Abstract
Autophagy is a critical factor in eukaryotic evolution. Cells provide nutrition and energy during autophagy by destroying non-essential components, thereby allowing intracellular material conversion and managing temporary survival stress. Autophagy is linked to a variety of oral disorders, including the type and extent of oral malignancies. Furthermore, autophagy is important in lymphocyte formation, innate immunity, and the regulation of acquired immune responses. It is also required for immunological responses in the oral cavity. Knowledge of autophagy has aided in the identification and treatment of common oral disorders, most notably cancers. The involvement of autophagy in the oral immune system may offer a new understanding of the immune mechanism and provide a novel approach to eliminating harmful bacteria in the body. This review focuses on autophagy creation, innate and acquired immunological responses to autophagy, and the status of autophagy in microbial infection research. Recent developments in the regulatory mechanisms of autophagy and therapeutic applications in oral illnesses, particularly oral cancers, are also discussed. Finally, the relationship between various natural substances that may be used as medications and autophagy is investigated.
Keywords: autophagy, natural substances, oral diseases, microbial infection, cancer
Introduction
Autophagy is an evolutionarily conserved cellular process that maintains energy homeostasis through the lysosomal machinery. It is responsible for transporting damaged organelles, misfolded proteins, and other macromolecular material to lysosomes for degradation and reuse (Wei et al., 2018). Autophagy can be divided into six main steps: initiation, vesicle nucleation, membrane elongation, closure, maturation, and degradation (Lapaquette et al., 2015). Several existing studies have revealed that autophagy is triggered to various degrees during the differentiation progression of many cells, such as angiogenesis (Yang et al., 2015), osteogenic differentiation (Damiati and El-Messeiry, 2021), adipogenesis (Deleyto-Seldas and Efeyan, 2021), neurogenesis, and other processes. The autophagy process was discovered in 1963 by the Belgian chemist Christian de Duve. Contemporary correlative research was fueled by studies of yeast in the 1990s through the identification of autophagy-related genes. One scientist, Yoshinori Ohsumi, was awarded the 2016 Nobel Prize in Physiology or Medicine for the discovery of cellular autophagy machinery (Chen et al., 2019). Autophagy is conducive to cellular catabolism and is also a vital defense mechanism against pathogen infection. Autophagosomes capture invading bacteria, deliver them to lysosomes for degradation, and limit the survival and reproduction of pathogens within the cell. However, certain microbes have also evolved various strategies to escape host cell autophagy. For example, some microbes can regulate autophagy, then use the autophagy machinery to form a replicative niche and promote intracellular growth (Brasil et al., 2017). With the progress of cell autophagy in pathogen infection, scholars have found that the interaction between microbes and host cell autophagy is complicated and varied (Lerner et al., 2017). Therefore, a comprehensive and systematic clarification of the molecular mechanisms by which autophagy develops and interacts with microbes will facilitate the understanding of the immune escape mechanisms of infection. Besides, it will provide a theoretical basis and effective therapeutic targets for the treatment of microbial infections. Recent studies have revealed that many oral diseases, such as periodontal disease, oral tumors, and oral mucosal conditions, are closely related to autophagy (Chen et al., 2017). Among them, oral tumors pose the greatest threat to human health. Relevant studies have found that autophagy affects the occurrence and development of oral tumors and regulates the progression of oral tumors through different autophagy signaling pathways. Also, multiple autophagy-related genes have dual regulatory effects on tumors (Chen et al., 2017). The differential expression of autophagy-related genes directly affects the progression of oral tumorigenesis and development (White et al., 2011), while advances in tumor therapeutics targeting the autophagy pathway also provide new ideas for treating oral cancer. Finally, several recent studies have also shown that many natural substances that are effective in disease treatment participate in autophagy regulation to varying degrees, which may exert therapeutic effects by regulating autophagy (Wang and Feng, 2015). After accounting for the various factors mentioned above, this article first reviews the links between oral diseases (especially tumors) and autophagy, then explores some associations between autophagy and microorganisms. Finally, it summarizes some of the possible therapeutic mechanisms of active ingredients from natural substances on diseases and their targets or pathways regulating cell autophagy and explores new targets with the drug candidates.
Cell autophagy
Autophagy classification
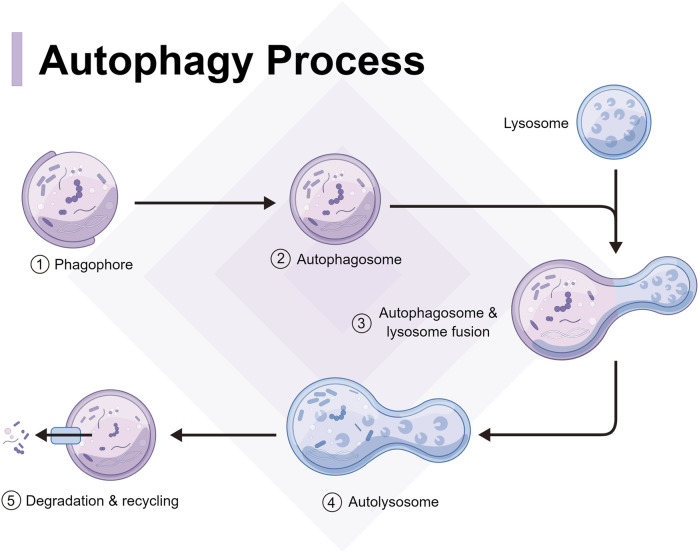
Autophagy can be categorized into two forms: classical and non-classical autophagy. Classical autophagy can be further divided into molecular chaperone-mediated autophagy, micro-autophagy, and macroautophagy, according to the substrate features, transport types, and regulatory mechanisms (Theofani and Xanthou, 2021). However, autophagy generally refers to macro-autophagy and in genetics, it is defined as an autophagosome characterized by a double membrane morphological structure in the cytoplasm that fuses with lysosomes (Acevo-Rodríguez et al., 2020). From yeast to mammals, autophagy has always been highly conserved, and the relevant process can be divided into several steps as follows (Figure 1): autophagy signal induction, precursor formation, phagocytic vesicle extension, autophagosome and lysosome fusion, and autophagolysosome degradation (Li et al., 2016a). Non-classical autophagy refers to LC3-related phagocytosis, which mainly targets intracellular macromolecules. Cells isolate pathogens after phagocytosis in an LC3-positive single membrane compartment called LAPosome. LAPosome then transports the pathogens to the lysosomes for degradation and clearance. The autophagy level is closely related to the occurrence and development of infectious diseases (Khandia et al., 2019), while macro-autophagy and LAP play an important role in resistance to pathogen infection.
FIGURE 1.
Process and mechanism of autophagy, including autophagy signal induction, precursor formation, phagocytic vesicle extension, autophagosome and lysosome fusion, and autophagolysosome degradation.
Autophagy occurrence and regulation
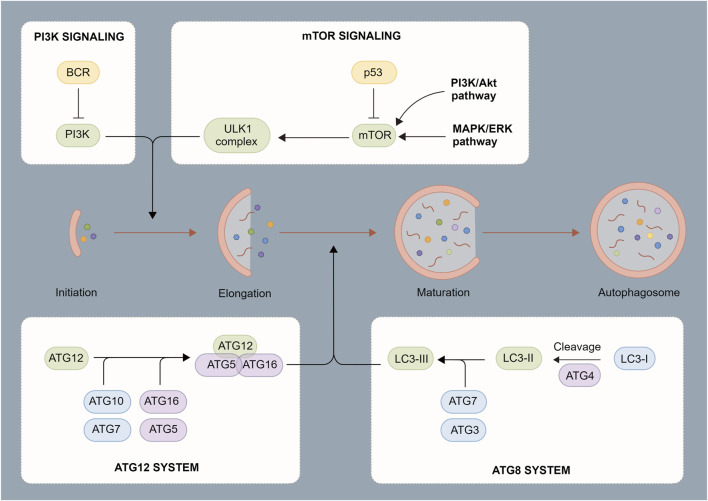
Autophagosome induction is strictly regulated by a series of signaling pathways (De Palma et al., 2014), including subcellular localization and the post-translational modification of the ATG protein. As an initiation complex of the autophagy signal, the Unc-51-like kinase 1 (ULK1) complex is mostly regulated by two upstream factors, mammalian target of rapamycin (mTOR) and AMP-activated protein kinase (AMPK). Under adequate intracellular nutrition, active mTOR inhibits autophagy through the phosphorylation of ULK1 Ser 757 (Li et al., 2019). When intracellular nutrition deficiency or pathogen infection occurs, mTOR becomes inactive and AMPK initiates autophagosome formation by phosphorylating ULK1 Ser 317 and Ser 777. Meanwhile, the ULK complex regulates phosphorylation and recruitment of Type III phosphatidylinositol 3-kinase complex, thereby promoting the production of phosphatidylinositol 3-phosphate (Wang et al., 2019a). At this point, phagocytic vacuoles begin to form at endoplasmic reticulum-mitochondrion contact sites to complete the synthesis of the phagosome precursor (Liu et al., 2017). Two independent ubiquitin-like connection systems control the production of the ATG12-ATG5-ATG16L1 complex and LC3-PE, which is critical for the elongation and closure of phagocytic vacuoles. The ATG16L complex promotes the formation of LC3-PE and anchors it to the autophagosome membrane. However, ATG4 may partially uncouple the LC3-PE complex and promote the recycling of LC3 to form new autophagosomes (Huang et al., 2021a). Finally, with the participation of multiple proteins such as Rab7, syntaxin 17, vesicle-associated membrane protein 8, and lysosome-associated membrane protein, autophagosomes fuse with lysosomes to form autophagolysosomes. After degradation with peptidase, lipase, and hydrolase, the autophagolysosome contents are recycled into the cytoplasm (Foroozandeh and Aziz, 2018).
Autophagy and human innate immunity
Autophagy is an essential element of natural immune response, but the signaling pathway for its activation remains unclear.
Monocytes/macrophages
Autophagy requires the differentiation and activation of monocytes/macrophages (Maiuri et al., 2013). Defects in autophagy may also compromise nitric oxide production, phagocytosis, and the bactericidal ability of macrophages. In macrophages without autophagy, mitochondria and reactive oxygen species (ROS) aggregate, leading to an increase in pro-inflammatory cytokine levels (Barbosa et al., 2018).
Dendritic cells
After infection, dendritic cells (DCs) induce autophagy through activated nod-like receptors and toll-like receptors. Autophagy provides lysosomes with cytoplasm and phagocytic extracellular antigens for further TLR activation, cytokine secretion, and antigen presentation (Allemailem et al., 2021). Mitochondria and reactive oxygen become uncontrolled due to a lack of autophagy, resulting in the aging of DCs (Figure 2).
FIGURE 2.
Process of autophagosome formation. Autophagosomes are composed of a double membrane enclosing a small portion of the cytoplasm. The cytoplasm contains digested material consisting of various components such as mitochondria, endoplasmic reticulum fragments, etc. The contents are degraded through fusion with lysosomes.
Neutrophils
Autophagy is induced by anti-neutrophil cytoplasmic Abs and promotes neutrophil extracellular trap formation (Bravo-Barrera et al., 2017). NETs are antimicrobial extracellular chromatin structures released by neutrophils that have undergone NETosis cell death. Most relevant studies assert that neutrophils in both humans and mice need autophagy to achieve optimal activation (Zhang et al., 2017). First, autophagy is induced when human neutrophils are activated. Also, autophagy defects lead to a series of functional deficiencies, including lower neutrophil degranulation in mice, reduced destruction of intracellular bacteria by human neutrophils, and compromised NET formation (Wang et al., 2019b). Since the function of aged neutrophils is reduced and autophagy is required for the optimal activation of neutrophils, it is necessary to consider whether the autophagy of aged neutrophils is compromised or whether autophagy improves the function of aged neutrophils (Jiang et al., 2019).
Autophagy and microbial infection
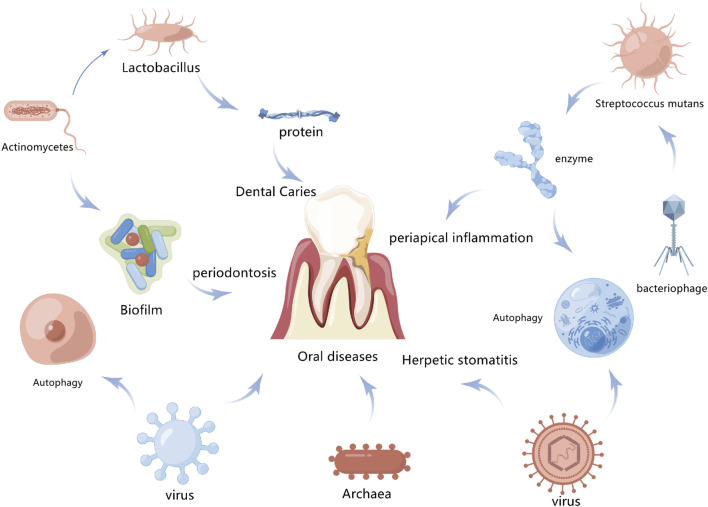
Autophagy acts on immune cells directly to regulate their function so that the body can resist the invasion of pathogens. However, as a “double-edged sword” (Figure 3), autophagy may be utilized by other pathogenic microorganisms to protect the body, thus promoting the infection of host cells and causing damage (Ke, 2018). Several types of pathogenic microorganisms have an intricate relationship with autophagy. Studying the relationship between autophagy and microbial infection may also provide a new direction for further exploring the infection mechanism of pathogenic microorganisms as well as novel methods for fighting infection.
FIGURE 3.
Relationship between autophagy and microbiota. Autophagy is a “double-edged sword” that is involved in host defense against pathogen infection, allowing invading bacteria to be transported to lysosomes for degradation via lipid raft mediated endocytosis. However, it can also be a unique strategy for certain pathogenic bacteria to evade immune surveillance, thereby promoting the survival, proliferation, and extent of infection in the incoming bacteria.
Autophagy for clearance of microbial infection
Resisting microbial invasion is one of the mechanisms for autophagy. Invading microorganisms are captured directly to form autophagosomes, which fuse with lysosomes (Huang et al., 2021b). Thus, microorganisms are degraded and eliminated successively. Studies have shown that a variety of mechanisms are involved in the regulation of this process, including pattern recognition receptors (PRRs), which are an important way to mediate the capture of invading microorganisms (Steimle and Frick, 2016). PRRs recognize the molecular signatures of microorganisms and then recruit intracellular autophagy devices to capture the invading microorganisms (Shibutani et al., 2015). Another path is based on invading microorganisms consuming nutrients in cells. In other words, autophagy activity may be promoted due to a lack of nutrients in cells, which is conducive to the elimination of invading microorganisms. The third pathway in regulating the activity of autophagy is the interaction of Beclin-1 with TNF receptor-associated factor 6 in the TLR signaling pathway (Wen et al., 2020). To initiate autophagy, the cells first detect whether microorganisms have entered the cytoplasm, then the corresponding locations with PRRs play an important role in microorganism identification (Wang et al., 2020). TLR and NOD-like receptors also sense microbial invasion and initiate autophagy in the early stages of infection. They induce autophagy when the cells engulf the bacteria or at different stages of replication after the cells are infected by viruses, thus facilitating the elimination of viruses. Autophagy is also involved in the resistance to toxoplasma gondii infection (Subauste, 2021).
Autophagy promoting microbial infection
Some bacteria may develop new mechanisms to evade or exploit autophagy through evolution. After invading host macrophages, Mycobacterium tuberculosis blocks the autophagy process by preventing autophagolysosomes from acidification and maturation. Therefore, it escapes the immune response of the body and poses a hidden danger for the incubation of Mycobacterium tuberculosis in organisms, thereby causing tuberculosis relapses (Kaleagasioglu et al., 2020). Shigella evades the autophagy of host cells through secretion of the IcsB protein by the Type III secretion system (Sasakawa, 2010). Salmonella and legionella interfere with the maturation of autophagosomes in host cells through the secretion system and survive by inducing the formation of autophagosome-like structures or causing the programmed death of macrophages. Helicobacter pylori stimulates macrophages to form autophagosomes and multiplies in double-membrane autophagosomes (Wang et al., 2021a). Autophagy may also be necessary for the growth of some parasites, such as Trypanosoma cruzi. By degrading organelles and some senescent proteins, autophagy provides nutrition for the development of Trypanosoma cruzi, thus playing an important role in its overall developmental cycle (Mesías et al., 2019). Some viruses may also escape clearance by either inhibiting or utilizing autophagy. Therefore, the resistance of autophagy against viral infection depends on the type and species of the cell. Additionally, autophagy-associated proteins are essential for the replication of the human immunodeficiency virus (HIV), which utilizes the formation of autophagosomes to promote its own replication (Ramesh et al., 2019).
Autophagy and oral diseases
Fluorine spot tooth
Fluorine spot tooth is also known as dental fluorosis or mottled enamel. It refers to incomplete enamel mineralization caused by damage to ameloblasts in the enamel mineralization stage due to excessive fluoride intake in the human tooth during its development and mineralization (Wei et al., 2019). It is widely accepted that endoplasmic reticulum stress (ERS) occurs when ameloblasts are stimulated by excessive fluoride, leading to protein expression errors and apoptosis, which are associated with the formation of fluorine spot tooth (Wei et al., 2019). When the mouse ameloblast-derived LS8 cell line is treated with fluoride at different concentrations, the expression levels of endoplasmic reticulum chaperone molecules GPR78 and XBP-1 rise with increasing fluoride concentrations, indicating that fluorosis induces ERS in ameloblasts (Yang et al., 2013). In one experiment, different concentrations of fluoride (0, 1, and 5 mmol/L) were used for 24 h of treatment of the first molars in Balb/c mice aged 1 day. As a result, the expression levels of pro-apoptotic proteins, e.g., Bcl-2 associated X protein (Bax), BH3 domain apoptosis protein, cysteine asparaginase-specific protease 8, and caspase-3, were up-regulated in a concentration-dependent manner. At the same time, the disintegration of ameloblasts and odontoblast-papillary area cells was observed. During tooth development, long-term or chronic fluoride intake allows ameloblasts to remain in ERS or even induces apoptosis. Thus, the early enamel matrix protein and late hydrolytic protease cannot be secreted normally, affecting the final enamel formation to a certain extent (Zhao et al., 2021). This may partially contribute to dental fluorosis. Additionally, Some scholars found that when ameloblasts were given excessive fluoride for intervention, the expression levels of LC3 and Beclin-1 increased with fluoride concentration, indicating that the intervention of excessive fluoride caused increased levels of autophagy. Some scholars have speculated that excessive fluoride may simultaneously cause ERS and autophagy in ameloblasts of developing teeth, thereby playing a role in the occurrence and development of dental fluorosis. However, the mechanism of mutual regulation between the two in the occurrence of dental fluorosis is yet to be fully explored (Tian et al., 2020).
Periodontal disease
Periodontal supporting tissue includes alveolar bone, gum, periodontal membrane, and cementum (Crotti et al., 2015). Periodontitis is a chronic disease that affects the integrity of the tissue that supports teeth. It is also the most common inflammatory disease in periodontal tissue, leading to progressive alveolar bone resorption. Inflammation, as a protective response of the body to injury or microbial infection, is closely related to both ERS and autophagy (Gilroy and Bishop-Bailey, 2019). In periodontitis, periodontal pathogens that carry or release virulence factors like LPS and bacterial DNA interact with toll-like receptors in periodontal cells to activate the autoimmune system, resulting in local inflammatory cell infiltration of tissue and releasing inflammatory factors (da Silva et al., 2019). TLR4 recruits Beclin-1 and competitively binds Beclin-1 with Bcl-2 to reduce the inhibition of Bcl-2. It also stimulates the aggregation of LC3 in the cytoplasm as a feature of autophagy induction. ERS also induces cell death and autophagy in human gingival cells via P38 mitogen-activated protein kinase (P38MAPK) (Wang et al., 2018). In mice, when autophagy genes Atg7 and Atg5 are knocked out, spontaneous aseptic pulmonary inflammation is observed (Arakawa et al., 2017). Inflammation is characterized by the mass recruitment of inflammatory cells associated with an increase in several pro-inflammatory cytokines in broncho-alveoli and serum, which is mainly mediated by interleukin-18 (IL-18). After LPS injection, the levels of pro-inflammatory cytokines in the lungs and serum as well as the mortality of autophagy-deficient mice are higher than in normal mice, indicating that autophagy negatively regulates the inflammatory response (Lei et al., 2019). However, the specific regulatory mechanism of autophagy in these inflammatory diseases is still unclear, and periodontitis is associated with many systemic diseases. Therefore, the relevant mechanism must be further explored (Figure 4).
FIGURE 4.
Association between autophagy and oral disease. Autophagy promotes the clearance of bacteria and toxins by infected cells and also contributes to the inhibition of inflammatory responses to maintain intracellular environment homeostasis, which is closely related to the occurrence and development of oral diseases.
Sjogren syndrome
As an autoimmune disease, Sjogren syndrome (SS) targets specific secretory epithelial tissue such as salivary and lacrimal glands, uncommon bronchioles, bile ducts, and distal renal tubules (Rizzo et al., 2019). Regarding the initiation and development of immune attacks, it has been hypothesized that secretory epithelial cells activated after chronic or acute external or internal injury cause the expression of SS-associated antigen A (SSA) and SS-associated antigen B (SSB) on the surface of cell membranes, resulting in an autoimmune response (Vletter et al., 2020). SS is a disease characterized by CD4+T cell infiltration, in which salivary gland epithelial cells present major histocompatibility complex II to CD4+T cells as antigen-presenting cells, thereby inducing the immune response (Vletter et al., 2020). An evaluation of protein levels in the saliva of SS patients revealed that lysosomal protease cathepsin D, a protein with relatively high concentrations, increases under autophagy. This suggests that many salivary epithelial cells in SS patients are in a state of autophagy. Autophagy has recently been identified as an antigen presentation causing polarization of T helper cell type 1 (Th1) and type 17 through the involvement of MHC II molecules in cytokine secretion (Münz, 2012). Clinically, non-specific autophagy-lysosomal inhibitors like chloroquine have been used to treat different autoimmune diseases. Understanding the complex interaction between autophagy and autoimmunity may help to develop more effective or specific therapeutic strategies (Jones et al., 2020).
Promotion of autophagy in oral tumorigenesis and its mechanism
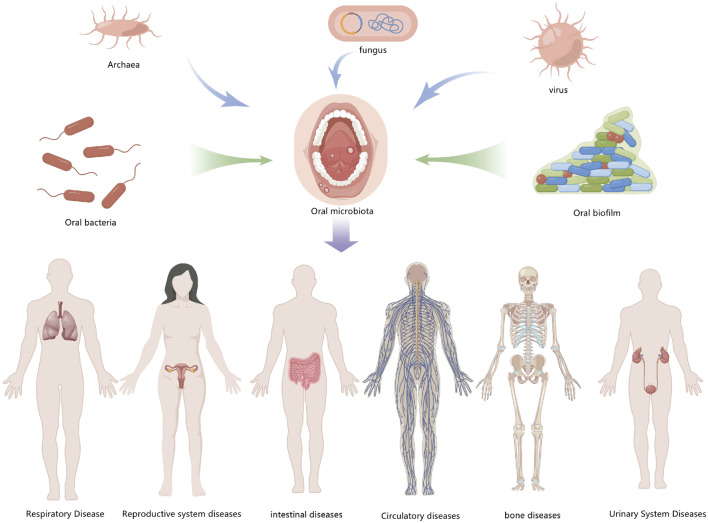
Oral health and systemic health are closely related, and modern medicine has demonstrated that periodontitis and other oral diseases are linked to cardiovascular disease, diabetes, cancer, respiratory disease, Alzheimer’s disease, and so on. Exploring the relationship between oral diseases and autophagy is therefore valuable for understanding other systemic diseases in the human body (Figure 5).
FIGURE 5.
Exploring oral diseases and autophagy may provide references for the study of diseases elsewhere in the body.
Promotive effect of autophagy on the development of oral carcinoma
The proteins involved in autophagy are known as autophagy-related genes (ATG). Besides, ATG8 (Fernández-Sanz et al., 2019), known as microtubule-associated protein 1 light chain 3 (MAP1LC3 or LC3 for short), is critical for autophagy. In cells of oral squamous cell carcinoma (OSCC), ATG7 participates in the covalent binding of ATG5 and ATG12 in the autophagosome membrane extension stage as an E1-like ubiquitination activase. As a result, the migration of autophagy tumor cells may be inhibited (Nitta et al., 2019). The mTOR protein kinase, a major negative regulator of autophagy (Viana et al., 2018), is involved in several signaling pathways that control cell growth, most of which are downstream of growth factor receptors with tyrosine kinase activity. Structural activation of these receptors, activation mutations of RAS, PI3K, AKT, and inactivation mutations of negative regulators such as PTEN often occur in the progression of cancer. This suggests that the inhibition of autophagy may promote tumor growth (Poillet-Perez and White, 2019). Some studies have emphasized that knocking out the neutrophil gelatinase-associated calcitonin (NGAL) gene induces mTOR activation, thereby inhibiting autophagy and promoting the development of oral tumors (Zhang et al., 2016). In the later stages of tumor evolution, activated autophagy promotes tumor growth by providing metabolically stressed tumor cells with nutrients. Besides, in the early stages of oral cancer, the long non-coding RNA FLJ22447 inhibits the autophagy of tumor-associated fibroblasts (CAFs) and impairs the autophagic degradation of IL33. Also, CAFs release a large amount of IL33 to promote the proliferation of OSCC cells (Saha et al., 2021).
ROS-dependent NUPR1-mediated autophagy and oral carcinoma
The stress-induced chromatin-related factor NUPR1 is overexpressed in tumor tissue, especially in cases with poor prognosis and high drug resistance (Emma et al., 2016). Also, NUPR1 is essential for the expression of genes associated with metabolic stress response, in particular for those involved in DNA repair, cell cycle regulation, apoptosis, dentate division, and autophagy (Mu et al., 2018). The migration and invasion of oral squamous cell carcinoma cells induced by repeated cadmium exposure may be prevented by inhibiting ROS/NURP1-dependent autophagy (Fan et al., 2019). Additionally, some studies have demonstrated that ROS-dependent NUPR1-mediated autophagy stimulates the proliferation of OSCC cells during repeated cadmium exposure.
MicroRNA-mediated autophagy and oral carcinoma
MicroRNAs are a group of endogenous RNAs without protein-coding capacity. They affect tumor growth through the action of oncogenes or tumor suppressors (Wozniak et al., 2016; Song et al., 2018; Gao et al., 2020) and inhibit autophagy by targeting autophagy-related genes, thereby promoting the occurrence of oral carcinoma. Pang et al. (2019) observed autophagy that was induced by the hypoxia of CAL-27 cells in human tongue squamous cell carcinoma (TSCC) with a transmission electron microscope. Results indicated that miR-17-5p overexpression limited the Beclin-1 mRNA and protein expression in CAL-27 cells, thereby inhibiting autophagy and promoting tumorigenesis.
Long Non-coding RNA and oral carcinoma
LncRNAs are divided into carcinogenic lncRNAs and anticancer lncRNAs, according to their effects on tumors (Li et al., 2016b; Dong et al., 2018). In autophagy regulation, lncRNAs form a network through interactions and connections between multiple pathways, thereby jointly regulating the growth and death of tumor cells. Yang et al. (Li et al., 2016b) found that overexpression of the lncRNA CASC9 activated the AKT/mTOR signaling pathway. As a result, autophagy and autophagy-mediated apoptosis are inhibited and the occurrence and development of oral carcinoma are promoted. This suggests that CASC9 is a potential biomarker for oral carcinoma diagnosis and also a potential site for targeted therapy.
Mechanism of the inhibitory effect of autophagy on oral tumorigenesis and development
Autophagy has a dual function in oral carcinoma (Yang et al., 2019). The inhibition of autophagy may promote the proliferation, invasion, and migration of oral cancer cells, thereby advancing the occurrence and development of oral cancer. Meanwhile, the autophagy of oral cancer cells can be enhanced by targeted drugs through chemotherapy or radiotherapy, preventing the manifestation or progression of oral cancer (Kardideh et al., 2019). Up-regulation or over-expression of the AKT/mTOR pathway leads to tumor growth and poor prognosis by affecting autophagy. Besides, the MAPK signaling pathway involved in the proliferation, differentiation, apoptosis, angiogenesis, invasion, and metastasis of tumor cells plays an important role in oral cancer (González et al., 2021). Therefore, by inhibiting activation of the MAPK signaling pathway, the occurrence of oral cancer may be suppressed. Studies have shown that fern diene induces autophagy in human OSCC cells by regulating the Akt and MAPK pathways. Besides, Bi-d1870, a specific inhibitor of downstream targets in the MAPK signaling pathway (Penisson et al., 2019), inhibits oral carcinoma by regulating the expression of cell cycle regulators like P21 and inducing cell cycle arrest in the G2/M phase. Studies have established that cancer suppressor genes controlling the autophagy pathway include PTEN, Beclin-1, and DAPK. The activated PTEN may promote downregulation of the PI3K/AKT pathway, affecting tumor growth by stimulating autophagy (Youn et al., 2021). As one of the most important autophagy regulators, Beclin-1 acts as a tumor suppressor mammalian cell. DAPK, a phosphorylated Beclin-1 kinase, is also known as a death-related protein kinase that disrupts the Beclin-1/Bcl-2 complex (Casterton et al., 2020; Kaushal et al., 2020). According to some studies, the DAPK gene provokes cancer development by inhibiting autophagy as an autophagy inducer inhibition in different types of human cancers.
Autophagy in oral cancer therapy
Autophagy is a dynamically balanced catabolic degradation process, in which cellular proteins and organelles are engulfed by autophagosomes, digested by lysosomes, and then recycled to maintain cellular metabolism (Watada and Fujitani, 2015). A lower OSCC cell autophagy ability reflects a higher degree of malignancy and chemotherapy resistance. Sensitivity to cell death may be improved by combining chemotherapy with autophagy inhibitors, suggesting that OSCC responds better to autophagy-based therapy. Additionally, OSCC cell autophagic death may occur during radiation. At the same time, some drugs induce the autophagy of OSCC cells, thereby treating OSCC. Several studies related to autophagy have provided a sound experimental basis for OSCC treatment, so utilizing autophagy to treat OSCC has become the focus of much current research (Adnan et al., 2019).
Using CerS6 to enhance sensitivity to cisplatin chemotherapy in OSCC treatment
To determine whether autophagy regulates drug resistance in oral cancer chemotherapy, some scholars (Li et al., 2018; Zhang et al., 2019; Krasikova et al., 2021) explored the effects of CerS6 on autophagy and mitochondrial fusion of chemotherapy-resistant cells using models of cisplatin-resistant OSCC cells and xenograft in nude mice. These studies revealed that high expression of CerS6 enhanced mitochondrial cleavage and apoptosis in cisplatin-resistant OSCC cells, and attenuated cisplatin-induced autophagy. Also, CerS6 may increase the sensitivity to cisplatin by altering the expression of calpain. The xenotransplantation of cisplatin-resistant OSCC cells into a nude mouse model demonstrated that CerS6 increased the sensitivity to cisplatin-resistant chemotherapy, and thus, the tumor volume was reduced.
Regulating autophagy-related genes to inhibit the development of oral carcinoma
As a histone methyltransferase, G9a is closely related to DNA methylation. In recent years, studies have shown that G9a affects oral carcinoma by regulating autophagy response. Also, G9a may combine transcription factors to participate in the occurrence of various diseases (Caffrey et al., 2016). Maeda et al. (Ahmad et al., 2016) discovered that G9a promoted the autophagy of OSCC cells and reduced the development of OSCC by inhibiting H3K9 methyltransferase. Sambandam et al. (Maeda et al., 2017) found that OSCC cells highly expressed with RANKL significantly increased the expression of autophagy-related genes LC3, ATG5, BECN1, and PI3KC3. A further study demonstrated that RANKL induced the formation of autophagosomes, providing evidence for the targeted therapy of oral carcinoma. Wang et al. (Sambandam et al., 2016) enhanced the autophagy of OSCC cells by treating OSCC cells with erianin. As a result, erianin reduced the vitality of OSCC cells, demonstrating its potential as an anticancer drug. Utaipan et al. (Wang et al., 2016) demonstrated that hordenine induced endoplasmic reticulum stress and triggered the p38MAPK-mediated apoptosis of drug-resistant cells and associated autophagic cell death in OSCC. With potential cytotoxic effects, isohordenine may provide potential antitumor effects against multi-drug-resistant oral carcinomas.
Regulating autophagy-related non-coding RNA to inhibit oral carcinoma
The development of non-coding miRNAs that bind to the 3′UTRs of target genes for translation inhibition or mRNA degradation suggests that it is possible to treat oral carcinoma using miRNAs with autophagy as the targeted pathway (Utaipan et al., 2017). Studies have shown that the low expression of miRNA-137 in OSCC is related to the degree of tumor differentiation, and miRNA-137 is expected to be a potential marker for the early diagnosis of OSCC (Shree Harini and Ezhilarasan, 2022). The study of Zhu et al. (Sun and Li, 2018) showed that miRNA-29b suppressed osteosarcoma cells by targeting CDK6 and inhibiting cell proliferation, migration, and invasion.
Autophagy induced by natural drugs
The therapeutic properties of chemicals isolated from natural products have been applied in the treatment of various diseases (Song et al., 2017; Zhu et al., 2021a; Wang et al., 2021b; Zhu et al., 2021b; Wen et al., 2021; Zhang et al., 2021; Bai et al., 2022). In recent years, the activation of autophagy by natural substances has gained increasing attention (Table 1; Figure 6). As an extract of Reynoutria japonica, Arachis hypogaea, Vitis vinifera, and Fructus mori, resveratrol is characterized by various biological activities and pharmacological effects, especially its anti-proliferation effect on tumors (Zhu et al., 2016). Research confirmed that low doses of resveratrol reduced colon tumor progression more effectively than high doses in subjects exposed to a high-fat diet (De Amicis et al., 2019). Besides, resveratrol suppresses the growth of cancer stem-like cells by inhibiting fatty acid synthase (Mouhid et al., 2017). An in vitro cell culture assay showed that increased uptake of resveratrol-modified liposomes by hepatocellular carcinoma (HepG2) cells led to better tumor cell killing ability compared with free resveratrol (Palomeras et al., 2018). Resveratrol induces autophagy by directly inhibiting mTOR through ATP competition (Kashapov et al., 2021), which may be related to its excellent anti-tumor effect. Curcumin is the primary curcuminoid in the rhizomes of Curcuma longa and is a constituent of the herbs and spices used in traditional Asian cooking (Kashapov et al., 2021). In the subcutaneous xenograft model of U87-MG cells, curcumin significantly inhibited tumor growth and induced autophagy (McBean et al., 2017; Wang et al., 2019c). Some scholars observed that curcumin induced caspase-3-dependent apoptosis and autophagy in Mtb-infected macrophages (Ravindran et al., 2009). While curcumin induces autophagic cell death in HCT116 human colon cancer cells by inducing ROS production (Gupta et al., 2017), it has also exhibited some defects in clinical applications. Higher concentrations of curcumin were observed in normal tissue than in malignant colorectal tissue in patients receiving 3.6 g/day curcumin, with trace levels of curcumin in peripheral blood circulation (Gupta et al., 2020). Bufalin is a key compound found in chansu and huachansu, as well as the toxins of other toad species such as Bufo marinus (Tomeh et al., 2019). It is an effective cardiotonic agent, anesthetic agent, blood pressure regulator, and anti-tumor drug, and it induces the autophagy of various human hepatoma cells (Qi et al., 2018). For instance, bufalin induces cell autophagy and inhibits the proliferation of liver cancer cells by influencing the expression of autophagy-related proteins including LC3-I, LC3-II, P62, and Beclin-1 in liver cancer cells. Furthermore, the autophagic state of liver cancer cells affects the inhibitory effect of bufalin in the proliferation of liver cancer cells (Sheng et al., 2021). Additionally, after constraining the autophagy of HCC-LM3 cells, bufalin significantly enhances inhibition of the adhesion, migration, and invasion of HCC-LM3 cells. Also, synergistic inhibition is strongest when different autophagy inhibitors are combined with 3 MA and CQ. After inhibiting autophagy, bufalin significantly inhibits the protein expression of P-AKT, Cyclin D1, MMP- 2, MMP-9, and VEGF in HCC-LM3 cells. Besides, the protein expression of PTEN and E-cadherin in HCC-LM3 cells increases significantly (Qi et al., 2018). Furthermore, bufalin simultaneously activates the autophagy and apoptosis of hepatoma and gastric cancer cells, and blocking bufalin-induced autophagy may aggravate the death of apoptotic cells (Sheng et al., 2018). Ginseng total saponin is a key ginseng extract with many effective components and strong anti-tumor activity, and it is the prime component responsible for the pharmacological effects of ginseng (Lv et al., 2021a). A previous study illustrated that Ginsenoside Rh4 triggered apoptosis and autophagy by activating the ROS/JNK/p53 pathway in colorectal cancer cells, providing evidence that Rh4 shows great potential as an anti-cancer agent (Liu et al., 2019). In another study, Ginsenosides (TGNs) were found to enhance autophagy by promoting acidic vacuole organelle formation, recruitment of enhanced green fluorescent protein-microtubule-associated protein light chain 3, and expression of autophagy-related factors in cervical cancer cell lines. These results prove that TGN has become a promising drug candidate for cancer therapy (Wu et al., 2018). Ursolic acid, a bioactive ingredient isolated from Radix Actinidiae chinensis, has a strong antitumor effect on osteosarcoma cells. It inhibits tumor cell proliferation and promotes the apoptosis of a variety of osteosarcoma cells. In a mouse osteosarcoma xenograft model, low-dose cisplatin combined with ursolic acid significantly reduced tumor growth, which is a process that may be associated with autophagy (Bian et al., 2021).
TABLE 1.
Summary of drugs and mechanisms by which active ingredients and compounds of TCMs activate autophagy.
| Drug | Cellular or animal models | dosage | Pathways that activate autophagy |
|---|---|---|---|
| Hyperthyroidism | M22 induces nthy - ori-3–1 in human thyroid follicular epithelial cells | Medicated serum was administered for 24 h in vitro | Inhibits the mTOR/P70S6K pathway, increases lc3ii, ATG5 expression, promotes autophagosome formation, and inhibits their degradation |
| Polysaccharide from Cordyceps pupae | High glucose induced podocytes in mice | 25 mg/L in vitro for 48 h | Inhibition of JAK/STAT pathway and increased lc3ii, Beclin1, ATG5, atg12 expression |
| Panax ginseng notoginseng Chuanxiong Hort | High glucose and high fat induced human aortic endothelial senescent cell HAEC | 200 mg/L in vitro for 24 h | Increased LC3, suppressed p62 expression and promoted autophagosome formation |
| Guben invigorating blood circulation complexing agents | High glucose stimulates podocyte MPC in podocytes | Medicated serum (compound gavage for 7 days) was administered for 48 h in vitro | Increased Atg7, atg12-atg5 expression and promoted autophagosome formation |
| Tongxin collaterals | Rat model of myocardial ischemia-reperfusion injury | 0.5 g/kg body weight for 7 days | Activates the PINK1/parkin mitophagy pathway, increases lc3ii/lc3i, suppresses p62 expression and increases autophagosome number |
| puerarin | Cadmium induced AML-12 in mouse hepatocytes | 200 μ Mol/L in vitro for 12 h | Inhibition of ROS, reduced lc3ii, p62 expression and reduced autophagosome accumulation |
| Formononetin | Human hepatoma HepG2 cells | 20 μ Mol/L in vitro for 24 h | The AMPK/TFEB pathway is activated to increase lc3ii and p62 expression and promote the fusion of autophagosomes with lysosomes |
| Tongluo Xingnao effervescent tablets | β Human neuroblastoma cells SH-SY5Y induced by 25–35 | 15 μ Mol/L in vitro for 24 h | Inhibition of mTOR/TFEB pathway, suppression of lc3ii/lc3i, p62 expression, and restoration of lysosomal function |
| Paeoniflorin and saikosaponin A | Corticosterone induced PC12 in human astrocytes | 400 μ Mol/L paeoniflorin and 10 μ Mol/L saikosaponin a for 24 h in vitro | Inhibition of mTOR pathway, reduced lc3ii/lc3i, p62 expression and decreased the number of autophagosomes |
| Yi Huang Tang | Human vaginal epithelial cells infected with herpes simplex virus type 2 VK2/E6E7 | 15 μ Mol/L in vitro for 24 h at 5 | Inhibition of PI3K/Akt/mTOR pathway, increased lc3ii, ATG5 expression and restored lysosomal function |
| berberine | High glucose induced H9c2 in rat cardiomyocytes | 100 μ Mol/L in vitro for 30 min | Activation of AMPK/mTOR pathway increased lc3ii, Beclin1, ATG5 expression |
| Tanshinone IIA | Doxorubicin induced H9c2 cells | 20 μ Mol/L in vitro for 24 h | Activation of Beclin1/lamp1 pathway reduces lc3ii and p62 expression and promotes autophagosome formation and lysosomal degradation |
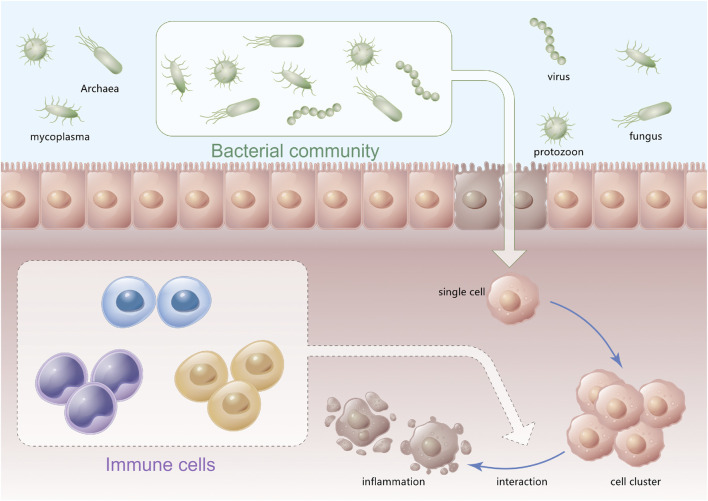
FIGURE 6.
Relationship between immune cells and microorganisms.
Inhibition of autophagy by natural substances
Certain natural substances may have an inhibitory effect on autophagy. In human glioblastoma LN229 and U251 cells, peiminine blocks the cell cycle by up-regulating the expression of LC3 and p62. It also inhibits autophagy through the AMPK/ULK1 pathway, which decreases the degradation of autophagosomes (Tang et al., 2021). ADCX is an extract from Cimicifugae Rhizoma. After treatment with metformin, an AMPK activator, the expression of p62 is down-regulated and autophagy is activated, which indicates that the AMPK target is vital for the inhibition of peiminine-induced autophagy (Zhao et al., 2018). Besides, ADCX promotes the apoptosis of HepG2/ADM cells by inhibiting autophagy flux. Additionally, ADCX is decisive in the inhibition of autophagic degradation by activating Akt and inhibiting CSTB (Sun et al., 2017). Astragaloside II is a natural substance that significantly reduces the expression of LC3-II and Beclin-1 in a dose-dependent manner. Astragaloside II (80 μm) further decreases the formation of LC3-II, Beclin-1, and GFP-LC3 puncta dots stimulated with 5-fluorouracil (0.2 mm) in Bel-7402/FU cells (p < 0.05). Moreover, Astragaloside II sensitizes cells to 5-fluorouracil-induced cell death via the inhibition of pro-survival autophagy involvement in the MAPK-mTOR pathway (Sanduja et al., 2016). Also, it inhibits autophagy by affecting lysosomal function and restores chemotherapy sensitivity to cisplatin through the PI3K/AKT/mTOR pathway. Furthermore, it reduces the survival rate of tumor cells, blocks them in phase S, and ultimately accelerates tumor cell death. Physakengose G (PG) is a new compound that was first isolated from Physalis alkekengi var. franchetii, an anticarcinogenic traditional Chinese medicine. PG showed promising anti-tumor effects in a previous study (Wang et al., 2017a), where it inhibited cell proliferation and induced apoptosis in human osteosarcoma cells. Also, PG treatment blocks EGFR phosphorylation and suppresses epidermal growth factor (EGF)-induced activation of downstream signaling molecules such as AKT and mTOR. PG treatment leads to lysosome dysfunction by altering lysosome acidification and LAMP1 levels, which causes autophagosome accumulation and autophagic flux inhibition. Besides, PG inhibits cell proliferation and EGFR/mTOR signaling in human osteosarcoma cells. Moreover, it induces apoptosis through the mitochondrial pathway and impedes autophagic flux via lysosome dysfunction (Wang et al., 2017a). Furthermore, PG changes lysosome acidification and LAMP1 levels to induce lysosome dysfunction, which causes autophagosome accumulation and autophagy inhibition, thereby inducing apoptosis. Sporoderm, which is the broken spores of Ganoderma lucidum water extract (BSGLWE), induces the autophagy activation of osteosarcoma cells and increases the number of autophagosomes blocking autophagy (Lin et al., 2018). It induces the apoptosis of osteosarcoma cells by inhibiting their proliferation and migration. In human colon cancer HT-29 and HCT116 cell lines, PG stimulates lysosome acidification and decreases cathepsin activity. This subsequently blocks the autophagosome-lysosome fusion process, thereby significantly inhibiting autophagy. Moreover, PG causes the accumulation of autophagosomes through the MAPK/ERK pathway and promotes the apoptosis of tumor cells.
Effect of mixed natural drugs on autophagy
Chinese herbal compounds are composed of more than two herbal medicines and are processed and administered according to regular methods (Wang et al., 2017b). These compounds for the treatment of diseases and syndromes constitute a major component of traditional Chinese medicine prescriptions (Shan et al., 2020). Tongluo Xingnao effervescent tablets restore the function of autophagolysosomes by inhibiting the mTOR/TFEB pathway in human neuroblastoma cells. They also activate autophagy to degrade autophagic vesicles and play a role in resisting Alzheimer’s disease (Wang and Wu, 2020). Additionally, Yi Huang Tang activates autophagy to resist HSV-2 infection by inhibiting the PI3K/AKT/mTOR pathway (Li et al., 2021). However, PI3K inhibitors prevent autophagy activation induced by Yi Huang Tang, indicating that this decoction activates PI3K targets related to autophagy flux. Guben Huayu Tongluo reduces urinary protein excretion and relieves renal pathological damage. Wnt4, p-GSK3β (S9), and β-catenin expression are decreased in the signaling pathway by Guben Huayu Tongluo, but GSK3β levels do not change in diabetic rats. Furthermore, the expression of TGF- and ILK decreases, but the level of E-cadherin increases in diabetic rats after treatment with HTH (Duan et al., 2020). Besides, Guben Huayu Tongluo activates autophagy by promoting the formation of autophagosomes, thereby improving the abnormal autophagy of podocytes under a high-glucose environment and alleviating the damage to podocytes induced by high glucose. In M22-induced human thyroid follicular epithelial cells (Nthy-ori 3-1), Jiakangning capsules containing serum stimulate autophagy and promote apoptosis by activating the AMPK/mTOR pathway (Bai et al., 2017). This indicates that AMPK targets play a vital role in activating autophagy and inhibiting the excessive proliferation of thyroid epithelial cells. In a hypoxia/reoxygenation model related to rat H9C2 cardiomyocytes, Tong Xin Luo activated autophagy flux through the PINK1/Parkin mitochondrial autophagy pathway. It eliminates damaged mitochondria and treats the degradation of misfolded protein through cooperation with the ubiquitin-proteasome system, exerting a protective effect on cardiomyocytes. Table 2 lists the active ingredients of herbal medicines and compounds that inhibit autophagy. Yiqi Huayu Jiedu (YQHYJD) blocks the autophagy of SMCC-7721 by inhibiting the PI3K/mTOR signaling pathway, increasing the number of autophagosomes, and promoting the death of tumor cells, thus playing a role in resisting tumors (Li et al., 2015). Congrong Tusizi Wan inhibits the activation of autophagy by preventing the expression of LC3 and Beclin-1 through the PI3K/Akt/mTOR pathway and reducing the number of autophagosomes and autophagolysosomes in rat ovarian granulosa cells. (Wu et al., 2017). In a lipopolysaccharide-induced macrophage activation model, the Buyang Huanwu decoction inhibited the autophagy of macrophages and the expression of LC3 and p62 by selectively blocking the PI3K/Akt/mTOR pathway (Lv et al., 2021b). This reduces macrophage autophagy and inflammatory reactions and stabilizes vulnerable atherosclerotic plaques. Also, in mouse glomerular podocytes induced by azithromycin, Modified Huangqi Chifeng Decoction (MHCD) inhibited the initiation of autophagy and the combination of autophagosomes and lysosomes through the ROS-autophagy pathway, thus blocking autophagy and protecting glomerular podocytes (Yu et al., 2018; Zheng et al., 2021).
TABLE 2.
Summary of drugs and mechanisms by which active ingredients and compound compounds of traditional Chinese medicine inhibit autophagy.
| Drug | Cellular or animal models | Dosage | Inhibition of autophagy signaling pathway |
|---|---|---|---|
| Astragaloside II | Cisplatin induced sgc-790 human gastric cancer cells, HepG2 human hepatocellular carcinoma cells, smc-7721 cells | 50 μ Mol/L in vitro for 30 min | Activates the PI3K/AKT/mTOR pathway, increases the expression of LC3II and p62, and inhibits lysosomal function |
| Physakengose G | Human osteosarcoma u-2os cells | 15 μ Mol/L in vitro for 12 h | Inhibition of EGFR/mTOR pathway, increased lc3ii, p62 expression and inhibited lysosomal degradation |
| Huangqi Chifeng Decoction | Doxorubicin induced podocytes in mouse glomeruli | Medicated serum (compound gavage for 7 days) was administered for 24 h in vitro | ROS pathway inhibited lc3ii/lc3i, Beclin1 expression, increased p62 expression and inhibited lysosomal function |
| buyang huanwu decoction | Lipopolysaccharide induced RAW264.7 macrophages | Medicated serum (compound gavage for 7 days) was administered for 24 h in vitro | Activated PI3K/Akt/mTOR pathway, inhibited lc3ii/lc3i, p62 expression and reduced autophagosome number |
| Yiqi invigorating and detoxifying formula | Human hepatoma cells smcc-7721 | 5 g/L in vitro for 24 h | Inhibition of PI3K/mTOR pathway increased lc3ii, Beclin1, p62 expression, leading to the impairment of autophagosome degradation |
| Beomyosin B | Human glioblastoma cells ln229 and U251 | 200 μ Mol/L in vitro for 7 days | Inhibition of AMPK/ulk1 pathway increased lc3ii/lc3i, p62 protein expression |
| curcumin | Gp120 induces BV2 in microglia | 15 μ Mol/L in vitro for 12 h | Inhibition of PI3K/AKT/IKK/NF-κB pathway, inhibition LC3II, Atg5 expression |
| Tanshinone IIA | Hypoxia model in primary rat cardiac myocytes | 5μ Mol/L in vitro for 24 h | Activation of mTOR pathway, inhibition of lc3ii, Beclin1, p62 expression, lysosomal dysfunction |
Discussion
Autophagy is a highly complex physiological process that plays a crucial role in the defense of the body against diseases. Additionally, the occurrence and development of multiple oral disorders are associated with abnormal autophagy. Therefore, the intervention of autophagic activity opens new avenues for the prevention and treatment of diseases. Autophagy has a dual impact on oral diseases. First, it suppresses lesions at an early stage by eliminating damaged organelles and proteins, alleviating cell damage, and ensuring metabolic stability. Besides, once the lesion has formed, autophagy improves the survival rate of cells under a stressful environment, thereby playing a disease-promoting role. The effect of autophagy on the occurrence and development of oral diseases is mainly expressed through autophagy-related genes. Besides, the autophagy pathway has been found to cure oral diseases and has become the focus of much research as a result. Therefore, exploring the mechanism of autophagy and elucidating the relationship between the signaling tracks of autophagy and oral diseases may lead to new developments in treating oral conditions. Autophagy plays an essential role in the defense against microbial infections by clearing pathogens and modulating innate and acquired immune responses. Thus, in-depth studies of the relationship between autophagy and microbial infection are essential for exploring effective therapeutic avenues for infectious diseases. In recent years, some progress has been made determining the molecular mechanism of cell autophagy. However, many issues, such as the signal transduction pathway of autophagy or the origin of autophagy, are still unclear. Therefore, more attention should be paid to these problems in future studies. Additionally, natural substances are now more widely used in the clinical treatment of diseases. Plants produce a variety of natural compounds that cope with various environmental pressures. Many of these chemicals have pharmacological and clinical characteristics that constitute a library of natural small molecules and macromolecules for the discovery of drugs. A focus of future research will be to isolate sugar-binding metabolites from traditional medicinal plants and study their effects on autophagy, to discover new mechanisms of phytochemicals in the treatment of diseases.
Acknowledgments
We acknowledge all the figure depicted by Figdraw (www.figdraw.com).
Author contributions
QC and PS conceived the idea. XC collected literature data, created the tables and figures, and wrote the first draft of the manuscript. QC reviewed and edited the manuscript. Both authors approved the final version of the manuscript.
Funding
The Public Welfare Technology Application Research Project of Zhejiang province (LGF22H140007); R&D program of the Stomatological Hospital of Zhejiang University school of Medicine (RD2022JXCK01).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Acevo-Rodríguez P. S., Maldonado G., Castro-Obregón S., Hernández G. (2020). Autophagy regulation by the translation machinery and its implications in cancer. Front. Oncol. 10, 322. 10.3389/fonc.2020.00322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adnan M., Islam W., Zhang J., Zheng W., Lu G. D. (2019). Diverse role of SNARE protein Sec22 in vesicle trafficking, membrane fusion, and autophagy. Cells 8, E337. 10.3390/cells8040337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad F., Dixit D., Joshi S. D., Sen E. (2016). G9a inhibition induced PKM2 regulates autophagic responses. Int. J. Biochem. Cell Biol. 78, 87–95. 10.1016/j.biocel.2016.07.009 [DOI] [PubMed] [Google Scholar]
- Allemailem K. S., Almatroudi A., Alrumaihi F., Almatroodi S. A., Alkurbi M. O., Basfar G. T., et al. (2021). Novel approaches of dysregulating lysosome functions in cancer cells by specific drugs and its nanoformulations: A smart approach of modern therapeutics. Int. J. Nanomedicine 16, 5065–5098. 10.2147/IJN.S321343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa S., Honda S., Yamaguchi H., Shimizu S. (2017). Molecular mechanisms and physiological roles of Atg5/Atg7-independent alternative autophagy. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 93, 378–385. 10.2183/pjab.93.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L., Huo B., Chen Z., Guo Q., Xu J., Fang J., et al. (2017). Effect of Huayu Tongluo herbs on reduction of proteinuria via inhibition of wnt/β-catenin signaling pathway in diabetic rats. Evid. Based. Complement. Altern. Med. 2017, 3054145. 10.1155/2017/3054145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X., Zhu Y., Jie J., Li D., Song L., Luo J. (2022). Maackiain protects against sepsis via activating AMPK/Nrf2/HO-1 pathway. Int. Immunopharmacol. 108, 108710. 10.1016/j.intimp.2022.108710 [DOI] [PubMed] [Google Scholar]
- Barbosa M. C., Grosso R. A., Fader C. M. (2018). Hallmarks of aging: An autophagic perspective. Front. Endocrinol. 9, 790. 10.3389/fendo.2018.00790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian S., Zhao Y., Li F., Lu S., He Z., Wang S., et al. (2021). Total ginsenosides induce autophagic cell death in cervical cancer cells accompanied by downregulation of bone marrow stromal antigen-2. Exp. Ther. Med. 22, 667. 10.3892/etm.2021.10099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasil T. R., Freire-de-Lima C. G., Morrot A., Veto Arnholdt A. C. (2017). Host-Toxoplasma gondii coadaptation leads to fine tuning of the immune response. Front. Immunol. 8, 1080. 10.3389/fimmu.2017.01080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Barrera J., Kourilovitch M., Galarza-Maldonado C. (2017). Neutrophil extracellular traps, antiphospholipid antibodies and treatment. Antibodies (Basel) 6, 6. 10.3390/antib6010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey P. B., Frenkel G. D., McAndrew K. L., Marks K. (2016). A model of the development of cisplatin resistance in human small cell lung cancer xenografts. Vivo 30, 745–749. 10.21873/invivo.10990 [DOI] [PubMed] [Google Scholar]
- Casterton R. L., Hunt R. J., Fanto M. (2020). Pathomechanism heterogeneity in the amyotrophic lateral sclerosis and frontotemporal dementia disease spectrum: Providing focus through the lens of autophagy. J. Mol. Biol. 432, 2692–2713. 10.1016/j.jmb.2020.02.018 [DOI] [PubMed] [Google Scholar]
- Chen P. H., Mahmood Q., Mariottini G. L., Chiang T. A., Lee K. W. (2017). Adverse health effects of betel quid and the risk of oral and pharyngeal cancers. Biomed. Res. Int. 2017, 3904098. 10.1155/2017/3904098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Shinozaki D., Luo J., Pottier M., Havé M., Marmagne A., et al. (2019). Autophagy and nutrients management in plants. Cells 8, 1426. 10.3390/cells8111426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti T. N., Dharmapatni A. A., Alias E., Haynes D. R. (2015). Osteoimmunology: Major and costimulatory pathway expression associated with chronic inflammatory induced bone loss. J. Immunol. Res. 2015, 281287. 10.1155/2015/281287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiati L. A., El-Messeiry S. (2021). An overview of RNA-based scaffolds for osteogenesis. Front. Mol. Biosci. 8, 682581. 10.3389/fmolb.2021.682581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva A. P. B., Alluri L., Bissada N. F., Gupta S. (2019). Association between oral pathogens and prostate cancer: Building the relationship. Am. J. Clin. Exp. Urol. 7 (1), 1–10. [PMC free article] [PubMed] [Google Scholar]
- De Amicis F., Chimento A., Montalto F. I., Casaburi I., Sirianni R., Pezzi V. (2019). Steroid receptor signallings as targets for resveratrol actions in breast and prostate cancer. Int. J. Mol. Sci. 20, E1087. 10.3390/ijms20051087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma C., Perrotta C., Pellegrino P., Clementi E., Cervia D. (2014). Skeletal muscle homeostasis in duchenne muscular dystrophy: Modulating autophagy as a promising therapeutic strategy. Front. Aging Neurosci. 6, 188. 10.3389/fnagi.2014.00188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleyto-Seldas N., Efeyan A. (2021). The mTOR-autophagy Axis and the control of metabolism. Front. Cell Dev. Biol. 9, 655731. 10.3389/fcell.2021.655731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P., Xiong Y., Yue J., Hanley S., Kobayashi N., Todo Y., et al. (2018). Long non-coding RNA NEAT1: A novel target for diagnosis and therapy in human tumors. Front. Genet. 9, 471. 10.3389/fgene.2018.00471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q., Liu T., Yuan P., Huang C., Shao Q., Xu L., et al. (2020). Antiviral effect of Chinese herbal prescription JieZe-1 on adhesion and penetration of VK2/E6E7 with herpes simplex viruses type 2. J. Ethnopharmacol. 249, 112405. 10.1016/j.jep.2019.112405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emma M. R., Iovanna J. L., Bachvarov D., Puleio R., Loria G. R., Augello G., et al. (2016). NUPR1, a new target in liver cancer: Implication in controlling cell growth, migration, invasion and sorafenib resistance. Cell Death Dis. 7, e2269. 10.1038/cddis.2016.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan T., Chen Y., He Z., Wang Q., Yang X., Ren Z., et al. (2019). Inhibition of ROS/NUPR1-dependent autophagy antagonises repeated cadmium exposure -induced oral squamous cell carcinoma cell migration and invasion. Toxicol. Lett. 314, 142–152. 10.1016/j.toxlet.2019.07.017 [DOI] [PubMed] [Google Scholar]
- Fernández-Sanz P., Ruiz-Gabarre D., García-Escudero V. (2019). Modulating effect of diet on Alzheimer's disease. Diseases 7, E12. 10.3390/diseases7010012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foroozandeh P., Aziz A. A. (2018). Insight into cellular uptake and intracellular trafficking of nanoparticles. Nanoscale Res. Lett. 13, 339. 10.1186/s11671-018-2728-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Song L., Xia H., Peng L., Wen Z. (2020). 6'-O-galloylpaeoniflorin regulates proliferation and metastasis of non-small cell lung cancer through AMPK/miR-299-5p/ATF2 axis. Respir. Res. 21 (1), 39. 10.1186/s12931-020-1277-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy D. W., Bishop-Bailey D. (2019). Lipid mediators in immune regulation and resolution. Br. J. Pharmacol. 176, 1009–1023. 10.1111/bph.14587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González A., Alonso-González C., González-González A., Menéndez-Menéndez J., Cos S., Martínez-Campa C. (2021). Melatonin as an adjuvant to antiangiogenic cancer treatments. Cancers (Basel) 13, 3263. 10.3390/cancers13133263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N., Verma K., Nalla S., Kulshreshtha A., Lall R., Prasad S. (2020). Free radicals as a double-edged sword: The cancer preventive and therapeutic roles of curcumin. Molecules 25, E5390. 10.3390/molecules25225390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V. K., Kumar M. M., Bisht D., Kaushik A. (2017). Plants in our combating strategies against Mycobacterium tuberculosis: Progress made and obstacles met. Pharm. Biol. 55, 1536–1544. 10.1080/13880209.2017.1309440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F. L., Yu S. J., Li C. L. (2021). Role of autophagy and apoptosis in acute lymphoblastic leukemia. Cancer Control. 28, 10732748211019138. 10.1177/10732748211019138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Li S., Feng H., Chen Y. (2021). Iron metabolism disorders for cognitive dysfunction after mild traumatic brain injury. Front. Neurosci. 15, 587197. 10.3389/fnins.2021.587197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G. M., Tan Y., Wang H., Peng L., Chen H. T., Meng X. J., et al. (2019). The relationship between autophagy and the immune system and its applications for tumor immunotherapy. Mol. Cancer 18, 17. 10.1186/s12943-019-0944-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. M., Carew J. S., Nawrocki S. T. (2020). Therapeutic targeting of autophagy for renal cell carcinoma therapy. Cancers (Basel) 12, 1185. 10.3390/cancers12051185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaleagasioglu F., Ali D. M., Berger M. R. (2020). Multiple facets of autophagy and the emerging role of alkylphosphocholines as autophagy modulators. Front. Pharmacol. 11, 547. 10.3389/fphar.2020.00547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardideh B., Samimi Z., Norooznezhad F., Kiani S., Mansouri K. (2019). Autophagy, cancer and angiogenesis: Where is the link. Cell Biosci. 9, 65. 10.1186/s13578-019-0327-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashapov R., Ibragimova A., Pavlov R., Gabdrakhmanov D., Kashapova N., Burilova E., et al. (2021). Nanocarriers for biomedicine: From lipid formulations to inorganic and hybrid nanoparticles. Int. J. Mol. Sci. 22, 7055. 10.3390/ijms22137055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal G. P., Chandrashekar K., Juncos L. A., Shah S. V. (2020). Autophagy function and regulation in kidney disease. Biomolecules 10, E100. 10.3390/biom10010100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke P. Y. (2018). The multifaceted roles of autophagy in flavivirus-host interactions. Int. J. Mol. Sci. 19, E3940. 10.3390/ijms19123940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandia R., Dadar M., Munjal A., Dhama K., Karthik K., Tiwari R., et al. (2019). A comprehensive review of autophagy and its various roles in infectious, non-infectious, and lifestyle diseases: Current knowledge and prospects for disease prevention, novel drug design, and therapy. Cells 8, E674. 10.3390/cells8070674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasikova Y., Rechkunova N., Lavrik O. (2021). Nucleotide excision repair: From molecular defects to neurological abnormalities. Int. J. Mol. Sci. 22, 6220. 10.3390/ijms22126220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapaquette P., Guzzo J., Bretillon L., Bringer M. A. (2015). Cellular and molecular connections between autophagy and inflammation. Mediat. Inflamm. 2015, 398483. 10.1155/2015/398483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M., Wang C. J., Yu F., Xie K., Lin S. H., Xu F. (2019). Different intensity of autophagy regulate interleukin-33 to control the uncontrolled inflammation of acute lung injury. Inflamm. Res. 68, 665–675. 10.1007/s00011-019-01250-y [DOI] [PubMed] [Google Scholar]
- Lerner A., Jeremias P., Matthias T. (2017). Gut-thyroid axis and celiac disease. Endocr. Connect. 6, R52–R58. 10.1530/EC-17-0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Song L., Wen Z., Li X., Jie J., Wang Y., et al. (2016). Strong evidence for LncRNA ZNRD1-AS1, and its functional Cis- eQTL locus contributing more to the susceptibility of lung cancer. Oncotarget 7 (24), 35813–35817. 10.18632/oncotarget.8411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. D., Meng X. M., Huang C., Zhang L., Lv X. W., Li J. (2019). Application of herbal traditional Chinese medicine in the treatment of acute kidney injury. Front. Pharmacol. 10, 376. 10.3389/fphar.2019.00376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yang Z., Li Y., Xia J., Li D., Li H., et al. (2016). Cell apoptosis, autophagy and necroptosis in osteosarcoma treatment. Oncotarget 7, 44763–44778. 10.18632/oncotarget.8206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. M., Wei J. P., Li M., Meng S. H. (2015). Effect of jiakangning capsule on thyroid function and akt/mTOR signal pathway of graves' disease mice: An experimental study. Zhongguo Zhong Xi Yi Jie He Za Zhi 35, 1119–1124. [PubMed] [Google Scholar]
- Li S., Wu Y., Ding Y., Yu M., Ai Z. (2018). CerS6 regulates cisplatin resistance in oral squamous cell carcinoma by altering mitochondrial fission and autophagy. J. Cell. Physiol. 233, 9416–9425. 10.1002/jcp.26815 [DOI] [PubMed] [Google Scholar]
- Li Y., Ren M., Wang J., Ma R., Chen H., Xie Q., et al. (2021). Progress in borneol intervention for ischemic stroke: A systematic review. Front. Pharmacol. 12, 606682. 10.3389/fphar.2021.606682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Zhang C., Zhang H., Xia Y. Z., Zhang C. Y., Luo J., et al. (2018). Physakengose G induces apoptosis via EGFR/mTOR signaling and inhibits autophagic flux in human osteosarcoma cells. Phytomedicine. 42, 190–198. 10.1016/j.phymed.2018.03.046 [DOI] [PubMed] [Google Scholar]
- Liu G., Pei F., Yang F., Li L., Amin A. D., Liu S., et al. (2017). Role of autophagy and apoptosis in non-small-cell lung cancer. Int. J. Mol. Sci. 18, 367. 10.3390/ijms18020367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Anderson G. A., Fernandez T. G., Doré S. (2019). Efficacy and mechanism of panax ginseng in experimental stroke. Front. Neurosci. 13, 294. 10.3389/fnins.2019.00294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L., Chang Y., Li Y., Chen H., Yao J., Xie Y., et al. (2021). Triptolide induces leydig cell apoptosis by disrupting mitochondrial dynamics in rats. Front. Pharmacol. 12, 616803. 10.3389/fphar.2021.616803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv S., Wang Z., Wang J., Wang H. (2021). Exogenous hydrogen sulfide plays an important role through regulating autophagy in ischemia/reperfusion injury. Front. Mol. Biosci. 8, 681676. 10.3389/fmolb.2021.681676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K., Doi S., Nakashima A., Nagai T., Irifuku T., Ueno T., et al. (2017). Inhibition of H3K9 methyltransferase G9a ameliorates methylglyoxal-induced peritoneal fibrosis. PLoS One 12, e0173706. 10.1371/journal.pone.0173706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri M. C., Grassia G., Platt A. M., Carnuccio R., Ialenti A., Maffia P. (2013). Macrophage autophagy in atherosclerosis. Mediat. Inflamm. 2013, 584715. 10.1155/2013/584715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBean G. J., López M. G., Wallner F. K. (2017). Redox-based therapeutics in neurodegenerative disease. Br. J. Pharmacol. 174, 1750–1770. 10.1111/bph.13551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesías A. C., Garg N. J., Zago M. P. (2019). Redox balance keepers and possible cell functions managed by redox homeostasis in trypanosoma cruzi. Front. Cell. Infect. Microbiol. 9, 435. 10.3389/fcimb.2019.00435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouhid L., Corzo-Martínez M., Torres C., Vázquez L., Reglero G., Fornari T., et al. (2017). Improving in vivo efficacy of bioactive molecules: An overview of potentially antitumor phytochemicals and currently available lipid-based delivery systems. J. Oncol. 2017, 7351976. 10.1155/2017/7351976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Y., Yan X., Li D., Zhao D., Wang L., Wang X., et al. (2018). NUPR1 maintains autolysosomal efflux by activating SNAP25 transcription in cancer cells. Autophagy 14, 654–670. 10.1080/15548627.2017.1338556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münz C. (2012). Antigen processing for MHC class II presentation via autophagy. Front. Immunol. 3, 9. 10.3389/fimmu.2012.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta A., Hori K., Tanida I., Igarashi A., Deyama Y., Ueno T., et al. (2019). Blocking LC3 lipidation and ATG12 conjugation reactions by ATG7 mutant protein containing C572S. Biochem. Biophys. Res. Commun. 508, 521–526. 10.1016/j.bbrc.2018.11.158 [DOI] [PubMed] [Google Scholar]
- Palomeras S., Ruiz-Martínez S., Puig T. (2018). Targeting breast cancer stem cells to overcome treatment resistance. Molecules 23, E2193. 10.3390/molecules23092193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang F., Liu C., Cui Y., Gong K., Liu G., Bian Y., et al. (2019). miR-17-5p promotes proliferation and migration of CAL-27 human tongue squamous cell carcinoma cells involved in autophagy inhibition under hypoxia. Int. J. Clin. Exp. Pathol. 12 (6), 2084–2091. [PMC free article] [PubMed] [Google Scholar]
- Penisson M., Ladewig J., Belvindrah R., Francis F. (2019). Corrigendum: Genes and mechanisms involved in the generation and amplification of basal radial glial cells. Front. Cell. Neurosci. 13, 462. 10.3389/fncel.2019.00462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poillet-Perez L., White E. (2019). Role of tumor and host autophagy in cancer metabolism. Genes Dev. 33, 610–619. 10.1101/gad.325514.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Zulfiker A., Li C., Good D., Wei M. Q. (2018). The development of toad toxins as potential therapeutic agents. Toxins (Basel) 10, E336. 10.3390/toxins10080336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh J., Ronsard L., Gao A., Venugopal B. (2019). Autophagy intertwines with different diseases-recent strategies for therapeutic approaches. Diseases 7, E15. 10.3390/diseases7010015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran J., Prasad S., Aggarwal B. B. (2009). Curcumin and cancer cells: How many ways can curry kill tumor cells selectively. AAPS J. 11, 495–510. 10.1208/s12248-009-9128-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo C., La Barbera L., Lo Pizzo M., Ciccia F., Sireci G., Guggino G. (2019). Invariant NKT cells and rheumatic disease: Focus on primary sjogren syndrome. Int. J. Mol. Sci. 20, E5435. 10.3390/ijms20215435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Zhang Y., Wilson B., Abounader R., Dutta A. (2021). The tumor-suppressive long noncoding RNA DRAIC inhibits protein translation and induces autophagy by activating AMPK. J. Cell Sci. 134, jcs259306. 10.1242/jcs.259306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandam Y., Sakamuri S., Balasubramanian S., Haque A. (2016). RANK ligand modulation of autophagy in oral squamous cell carcinoma tumor cells. J. Cell. Biochem. 117, 118–125. 10.1002/jcb.25255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanduja S., Feng Y., Mathis R. A., Sokol E. S., Reinhardt F., Halaban R., et al. (2016). AMPK promotes tolerance to Ras pathway inhibition by activating autophagy. Oncogene 35, 5295–5303. 10.1038/onc.2016.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakawa C. (2010). A new paradigm of bacteria-gut interplay brought through the study of Shigella. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 86, 229–243. 10.2183/pjab.86.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q. Y., Sang X. N., Hui H., Shou Q. Y., Fu H. Y., Hao M., et al. (2020). Processing and polyherbal formulation of tetradium ruticarpum (A. Juss.) hartley: Phytochemistry, pharmacokinetics, and toxicity. Front. Pharmacol. 11, 133. 10.3389/fphar.2020.00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng X., Zhu P., Qin J., Li Q. (2018). The biological role of autophagy in regulating and controlling the proliferation of liver cancer cells induced by bufalin. Oncol. Rep. 39, 2931–2941. 10.3892/or.2018.6365 [DOI] [PubMed] [Google Scholar]
- Sheng X., Zhu P., Zhao Y., Zhang J., Li H., Zhao H., et al. (2021). Effect of PI3K/AKT/mTOR signaling pathway on regulating and controlling the anti-invasion and metastasis of hepatoma cells by bufalin. Recent Pat. anticancer. Drug Discov. 16, 54–65. 10.2174/1574892816666210201120324 [DOI] [PubMed] [Google Scholar]
- Shibutani S. T., Saitoh T., Nowag H., Münz C., Yoshimori T. (2015). Autophagy and autophagy-related proteins in the immune system. Nat. Immunol. 16, 1014–1024. 10.1038/ni.3273 [DOI] [PubMed] [Google Scholar]
- Shree Harini K., Ezhilarasan D. (2022). Promising autophagy inhibitors: Therapeutic implications in oral cancer. Oral Oncol. 131, 105948. 10.1016/j.oraloncology.2022.105948 [DOI] [PubMed] [Google Scholar]
- Song L., Li X., Bai X. X., Gao J., Wang C. Y. (2017). Calycosin improves cognitive function in a transgenic mouse model of Alzheimer's disease by activating the protein kinase C pathway. Neural Regen. Res. 12 (11), 1870–1876. 10.4103/1673-5374.219049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Peng L., Hua S., Li X., Ma L., Jie J., et al. (2018). miR-144-5p enhances the radiosensitivity of non-small-cell lung cancer cells via targeting ATF2. Biomed. Res. Int. 2018, 5109497. 10.1155/2018/5109497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimle A., Frick J. S. (2016). Molecular mechanisms of induction of tolerant and tolerogenic intestinal dendritic cells in mice. J. Immunol. Res. 2016, 1958650. 10.1155/2016/1958650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subauste C. S. (2021). Recent advances in the roles of autophagy and autophagy proteins in host cells during toxoplasma gondii infection and potential therapeutic implications. Front. Cell Dev. Biol. 9, 673813. 10.3389/fcell.2021.673813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Li J. (2018). Expression of MiRNA-137 in oral squamous cell carcinoma and its clinical significance. J. BUON 23 (1), 167–172. [PubMed] [Google Scholar]
- Sun H., Huang M., Yao N., Hu J., Li Y., Chen L., et al. (2017). The cycloartane triterpenoid ADCX impairs autophagic degradation through Akt overactivation and promotes apoptotic cell death in multidrug-resistant HepG2/ADM cells. Biochem. Pharmacol. 146, 87–100. 10.1016/j.bcp.2017.10.012 [DOI] [PubMed] [Google Scholar]
- Tang Z., Dong H., Li T., Wang N., Wei X., Wu H., et al. (2021). The synergistic reducing drug resistance effect of cisplatin and ursolic acid on osteosarcoma through a multistep mechanism involving ferritinophagy. Oxid. Med. Cell. Longev. 2021, 5192271. 10.1155/2021/5192271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theofani E., Xanthou G. (2021). Autophagy: A friend or foe in allergic asthma. Int. J. Mol. Sci. 22, 6314. 10.3390/ijms22126314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Xie J., Chen X., Dong N., Feng J., Gao Y., et al. (2020). Deregulation of autophagy is involved in nephrotoxicity of arsenite and fluoride exposure during gestation to puberty in rat offspring. Arch. Toxicol. 94, 749–760. 10.1007/s00204-019-02651-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomeh M. A., Hadianamrei R., Zhao X. (2019). A review of curcumin and its derivatives as anticancer agents. Int. J. Mol. Sci. 20, E1033. 10.3390/ijms20051033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utaipan T., Athipornchai A., Suksamrarn A., Chunsrivirot S., Chunglok W. (2017). Isomahanine induces endoplasmic reticulum stress and simultaneously triggers p38?MAPK-mediated apoptosis and autophagy in multidrug-resistant human oral squamous cell carcinoma cells. Oncol. Rep. 37, 1243–1252. 10.3892/or.2017.5352 [DOI] [PubMed] [Google Scholar]
- Viana S. D., Reis F., Alves R. (2018). Therapeutic use of mTOR inhibitors in renal diseases: Advances, drawbacks, and challenges. Oxid. Med. Cell. Longev. 2018, 3693625. 10.1155/2018/3693625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vletter E. M., Koning M. T., Scherer H. U., Veelken H., Toes R. (2020). A comparison of immunoglobulin variable region N-linked glycosylation in healthy donors, autoimmune disease and lymphoma. Front. Immunol. 11, 241. 10.3389/fimmu.2020.00241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Hu Y., Yang H., Wang S., Zhou B., Bao Y., et al. (2021). Function of non-coding RNA in Helicobacter pylori-infected gastric cancer. Front. Mol. Biosci. 8, 649105. 10.3389/fmolb.2021.649105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Luo J., Bai X., Hua S., Jie J., Liu H., et al. (2021). Calycosin alleviates injury in airway epithelial cells caused by PM 2.5 exposure via activation of AMPK signalling. Evid. Based. Complement. Altern. Med. 2021, 8885716. 10.1155/2021/8885716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhang T., Sun W., Wang Z., Zuo D., Zhou Z., et al. (2016). Erianin induces G2/M-phase arrest, apoptosis, and autophagy via the ROS/JNK signaling pathway in human osteosarcoma cells in vitro and in vivo . Cell Death Dis. 7, e2247. 10.1038/cddis.2016.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Cao B., Zhao H., Feng J. (2017). Emerging roles of Ganoderma lucidum in anti-aging. Aging Dis. 8, 691–707. 10.14336/AD.2017.0410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Ran Q., Zeng H. R., Wang L., Hu C. J., Huang Q. W. (2018). Cellular stress response mechanisms of rhizoma coptidis: A systematic review. Chin. Med. 13, 27. 10.1186/s13020-018-0184-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wu X. (2020). Traditional Chinese medicine jiuwei zhenxin granules in treating depression: An overview. Neuropsychiatr. Dis. Treat. 16, 2237–2255. 10.2147/NDT.S273324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Chen Y., Zhang P., Lin P., Xie N., Wu M. (2019). Protective features of autophagy in pulmonary infection and inflammatory diseases. Cells 8, E123. 10.3390/cells8020123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Huang C., Su Y., Yang C., Xia Q., Xu D. J. (2017). Astragaloside II sensitizes human hepatocellular carcinoma cells to 5-fluorouracil via suppression of autophagy. J. Pharm. Pharmacol. 69, 743–752. 10.1111/jphp.12706 [DOI] [PubMed] [Google Scholar]
- Wang M., Jiang S., Zhou L., Yu F., Ding H., Li P., et al. (2019). Potential mechanisms of action of curcumin for cancer prevention: Focus on cellular signaling pathways and miRNAs. Int. J. Biol. Sci. 15, 1200–1214. 10.7150/ijbs.33710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. M., Feng Y. S., Yang S. D., Xing Y., Zhang J., Dong F., et al. (2019). The relationship between autophagy and brain plasticity in neurological diseases. Front. Cell. Neurosci. 13, 228. 10.3389/fncel.2019.00228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Feng Y. (2015). Elaborating the role of natural products-induced autophagy in cancer treatment: Achievements and artifacts in the state of the art. Biomed. Res. Int. 2015, 934207. 10.1155/2015/934207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang D., Hou Y., Shen S., Wang T. (2020). The adaptor protein CARD9, from fungal immunity to tumorigenesis. Am. J. Cancer Res. 10 (8), 2203–2225. [PMC free article] [PubMed] [Google Scholar]
- Watada H., Fujitani Y. (2015). Minireview: Autophagy in pancreatic β-cells and its implication in diabetes. Mol. Endocrinol. 29, 338–348. 10.1210/me.2014-1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Pang S., Sun D. (2019). The pathogenesis of endemic fluorosis: Research progress in the last 5?years. J. Cell. Mol. Med. 23, 2333–2342. 10.1111/jcmm.14185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Liu M., Li X., Liu J., Li H. (2018). Origin of the autophagosome membrane in mammals. Biomed. Res. Int. 2018, 1012789. 10.1155/2018/1012789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen N., Lv Q., Du Z. G. (2020). MicroRNAs involved in drug resistance of breast cancer by regulating autophagy. J. Zhejiang Univ. Sci. B 21, 690–702. 10.1631/jzus.B2000076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen T., Song L., Hua S. (2021). Perspectives and controversies regarding the use of natural products for the treatment of lung cancer. Cancer Med. 10 (7), 2396–2422. 10.1002/cam4.3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. J., Martin V., Liu J. L., Klein S. R., Piya S., Gomez-Manzano C., et al. (2011). Autophagy regulation in cancer development and therapy. Am. J. Cancer Res. 1, 362–372. [PMC free article] [PubMed] [Google Scholar]
- Wozniak M., Mielczarek A., Czyz M. (2016). miRNAs in melanoma: Tumor suppressors and oncogenes with prognostic potential. Curr. Med. Chem. 23, 3136–3153. 10.2174/1389557516666160831164544 [DOI] [PubMed] [Google Scholar]
- Wu Q., Deng J., Fan D., Duan Z., Zhu C., Fu R., et al. (2018). Ginsenoside Rh4 induces apoptosis and autophagic cell death through activation of the ROS/JNK/p53 pathway in colorectal cancer cells. Biochem. Pharmacol. 148, 64–74. 10.1016/j.bcp.2017.12.004 [DOI] [PubMed] [Google Scholar]
- Wu T. T., Lu J., Zheng P. Q., Liu S. L., Wu J., Sun W., et al. (2017). Yiqi Huayu Jiedu decoction inhibits the invasion and metastasis of gastric cancer cells through TGF-β/smad pathway. Evid. Based. Complement. Altern. Med. 2017, 1871298. 10.1155/2017/1871298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Zhang Y., Li Y., Hao Y., Zhou M., Dong N., et al. (2013). High amounts of fluoride induce apoptosis/cell death in matured ameloblast-like LS8 cells by downregulating Bcl-2. Arch. Oral Biol. 58, 1165–1173. 10.1016/j.archoralbio.2013.03.016 [DOI] [PubMed] [Google Scholar]
- Yang X., Yu D. D., Yan F., Jing Y. Y., Han Z. P., Sun K., et al. (2015). The role of autophagy induced by tumor microenvironment in different cells and stages of cancer. Cell Biosci. 5, 14. 10.1186/s13578-015-0005-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Chen D., Liu H., Yang K. (2019). Increased expression of lncRNA CASC9 promotes tumor progression by suppressing autophagy-mediated cell apoptosis via the AKT/mTOR pathway in oral squamous cell carcinoma. Cell Death Dis. 10, 41. 10.1038/s41419-018-1280-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn M., Gomez J. O., Mark K., Sakamoto K. M. (2021). RSK isoforms in acute myeloid leukemia. Biomedicines 9, 726. 10.3390/biomedicines9070726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z. K., Yang B., Zhang Y., Li L. S., Zhao J. N., Hao W. (2018). Modified Huangqi Chifeng decoction inhibits excessive autophagy to protect against Doxorubicin-induced nephrotic syndrome in rats via the PI3K/mTOR signaling pathway. Exp. Ther. Med. 16, 2490–2498. 10.3892/etm.2018.6492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Gu Y., Chen B. (2019). Mechanisms of drug resistance in acute myeloid leukemia. Onco. Targets. Ther. 12, 1937–1945. 10.2147/OTT.S191621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wang P., Wan L., Xu S., Pang D. (2017). The emergence of noncoding RNAs as Heracles in autophagy. Autophagy 13, 1004–1024. 10.1080/15548627.2017.1312041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Yang S., Cui L., Zhang J. (2016). Neutrophil gelatinase-associated lipocalin worsens ischemia/reperfusion damage of kidney cells by autophagy. Ren. Fail. 38, 1136–1140. 10.3109/0886022X.2016.1158041 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu Y., Luo J., Jie J., Deng X., Song L. (2021). The herbal compound thymol targets multiple Salmonella typhimurium virulence factors for lon protease degradation. Front. Pharmacol. 12, 674955. 10.3389/fphar.2021.674955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Shen C., Zheng Z., Wang X., Zhao W., Chen X., et al. (2018). Peiminine inhibits glioblastoma in vitro and in vivo through cell cycle arrest and autophagic flux blocking. Cell. Physiol. biochem. 51, 1566–1583. 10.1159/000495646 [DOI] [PubMed] [Google Scholar]
- Zhao L., Su J., Liu S., Li Y., Xi T., Ruan J., et al. (2021). MAP kinase phosphatase MKP-1 regulates p-ERK1/2 signaling pathway with fluoride treatment. Biochem. Biophys. Res. Commun. 542, 65–72. 10.1016/j.bbrc.2020.12.100 [DOI] [PubMed] [Google Scholar]
- Zheng X. Y., Zhang Y. H., Song W. T., Cao H., Liu J. X. (2021). Effects of Buyang Huanwu decoction on neurovascular units after cerebral ischemia: A review. Zhongguo Zhong Yao Za Zhi 46, 5226–5232. 10.19540/j.cnki.cjcmm.20210610.706 [DOI] [PubMed] [Google Scholar]
- Zhu K., Liu L., Zhang J., Wang Y., Liang H., Fan G., et al. (2016). MiR-29b suppresses the proliferation and migration of osteosarcoma cells by targeting CDK6. Protein Cell 7, 434–444. 10.1007/s13238-016-0277-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Sun D., Liu H., Sun L., Jie J., Luo J., et al. (2021). Bixin protects mice against bronchial asthma though modulating PI3K/Akt pathway. Int. Immunopharmacol. 101, 108266. 10.1016/j.intimp.2021.108266 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Wang C., Luo J., Hua S., Li D., Peng L., et al. (2021). The protective role of Zingerone in a murine asthma model via activation of the AMPK/Nrf2/HO-1 pathway. Food Funct. 12 (7), 3120–3131. 10.1039/d0fo01583k [DOI] [PubMed] [Google Scholar]