (13), and protein concentrations were determined by the method of Bradford (1), using bovine serum albumin as standard.
Purification of the His6-tagged GlmM enzyme.
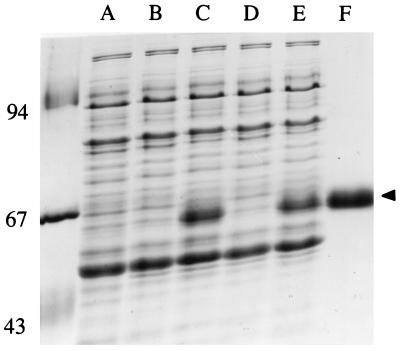
The one-step purification procedure was carried out under native conditions, basically following the recommendations of the manufacturer (Qiagen, Santa Clarita, Calif.): binding of His6-GlmM on Ni2+-nitrilotriacetate agarose and washing with buffer A containing 200 mM KCl and 20 mM imidazole to remove impurities, elution of the protein with increasing concentrations of imidazole added to buffer A (from 50 to 300 mM; the His6-GlmM protein was eluted between 200 and 300 mM), and dialysis of eluted His6-GlmM against buffer A containing 10% glycerol. The His6-tagged enzyme prepared in this manner was more than 90% pure, as estimated by SDS-PAGE (Fig. 1).
FIG. 1.
Overproduction and purification of P. aeruginosa phosphoglucosamine mutase. Lane A, crude extract from parental JM83 cells; lanes B and C, crude extracts from PAE831 cells grown in the absence or presence of IPTG, respectively; lanes D and E, crude extracts from PAE832 cells grown in the absence or presence of IPTG, respectively. A high-level overproduction of the wild-type (lane C) and His6-tagged (lane E) forms of GlmM enzyme (arrowhead) is observed following induction of gene expression with IPTG. Lane F, purified His6-tagged GlmM enzyme. Molecular mass standards, indicated on the left, are as follows: phosphorylase b, 94 kDa; bovine serum albumin, 67 kDa; and ovalbumin, 43 kDa.
Enzymatic assays. (i) Biochemicals.
14C-labeled acetyl coenzyme A (acetyl-CoA) (1.9 GBq mmol−1) was from ICN (Irvine, Calif.). GlcN-6-P, acetyl-CoA, UTP, Glc-1-P, glucose-1,6-diphosphate (Glc-1,6-diP), Man-1-P, NAD, Glc-6-P dehydrogenase, and phosphomannose isomerase were bought from Sigma. Pure GlmU enzyme was prepared as previously described (24).
(ii) GlmM assay.
The coupled assay in which the GlcN-1-P synthesized from GlcN-6-P by the mutase was quantitatively converted to UDP-N-acetylglucosamine in the presence of bifunctional GlmU enzyme (20, 24) was used. The standard assay mixture (100 μl) contained 50 mM Tris-HCl buffer (pH 8), 3 mM MgCl2, 1 mM GlcN-6-P, 0.4 mM [14C]acetyl-CoA (700 Bq), 10 mM UTP, 0.7 mM Glc-1,6-diP, pure GlmU enzyme (1 μg), and GlmM enzyme (0.1 to 10 μg of protein, depending on overexpression or purification factors). Appropriate dilutions of the enzyme prior to assays were performed in buffer A supplemented with 100 μg of bovine serum albumin per ml. Reaction mixtures were incubated at 37°C for 30 min, and the reaction was stopped by the addition of 10 μl of glacial acetic acid. Reaction products were separated with high-voltage electrophoresis 3469 filter paper (Schleicher & Schuell, Dassel, Germany) in 2% formic acid (pH 1.9) for 90 min at 40 V · cm−1 using an LT36 apparatus (Savant Instruments, Hicksville, N.Y.). The radioactive spots were located and quantified with a radioactivity scanner (model Multi-Tracermaster LB285; EG&G Wallac/Berthold). One unit of enzyme activity was defined as the amount which catalyzed the synthesis of 1 μmol of product in 1 min.
(iii) PGM and PMM assays.
The PGM activity of the GlmM enzyme was assayed at 37°C in a reaction mixture (100 μl) containing 50 mM Tris-HCl (pH 8), 5 mM MgCl2, 1 mM Glc-1-P, 0.7 mM Glc-1,6-diP, 1 mM NAD, Glc-6-P dehydrogenase (2 μg), and pure GlmM enzyme (10 μg). The PMM activity of the GlmM enzyme was assayed at 37°C in a reaction mixture (100 μl) containing 50 mM Tris-HCl (pH 8), 5 mM MgCl2, 1 mM Man-1-P, 0.7 mM Glc-1,6-diP, 1 mM NAD, Glc-6-P dehydrogenase (2 μg), phosphomannose isomerase (1 μg), and pure GlmM enzyme (1 μg). In both cases, the formation of NADH was monitored at 340 nm during a 2-h period with a Shimadzu UV-1601 spectrophotometer. One unit of enzyme activity was defined as the amount of enzyme that reduced 1 μmol of NAD per min under the assay conditions.
All of the enzyme assays were performed in triplicate; results were concordant, and mean values are shown.
Isolation of sacculi and quantification of peptidoglycan.
Exponential-phase cells (0.5-liter cultures) were grown at 37°C in 2YT medium supplemented with ampicillin. When the optical density of the culture reached 0.7 (about 3 × 108 cells · ml−1), cells were harvested in the cold, washed with a cold 0.9% NaCl solution, and rapidly suspended by vigorous stirring in 20 ml of a hot (95 to 100°C) aqueous 4% SDS solution for 30 min. After standing overnight at room temperature, the suspensions were centrifuged for 30 min at 200,000 × g in a Beckman TL100 centrifuge, and the pellets were washed several times with water. Final suspensions made in 2 ml of water were homogenized by brief sonication. Aliquots were hydrolyzed and analyzed as previously described, and the peptidoglycan content of the sacculi was expressed in terms of its muramic acid content (19).
RESULTS AND DISCUSSION
The P. aeruginosa glmM gene complements a E. coli glmM mutation.
A recent search for a potential algC homologue within the P. aeruginosa PAO1 genome database (http://www.pseudomonas.com) revealed a putative gene whose product showed 26% sequence identity with AlgC (28). A search within GenBank database showed significant homology of the AlgC homologue with enzymes belonging to the hexosephosphate mutase family. The highest degree of similarity was observed with recently characterized phosphoglucosamine mutases (GlmM) from E. coli, S. aureus, and H. pylori (40 to 60% identity for 445 amino acids). These enzymes catalyze the formation of GlcN-1-P from GlcN-6-P, an essential step in the pathway for UDP-N-acetylglucosamine synthesis (21, 29).
To see whether the putative P. aeruginosa gene could compensate a GlmM deficiency, pIT352, a broad-host-range plasmid carrying a 1.5-kb chromosomal insert from P. aeruginosa with this gene expressed under control of the tac promoter, was transformed into the thermosensitive E. coli glmM mutant. Unfortunately, transformants failed to grow at the restrictive temperature of 42°C, suggesting either that the gene did not code for a phosphoglucosamine mutase or that its expression from the plasmid was too low to meet specific cell requirements. The gene sequence was then amplified by PCR and cloned into vectors allowing its expression under the control of the strong IPTG-inducible trc promoter, pTrc99A and pTrcHis60, for expression of wild-type and His6-tagged enzyme forms, respectively. The two resulting plasmids, pMLD136 and pMLD137, restored the capability of strain GPM83 to grow at 42°C. Complementation occurred even in the absence of IPTG, indicating that the basal expression from these vectors was enough to ensure normal cell growth. As judged by their sensitivity to chloramphenicol, these transformants, named PAE831 and PAE832, respectively, had effectively been cured of the thermosensitive pGMM plasmid originally present in strain GPM83, and they consequently expressed the P. aeruginosa enzyme as the sole source of phosphoglucosamine mutase activity. The peptidoglycan contents of exponentially growing JM83 and PAE831 cells were determined and appeared to be quite similar (9,000 and 11,000 nmol per g [dry weight] of bacteria, respectively), further demonstrating that the P. aeruginosa glmM gene fully complemented the glmM defect of E. coli strain GPM83.
Overproduction and purification of P. aeruginosa phosphoglucosamine mutase in E. coli.
When PAE831 and PAE832 cells were induced with 1 mM IPTG for 3 h, the accumulation of a protein species was observed (Fig. 1), whose molecular mass, about 50 kDa, was consistent with that calculated from the gene sequence (47.8 kDa). The overproduced protein represented about 10 to 15% of total cell proteins, and most of it (about 90%) was recovered in the soluble fraction after a typical cell fractionation. As shown in Table 2, the overproduction of this protein was correlated with an increase of phosphoglucosamine mutase activity in cell extracts. As the two E. coli strains PAE831 and PAE832 carried an inactivated copy of the glmM gene on the chromosome, they consequently expressed the P. aeruginosa gene present on plasmids as the sole source of phosphoglucosamine mutase. In the absence of IPTG, the GlmM activity that could be detected in these cells represented approximately 20% of the activity normally detected in a wild-type E. coli strain. It was sufficient, however, to complement a thermosensitive glmM mutant strain, suggesting that the E. coli GlmM enzyme is normally in great excess in wild-type cells with respect to specific cell requirements (21). When induced with IPTG, PAE831 and PAE832 cells contained 40-fold more phosphoglucosamine mutase activity than noninduced cells (Table 2). It should be mentioned that the levels of GlmM activity detected in these two strains were quite similar, suggesting that the presence of the His6 tag extension at the C terminus of the protein had no significant effect on its enzymatic activity.
TABLE 2.
Phosphoglucosamine mutase activity in E. coli cells expressing the P. aeruginosa glmM gene
| Enzyme source | Growth temp (°C) | IPTG (1 mM) | Sp act (μmol · min−1 · mg of protein−1) |
|---|---|---|---|
| Crude extracts from: | |||
| JM83 | 30, 37, or 42 | − | 0.04 |
| GPM83 | 30 | − | 0.08 |
| 42 | − | 0.001 | |
| PAE831 | 37 | − | 0.008 |
| 37 | + | 0.35 | |
| PAE832 | 37 | − | 0.007 |
| 37 | + | 0.30 | |
| Pure P. aeruginosa GlmM enzyme (His6-tagged form) | 2.5 |
The purification of the His6-tagged P. aeruginosa enzyme was easily achieved in one step by chromatography of crude extracts from induced PAE832 cells on Ni2+-nitrilotriacetate agarose. Two to three milligrams of protein was routinely obtained from 0.5 liter of culture, which appeared to be at least 90% pure as judged by SDS-PAGE (Fig. 1). The specific activity of this pure enzyme preparation was 2.5 U · mg of protein−1 (Table 2), a value quite similar to that recently estimated for the E. coli enzyme, i.e., 3 U · mg of protein−1 for the His6-tagged form (11).
All of these results taken together support the conclusion that the GlmM protein from P. aeruginosa is a new member of the family of confirmed bacterial phosphoglucosamine mutases, which to date included only the glmM gene products from E. coli (21), S. aureus (12, 31), and H. pylori (5). All of them contain in their amino acid sequence the characteristic signature of hexosephosphate mutases (appearing as G95VVISAS*HNPHDDN108 in the sequence of the P. aeruginosa GlmM enzyme), which includes the serine residue (S*) whose phosphorylation is a prerequisite for enzyme activity. The reaction mechanism of the E. coli GlmM enzyme was recently identified as a ping-pong-type mechanism in which glucosamine-1,6-diphosphate, the reaction intermediate, acts as both the first product and the second substrate (11). It is at present unknown whether this compound or another hexose-1,6-diphosphate also ensures the initial activation (phosphorylation) of the enzyme in vivo, a question still open considering that Glc-1,6-diP (the only hexose diphosphate commercially available) could activate GlmM enzymes in vitro (21). As previously observed with the other GlmM enzymes (5, 11, 12), the pure P. aeruginosa enzyme is active in vitro when assayed in the absence of sugar diphosphate, and this basal activity (0.12 U · mg of protein−1) is greatly enhanced (about 20-fold) in the presence of this compound. This suggests that this enzyme that has been expressed in E. coli cells is already partially phosphorylated (active) but that full expression requires further activation by the sugar diphosphate.
Additional PGM and PMM activities of the P. aeruginosa phosphoglucosamine mutase.
The ability of the P. aeruginosa GlmM enzyme to catalyze the interconversion of other sugar phosphates was investigated. The pure enzyme was shown to exhibit both PGM and PMM activities, which were 50- and 5-fold lower than its GlmM activity, respectively (Table 3). As reported recently (11), the E. coli GlmM enzyme also exhibits a PGM activity, which is 1,400-fold lower than its GlmM activity. The PMM activity of the E. coli enzyme was now tested and also appeared to be relatively high (1.75 U · mg of protein−1), representing almost 20% of its GlmM activity. The PMM activities of the two GlmM enzymes were thus much higher than their PGM activities and only slightly lower than their GlmM activities (Table 3). These results suggest that possibly all members of the phosphoglucosamine mutase family can use both mannose and glucose phosphates as substrates, besides glucosamine phosphate, although they can exhibit a preference for one substrate over another.
TABLE 3.
PGM and PMM activities of GlmM enzymes from P. aeruginosa and E. coli
| Enzyme source | Sp act (μmol · min−1 · mg of protein−1)
|
||
|---|---|---|---|
| PGM | PMM | GlmM | |
| Pure His6-tagged GlmM from P. aeruginosa | 0.06 | 0.55 | 2.5 |
| Pure wild-type GlmM from E. coli | 0.007 | 1.75 | 10 |
The main physiological role of the P. aeruginosa GlmM protein is clearly the catalysis of the first step in the biosynthetic pathway leading to the essential peptidoglycan precursor UDP-N-acetylglucosamine. However, the detection of the additional PMM (relatively high) and PGM (low) activities of this protein raises the question of its eventual involvement, under specific conditions, in LPS and alginate biosynthesis. Nevertheless, when plasmid pIT352 was introduced by triparental mating into the algC mutant strain P. aeruginosa 8858, alginate synthesis was not restored, at either 37 or 30°C (14), with or without IPTG induction. Under identical conditions, plasmid pNZ49, carrying the P. aeruginosa algC gene into the same cloning vector pMMB66(HE), restored the mucoid phenotype. These observations suggest either that the expression of PMM activity from the plasmid pIT352 is too low to fulfill cell requirements for alginate synthesis or that GlmM protein cannot convert Man-6-P into Man-1-P in vivo. This conclusion is consistent with previous observations leading to the concept that the P. aeruginosa algC gene provides the only source of PMM and PGM activities participating in the synthesis of various sugar nucleotides required for the production of a number of cell surface and extracellular polysaccharides (2, 26, 33).
ACKNOWLEDGMENTS
This work was supported by the FCT, FEDER, and PRAXIS XXI program (grant PRAXIS/PSAU/P/SAU/59/96 and Ph.D. scholarship BD/13496/97 to I.M.T.) and by a grant from the Centre National de la Recherche Scientifique (EP1088). Financial support from Hoechst Marion Roussel to L.J. and F.P. is acknowledged.
REFERENCES
- 1.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 2.Coyne M J, Jr, Russell K S, Coyle C L, Goldberg J B. The Pseudomonas aeruginosa algC gene encodes phosphoglucomutase, required for the synthesis of a complete lipopolysaccharide core. J Bacteriol. 1994;176:3500–3507. doi: 10.1128/jb.176.12.3500-3507.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cryz S J, Jr, Pitt T L, Furer E, Germanier R. Role of lipopolysaccharide in virulence of Pseudomonas aeruginosa. Infect Immun. 1984;44:508–513. doi: 10.1128/iai.44.2.508-513.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dagert M, Ehrlich S D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979;6:23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- 5.de Reuse H, Labigne A, Mengin-Lecreulx D. The Helicobacter pylori ureC gene codes for a phosphoglucosamine mutase. J Bacteriol. 1997;179:3488–3493. doi: 10.1128/jb.179.11.3488-3493.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furste J P, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 8.Goldberg J B, Ohman D E. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J Bacteriol. 1984;158:1115–1121. doi: 10.1128/jb.158.3.1115-1121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 11.Jolly L, Ferrari P, Blanot D, van Heijenoort J, Fassy F, Mengin-Lecreulx D. Reaction mechanism of phosphoglucosamine mutase from Escherichia coli. Eur J Biochem. 1999;262:202–210. doi: 10.1046/j.1432-1327.1999.00373.x. [DOI] [PubMed] [Google Scholar]
- 12.Jolly L, Wu S, van Heijenoort J, de Lencastre H, Mengin-Lecreulx D, Tomasz A. The femR315 gene from Staphylococcus aureus, the interruption of which results in reduced methicillin resistance, encodes a phosphoglucosamine mutase. J Bacteriol. 1997;179:5321–5325. doi: 10.1128/jb.179.17.5321-5325.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laemmli U K, Favre M. Maturation of the head of bacteriophage T4. J Mol Biol. 1973;80:575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- 14.Leitão J H, Fialho A M, Correia I S. Effects of growth temperature on alginate synthesis and enzymes in Pseudomonas aeruginosa variants. J Gen Microbiol. 1992;138:605–610. doi: 10.1099/00221287-138-3-605. [DOI] [PubMed] [Google Scholar]
- 15.Leitão J H, Correia I S. Manipulation of Pseudomonas aeruginosa alginate pathway by varying the level of biosynthetic enzymes and growth temperature. J Appl Bacteriol. 1993;74:452–459. doi: 10.1111/j.1365-2672.1993.tb05153.x. [DOI] [PubMed] [Google Scholar]
- 16.Leitão J H, Alvim T, Correia I S. Ribotyping of Pseudomonas aeruginosa isolates from patients and water springs and genome fingerprinting of variants concerning mucoidy. FEMS Immunol Med Microbiol. 1996;13:287–292. doi: 10.1111/j.1574-695X.1996.tb00253.x. [DOI] [PubMed] [Google Scholar]
- 17.Lightfoot J, Lam J S. Chromosomal mapping, expression and synthesis of lipopolysaccharide in Pseudomonas aeruginosa: a role for guanosine diphospho-(GDP)-d-mannose. Mol Microbiol. 1993;8:771–782. doi: 10.1111/j.1365-2958.1993.tb01620.x. [DOI] [PubMed] [Google Scholar]
- 18.May T B, Chakrabarty A M. Pseudomonas aeruginosa: genes and enzymes of alginate synthesis. Trends Microbiol. 1994;2:151–157. doi: 10.1016/0966-842x(94)90664-5. [DOI] [PubMed] [Google Scholar]
- 19.Mengin-Lecreulx D, van Heijenoort J. Effect of growth conditions on peptidoglycan content and cytoplasmic steps of its biosynthesis in Escherichia coli. J Bacteriol. 1985;163:208–212. doi: 10.1128/jb.163.1.208-212.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mengin-Lecreulx D, van Heijenoort J. Copurification of glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-phosphate uridyltransferase activities of Escherichia coli: characterization of the glmU gene product as a bifunctional enzyme catalyzing two subsequent steps in the pathway for UDP-N-acetylglucosamine synthesis. J Bacteriol. 1994;176:5788–5795. doi: 10.1128/jb.176.18.5788-5795.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mengin-Lecreulx D, van Heijenoort J. Characterization of the essential gene glmM encoding phosphoglucosamine mutase in Escherichia coli. J Biol Chem. 1996;271:32–39. doi: 10.1074/jbc.271.1.32. [DOI] [PubMed] [Google Scholar]
- 22.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 23.Olvera C, Goldberg J B, Sánchez R, Soberón-Chávez G. The Pseudomonas aeruginosa algC gene product participates in rhamnolipid biosynthesis. FEMS Microbiol Lett. 1999;71:85–90. doi: 10.1111/j.1574-6968.1999.tb08712.x. [DOI] [PubMed] [Google Scholar]
- 24.Pompeo F, van Heijenoort J, Mengin-Lecreulx D. Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the bifunctional GlmU protein from Escherichia coli: site-directed mutagenesis and characterization of the mutant enzymes. J Bacteriol. 1998;180:4799–4803. doi: 10.1128/jb.180.18.4799-4803.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rochetta H L, Burrows L L, Lam J S. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol Mol Biol Rev. 1999;63:523–553. doi: 10.1128/mmbr.63.3.523-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rochetta H L, Pacan J C, Lam J S. Synthesis of the A-band polysaccharide sugar d-rhamnose requires Rmd and WbpW: identification of multiple AlgA homologues, WbpW and ORF488, in Pseudomonas aeruginosa. Mol Microbiol. 1998;29:1419–1434. doi: 10.1046/j.1365-2958.1998.01024.x. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Tavares I M, Leitão J H, Fialho A M, Correia I S. Pattern of changes in the activity of enzymes of GDP-d-mannuronic acid synthesis and in the level of transcription of algA, algC and algD genes accompanying the loss and emergence of mucoidy in Pseudomonas aeruginosa. Res Microbiol. 1999;150:105–116. doi: 10.1016/s0923-2508(99)80028-x. [DOI] [PubMed] [Google Scholar]
- 29.van Heijenoort J. Murein synthesis. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1025–1034. [Google Scholar]
- 30.Woods D E, Vasil M L. Pathogenesis of Pseudomonas aeruginosa infections. In: Baltch A L, Smith R P, editors. Pseudomonas aeruginosa infections and treatment. New York, N.Y: Marcel Dekker Inc.; 1994. pp. 21–50. [Google Scholar]
- 31.Wu S, de Lencastre H, Sali A, Tomasz A. A phosphoglucomutase-like gene essential for the optimal expression of methicillin resistance in Staphylococcus aureus: molecular cloning and DNA sequencing. Microb Drug Resist. 1996;2:277–286. doi: 10.1089/mdr.1996.2.277. [DOI] [PubMed] [Google Scholar]
- 32.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 33.Ye R W, Zielinski N A, Chakrabarty A M. Purification and characterization of phosphomannomutase/phosphoglucomutase from Pseudomonas aeruginosa involved in biosynthesis of both alginate and lipopolysaccharide. J Bacteriol. 1994;176:4851–4857. doi: 10.1128/jb.176.16.4851-4857.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zielinski N A, Chakrabarty A M, Berry A. Characterization and regulation of the Pseudomonas aeruginosa algC encoding phosphomannomutase. J Biol Chem. 1991;266:9754–9763. [PubMed] [Google Scholar]