The structure of the trans-Pt(NH3)2(mnt)2 complex has Pt—N and Pt—S distances that are are consistent with those in other platinum(IV) complexes. The nitrile nitrogen atoms are positioned suitably to hydrogen bond with adjacent ammines.
Keywords: crystal structure, platinum(IV), dithiolene
Abstract
The title compound, [Pt(C4N2S2)2(NH3)2], represents an octahedral platinum(IV) complex with two trans-ammine and two mnt (mnt = 1,2-dicyanoethene-1,2-dithiolato) ligands. The Pt—N and Pt—S distances are consistent with those in other platinum(IV) complexes. As a result of a slight canting of the coordination of the mnt ligand to the platinum(IV) atom, the nitrile nitrogen atoms are positioned suitably to hydrogen-bond with adjacent ammines.

Structure description
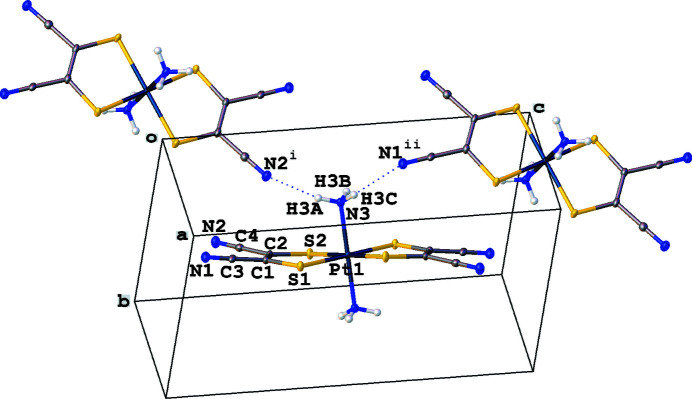
The neutral title complex contains two ammine and two mnt ligands forming an octahedral platinum(IV) complex. The C—S distances of 1.747 (3) and 1.744 (3) Å and the C=C distance of 1.358 (4) Å support the ene-1,2-dithiolate form of the mnt ligand (Güntner et al., 1989 ▸; Chandrasekaran et al., 2014 ▸). The two ammine ligands are trans with a Pt—N bond length of 2.055 (2) Å, which is consistent with the Pt—N distances in other platinum(IV) complexes of 2.056 (9) (Fanwick & Huckaby, 1982 ▸) and 2.053 (5) Å (Brawner et al., 1978 ▸). The Pt—S distances of 2.3434 (8) and 2.3461 (7) Å are longer than in square-planar platinum complexes with mnt such as the PtII—S distances of 2.290 and 2.282 Å in [Pt(mnt)2]2− (Günter et al., 1989 ▸) or the PtIII—S distance of 2.262 Å in [Pt(mnt)2]− (Mochida et al., 2010 ▸). This longer Pt—S bond is comparable, however, with the Pt—S distance of 2.3619 Å found in a similar octahedral platinum(IV) complex with two dithiolene and two trans phosphine ligands (Chandrasekaran et al., 2014 ▸). The coordination of the mnt ligands is slightly canted from the platinum(IV) atom, which allows for hydrogen bonding between the nitrile nitrogen atoms and adjacent ammines (Fig. 1 ▸, Table 1 ▸). These interactions lead to the formation of a three-dimensional network.
Figure 1.
Displacement ellipsoid plot 50% probability of all non-H atoms showing N—H hydrogen bonding between nitrile nitrogen atoms and hydrogen atoms on adjacent ammines. Symmetry codes: (i) −x, y −
 , −z +
, −z +
 ; (ii) x, −y +
; (ii) x, −y +
 , z +
, z +
 .
.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N3—H3A⋯N2i | 0.89 | 2.16 | 3.016 (3) | 160 |
| N3—H3B⋯S2ii | 0.89 | 2.73 | 3.610 (3) | 171 |
| N3—H3C⋯N1iii | 0.89 | 2.26 | 3.011 (3) | 142 |
Symmetry codes: (i)
 ; (ii)
; (ii)
 ; (iii)
; (iii)
 .
.
Synthesis and crystallization
A solution of 13.9 mg (7.46 × 10 −5mol) of Na2mnt dissolved in 10 mL of water was combined with a solution of 25 mg (7.48 × 10−5 mol) of tetraammineplatinum(II) chloride dissolved in 25 mL of water, and stirred for 2 h in air. The solvent was removed using a vacuum oven to give 26.5 mg of a brown product isolated [1H NMR (d-DMSO) 4.23ppm]. Light-orange crystals of the title compound were grown by liquid diffusion of diethyl ether into a methanol solution of the synthesized product in a tall, narrow tube that was covered with parafilm. The platinum(II) dithiolene complex is presumed to oxidize to the ammine-stabilized octahedral platinum(IV) dithiolene compound via air, demonstrating a synthetic route toward stable neutral PtIV dithiolene complexes (Geiger et al., 2001 ▸).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | [Pt(C4N2S2)2(NH3)2] |
| M r | 509.52 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 100 |
| a, b, c (Å) | 6.1778 (3), 7.7700 (4), 14.8862 (7) |
| β (°) | 95.935 (4) |
| V (Å3) | 710.73 (6) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 10.45 |
| Crystal size (mm) | 0.05 × 0.02 × 0.01 |
| Data collection | |
| Diffractometer | Rigaku XtaLAB Synergy, Dualflex, HyPix |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku OD, 2019 ▸) |
| T min, T max | 0.509, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 8027, 1556, 1375 |
| R int | 0.034 |
| (sin θ/λ)max (Å−1) | 0.641 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.017, 0.038, 1.05 |
| No. of reflections | 1556 |
| No. of parameters | 89 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.62, −0.54 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2414314620009803/bv4032sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314620009803/bv4032Isup2.hkl
CCDC reference: 2017151
Additional supporting information: crystallographic information; 3D view; checkCIF report
full crystallographic data
Crystal data
| [Pt(C4N2S2)2(NH3)2] | F(000) = 476 |
| Mr = 509.52 | Dx = 2.381 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 6.1778 (3) Å | Cell parameters from 5207 reflections |
| b = 7.7700 (4) Å | θ = 2.7–29.7° |
| c = 14.8862 (7) Å | µ = 10.45 mm−1 |
| β = 95.935 (4)° | T = 100 K |
| V = 710.73 (6) Å3 | Plate, clear light orange |
| Z = 2 | 0.05 × 0.02 × 0.01 mm |
Data collection
| Rigaku XtaLAB Synergy, Dualflex, HyPix diffractometer | 1556 independent reflections |
| Radiation source: micro-focus sealed X-ray tube, PhotonJet (Mo) X-ray Source | 1375 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.034 |
| Detector resolution: 10.0000 pixels mm-1 | θmax = 27.1°, θmin = 2.8° |
| ω scans | h = −7→7 |
| Absorption correction: multi-scan (CrysAlisPro; Rigaku OD, 2019) | k = −9→9 |
| Tmin = 0.509, Tmax = 1.000 | l = −18→19 |
| 8027 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.017 | H-atom parameters constrained |
| wR(F2) = 0.038 | w = 1/[σ2(Fo2) + (0.0154P)2 + 0.3678P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max < 0.001 |
| 1556 reflections | Δρmax = 0.62 e Å−3 |
| 89 parameters | Δρmin = −0.54 e Å−3 |
| 0 restraints |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Pt1 | 0.500000 | 0.500000 | 0.500000 | 0.00819 (6) | |
| S1 | 0.62740 (13) | 0.46855 (9) | 0.35800 (5) | 0.01208 (16) | |
| S2 | 0.20123 (12) | 0.65846 (10) | 0.43437 (5) | 0.01294 (16) | |
| N1 | 0.4195 (5) | 0.4964 (3) | 0.1195 (2) | 0.0179 (6) | |
| N2 | −0.0905 (4) | 0.7217 (3) | 0.20727 (17) | 0.0182 (6) | |
| N3 | 0.3182 (4) | 0.2794 (3) | 0.47944 (16) | 0.0125 (5) | |
| H3A | 0.283811 | 0.263528 | 0.420476 | 0.015* | |
| H3B | 0.197076 | 0.289137 | 0.506596 | 0.015* | |
| H3C | 0.395109 | 0.189825 | 0.502333 | 0.015* | |
| C1 | 0.4056 (5) | 0.5506 (4) | 0.2889 (2) | 0.0110 (6) | |
| C2 | 0.2301 (5) | 0.6263 (4) | 0.32026 (18) | 0.0116 (6) | |
| C3 | 0.4129 (5) | 0.5226 (3) | 0.1948 (2) | 0.0120 (6) | |
| C4 | 0.0516 (5) | 0.6804 (4) | 0.25742 (19) | 0.0124 (6) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Pt1 | 0.00671 (9) | 0.01158 (10) | 0.00598 (9) | 0.00083 (6) | −0.00073 (6) | 0.00050 (6) |
| S1 | 0.0099 (4) | 0.0183 (4) | 0.0079 (3) | 0.0023 (3) | 0.0005 (3) | 0.0006 (3) |
| S2 | 0.0104 (3) | 0.0193 (4) | 0.0088 (3) | 0.0045 (3) | −0.0006 (3) | 0.0021 (3) |
| N1 | 0.0231 (16) | 0.0163 (15) | 0.0136 (14) | 0.0000 (11) | −0.0009 (12) | 0.0000 (10) |
| N2 | 0.0171 (14) | 0.0228 (15) | 0.0143 (13) | 0.0030 (12) | −0.0008 (11) | 0.0019 (11) |
| N3 | 0.0099 (13) | 0.0157 (13) | 0.0117 (12) | 0.0002 (10) | −0.0008 (10) | 0.0027 (10) |
| C1 | 0.0123 (15) | 0.0119 (14) | 0.0085 (14) | −0.0017 (12) | −0.0010 (12) | 0.0009 (11) |
| C2 | 0.0136 (15) | 0.0119 (15) | 0.0086 (14) | −0.0020 (12) | −0.0028 (11) | 0.0024 (11) |
| C3 | 0.0149 (16) | 0.0076 (15) | 0.0124 (16) | 0.0001 (11) | −0.0035 (12) | 0.0008 (11) |
| C4 | 0.0135 (15) | 0.0111 (15) | 0.0127 (14) | −0.0019 (12) | 0.0014 (12) | −0.0002 (12) |
Geometric parameters (Å, º)
| Pt1—S1i | 2.3434 (8) | N1—C3 | 1.143 (4) |
| Pt1—S1 | 2.3434 (8) | N2—C4 | 1.138 (4) |
| Pt1—S2 | 2.3461 (7) | N3—H3A | 0.8900 |
| Pt1—S2i | 2.3461 (7) | N3—H3B | 0.8900 |
| Pt1—N3i | 2.055 (2) | N3—H3C | 0.8900 |
| Pt1—N3 | 2.055 (2) | C1—C2 | 1.358 (4) |
| S1—C1 | 1.747 (3) | C1—C3 | 1.423 (4) |
| S2—C2 | 1.744 (3) | C2—C4 | 1.433 (4) |
| S1i—Pt1—S1 | 180.0 | C2—S2—Pt1 | 100.09 (10) |
| S1i—Pt1—S2 | 89.87 (3) | Pt1—N3—H3A | 109.5 |
| S1—Pt1—S2i | 89.87 (3) | Pt1—N3—H3B | 109.5 |
| S1—Pt1—S2 | 90.13 (3) | Pt1—N3—H3C | 109.5 |
| S1i—Pt1—S2i | 90.13 (3) | H3A—N3—H3B | 109.5 |
| S2i—Pt1—S2 | 180.0 | H3A—N3—H3C | 109.5 |
| N3i—Pt1—S1i | 90.46 (7) | H3B—N3—H3C | 109.5 |
| N3—Pt1—S1i | 89.54 (7) | C2—C1—S1 | 124.1 (2) |
| N3—Pt1—S1 | 90.46 (7) | C2—C1—C3 | 120.8 (3) |
| N3i—Pt1—S1 | 89.54 (7) | C3—C1—S1 | 114.9 (2) |
| N3i—Pt1—S2 | 91.06 (7) | C1—C2—S2 | 124.2 (2) |
| N3—Pt1—S2i | 91.06 (7) | C1—C2—C4 | 119.4 (3) |
| N3—Pt1—S2 | 88.94 (7) | C4—C2—S2 | 116.4 (2) |
| N3i—Pt1—S2i | 88.94 (7) | N1—C3—C1 | 178.5 (3) |
| N3—Pt1—N3i | 180.0 | N2—C4—C2 | 179.3 (3) |
| C1—S1—Pt1 | 100.20 (11) | ||
| Pt1—S1—C1—C2 | 6.6 (3) | S1—C1—C2—S2 | 1.7 (4) |
| Pt1—S1—C1—C3 | −169.3 (2) | S1—C1—C2—C4 | −176.5 (2) |
| Pt1—S2—C2—C1 | −8.8 (3) | C3—C1—C2—S2 | 177.4 (2) |
| Pt1—S2—C2—C4 | 169.4 (2) | C3—C1—C2—C4 | −0.8 (5) |
Symmetry code: (i) −x+1, −y+1, −z+1.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N3—H3A···N2ii | 0.89 | 2.16 | 3.016 (3) | 160 |
| N3—H3B···S2iii | 0.89 | 2.73 | 3.610 (3) | 171 |
| N3—H3C···N1iv | 0.89 | 2.26 | 3.011 (3) | 142 |
Symmetry codes: (ii) −x, y−1/2, −z+1/2; (iii) −x, −y+1, −z+1; (iv) x, −y+1/2, z+1/2.
Funding Statement
We are grateful of the Welch Foundation (AD-0007) for a department grant supporting undergraduate research and the NSF MRI for a Jeol ECZ-400 NMR at Austin College (CHE-1725651) and a Rigaku XtaLAB Synergy-S X-ray diffractometer at UNT (CHE-1726652).
References
- Brawner, S. A., Lin, I. J. B., Kim, J.-H. & Everett, G. W. (1978). Inorg. Chem. 17, 1304–1308.
- Chandrasekaran, P., Greene, A. F., Lillich, K., Capone, S., Mague, J. T., DeBeer, S. & Donahue, J. P. (2014). Inorg. Chem. 53, 9192–9205. [DOI] [PubMed]
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Fanwick, P. E. & Huckaby, J. L. (1982). Inorg. Chem. 21, 3067–3071.
- Geiger, W. E., Barrière, F., LeSuer, R. J. & Trupia, S. (2001). Inorg. Chem. 40, 2472–2473. [DOI] [PubMed]
- Güntner, W., Gliemann, G., Klement, U. & Zabel, M. (1989). Inorg. Chim. Acta, 165, 51–56.
- Mochida, T., Nagabuchi, E. & Ueda, M. (2010). Inorg. Chim. Acta, 363, 4108–4111.
- Rigaku OD (2019). CrysAlis PRO. Rigaku Oxford Diffraction, Yarnton, England.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2414314620009803/bv4032sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314620009803/bv4032Isup2.hkl
CCDC reference: 2017151
Additional supporting information: crystallographic information; 3D view; checkCIF report