In the title compound, the pendant groups differ in their divergence from coplanarity with the central ring.
Keywords: crystal structure, hydrogen bonding
Abstract
In the title compound, C9H10N2O4, the three substituents vary in the degree of lack of planarity with the central phenyl ring. The methoxy group is nearest to being coplanar, with a C—C—O—C torsion angle of 6.1 (5)°. The nitro group is less coplanar, with a 12.8 (5)° twist about the C—N bond and the acetamido group is considerably less coplanar with the central ring, having a 25.4 (5)° twist about the C—N bond to the ring. The NH group forms an intramolecular N—H⋯O hydrogen bond to a nitro-group O atom.
Structure description
The analgesic use of 4-alkoxyacetanilides, in particular 4-ethoxyacetanilide or 4-EA, predates the First World War. 4-Hydroxyacetanilide (popularly known as Tylenol or acetaminophen) and 4-EA were introduced into the markets at around the same time; however, 4-EA was withdrawn from sale some decades ago due to its carcinogenic and kidney-damaging properties (Dubach et al., 1983 ▸; Nakanishi et al., 1982 ▸). Although there has been extensive information on phase I and phase II biotransformation of 4-alkoxyacetanilides (Hinson, 1983 ▸; Kapetanović et al., 1979 ▸; Mulder et al., 1984 ▸; Veronese et al., 1985 ▸), little or no information is available on nitrated or other oxidation products that could be formed in reactions with cellular oxidants, such as hypochlorite (−OCl)/hypochlorous acid (HOCl; pK a ≃ 7.53) and peroxynitrite (ONOO−)/peroxynitrous acid (ONOOH; pK a ≃ 6.2; ONOOH and ONOO− are collectively referred to as peroxynitrite or PN). We have shown, for instance, that 4-hydroxyacetanilide forms nitrated and chlorinated products along with varying amounts of dimers when reacted with HOCl/−OCl and PN/CO2 under physiologically relevant conditions (Uppu & Martin, 2005 ▸; Deere et al., 2022 ▸). We suspect that similar products (or their positional isomers) may be formed in the reactions of 4-alkoxyacetanilides with the cellular oxidants referenced above. Towards a better understanding of this and to shed light on molecular targets (Bertolini et al., 2006 ▸), we have synthesized the title compound, C9H10N2O4: single crystals grown from aqueous solution were analyzed by X-ray diffraction.
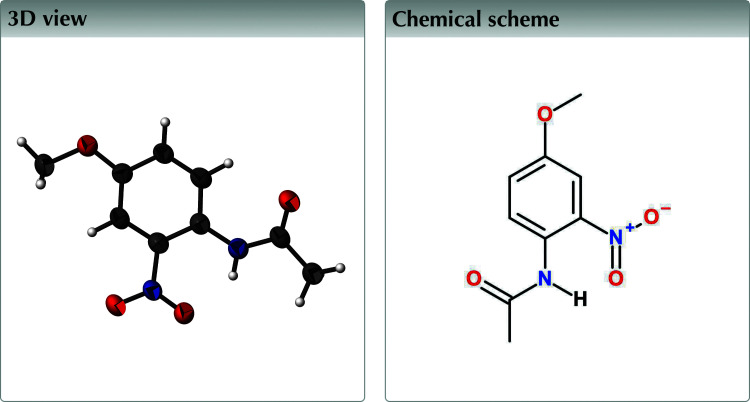
The title compound is shown in Fig. 1 ▸. It is significantly non-planar, and its deviation from planarity may be described by torsion angles about bonds from the central C1–C6 phenyl ring to the three substituents. The methoxy group is nearest to being coplanar, with a C9—O2—C4–C3 torsion angle of 6.1 (5)°. The nitro group deviates more from coplanarity with the central ring, with the O3—N2—C2—C1 torsion angle being −12.8 (5)°. The acetamido group is considerably less coplanar with the central ring, with a C7—N1—C1—C6 torsion angle of 25.4 (5)°. These deviations are similar to those seen in the analogous 4-ethoxy compound (Uppu et al., 2020 ▸), in which the corresponding torsion angles are 0.56 (12), −14.94 (13) and 18.23 (15)°, respectively. N-(4-Hydroxy-2-nitrophenyl)acetamide (Hines et al., 2022 ▸) is considerably more planar, with torsion angles to the nitro group and to the acetamido group being −0.79 (19) and 3.1 (2)°, respectively, likely as a result of intermolecular hydrogen bonding by the OH group. The structure of N-(4-hydroxy-3-nitrophenyl)acetamide, in which the OH group likewise participates in intermolecular hydrogen bonding, has also been reported (Salahifar et al., 2015 ▸; Deere et al., 2019 ▸). It is also more planar than the title compound, with a torsion angle of −11.8 (2)° for the nitro group and 9.0 (2)° for the acetamido group. An intramolecular N1—H1N⋯O3 hydrogen bond (Table 1 ▸) is observed in the title compound.
Figure 1.

The title molecule with 50% displacement ellipsoids with the intramolecular N—H⋯O hydrogen bond shown as a blue dashed line.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N⋯O3 | 0.87 (5) | 1.92 (5) | 2.632 (4) | 137 (4) |
| C5—H5⋯O2i | 0.95 | 2.48 | 3.418 (4) | 171 |
| C6—H6⋯O1 | 0.95 | 2.30 | 2.864 (4) | 117 |
| C8—H8B⋯O3ii | 0.98 | 2.64 | 3.578 (4) | 160 |
| C8—H8C⋯O4iii | 0.98 | 2.63 | 3.546 (4) | 156 |
Symmetry codes: (i)
 ; (ii)
; (ii)
 ; (iii)
; (iii)
 .
.
The unit cell of the title compound is shown in Figs. 2 ▸ and 3 ▸. The closest intermolecular contact is C5—H5⋯O2 (at 1 − x, −y, 1 − z), forming dimers about inversion centers with a C⋯O distance of 3.418 (4) Å and 171° angle about H. Molecules form a herringbone pattern in the [101] direction with alternate phenyl rings forming a dihedral angle of 65.7 (2)°.
Figure 2.
The unit cell, viewed down the [010] direction, showing intramolecular hydrogen bonds.
Figure 3.
The unit cell, viewed down the [101] direction. H atoms are not shown.
Synthesis and crystallization
N-(4-Methoxy-2-nitrophenyl)acetamide was synthesized by acetylation of 4-methoxy-2-nitroaniline using acetic anhydride in acetic acid solvent: 3.36 g (20 mmol) of 4-methoxy-2-nitroaniline in 30 ml of glacial acetic was allowed to react with 2.46 g (24 mmol) of acetic anhydride for 18 h at room temperature. The reaction mixture was stirred continuously during the reaction. In the end, the mixture was dried under vacuum, and the N-(4-methoxy-2-nitrophenyl)acetamide in the residue was purified by recrystallization twice from aqueous solution. Single crystals in the form of yellow laths were grown in water by slow cooling of a hot and nearly saturated solution of the title compound.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C9H10N2O4 |
| M r | 210.19 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 90 |
| a, b, c (Å) | 14.8713 (7), 3.9563 (2), 17.2057 (9) |
| β (°) | 114.051 (3) |
| V (Å3) | 924.42 (8) |
| Z | 4 |
| Radiation type | Cu Kα |
| μ (mm−1) | 1.03 |
| Crystal size (mm) | 0.42 × 0.06 × 0.01 |
| Data collection | |
| Diffractometer | Bruker Kappa APEXII DUO CCD |
| Absorption correction | Multi-scan (SADABS; Krause et al., 2015 ▸) |
| T min, T max | 0.692, 0.990 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 11516, 1638, 1211 |
| R int | 0.122 |
| (sin θ/λ)max (Å−1) | 0.595 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.071, 0.203, 1.09 |
| No. of reflections | 1638 |
| No. of parameters | 141 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.24, −0.27 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2414314622002772/hb4403sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314622002772/hb4403Isup2.hkl
Supporting information file. DOI: 10.1107/S2414314622002772/hb4403Isup3.cml
CCDC reference: 2157748
Additional supporting information: crystallographic information; 3D view; checkCIF report
full crystallographic data
Crystal data
| C9H10N2O4 | F(000) = 440 |
| Mr = 210.19 | Dx = 1.510 Mg m−3 |
| Monoclinic, P21/n | Cu Kα radiation, λ = 1.54184 Å |
| a = 14.8713 (7) Å | Cell parameters from 2102 reflections |
| b = 3.9563 (2) Å | θ = 3.3–66.3° |
| c = 17.2057 (9) Å | µ = 1.03 mm−1 |
| β = 114.051 (3)° | T = 90 K |
| V = 924.42 (8) Å3 | Lath, yellow |
| Z = 4 | 0.42 × 0.06 × 0.01 mm |
Data collection
| Bruker Kappa APEXII DUO CCD diffractometer | 1638 independent reflections |
| Radiation source: IµS microfocus | 1211 reflections with I > 2σ(I) |
| QUAZAR multilayer optics monochromator | Rint = 0.122 |
| φ and ω scans | θmax = 66.7°, θmin = 3.3° |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | h = −17→17 |
| Tmin = 0.692, Tmax = 0.990 | k = −4→4 |
| 11516 measured reflections | l = −20→20 |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.071 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.203 | w = 1/[σ2(Fo2) + 0.298P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.09 | (Δ/σ)max < 0.001 |
| 1638 reflections | Δρmax = 0.24 e Å−3 |
| 141 parameters | Δρmin = −0.27 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. All H atoms were located in difference maps and those on C were thereafter treated as riding in geometrically idealized positions with C—H distances 0.95 Å for phenyl and 0.98 Å for methyl. Coordinates of the N—H hydrogen atom were refined. Uiso(H) values were assigned as 1.2Ueq for the attached atom (1.5 for methyl). |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.58485 (18) | 0.7604 (7) | 0.81731 (16) | 0.0512 (7) | |
| O2 | 0.36866 (18) | 0.2368 (6) | 0.43490 (15) | 0.0455 (6) | |
| O3 | 0.23210 (18) | 0.9412 (7) | 0.67288 (16) | 0.0529 (7) | |
| O4 | 0.16683 (18) | 0.9595 (6) | 0.53559 (16) | 0.0480 (7) | |
| N1 | 0.4165 (2) | 0.7462 (7) | 0.75182 (18) | 0.0447 (7) | |
| H1N | 0.363 (4) | 0.830 (11) | 0.753 (3) | 0.054* | |
| N2 | 0.2326 (2) | 0.8718 (7) | 0.60286 (18) | 0.0416 (7) | |
| C1 | 0.4018 (3) | 0.6185 (9) | 0.6716 (2) | 0.0428 (8) | |
| C2 | 0.3144 (2) | 0.6751 (8) | 0.5992 (2) | 0.0420 (8) | |
| C3 | 0.3005 (2) | 0.5555 (8) | 0.5182 (2) | 0.0415 (8) | |
| H3A | 0.241073 | 0.602472 | 0.470197 | 0.050* | |
| C4 | 0.3736 (3) | 0.3699 (8) | 0.5092 (2) | 0.0417 (8) | |
| C5 | 0.4605 (3) | 0.3057 (8) | 0.5806 (2) | 0.0431 (8) | |
| H5 | 0.511084 | 0.176401 | 0.574482 | 0.052* | |
| C6 | 0.4740 (3) | 0.4264 (9) | 0.6593 (2) | 0.0442 (8) | |
| H6 | 0.533915 | 0.378381 | 0.706654 | 0.053* | |
| C7 | 0.5054 (3) | 0.8178 (8) | 0.8187 (2) | 0.0435 (8) | |
| C8 | 0.4928 (3) | 0.9743 (9) | 0.8926 (2) | 0.0475 (8) | |
| H8A | 0.473964 | 0.799894 | 0.923540 | 0.071* | |
| H8B | 0.441299 | 1.147414 | 0.872021 | 0.071* | |
| H8C | 0.554993 | 1.078496 | 0.930706 | 0.071* | |
| C9 | 0.2843 (3) | 0.3245 (9) | 0.3589 (2) | 0.0488 (9) | |
| H9A | 0.224837 | 0.228708 | 0.361637 | 0.073* | |
| H9B | 0.292327 | 0.233462 | 0.309172 | 0.073* | |
| H9C | 0.278101 | 0.571021 | 0.354165 | 0.073* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0331 (14) | 0.0634 (15) | 0.0497 (14) | 0.0015 (10) | 0.0093 (11) | −0.0066 (11) |
| O2 | 0.0413 (14) | 0.0494 (13) | 0.0439 (13) | −0.0005 (10) | 0.0154 (10) | −0.0013 (10) |
| O3 | 0.0419 (14) | 0.0706 (16) | 0.0434 (14) | 0.0026 (12) | 0.0145 (11) | −0.0053 (11) |
| O4 | 0.0361 (13) | 0.0553 (14) | 0.0444 (13) | 0.0047 (10) | 0.0079 (10) | 0.0026 (10) |
| N1 | 0.0398 (17) | 0.0513 (16) | 0.0424 (15) | 0.0000 (12) | 0.0162 (13) | 0.0006 (12) |
| N2 | 0.0308 (15) | 0.0463 (15) | 0.0425 (16) | −0.0030 (11) | 0.0097 (13) | −0.0028 (12) |
| C1 | 0.0376 (18) | 0.0453 (17) | 0.0429 (18) | −0.0019 (13) | 0.0140 (15) | 0.0030 (13) |
| C2 | 0.0354 (18) | 0.0430 (17) | 0.0458 (19) | −0.0024 (13) | 0.0148 (15) | 0.0032 (13) |
| C3 | 0.0342 (17) | 0.0411 (17) | 0.0432 (17) | −0.0037 (13) | 0.0095 (14) | 0.0033 (13) |
| C4 | 0.0381 (18) | 0.0425 (17) | 0.0446 (18) | −0.0025 (13) | 0.0170 (15) | −0.0011 (13) |
| C5 | 0.0331 (18) | 0.0454 (17) | 0.0469 (19) | −0.0008 (13) | 0.0125 (15) | −0.0009 (14) |
| C6 | 0.0366 (18) | 0.0437 (17) | 0.0495 (19) | 0.0007 (13) | 0.0146 (15) | 0.0033 (14) |
| C7 | 0.0342 (19) | 0.0449 (17) | 0.0452 (18) | −0.0005 (13) | 0.0099 (15) | 0.0039 (13) |
| C8 | 0.0412 (19) | 0.0501 (19) | 0.0462 (18) | 0.0005 (15) | 0.0126 (15) | −0.0017 (15) |
| C9 | 0.045 (2) | 0.0517 (19) | 0.0416 (18) | 0.0030 (15) | 0.0089 (16) | −0.0008 (14) |
Geometric parameters (Å, º)
| O1—C7 | 1.214 (4) | C3—H3A | 0.9500 |
| O2—C4 | 1.357 (4) | C4—C5 | 1.395 (5) |
| O2—C9 | 1.438 (4) | C5—C6 | 1.371 (5) |
| O3—N2 | 1.239 (4) | C5—H5 | 0.9500 |
| O4—N2 | 1.222 (4) | C6—H6 | 0.9500 |
| N1—C7 | 1.383 (5) | C7—C8 | 1.492 (5) |
| N1—C1 | 1.401 (5) | C8—H8A | 0.9800 |
| N1—H1N | 0.87 (5) | C8—H8B | 0.9800 |
| N2—C2 | 1.467 (4) | C8—H8C | 0.9800 |
| C1—C6 | 1.400 (5) | C9—H9A | 0.9800 |
| C1—C2 | 1.405 (5) | C9—H9B | 0.9800 |
| C2—C3 | 1.404 (5) | C9—H9C | 0.9800 |
| C3—C4 | 1.372 (5) | ||
| C4—O2—C9 | 117.1 (3) | C6—C5—H5 | 119.5 |
| C7—N1—C1 | 127.4 (3) | C4—C5—H5 | 119.5 |
| C7—N1—H1N | 118 (3) | C5—C6—C1 | 121.7 (3) |
| C1—N1—H1N | 113 (3) | C5—C6—H6 | 119.1 |
| O4—N2—O3 | 122.6 (3) | C1—C6—H6 | 119.1 |
| O4—N2—C2 | 117.9 (3) | O1—C7—N1 | 123.6 (3) |
| O3—N2—C2 | 119.6 (3) | O1—C7—C8 | 123.8 (3) |
| C6—C1—N1 | 121.6 (3) | N1—C7—C8 | 112.7 (3) |
| C6—C1—C2 | 116.2 (3) | C7—C8—H8A | 109.5 |
| N1—C1—C2 | 122.2 (3) | C7—C8—H8B | 109.5 |
| C3—C2—C1 | 122.4 (3) | H8A—C8—H8B | 109.5 |
| C3—C2—N2 | 115.6 (3) | C7—C8—H8C | 109.5 |
| C1—C2—N2 | 122.0 (3) | H8A—C8—H8C | 109.5 |
| C4—C3—C2 | 119.2 (3) | H8B—C8—H8C | 109.5 |
| C4—C3—H3A | 120.4 | O2—C9—H9A | 109.5 |
| C2—C3—H3A | 120.4 | O2—C9—H9B | 109.5 |
| O2—C4—C3 | 124.9 (3) | H9A—C9—H9B | 109.5 |
| O2—C4—C5 | 115.7 (3) | O2—C9—H9C | 109.5 |
| C3—C4—C5 | 119.4 (3) | H9A—C9—H9C | 109.5 |
| C6—C5—C4 | 121.0 (3) | H9B—C9—H9C | 109.5 |
| C7—N1—C1—C6 | 25.4 (5) | C9—O2—C4—C3 | 6.1 (5) |
| C7—N1—C1—C2 | −154.6 (3) | C9—O2—C4—C5 | −174.3 (3) |
| C6—C1—C2—C3 | −1.8 (5) | C2—C3—C4—O2 | 179.1 (3) |
| N1—C1—C2—C3 | 178.2 (3) | C2—C3—C4—C5 | −0.4 (5) |
| C6—C1—C2—N2 | 180.0 (3) | O2—C4—C5—C6 | −179.9 (3) |
| N1—C1—C2—N2 | −0.1 (5) | C3—C4—C5—C6 | −0.3 (5) |
| O4—N2—C2—C3 | −10.9 (4) | C4—C5—C6—C1 | −0.1 (5) |
| O3—N2—C2—C3 | 168.8 (3) | N1—C1—C6—C5 | −178.9 (3) |
| O4—N2—C2—C1 | 167.5 (3) | C2—C1—C6—C5 | 1.1 (5) |
| O3—N2—C2—C1 | −12.8 (5) | C1—N1—C7—O1 | −3.9 (6) |
| C1—C2—C3—C4 | 1.5 (5) | C1—N1—C7—C8 | 175.8 (3) |
| N2—C2—C3—C4 | 179.9 (3) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O3 | 0.87 (5) | 1.92 (5) | 2.632 (4) | 137 (4) |
| C5—H5···O2i | 0.95 | 2.48 | 3.418 (4) | 171 |
| C6—H6···O1 | 0.95 | 2.30 | 2.864 (4) | 117 |
| C8—H8B···O3ii | 0.98 | 2.64 | 3.578 (4) | 160 |
| C8—H8C···O4iii | 0.98 | 2.63 | 3.546 (4) | 156 |
Symmetry codes: (i) −x+1, −y, −z+1; (ii) −x+1/2, y+1/2, −z+3/2; (iii) x+1/2, −y+5/2, z+1/2.
Funding Statement
The authors acknowledge the support from the National Institutes of Health (NIH) through the National Institute of General Medical Science (NIGMS) grant No. 5 P2O GM103424–17 and the US Department of Education (US DoE; Title III, HBGI Part B grant No. P031B040030). Its contents are solely the responsibility of authors and do not represent the official views of NIH, NIGMS, or US DoE. The upgrade of the diffractometer was made possible by grant No. LEQSF(2011–12)-ENH-TR-01, administered by the Louisiana Board of Regents
References
- Bertolini, A., Ferrari, A., Ottani, A., Guerzoni, S., Tacchi, R. & Leone, S. (2006). CNS Drug Rev. 12, 250–275. [DOI] [PMC free article] [PubMed]
- Bruker (2016). APEX2 and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Deere, C. J., Hines, J. E. III, Agu, O. A., Fronczek, F. R. & Uppu, R. M. (2019). CSD Communication (CCDC 1910293). CCDC, Cambridge, England. https://doi.org/10.5517/ccdc.csd.cc223tcd.
- Deere, C. J., Hines, J. E. III & Uppu, R. M. (2022). Unpublished.
- Dubach, U. C., Rosner, B. & Pfister, E. (1983). New Engl. J. Med. 308, 357–362. [DOI] [PubMed]
- Hines, J. E. III, Deere, C. J., Fronczek, F. R. & Uppu, R. M. (2022). IUCr Data, 7, x220201. [DOI] [PMC free article] [PubMed]
- Hinson, J. A. (1983). Environ. Health Perspect. 49, 71–79. [DOI] [PMC free article] [PubMed]
- Kapetanović, I. M., Strong, J. M. & Mieyal, J. J. (1979). J. Pharmacol. Exp. Ther. 209, 20–24. [PubMed]
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Macrae, C. F., Sovago, I., Cottrell, S. J., Galek, P. T. A., McCabe, P., Pidcock, E., Platings, M., Shields, G. P., Stevens, J. S., Towler, M. & Wood, P. A. (2020). J. Appl. Cryst. 53, 226–235. [DOI] [PMC free article] [PubMed]
- Mulder, G. J., Kadlubar, F. F., Mays, J. B. & Hinson, J. A. (1984). Mol. Pharmacol. 26, 342–347. [PubMed]
- Nakanishi, K., Kurata, Y., Oshima, M., Fukushima, S. & Ito, N. (1982). Int. J. Cancer, 29, 439–444. [DOI] [PubMed]
- Salahifar, E., Nematollahi, D., Bayat, M., Mahyari, A. & Amiri Rudbari, H. (2015). Org. Lett. 17, 4666–4669. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Uppu, R. M. & Martin, R. J. (2005). Toxicologist (supplement to Toxicol. Sci), p. 319. https://www.toxicology.org/pubs/docs/Tox/2005Tox.pdf
- Uppu, S. N., Agu, O. A., Deere, C. J. & Fronczek, F. R. (2020). CSD Communication (CCDC 2021362). CCDC, Cambridge, England. https://doi.org/10.5517/ccdc.csd.cc25vd7p.
- Veronese, M. E., McLean, S., D’Souza, C. A. & Davies, N. W. (1985). Xenobiotica, 15, 929–940. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2414314622002772/hb4403sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314622002772/hb4403Isup2.hkl
Supporting information file. DOI: 10.1107/S2414314622002772/hb4403Isup3.cml
CCDC reference: 2157748
Additional supporting information: crystallographic information; 3D view; checkCIF report




