The crystal structure of the nickel(II) dichloride complex of the ligand 2-(pyridin-2-yl)-1H-benzimidazole is described.
Keywords: crystal structure, nickel(II) complex, 2-(pyridin-2-yl)-1H-benzimidazole, hydrogen bonding, C—H⋯π interactions, transfer hydrogenation
Abstract
In the title complex, [NiCl2(C12H9N3)2]·H2O, a divalent nickel atom is coordinated by two 2-(pyridin-2-yl)-1H-benzimidazole ligands in a slightly distorted octahedral environment defined by four N donors of two N,N′-chelating ligands, along with two cis-oriented anionic chloride donors. The title complex crystallized with a water molecule disordered over two positions. In the crystal, a combination of O—H⋯Cl, O—H.·O and N—H⋯Cl hydrogen bonds, together with C—H⋯O, C—H⋯Cl and C—H⋯π interactions, links the complex molecules and the water molecules to form a supramolecular three-dimensional framework. The title complex is isostructural with the cobalt(II) dichloride complex reported previously [Das et al. (2011 ▸). Org. Biomol. Chem. 9, 7097–7107].
Structure description
Transition-metal-catalyzed transfer hydrogenation (TH) is an effective method of reducing ketones to the corresponding secondary alcohols (Zhu et al., 2014 ▸). Generally, the method is operationally simple, selective, and sources hydrogen from alcohols, thus avoiding high pressures of H2 gas (Zhu et al., 2014 ▸). Several transition-metal complexes have been studied in catalytic TH and have been used on laboratory and industrial scales. Complexes of precious metals (Rh, Ir, and Ru) have been the preferred catalysts for TH owing to their high activity and commercial availability (Raja et al., 2012 ▸; Wang et al., 2015 ▸; Li et al., 2015 ▸). With growing concern surrounding the economic and environmental impact of using precious metals in chemistry, a renewed interest in Earth-abundant metal catalysis has prompted our research into TH catalysts featuring first-row transition metals, such as iron, cobalt, or nickel (Morris, 2009 ▸; Garduño & García, 2017 ▸; Abubakar et al., 2018 ▸; Chen et al., 2010 ▸). Recognizing that nickel(II) complexes of chiral bis(phosphines) have been utilized in asymmetric TH, we turned our attention to nickel(II) complexes of the commercially available ligand 2-(pyridin-2-yl)-1H-benzimidazole.
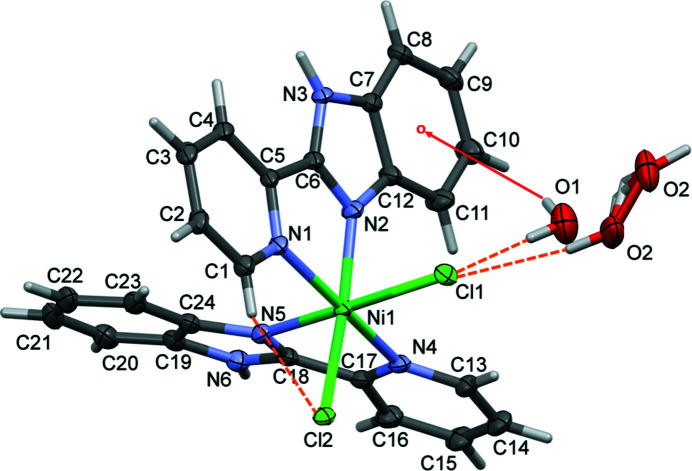
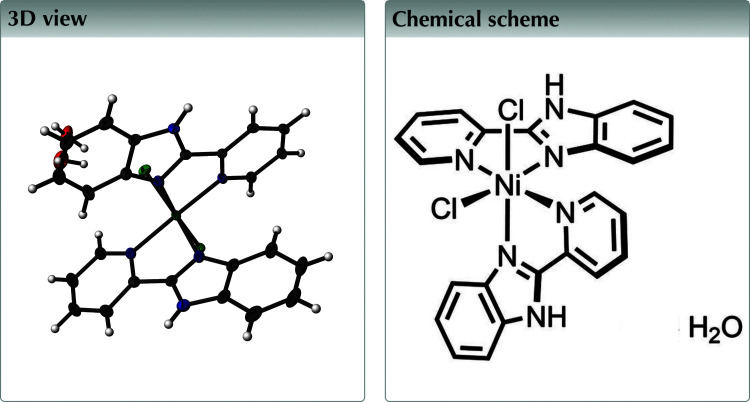
The asymmetric unit of the title complex consists of a NiII ion coordinated by two 2-(pyridin-2-yl)-1H-benzimidazole ligands bound in a κ2-N,N arrangement, along with two cis-oriented anionic chloride donors (Fig. 1 ▸). The complex crystallized as a monohydrate with the water molecule disordered over two sites (Fig. 1 ▸). The metal center adopts a slightly distorted octahedral geometry. The pyridyl N-donor atoms are trans-disposed [N1—Ni1—N4 = 170.66 (8)°], while the chloride ligands are cis-disposed [Cl2—Ni1—Cl1 = 93.04 (2)°]. The disordered water molecules are linked to the complex molecule by O—H⋯Cl hydrogen bonds, and water H atom H2B is directed to the centroid of the C7–C12 ring (Fig. 1 ▸, Table 1 ▸).
Figure 1.
The molecular structure of the title complex, with atom labeling. Displacement ellipsoids are drawn at the 50% probability level. Hydrogen bonds are shown as orange dashed lines and the O—H⋯π interaction as a red arrow (Table 1 ▸).
Table 1. Hydrogen-bond geometry (Å, °).
Cg1, Cg2, Cg3, Cg4 and Cg5 are the centroids of the C7–C12, N5/N6/C18/C19/C24, N1/C1–C5, N4/C13–C17 and C19–C24 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C1—H1⋯Cl2 | 0.95 | 2.75 | 3.378 (2) | 124 |
| O1—H1A⋯Cl1 | 0.85 | 2.37 | 3.221 (4) | 174 |
| O2—H2B⋯Cl1 | 0.85 | 2.41 | 3.239 (4) | 165 |
| O2—H2A⋯O1i | 0.85 | 1.97 | 2.806 (6) | 167 |
| N3—H3⋯Cl2ii | 0.88 | 2.29 | 3.162 (2) | 171 |
| N6—H6⋯Cl1iii | 0.88 | 2.23 | 3.069 (2) | 160 |
| C2—H2⋯O2iv | 0.95 | 2.56 | 3.400 (5) | 147 |
| C20—H20⋯O2iii | 0.95 | 2.54 | 3.317 (5) | 139 |
| O1—H1B⋯Cg1 | 0.85 | 3.11 | 3.869 (3) | 150 |
| C3—H3A⋯Cg5ii | 0.95 | 2.97 | 3.738 (3) | 139 |
| C8—H8⋯Cg2v | 0.95 | 2.69 | 3.579 (3) | 155 |
| C9—H9⋯Cg5v | 0.95 | 2.88 | 3.542 (3) | 128 |
| C11—H11⋯Cg4 | 0.95 | 2.93 | 3.810 (3) | 155 |
| C23—H23⋯Cg3 | 0.95 | 2.94 | 3.733 (3) | 142 |
Symmetry codes: (i)
 ; (ii)
; (ii)
 ; (iii)
; (iii)
 ; (iv)
; (iv)
 ; (v)
; (v)
 .
.
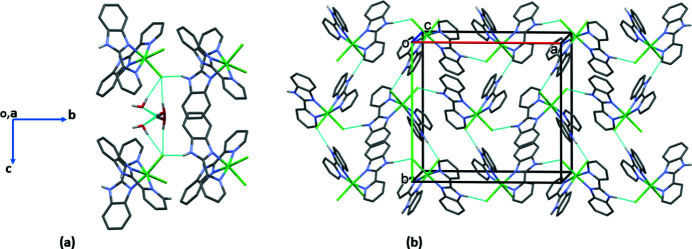
In the crystal, extensive hydrogen bonding is observed involving the disordered water molecule, the ligand NH groups and the chloride ions (Fig. 2 ▸ a and 2b and Table 1 ▸). The result is the formation of a supramolecular three-dimensional network (Fig. 3 ▸). There are also C—H⋯O and C—H⋯π interactions present (Table 1 ▸) consolidating the packing.
Figure 2.
Hydrogen-bonding networks involving, (a) the discorded water molecule, and (b) the N—H⋯Cl hydrogen bonds. For clarity, only the H atoms involved in hydrogen bonding (dashed lines; Table 1 ▸) have been included.
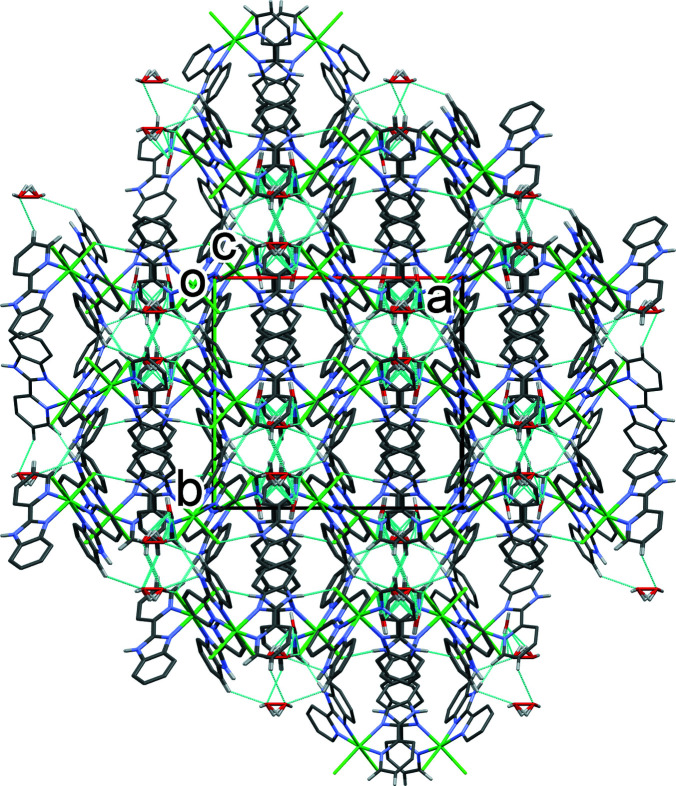
Figure 3.
A view along the c axis of the crystal packing of the title complex. For clarity, only the H atoms involved in hydrogen bonding (dashed lines; Table 1 ▸) have been included.
A search of the Cambridge Structural Database (CSD, Version 5.40, May 2019; Groom et al., 2016 ▸) revealed that the title compound is isostructural with the cobalt(II) complex dichloridobis-[2-(pyridin-2-yl)-1H-benzimidazole]cobalt(II) monohydrate (CSD refcode DACRIK; Das et al., 2011 ▸). The later was reported in space group C2/c but transformation of the unit cell gives space group I2/a (ADDSYMM in PLATON; Spek, 2020 ▸) with almost identical cell parameters to those of the title complex – see Fig. 4 ▸.
Figure 4.
A view of the ADDSYM (PLATON; Spek, 2020 ▸) transformation of the cell dimensions of the isostructural compound dichloridobis[2-(pyridin-2-yl)-1H-benzimidazole]cobalt(II) monohydrate (CSD refcode DACRIK; Das et al., 2011 ▸).
Synthesis and crystallization
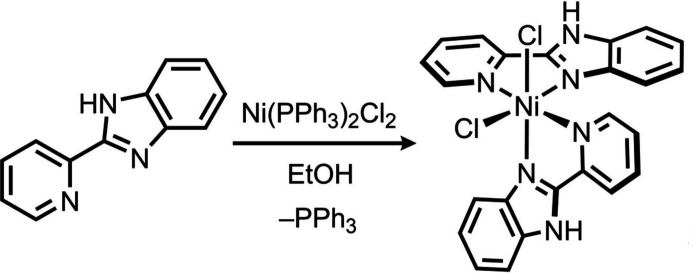
The reaction scheme for the synthesis of the title complex is given in Fig. 5 ▸. A solution of 2-(pyridin-2-yl)-1H-benzimidazole (0.15 g, 0.78 mmol) in ethanol (5 ml) was added dropwise to a stirring ethanolic solution of bis(triphenylphosphine)nickel(II) dichloride (0.50 g, 0.76 mmol). The mixture was stirred at room temperature for 24 h. The resulting mixture was concentrated and the product isolated by addition of diethyl ether (5 ml) giving a light-brown solid. Yield: 0.27 g (68%). Analysis calculated for C24H18Cl2N6Ni: C, 55.43; H, 3.49; N, 16.16%. Found: C, 55.23; H, 3.59; N, 16.25%. Light-blue plate-like crystals, suitable for X-ray diffraction analysis, were obtained by slow evaporation of a concentrated ethanol solution.
Figure 5.
Reaction scheme for the synthesis of the title complex.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. The complex crystallized as a monohydrate with the water molecule disordered over two sites (O1 and O2); occupancies fixed at 0.5 each.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | [NiCl2(C12H9N3)2]·H2O |
| M r | 538.07 |
| Crystal system, space group | Monoclinic, I2/a |
| Temperature (K) | 100 |
| a, b, c (Å) | 15.9019 (6), 14.7008 (7), 20.0039 (7) |
| β (°) | 95.924 (4) |
| V (Å3) | 4651.4 (3) |
| Z | 8 |
| Radiation type | Mo Kα |
| μ (mm−1) | 1.10 |
| Crystal size (mm) | 0.21 × 0.15 × 0.1 |
| Data collection | |
| Diffractometer | Rigaku Oxford Diffraction SuperNova, Dual, Cu at zero, Pilatus 200/300K |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku OD, 2015 ▸) |
| T min, T max | 0.785, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 29773, 6268, 5296 |
| R int | 0.046 |
| (sin θ/λ)max (Å−1) | 0.734 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.043, 0.113, 1.06 |
| No. of reflections | 6268 |
| No. of parameters | 322 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 1.16, −0.64 |
Supplementary Material
Crystal structure: contains datablock(s) Global, I. DOI: 10.1107/S2414314620000401/su4175sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314620000401/su4175Isup2.hkl
CCDC reference: 1946553
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
CSM is grateful to Professor Paul J. Chirik of Princeton University for hosting during the submission of the manuscript.
full crystallographic data
Crystal data
| [NiCl2(C12H9N3)2]·H2O | F(000) = 2208 |
| Mr = 538.07 | Dx = 1.537 Mg m−3 |
| Monoclinic, I2/a | Mo Kα radiation, λ = 0.71073 Å |
| a = 15.9019 (6) Å | Cell parameters from 13739 reflections |
| b = 14.7008 (7) Å | θ = 3.8–30.9° |
| c = 20.0039 (7) Å | µ = 1.10 mm−1 |
| β = 95.924 (4)° | T = 100 K |
| V = 4651.4 (3) Å3 | Plate, clear light blue |
| Z = 8 | 0.21 × 0.15 × 0.1 mm |
Data collection
| Rigaku Oxford Diffraction SuperNova, Dual, Cu at zero, Pilatus 200/300K diffractometer | 5296 reflections with I > 2σ(I) |
| ω scans | Rint = 0.046 |
| Absorption correction: multi-scan (CrysAlisPro; Rigaku OD, 2015) | θmax = 31.4°, θmin = 3.4° |
| Tmin = 0.785, Tmax = 1.000 | h = −22→21 |
| 29773 measured reflections | k = −19→19 |
| 6268 independent reflections | l = −29→26 |
Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.043 | Hydrogen site location: mixed |
| wR(F2) = 0.113 | H-atom parameters constrained |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0511P)2 + 11.2375P] where P = (Fo2 + 2Fc2)/3 |
| 6268 reflections | (Δ/σ)max = 0.001 |
| 322 parameters | Δρmax = 1.16 e Å−3 |
| 0 restraints | Δρmin = −0.64 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. The O-, N- and C-bound H atoms were included in calculated positions and treated as riding on the parent atom: O—H = 0.85 Å, N—H = 0.88 Å, C—H = 0.95 Å with Uiso(H) = 1.5Ueq(O) and 1.2Ueq(N,C). |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Ni1 | 0.41587 (2) | 0.53396 (2) | 0.26342 (2) | 0.01946 (9) | |
| Cl2 | 0.51535 (3) | 0.65004 (4) | 0.30400 (2) | 0.02048 (12) | |
| Cl1 | 0.34436 (3) | 0.63357 (5) | 0.17809 (3) | 0.02851 (14) | |
| N1 | 0.32601 (11) | 0.56738 (14) | 0.32888 (9) | 0.0206 (4) | |
| N4 | 0.50259 (11) | 0.47857 (14) | 0.20265 (9) | 0.0223 (4) | |
| N3 | 0.19362 (11) | 0.38608 (14) | 0.25224 (9) | 0.0214 (4) | |
| H3 | 0.1459 | 0.3800 | 0.2704 | 0.026* | |
| N5 | 0.47605 (11) | 0.43649 (14) | 0.32845 (9) | 0.0216 (4) | |
| N2 | 0.32190 (11) | 0.43767 (14) | 0.23554 (9) | 0.0230 (4) | |
| N6 | 0.54931 (12) | 0.30631 (15) | 0.33216 (10) | 0.0251 (4) | |
| H6 | 0.5796 | 0.2610 | 0.3188 | 0.030* | |
| C1 | 0.33068 (14) | 0.63587 (16) | 0.37299 (11) | 0.0220 (4) | |
| H1 | 0.3771 | 0.6767 | 0.3740 | 0.026* | |
| C6 | 0.25743 (13) | 0.44376 (16) | 0.27227 (10) | 0.0197 (4) | |
| C2 | 0.27009 (14) | 0.64957 (17) | 0.41747 (11) | 0.0239 (5) | |
| H2 | 0.2741 | 0.6996 | 0.4477 | 0.029* | |
| C13 | 0.52147 (14) | 0.50999 (18) | 0.14323 (12) | 0.0260 (5) | |
| H13 | 0.4931 | 0.5627 | 0.1250 | 0.031* | |
| C7 | 0.21767 (13) | 0.33846 (16) | 0.19770 (11) | 0.0220 (4) | |
| C5 | 0.25879 (13) | 0.51099 (16) | 0.32589 (10) | 0.0200 (4) | |
| C3 | 0.20350 (14) | 0.58805 (18) | 0.41653 (11) | 0.0251 (5) | |
| H3A | 0.1628 | 0.5939 | 0.4479 | 0.030* | |
| C4 | 0.19672 (13) | 0.51814 (17) | 0.36966 (11) | 0.0232 (5) | |
| H4 | 0.1509 | 0.4764 | 0.3676 | 0.028* | |
| C12 | 0.29871 (14) | 0.37186 (17) | 0.18758 (11) | 0.0239 (5) | |
| C17 | 0.54399 (13) | 0.40452 (16) | 0.22848 (11) | 0.0229 (4) | |
| C19 | 0.52027 (14) | 0.31581 (18) | 0.39437 (11) | 0.0266 (5) | |
| C18 | 0.52220 (13) | 0.38021 (17) | 0.29551 (11) | 0.0229 (4) | |
| C24 | 0.47421 (14) | 0.39799 (17) | 0.39175 (11) | 0.0247 (5) | |
| C8 | 0.17716 (14) | 0.27144 (18) | 0.15768 (12) | 0.0273 (5) | |
| H8 | 0.1232 | 0.2488 | 0.1658 | 0.033* | |
| C9 | 0.21903 (16) | 0.23914 (19) | 0.10533 (12) | 0.0305 (5) | |
| H9 | 0.1928 | 0.1942 | 0.0761 | 0.037* | |
| C16 | 0.60480 (15) | 0.35909 (18) | 0.19622 (12) | 0.0285 (5) | |
| H16 | 0.6329 | 0.3071 | 0.2159 | 0.034* | |
| C10 | 0.29967 (16) | 0.2715 (2) | 0.09461 (13) | 0.0335 (6) | |
| H10 | 0.3266 | 0.2477 | 0.0582 | 0.040* | |
| C14 | 0.58124 (16) | 0.46814 (19) | 0.10720 (13) | 0.0320 (6) | |
| H14 | 0.5929 | 0.4915 | 0.0648 | 0.038* | |
| C11 | 0.34100 (15) | 0.3368 (2) | 0.13516 (13) | 0.0319 (6) | |
| H11 | 0.3960 | 0.3572 | 0.1279 | 0.038* | |
| C23 | 0.43824 (16) | 0.42902 (19) | 0.44841 (11) | 0.0290 (5) | |
| H23 | 0.4070 | 0.4842 | 0.4476 | 0.035* | |
| C22 | 0.45004 (18) | 0.3760 (2) | 0.50589 (12) | 0.0345 (6) | |
| H22 | 0.4270 | 0.3959 | 0.5453 | 0.041* | |
| C20 | 0.53109 (16) | 0.2614 (2) | 0.45225 (13) | 0.0335 (6) | |
| H20 | 0.5614 | 0.2057 | 0.4534 | 0.040* | |
| C21 | 0.49486 (17) | 0.2940 (2) | 0.50755 (12) | 0.0362 (6) | |
| H21 | 0.5006 | 0.2596 | 0.5479 | 0.043* | |
| C15 | 0.62325 (16) | 0.3922 (2) | 0.13411 (13) | 0.0320 (5) | |
| H15 | 0.6644 | 0.3627 | 0.1105 | 0.038* | |
| O1 | 0.3180 (3) | 0.5134 (3) | 0.0429 (2) | 0.0440 (10) | 0.5 |
| H1A | 0.3268 | 0.5413 | 0.0802 | 0.066* | 0.5 |
| H1B | 0.3085 | 0.4575 | 0.0502 | 0.066* | 0.5 |
| O2 | 0.2882 (3) | 0.6487 (3) | 0.01818 (18) | 0.0429 (10) | 0.5 |
| H2A | 0.2602 | 0.6019 | 0.0042 | 0.064* | 0.5 |
| H2B | 0.3113 | 0.6391 | 0.0578 | 0.064* | 0.5 |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Ni1 | 0.01005 (13) | 0.03529 (17) | 0.01374 (13) | −0.00391 (10) | 0.00448 (10) | −0.00355 (11) |
| Cl2 | 0.0109 (2) | 0.0340 (3) | 0.0169 (2) | −0.00172 (18) | 0.00350 (17) | −0.00279 (19) |
| Cl1 | 0.0169 (2) | 0.0491 (4) | 0.0188 (2) | −0.0029 (2) | −0.00126 (19) | 0.0018 (2) |
| N1 | 0.0130 (8) | 0.0355 (10) | 0.0141 (8) | −0.0013 (7) | 0.0049 (6) | −0.0016 (7) |
| N4 | 0.0129 (8) | 0.0379 (11) | 0.0165 (8) | −0.0066 (7) | 0.0038 (7) | −0.0047 (8) |
| N3 | 0.0098 (7) | 0.0387 (11) | 0.0162 (8) | −0.0036 (7) | 0.0033 (6) | 0.0001 (8) |
| N5 | 0.0154 (8) | 0.0346 (10) | 0.0153 (8) | −0.0064 (7) | 0.0036 (7) | −0.0023 (7) |
| N2 | 0.0133 (8) | 0.0401 (11) | 0.0166 (8) | −0.0052 (8) | 0.0059 (7) | −0.0046 (8) |
| N6 | 0.0167 (8) | 0.0376 (11) | 0.0208 (9) | −0.0022 (8) | 0.0007 (7) | −0.0010 (8) |
| C1 | 0.0172 (10) | 0.0334 (12) | 0.0158 (9) | −0.0009 (8) | 0.0035 (8) | −0.0014 (8) |
| C6 | 0.0114 (9) | 0.0319 (11) | 0.0159 (9) | −0.0005 (8) | 0.0020 (7) | 0.0000 (8) |
| C2 | 0.0196 (10) | 0.0384 (13) | 0.0143 (9) | 0.0055 (9) | 0.0048 (8) | −0.0004 (9) |
| C13 | 0.0179 (10) | 0.0420 (13) | 0.0192 (10) | −0.0054 (9) | 0.0064 (8) | −0.0015 (9) |
| C7 | 0.0141 (9) | 0.0351 (12) | 0.0163 (9) | −0.0028 (8) | 0.0000 (8) | 0.0011 (9) |
| C5 | 0.0113 (9) | 0.0368 (12) | 0.0120 (8) | 0.0018 (8) | 0.0020 (7) | 0.0007 (8) |
| C3 | 0.0149 (9) | 0.0445 (14) | 0.0166 (9) | 0.0053 (9) | 0.0049 (8) | 0.0013 (9) |
| C4 | 0.0111 (9) | 0.0423 (13) | 0.0163 (9) | 0.0005 (8) | 0.0022 (8) | 0.0036 (9) |
| C12 | 0.0160 (10) | 0.0385 (13) | 0.0172 (9) | −0.0054 (9) | 0.0025 (8) | −0.0057 (9) |
| C17 | 0.0129 (9) | 0.0362 (12) | 0.0196 (10) | −0.0059 (8) | 0.0021 (8) | −0.0051 (9) |
| C19 | 0.0190 (10) | 0.0422 (13) | 0.0178 (10) | −0.0096 (9) | −0.0019 (8) | −0.0011 (9) |
| C18 | 0.0128 (9) | 0.0383 (12) | 0.0173 (9) | −0.0046 (8) | −0.0002 (8) | −0.0024 (9) |
| C24 | 0.0174 (10) | 0.0386 (13) | 0.0176 (10) | −0.0094 (9) | −0.0009 (8) | −0.0009 (9) |
| C8 | 0.0180 (10) | 0.0414 (14) | 0.0216 (10) | −0.0077 (9) | −0.0025 (9) | −0.0010 (10) |
| C9 | 0.0255 (11) | 0.0430 (14) | 0.0219 (11) | −0.0073 (10) | −0.0036 (9) | −0.0077 (10) |
| C16 | 0.0179 (10) | 0.0418 (14) | 0.0264 (11) | −0.0009 (9) | 0.0053 (9) | −0.0049 (10) |
| C10 | 0.0252 (12) | 0.0514 (16) | 0.0245 (11) | −0.0056 (11) | 0.0050 (9) | −0.0140 (11) |
| C14 | 0.0236 (12) | 0.0501 (16) | 0.0242 (11) | −0.0064 (11) | 0.0123 (10) | −0.0039 (11) |
| C11 | 0.0204 (11) | 0.0509 (15) | 0.0260 (12) | −0.0089 (10) | 0.0098 (9) | −0.0165 (11) |
| C23 | 0.0253 (11) | 0.0434 (14) | 0.0182 (10) | −0.0138 (10) | 0.0021 (9) | −0.0041 (10) |
| C22 | 0.0358 (14) | 0.0520 (16) | 0.0158 (10) | −0.0193 (12) | 0.0023 (10) | −0.0037 (10) |
| C20 | 0.0270 (12) | 0.0454 (15) | 0.0263 (12) | −0.0106 (11) | −0.0057 (10) | 0.0043 (11) |
| C21 | 0.0362 (14) | 0.0533 (17) | 0.0174 (10) | −0.0191 (12) | −0.0048 (10) | 0.0042 (11) |
| C15 | 0.0201 (11) | 0.0511 (15) | 0.0268 (12) | −0.0027 (10) | 0.0114 (9) | −0.0093 (11) |
| O1 | 0.057 (3) | 0.048 (2) | 0.0260 (19) | 0.009 (2) | −0.0008 (18) | 0.0042 (17) |
| O2 | 0.052 (3) | 0.057 (3) | 0.0188 (17) | −0.022 (2) | 0.0026 (17) | −0.0022 (16) |
Geometric parameters (Å, º)
| Ni1—Cl2 | 2.4101 (6) | C3—C4 | 1.388 (3) |
| Ni1—Cl1 | 2.4394 (6) | C4—H4 | 0.9500 |
| Ni1—N1 | 2.0937 (18) | C12—C11 | 1.401 (3) |
| Ni1—N4 | 2.0949 (19) | C17—C18 | 1.463 (3) |
| Ni1—N5 | 2.099 (2) | C17—C16 | 1.388 (3) |
| Ni1—N2 | 2.0918 (19) | C19—C24 | 1.411 (4) |
| N1—C1 | 1.336 (3) | C19—C20 | 1.403 (3) |
| N1—C5 | 1.349 (3) | C24—C23 | 1.398 (3) |
| N4—C13 | 1.338 (3) | C8—H8 | 0.9500 |
| N4—C17 | 1.347 (3) | C8—C9 | 1.382 (4) |
| N3—H3 | 0.8800 | C9—H9 | 0.9500 |
| N3—C6 | 1.351 (3) | C9—C10 | 1.405 (4) |
| N3—C7 | 1.383 (3) | C16—H16 | 0.9500 |
| N5—C18 | 1.326 (3) | C16—C15 | 1.393 (4) |
| N5—C24 | 1.390 (3) | C10—H10 | 0.9500 |
| N2—C6 | 1.325 (3) | C10—C11 | 1.378 (3) |
| N2—C12 | 1.385 (3) | C14—H14 | 0.9500 |
| N6—H6 | 0.8800 | C14—C15 | 1.382 (4) |
| N6—C19 | 1.378 (3) | C11—H11 | 0.9500 |
| N6—C18 | 1.356 (3) | C23—H23 | 0.9500 |
| C1—H1 | 0.9500 | C23—C22 | 1.386 (4) |
| C1—C2 | 1.392 (3) | C22—H22 | 0.9500 |
| C6—C5 | 1.457 (3) | C22—C21 | 1.398 (4) |
| C2—H2 | 0.9500 | C20—H20 | 0.9500 |
| C2—C3 | 1.391 (3) | C20—C21 | 1.385 (4) |
| C13—H13 | 0.9500 | C21—H21 | 0.9500 |
| C13—C14 | 1.394 (3) | C15—H15 | 0.9500 |
| C7—C12 | 1.413 (3) | O1—H1A | 0.8505 |
| C7—C8 | 1.386 (3) | O1—H1B | 0.8503 |
| C5—C4 | 1.389 (3) | O2—H2A | 0.8507 |
| C3—H3A | 0.9500 | O2—H2B | 0.8498 |
| Cl2—Ni1—Cl1 | 93.04 (2) | C5—C4—H4 | 120.9 |
| N1—Ni1—Cl2 | 95.15 (5) | C3—C4—C5 | 118.1 (2) |
| N1—Ni1—Cl1 | 89.87 (5) | C3—C4—H4 | 120.9 |
| N1—Ni1—N4 | 170.66 (8) | N2—C12—C7 | 108.96 (19) |
| N1—Ni1—N5 | 93.99 (7) | N2—C12—C11 | 131.4 (2) |
| N4—Ni1—Cl2 | 91.27 (5) | C11—C12—C7 | 119.6 (2) |
| N4—Ni1—Cl1 | 96.57 (6) | N4—C17—C18 | 113.3 (2) |
| N4—Ni1—N5 | 78.99 (8) | N4—C17—C16 | 123.1 (2) |
| N5—Ni1—Cl2 | 91.90 (5) | C16—C17—C18 | 123.4 (2) |
| N5—Ni1—Cl1 | 173.44 (6) | N6—C19—C24 | 105.9 (2) |
| N2—Ni1—Cl2 | 174.23 (5) | N6—C19—C20 | 131.6 (3) |
| N2—Ni1—Cl1 | 87.19 (6) | C20—C19—C24 | 122.5 (2) |
| N2—Ni1—N1 | 79.09 (7) | N5—C18—N6 | 113.1 (2) |
| N2—Ni1—N4 | 94.43 (7) | N5—C18—C17 | 119.9 (2) |
| N2—Ni1—N5 | 88.32 (8) | N6—C18—C17 | 126.8 (2) |
| C1—N1—Ni1 | 126.52 (15) | N5—C24—C19 | 108.8 (2) |
| C1—N1—C5 | 118.82 (18) | N5—C24—C23 | 130.9 (2) |
| C5—N1—Ni1 | 114.63 (15) | C23—C24—C19 | 120.2 (2) |
| C13—N4—Ni1 | 127.08 (17) | C7—C8—H8 | 121.6 |
| C13—N4—C17 | 118.3 (2) | C9—C8—C7 | 116.8 (2) |
| C17—N4—Ni1 | 114.60 (15) | C9—C8—H8 | 121.6 |
| C6—N3—H3 | 126.5 | C8—C9—H9 | 119.4 |
| C6—N3—C7 | 106.94 (17) | C8—C9—C10 | 121.2 (2) |
| C7—N3—H3 | 126.5 | C10—C9—H9 | 119.4 |
| C18—N5—Ni1 | 111.00 (15) | C17—C16—H16 | 121.1 |
| C18—N5—C24 | 105.3 (2) | C17—C16—C15 | 117.9 (2) |
| C24—N5—Ni1 | 142.33 (16) | C15—C16—H16 | 121.1 |
| C6—N2—Ni1 | 112.26 (15) | C9—C10—H10 | 118.9 |
| C6—N2—C12 | 105.40 (18) | C11—C10—C9 | 122.2 (2) |
| C12—N2—Ni1 | 142.08 (15) | C11—C10—H10 | 118.9 |
| C19—N6—H6 | 126.6 | C13—C14—H14 | 120.6 |
| C18—N6—H6 | 126.6 | C15—C14—C13 | 118.9 (2) |
| C18—N6—C19 | 106.8 (2) | C15—C14—H14 | 120.6 |
| N1—C1—H1 | 118.8 | C12—C11—H11 | 121.3 |
| N1—C1—C2 | 122.4 (2) | C10—C11—C12 | 117.4 (2) |
| C2—C1—H1 | 118.8 | C10—C11—H11 | 121.3 |
| N3—C6—C5 | 126.71 (19) | C24—C23—H23 | 121.4 |
| N2—C6—N3 | 113.18 (19) | C22—C23—C24 | 117.2 (3) |
| N2—C6—C5 | 120.04 (19) | C22—C23—H23 | 121.4 |
| C1—C2—H2 | 120.9 | C23—C22—H22 | 119.0 |
| C3—C2—C1 | 118.3 (2) | C23—C22—C21 | 122.0 (2) |
| C3—C2—H2 | 120.9 | C21—C22—H22 | 119.0 |
| N4—C13—H13 | 118.8 | C19—C20—H20 | 122.1 |
| N4—C13—C14 | 122.3 (2) | C21—C20—C19 | 115.9 (3) |
| C14—C13—H13 | 118.8 | C21—C20—H20 | 122.1 |
| N3—C7—C12 | 105.53 (19) | C22—C21—H21 | 118.9 |
| N3—C7—C8 | 131.7 (2) | C20—C21—C22 | 122.1 (2) |
| C8—C7—C12 | 122.7 (2) | C20—C21—H21 | 118.9 |
| N1—C5—C6 | 113.64 (18) | C16—C15—H15 | 120.3 |
| N1—C5—C4 | 122.5 (2) | C14—C15—C16 | 119.5 (2) |
| C4—C5—C6 | 123.9 (2) | C14—C15—H15 | 120.3 |
| C2—C3—H3A | 120.1 | H1A—O1—H1B | 109.4 |
| C4—C3—C2 | 119.8 (2) | H2A—O2—H2B | 109.5 |
| C4—C3—H3A | 120.1 |
Hydrogen-bond geometry (Å, º)
Cg1, Cg2, Cg3, Cg4 and Cg5 are the centroids of the C7–C12, N5/N6/C18/C19/C24, N1/C1–C5, N4/C13–C17 and C19–C24 rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C1—H1···Cl2 | 0.95 | 2.75 | 3.378 (2) | 124 |
| O1—H1A···Cl1 | 0.85 | 2.37 | 3.221 (4) | 174 |
| O2—H2B···Cl1 | 0.85 | 2.41 | 3.239 (4) | 165 |
| O2—H2A···O1i | 0.85 | 1.97 | 2.806 (6) | 167 |
| N3—H3···Cl2ii | 0.88 | 2.29 | 3.162 (2) | 171 |
| N6—H6···Cl1iii | 0.88 | 2.23 | 3.069 (2) | 160 |
| C2—H2···O2iv | 0.95 | 2.56 | 3.400 (5) | 147 |
| C20—H20···O2iii | 0.95 | 2.54 | 3.317 (5) | 139 |
| O1—H1B···Cg1 | 0.85 | 3.11 | 3.869 (3) | 150 |
| C3—H3A···Cg5ii | 0.95 | 2.97 | 3.738 (3) | 139 |
| C8—H8···Cg2v | 0.95 | 2.69 | 3.579 (3) | 155 |
| C9—H9···Cg5v | 0.95 | 2.88 | 3.542 (3) | 128 |
| C11—H11···Cg4 | 0.95 | 2.93 | 3.810 (3) | 155 |
| C23—H23···Cg3 | 0.95 | 2.94 | 3.733 (3) | 142 |
Symmetry codes: (i) −x+1/2, y, −z; (ii) x−1/2, −y+1, z; (iii) −x+1, y−1/2, −z+1/2; (iv) −x+1/2, −y+3/2, −z+1/2; (v) −x+1/2, −y+1/2, −z+1/2.
Funding Statement
The authors thank the NSERC of Canada for funding. CSM is grateful to NSERC for a CGS-D fellowship. PGH thanks NSERC for a Discovery Grant and the University of Lethbridge for a Tier I Board of Governors Research Chair in Organometallic Chemistry. SOO is grateful to the University of KwaZulu-Natal and National Research Foundation–South Africa (grant No. CPRR98938) for financial support.
References
- Abubakar, S. & Bala, M. D. (2018). J. Coord. Chem. 71, 2913–2923.
- Chen, Z., Zeng, M., Zhang, Y., Zhang, Z. & Liang, F. (2010). Appl. Organomet. Chem. 24, 625–630.
- Das, S., Guha, S., Banerjee, A., Lohar, S., Sahana, A. & Das, D. (2011). Org. Biomol. Chem. 9, 7097–7104. [DOI] [PubMed]
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Garduño, J. A. & García, J. J. (2017). ACS Omega, 2, 2337–2343. [DOI] [PMC free article] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Li, Y. Y., Yu, S. L., Shen, W. Y. & Gao, J. X. (2015). Acc. Chem. Res. 48, 2587–2598. [DOI] [PubMed]
- Morris, R. H. (2009). Chem. Soc. Rev. 38, 2282–2291.
- Raja, N. & Ramesh, R. (2012). Tetrahedron Lett. 53, 4770–4774.
- Rigaku OD (2015). CrysAlis PRO. Rigaku Oxford Diffraction Ltd., Yarnton, England.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Spek, A. L. (2020). Acta Cryst. E76, 1–11. [DOI] [PMC free article] [PubMed]
- Wang, D. & Astruc, D. (2015). Chem. Rev. 115, 6621–6686. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Zhu, M. (2014). Appl. Catal. Gen. 479, 45–48.
- Zhu, X.-H., Cai, L., Wang, C., Wang, Y., Guo, X. & Hou, X. (2014). J. Mol. Catal. A Chem. 393, 134–141.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) Global, I. DOI: 10.1107/S2414314620000401/su4175sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314620000401/su4175Isup2.hkl
CCDC reference: 1946553
Additional supporting information: crystallographic information; 3D view; checkCIF report