Abstract
Streptococcus agalactiae is a primary cause of neonatal morbidity and mortality. Essential to the virulence of this pathogen is the production of a type-specific capsular polysaccharide (CPS) that enables the bacteria to evade host immune defenses. The identification, cloning, sequencing, and functional characterization of seven genes involved in type III capsule production have been previously reported. Here, we describe the cloning and sequencing of nine additional adjacent genes, cpsIIIFGHIJKL, neuIIIB, and neuIIIC. Sequence comparisons suggested that these genes are involved in sialic acid synthesis, pentasaccharide repeating unit formation, and oligosaccharide transport and polymerization. The type III CPS (cpsIII) locus was comprised of 16 genes within 15.5 kb of contiguous chromosomal DNA. Primer extension analysis and investigation of mRNA from mutants with polar insertions in their cpsIII loci supported the hypothesis that the operon is transcribed as a single polycistronic message. The translated cpsIII sequences were compared to those of the S. agalactiae cpsIa locus, and the primary difference between the operons was found to reside in cpsIIIH, the putative CPS polymerase gene. Expression of cpsIIIH in a type Ia strain resulted in suppression of CPS Ia synthesis and in production of a CPS which reacted with type III-specific polyclonal antibody. Likewise, expression of the putative type Ia polymerase gene in a type III strain reduced synthesis of type III CPS with production of a type Ia immunoreactive capsule. Based on the similar structures of the oligosaccharide repeating units of the type Ia and III capsules, our observations demonstrated that cpsIaH and cpsIIIH encoded the type Ia and III CPS polymerases, respectively. Additionally, these findings suggested that a single gene can confer serotype specificity in organisms that produce complex polysaccharides.
Group B streptococci (GBS) (Streptococcus agalactiae) are the leading cause of serious bacterial infections (bacteremia, pneumonia, and meningitis) in newborns, causing two to three cases per 1,000 live births (47). An indispensable GBS virulence determinant is the production of a type-specific capsular polysaccharide (CPS), which prevents the deposition of host complement factor C3b and inhibits opsonophagocytosis (45). Nine distinct capsular serotypes, Ia, Ib, and II to VIII, have been identified (54), and their chemical compositions and structures have been determined (16–19, 51, 55, 56, 58). Type Ia, Ib, II to V, and VII CPS consist of the monosaccharides glucose, galactose, N-acetylglucosamine, and N-acetylneuraminic acid. Serotypes VI and VIII lack N-acetylglucosamine, and type VIII contains rhamnose (19). Although serotypes Ia, III, and V are currently the most common isolates from the United States associated with early-onset disease (within 1 week of birth), comprising 82% of isolates (27), type III GBS are the most prevalent isolates associated with neonatal disease (5).
We previously identified a region of the GBS chromosome encoding genes involved in type III capsule production (the cpsIII locus) by screening genomic transposon libraries for CPS mutants (45, 59). DNA sequence analysis of the acapsular type III mutants led to the identification of four genes designated cpsA, cpsB, cpsC, and cpsD (43). To conform to the emerging consensus nomenclature in the CPS literature, these genes have been designated cpsIIIB, cpsIIIC, cpsIIID, and cpsIIIE, respectively. CpsIIIE was assigned a function as a galactosyltransferase, but due to potential endogenous C-4 epimerase activity, the possibility that CpsIIIE may act as a glucosyltransferase could not be ruled out (43). Mutants which produced a capsule lacking sialic acid were also identified (25, 57, 59). The asialo mutants had a common transposon insertion site in a gene approximately 9 kb downstream of cpsIIIE, which was designated cpsF (now referred to as neuIIIA). The neuIIIA gene was shown to encode a CMP-N-acetylneuraminic acid synthetase (15, 46) which complemented an Escherichia coli K1 neuA mutant, restoring synthesis of the polysialic acid capsule (15). A gene homologous to E. coli K1 neuD (formerly designated cpsE and now called neuIIID) was found adjacent to the 5′ end of neuIIIA (8). Additionally, upstream of cpsIIIB, two genes similar to regulatory proteins, designated cpsY and cpsX (referred to here as cpsIIIA to conform to the usage for the S. agalactiae type Ia homologue), have been identified, although their functions have not been confirmed (24). Thus, seven genes of the cpsIII locus have previously been described, along with the divergently transcribed cpsY, whose role in CPS synthesis is not known.
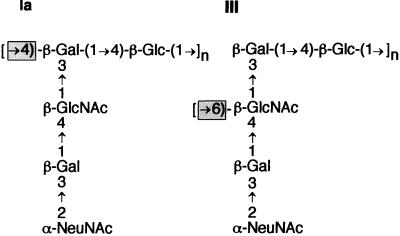
The type III CPS produced under the direction of the cpsIII operon possesses a backbone of repeating [→6)-β-d-N-acetylglucosamine-(1→3)-β-d-galactose-(1→4)-β-d-glucose-(1→]n trisaccharide units. Each repeating unit carries a disaccharide side chain of α-d-N-acetylneuraminic acid-(2→3)-β-d-galactose, 1→4 linked via the galactose to the backbone N-acetylglucosamine. The type Ia CPS is composed of a polymer of β-1,4-linked lactose to which a trisaccharide side chain of α-d-N-acetylneuraminic acid-(2→3)-β-d-galactose-(1→4)-β-d-N-acetylglucosamine is attached to each repeating unit galactose by a β-d-N-acetylglucosaminyl-(1→3) linkage. Traditionally, the GBS CPS repeating unit structures have been depicted to emphasize these differences and to reflect their conformational structure in the native polysaccharide configuration (16, 17). However, if the repeating unit structures are redrawn as in Fig. 1, it can be seen that the type Ia and type III CPS oligosaccharide structures differ only by the linkage between repeating units. Therefore, the essential difference between the two capsules is due to the glycosidic bond formed during CPS polymerization. The structural similarity of these capsule oligosaccharides has led us to hypothesize that there is a high degree of genetic relatedness between their capsule synthesis loci. This hypothesis is supported by two further observations: first, the CPS structures of S. pneumoniae type 14 and type III GBS are identical except for the lack of the terminal sialic acid side chain residue of the former (Fig. 1); second, the cps loci of S. pneumoniae type 14 and GBS type Ia are highly homologous within their regulatory, repeat unit, and oligosaccharide transport genes (60).
FIG. 1.
GBS type Ia and III CPS repeating unit structures. Ia, S. agalactiae type Ia CPS subunit structure; III, S. agalactiae type III CPS subunit structure; Gal, galactose; Glc, glucose; GlcNAc, N-acetylglucosamine; NeuNAc, N-acetylneuraminic (sialic) acid. The critical linkages differentiating the type Ia and III CPS are shown in the shaded boxes.
In this study, we have completed the molecular characterization of nine additional genes that make up the remainder of the cpsIII operon. We present the organization of the 15.5 kb of DNA containing the 16 genes involved in CPS III synthesis, describe its similarity to other complex polysaccharide synthesis loci, and demonstrate its expression as a polycistronic operon. Last, we provide evidence that a single polymerase gene determines the unique bond formed during polymerization of the type Ia or type III repeating units and thus confers CPS serotype specificity.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used for this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Genotype/phenotypea | Reference or source |
|---|---|---|
| S. agalactiae | ||
| A909 | Wild type/cpsIa | 29 |
| COH1 | Wild type/cpsIII, Tetr | 32 |
| COH1-13 | COH1, Tn916ΔE cpsE/CPS− Tetr Emr | 43 |
| COHY-104 | COH1, cpsIIIB, ΩKm-2 via pHY108/CPS− Tetr Kanr Cms | This study |
| COHY-105 | COH1, cpsIIIC, ΩKm-2 via pHY113/CPS− Tetr Kanr Cms | This study |
| COHY-106 | COH1, cpsIIID, ΩKm-2 via pHY109/CPS− Tetr Kanr Cms | This study |
| COHY-107 | COH1, cpsIIIA, ΩKm-2 via pHY120/CPS− Tetr Kanr Cms | This study |
| E. coli | ||
| DH5α | F−endA1 (rK− mK+) supE44 thi-1 hsdR17 gyrA (Nalr) recA1 Δ(lacZYA-argF)U169 deoR relA1 [φ80Δlac(lacZ)M15] | BRL, Inc. |
| MC1061 | F−araD139 Δ(ara-leu)7696 Δ(lac)X74 galU galK hadR2 (rK− mK+) mcrB1 rpsL (Strr) | 53 |
| Plasmids | ||
| pBS | Cloning vector, Apr 2.8 kb | Stratagene |
| pBS9.0 | pBS, cpsIIIIJKLneuBCDA/Apr 11.8 kb | This study |
| pCER107 | pBS, cpsIIIABCDE/Apr 5.7 kb | 25 |
| pCER108 | pBS, cpsIIIEFGHI/Apr 6.1 kb | 25 |
| pDC128 | Cmr, pDC123 with a 7,122-bp XbaI fragment containing cpsIIIKLneuBCDA; 11.6 kb | This study |
| pCIV2 | Kanr/ΩKm-2 5.9 kb | 38 |
| pHY101 | Cmr, 597 bp ‘cpsIIIB’ fragment cloned from pCER107 via ApaI/HindIII ligation into pVE6007 | This study |
| pHY104 | Cmr, 412-bp ‘cpsIIID’ fragment cloned from pHY204 via KpnI/HindIII ligation into pVE6007 | This study |
| pHY105 | Cmr, 537-bp ‘cpsIIIC’ fragment cloned from pHY205 via KpnI/HindIII ligation into pVE6007 | This study |
| pHY108 | pHY101 with ΩKm-2 from pCIV2 cloned into FspI site of ‘cpsIIIB’/Kanr | This study |
| pHY109 | pHY104 with ΩKm-2 from pCIV2 cloned into BglII site of ‘cpsIIID’/Kanr | This study |
| pHY113 | pHY105 with ΩKm-2 from pCIV2 cloned into BglII site of ‘cpsIIIC’/Kanr | This study |
| pHY115 | Cmr, 648-bp ‘cpsIIIA’ fragment cloned into pVE6007 from pHY208 via KpnI/HindIII ligation | This study |
| pHY120 | pHY115 with ΩKm-2 from pCIV2 cloned into NsiI site of cpsIIIA using a BamHI-NsiI adapter/Kanr | This study |
| pHY204 | Apr, 345-bp ‘cpsIIID’ amplicon cloned into pT7Blue vector | This study |
| pHY205 | Apr, 470-bp ‘cpsIIIC’ amplicon cloned into pT7Blue vector | This study |
| pHY208 | Apr, 583-bp ‘cpsIIIA’ amplicon cloned into pT7Blue vector | This study |
| pSH101 | Apr, 541-bp ‘neuIIIA’ amplicon cloned into pT7Blue vector | This study |
| pSH102 | Cmr, 525-bp ‘neuIIIA’ cloned from pSH101 via ApaI/EcoRV ligation into pVE6007 | This study |
| pSH104 | Cmr, plasmid rescue construct containing pSH102 and sequences downstream of neuIIIA, 7.3 kb | This study |
| pT7Blue | Apr 2.9-kb T-overhang PCR cloning vector | Novagen |
| pVE6007 | Cmr temperature-sensitive shuttle vector, 3.4 kb, ori[Ts] | 30 |
| pDC123 | Cmr, phoZ, gram-positive/negative blue/white screening expression vector | 8 |
| pDC123(GHIa) | Cmr, 2,350-bp PCR product from type Ia GBS strain A909 containing cpsIaGH cloned into AflII/ApaI-cut pDC123 | This study |
| pDC123(HIa) | Cmr, 1,767-bp XbaI-cut PCR product containing cpsIaH from type Ia GBS strain A909 cloned into SmaI/XbaI-cut pDC123 | This study |
| pDC123(GHIII) | Cmr, 2,244-bp PCR product containing cpsIIIGH from type III GBS strain COH1 cloned into AflII/EcoRV-cut pDC123 | This study |
| pDC123(HIII) | Cmr, 209-bp BclI deletion of pDC123(GHIII) | This study |
Tetr, tetracycline resistance; Emr, erythromycin resistance; Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Cms, chloramphenicol susceptible; Kanr, kanamycin resistance; Nalr, nalidixic acid resistance; CPS−, capsule negative.
Nucleotide sequencing and DNA analysis.
Sequencing was performed on plasmid subclones containing COH1 genomic DNA (25), purified PCR products, and a subclone generated by plasmid insertion/rescue (Fig. 2). Fluorescent dye terminator sequencing reactions were performed using a Ready Reaction sequencing kit (Perkin-Elmer Cetus, Foster City, Calif.) according to the manufacturer's specifications and analyzed on an Applied Biosystems 373A automated sequencer. Both strands of each DNA template were sequenced.
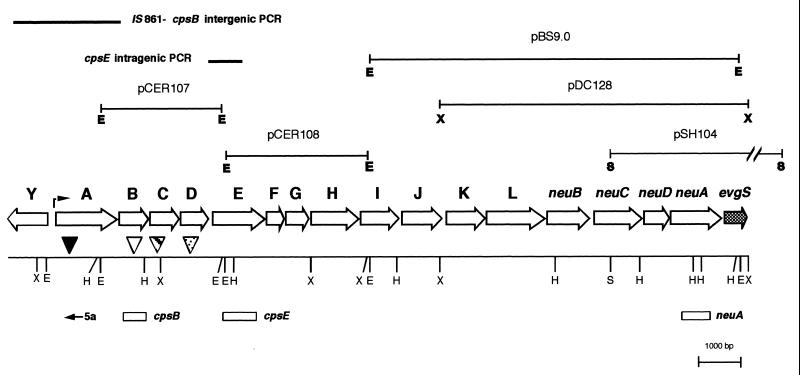
FIG. 2.
Organizational map of the type III GBS cps operon. ORFs within the operon, with the direction of transcription, are indicated by the open arrows. The corresponding gene designation is shown above each ORF. Restriction site designations: E, EcoRI; H, HindIII; S, SacI; X, XbaI. Plasmids used in sequencing the locus are depicted above the operon map. PCR products used to derive additional sequence are shown as solid bars above the operon map. The cpsIII transcriptional start site is indicated with an arrow in the cpsY-cpsA intergenic region. The ΩKm-2 insertion sites are indicated with triangles: filled triangle, cpsIIIA insertion; open triangle, cpsIIIB insertion; striped triangle, cpsIIIC insertion; dotted triangle, cpsIIID insertion. Sequences used in generating RNA probes are marked with open boxes. The location and orientation of the oligonucleotide used to initiate the primer extension reaction (primer 5a) is also shown.
cpsIIID-cpsIIIE and IS861-cpsIIIB intergenic regions were amplified from the COH1 chromosome by PCR. Primers used for amplifying the IS861-cpsIIIB intergenic region hybridized to a region downstream of IS861 (5′-GAAAGGTTTGCTTTGTCGTGTCGGATATAG-3′) (44) and to the 5′ end of cpsIIIB (5′-CTTCAACGCTTTTGGGCCCATCATCTACATC-3′) (43). The second PCR product was an amplified region of cpsIIID-cpsIIIE that was produced by using primers 5′-GAAGTAAGGGACTCTGGTATTGA-3′, which hybridizes to the 3′ end of cpsIIID, and 5′-GAGCAAACCTATAATAGCACCCGT-3′, which hybridizes to cpsIIIE. The PCR products were prepared for sequencing by purification using a QiaQuick PCR purification kit (Qiagen Inc., Valencia, Calif.).
DNA sequence analysis and FASTA homology searches were performed using the Genetics Computing Group (University of Wisconsin) software on a UNIX computer at the University of Washington. ENTREZ and BLAST homology searches were performed using the National Center for Biotechnology Information Internet server.
Plasmid insertion/rescue.
To clone and sequence chromosomal DNA 3′ of neuIIIA, an intragenic ‘neuIIIA’ fragment was amplified and cloned into the pT7Blue vector (Novagen Inc., Madison, Wis.) to create pSH101. An ApaI/EcoRV fragment containing the ‘neuIIIA’ sequence was isolated from pSH101 and ligated into the temperature-sensitive vector pVE6007 (30) previously digested with the same restriction enzymes. This new plasmid was designated pSH102 and transformed into competent COH1 as described elsewhere (13). Transformants were screened for plasmid integration within the chromosomal wild-type (wt) neuIIIA as described elsewhere (61). Subsequent digestion of integrant chromosomal DNA with SacI, followed by intramolecular ligation, rescued the plasmid and flanking chromosomal sequences. The ligated DNA was transformed into E. coli DH5α, and chloramphenicol-resistant clones were isolated by growth at 30°C on L agar containing 10 μg of chloramphenicol per ml.
Generation of cps gene allelic exchange plasmids.
Intragenic fragments of cpsIIIA, cpsIIIC, and cpsIIID were amplified from COH1 chromosomal DNA by PCR and directly ligated into the vector pT7Blue. The intragenic fragments were excised from pT7Blue with KpnI and HindIII and ligated into KpnI- and HindIII-digested pVE6007. To clone the intragenic fragment of cpsIIIB, pCER107 (43) was digested with ApaI and HindIII, removing 47 nucleotides (nt) from the 5′ end and 425 nt from the 3′ end of cpsIIIB. This 616-bp intragenic fragment was separated by agarose gel electrophoresis, extracted (QiaEx gel purification kit; Qiagen), and ligated into ApaI- and HindIII-digested pVE6007.
The ΩKm-2 fragment (38) from pCIV2 was subsequently cloned into sites within each of the intragenic cps gene fragments in the pVE6007-based vectors described above. For the cpsIIIC and cpsIIID constructs, the ΩKm-2 fragment was digested from pCIV2 with BamHI and cloned directly into the BglII site within each gene fragment. To insert ΩKm-2 within the cpsIIIB construct, an FspI site in cpsIIIB was ligated to the ΩKm-2 cassette previously prepared by BamHI digestion and end filled with Klenow fragment. The ΩKm-2 fragment was ligated into the NsiI site in the cpsIIIA construct via a BamHI-NsiI adapter, 5′-GATCAGCGGCCGCTTGCA-3′. Plasmid constructs containing ΩKm-2 cassettes in the same orientation relative to the chromosomal cps genes were chosen for deriving the allelic exchange mutants in COH1. After transformation of the allelic exchange vectors into GBS, the cells were grown under conditions promoting homologous recombination as described previously (61). Recombination occurring on both sides of the ΩKm-2 cassette resulted in a double-crossover recombination event and an allelic exchange mutation within the recipient gene.
PCR amplification.
PCRs were performed using standard conditions (4) and Taq polymerase (Promega Corp., Madison, Wis.). To amplify larger (>2.0-kb) PCR products, Taq extender (5 U; Stratagene Cloning Systems, La Jolla, Calif.) was added to a standard 100-μl reaction according to the manufacturer's instructions.
RNA analysis.
Total bacterial cell RNA was extracted from GBS using a rapid cell disruption method with a dental amalgamator and glass beads as described previously (62). Dot blots were generated as follows. A 2.5-μg aliquot of total cellular RNA was applied to Magnagraph nylon membrane (Micron Separations Inc., Westborough, Mass.) using a Minifold I dot blot vacuum apparatus (Schleicher & Schuell, Keene, N.H.) according to the manufacturer's recommendations and UV cross-linked. Blots were hybridized using standard 50% formamide buffers (31) and 32P-labeled RNA gene probes. After hybridization, the blot was washed twice for 15 min with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS) at room temperature and twice with 0.1× SSC–0.1% SDS at 65°C, excess buffer was removed, and the filter was subjected to autoradiography.
Primer extension reactions were used to identify the 5′ end of cps-specific mRNA. Primer 5a (5′-CACATACACCTCTACGG-3′) was end labeled using [γ-32P]ATP and T4 polynucleotide kinase, hybridized to 50 μg of total cellular RNA, and extended, using displayTHERMO-RT RNA polymerase (Display Systems Biotech, Inc., Vista, Calif.). The sample was ethanol precipitated, resuspended in 8 μl of RNase A (200 μg/ml), and incubated for 20 min at 23°C. A reference standard sequencing ladder was generated using [γ-32P]ATP-labeled primer 5a with a cycleSEQ manual cycle sequencing kit (Display Systems Biotech) and loaded next to the primer extension reaction on a 7% acrylamide–8 M urea–1× Tris-borate-EDTA gel. The samples were separated by electrophoresis at 45 W for 2.5 h; the gel was transferred to Whatman 3MM filter paper, dried, and exposed to X-ray film.
Generation of cps type Ia and III gene expression constructs.
Plasmid constructs were generated to test the effect on capsule production of heterologous cps type Ia and III gene expression. The cpsIaGH and cpsIIIGH genes were PCR amplified from strain A909 and COH1 chromosomal DNA, respectively, using primer Pol1f, which contained a 5′ ApaI adapter (5′-CCCGGGCCCAGATGTTATCATATCA-3′), and Pol1r, containing a 5′ AflII adapter (5′-CCAGATCTTAAGTTTCGTCTTTTCTTC-3′) (adapters are underlined). The 2.35-kb cpsIaGH amplicon was digested with ApaI and AflII and ligated to the ApaI/AflII-cut pDC123 expression vector, forming pDC123(GHIa). The 2.24-kb cpsIIIGH amplicon was digested with AflII and ligated with EcoRV/AflII-cut pDC123, forming pDC123(GHIII). The cpsIaH gene was amplified from the GBS A909 chromosome using primers Pol2f (5′-CTGAGATTGTTATCACAC-3′) and Pol1r. The cpsIaH amplicon (1.95 kb) was digested with XbaI and cloned into SmaI/XbaI-cut pDC123, forming pDC123(HIa). Constructs for the expression of cpsIIIH were derived from pDC123(HIIII) by excision of a 0.21-kb BclI fragment intragenic to cpsIIIG, forming pDC123(IIII). The constructs were transferred to the appropriate GBS hosts as described (13), and their identity was subsequently confirmed by PCR and restriction analyses.
Serotype determination of cell-associated CPS.
A modification of the colony immunoblot method described by Rubens et al. (45) was used to identify the type of CPS produced on recombinant strains compared to the wt strains. Aliquots (3 μl) of stationary-phase cultures were spotted onto a NitroPlus membrane (Micron Separations), fixed for 5 min with 70% ethanol, air dried, and blocked with BLOTTO (5% nonfat dry milk [NFDM] in phosphate-buffered saline [PBS]) for 30 min. The blots were washed three times for 5 min each with PBS and then incubated for 1 h with either type Ia or III rabbit anti-CPS antiserum (kindly provided by Michael Wessels, Channing Laboratory, Boston, Mass.) diluted 1:20,000 (type Ia) or 1:30,000 (type III) in BLOTTO. The blots were washed three times (5 min each in PBS) and then incubated for 1 h with horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody, diluted 1:20,000 in BLOTTO (Sigma, St. Louis, Mo.). After three 5-min PBS washes, 1 ml of SuperSignal chemiluminescent substrate for HRP (Pierce, Rockford, Ill.) was added to the surface of the filter according to the manufacturer's directions. Chemiluminescence was detected by exposing the blot to X-ray film (Kodak, Rochester, N.Y.).
Quantification of cell-associated CPS.
Mutanolysin digests of bacterial cells for extraction of CPS were carried out as described by Paoletti et al. (39), with minor modifications as follows. GBS were grown overnight in 10 ml of Todd-Hewitt broth, with antibiotic selection if necessary, harvested by centrifugation at 10,000 × g for 10 min at 4°C, and washed twice with 1.5 ml of 50 mM sodium phosphate buffer (pH 7.0). The cell pellets were resuspended in a final volume of 0.6 ml of 50 mM phosphate buffer (pH 7.0); 200 μl of the cell solution was transferred in duplicate to fresh microcentrifuge tubes; the cells were pelleted at 12,000 × g for 10 min at 4°C and then resuspended in 750 μl of 40% sucrose (wt/vol) in 50 mM sodium phosphate buffer (pH 7.0). To each tube, 250 μl of mutanolysin (1 mg/ml in 50 mM sodium phosphate buffer [pH 7.0]) was added. The samples were incubated at 37°C with end-over-end mixing for 1 h. Protoplasts and cell debris were removed by centrifugation at 13,000 × g for 4 min at 4°C, and the supernatants were transferred to fresh tubes and stored at −20°C.
Plates for quantitative enzyme-linked immunosorbent assay (ELISA) were prepared by coupling poly-l-lysine to purified type Ia or III CPS (kindly provided by Lawrence Paoletti, Channing Laboratory, Brigham and Women's Hospital, Harvard Medical School, Boston, Mass.), which was used to coat 96-well microtiter plates as described elsewhere (45). Coating solution (CPS [1.0 μg/ml] coupled to poly-l-lysine in 40 mM sodium phosphate buffer [pH 7.0]; 10 ml/plate) was prepared. To each well of the microtiter plate, 100 μl of the coating solution was dispensed, and the plates were incubated at 37°C for 1 h. The coating solution was discarded, and the plates were washed three times with 40 mM sodium phosphate (pH 7.0)–0.05% Tween 20 (PBT; 200 μl/well) and then blocked with 0.5% (wt/vol) NFDM in 40 mM sodium phosphate buffer (pH 7.0). The blocking solution was decanted, the wells were washed three times with 200 μl of PBT; and the plates were stored with 200 μl of PBT per well for up to 48 h before use at 4°C.
The amount of cell-associated, serotype-specific CPS produced by each strain was determined by competitive inhibition ELISA assay as described elsewhere (39). Assays were performed with duplicate plates in at least three independent experiments for each strain and serotype combination. CPS standards were prepared by dissolving purified type Ia or III CPS in H2O to a final concentration of 10 ng/μl. Then 160 μl of CPS standard or CPS mutanolysin extract was transferred to the first column of the plate, and serial fourfold dilutions were performed across the plate from left to right into 40 mM sodium phosphate (pH 7.0). Rabbit anti-CPS antiserum (diluted 1:10,000 in 5% NFDM–50 mM sodium phosphate [pH 7.0]) was dispensed into the plate (120 μl/well), which was then allowed to stand at room temperature for 30 min, allowing the antibody to bind. From each well of the dilution series plate, 200 μl was transferred to the corresponding well (i.e., A1 to A1) of a CPS–poly-l-lysine-coated microtiter plate. One well of the plate, chosen as a blank, received 5% NFDM–50 mM Na phosphate (pH 7.0) instead of an antibody-CPS dilution. The ELISA was then developed as described elsewhere (39). The ELISA data were fitted to the four-parameter model of Rodbard (42), and the concentration of type-specific CPS in the undiluted extracts was calculated (50% inhibitory concentration of standard/50% inhibitory concentration of unknown) × concentration of undiluted standard).
Nucleotide sequence accession number.
The cpsIII DNA sequence has been deposited in GenBank with accession no. AF163833.
RESULTS
Organization of the type III GBS cps locus.
We have completed the sequencing of the entire GBS type III capsule synthesis locus by subcloning or amplifying the genomic DNA 5′ and 3′ of the cps genes identified previously (Fig. 2). Analysis of the compiled sequence revealed a total of 16 cps-related open reading frames (ORFs), of which 9 were previously undescribed. Additionally, a divergently transcribed monocistronic ORF 5′ of the type III locus containing cpsIIIY was observed. To facilitate comparison with the type Ia S. agalactiae and type 14 S. pneumoniae cps loci, we have changed the gene designations for cpsABCDEF, and thus the gene order for the CPS synthesis locus is cpsIIIABCDEFGHIJKL neuIIIBCDA (Fig. 2). All of the ORFs except cpsIIIY are preceded by recognizable ribosome binding sites. Most of the ORFs are closely linked, with stop codons that overlap the translational initiation sites of the adjacent ORFs. There are significant gaps, however, between cpsIIID and cpsIIIE (51 bp), cpsIIII and cpsIIIJ (33 bp), cpsIIIJ and cpsIIIK (84 bp), and neuIIIB and neuIIIC (75 bp). We have examined these intergenic gaps and found that they do not contain potential regions of dyad symmetry, transcriptional terminators, or consensus promoter sequences. If we allow the ORFs to initiate with alternative start codons such as CUG or UUG, the intragenic gaps between cpsIIID and cpsIIIE, cpsIIII and cpsIIIJ, and neuIIID and neuIIIA decrease in size from 51 to 7, 33 to 0, and 69 to 21 bp, respectively. The alternative start codons for these putative ORFs also contain upstream ribosome binding sites.
The cpsIII locus is nearly identical to the cpsIa operon of S. agalactiae type Ia (Fig. 3 and Table 2). Identity between translated ORFs ranges between 94 and 100% with the exception of CpsIIIG and CpsIIIH, where it falls to 72 and 24%, respectively. A homologue for CpsIIIY was not reported for type Ia S. agalactiae. The arrangement of genes from cpsIIIA to cpsIIIL of the GBS type III cps operon is also strikingly similar to that of the entire capsule locus from S. pneumoniae type 14 (Fig. 3). The nucleotide sequence similarity over the length of the common genes is 52% (7,784 identical nt out of 15,133 nt, with 215 gaps) when the type 14 capsule gene sequence is aligned with the GBS type III locus. The resemblance between the genes when translated is also high at approximately 50% identity and 60% similarity as anticipated, based on their identical capsular structures if the terminal sialic acid residue present on the type III CPS is excluded.
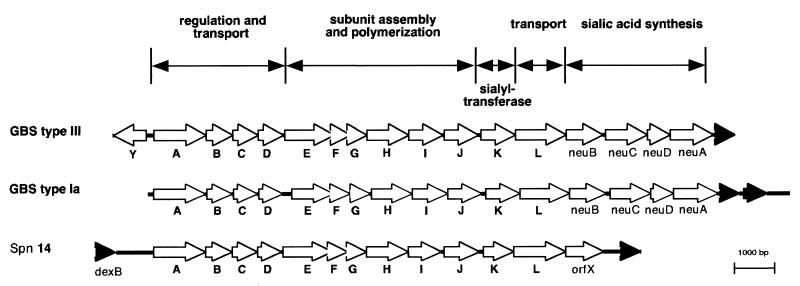
FIG. 3.
Schematic representation of the organization of the CPS synthesis loci of S. agalactiae type III (GBS type III), S. agalactiae type Ia (GBS type Ia) (60), and S. pneumoniae type 14 (Spn 14) (23). The unfilled arrows indicate ORFs within the operons and the direction of transcription. Filled arrows indicate ORFs not involved in CPS synthesis that flank the cps gene regions. Letters below the arrows indicate the gene designations. The organization of the cps loci into regions of associated gene function is indicated above the ORF diagrams.
TABLE 2.
Sequence comparisons to the NCBI databasea
| Gene product | Proposed function | Similar gene products | % Identity | Range (amino acid) | Accession no. |
|---|---|---|---|---|---|
| CpsIIIY | Transcriptional regulatorb | Azorhizobium caulinodans, NACc | 42 | 1–80 | AJ006238 |
| Klebsiella pneumoniae, NACc | 44 | 1–74 | L01114 | ||
| Comamonas testosteroni, TsaRf | 37 | 1–80 | U32622 | ||
| H. influenzae Rd KW20, OxyRc | 27 | 1–205 | U49355 | ||
| E. coli, NACc | 43 | 1–74 | U56736 | ||
| CpsIIIA | Transcriptional regulatorb | S. agalactiae type Ia, CpsIaAf | 95 | 1–486 | AB028896 |
| S. thermophilus Sfi6, EpsAf | 47 | 1–486 | U40830 | ||
| S. thermophilus, EpsAf | 52 | 78–486 | AF053346 | ||
| S. pneumoniae, Cps14fAf | 42 | 5–485 | X85787 | ||
| S. pneumoniae, Cps23fAf | 42 | 5–485 | AF030373 | ||
| S. pneumoniae, Cps33fAf | 42 | 5–485 | AJ006986 | ||
| CpsIIIB | Tyrosine phosphataseb | S. agalactiae type Ia, CpsIaBg | 99 | 1–243 | AB028896 |
| S. thermophilus, EpsBg | 65 | 1–243 | AF053346 | ||
| S. thermophilus Sfi6, EpsBg | 64 | 1–243 | U40830 | ||
| S. salivarius, CpsBg | 64 | 1–243 | X94980 | ||
| S. pneumoniae, Cps19fBc | 60 | 1–243 | AF030367 | ||
| CpsIIIC | Chain length regulatorb | S. agalactiae type Ia, CpsIaCf | 99 | 1–230 | AB028896 |
| S. thermophilus MR-1C, EpsCf | 50 | 9–230 | AF053348 | ||
| S. salivarius, CpsCf | 50 | 1–230 | X94980 | ||
| S. thermophilus Sfi6, EpsCf | 49 | 1–230 | U40830 | ||
| S. pneumoniae, Cap33fCf | 45 | 1–230 | AJ006986 | ||
| S. pneumoniae, Cps23fCf | 44 | 9–230 | AF030373 | ||
| CpsIIID | Tyrosine kinase, CPS chain length regulator/exporterb | S. agalactiae type Ia, CpsIaCf | 99 | 1–229 | AB028896 |
| S. salivarius, CpsDf | 57 | 1–222 | X94980 | ||
| S. thermophilus Sfi6, EpsDf | 56 | 1–222 | U40830 | ||
| S. pneumoniae, Cps23fDf | 51 | 1–229 | AF030373 | ||
| S. pneumoniae, Cps19fDc | 53 | 1–224 | U09239 | ||
| CpsIIIE | Glc-1-P transferasec | S. agalactiae Ia, CpsIaEc | 99 | 1–449 | AB028896 |
| S. salivarius, CpsEf | 47 | 1–432 | X94980 | ||
| S. pneumoniae, Cap33fEf | 48 | 33–432 | AJ006986 | ||
| S. pneumoniae, Cps19fEf | 44 | 1–432 | U09239 | ||
| S. pneumoniae, Cps14fEc | 44 | 1–432 | X85787 | ||
| CpsIIIF | β-1,4-Gal transferase enhancerb | S. agalactiae Ia, CpsIaFg | 96 | 1–149 | AB028896 |
| S. pneumoniae, Cps14fFf | 83 | 1–149 | X85787 | ||
| Lactococcus lactis NIZO B40, EpsEc | 38 | 2–149 | U93364 | ||
| Sphingomonas strain s88, SpsKc | 32 | 1–142 | U51197 | ||
| E. coli, N-GlcAc | 30 | 3–146 | AF013583 | ||
| CpsIIIG | β-1,4-Gal transferased | S. agalactiae Ia, CpsIaGc | 72 | 1–147 | AB028896 |
| S. pneumoniae, Cps14fGc | 63 | 1–126 | X85787 | ||
| L. lactis NIZO B40, EpsFc | 32 | 1–157 | U93364 | ||
| Rhizobium leguminosarum, PssEc | 34 | 1–111 | X99850 | ||
| R. leguminosarum, PssEc | 34 | 1–111 | AF014054 | ||
| CpsIIIH | CPS polymerasee | S. pneumoniae, Cps14fHf | 25 | 1–375 | X85787 |
| S. agalactiae Ia, CpsIaHf | 24 | 137–363 | AB028896 | ||
| S. suis Cps1Hf | 23 | 6–361 | AF155804 | ||
| CpsIIII | β-1,3-GlcNAc transferased | S. agalactiae Ia, CpsIaIc | 98 | 5–320 | AB028896 |
| S. pneumoniae, Cps23fEf | 53 | 1–276 | AF030373 | ||
| S. thermophilus Sfi6, EpsIf | 35 | 1–255 | U40830 | ||
| S. pneumoniae, Cap33ff | 36 | 5–237 | AJ006986 | ||
| S. pneumoniae, Cps14fEc | 27 | 1–290 | X85787 | ||
| CpsIIIJ | β-1,4-Gal transferased | S. agalactiae Ia, CpsIaJc | 99 | 1–315 | AB028896 |
| S. pneumoniae, Cps14fJc | 38 | 7–224 | X85787 | ||
| S. thermophilus Sfi6, EpsIf | 37 | 7–242 | U40830 | ||
| S. pneumoniae, Cps23fEf | 37 | 9–237 | AF030373 | ||
| S. pneumoniae, Cap33ff | 27 | 9–247 | AJ006986 | ||
| CpsIIIK | Sialyltransferaseb | S. agalactiae Ia, CpsIaKg | 99 | 1–460 | AB028896 |
| H. ducreyi Lstc | 27 | 143–299 | AF101047 | ||
| H. influenzae Rd KW20, OrfYg | 26 | 102–298 | HI0871 | ||
| CpsIIIL | Repeating unit transporterb | S. agalactiae Ia, CpsIaL | 100 | 1–460 | AB028896 |
| H. influenzae Rd KW20g | 26 | 102–298 | M94855 | ||
| Staphylococcus aureus, Cap1Ff | 24 | 183–400 | U10927 | ||
| Streptococcus pneumoniae, Cps23ff | 19 | 4–456 | AF030373 | ||
| NeuIIIB | NeuNAc synthetasec | S. agalactiae Ia, NeuBf | 99 | 1–341 | AB028896 |
| E. coli, NeuBc | 56 | 2–337 | U05248 | ||
| Methanococcus jannaschiif | 47 | 3–329 | H64432 | ||
| N. meningitidis, SiaCc | 38 | 4–315 | M95053 | ||
| Bacillus subtilis, SpsEf | 34 | 2–334 | X73124 | ||
| NeuIIIC | UDP-GlcNAc 2-epimerasec | S. agalactiae Ia, NeuCf | 94 | 1–320 | AB028896 |
| E. coli, NeuCc | 39 | 1–380 | M84026 | ||
| N. meningitidis, SiaAf | 38 | 4–315 | M95093 | ||
| Rattus norvegicusc | 29 | 1–318 | Y07744 | ||
| Methanococcus thermoautotrophicumf | 47 | 3–329 | AE000860 | ||
| NeuIIID | Sialic acid synthesisc | S. agalactiae type Ia, NeuDf | 100 | 1–209 | AB028896 |
| E. coli, NeuDc | 30 | 1–209 | U05248 | ||
| B. subtilis, YvfDg | 25 | 3–208 | Z71928 | ||
| Caulobacter crescentus, LpsBg | 27 | 56–206 | AF062345 | ||
| Campylobacter jejuni, WlaIg | 27 | 65–207 | Y11648 | ||
| NeuIIIA | CMP-NeuNAc synthetasec | S. agalactiae type Ia, NeuAb | 94 | 1–370 | AB028896 |
| E. coli, NeuAc | 37 | 2–395 | J05023 | ||
| N. meningitidis, SiaBc | 35 | 4–213 | U04328 | ||
| H. influenzae, NeuAc | 33 | 1–213 | U32807 | ||
| H. ducreyi, NeuAc | 29 | 1–209 | U54496 |
Translations of cpsIII ORFs were aligned using BLAST 2.0 with the BLASTP algorithm using the nonredundant database and the BLOSUM62 matrix at the National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, Md. The extent of the alignment returned by BLAST is given as the range.
Predicted by sequence similarity.
Enzymatic activity determined.
Predicted based on similarity to S. agalactiae type Ia.
Determined in this study.
Proposed function.
Unknown function.
Upstream of the GBS CPS synthesis locus, but in the opposite orientation, is cpsIIIY. It is separated by an intergenic span of 189 nt from the cpsABCDEFGHIJKL neuIIIBCDA (cpsIII) operon. At the 3′ end of the operon, downstream of neuIIIA is a gap of 170 nt before another ORF is detected. This putative ORF has amino acid sequence homology with the central portion of the E. coli gene evgS, a putative sensor protein from a two-component regulatory system, and is therefore probably not a component of the capsule synthesis genes.
Sequence similarities of the CPS ORFs to other genes.
All of the cps gene sequences were translated, and the amino acid sequences were compared to protein sequence databases at the National Center for Biotechnology Information using the gapped BLAST version 2.0 program (1). Similarities to proteins in the sequence databases are presented in Table 2. The translated sequences of cpsIIIA through cpsIIIJ are similar to those of a number of genes from gram-positive organisms involved in CPS or exopolysaccharide synthesis.
The cpsIIIY and cpsIIIA (previously cpsX) genes coding for homologues of LysR and LytR, respectively, were previously described (24). The genes cpsIIIB, cpsIIIC, cpsIIID, and cpsIIIE have also been described (43). CpsIIIB is homologus to S. pneumoniae CpsB, a tyrosine phosphatase involved in CPS regulation (36). Capsule expression is reduced when CpsB dephosphoryates CpsD, an autophosphorylating protein kinase involved in CPS chain length determination and transport. CpsIIID is homologous to the S. pneumoniae CpsD and also shares amino acid similarity with the carboxy terminus of Wzc, an autophosphorylating tyrosine kinase for lipopolysaccharide synthesis in E. coli. CpsIIIC, transcribed from the ORF immediately 5′ of cpsIIID, is homologous to the amino-terminal end of Wzc (40). CpsIIIE is a glycosyl-1-phosphate transferase (60). The function of CpsIIIF is unknown, but its S. pneumoniae type 14 homologue, Cps14F, is thought to enhance the activity of Cps14G. CpsIIIG is homologous to Cps14G of S. pneumoniae and CpsIaG from S. agalactiae type Ia, β-1,4-galactosyltransferases that catalyze the addition of the second monosaccharide in the formation of the CPS repeating unit.
The cpsIIIH gene is predicted to encode a protein with eight transmembrane domains which is homologous to the putative polysaccharide polymerase encoded by cps14H from S. pneumoniae type 14 (22). This homology led us to speculate that CpsIIIH was responsible for polymerization of the oligosaccharide precursors in the type III CPS and thereby introduction of the structural determinant distinguishing type III from type Ia capsule. The aligned sequences of the cpsIIIH gene product, Cps14H from S. pneumoniae, and CpsIaH, a GBS type Ia homologue (60), are depicted in Fig. 4. The translated type Ia and type III gene products are strikingly similar outside the CpsIIIH region (99.3% similarity and 99.2% identity) but diverge markedly from the carboxy-terminal end of CpsIIIG to the amino-terminal end of CpsIIII (41% similarity and 28% identity). Further analysis of this region is discussed below. The CpsIIII and CpsIIIJ protein sequences are highly homologous to sequences of several glycosyltransferases, including Cps14I and Cps14J from S. pneumoniae type 14 (20, 22, 23).
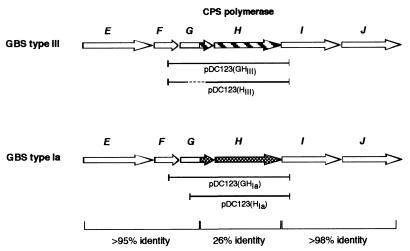
FIG. 4.
Capsule subunit assembly locus. Regions of divergent sequence between type Ia and III strains are shown either striped (type III) or checked (type Ia). The degree of homology between the conserved and nonconserved regions is shown at the bottom. Gene designations are shown as single letters above each ORF map, and the CPS polymerase (H) is also indicated. The plasmids used in the polymerase expression constructs are shown beneath each map. Sequences deleted in the construction of pDC123(HIII) are shown with dashes.
The cpsIIIK gene shares homology with its type Ia GBS homologue, cpsIaK, with the lipooligosaccharide synthetase orfY (HI0871) from Haemophilus influenzae, and lst, which encodes a novel sialyltransferase in H. ducreyi (7). Lst, like CpsIIIK, lacks homology to sialyltransferases in E. coli, H. influenzae, Neisseria gonorrhoeae, N. meningitidis, and mammals but shows significant homology to the H. influenzae OrfY. Interestingly, Lst catalyzes an α(2→3) linkage between the terminal sialic acid and a galactose residue in the lipooligosaccharide of H. ducreyi, the same specific glycosidic bond between these residues in the GBS capsular polysaccharides. These observations suggest that CpsIIIK is the cpsIII sialyltransferase.
The cpsIIIL gene encodes a protein which is weakly similar to several known and presumed oligosaccharide transport proteins. The genes neuIIIB, neuIIIC, neuIIID, and neuIIIA encode the final four proteins in the locus. NeuIIID (formerly called CpsE) and NeuIIIA have been previously described (8, 15). These four genes are most similar to the neuABCD genes from the gram-negative organism E. coli K1 (6) and the siaCAB genes of N. meningitidis serogroup B (11), which are responsible for the synthesis and activation of sialic acid for capsule production in these species. We previously demonstrated that neuIIIA encodes a CMP-NeuNAc synthetase, catalyzing the activation of sialic acid with CMP (15). Homologues of NeuIIIB are NeuNAc synthetases (2). neuC of E. coli K1 has been characterized as a UDP-N-acetylglucosamine→ManNAc epimerase (R. P. Silver et al., submitted for publication). In this study, complementation of an E. coli K1 neuC mutant by neuIIIC indicated that NeuIIIC is functionally equivalent to the E. coli epimerase. Although the exact function of neuIIID is unknown, a role in sialic acid synthesis has recently been established, since a nonpolar mutation in the E. coli K1 homologue neuD can be complemented by the addition of exogenous sialic acid or expression of neuIIID in trans (9a).
Identification of the cpsIII transcriptional start site.
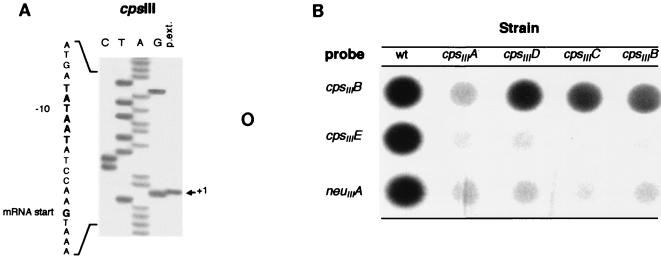
We hypothesized that the start of transcription occurred proximal to the cpsIIIA gene. To investigate this possibility, we used COH1 mRNA and a radiolabeled primer complementary to the 5′ end of cpsIIIA to identify the 5′ transcriptional start site of the cpsIII locus by primer extension analysis. The products of the primer extension reaction are shown in Fig. 5A. We mapped the 5′ end of the transcript (indicated by the arrow) to a G nucleotide on the DNA template 38 bp upstream of the cpsIIIA start codon. We examined the sequence directly upstream of the G nucleotide to identify canonical consensus promoter sequences. A sequence (TATAAT) identical to the −10 region of the consensus ς70 E. coli promoter sequence was detected 7 bp above the 5′ end of the cpsIII transcript. A sequence 17 bp 5′ of the −10 region (TTGAAT) matched the canonical −35 region from E. coli at four of six positions. It also matched a consensus sequence for streptococcal promoters which is similar to the E. coli promoter, except that the −10 region is extended on the 5′ side to include the sequence TGN (with N being any nucleotide) (12, 52). We concluded this sequence is the cpsIII promoter.
FIG. 5.
(A) Primer extension reaction. Primer extension reactions were carried out on total cellular RNA derived from GBS strain COH1, using an antisense primer that hybridized to cpsIIIA mRNA. A sequencing ladder was generated from cpsIIIA double-stranded DNA using the same primer and was separated on a sequencing gel in lanes adjacent (lanes CTAG) to the primer extension products (p.ext.). The position of the apparent first ribonucleotide of the primer extension product (+1) is indicated to the right. The DNA sequence surrounding the start position of the mRNA is depicted vertically on the left, with the consensus −10 promoter sequence and the mRNA start site highlighted in bold. (B) RNA dot blots of total cellular RNA derived from the wt GBS strain COH1 and the ΩKm-2 cassette allelic exchange mutant strains COHY-107 (cpsIIIA), COHY-106 (cpsIIID), COHY-105 (cpsIIIC), and COHY-104 (cpsIIIB). The blots were hybridized to intragenic probes derived from the wt COH1 cpsIIIB, cpsIIIE, and neuIIIA genes and exposed to X-ray film.
Polar effects of allelic exchange mutagenesis within the cpsIII locus.
To determine if there were additional transcriptional start sites within the cpsIII locus, we developed strains incorporating transcriptional terminators within their cps loci by allelic exchange mutagenesis (26, 61). To ascertain if these mutations were polar, we tested each clone for type III capsule production by immunoblot analysis as described. The type III antiserum recognizes both sialylated and asialo forms of the type III CPS. The wt strain, COH1, reacted with the antibody as expected; however, each of the CPS mutants failed to react, indicating they were acapsular (data not shown). These data demonstrated that mutations generated by homologous recombination in the 5′ region of the CPS synthesis locus prevented expression of capsule synthesis. These results also suggested that mutations in these upstream genes were polar and altered expression of downstream cps genes. This was expected, as a transposon mutant in cpsIIIB was previously shown to be acapsular (43).
Since each of the allelic exchange mutations above abolished capsule production, we sought to determine the effect of the mutations on the expression of downstream cps genes. RNA dot blot analysis was performed on total cellular RNA isolated from COH1 and each of the isogenic mutants. Equal amounts of RNA from each strain were hybridized to three different cps gene probes independently. The probes were specific for different regions of the CPS synthesis locus: the 5′ end (cpsIIIB), the central portion (cpsIIID), and the 3′ end (neuIIIA). As shown in Fig. 5B, each of the probes hybridized to the RNA isolated from COH1 (wt) as expected and demonstrated comparable levels of mRNA transcribed from cpsIIIB, cpsIIIE, and neuIIIA. In contrast, virtually no hybridization was observed between the three probes and the RNA from the cpsIIIA mutant. Since all of the probes are specific for genes 3′ of the ΩKm-2 insertion in cpsIIIA, these data suggest that this mutation was polar and blocked expression of all downstream genes.
RNA dot blot analysis using cpsIIIB-, cpsIIIC-, and cpsIIID-derived probes demonstrated hybridization proximal but not distal to each mutation as far downstream as neuIIIA (data not shown). These results confirmed the observation that the cps genes are transcribed up to the site of the ΩKm-2 mutation but not distal to the site of the ΩKm-2 insertion for any of the mutants. Therefore, the ΩKm-2 cassette mutations interrupted transcription of the distal genes, providing further evidence that the CPS synthesis locus is transcribed as a large polycistronic message from cpsIIIA to neuIIIA.
Development of strains expressing heterologous CPS.
Based on the observation that the GBS type Ia and type III CPS oligosaccharide repeating units are structurally identical in vivo, the serotype specificity of these two polysaccharides would be determined by the unique linkage introduced during polymerization of the repeating units into high-molecular-weight CPS (Fig. 1). Homology analysis between the CPS type Ia and III loci revealed a striking divergence between CpsIaH and CpsIIIH (Fig. 4), both of which share homology with repeating unit polymerase genes in other polysaccharide operons. In addition, we observed that the homology between the cpsIa and cpsIII loci diverged at amino acid 110 of the upstream glycosyltransferase gene cpsIaG (Fig. 4). We hypothesized that the putative polymerases are responsible for catalyzing the linkage between the type Ia or III subunits, which confer structural and therefore serotype specificity.
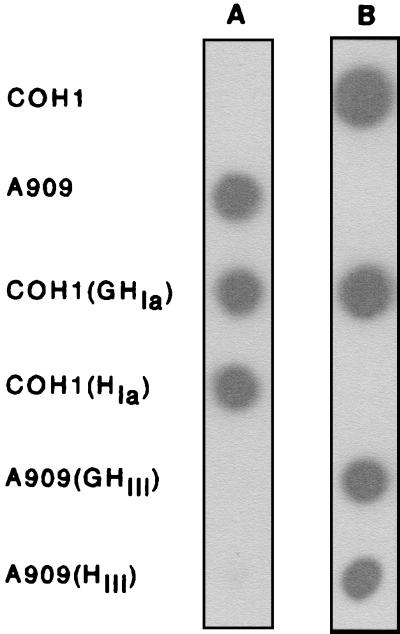
To address this hypothesis, we investigated how expression of the cpsIaH and cpsIIIH genes in the heterologous serotype strain would affect the type of CPS produced. cpsIaH and cpsIIIH were cloned independently into the gram-positive expression vector pDC123 (Fig. 4; Table 1). The cpsIaH construct, pDC123(HIa), was transformed into the type III capsule-producing strain, COH1. The type Ia-producing strain, A909, was transformed with pDC123(HIII) containing cpsIIIH. Due to the divergence in their 3′ termini, we also explored the contribution of the glycosyltransferase genes, cpsIaG and cpsIIIG, to the specific serotype of CPS synthesized. Therefore, additional constructs containing either cpsIaGH [pDC123(GHIa)] or cpsIIIGH [pDC123(GHIII)] were generated and transformed into the respective heterologous wt hosts. Immunoblot analysis was performed using rabbit type Ia or type III CPS polyclonal antisera to determine the nature of the CPS being expressed by the recombinant strains compared to the wt strains as controls. As seen in Fig. 6, cross-reactivity was not observed between A909 and the type III antisera or COH1 and antisera against type Ia CPS. When cpsIIIGH or cpsIIIH was expressed in A909, however, reactivity with the type III antiserum was observed, but reactivity with the type Ia antiserum was absent. Similarly, expression of cpsIaGH or cpsIaH alone in COH1 resulted in reactivity with the type Ia antisera (in addition to reactivity to the type III antisera for cpsIaGH). These data suggested that expression of cpsIIIH in A909 alone was capable of altering the type of CPS produced from Ia to III. Likewise, expression of cpsIaH in COH1 converted the CPS produced from type III to type Ia. Inclusion of the glycosyltransferase gene(s) (cpsIaG or cpsIIIG) just 5′ of the putative polymerase gene did not influence the nature of the CPS produced, as was anticipated from their roles as glycosyltransferases (60).
FIG. 6.
Immunoblots directed against immobilized whole GBS utilizing rabbit polyclonal anti-CPS antisera, detected with HRP-conjugated, goat anti-rabbit secondary antibody and chemiluminescent HRP substrate. Strains used: COH1, GBS type III strain; A909, GBS type Ia strain; COH1(GHIa), strain COH1 containing pDC123(GHIa); COH1(HIa), strain COH1 containing pDC123(HIa); A909(GHIII), strain A909 containing pDC123(GHIII); A909(HIII), strain A909 containing pDC123(HIII). (A) Blot exposed to type Ia antisera; (B) blot exposed to type III antisera.
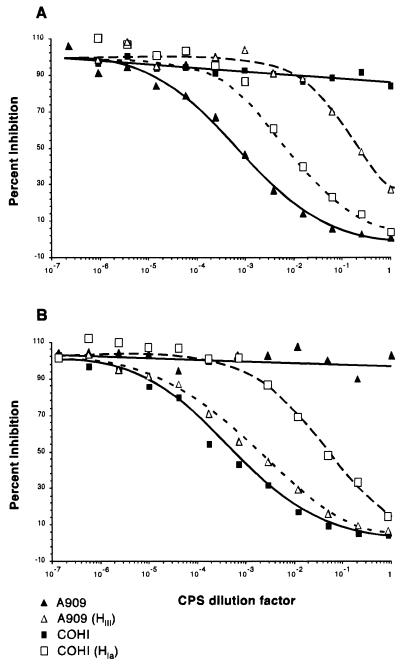
The amount of CPS produced by the wt strain and the strains carrying the heterologous polymerase genes was quantified by competitive ELISA to determine if any native CPS in the latter strains was produced. Mutanolysin CPS extracts from the wt and recombinant strains were used to competitively inhibit the binding of serotype-specific polyclonal antisera to purified type Ia or type III CPS immobilized on microtiter plates. The degree of inhibition was compared to that of purified CPS similarly preincubated with the antibody, prior to transfer to the coated plates (Fig. 7). The CPS extracts from the wt strains A909 and COH1 inhibited type Ia and III antibody binding, respectively, as expected, and failed to inhibit reactivity of the heterologous CPS serotype antisera. For A909 containing pDC123(GHIII) or pDC123(HIII), production of type Ia CPS was reduced (1.7 and 1.2 ng/μl, respectively, versus 63.5 ng/μl for A909 alone). Both of these strains demonstrated a >37-fold decrease in type Ia-reactive CPS material compared to wt but produced a substantial amount of CPS which reacted with the type III antisera (20.8 and 26.1 ng/μl). COH1 containing pDC123(HIa), expressing the type Ia CPS polymerase, produced less type III CPS (10.5 ng/μl) and synthesized a quantity of anti-type Ia-reactive CPS equal to 72% of the wt A909 levels (45.4 ng/μl). A less dramatic effect was seen with COH1 containing pDC123(GHIa), expressing both the cpsIaG and cpsIaH alleles. The reason for this disparity is unclear but could be due to decreased expression of the CPS polymerase in this construct. Nevertheless, an anti-type Ia-reactive CPS was produced (3.2 ng/μl) with a concomitant decrease in type III CPS production (61.9 versus 91.0 ng/μl). These data demonstrated that expression of the heterologous CPS polymerase gene alone, in A909 or COH1, is sufficient to confer binary synthesis of CPS that are immunoreactive to both type Ia and type III antisera. Furthermore, inclusion of cpsIaG or its type III homologue cpsIIIG in the expression constructs was unnecessary for production of the altered CPS phenotype. These data also support the hypothesis that the putative polymerase gene is responsible for the glycosidic linkage between the repeating units of type Ia or III CPS and ultimately confers the serotype specificity of the CPS.
FIG. 7.
Competitive ELISA inhibition curves produced from mutanolysin extracts of wt and heterologous CPS polymerase-expressing strains. CPS extracts were used to compete with immobilized purified type Ia CPS for anti-type Ia polyclonal antibody (A) or with immobilized purified type III CPS for anti-type III polyclonal antibody (B). Strains A909 and COH1 produce native type Ia and type III CPS, respectively. Strain A909(HIII) expresses the cpsIIIH allele from COH1, and strain COH1(HIa) expresses the cpsIaH allele from strain A909.
DISCUSSION
The complete DNA sequences of several bacterial CPS and exopolysaccharide synthesis operons have been reported (10, 23, 28, 33, 35, 41, 48, 49). The organization of the regulatory and structural genes within these loci is remarkably well conserved across CPS and exopolysaccharide-producing species. The typical arrangement begins with regulatory determinants, then genes coding for control of polymer chain length and export, followed by structural genes for repeating unit assembly (glycosyltransferases) and polymerization (polymerases), and finally additional gene(s) involved in subunit transport and/or activated monosaccharide biosynthesis. The basic organization of the GBS type III operon reported here is similar to that of polysaccharide operons in other species and parallels that reported for the S. agalactiae type Ia operon (60).
Identification and characterization of the glycosyltransferases responsible for the structural organization of the oligosaccharide repeating units of S. pneumoniae types 3 (3, 10) and 14 (20, 22, 23) and the eps genes of Lactococcus lactis (49, 50) have recently been reported (Table 2). In addition, the S. agalactiae type Ia glycosyltransferases have recently been characterized and found to be functionally equivalent to their S. pneumoniae type 14 homologues Cps14E, Cps14G, Cps14I, and Cps14J (22, 60). In both species, these enzymes are responsible for the assembly of a tetrasaccharide that is chemically identical to the asialo type III GBS CPS repeating unit. We previously reported that cpsIIIE most likely encodes a galactosyltransferase. We could not, however, rule out the possibility that it may encode a glucosyltransferase (43). Given the homology of the type Ia glycosyltransferase genes to those in the cpsIII operon, and the identical structure of the oligosaccharide repeating unit, it is likely that these genes encode the same functions in both serotypes. Subsequent biochemical analysis of CpsIaE and Cps14E in E. coli has shown they are glucosyl-1-phosphate transferases (22, 60), which catalyze the transfer of glucose to a receptor lipid intermediate to begin synthesis of the repeating unit. Based on these observations collectively, cpsIIIE should also code for a glucosyltransferase, and the assignment of cpsIIIE as a probable galactosyltransferase must be reevaluated.
Since the glycosidic linkage between the Ia and III pentasaccharide repeating units is the only apparent difference between the two CPS structures, and the type Ia and type III operons are virtually identical except for the region encoding the CPS polymerase, we hypothesized that the cpsH gene in both serotypes encoded the polymerase responsible for the repeating unit linkage. Although the carboxy-terminal domain of the upstream-encoded β-1,4-galactosyltransferases, CpsIIIG and CpsIaG diverged, our results confirm that these enzymes are not involved in linking the repeating units. Overexpressing the cpsIaH and cpsIIIH alleles in the heterologous serotype strains resulted in decreased expression of the native CPS and promoted synthesis of the alternative serotype CPS. We postulate that since plasmid pDC123 replicates at ∼90 copies/cell (8), the cps gene dosage of the recombinant plasmids was significantly higher than that of the chromosomal copy. Hence, depletion of the CPS precursor pool by the episomal polymerase gene product redirected polymerization of the repeating units into the alternative CPS structure. These results demonstrated that CpsIaH and CpsIIIH were responsible for the linkage between the repeating units of each CPS polymer.
Despite a number of recent publications characterizing the CPS glycosyltransferases in various gram-positive species, there is a lack of studies describing the CPS polymerases. The identification of genes encoding CPS polymerases within gram-positive cps loci has been based on the weak homology of the translated genes to the O-antigen polymerases of Shigella flexneri (37) and Salmonella enterica serovar Typhimurium (9). Direct evidence of polymerase function has been difficult to obtain presumably because biochemical approaches require significant amounts of the precursor oligosaccharides, which are difficult to isolate or synthesize. The need to reconstitute the polymerase with potential membrane-bound accessory proteins in a host suitable for carrying out the assays, such as E. coli, presents another difficulty. As a result, direct in vitro evidence of CPS polymerase activity has not been reported for gram-positive hosts producing complex, branched CPS. Only one other gene product from any gram-positive CPS- or exopolysaccharide-producing species has been unambiguously assigned CPS polymerase function, cap3B of type 3 S. pneumoniae (14). This enzyme does not share homology with CpsIIIH and differs in activity, as it processively polymerizes an unbranched β(1→3)-linked glucuronic acid-(1→4)-β-d-glucose heteropolymer (7a). Examining CPS polymerase function by heterologous gene expression, as done here, provides the advantage of using the original host's native machinery in an intact cell for oligosaccharide production and provides a novel approach to assigning CPS gene function.
Divergent CPS polymerase genes have been detected in several pneumococcal serotypes and may be responsible for altered CPS structures in this species. In a study examining the genetic diversity of pneumococcal CPS synthesis loci (21), DNA from 26 different S. pneumoniae serotypes was probed with cps14H, the gene encoding the putative CPS polymerase. Only serotypes 15b and 15c, which have the same CPS core structure as type 14, hybridized to this probe. The structural differences between S. pneumoniae type 19A and type 19F CPS have been attributed to the potential activities of their respective CPS polymerases (34). Transformation of a type 19F strain with DNA containing the cps19AH and cps19AI genes resulted in a strain that produced a type 19A serotype. These data suggest that although CPS structural diversity within pneumococci is generally due to modification of glycosyltransferase and monosaccharide precursor usage, it can also be achieved through changes in the specificity of the polysaccharide polymerases. The S. agalactiae type Ia and type III data presented here provides compelling evidence that the polymerase gene is responsible for generating the linkage which confers the type-specific glycosidic bond between the oligosaccharide subunits and hence the serotype CPS produced.
Mutagenesis of the first four genes in the cpsIII operon and RNA dot blot analysis failed to show transcription starting distal to the promoter upstream of cpsIIIA, suggesting the absence of additional active promoters in the rest of the operon. These data indicated that the cpsIII operon is transcribed as a large polycistronic message. In contrast, Yamamoto et al. (60) identified an additional promoter in the type Ia locus between cpsIaD and cpsIaE (homologues of cpsIIID and cpsIIIE, respectively). This promoter may arise from differences between the type Ia and type III nucleotide sequences within the region where the type Ia secondary promoter was located. The most significant of these is the insertion of an additional A at base 3894 in the Ia nucleotide sequence that introduces a frameshift and subsequent stop codon. This results in the truncation of CpsIaE compared to the type III homologue (the type III cpsIIIE ORF extends an additional 58 codons 5′ of the start site of the homologue type Ia cpsIaE gene). The additional adenine is located within the 17-bp spacer between the −35 and −10 regions of the type Ia promoter sequence. The shorter spacer in the type III locus may account for the lack of secondary promoter activity in our transcriptional analysis.
We are continuing to investigate the cps loci of the other GBS serotypes by analyzing the sequence differences of their operons. As might be expected from their CPS structure, we have obtained preliminary evidence, using the same genetic approach, that the CPS of serotype Ib S. agalactiae may be converted to type Ia by transferring and overexpressing a single type III galactosyltransferase gene. Similarly, an antigenically indistinguishable type III CPS can be produced from a type VI strain by transfer of a single type III glycosyltransferase gene. This approach promises to enable a rapid means of assigning CPS gene function and investigating polysaccharide biosynthesis in GBS and other gram-positive organisms.
ACKNOWLEDGMENTS
This study was supported by NIH grants AI22498 and The Group B Streptococcal Initiative AI22152 to C.E.R.
We thank Glen Tamura and Dan Shelver for thoughtful comments on the manuscript and Erin Sweet and Aphakorn Nittayajarn for technical assistance.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Annunziato P W, Wright L F, Vann W F, Silver R P. Nucleotide sequence and genetic analysis of the neuD and neuB genes in region 2 of the polysialic acid gene cluster of Escherichia coli K1. J Bacteriol. 1995;177:312–319. doi: 10.1128/jb.177.2.312-319.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrecubieta C, Garcia E, Lopez R. Demonstration of UDP-glucose dehydrogenase activity in cell extracts of Escherichia coli expressing the pneumococcal cap3A gene required for the synthesis of type 3 capsular polysaccharide. J Bacteriol. 1996;178:2971–2974. doi: 10.1128/jb.178.10.2971-2974.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons; 1995. [Google Scholar]
- 5.Baker C J, Edwards M S. Group B streptococcal infections. In: Remington J S, Klein J O, editors. Infectious diseases of the fetus and newborn infant. 4th ed. Philadelphia, Pa: W. B. Saunders Co.; 1995. pp. 980–1054. [Google Scholar]
- 6.Bliss J M, Garon C F, Silver R P. Polysialic acid export in Escherichia coli K1: the role of KpsT, the ATP-binding component of an ABC transporter, in chain translocation. Glycobiology. 1996;6:445–452. doi: 10.1093/glycob/6.4.445. [DOI] [PubMed] [Google Scholar]
- 7.Bozue J A, Tullius M V, Wang J, Gibson B W, Munson R S., Jr Haemophilus ducreyi produces a novel sialyltransferase. Identification of the sialyltransferase gene and construction of mutants deficient in the production of the sialic acid-containing glycoform of the lipooligosaccharide. J Biol Chem. 1999;274:4106–4114. doi: 10.1074/jbc.274.7.4106. [DOI] [PubMed] [Google Scholar]
- 7a.Cartee R T, Forsee T W, Schutzbach J S, Yother J. Mechanism of type 3 capsular polysaccharide synthesis in Streptococcus pneumoniae. J Biol Chem. 2000;275:3907–3914. doi: 10.1074/jbc.275.6.3907. [DOI] [PubMed] [Google Scholar]
- 8.Chaffin D O, Rubens C E. Blue/white screening of recombinant plasmids in Gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene. 1998;219:91–99. doi: 10.1016/s0378-1119(98)00396-5. [DOI] [PubMed] [Google Scholar]
- 9.Collins L V, Hackett J. Molecular cloning, characterization, and nucleotide sequence of the rfc gene, which encodes an O-antigen polymerase of Salmonella typhimurium. J Bacteriol. 1991;173:2521–2529. doi: 10.1128/jb.173.8.2521-2529.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Dains, D. A., L. F. Wright, D. O. Chaffin, C. E. Rubens, and R. P. Silver. NeuD plays a role in the synthesis of sialic acid in Escherichia coli K1. FEMS Microbiol. Lett., in press. [DOI] [PubMed]
- 10.Dillard J P, Vandersea M W, Yother J. Characterization of the cassette containing genes for the type 3 capsular polysaccharide biosynthesis in Streptococcus pneumoniae. J Exp Med. 1995;181:973–983. doi: 10.1084/jem.181.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards U, Muller A, Hammerschmidt S, Gerardy-Schahn R, Frosch M. Molecular analysis of the biosynthesis pathway of the α-2,8 polysialic acid capsule by Neisseria meningitidis serogroup B. Mol Microbiol. 1994;14:141–149. doi: 10.1111/j.1365-2958.1994.tb01274.x. [DOI] [PubMed] [Google Scholar]
- 12.Ferretti J J, Curtiss R, editors. Streptococcal genetics. Washington, D.C.: American Society for Microbiology; 1987. [Google Scholar]
- 13.Framson P E, Nittayajarn A, Merry J, Youngman P, Rubens C E. New genetic techniques for group B streptococcus: high-efficiency transformation, maintenance of temperature-sensitive pWV01 plasmids, and mutagenesis with Tn917. Appl Environ Microbiol. 1997;63:3539–3547. doi: 10.1128/aem.63.9.3539-3547.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia E, Arrecubieta C, Munoz R, Mollerach M, Lopez R. A functional analysis of the Streptococcus pneumoniae genes involved in the synthesis of type 1 and type 3 capsular polysaccharides. Microb Drug Resist. 1997;3:73–88. doi: 10.1089/mdr.1997.3.73. [DOI] [PubMed] [Google Scholar]
- 15.Haft R F, Wessels M R, Mebane M F, Conaty N, Rubens C E. Characterization of cpsF and its product CMP-N-acetylneuraminic acid synthetase, a group B streptococcal enzyme that can function in K1 capsular polysaccharide biosynthesis in Escherichia coli. Mol Microbiol. 1996;19:555–563. doi: 10.1046/j.1365-2958.1996.395931.x. [DOI] [PubMed] [Google Scholar]
- 16.Jennings H J, Katzenellenbogen E, Lugowski C, Kasper D L. Structure of native polysaccharide antigens of type Ia and type Ib group B streptococcus. Biochemistry. 1983;22:1258–1264. doi: 10.1021/bi00274a042. [DOI] [PubMed] [Google Scholar]
- 17.Jennings H J, Rosell K G, Katzenellenbogen E, Kasper D L. Structural determination of the capsular polysaccharide antigen of type II group B streptococcus. J Biol Chem. 1983;258:1793–1798. [PubMed] [Google Scholar]
- 18.Kogan G, Brisson J R, Kasper D L, von Hunolstein C, Orefici G, Jennings H J. Structural elucidation of the novel type VII group B streptococcus capsular polysaccharide by high resolution NMR spectroscopy. Carbohydr Res. 1995;277:1–9. doi: 10.1016/0008-6215(95)00195-y. [DOI] [PubMed] [Google Scholar]
- 19.Kogan G, Uhrin D, Brisson J R, Paoletti L C, Blodgett A E, Kasper D L, Jennings H J. Structural and immunochemical characterization of the type VIII group B streptococcus capsular polysaccharide. J Biol Chem. 1996;271:8786–8790. doi: 10.1074/jbc.271.15.8786. [DOI] [PubMed] [Google Scholar]
- 20.Kolkman M A, Morrison D A, van der Zeijst B A M, Nuijten P J M. The capsule polysaccharide synthesis locus of Streptococcus pneumoniae serotype 14: identification of the glycosyl transferase gene cps14E. J Bacteriol. 1996;178:3736–3741. doi: 10.1128/jb.178.13.3736-3741.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolkman M A, van der Zeijst B A, Nuijten P J. Diversity of capsular polysaccharide synthesis gene clusters in Streptococcus pneumoniae. J Biochem (Tokyo) 1998;123:937–945. doi: 10.1093/oxfordjournals.jbchem.a022028. [DOI] [PubMed] [Google Scholar]
- 22.Kolkman M A, van der Zeijst B A, Nuijten P J. Functional analysis of glycosyltransferases encoded by the capsular polysaccharide biosynthesis locus of Streptococcus pneumoniae serotype 14. J Biol Chem. 1997;272:19502–19508. doi: 10.1074/jbc.272.31.19502. [DOI] [PubMed] [Google Scholar]
- 23.Kolkman M A B, Wakarchuk W, Nuijten P J M, van der Zeijst B A M. Capsular polysaccharide synthesis in Streptococcus pneumoniae type 14: molecular analysis of the complete cps locus and identification of genes encoding glycosyltransferases required for the biosynthesis of the tetrasaccaride subunit. Mol Microbiol. 1997;26:197–208. doi: 10.1046/j.1365-2958.1997.5791940.x. [DOI] [PubMed] [Google Scholar]
- 24.Koskiniemi S, Sellin M, Norgren M. Identification of two genes, cpsX and cpsY, with putative regulatory function on capsule expression in group B streptococci. FEMS Immunol Med Microbiol. 1998;21:159–168. doi: 10.1111/j.1574-695X.1998.tb01162.x. [DOI] [PubMed] [Google Scholar]
- 25.Kuypers J M, Heggen L M, Rubens C E. Molecular analysis of a region of the group B streptococcus chromosome involved in type III capsule expression. Infect Immun. 1989;57:3058–3065. doi: 10.1128/iai.57.10.3058-3065.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Kasper D L, Ausubel F M, Rosner B, Michel J L. Inactivation of the alpha C protein antigen gene, bca, by a novel shuttle/suicide vector results in attenuation of virulence and immunity in group B streptococcus. Proc Natl Acad Sci USA. 1997;94:13251–13256. doi: 10.1073/pnas.94.24.13251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin F Y, Clemens J D, Azimi P H, Regan J A, Weisman L E, Philips III J B, Rhoads G G, Clark P, Brenner R A, Ferrieri P. Capsular polysaccharide types of group B streptococcal isolates from neonates with early-onset systemic infection. J Infect Dis. 1998;177:790–792. doi: 10.1086/517810. [DOI] [PubMed] [Google Scholar]
- 28.Lin W S, Cunneen T, Lee C Y. Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J Bacteriol. 1994;176:7005–7016. doi: 10.1128/jb.176.22.7005-7016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madoff L C, Michel J L, Kasper D L. A monoclonal antibody identifies a protective C-protein alpha-antigen epitope in group B streptococci. Infect Immun. 1991;59:204–210. doi: 10.1128/iai.59.1.204-210.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maguin E, Duwaat P, Hege T, Ehrlich D, Gruss A. New thermosensitive plasmid for gram-positive bacteria. J Bacteriol. 1992;174:5633–5638. doi: 10.1128/jb.174.17.5633-5638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 32.Martin T R, Rubens C E, Wilson C B. Lung antibacterial defense mechanisms in infant and adult rats: implications for the pathogenesis of group B streptococcal infections in neonatal lung. J Infect Dis. 1988;157:91–100. doi: 10.1093/infdis/157.1.91. [DOI] [PubMed] [Google Scholar]
- 33.Morona J K, Morona R, Paton J C. Characterization of the locus encoding the Streptococcus pneumoniae type 19F capsular polysaccharide biosynthetic pathway. Mol Microbiol. 1997;23:751–763. doi: 10.1046/j.1365-2958.1997.2551624.x. [DOI] [PubMed] [Google Scholar]
- 34.Morona J K, Morona R, Paton J C. Comparative genetics of capsular polysaccharide biosynthesis in Streptococcus pneumoniae types belonging to serogroup 19. J Bacteriol. 1999;181:5355–5364. doi: 10.1128/jb.181.17.5355-5364.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morona J K, Morona R, Paton J C. Molecular and genetic characterization of the capsule biosynthesis locus of Streptococcus pneumoniae type 19B. J Bacteriol. 1997;179:4953–4958. doi: 10.1128/jb.179.15.4953-4958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morona J K, Paton J C, Miller D C, Morona R. Tyrosine phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in streptococcus pneumoniae. Mol Microbiol. 2000;35:1431–1442. doi: 10.1046/j.1365-2958.2000.01808.x. [DOI] [PubMed] [Google Scholar]
- 37.Morona R, Mavris M, Fallarino A, Manning P A. Characterization of the rfc region of Shigella flexneri. J Bacteriol. 1994;176:733–747. doi: 10.1128/jb.176.3.733-747.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okada N, Geist R T, Caparon M G. Positive transcriptional control of mry regulates virulence in the group A streptococcus. Mol Microbiol. 1993;7:893–903. doi: 10.1111/j.1365-2958.1993.tb01180.x. [DOI] [PubMed] [Google Scholar]
- 39.Paoletti L C, Ross R A, Johnson K D. Cell growth rate regulates expression of group B streptococcus type III capsular polysaccharide. Infect Immun. 1996;64:1220–1226. doi: 10.1128/iai.64.4.1220-1226.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paulsen I T, Beness A M, Saier M H., Jr Computer-based analyses of the protein constituents of transport systems catalysing export of complex carbohydrates in bacteria. Microbiology. 1997;143:2685–2699. doi: 10.1099/00221287-143-8-2685. [DOI] [PubMed] [Google Scholar]
- 41.Ramirez M, Tomasz A. Molecular characterization of the complete 23F capsular polysaccharide locus of Streptococcus pneumoniae. J Bacteriol. 1998;180:5273–5278. doi: 10.1128/jb.180.19.5273-5278.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodbard D. Apparent positive cooperative effects in cyclic AMP and corticosterone production by isolated adrenal cells in response to ACTH analogues. Endocrinology. 1974;94:1427–1437. doi: 10.1210/endo-94-5-1427. [DOI] [PubMed] [Google Scholar]
- 43.Rubens C E, Heggen L M, Haft R F, Wessels M R. Identification of cpsD, a gene essential for type III capsule expression in group B streptococci. Mol Microbiol. 1993;8:843–855. doi: 10.1111/j.1365-2958.1993.tb01631.x. [DOI] [PubMed] [Google Scholar]
- 44.Rubens C E, Heggen L M, Kuypers J M. IS861, a group B streptococcal insertion sequence related to IS150 and IS3 of Escherichia coli. J Bacteriol. 1989;171:5531–5535. doi: 10.1128/jb.171.10.5531-5535.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rubens C E, Wessels M R, Heggen L M, Kasper D L. Transposon mutagenesis of type III group B streptococcus: correlation of capsule expression with virulence. Proc Natl Acad Sci USA. 1987;84:7208–7212. doi: 10.1073/pnas.84.20.7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rubens C E, Wessels M R, Kuypers J M, Kasper D L, Weiser J N. Molecular analysis of two group B streptococcal virulence factors. Semin Perinatol. 1990;14(4 Suppl. 1):22–29. [PubMed] [Google Scholar]
- 47.Schuchat A. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin Microbiol Rev. 1998;11:497–513. doi: 10.1128/cmr.11.3.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stingele F, Neeser J R, Mollet B. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J Bacteriol. 1996;178:1680–1690. doi: 10.1128/jb.178.6.1680-1690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Kranenburg R, Marugg J D, van Swam I I, Willem N J, de Vos W M. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol Microbiol. 1997;24:387–397. doi: 10.1046/j.1365-2958.1997.3521720.x. [DOI] [PubMed] [Google Scholar]
- 50.van Kranenburg R, Vos H R, van Swan I I, Kleerebezem M, de Vos W M. Functional analysis of glycosyltransferase genes from Lactococcus lactis and other gram-positive cocci: complementation, expression, and diversity. J Bacteriol. 1999;181:6347–6353. doi: 10.1128/jb.181.20.6347-6353.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Hunolstein C, D'Ascenzi S, Wagner B, Jelinkova J, Alfarone G, Recchia S, Wagner M, Orefici G. Immunochemistry of capsular type polysaccharide and virulence properties of type VI Streptococcus agalactiae (group B streptococci) Infect Immun. 1993;61:1272–1280. doi: 10.1128/iai.61.4.1272-1280.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voskuil M I, Chambliss G H. The −16 region of Bacillus subtilis and other gram-positive bacterial promoters. Nucleic Acids Res. 1998;26:3584–3590. doi: 10.1093/nar/26.15.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wertman K F, Wyman A R, Botstein D. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene. 1986;49:253–262. doi: 10.1016/0378-1119(86)90286-6. [DOI] [PubMed] [Google Scholar]
- 54.Wessels M R. Biology of streptococcal capsular polysaccharides. Soc Appl Bacteriol Symp Ser. 1997;26:20S–31S. [PubMed] [Google Scholar]
- 55.Wessels M R, Benedi W J, Jennings H J, Michon F, DiFabio J L, Kasper D L. Isolation and characterization of type IV group B streptococcus capsular polysaccharide. Infect Immun. 1989;57:1089–1094. doi: 10.1128/iai.57.4.1089-1094.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wessels M R, DiFabio J L, Benedi V J, Kasper D L, Michon F, Brisson J R, Jel'inkov'a J, Jennings H J. Structural determination and immunochemical characterization of the type V group B streptococcus capsular polysaccharide. J Biol Chem. 1991;266:6714–6719. [PubMed] [Google Scholar]
- 57.Wessels M R, Haft R F, Heggen L M, Rubens C E. Identification of a genetic locus essential for capsule sialylation in type III group B streptococci. Infect Immun. 1992;60:392–400. doi: 10.1128/iai.60.2.392-400.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wessels M R, Pozsgay V, Kasper D L, Jennings H J. Structure and immunochemistry of an oligosaccharide repeating unit of the capsular polysaccharide of type III group B streptococcus. A revised structure for the type III group B streptococcal polysaccharide antigen. J Biol Chem. 1987;262:8262–8267. [PubMed] [Google Scholar]
- 59.Wessels M R, Rubens C E, Bened I V J, Kasper D L. Definition of a bacterial virulence factor: sialylation of the group B streptococcal capsule. Proc Natl Acad Sci USA. 1989;86:8983–8987. doi: 10.1073/pnas.86.22.8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamamoto S, Miyake K, Koike Y, Watanabe M, Machida Y, Ohta M, Iijima S. Molecular characterization of type-specific capsular polysaccharide biosynthesis genes of Streptococcus agalactiae type Ia. J Bacteriol. 1999;181:5176–5184. doi: 10.1128/jb.181.17.5176-5184.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yim H H, Rubens C E. Site-specific homologous recombination mutagenesis in group B streptococci. Methods Cell Sci. 1998;20:13–20. [Google Scholar]
- 62.Yim H H, Rubens C E. Use of a dental amalgamator to extract RNA from the gram-positive bacterium Streptococcus agalactiae. BioTechniques. 1997;23:229–231. doi: 10.2144/97232bm11. [DOI] [PubMed] [Google Scholar]