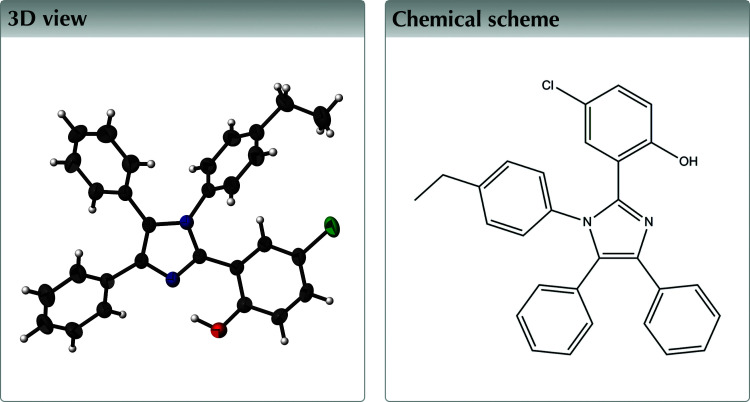
In the title compound, the 5-chlorophenol ring and the imidazole ring are nearly coplanar, with a dihedral angle of 15.76 (9)°. The ethylphenyl ring and the two phenyl rings subtend at angles of 71.09 (7), 43.95 (5) and 36.53 (9)°, respectively, with the imidazole plane.
Keywords: crystal structure, imidazole, hydrogen bonding
Abstract
In the title compound, C29H23ClN2O, the 5-chlorophenol ring and the imidazole ring are nearly coplanar, with a dihedral angle of 15.76 (9)° between them. The ethylphenyl ring and the two phenyl rings subtend angles of 71.09 (7), 43.95 (5) and 36.53 (9)°, respectively, with the imidazole plane. An intramolecular O—H⋯N hydrogen bond supports the molecular conformation, and an intermolecular C—H⋯O interaction, originating from an ortho-phenyl H atom, stabilizes the packing arrangement. In addition, a weak C—H⋯π interaction, also involving an ortho-phenyl H atom, is observed.
Structure description
The imidazole moiety is known to play an important role in biological systems being a part of the histidyl residue in peptides and proteins (Sigel et al., 2000 ▸). Multi-substituted imidazoles are an important class of heterocyclic compounds that exhibit diverse biological activities such as anti-inflammatory (Gaonkar et al., 2009 ▸), antileishmanial (Bhandari et al., 2010 ▸) and anticancer (Ozkay et al., 2010 ▸) activities. As part of our ongoing studies in this area, we herein report the synthesis and crystal structure of the title compound, 4-chloro-2-(1-(4-ethylphenyl)-4,5-diphenyl-1H-imidazol-2-yl)phenol (Fig. 1 ▸). The 5-chlorophenol ring, two phenyl rings and the ethylphenyl ring are substituents on the central five-membered imidazole ring (C1/N2/C3/C2/N1). The imidazole and the 5-chlorophenol rings are close to coplanar with a dihedral angle of 15.76 (9)° between them. The imidazole ring subtends at dihedral angles of 71.09 (7), 43.95 (5) and 36.53 (9)° with the ethylphenyl ring and the two phenyl rings (C18–C23 and C24–C29), respectively. A strong intramolecular O1—H1⋯N1 hydrogen bond is formed between the O1 atom of the 5-chlorophenol group and atom N1 of the imidazole ring (Fig. 2 ▸), forming an
 (6) graph-set motif, which stabilizes the close to coplanar arrangement of the imidazole and phenol rings.
(6) graph-set motif, which stabilizes the close to coplanar arrangement of the imidazole and phenol rings.
Figure 1.
The molecular structure of the title compound with the atom-numbering scheme. Displacement ellipsoids are drawn at the 50% probability level. H atoms are represented as small spheres of arbitrary radius.
Figure 2.
Unit-cell packing of the title compound showing the intramolecular O—H⋯N interactions, intermolecular C—H⋯O interactions and intermolecular C—H⋯π interactions as dotted lines. H atoms not involved in hydrogen bonding have been excluded.
In the crystal, atom C19 of the phenyl ring and the hydroxyl O1 atom of the phenol group are involved in a weak C19—H19⋯O1i interaction that links the molecules along the a-axis direction (Fig. 2 ▸). Thus the hydroxyl O atom acts as both a hydrogen-bond donor and an acceptor. The crystal structure is further consolidated by a C15—H15⋯Cg ii interaction with the aryl ring (Table 1 ▸, Fig. 2 ▸).
Table 1. Hydrogen-bond geometry (Å, °).
Cg is the centroid of the C4–C9 aryl ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯N1 | 1.00 | 1.66 | 2.549 (1) | 145 |
| C19—H19⋯O1i | 0.93 | 2.57 | 3.242 (3) | 129 |
| C15—H15⋯Cg ii | 0.93 | 2.72 | 3.527 (2) | 146 |
Symmetry codes: (i)
 ; (ii)
; (ii)
 .
.
Synthesis and crystallization
The title compound was synthesized by the one-pot reaction of benzil (10 mmol), 4-ethylaniline (10 mmol) and 5-chloro-2-hydroxybenzaldehyde (10 mmol) with ammonium acetate (10 mmol) in a glacial acetic acid (20 ml) medium. The mixture was refluxed for 5 h at 343 K, the progress of the reaction being monitored by TLC. After completion of the reaction, the mixture was cooled to room temperature and poured into 100 ml of ice-cold water. The resulting precipitate was filtered, dried and further purified by column chromatography (7:3 petroleum ether:ethyl acetate) and isolated in good yield (85%). The product was recrystallized from ethanol solution. IR (KBr) (cm−1): 3448.63 (OH), 1947.51 (C=C), 1601.84 (C=N). 1H NMR (CDCl3): δ 1.242 (t, J = 7.2 Hz, 2H), 2.64–2.68 (q, J = 7.2 Hz, 3H), 7.09–7.26 (m, 13H), 7.50–7.52 (m, 4H), 6.34–6.35 (s, 1H). GC–MS (EI, 70 eV): m/z: 450.95.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C29H23ClN2O |
| M r | 450.94 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 297 |
| a, b, c (Å) | 9.0627 (6), 10.7595 (8), 24.4636 (19) |
| β (°) | 100.599 (3) |
| V (Å3) | 2344.7 (3) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.19 |
| Crystal size (mm) | 0.45 × 0.38 × 0.35 |
| Data collection | |
| Diffractometer | Bruker SMART APEX CCD |
| Absorption correction | Multi-scan (SADABS; Bruker, 1998 ▸) |
| T min, T max | 0.821, 0.928 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 34596, 4807, 3593 |
| R int | 0.044 |
| (sin θ/λ)max (Å−1) | 0.627 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.047, 0.129, 1.03 |
| No. of reflections | 4807 |
| No. of parameters | 301 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.26, −0.29 |
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2414314619016900/zl4037sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314619016900/zl4037Isup2.hkl
Supporting information file. DOI: 10.1107/S2414314619016900/zl4037sup3.tif
Supporting information file. DOI: 10.1107/S2414314619016900/zl4037sup4.pdf
Proton-NMR. DOI: 10.1107/S2414314619016900/zl4037sup5.pdf
Supporting information file. DOI: 10.1107/S2414314619016900/zl4037Isup6.cml
CCDC reference: 1950453
Additional supporting information: crystallographic information; 3D view; checkCIF report
full crystallographic data
Crystal data
| C29H23ClN2O | F(000) = 944 |
| Mr = 450.94 | Dx = 1.277 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 9.0627 (6) Å | Cell parameters from 4807 reflections |
| b = 10.7595 (8) Å | θ = 2.6–26.5° |
| c = 24.4636 (19) Å | µ = 0.19 mm−1 |
| β = 100.599 (3)° | T = 297 K |
| V = 2344.7 (3) Å3 | Block, colorless |
| Z = 4 | 0.45 × 0.38 × 0.35 mm |
Data collection
| Bruker SMART APEX CCD diffractometer | 3593 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.044 |
| ω scans | θmax = 26.5°, θmin = 2.6° |
| Absorption correction: multi-scan (SADABS; Bruker, 1998) | h = −11→11 |
| Tmin = 0.821, Tmax = 0.928 | k = −13→13 |
| 34596 measured reflections | l = −30→30 |
| 4807 independent reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.047 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.129 | w = 1/[σ2(Fo2) + (0.0461P)2 + 1.5828P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max = 0.001 |
| 4807 reflections | Δρmax = 0.26 e Å−3 |
| 301 parameters | Δρmin = −0.29 e Å−3 |
| 0 restraints | Extinction correction: SHELXL2018 (Sheldrick, 2015), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0135 (11) |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. H atoms were placed at calculated positions in the riding-model approximation, with O—H = 1.00 Å and C—H = 0.93, 0.96 and 0.97 Å for aromatic, methyl and methine H atoms, respectively, and with Uiso(H) = 1.5Ueq(C) for methyl H atoms and 1.2Ueq(C) otherwise. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 1.10374 (7) | 0.46682 (6) | 1.17331 (2) | 0.0588 (2) | |
| O1 | 0.67555 (18) | 0.23218 (14) | 0.99506 (7) | 0.0538 (4) | |
| H1 | 0.647 (3) | 0.2890 (19) | 0.9625 (11) | 0.081* | |
| N1 | 0.65078 (19) | 0.43455 (15) | 0.93998 (7) | 0.0412 (4) | |
| N2 | 0.71390 (18) | 0.61739 (15) | 0.97832 (6) | 0.0367 (4) | |
| C1 | 0.7255 (2) | 0.49090 (18) | 0.98494 (8) | 0.0374 (4) | |
| C2 | 0.6291 (2) | 0.64067 (18) | 0.92560 (7) | 0.0365 (4) | |
| C3 | 0.5904 (2) | 0.52542 (18) | 0.90324 (8) | 0.0372 (4) | |
| C4 | 0.8032 (2) | 0.42008 (18) | 1.03298 (8) | 0.0369 (4) | |
| C5 | 0.7687 (2) | 0.29210 (19) | 1.03599 (8) | 0.0404 (5) | |
| C6 | 0.8346 (2) | 0.2226 (2) | 1.08197 (9) | 0.0451 (5) | |
| H6 | 0.808858 | 0.139367 | 1.084268 | 0.054* | |
| C7 | 0.9371 (2) | 0.2747 (2) | 1.12404 (8) | 0.0453 (5) | |
| H7 | 0.980055 | 0.227497 | 1.154686 | 0.054* | |
| C8 | 0.9756 (2) | 0.3984 (2) | 1.12027 (8) | 0.0407 (5) | |
| C9 | 0.9117 (2) | 0.47049 (19) | 1.07553 (8) | 0.0401 (4) | |
| H9 | 0.940636 | 0.553051 | 1.073556 | 0.048* | |
| C10 | 0.7779 (2) | 0.70811 (17) | 1.01924 (7) | 0.0343 (4) | |
| C11 | 0.7207 (2) | 0.7207 (2) | 1.06745 (8) | 0.0410 (5) | |
| H11 | 0.634020 | 0.678509 | 1.071702 | 0.049* | |
| C12 | 0.7936 (2) | 0.7967 (2) | 1.10933 (8) | 0.0465 (5) | |
| H12 | 0.755755 | 0.804476 | 1.142025 | 0.056* | |
| C13 | 0.9217 (2) | 0.86161 (19) | 1.10368 (8) | 0.0431 (5) | |
| C14 | 0.9724 (2) | 0.8518 (2) | 1.05376 (9) | 0.0483 (5) | |
| H14 | 1.055933 | 0.897377 | 1.048605 | 0.058* | |
| C15 | 0.9012 (2) | 0.7755 (2) | 1.01134 (8) | 0.0428 (5) | |
| H15 | 0.936377 | 0.769909 | 0.978027 | 0.051* | |
| C16 | 1.0063 (3) | 0.9367 (2) | 1.15138 (10) | 0.0616 (7) | |
| H16A | 0.936247 | 0.989876 | 1.165958 | 0.074* | |
| H16B | 1.078251 | 0.989614 | 1.137768 | 0.074* | |
| C17 | 1.0880 (4) | 0.8561 (3) | 1.19769 (13) | 0.0940 (11) | |
| H17A | 1.139962 | 0.907784 | 1.227008 | 0.141* | |
| H17B | 1.017021 | 0.804672 | 1.211867 | 0.141* | |
| H17C | 1.159005 | 0.804410 | 1.183681 | 0.141* | |
| C18 | 0.6046 (2) | 0.76627 (18) | 0.90155 (8) | 0.0372 (4) | |
| C19 | 0.5629 (3) | 0.8667 (2) | 0.93102 (9) | 0.0504 (5) | |
| H19 | 0.549954 | 0.855868 | 0.967564 | 0.061* | |
| C20 | 0.5404 (3) | 0.9826 (2) | 0.90656 (11) | 0.0641 (7) | |
| H20 | 0.513916 | 1.049426 | 0.926859 | 0.077* | |
| C21 | 0.5569 (3) | 0.9994 (2) | 0.85245 (12) | 0.0686 (8) | |
| H21 | 0.540690 | 1.077261 | 0.835939 | 0.082* | |
| C22 | 0.5975 (3) | 0.9009 (2) | 0.82270 (10) | 0.0639 (7) | |
| H22 | 0.608119 | 0.912305 | 0.785941 | 0.077* | |
| C23 | 0.6226 (2) | 0.7851 (2) | 0.84696 (9) | 0.0475 (5) | |
| H23 | 0.651751 | 0.719374 | 0.826657 | 0.057* | |
| C24 | 0.4960 (2) | 0.48681 (18) | 0.85017 (8) | 0.0376 (4) | |
| C25 | 0.3671 (2) | 0.5502 (2) | 0.82669 (9) | 0.0490 (5) | |
| H25 | 0.341256 | 0.623145 | 0.843045 | 0.059* | |
| C26 | 0.2763 (3) | 0.5057 (3) | 0.77900 (10) | 0.0602 (6) | |
| H26 | 0.190035 | 0.549194 | 0.763527 | 0.072* | |
| C27 | 0.3123 (3) | 0.3978 (3) | 0.75417 (9) | 0.0599 (6) | |
| H27 | 0.250298 | 0.368055 | 0.722302 | 0.072* | |
| C28 | 0.4405 (3) | 0.3347 (2) | 0.77687 (9) | 0.0568 (6) | |
| H28 | 0.466097 | 0.262341 | 0.760037 | 0.068* | |
| C29 | 0.5316 (3) | 0.3779 (2) | 0.82458 (8) | 0.0471 (5) | |
| H29 | 0.617670 | 0.333929 | 0.839817 | 0.057* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0561 (3) | 0.0675 (4) | 0.0446 (3) | 0.0094 (3) | −0.0121 (2) | −0.0065 (3) |
| O1 | 0.0686 (10) | 0.0383 (8) | 0.0474 (9) | −0.0101 (7) | −0.0079 (7) | 0.0039 (7) |
| N1 | 0.0487 (10) | 0.0358 (9) | 0.0348 (8) | −0.0001 (7) | −0.0034 (7) | 0.0022 (7) |
| N2 | 0.0440 (9) | 0.0315 (8) | 0.0318 (8) | −0.0011 (7) | −0.0006 (7) | 0.0009 (6) |
| C1 | 0.0424 (10) | 0.0343 (10) | 0.0326 (9) | 0.0000 (8) | −0.0008 (8) | 0.0025 (8) |
| C2 | 0.0400 (10) | 0.0363 (10) | 0.0311 (9) | 0.0031 (8) | 0.0011 (8) | 0.0010 (8) |
| C3 | 0.0414 (10) | 0.0362 (10) | 0.0316 (9) | 0.0025 (8) | 0.0006 (8) | 0.0026 (8) |
| C4 | 0.0421 (10) | 0.0335 (10) | 0.0336 (9) | 0.0027 (8) | 0.0027 (8) | 0.0030 (8) |
| C5 | 0.0450 (11) | 0.0380 (10) | 0.0372 (10) | 0.0005 (9) | 0.0043 (8) | 0.0021 (8) |
| C6 | 0.0543 (12) | 0.0377 (11) | 0.0433 (11) | 0.0023 (9) | 0.0086 (9) | 0.0089 (9) |
| C7 | 0.0515 (12) | 0.0488 (12) | 0.0347 (10) | 0.0112 (10) | 0.0051 (9) | 0.0110 (9) |
| C8 | 0.0416 (10) | 0.0466 (12) | 0.0320 (10) | 0.0096 (9) | 0.0020 (8) | −0.0016 (9) |
| C9 | 0.0441 (11) | 0.0364 (10) | 0.0370 (10) | 0.0052 (8) | −0.0001 (8) | 0.0020 (8) |
| C10 | 0.0372 (9) | 0.0331 (9) | 0.0304 (9) | 0.0013 (8) | 0.0004 (7) | −0.0011 (7) |
| C11 | 0.0377 (10) | 0.0483 (12) | 0.0373 (10) | −0.0013 (9) | 0.0076 (8) | 0.0021 (9) |
| C12 | 0.0518 (12) | 0.0558 (13) | 0.0321 (10) | 0.0068 (10) | 0.0083 (9) | −0.0040 (9) |
| C13 | 0.0497 (12) | 0.0388 (11) | 0.0368 (10) | 0.0045 (9) | −0.0024 (9) | −0.0023 (9) |
| C14 | 0.0479 (12) | 0.0486 (12) | 0.0478 (12) | −0.0126 (10) | 0.0073 (9) | −0.0018 (10) |
| C15 | 0.0477 (11) | 0.0466 (12) | 0.0355 (10) | −0.0050 (9) | 0.0109 (9) | −0.0023 (9) |
| C16 | 0.0747 (16) | 0.0534 (14) | 0.0492 (13) | −0.0026 (12) | −0.0083 (12) | −0.0121 (11) |
| C17 | 0.113 (3) | 0.083 (2) | 0.0662 (18) | −0.0077 (19) | −0.0367 (17) | 0.0015 (16) |
| C18 | 0.0393 (10) | 0.0357 (10) | 0.0337 (9) | 0.0006 (8) | −0.0007 (8) | 0.0013 (8) |
| C19 | 0.0603 (14) | 0.0431 (12) | 0.0457 (12) | 0.0093 (10) | 0.0038 (10) | −0.0019 (10) |
| C20 | 0.0767 (17) | 0.0389 (12) | 0.0701 (17) | 0.0139 (12) | −0.0044 (13) | −0.0034 (12) |
| C21 | 0.0850 (19) | 0.0389 (13) | 0.0733 (18) | 0.0027 (12) | −0.0085 (14) | 0.0181 (12) |
| C22 | 0.0799 (17) | 0.0586 (15) | 0.0495 (14) | −0.0013 (13) | 0.0021 (12) | 0.0194 (12) |
| C23 | 0.0583 (13) | 0.0450 (12) | 0.0377 (11) | 0.0030 (10) | 0.0047 (9) | 0.0056 (9) |
| C24 | 0.0414 (10) | 0.0391 (10) | 0.0307 (9) | −0.0038 (8) | 0.0025 (8) | 0.0006 (8) |
| C25 | 0.0476 (12) | 0.0532 (13) | 0.0431 (11) | 0.0037 (10) | 0.0000 (9) | −0.0028 (10) |
| C26 | 0.0478 (13) | 0.0774 (18) | 0.0484 (13) | 0.0038 (12) | −0.0092 (10) | −0.0020 (12) |
| C27 | 0.0625 (15) | 0.0707 (17) | 0.0408 (12) | −0.0156 (13) | −0.0060 (11) | −0.0064 (12) |
| C28 | 0.0792 (17) | 0.0480 (13) | 0.0403 (12) | −0.0074 (12) | 0.0030 (11) | −0.0107 (10) |
| C29 | 0.0577 (13) | 0.0409 (11) | 0.0391 (11) | 0.0029 (10) | −0.0009 (9) | −0.0013 (9) |
Geometric parameters (Å, º)
| Cl1—C8 | 1.738 (2) | C14—H14 | 0.9300 |
| O1—C5 | 1.349 (2) | C15—H15 | 0.9300 |
| O1—H1 | 1.00 (3) | C16—C17 | 1.509 (4) |
| N1—C1 | 1.327 (2) | C16—H16A | 0.9700 |
| N1—C3 | 1.372 (2) | C16—H16B | 0.9700 |
| N2—C1 | 1.372 (2) | C17—H17A | 0.9600 |
| N2—C2 | 1.397 (2) | C17—H17B | 0.9600 |
| N2—C10 | 1.441 (2) | C17—H17C | 0.9600 |
| C1—C4 | 1.468 (3) | C18—C23 | 1.390 (3) |
| C2—C3 | 1.375 (3) | C18—C19 | 1.390 (3) |
| C2—C18 | 1.474 (3) | C19—C20 | 1.382 (3) |
| C3—C24 | 1.477 (3) | C19—H19 | 0.9300 |
| C4—C9 | 1.403 (3) | C20—C21 | 1.371 (4) |
| C4—C5 | 1.417 (3) | C20—H20 | 0.9300 |
| C5—C6 | 1.391 (3) | C21—C22 | 1.373 (4) |
| C6—C7 | 1.373 (3) | C21—H21 | 0.9300 |
| C6—H6 | 0.9300 | C22—C23 | 1.381 (3) |
| C7—C8 | 1.383 (3) | C22—H22 | 0.9300 |
| C7—H7 | 0.9300 | C23—H23 | 0.9300 |
| C8—C9 | 1.379 (3) | C24—C25 | 1.383 (3) |
| C9—H9 | 0.9300 | C24—C29 | 1.394 (3) |
| C10—C15 | 1.376 (3) | C25—C26 | 1.383 (3) |
| C10—C11 | 1.379 (3) | C25—H25 | 0.9300 |
| C11—C12 | 1.380 (3) | C26—C27 | 1.377 (4) |
| C11—H11 | 0.9300 | C26—H26 | 0.9300 |
| C12—C13 | 1.384 (3) | C27—C28 | 1.372 (4) |
| C12—H12 | 0.9300 | C27—H27 | 0.9300 |
| C13—C14 | 1.386 (3) | C28—C29 | 1.381 (3) |
| C13—C16 | 1.507 (3) | C28—H28 | 0.9300 |
| C14—C15 | 1.386 (3) | C29—H29 | 0.9300 |
| C5—O1—H1 | 109.5 | C14—C15—H15 | 120.5 |
| C1—N1—C3 | 107.38 (16) | C13—C16—C17 | 112.5 (2) |
| C1—N2—C2 | 107.68 (15) | C13—C16—H16A | 109.1 |
| C1—N2—C10 | 125.28 (15) | C17—C16—H16A | 109.1 |
| C2—N2—C10 | 127.04 (16) | C13—C16—H16B | 109.1 |
| N1—C1—N2 | 109.82 (16) | C17—C16—H16B | 109.1 |
| N1—C1—C4 | 121.54 (17) | H16A—C16—H16B | 107.8 |
| N2—C1—C4 | 128.64 (17) | C16—C17—H17A | 109.5 |
| C3—C2—N2 | 105.22 (16) | C16—C17—H17B | 109.5 |
| C3—C2—C18 | 131.25 (17) | H17A—C17—H17B | 109.5 |
| N2—C2—C18 | 123.34 (17) | C16—C17—H17C | 109.5 |
| N1—C3—C2 | 109.88 (16) | H17A—C17—H17C | 109.5 |
| N1—C3—C24 | 118.18 (17) | H17B—C17—H17C | 109.5 |
| C2—C3—C24 | 131.90 (18) | C23—C18—C19 | 118.45 (19) |
| C9—C4—C5 | 117.94 (17) | C23—C18—C2 | 118.78 (18) |
| C9—C4—C1 | 124.14 (17) | C19—C18—C2 | 122.77 (18) |
| C5—C4—C1 | 117.91 (17) | C20—C19—C18 | 120.6 (2) |
| O1—C5—C6 | 117.42 (18) | C20—C19—H19 | 119.7 |
| O1—C5—C4 | 122.68 (17) | C18—C19—H19 | 119.7 |
| C6—C5—C4 | 119.88 (19) | C21—C20—C19 | 120.2 (2) |
| C7—C6—C5 | 121.2 (2) | C21—C20—H20 | 119.9 |
| C7—C6—H6 | 119.4 | C19—C20—H20 | 119.9 |
| C5—C6—H6 | 119.4 | C20—C21—C22 | 119.8 (2) |
| C6—C7—C8 | 119.18 (18) | C20—C21—H21 | 120.1 |
| C6—C7—H7 | 120.4 | C22—C21—H21 | 120.1 |
| C8—C7—H7 | 120.4 | C21—C22—C23 | 120.5 (2) |
| C9—C8—C7 | 121.29 (19) | C21—C22—H22 | 119.8 |
| C9—C8—Cl1 | 118.69 (17) | C23—C22—H22 | 119.8 |
| C7—C8—Cl1 | 120.00 (15) | C22—C23—C18 | 120.4 (2) |
| C8—C9—C4 | 120.42 (19) | C22—C23—H23 | 119.8 |
| C8—C9—H9 | 119.8 | C18—C23—H23 | 119.8 |
| C4—C9—H9 | 119.8 | C25—C24—C29 | 118.41 (19) |
| C15—C10—C11 | 120.72 (18) | C25—C24—C3 | 122.32 (19) |
| C15—C10—N2 | 119.52 (17) | C29—C24—C3 | 119.12 (18) |
| C11—C10—N2 | 119.63 (17) | C26—C25—C24 | 120.3 (2) |
| C10—C11—C12 | 119.32 (19) | C26—C25—H25 | 119.8 |
| C10—C11—H11 | 120.3 | C24—C25—H25 | 119.8 |
| C12—C11—H11 | 120.3 | C27—C26—C25 | 120.8 (2) |
| C11—C12—C13 | 121.36 (19) | C27—C26—H26 | 119.6 |
| C11—C12—H12 | 119.3 | C25—C26—H26 | 119.6 |
| C13—C12—H12 | 119.3 | C28—C27—C26 | 119.4 (2) |
| C12—C13—C14 | 118.03 (19) | C28—C27—H27 | 120.3 |
| C12—C13—C16 | 120.6 (2) | C26—C27—H27 | 120.3 |
| C14—C13—C16 | 121.3 (2) | C27—C28—C29 | 120.4 (2) |
| C13—C14—C15 | 121.4 (2) | C27—C28—H28 | 119.8 |
| C13—C14—H14 | 119.3 | C29—C28—H28 | 119.8 |
| C15—C14—H14 | 119.3 | C28—C29—C24 | 120.7 (2) |
| C10—C15—C14 | 119.03 (19) | C28—C29—H29 | 119.6 |
| C10—C15—H15 | 120.5 | C24—C29—H29 | 119.6 |
| C3—N1—C1—N2 | 0.4 (2) | C15—C10—C11—C12 | −3.6 (3) |
| C3—N1—C1—C4 | 179.45 (17) | N2—C10—C11—C12 | 172.22 (18) |
| C2—N2—C1—N1 | −1.1 (2) | C10—C11—C12—C13 | 0.8 (3) |
| C10—N2—C1—N1 | 178.44 (17) | C11—C12—C13—C14 | 2.3 (3) |
| C2—N2—C1—C4 | 179.94 (19) | C11—C12—C13—C16 | −175.5 (2) |
| C10—N2—C1—C4 | −0.5 (3) | C12—C13—C14—C15 | −2.6 (3) |
| C1—N2—C2—C3 | 1.3 (2) | C16—C13—C14—C15 | 175.1 (2) |
| C10—N2—C2—C3 | −178.22 (17) | C11—C10—C15—C14 | 3.3 (3) |
| C1—N2—C2—C18 | −174.15 (18) | N2—C10—C15—C14 | −172.56 (18) |
| C10—N2—C2—C18 | 6.3 (3) | C13—C14—C15—C10 | −0.1 (3) |
| C1—N1—C3—C2 | 0.4 (2) | C12—C13—C16—C17 | 71.3 (3) |
| C1—N1—C3—C24 | −177.67 (17) | C14—C13—C16—C17 | −106.4 (3) |
| N2—C2—C3—N1 | −1.1 (2) | C3—C2—C18—C23 | −40.4 (3) |
| C18—C2—C3—N1 | 173.9 (2) | N2—C2—C18—C23 | 133.8 (2) |
| N2—C2—C3—C24 | 176.7 (2) | C3—C2—C18—C19 | 139.0 (2) |
| C18—C2—C3—C24 | −8.4 (4) | N2—C2—C18—C19 | −46.8 (3) |
| N1—C1—C4—C9 | 164.28 (19) | C23—C18—C19—C20 | −0.2 (3) |
| N2—C1—C4—C9 | −16.9 (3) | C2—C18—C19—C20 | −179.6 (2) |
| N1—C1—C4—C5 | −14.4 (3) | C18—C19—C20—C21 | 1.0 (4) |
| N2—C1—C4—C5 | 164.4 (2) | C19—C20—C21—C22 | −0.7 (4) |
| C9—C4—C5—O1 | −174.41 (19) | C20—C21—C22—C23 | −0.4 (4) |
| C1—C4—C5—O1 | 4.4 (3) | C21—C22—C23—C18 | 1.1 (4) |
| C9—C4—C5—C6 | 4.1 (3) | C19—C18—C23—C22 | −0.8 (3) |
| C1—C4—C5—C6 | −177.12 (19) | C2—C18—C23—C22 | 178.6 (2) |
| O1—C5—C6—C7 | 176.40 (19) | N1—C3—C24—C25 | 140.6 (2) |
| C4—C5—C6—C7 | −2.2 (3) | C2—C3—C24—C25 | −37.0 (3) |
| C5—C6—C7—C8 | −0.3 (3) | N1—C3—C24—C29 | −34.8 (3) |
| C6—C7—C8—C9 | 0.9 (3) | C2—C3—C24—C29 | 147.5 (2) |
| C6—C7—C8—Cl1 | 179.40 (16) | C29—C24—C25—C26 | 0.1 (3) |
| C7—C8—C9—C4 | 1.1 (3) | C3—C24—C25—C26 | −175.4 (2) |
| Cl1—C8—C9—C4 | −177.38 (15) | C24—C25—C26—C27 | 0.1 (4) |
| C5—C4—C9—C8 | −3.6 (3) | C25—C26—C27—C28 | −0.6 (4) |
| C1—C4—C9—C8 | 177.71 (19) | C26—C27—C28—C29 | 0.8 (4) |
| C1—N2—C10—C15 | 107.0 (2) | C27—C28—C29—C24 | −0.6 (4) |
| C2—N2—C10—C15 | −73.5 (3) | C25—C24—C29—C28 | 0.1 (3) |
| C1—N2—C10—C11 | −68.8 (3) | C3—C24—C29—C28 | 175.8 (2) |
| C2—N2—C10—C11 | 110.7 (2) |
Hydrogen-bond geometry (Å, º)
Cg is the centroid of the C4–C9 aryl ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···N1 | 1.00 | 1.66 | 2.549 (1) | 145 |
| C19—H19···O1i | 0.93 | 2.57 | 3.242 (3) | 129 |
| C15—H15···Cgii | 0.93 | 2.72 | 3.527 (2) | 146 |
Symmetry codes: (i) −x+1, −y+1, −z+2; (ii) −x+2, −y+1, −z+2.
References
- Bhandari, K., Srinivas, N., Marrapu, V. K., Verma, A., Srivastava, S. & Gupta, S. (2010). Bioorg. Med. Chem. Lett. 20, 291–293. [DOI] [PubMed]
- Bruker. (1998). SMART, SAINT-Plus and SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Gaonkar, S. L., Rai, K. M. L. & Shetty, N. S. (2009). Med. Chem. Res. 18, 221–230.
- Özkay, Y., Işıkdağ, , İncesu, Z. & Akalın, G. (2010). Eur. J. Med. Chem. 45, 3320–3328. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Sigel, H., Saha, A., Saha, N., Carloni, P., Kapinos, L. E. & Griesser, R. (2000). J. Inorg. Biochem. 78, 129–137. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2414314619016900/zl4037sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314619016900/zl4037Isup2.hkl
Supporting information file. DOI: 10.1107/S2414314619016900/zl4037sup3.tif
Supporting information file. DOI: 10.1107/S2414314619016900/zl4037sup4.pdf
Proton-NMR. DOI: 10.1107/S2414314619016900/zl4037sup5.pdf
Supporting information file. DOI: 10.1107/S2414314619016900/zl4037Isup6.cml
CCDC reference: 1950453
Additional supporting information: crystallographic information; 3D view; checkCIF report




