Cations of the title compound interact via π–π interactions and hydrogen bond with water to form a zigzag ribbon.
Keywords: crystal structure, pyridinium, hydrogen bonding, π–π interactions
Abstract
The chevron-shaped cations of the title hydrated salt, C25H22N4
2+·2Br−·2H2O, are arranged in back-to-back alternating directions to form a zigzag ribbon propagating along the [010] direction. Intermolecular interactions comprising these ribbons are π–π interactions between the pyridinium and adjacent pyridyl rings, as well as O—H⋯O hydrogen bonding between water molecules and two adjacent pyridyl N atoms. Half of the cation is generated by the mirror plane. The water O atoms, the central C atom and one Br atom are located on this mirror plane while the other Br atom is on a twofold screw axis.
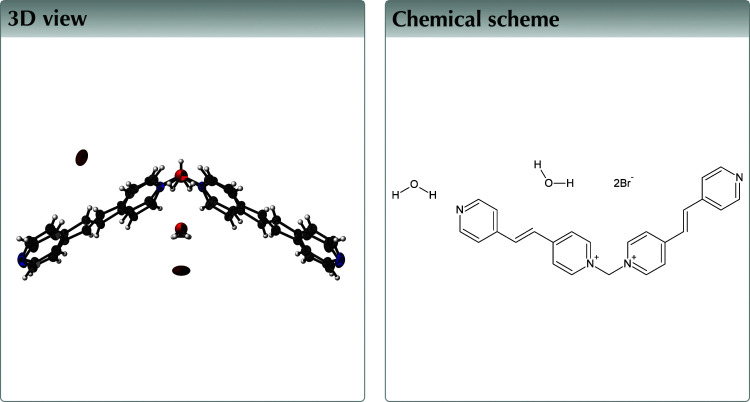
Structure description
Half of the cation is generated by the mirror plane (x,
 − y, z). The O1, O2, Br1, and C1 atoms are located on this mirror plane and the Br2 atom is on a twofold screw axis (−x,
− y, z). The O1, O2, Br1, and C1 atoms are located on this mirror plane and the Br2 atom is on a twofold screw axis (−x,
 + y, −z). The pyridyl–vinyl–pyridinium moiety (Fig. 1 ▸) is essentially planar with a 1.7 (3)° dihedral angle between the planes of the pyridinium (N1/C2–C6) and pyridyl (N2/C9–C13) rings. The N1—C1—N1(x,
+ y, −z). The pyridyl–vinyl–pyridinium moiety (Fig. 1 ▸) is essentially planar with a 1.7 (3)° dihedral angle between the planes of the pyridinium (N1/C2–C6) and pyridyl (N2/C9–C13) rings. The N1—C1—N1(x,
 − y, z) angle is 110.9 (10)°, which is similar to the N—C—N angles of 111.1 (4) or 112.3 (4)° found in the bromide (Schuster et al. 2022 ▸) or PF6
− (Blanco et al., 2007 ▸) salts, respectively, of the 1,1′-methylenebis-4,4′-bipyridinium cation. When two of the title cations are used in a supramolecular cyclic compound with two Pd(ethylenediamine) moieties, the crystal structure had this same N—C—N angle remaining relatively unchanged at 109.1 (19) and 111.2 (11)° (Blanco et al., 2009 ▸).
− y, z) angle is 110.9 (10)°, which is similar to the N—C—N angles of 111.1 (4) or 112.3 (4)° found in the bromide (Schuster et al. 2022 ▸) or PF6
− (Blanco et al., 2007 ▸) salts, respectively, of the 1,1′-methylenebis-4,4′-bipyridinium cation. When two of the title cations are used in a supramolecular cyclic compound with two Pd(ethylenediamine) moieties, the crystal structure had this same N—C—N angle remaining relatively unchanged at 109.1 (19) and 111.2 (11)° (Blanco et al., 2009 ▸).
Figure 1.
Ellipsoid (50%) representation of the title complex with disorder omitted for clarity.
In the extended structure, the chevron-shaped cations of the title compound arrange in back-to-back alternating directions to form a zigzag ribbon (Fig. 2 ▸) propagating along the [010] direction. Water molecules are positioned to interact with the terminal pyridyl nitrogen atom, N2, with an N2—H1D(
 − x, 1 − y,
− x, 1 − y,
 + z) distance of 2.01 Å (Table 1 ▸). The distance between back-to-back pyridinium and pyridyl rings [the closest distance between carbon atoms, C6 of the pyridinium and C13(1 − x, 1 − y, 1 − z) of a pyridyl ring, being 3.46 (1) Å (Fig. 2 ▸)] is suitable for π–π interactions (Sinnokrot et al., 2002 ▸), which further consolidate these zigzag ribbons. Water molecules and bromide ions pack between the ribbons (Fig. 3 ▸). Other hydrogen-bonded zigzag ribbon structures are observed in 1,3-bis[(tetrahydrofuran-2-yl)methyl]thiourea (Peña et al., 2009 ▸) or 1-(4-bromophenyl)-3-(4-ethoxyphenyl)prop-2-en-1-one (Fun et al., 2008 ▸).
+ z) distance of 2.01 Å (Table 1 ▸). The distance between back-to-back pyridinium and pyridyl rings [the closest distance between carbon atoms, C6 of the pyridinium and C13(1 − x, 1 − y, 1 − z) of a pyridyl ring, being 3.46 (1) Å (Fig. 2 ▸)] is suitable for π–π interactions (Sinnokrot et al., 2002 ▸), which further consolidate these zigzag ribbons. Water molecules and bromide ions pack between the ribbons (Fig. 3 ▸). Other hydrogen-bonded zigzag ribbon structures are observed in 1,3-bis[(tetrahydrofuran-2-yl)methyl]thiourea (Peña et al., 2009 ▸) or 1-(4-bromophenyl)-3-(4-ethoxyphenyl)prop-2-en-1-one (Fun et al., 2008 ▸).
Figure 2.
Zigzag ribbons composed of back-to-back chevron-shaped cations of the title complex. The distance between N2 and H1D(
 − x, 1 − y,
− x, 1 − y,
 + z) is shown. Ellipsoids at 50% with disorder, bromide ions and some water molecules omitted for clarity.
+ z) is shown. Ellipsoids at 50% with disorder, bromide ions and some water molecules omitted for clarity.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1C⋯N2i | 0.88 | 2.26 | 2.880 (11) | 128 |
| O1—H1D⋯N2ii | 0.88 | 2.01 | 2.880 (11) | 171 |
Symmetry codes: (i)
 ; (ii)
; (ii)
 .
.
Figure 3.
Ellipsoid (50%) representation of ribbons of cations with bromide ions (brown) and water molecules positioned between them. Ellipsoids at 50% with disorder omitted for clarity.
Synthesis and crystallization
The title compound was synthesized according to published procedures (Blanco et al., 2009 ▸). Colorless plates were grown from liquid diffusion of tetrahydrofuran into a dimethylformamide solution of the pyridinium bromide salt.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. Disorder of the 4-[(E)-2-(pyridin-4-yl)ethenyl]pyridinium moiety was refined using ‘PART 1’ and ‘PART 2’ with the ratio of occupancies at 47 and 53%. All our attempts to refine the structure to achieve equal occupancies led to a drastic worsening of R1 and wR2 values.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C25H22N4 2+·2Br−·2H2O |
| M r | 574.32 |
| Crystal system, space group | Orthorhombic, P n m a |
| Temperature (K) | 220 |
| a, b, c (Å) | 15.4863 (2), 22.2936 (3), 7.2100 (1) |
| V (Å3) | 2489.22 (6) |
| Z | 4 |
| Radiation type | Cu Kα |
| μ (mm−1) | 4.37 |
| Crystal size (mm) | 0.04 × 0.03 × 0.02 |
| Data collection | |
| Diffractometer | XtaLAB Synergy, Dualflex, HyPix |
| Absorption correction | Multi-scan CrysAlis PRO (Rigaku OD, 2021 ▸) |
| T min, T max | 0.671, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 25704, 2780, 2439 |
| R int | 0.030 |
| (sin θ/λ)max (Å−1) | 0.639 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.036, 0.104, 1.09 |
| No. of reflections | 2780 |
| No. of parameters | 244 |
| No. of restraints | 8 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.99, −0.86 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2414314622005259/bx4021sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314622005259/bx4021Isup2.hkl
Supporting information file. DOI: 10.1107/S2414314622005259/bx4021Isup3.mol
Supporting information file. DOI: 10.1107/S2414314622005259/bx4021Isup4.cml
CCDC reference: 2173317
Additional supporting information: crystallographic information; 3D view; checkCIF report
full crystallographic data
Crystal data
| C25H22N42+·2Br−·2H2O | Dx = 1.532 Mg m−3 |
| Mr = 574.32 | Cu Kα radiation, λ = 1.54184 Å |
| Orthorhombic, Pnma | Cell parameters from 14220 reflections |
| a = 15.4863 (2) Å | θ = 6.1–79.8° |
| b = 22.2936 (3) Å | µ = 4.37 mm−1 |
| c = 7.2100 (1) Å | T = 220 K |
| V = 2489.22 (6) Å3 | Plate, clear light colourless |
| Z = 4 | 0.04 × 0.03 × 0.02 mm |
| F(000) = 1160 |
Data collection
| XtaLAB Synergy, Dualflex, HyPix diffractometer | 2780 independent reflections |
| Radiation source: micro-focus sealed X-ray tube, PhotonJet (Cu) X-ray Source | 2439 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.030 |
| Detector resolution: 10.0000 pixels mm-1 | θmax = 80.3°, θmin = 4.0° |
| ω scans | h = −17→19 |
| Absorption correction: multi-scan CrysAlisPro (Rigaku OD, 2021) | k = −28→27 |
| Tmin = 0.671, Tmax = 1.000 | l = −9→9 |
| 25704 measured reflections |
Refinement
| Refinement on F2 | 8 restraints |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.036 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.104 | w = 1/[σ2(Fo2) + (0.0464P)2 + 1.8748P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.09 | (Δ/σ)max < 0.001 |
| 2780 reflections | Δρmax = 0.99 e Å−3 |
| 244 parameters | Δρmin = −0.86 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Br1 | 0.83013 (2) | 0.250000 | 0.51496 (5) | 0.05780 (16) | |
| Br2 | 0.500000 | 0.500000 | 0.000000 | 0.06787 (19) | |
| O1 | 0.59949 (18) | 0.250000 | −0.0276 (5) | 0.0644 (5) | |
| H1C | 0.628097 | 0.228712 | 0.053830 | 0.097* | 0.5 |
| H1D | 0.637353 | 0.279127 | −0.030494 | 0.097* | 0.5 |
| O2 | 0.61749 (17) | 0.250000 | 0.5693 (5) | 0.0644 (5) | |
| H2B | 0.666046 | 0.234075 | 0.534382 | 0.097* | 0.5 |
| H2C | 0.616243 | 0.229500 | 0.676421 | 0.097* | 0.5 |
| N1 | 0.4441 (7) | 0.3047 (4) | 0.4944 (11) | 0.0280 (19) | 0.471 (7) |
| C2 | 0.4622 (7) | 0.3261 (4) | 0.6635 (9) | 0.0336 (16) | 0.471 (7) |
| H2 | 0.441055 | 0.306491 | 0.769534 | 0.040* | 0.471 (7) |
| C3 | 0.5120 (5) | 0.3769 (3) | 0.6814 (7) | 0.0338 (14) | 0.471 (7) |
| H3 | 0.523534 | 0.392117 | 0.800374 | 0.041* | 0.471 (7) |
| C4 | 0.5451 (3) | 0.4059 (2) | 0.5289 (9) | 0.0259 (12) | 0.471 (7) |
| C5 | 0.5254 (4) | 0.3821 (3) | 0.3578 (7) | 0.0355 (15) | 0.471 (7) |
| H5 | 0.546674 | 0.400906 | 0.250362 | 0.043* | 0.471 (7) |
| C6 | 0.4753 (6) | 0.3316 (4) | 0.3410 (8) | 0.0375 (17) | 0.471 (7) |
| H6 | 0.462840 | 0.315841 | 0.223042 | 0.045* | 0.471 (7) |
| N2 | 0.7822 (9) | 0.6508 (4) | 0.5018 (14) | 0.048 (3) | 0.471 (7) |
| C11 | 0.7660 (7) | 0.6282 (4) | 0.3350 (11) | 0.0430 (17) | 0.471 (7) |
| H11 | 0.788849 | 0.647826 | 0.230565 | 0.052* | 0.471 (7) |
| C10 | 0.7170 (5) | 0.5769 (3) | 0.3083 (8) | 0.0383 (15) | 0.471 (7) |
| H10 | 0.707421 | 0.562561 | 0.187380 | 0.046* | 0.471 (7) |
| C9 | 0.6822 (3) | 0.54692 (19) | 0.4566 (10) | 0.0301 (12) | 0.471 (7) |
| C13 | 0.6980 (5) | 0.5710 (3) | 0.6290 (8) | 0.0394 (16) | 0.471 (7) |
| H13 | 0.674916 | 0.552711 | 0.735506 | 0.047* | 0.471 (7) |
| C12 | 0.7480 (8) | 0.6224 (4) | 0.6452 (10) | 0.050 (2) | 0.471 (7) |
| H12 | 0.758017 | 0.637927 | 0.764548 | 0.060* | 0.471 (7) |
| C1 | 0.39067 (17) | 0.250000 | 0.4756 (4) | 0.0281 (5) | |
| C7 | 0.5974 (3) | 0.4592 (2) | 0.5575 (7) | 0.0328 (12) | 0.471 (7) |
| H7 | 0.608461 | 0.470689 | 0.680642 | 0.039* | 0.471 (7) |
| C8 | 0.6307 (3) | 0.4929 (2) | 0.4239 (6) | 0.0321 (13) | 0.471 (7) |
| H8 | 0.620576 | 0.481388 | 0.300419 | 0.038* | 0.471 (7) |
| C8A | 0.6252 (3) | 0.4946 (2) | 0.5660 (6) | 0.0352 (12) | 0.529 (7) |
| H8A | 0.604322 | 0.485675 | 0.685211 | 0.042* | 0.529 (7) |
| C7A | 0.6033 (3) | 0.4578 (2) | 0.4290 (6) | 0.0330 (11) | 0.529 (7) |
| H7A | 0.624114 | 0.466670 | 0.309581 | 0.040* | 0.529 (7) |
| N1A | 0.4453 (6) | 0.3041 (3) | 0.4673 (9) | 0.0244 (15) | 0.529 (7) |
| C2A | 0.4791 (5) | 0.3212 (3) | 0.3033 (8) | 0.0290 (12) | 0.529 (7) |
| H2A | 0.467537 | 0.298861 | 0.195459 | 0.035* | 0.529 (7) |
| C3A | 0.5305 (3) | 0.3713 (2) | 0.2945 (6) | 0.0307 (12) | 0.529 (7) |
| H3A | 0.553479 | 0.383222 | 0.179661 | 0.037* | 0.529 (7) |
| C4A | 0.5489 (3) | 0.40443 (19) | 0.4506 (8) | 0.0264 (11) | 0.529 (7) |
| C5A | 0.5131 (4) | 0.3853 (3) | 0.6147 (7) | 0.0341 (13) | 0.529 (7) |
| H5A | 0.523793 | 0.407169 | 0.723830 | 0.041* | 0.529 (7) |
| C6A | 0.4621 (6) | 0.3349 (3) | 0.6226 (7) | 0.0323 (13) | 0.529 (7) |
| H6A | 0.439049 | 0.322149 | 0.736540 | 0.039* | 0.529 (7) |
| N2A | 0.7859 (8) | 0.6497 (4) | 0.5226 (13) | 0.052 (3) | 0.529 (7) |
| C11A | 0.7539 (7) | 0.6311 (4) | 0.6829 (10) | 0.0468 (16) | 0.529 (7) |
| H11A | 0.767754 | 0.652790 | 0.790561 | 0.056* | 0.529 (7) |
| C10A | 0.7011 (4) | 0.5811 (3) | 0.7003 (7) | 0.0401 (14) | 0.529 (7) |
| H10A | 0.680077 | 0.569719 | 0.817495 | 0.048* | 0.529 (7) |
| C9A | 0.6796 (3) | 0.54816 (18) | 0.5456 (9) | 0.0325 (11) | 0.529 (7) |
| C13A | 0.7118 (4) | 0.5682 (3) | 0.3798 (8) | 0.0442 (15) | 0.529 (7) |
| H13A | 0.698343 | 0.547622 | 0.269787 | 0.053* | 0.529 (7) |
| C12A | 0.7641 (7) | 0.6187 (4) | 0.3728 (10) | 0.054 (2) | 0.529 (7) |
| H12A | 0.785018 | 0.631399 | 0.256891 | 0.065* | 0.529 (7) |
| H1A | 0.359 (2) | 0.250000 | 0.584 (4) | 0.025 (7)* | |
| H1B | 0.363 (2) | 0.250000 | 0.372 (5) | 0.036 (9)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.0341 (2) | 0.1080 (4) | 0.03128 (19) | 0.000 | 0.00029 (12) | 0.000 |
| Br2 | 0.1255 (5) | 0.0507 (2) | 0.0274 (2) | 0.0236 (2) | 0.00356 (18) | 0.00004 (13) |
| O1 | 0.0401 (9) | 0.0554 (10) | 0.0978 (16) | 0.000 | −0.0060 (10) | 0.000 |
| O2 | 0.0401 (9) | 0.0554 (10) | 0.0978 (16) | 0.000 | −0.0060 (10) | 0.000 |
| N1 | 0.023 (3) | 0.025 (4) | 0.036 (3) | 0.001 (3) | 0.004 (2) | 0.007 (3) |
| C2 | 0.045 (3) | 0.030 (3) | 0.025 (3) | −0.006 (3) | 0.007 (3) | −0.003 (2) |
| C3 | 0.046 (3) | 0.033 (3) | 0.022 (3) | −0.006 (2) | 0.003 (3) | −0.008 (3) |
| C4 | 0.029 (2) | 0.023 (2) | 0.026 (3) | 0.0000 (16) | 0.002 (2) | −0.004 (3) |
| C5 | 0.048 (3) | 0.033 (3) | 0.025 (4) | −0.002 (2) | 0.008 (3) | 0.008 (3) |
| C6 | 0.051 (4) | 0.036 (3) | 0.026 (3) | −0.003 (3) | −0.010 (3) | −0.004 (3) |
| N2 | 0.039 (6) | 0.025 (6) | 0.078 (6) | 0.000 (5) | 0.003 (5) | 0.007 (5) |
| C11 | 0.042 (3) | 0.028 (3) | 0.060 (4) | −0.005 (2) | 0.005 (3) | 0.006 (3) |
| C10 | 0.040 (3) | 0.035 (3) | 0.040 (4) | −0.002 (2) | 0.004 (3) | −0.001 (3) |
| C9 | 0.028 (2) | 0.023 (2) | 0.039 (4) | 0.0008 (16) | 0.001 (3) | 0.001 (3) |
| C13 | 0.040 (3) | 0.036 (4) | 0.042 (4) | −0.004 (2) | 0.000 (3) | 0.001 (3) |
| C12 | 0.052 (4) | 0.032 (3) | 0.065 (5) | −0.001 (3) | −0.014 (4) | −0.011 (3) |
| C1 | 0.0243 (12) | 0.0240 (11) | 0.0360 (14) | 0.000 | −0.0007 (11) | 0.000 |
| C7 | 0.033 (2) | 0.027 (3) | 0.038 (3) | −0.0030 (19) | 0.0006 (18) | −0.0038 (17) |
| C8 | 0.032 (2) | 0.028 (3) | 0.035 (3) | −0.0021 (19) | −0.0027 (17) | −0.0009 (16) |
| C8A | 0.0346 (19) | 0.032 (2) | 0.039 (3) | −0.0033 (18) | 0.0025 (16) | 0.0023 (16) |
| C7A | 0.0326 (19) | 0.029 (2) | 0.037 (2) | −0.0024 (17) | 0.0024 (15) | 0.0007 (16) |
| N1A | 0.026 (3) | 0.022 (3) | 0.026 (2) | 0.000 (2) | −0.004 (2) | −0.004 (2) |
| C2A | 0.034 (2) | 0.032 (3) | 0.022 (2) | −0.008 (2) | 0.0000 (18) | 0.0004 (19) |
| C3A | 0.032 (2) | 0.034 (2) | 0.026 (3) | −0.0048 (18) | 0.004 (2) | 0.003 (2) |
| C4A | 0.0276 (18) | 0.027 (2) | 0.024 (3) | 0.0010 (14) | −0.001 (2) | −0.008 (2) |
| C5A | 0.047 (3) | 0.032 (3) | 0.024 (3) | −0.002 (2) | 0.000 (3) | −0.008 (3) |
| C6A | 0.039 (3) | 0.034 (3) | 0.023 (3) | 0.001 (2) | 0.006 (2) | −0.002 (2) |
| N2A | 0.041 (5) | 0.033 (6) | 0.081 (5) | −0.010 (5) | −0.001 (5) | −0.007 (5) |
| C11A | 0.045 (3) | 0.035 (3) | 0.061 (4) | −0.006 (2) | −0.007 (3) | −0.007 (3) |
| C10A | 0.042 (2) | 0.034 (3) | 0.044 (3) | −0.001 (2) | −0.006 (3) | 0.000 (3) |
| C9A | 0.0290 (19) | 0.028 (2) | 0.041 (3) | 0.0021 (16) | −0.001 (2) | −0.003 (3) |
| C13A | 0.053 (3) | 0.037 (3) | 0.042 (4) | −0.007 (2) | 0.005 (3) | −0.005 (3) |
| C12A | 0.058 (4) | 0.036 (3) | 0.068 (5) | −0.005 (3) | 0.017 (4) | 0.003 (3) |
Geometric parameters (Å, º)
| O1—H1C | 0.8753 | C1—N1Ai | 1.474 (4) |
| O1—H1Ci | 0.88 (6) | C1—H1A | 0.92 (3) |
| O1—H1D | 0.8752 | C1—H1B | 0.86 (4) |
| O1—H1Di | 0.88 (8) | C7—H7 | 0.9400 |
| O2—H2B | 0.8688 | C7—C8 | 1.326 (7) |
| O2—H2Bi | 0.87 (5) | C8—H8 | 0.9400 |
| O2—H2C | 0.8975 | C8A—H8A | 0.9400 |
| O2—H2Ci | 0.90 (6) | C8A—C7A | 1.328 (6) |
| N1—C2 | 1.3385 | C8A—C9A | 1.468 (6) |
| N1—C6 | 1.3481 | C7A—H7A | 0.9400 |
| N1—C1 | 1.480 (5) | C7A—C4A | 1.467 (7) |
| C2—H2 | 0.9400 | N1A—C2A | 1.3482 |
| C2—C3 | 1.3755 | N1A—C6A | 1.3387 |
| C3—H3 | 0.9400 | C2A—H2A | 0.9400 |
| C3—C4 | 1.3742 | C2A—C3A | 1.3735 |
| C4—C5 | 1.3759 | C3A—H3A | 0.9400 |
| C4—C7 | 1.453 (7) | C3A—C4A | 1.3761 |
| C5—H5 | 0.9400 | C4A—C5A | 1.3737 |
| C5—C6 | 1.3734 | C5A—H5A | 0.9400 |
| C6—H6 | 0.9400 | C5A—C6A | 1.3753 |
| N2—C11 | 1.3275 | C6A—H6A | 0.9400 |
| N2—C12 | 1.3235 | N2A—C11A | 1.3241 |
| C11—H11 | 0.9400 | N2A—C12A | 1.3271 |
| C11—C10 | 1.3868 | C11A—H11A | 0.9400 |
| C10—H10 | 0.9400 | C11A—C10A | 1.3883 |
| C10—C9 | 1.3715 | C10A—H10A | 0.9400 |
| C9—C13 | 1.3756 | C10A—C9A | 1.3758 |
| C9—C8 | 1.464 (7) | C9A—C13A | 1.3709 |
| C13—H13 | 0.9400 | C13A—H13A | 0.9400 |
| C13—C12 | 1.3883 | C13A—C12A | 1.3864 |
| C12—H12 | 0.9400 | C12A—H12A | 0.9400 |
| C1—N1A | 1.474 (4) | ||
| H1C—O1—H1Ci | 65.7 | N1A—C1—H1A | 110.0 (10) |
| H1Ci—O1—H1Di | 94.5 | N1Ai—C1—H1A | 110.0 (10) |
| H1C—O1—H1Di | 43.5 | N1Ai—C1—H1B | 104.5 (13) |
| H1C—O1—H1D | 94.5 | N1A—C1—H1B | 104.5 (13) |
| H1D—O1—H1Ci | 43.5 | H1A—C1—H1B | 118 (3) |
| H1D—O1—H1Di | 95.8 | C4—C7—H7 | 117.4 |
| H2B—O2—H2Bi | 48.2 | C8—C7—C4 | 125.3 (6) |
| H2Bi—O2—H2Ci | 93.4 | C8—C7—H7 | 117.4 |
| H2B—O2—H2Ci | 118.4 | C9—C8—H8 | 117.9 |
| H2B—O2—H2C | 93.4 | C7—C8—C9 | 124.2 (5) |
| H2C—O2—H2Bi | 118.4 | C7—C8—H8 | 117.9 |
| H2C—O2—H2Ci | 61.2 | C7A—C8A—H8A | 117.5 |
| C2—N1—C6 | 120.9 | C7A—C8A—C9A | 125.1 (5) |
| C2—N1—C1 | 119.6 (5) | C9A—C8A—H8A | 117.5 |
| C6—N1—C1 | 119.4 (5) | C8A—C7A—H7A | 117.7 |
| N1—C2—H2 | 120.1 | C8A—C7A—C4A | 124.7 (5) |
| N1—C2—C3 | 119.8 | C4A—C7A—H7A | 117.7 |
| C3—C2—H2 | 120.1 | C2A—N1A—C1 | 119.3 (4) |
| C2—C3—H3 | 119.3 | C6A—N1A—C1 | 119.8 (4) |
| C4—C3—C2 | 121.4 | C6A—N1A—C2A | 120.9 |
| C4—C3—H3 | 119.3 | N1A—C2A—H2A | 120.2 |
| C3—C4—C5 | 117.0 | N1A—C2A—C3A | 119.7 |
| C3—C4—C7 | 118.6 (4) | C3A—C2A—H2A | 120.2 |
| C5—C4—C7 | 124.4 (4) | C2A—C3A—H3A | 119.4 |
| C4—C5—H5 | 119.4 | C2A—C3A—C4A | 121.3 |
| C6—C5—C4 | 121.3 | C4A—C3A—H3A | 119.4 |
| C6—C5—H5 | 119.4 | C3A—C4A—C7A | 117.9 (4) |
| N1—C6—C5 | 119.7 | C5A—C4A—C7A | 125.1 (4) |
| N1—C6—H6 | 120.2 | C5A—C4A—C3A | 117.0 |
| C5—C6—H6 | 120.2 | C4A—C5A—H5A | 119.3 |
| C12—N2—C11 | 116.8 | C4A—C5A—C6A | 121.4 |
| N2—C11—H11 | 118.6 | C6A—C5A—H5A | 119.3 |
| N2—C11—C10 | 122.9 | N1A—C6A—C5A | 119.8 |
| C10—C11—H11 | 118.6 | N1A—C6A—H6A | 120.1 |
| C11—C10—H10 | 119.7 | C5A—C6A—H6A | 120.1 |
| C9—C10—C11 | 120.6 | C11A—N2A—C12A | 116.8 |
| C9—C10—H10 | 119.7 | N2A—C11A—H11A | 118.2 |
| C10—C9—C13 | 116.4 | N2A—C11A—C10A | 123.5 |
| C10—C9—C8 | 119.3 (5) | C10A—C11A—H11A | 118.2 |
| C13—C9—C8 | 124.3 (5) | C11A—C10A—H10A | 120.1 |
| C9—C13—H13 | 120.1 | C9A—C10A—C11A | 119.8 |
| C9—C13—C12 | 119.8 | C9A—C10A—H10A | 120.1 |
| C12—C13—H13 | 120.1 | C10A—C9A—C8A | 119.4 (4) |
| N2—C12—C13 | 123.5 | C13A—C9A—C8A | 124.2 (4) |
| N2—C12—H12 | 118.2 | C13A—C9A—C10A | 116.4 |
| C13—C12—H12 | 118.2 | C9A—C13A—H13A | 119.7 |
| N1i—C1—N1 | 110.9 (10) | C9A—C13A—C12A | 120.6 |
| N1—C1—H1A | 102.9 (11) | C12A—C13A—H13A | 119.7 |
| N1i—C1—H1A | 102.9 (11) | N2A—C12A—C13A | 122.8 |
| N1—C1—H1B | 110.9 (11) | N2A—C12A—H12A | 118.6 |
| N1i—C1—H1B | 110.9 (11) | C13A—C12A—H12A | 118.6 |
| N1A—C1—N1Ai | 109.8 (9) | ||
| N1—C2—C3—C4 | −1.2 | C7—C4—C5—C6 | −179.8 (6) |
| C2—N1—C6—C5 | −0.9 | C8—C9—C13—C12 | 179.8 (6) |
| C2—N1—C1—N1i | −84.1 (7) | C8A—C7A—C4A—C3A | 178.8 (4) |
| C2—C3—C4—C5 | 0.7 | C8A—C7A—C4A—C5A | 0.2 (6) |
| C2—C3—C4—C7 | −179.8 (6) | C8A—C9A—C13A—C12A | −178.8 (6) |
| C3—C4—C5—C6 | −0.3 | C7A—C8A—C9A—C10A | −176.9 (4) |
| C3—C4—C7—C8 | −177.3 (5) | C7A—C8A—C9A—C13A | 2.7 (7) |
| C4—C5—C6—N1 | 0.4 | C7A—C4A—C5A—C6A | 179.4 (5) |
| C4—C7—C8—C9 | 179.1 (4) | N1Ai—C1—N1A—C2A | 76.4 (7) |
| C5—C4—C7—C8 | 2.2 (7) | N1Ai—C1—N1A—C6A | −102.3 (5) |
| C6—N1—C2—C3 | 1.3 | N1A—C2A—C3A—C4A | 0.5 |
| C6—N1—C1—N1i | 94.7 (7) | C2A—N1A—C6A—C5A | 1.2 |
| N2—C11—C10—C9 | 0.2 | C2A—C3A—C4A—C7A | −179.2 (5) |
| C11—N2—C12—C13 | 0.7 | C2A—C3A—C4A—C5A | −0.5 |
| C11—C10—C9—C13 | 0.8 | C3A—C4A—C5A—C6A | 0.8 |
| C11—C10—C9—C8 | 180.0 (6) | C4A—C5A—C6A—N1A | −1.2 |
| C10—C9—C13—C12 | −1.1 | C6A—N1A—C2A—C3A | −0.9 |
| C10—C9—C8—C7 | 177.0 (5) | N2A—C11A—C10A—C9A | 0.0 |
| C9—C13—C12—N2 | 0.3 | C11A—N2A—C12A—C13A | −1.1 |
| C13—C9—C8—C7 | −3.9 (7) | C11A—C10A—C9A—C8A | 178.7 (6) |
| C12—N2—C11—C10 | −1.0 | C11A—C10A—C9A—C13A | −1.0 |
| C1—N1—C2—C3 | 180.0 (10) | C10A—C9A—C13A—C12A | 0.8 |
| C1—N1—C6—C5 | −179.6 (9) | C9A—C8A—C7A—C4A | −180.0 (4) |
| C1—N1A—C2A—C3A | −179.6 (8) | C9A—C13A—C12A—N2A | 0.2 |
| C1—N1A—C6A—C5A | 179.9 (8) | C12A—N2A—C11A—C10A | 1.0 |
Symmetry code: (i) x, −y+1/2, z.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1C···N2ii | 0.88 | 2.26 | 2.880 (11) | 128 |
| O1—H1D···N2iii | 0.88 | 2.01 | 2.880 (11) | 171 |
Symmetry codes: (ii) −x+3/2, y−1/2, z−1/2; (iii) −x+3/2, −y+1, z−1/2.
Funding Statement
Funding for this research was provided by: National Science Foundation (grant No. 1726652 to UNT; grant No. 1712066 to Austin College); Welch Foundation (grant No. AD-0007 to Austin College).
References
- Blanco, V., Chas, M., Abella, D., Peinador, C. & Quintela, J. M. (2007). J. Am. Chem. Soc. 129, 13978–13986. [DOI] [PubMed]
- Blanco, V., Gutiérrez, A., Platas-Iglesias, C., Peinador, C. & Quintela, J. M. (2009). J. Org. Chem. 74, 6577–6583. [DOI] [PubMed]
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Fun, H.-K., Patil, P. S., Dharmaprakash, S. M. & Chantrapromma, S. (2008). Acta Cryst. E64, o1540–o1541. [DOI] [PMC free article] [PubMed]
- Macrae, C. F., Sovago, I., Cottrell, S. J., Galek, P. T. A., McCabe, P., Pidcock, E., Platings, M., Shields, G. P., Stevens, J. S., Towler, M. & Wood, P. A. (2020). J. Appl. Cryst. 53, 226–235. [DOI] [PMC free article] [PubMed]
- Peña, Ú., Bernès, S. & Gutiérrez, R. (2009). Acta Cryst. E65, o96. [DOI] [PMC free article] [PubMed]
- Rigaku OD (2021). CrysAlis PRO. Rigaku Oxford Diffraction, Yarnton, England.
- Schuster, S. A., Nesterov, V. V. & Smucker, B. W. (2022). IUCrData, 7, x220526. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Sinnokrot, M. O., Valeev, E. F. & Sherrill, C. D. (2002). J. Am. Chem. Soc. 124, 10887–10893. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2414314622005259/bx4021sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314622005259/bx4021Isup2.hkl
Supporting information file. DOI: 10.1107/S2414314622005259/bx4021Isup3.mol
Supporting information file. DOI: 10.1107/S2414314622005259/bx4021Isup4.cml
CCDC reference: 2173317
Additional supporting information: crystallographic information; 3D view; checkCIF report





