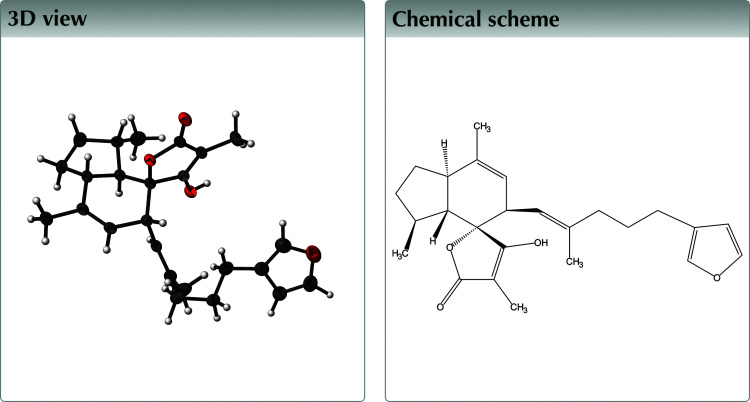
The title compound ircinianin belongs to the sesterterpene tetronic acid compound family and was isolated from the marine sponge Ircinia wistarii. These chemical scaffolds are pharmacologically relevant, since they represent a new class of glycine receptor modulators.
Keywords: crystal structure, natural product, ircinianin
Abstract
The title compound, ircinianin, C25H32O4, belongs to the sesterterpene tetronic acid compound family and was isolated from the marine sponge Ircinia wistarii. These chemical scaffolds are pharmacologically relevant, since they represent a new class of glycine receptor modulators. The furan ring makes a dihedral angle of 35.14 (12)° to the 4-hydroxy-3-methylfuran-2(5H)-one ring. The crystal packing is characterized by intermolecular O—H⋯O hydrogen bonds, which generate [010] chains.
Structure description
The genus Ircinia of the sea sponge family Irciniidae is a prolific source of natural products with a huge variety of different natural product classes like macrolides, alkaloids, steroids, peptides and terpenes (Coll et al., 1997 ▸; Kondo et al., 1992 ▸; Kobayashi et al., 1995 ▸; Mau et al., 1996 ▸; Chevallier et al., 2006 ▸). Particularly, regarding the latter compound class, Ircinia spp. are known to produce unusual and rare terpenoids, especially sesterterpene tetronic acids in a linear and cyclic form, like ircinianin and its structural congeners (Hofheinz & Schönholzer, 1977 ▸; Barrow et al., 1988 ▸; Coll et al., 1997 ▸; Höller et al., 1997 ▸; Balansa et al., 2013 ▸; Balansa et al., 2010 ▸).
Balansa et al. (2013 ▸) showed that these analogues exhibit a significant isoform-selective potentiation of glycine-gated chloride channel receptors (GlyRs). The compounds have therefore the potential to be developed either as molecular tools to probe GlyR function or can serve as lead structures to treat GlyR-mediated neural disorders.
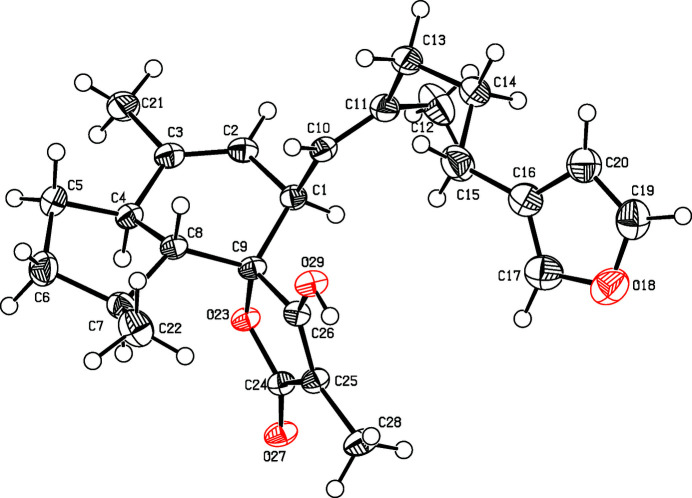
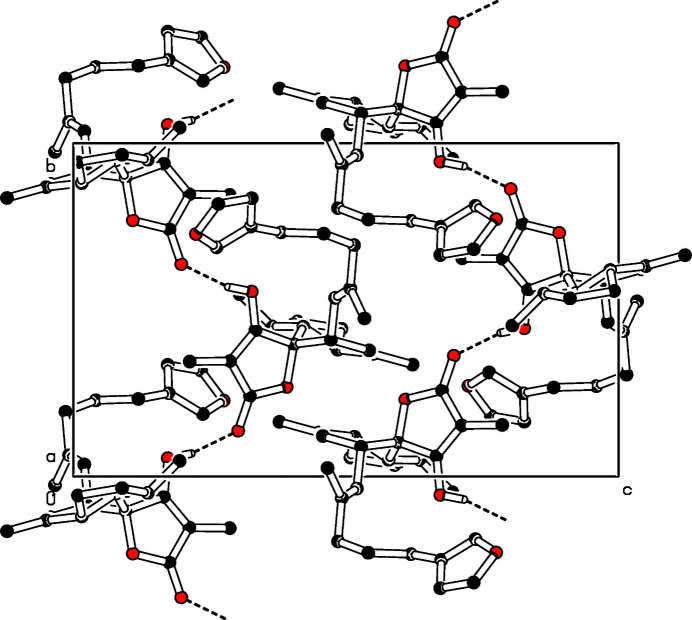
The title compound (Fig. 1 ▸) is a polycyclic sesterterpene tetronic acid with a furan moiety. The furan ring makes a dihedral angle of 35.14 (12)° to the 4-hydroxy-3-methylfuran-2(5H)-one ring. In the crystal, the molecules are linked by O—H⋯O hydrogen bonds (Table 1 ▸, Fig. 2 ▸), forming chains parallel to the b axis. The crystal structure of the title compound has already been reported in 1977 by researchers from the pharmaceutical company Hoffmann La Roche (Hofheinz & Schönholzer, 1977 ▸; CCDC reference: 1180878). However, in this study the hydrogen atoms were not refined, and only the relative stereochemistry could be deduced. The absolute structure was so far solely determined by asymmetric total synthesis in 1997 (Uenishi et al., 1997 ▸).
Figure 1.
Perspective view of the title compound. Displacement ellipsoids are drawn at the 50% probability level.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O29—H29⋯O27i | 0.84 (3) | 1.80 (3) | 2.6207 (18) | 166 (3) |
Symmetry code: (i)
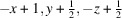 .
.
Figure 2.
Partial packing diagram of the title compound. View along the a-axis.
Synthesis and crystallization
The title compound C25H32O4 was isolated from the marine sponge Ircinia wistarii. The sample (voucher number HER6) was collected from Wistarii Reef, Heron Island, Great Barrier Reef, Australia in July 1998 from a depth of 20 m. After collection, the material was stored in EtOH and kept frozen at 253 K until use.
The sponge material (800 g, wet weight) was cut into smaller pieces (2 × 2 cm) and was extracted with a solvent mixture of CHCl3/MeOH (1:1, v/v; 2 l of volume per extraction step) for three times (after 4, 8 and 20 h). The extraction solvent of each step was collected and combined. After filtration and evaporation to dryness, 25.46 g crude extract was obtained. The crude extract was redissolved in MeOH and fractioned by preparative reversed phase open column chromatography [Polygoprep 60–50 C18 (Macherey-Nagel) as stationary phase] using gravity and stepwise MeOH/H2O gradients with increasing lipophilicity and DCM. In total, eleven fractions were gained, and the ircinianin-enriched fraction (MeOH/H2O – 90:10) was identified by LC–MS. This fraction was then purified by reversed phase HPLC [Luna Omega 5 µm Polar C18 100 Å column, 250 × 4.6 mm, at 1.2 ml min−1 and UV detection at 215 nm with a 3 min gradient elution, from 20:80 to 55:45 ACN/H2O + 0.1% TFA, followed by ramping over 27 min to 90:10], yielding 140 mg of ircinianin, judged as pure based on total ion current profiles, ESI–MS and NMR spectrometry. Suitable crystals were prepared by slow evaporation at room temperature from a ACN/H2O (65:35) solution under atmospheric pressure.
Spectroscopic data of the title compound were in accordance with literature data (Balansa et al., 2013 ▸). For ease of comparison with related compounds, the title compound was given in the NMR section the same numbering scheme as previously used in the literature (Balansa et al., 2013 ▸):
1H NMR (400 MHz, MeOH-d4 ): δ 7.38 (H-1, t, 1.6), 7.26 (H-4, m), 6.30 (H-2, m), 5.11 (H-10, dd, 10.3, 1.1), 5.03 (H-12, m), 3.08 (H-11, dm, 10.3), 2.42 (H-15, m)A, 2.41 (H-5 br t, 7.5)A, 2.04 (H-7, m), 2.00 (H-17a, m), 1.89 (H-16a, m), 1.71 (H-14, m), 1.68 (H-6, m), 1.65 (H-18, m), 1.64 (H-25, s), 1.60 (H-20, m), 1.57 (H-9, d, 1.3), 1.33 (H-16b, m), 1.31 (H-17b, m), 0.92 (H-19, d, 6.3).
13C NMR (100 MHz, MeOH-d4 ): δ 179.2 (C-22, s C)B, 177.7 (C-24, s)B, 144.0 (C-1, d), 140.3 (C-4, d), 137.1 (C-13, s), 136.6 (C-8, s), 126.5 (C-3, s), 125.0 (C-10, d), 123.6 (C-12, d), 112.1 (C-2, d), 97.5 (C-23, s), 86.9 (C-21, s), 52.0 (C-20, d), 48.7 (C-11, d), 46.2 (C-15, d), 40.5 (C-7, t), 33.6 (C-17, t), 33.2 (C-18, d), 29.5 (C-6, t), 27.3 (C-16, t), 25.3 (C-5, t), 20.8 (C-14, q), 20.7 (C-19, q), 16.3 (C-9, q), 6.1 (C-25, q). [A Overlapping signals; B assignments interchangeable; C implied multiplicities determined by DEPT (qC = s; CH = d; CH2 = t; CH3 = q).]
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. All hydrogen atoms were located in difference Fourier maps and were refined with isotropic displacement parameters.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C25H32O4 |
| M r | 396.50 |
| Crystal system, space group | Orthorhombic, P212121 |
| Temperature (K) | 120 |
| a, b, c (Å) | 10.8217 (2), 11.1644 (2), 18.2804 (5) |
| V (Å3) | 2208.60 (8) |
| Z | 4 |
| Radiation type | Cu Kα |
| μ (mm−1) | 0.63 |
| Crystal size (mm) | 0.91 × 0.08 × 0.08 |
| Data collection | |
| Diffractometer | Stoe IPDS 2T |
| Absorption correction | Integration |
| T min, T max | 0.914, 0.990 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 18913, 3945, 3849 |
| R int | 0.018 |
| (sin θ/λ)max (Å−1) | 0.600 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.032, 0.086, 1.08 |
| No. of reflections | 3945 |
| No. of parameters | 377 |
| H-atom treatment | All H-atom parameters refined |
| Δρmax, Δρmin (e Å−3) | 0.19, −0.20 |
| Absolute structure | Flack x determined using 1639 quotients [(I +)−(I −)]/[(I +)+(I −)] (Parsons et al., 2013 ▸) |
| Absolute structure parameter | 0.03 (9) |
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2414314620015783/bt4103sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314620015783/bt4103Isup2.hkl
CCDC reference: 2047802
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
We gratefully thank Anthony D. Wright for collecting the animal material by SCUBA diving and for providing the sponge sample to the Department of Pharmaceutical Biology, Eberhard Karls University, Tübingen.
full crystallographic data
Crystal data
| C25H32O4 | Dx = 1.192 Mg m−3 |
| Mr = 396.50 | Cu Kα radiation, λ = 1.54186 Å |
| Orthorhombic, P212121 | Cell parameters from 58680 reflections |
| a = 10.8217 (2) Å | θ = 2.4–68.0° |
| b = 11.1644 (2) Å | µ = 0.63 mm−1 |
| c = 18.2804 (5) Å | T = 120 K |
| V = 2208.60 (8) Å3 | Coloumn, colourless |
| Z = 4 | 0.91 × 0.08 × 0.08 mm |
| F(000) = 856 |
Data collection
| Stoe IPDS 2T diffractometer | 3945 independent reflections |
| Radiation source: Incoatec microSource Cu | 3849 reflections with I > 2σ(I) |
| Detector resolution: 6.67 pixels mm-1 | Rint = 0.018 |
| rotation method, ω scans | θmax = 67.8°, θmin = 4.6° |
| Absorption correction: integration | h = −12→12 |
| Tmin = 0.914, Tmax = 0.990 | k = −13→13 |
| 18913 measured reflections | l = −19→21 |
Refinement
| Refinement on F2 | Hydrogen site location: difference Fourier map |
| Least-squares matrix: full | All H-atom parameters refined |
| R[F2 > 2σ(F2)] = 0.032 | w = 1/[σ2(Fo2) + (0.0583P)2 + 0.3312P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.086 | (Δ/σ)max < 0.001 |
| S = 1.08 | Δρmax = 0.19 e Å−3 |
| 3945 reflections | Δρmin = −0.20 e Å−3 |
| 377 parameters | Absolute structure: Flack x determined using 1639 quotients [(I+)-(I-)]/[(I+)+(I-)] (Parsons et al., 2013) |
| 0 restraints | Absolute structure parameter: 0.03 (9) |
| Primary atom site location: dual |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.49694 (18) | 0.41060 (15) | 0.47235 (10) | 0.0219 (4) | |
| H1 | 0.428 (2) | 0.357 (2) | 0.4651 (13) | 0.030 (6)* | |
| C2 | 0.56472 (18) | 0.37841 (17) | 0.54252 (10) | 0.0253 (4) | |
| H2 | 0.513 (2) | 0.368 (2) | 0.5861 (13) | 0.025 (5)* | |
| C3 | 0.68627 (19) | 0.36830 (17) | 0.55010 (10) | 0.0259 (4) | |
| C4 | 0.76937 (17) | 0.38055 (17) | 0.48472 (10) | 0.0236 (4) | |
| H4 | 0.786 (2) | 0.301 (2) | 0.4652 (13) | 0.030 (6)* | |
| C5 | 0.89455 (18) | 0.4432 (2) | 0.49168 (11) | 0.0301 (4) | |
| H5A | 0.954 (2) | 0.393 (2) | 0.5169 (13) | 0.029 (4)* | |
| H5AB | 0.883 (2) | 0.521 (2) | 0.5200 (13) | 0.029 (4)* | |
| C6 | 0.9310 (2) | 0.4654 (2) | 0.41093 (12) | 0.0380 (5) | |
| H6A | 0.993 (3) | 0.406 (3) | 0.3941 (16) | 0.052 (5)* | |
| H6AB | 0.967 (3) | 0.552 (3) | 0.4041 (16) | 0.052 (5)* | |
| C7 | 0.81215 (17) | 0.45092 (17) | 0.36372 (10) | 0.0259 (4) | |
| H7 | 0.815 (3) | 0.373 (2) | 0.3362 (15) | 0.042 (7)* | |
| C8 | 0.70955 (17) | 0.44944 (16) | 0.42160 (10) | 0.0214 (4) | |
| H8 | 0.694 (2) | 0.535 (2) | 0.4385 (12) | 0.024 (5)* | |
| C9 | 0.58241 (17) | 0.39703 (15) | 0.40444 (10) | 0.0205 (4) | |
| C10 | 0.44429 (17) | 0.53607 (16) | 0.48011 (10) | 0.0235 (4) | |
| H10 | 0.500 (2) | 0.602 (2) | 0.4651 (13) | 0.030 (6)* | |
| C11 | 0.33472 (19) | 0.56473 (18) | 0.50782 (10) | 0.0284 (4) | |
| C12 | 0.2416 (2) | 0.4744 (2) | 0.53387 (18) | 0.0485 (6) | |
| H12A | 0.181 (4) | 0.508 (5) | 0.553 (3) | 0.111 (9)* | |
| H12B | 0.277 (5) | 0.402 (4) | 0.551 (3) | 0.111 (9)* | |
| H12C | 0.208 (5) | 0.437 (4) | 0.482 (3) | 0.111 (9)* | |
| C13 | 0.2955 (2) | 0.69443 (19) | 0.51459 (12) | 0.0333 (5) | |
| H13A | 0.368 (3) | 0.746 (3) | 0.5061 (17) | 0.053 (6)* | |
| H13B | 0.260 (3) | 0.702 (3) | 0.5669 (18) | 0.053 (6)* | |
| C14 | 0.1950 (2) | 0.7288 (2) | 0.45910 (12) | 0.0347 (5) | |
| H14A | 0.122 (3) | 0.665 (3) | 0.4642 (16) | 0.054 (6)* | |
| H14B | 0.158 (3) | 0.819 (3) | 0.4699 (17) | 0.054 (6)* | |
| C15 | 0.2427 (2) | 0.7328 (2) | 0.38087 (13) | 0.0426 (5) | |
| H15A | 0.304 (3) | 0.814 (3) | 0.3804 (18) | 0.066 (6)* | |
| H15B | 0.280 (3) | 0.652 (3) | 0.3716 (18) | 0.066 (6)* | |
| C16 | 0.1423 (2) | 0.7499 (2) | 0.32518 (12) | 0.0363 (5) | |
| C17 | 0.1271 (2) | 0.6877 (2) | 0.26257 (13) | 0.0422 (5) | |
| H17 | 0.169 (2) | 0.617 (2) | 0.2377 (13) | 0.033 (6)* | |
| O18 | 0.02589 (17) | 0.72790 (16) | 0.22460 (9) | 0.0467 (4) | |
| C19 | −0.0236 (2) | 0.8178 (2) | 0.26580 (14) | 0.0444 (6) | |
| H19 | −0.103 (3) | 0.863 (3) | 0.2478 (16) | 0.049 (8)* | |
| C20 | 0.0435 (2) | 0.8351 (2) | 0.32708 (13) | 0.0404 (5) | |
| H20 | 0.029 (3) | 0.894 (3) | 0.3654 (16) | 0.044 (7)* | |
| C21 | 0.7463 (2) | 0.3359 (2) | 0.62195 (12) | 0.0371 (5) | |
| H21A | 0.810 (3) | 0.393 (3) | 0.6351 (16) | 0.052 (5)* | |
| H21B | 0.791 (3) | 0.266 (3) | 0.6173 (16) | 0.052 (5)* | |
| H21C | 0.686 (3) | 0.331 (3) | 0.6612 (17) | 0.052 (5)* | |
| C22 | 0.8012 (2) | 0.5468 (2) | 0.30527 (13) | 0.0395 (5) | |
| H22A | 0.727 (3) | 0.533 (3) | 0.2718 (17) | 0.053 (4)* | |
| H22B | 0.881 (3) | 0.544 (3) | 0.2759 (17) | 0.053 (4)* | |
| H22C | 0.792 (3) | 0.626 (3) | 0.3312 (17) | 0.053 (4)* | |
| O23 | 0.59733 (12) | 0.26878 (10) | 0.39111 (7) | 0.0211 (3) | |
| C24 | 0.55392 (17) | 0.24167 (15) | 0.32376 (9) | 0.0217 (4) | |
| C25 | 0.50545 (18) | 0.34673 (15) | 0.28756 (9) | 0.0228 (4) | |
| C26 | 0.52443 (16) | 0.43886 (16) | 0.33396 (9) | 0.0209 (4) | |
| O27 | 0.56032 (13) | 0.13752 (11) | 0.30220 (7) | 0.0271 (3) | |
| C28 | 0.4465 (2) | 0.34514 (19) | 0.21335 (11) | 0.0321 (5) | |
| H28A | 0.434 (4) | 0.417 (4) | 0.195 (2) | 0.077 (6)* | |
| H28B | 0.375 (4) | 0.296 (3) | 0.215 (2) | 0.077 (6)* | |
| H28C | 0.499 (4) | 0.307 (3) | 0.178 (2) | 0.077 (6)* | |
| O29 | 0.50180 (13) | 0.55511 (11) | 0.32690 (7) | 0.0258 (3) | |
| H29 | 0.481 (3) | 0.569 (3) | 0.2837 (18) | 0.048 (8)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0240 (8) | 0.0201 (8) | 0.0215 (9) | −0.0012 (7) | 0.0001 (7) | −0.0006 (6) |
| C2 | 0.0310 (10) | 0.0253 (9) | 0.0196 (8) | 0.0011 (8) | 0.0014 (8) | −0.0005 (7) |
| C3 | 0.0316 (10) | 0.0257 (9) | 0.0205 (9) | 0.0010 (8) | −0.0017 (8) | −0.0013 (7) |
| C4 | 0.0266 (9) | 0.0231 (9) | 0.0212 (9) | 0.0005 (7) | −0.0032 (7) | −0.0007 (7) |
| C5 | 0.0255 (10) | 0.0339 (10) | 0.0310 (10) | −0.0016 (8) | −0.0049 (8) | −0.0019 (9) |
| C6 | 0.0274 (11) | 0.0511 (14) | 0.0356 (11) | −0.0073 (10) | 0.0008 (9) | 0.0015 (10) |
| C7 | 0.0269 (9) | 0.0252 (9) | 0.0255 (9) | −0.0016 (7) | 0.0036 (7) | −0.0008 (7) |
| C8 | 0.0256 (9) | 0.0180 (8) | 0.0206 (8) | −0.0008 (7) | −0.0006 (7) | −0.0005 (7) |
| C9 | 0.0273 (9) | 0.0147 (8) | 0.0195 (8) | 0.0010 (6) | −0.0015 (7) | −0.0012 (6) |
| C10 | 0.0263 (9) | 0.0218 (8) | 0.0224 (8) | −0.0006 (7) | −0.0033 (7) | −0.0026 (7) |
| C11 | 0.0295 (10) | 0.0281 (9) | 0.0276 (9) | 0.0021 (8) | −0.0004 (7) | −0.0030 (8) |
| C12 | 0.0376 (12) | 0.0383 (13) | 0.0695 (18) | 0.0016 (10) | 0.0184 (12) | 0.0015 (12) |
| C13 | 0.0331 (10) | 0.0309 (11) | 0.0358 (11) | 0.0066 (9) | −0.0017 (9) | −0.0085 (8) |
| C14 | 0.0321 (10) | 0.0346 (11) | 0.0374 (11) | 0.0067 (9) | −0.0001 (9) | −0.0071 (9) |
| C15 | 0.0372 (12) | 0.0515 (14) | 0.0393 (12) | 0.0121 (11) | 0.0019 (10) | 0.0029 (11) |
| C16 | 0.0388 (12) | 0.0351 (10) | 0.0351 (11) | −0.0006 (9) | 0.0030 (9) | 0.0030 (9) |
| C17 | 0.0490 (13) | 0.0400 (12) | 0.0377 (12) | −0.0029 (11) | 0.0054 (10) | 0.0032 (10) |
| O18 | 0.0504 (10) | 0.0526 (10) | 0.0371 (8) | −0.0140 (8) | −0.0016 (7) | 0.0023 (7) |
| C19 | 0.0394 (13) | 0.0498 (14) | 0.0439 (13) | −0.0018 (10) | −0.0021 (10) | 0.0103 (11) |
| C20 | 0.0414 (12) | 0.0399 (12) | 0.0398 (12) | 0.0038 (10) | −0.0014 (10) | 0.0006 (10) |
| C21 | 0.0370 (12) | 0.0530 (14) | 0.0214 (10) | 0.0089 (11) | −0.0025 (9) | 0.0007 (9) |
| C22 | 0.0391 (12) | 0.0416 (12) | 0.0378 (12) | −0.0005 (10) | 0.0090 (9) | 0.0124 (10) |
| O23 | 0.0291 (7) | 0.0148 (6) | 0.0192 (6) | 0.0009 (5) | −0.0029 (5) | −0.0005 (4) |
| C24 | 0.0255 (9) | 0.0200 (8) | 0.0196 (8) | −0.0018 (7) | 0.0004 (7) | −0.0013 (6) |
| C25 | 0.0275 (9) | 0.0202 (9) | 0.0207 (8) | 0.0004 (7) | −0.0027 (7) | 0.0003 (7) |
| C26 | 0.0240 (8) | 0.0180 (8) | 0.0208 (8) | 0.0007 (7) | 0.0008 (7) | 0.0011 (7) |
| O27 | 0.0388 (7) | 0.0178 (6) | 0.0247 (6) | 0.0005 (5) | −0.0051 (6) | −0.0038 (5) |
| C28 | 0.0453 (12) | 0.0258 (10) | 0.0251 (9) | 0.0035 (9) | −0.0114 (9) | −0.0027 (8) |
| O29 | 0.0379 (7) | 0.0177 (6) | 0.0217 (6) | 0.0035 (5) | −0.0040 (6) | 0.0013 (5) |
Geometric parameters (Å, º)
| C1—C10 | 1.519 (2) | C13—H13A | 0.98 (3) |
| C1—C2 | 1.521 (2) | C13—H13B | 1.03 (3) |
| C1—C9 | 1.555 (2) | C14—C15 | 1.521 (3) |
| C1—H1 | 0.96 (2) | C14—H14A | 1.07 (3) |
| C2—C3 | 1.327 (3) | C14—H14B | 1.10 (3) |
| C2—H2 | 0.98 (2) | C15—C16 | 1.502 (3) |
| C3—C4 | 1.502 (3) | C15—H15A | 1.12 (3) |
| C3—C21 | 1.509 (3) | C15—H15B | 1.01 (3) |
| C4—C5 | 1.530 (3) | C16—C17 | 1.349 (3) |
| C4—C8 | 1.530 (2) | C16—C20 | 1.431 (3) |
| C4—H4 | 0.97 (2) | C17—O18 | 1.372 (3) |
| C5—C6 | 1.548 (3) | C17—H17 | 1.01 (3) |
| C5—H5A | 0.97 (2) | O18—C19 | 1.365 (3) |
| C5—H5AB | 1.02 (3) | C19—C20 | 1.349 (3) |
| C6—C7 | 1.557 (3) | C19—H19 | 1.05 (3) |
| C6—H6A | 0.99 (3) | C20—H20 | 0.98 (3) |
| C6—H6AB | 1.05 (3) | C21—H21A | 0.97 (3) |
| C7—C22 | 1.517 (3) | C21—H21B | 0.92 (3) |
| C7—C8 | 1.534 (2) | C21—H21C | 0.97 (3) |
| C7—H7 | 1.00 (3) | C22—H22A | 1.02 (3) |
| C8—C9 | 1.528 (3) | C22—H22B | 1.02 (3) |
| C8—H8 | 1.02 (2) | C22—H22C | 1.01 (3) |
| C9—O23 | 1.4613 (19) | O23—C24 | 1.352 (2) |
| C9—C26 | 1.507 (2) | C24—O27 | 1.230 (2) |
| C10—C11 | 1.329 (3) | C24—C25 | 1.445 (2) |
| C10—H10 | 0.99 (3) | C25—C26 | 1.349 (3) |
| C11—C12 | 1.503 (3) | C25—C28 | 1.499 (2) |
| C11—C13 | 1.514 (3) | C26—O29 | 1.327 (2) |
| C12—H12A | 0.83 (5) | C28—H28A | 0.88 (4) |
| C12—H12B | 0.95 (5) | C28—H28B | 0.95 (4) |
| C12—H12C | 1.10 (5) | C28—H28C | 0.96 (4) |
| C13—C14 | 1.536 (3) | O29—H29 | 0.84 (3) |
| C10—C1—C2 | 108.67 (15) | C11—C13—C14 | 112.56 (17) |
| C10—C1—C9 | 112.79 (14) | C11—C13—H13A | 108.7 (19) |
| C2—C1—C9 | 111.31 (15) | C14—C13—H13A | 108.3 (19) |
| C10—C1—H1 | 107.0 (14) | C11—C13—H13B | 105.1 (17) |
| C2—C1—H1 | 110.1 (14) | C14—C13—H13B | 108.9 (17) |
| C9—C1—H1 | 106.9 (14) | H13A—C13—H13B | 113 (3) |
| C3—C2—C1 | 125.88 (17) | C15—C14—C13 | 112.82 (19) |
| C3—C2—H2 | 117.9 (14) | C15—C14—H14A | 110.7 (16) |
| C1—C2—H2 | 116.2 (14) | C13—C14—H14A | 107.4 (17) |
| C2—C3—C4 | 120.16 (17) | C15—C14—H14B | 105.3 (16) |
| C2—C3—C21 | 122.54 (19) | C13—C14—H14B | 111.4 (16) |
| C4—C3—C21 | 117.16 (17) | H14A—C14—H14B | 109 (2) |
| C3—C4—C5 | 120.37 (16) | C16—C15—C14 | 113.3 (2) |
| C3—C4—C8 | 113.11 (16) | C16—C15—H15A | 108.9 (17) |
| C5—C4—C8 | 101.99 (15) | C14—C15—H15A | 103.4 (17) |
| C3—C4—H4 | 108.6 (14) | C16—C15—H15B | 107 (2) |
| C5—C4—H4 | 106.4 (14) | C14—C15—H15B | 105.6 (19) |
| C8—C4—H4 | 105.2 (14) | H15A—C15—H15B | 119 (3) |
| C4—C5—C6 | 102.69 (16) | C17—C16—C20 | 105.8 (2) |
| C4—C5—H5A | 111.3 (14) | C17—C16—C15 | 126.7 (2) |
| C6—C5—H5A | 112.2 (14) | C20—C16—C15 | 127.5 (2) |
| C4—C5—H5AB | 108.8 (13) | C16—C17—O18 | 111.0 (2) |
| C6—C5—H5AB | 112.4 (13) | C16—C17—H17 | 136.5 (14) |
| H5A—C5—H5AB | 109.3 (19) | O18—C17—H17 | 112.5 (14) |
| C5—C6—C7 | 107.52 (16) | C19—O18—C17 | 105.96 (18) |
| C5—C6—H6A | 111.3 (17) | C20—C19—O18 | 110.6 (2) |
| C7—C6—H6A | 108.5 (18) | C20—C19—H19 | 129.1 (17) |
| C5—C6—H6AB | 110.9 (16) | O18—C19—H19 | 120.3 (16) |
| C7—C6—H6AB | 109.8 (16) | C19—C20—C16 | 106.7 (2) |
| H6A—C6—H6AB | 109 (2) | C19—C20—H20 | 127.2 (17) |
| C22—C7—C8 | 115.92 (17) | C16—C20—H20 | 126.1 (17) |
| C22—C7—C6 | 112.42 (18) | C3—C21—H21A | 111.3 (18) |
| C8—C7—C6 | 102.52 (15) | C3—C21—H21B | 110.5 (19) |
| C22—C7—H7 | 104.9 (16) | H21A—C21—H21B | 102 (3) |
| C8—C7—H7 | 111.4 (16) | C3—C21—H21C | 111.6 (18) |
| C6—C7—H7 | 109.8 (16) | H21A—C21—H21C | 109 (2) |
| C9—C8—C4 | 110.08 (15) | H21B—C21—H21C | 112 (3) |
| C9—C8—C7 | 120.96 (15) | C7—C22—H22A | 111.9 (17) |
| C4—C8—C7 | 102.67 (15) | C7—C22—H22B | 106.6 (17) |
| C9—C8—H8 | 105.9 (12) | H22A—C22—H22B | 110 (2) |
| C4—C8—H8 | 108.2 (12) | C7—C22—H22C | 107.2 (18) |
| C7—C8—H8 | 108.6 (12) | H22A—C22—H22C | 110 (2) |
| O23—C9—C26 | 101.95 (13) | H22B—C22—H22C | 111 (3) |
| O23—C9—C8 | 108.06 (14) | C24—O23—C9 | 109.44 (13) |
| C26—C9—C8 | 115.58 (15) | O27—C24—O23 | 118.94 (16) |
| O23—C9—C1 | 107.11 (13) | O27—C24—C25 | 129.87 (17) |
| C26—C9—C1 | 113.87 (14) | O23—C24—C25 | 111.19 (14) |
| C8—C9—C1 | 109.53 (14) | C26—C25—C24 | 106.00 (15) |
| C11—C10—C1 | 126.34 (18) | C26—C25—C28 | 130.00 (17) |
| C11—C10—H10 | 118.1 (14) | C24—C25—C28 | 124.01 (16) |
| C1—C10—H10 | 115.5 (14) | O29—C26—C25 | 131.03 (16) |
| C10—C11—C12 | 123.88 (19) | O29—C26—C9 | 117.58 (15) |
| C10—C11—C13 | 120.76 (19) | C25—C26—C9 | 111.38 (15) |
| C12—C11—C13 | 115.36 (19) | C25—C28—H28A | 114 (3) |
| C11—C12—H12A | 111 (3) | C25—C28—H28B | 109 (2) |
| C11—C12—H12B | 114 (3) | H28A—C28—H28B | 114 (3) |
| H12A—C12—H12B | 125 (4) | C25—C28—H28C | 111 (2) |
| C11—C12—H12C | 102 (3) | H28A—C28—H28C | 104 (3) |
| H12A—C12—H12C | 106 (4) | H28B—C28—H28C | 103 (3) |
| H12B—C12—H12C | 95 (3) | C26—O29—H29 | 109 (2) |
| C10—C1—C2—C3 | −109.2 (2) | C1—C10—C11—C12 | −2.5 (3) |
| C9—C1—C2—C3 | 15.6 (3) | C1—C10—C11—C13 | 177.95 (18) |
| C1—C2—C3—C4 | −4.4 (3) | C10—C11—C13—C14 | 107.9 (2) |
| C1—C2—C3—C21 | 180.00 (19) | C12—C11—C13—C14 | −71.7 (3) |
| C2—C3—C4—C5 | 142.7 (2) | C11—C13—C14—C15 | −69.9 (3) |
| C21—C3—C4—C5 | −41.5 (3) | C13—C14—C15—C16 | 172.2 (2) |
| C2—C3—C4—C8 | 21.9 (3) | C14—C15—C16—C17 | −133.3 (2) |
| C21—C3—C4—C8 | −162.28 (18) | C14—C15—C16—C20 | 46.9 (3) |
| C3—C4—C5—C6 | −165.52 (18) | C20—C16—C17—O18 | 0.3 (3) |
| C8—C4—C5—C6 | −39.4 (2) | C15—C16—C17—O18 | −179.6 (2) |
| C4—C5—C6—C7 | 16.7 (2) | C16—C17—O18—C19 | −0.5 (3) |
| C5—C6—C7—C22 | 137.51 (19) | C17—O18—C19—C20 | 0.5 (3) |
| C5—C6—C7—C8 | 12.3 (2) | O18—C19—C20—C16 | −0.4 (3) |
| C3—C4—C8—C9 | −50.8 (2) | C17—C16—C20—C19 | 0.1 (3) |
| C5—C4—C8—C9 | 178.42 (14) | C15—C16—C20—C19 | 179.9 (2) |
| C3—C4—C8—C7 | 179.09 (15) | C26—C9—O23—C24 | −0.34 (18) |
| C5—C4—C8—C7 | 48.36 (17) | C8—C9—O23—C24 | −122.55 (15) |
| C22—C7—C8—C9 | 77.3 (2) | C1—C9—O23—C24 | 119.52 (15) |
| C6—C7—C8—C9 | −159.85 (17) | C9—O23—C24—O27 | 178.94 (16) |
| C22—C7—C8—C4 | −159.65 (18) | C9—O23—C24—C25 | −0.9 (2) |
| C6—C7—C8—C4 | −36.81 (18) | O27—C24—C25—C26 | −177.91 (19) |
| C4—C8—C9—O23 | −54.40 (18) | O23—C24—C25—C26 | 2.0 (2) |
| C7—C8—C9—O23 | 65.0 (2) | O27—C24—C25—C28 | 2.5 (3) |
| C4—C8—C9—C26 | −167.82 (14) | O23—C24—C25—C28 | −177.65 (18) |
| C7—C8—C9—C26 | −48.4 (2) | C24—C25—C26—O29 | 177.21 (18) |
| C4—C8—C9—C1 | 61.96 (18) | C28—C25—C26—O29 | −3.2 (4) |
| C7—C8—C9—C1 | −178.59 (15) | C24—C25—C26—C9 | −2.2 (2) |
| C10—C1—C9—O23 | −163.92 (14) | C28—C25—C26—C9 | 177.4 (2) |
| C2—C1—C9—O23 | 73.62 (17) | O23—C9—C26—O29 | −177.86 (14) |
| C10—C1—C9—C26 | −52.0 (2) | C8—C9—C26—O29 | −61.0 (2) |
| C2—C1—C9—C26 | −174.48 (14) | C1—C9—C26—O29 | 67.2 (2) |
| C10—C1—C9—C8 | 79.12 (18) | O23—C9—C26—C25 | 1.60 (19) |
| C2—C1—C9—C8 | −43.34 (18) | C8—C9—C26—C25 | 118.51 (17) |
| C2—C1—C10—C11 | −88.2 (2) | C1—C9—C26—C25 | −113.39 (17) |
| C9—C1—C10—C11 | 147.88 (19) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O29—H29···O27i | 0.84 (3) | 1.80 (3) | 2.6207 (18) | 166 (3) |
Symmetry code: (i) −x+1, y+1/2, −z+1/2.
References
- Balansa, W., Islam, R., Fontaine, F., Piggott, A. M., Zhang, H., Webb, T. I., Gilbert, D. F., Lynch, J. W. & Capon, R. J. (2010). Bioorg. Med. Chem. 18, 2912–2919. [DOI] [PubMed]
- Balansa, W., Islam, R., Fontaine, F., Piggott, A. M., Zhang, H., Xiao, X., Webb, T. I., Gilbert, D. F., Lynch, J. W. & Capon, R. J. (2013). Org. Biomol. Chem. 11, 4695–4701. [DOI] [PubMed]
- Barrow, C. J., Blunt, J. W., Munro, M. H. G. & Perry, N. B. (1988). J. Nat. Prod. 51, 1294–1298.
- Burla, M. C., Caliandro, R., Camalli, M., Carrozzini, B., Cascarano, G. L., De Caro, L., Giacovazzo, C., Polidori, G. & Spagna, R. (2005). J. Appl. Cryst. 38, 381–388.
- Chevallier, C., Bugni, T. S., Feng, X., Harper, M. K., Orendt, A. M. & Ireland, C. M. (2006). J. Org. Chem. 71, 2510–2513. [DOI] [PMC free article] [PubMed]
- Coll, J. C., Kearns, P. S., Rideout, J. A. & Hooper, J. (1997). J. Nat. Prod. 60, 1178–1179.
- Hofheinz, W. & Schönholzer, P. (1977). Helv. Chim. Acta, 60, 1367–1370. [DOI] [PubMed]
- Höller, U., König, G. M. & Wright, A. D. (1997). J. Nat. Prod. 60, 832–835.
- Kobayashi, J., Shinonaga, H., Shigemori, H., Umeyama, A., Shoji, N. & Arihara, S. (1995). J. Nat. Prod. 58, 312–318. [DOI] [PubMed]
- Kondo, K., Shigemori, H., Kikuchi, Y., Ishibashi, M., Sasaki, T. & Kobayashi, J. (1992). J. Org. Chem. 57, 2480–2483.
- Mau, C. M. S., Nakao, Y., Yoshida, W. Y., Scheuer, P. J. & Kelly-Borges, M. (1996). J. Org. Chem. 61, 6302–6304. [DOI] [PubMed]
- Parsons, S., Flack, H. D. & Wagner, T. (2013). Acta Cryst. B69, 249–259. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Spek, A. L. (2020). Acta Cryst. E76, 1–11. [DOI] [PMC free article] [PubMed]
- Stoe & Cie (2019). X-RED32 and X-AREA. Stoe & Cie, Darmstadt, Germany.
- Uenishi, J., Kawahama, R. & Yonemitsu, O. (1997). J. Org. Chem. 62, 1691–1701.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2414314620015783/bt4103sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314620015783/bt4103Isup2.hkl
CCDC reference: 2047802
Additional supporting information: crystallographic information; 3D view; checkCIF report