The title molecule is built by annulation of a half-chair cyclohexenone and a twist-cyclohexenone to a flat 4-H-pyrane boat. In the crystal, molecules are connected via van der Waals interactions and C—H⋯O hydrogen bonds.
Keywords: crystal structure, heterocycles, polycyclic system
Abstract
The title molecule, C19H24O5, is built by annulation of a half-chair cyclohexenone and a twist-cyclohexenone to a flat 4-H-pyrane boat. In the crystal, molecules are connected via van der Waals interactions and C—H⋯O hydrogen bonds.
Structure description
The title compound was obtained as a side product during the formation of methyl methoxy(2,6-dioxo-4,4-dimethylcyclohexyl)acetate according to the procedure of Grosz & Freiberg (1966 ▸). A similar product (1,2,3,4,5,6,7,8-octahydro-3,3,6,6-tetramethyl-1,8-dioxo-9-xanthenyl acetic acid) was obtained by Gustafsson (1948 ▸) in the condensation of dimedone and glyoxalic acid. The free acid is an isomer of the title compound with a methylene group connecting the heterocyclic unit and carboxylic acid group. A short route to these compounds is the uncatalysed tandem aldol condensation/elimination/Michael addition/condensation, as discovered by Rohr & Mahrwald (2009 ▸).
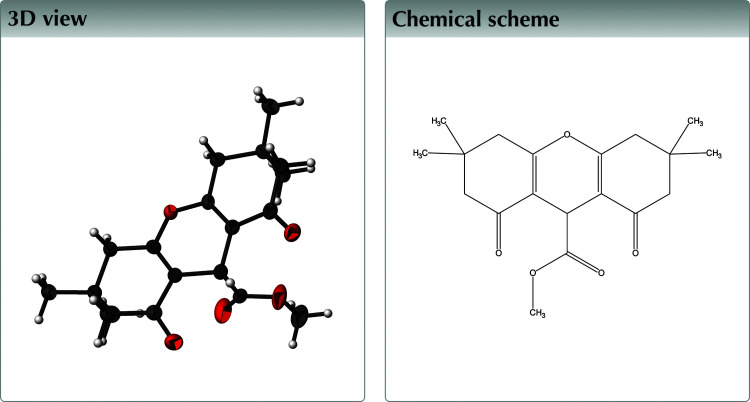
The molecule is composed of two dimethylcyclohexenone units annulated to a central 4H-pyrane (Fig. 1 ▸). While the conformation of the latter is a flat boat, one cyclohexenone (C2–C7) forms a half-chair and the other (C9–C14) has a twist form. The pyrane boat promotes a folded shape of the molecule, the angle between the mean planes through atoms C1–C3/C6/C7/O8 and O8/C9/C10/C13//C14 being 22.42 (11)°, with maximum deviations from the mean planes at O8 [−0.1046 (18) Å] and C1 [0.051 (3) Å]. The torsion angle of the ester group (O17—C15—C1—C2) is 66.4 (3)°.
Figure 1.
Perspective view of the title compound. Displacement ellipsoids are drawn at the 50% probability level.
Four molecules occupy the monoclinic unit cell, the packing in the cell being dominated by van der Waals interactions and hydrogen-bonding interactions (Table 1 ▸ and Fig. 2 ▸). The C—H⋯O hydrogen bonds (Steiner, 1996 ▸) C18—H18A⋯O17 and C18—H18A⋯O19 form a hydrogen-bonded dimer while the C6—H6B⋯O24 interaction connects two molecules related by the c-glide plane.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C6—H6B⋯O24i | 0.99 | 2.50 | 3.324 (4) | 140 |
| C18—H18A⋯O17ii | 0.98 | 2.53 | 3.451 (5) | 156 |
| C18—H18A⋯O19ii | 0.98 | 2.41 | 3.093 (4) | 126 |
Symmetry codes: (i)
 ; (ii)
; (ii)
 .
.
Figure 2.
Partial packing diagram of the title compound with a view along the b axis. Most of the hydrogen atoms omitted for clarity. Hydrogen bonds are depicted with dashed lines.
Synthesis and crystallization
Dimedone (7.01 g, 0.05 mol, 1 eq.) and triethylamine (5.05 g, 50 mmol, 6.9 ml, 1 eq.) were dissolved in dichloromethane (25 ml) in a 250 ml flask under nitrogen. Methyl chloromethoxyacetate (5.9 ml, 6.49 g, 0.525 mol, 1.05 eq.) was added dropwise to the ice-cooled mixture under stirring and the stirring was continued for 75 min at room temperature and a further 3 h under reflux conditions. The solvent was evaporated, methyl tert-butyl ether was added to the suspension and triethyl ammonium chloride was removed via filtration. The etheral layer was washed with aqueous sodium carbonate and brine, and dried over sodium sulfate. Evaporation of the solvent and chromatography (silica gel, petroleum ether/ethyl acetate = 3/1, R f = 3:1) yielded 0.83 g (2.5 mmol, 5%) of the title compound as colourless crystals with m.p. = 474–478 K. The main product yield was 83%. Crystals of the title compound were obtained from a solution in ethyl acetate. IR: 2959, 2875, 1728, 1663, 1368, 1193, 995. 1H NMR (300 MHz, CDCl3) δ/p.p.m.: 4.47–4.46 (s, 1H), 3.68 (s, 3H), 2.43 (2*d, 2*2 gem H, J = 18 Hz), 4H), 2.27 (2*d, 2*2 gem H, J = 18 Hz), 1.11 (s, 12H).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C19H24O5 |
| M r | 332.38 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 120 |
| a, b, c (Å) | 13.1494 (10), 9.6899 (6), 14.8185 (13) |
| β (°) | 113.295 (6) |
| V (Å3) | 1734.2 (2) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.09 |
| Crystal size (mm) | 0.22 × 0.11 × 0.06 |
| Data collection | |
| Diffractometer | Stoe IPDS 2T |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 8583, 4123, 2784 |
| R int | 0.036 |
| (sin θ/λ)max (Å−1) | 0.659 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.072, 0.185, 1.05 |
| No. of reflections | 4123 |
| No. of parameters | 222 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.24, −0.30 |
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2414314620010184/gg4005sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314620010184/gg4005Isup2.hkl
schematic diagram of the reaction. DOI: 10.1107/S2414314620010184/gg4005sup3.tif
Supporting information file. DOI: 10.1107/S2414314620010184/gg4005Isup4.cml
CCDC reference: 2018468
Additional supporting information: crystallographic information; 3D view; checkCIF report
full crystallographic data
Crystal data
| C19H24O5 | F(000) = 712 |
| Mr = 332.38 | Dx = 1.273 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 13.1494 (10) Å | Cell parameters from 9146 reflections |
| b = 9.6899 (6) Å | θ = 2.6–28.2° |
| c = 14.8185 (13) Å | µ = 0.09 mm−1 |
| β = 113.295 (6)° | T = 120 K |
| V = 1734.2 (2) Å3 | Plate, colourless |
| Z = 4 | 0.22 × 0.11 × 0.06 mm |
Data collection
| STOE IPDS 2T diffractometer | 2784 reflections with I > 2σ(I) |
| Radiation source: sealed X-ray tube, 12 x 0.4 mm long-fine focus | Rint = 0.036 |
| Detector resolution: 6.67 pixels mm-1 | θmax = 28.0°, θmin = 2.6° |
| rotation method, ω scans | h = −16→17 |
| 8583 measured reflections | k = −12→12 |
| 4123 independent reflections | l = −19→19 |
Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.072 | H-atom parameters constrained |
| wR(F2) = 0.185 | w = 1/[σ2(Fo2) + (0.0577P)2 + 2.4014P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max = 0.002 |
| 4123 reflections | Δρmax = 0.24 e Å−3 |
| 222 parameters | Δρmin = −0.30 e Å−3 |
| 0 restraints |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Hydrogen atoms attached to carbons were placed at calculated positions and were refined in the riding-model approximation with C–H = 0.95 Å, and with Uiso(H) = 1.2 Ueq(C). |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.6910 (2) | 0.3522 (3) | 0.3759 (2) | 0.0290 (5) | |
| H1 | 0.715526 | 0.337644 | 0.320767 | 0.035* | |
| C2 | 0.7343 (2) | 0.2359 (3) | 0.4490 (2) | 0.0284 (5) | |
| C3 | 0.8403 (2) | 0.1694 (3) | 0.4650 (2) | 0.0292 (5) | |
| C4 | 0.8801 (2) | 0.0564 (3) | 0.5415 (2) | 0.0327 (6) | |
| H4A | 0.962100 | 0.056545 | 0.570806 | 0.039* | |
| H4B | 0.855649 | −0.033835 | 0.508675 | 0.039* | |
| C5 | 0.8385 (2) | 0.0705 (3) | 0.6237 (2) | 0.0302 (6) | |
| C6 | 0.7113 (2) | 0.0808 (3) | 0.5759 (2) | 0.0313 (6) | |
| H6A | 0.679951 | −0.010095 | 0.548221 | 0.038* | |
| H6B | 0.683107 | 0.105346 | 0.626632 | 0.038* | |
| C7 | 0.67398 (19) | 0.1865 (3) | 0.49633 (19) | 0.0270 (5) | |
| O8 | 0.56625 (14) | 0.22547 (18) | 0.47408 (13) | 0.0293 (4) | |
| C9 | 0.5126 (2) | 0.2955 (3) | 0.38683 (19) | 0.0283 (5) | |
| C10 | 0.3910 (2) | 0.3002 (3) | 0.3590 (2) | 0.0301 (6) | |
| H10A | 0.375949 | 0.313286 | 0.418829 | 0.036* | |
| H10B | 0.358002 | 0.211094 | 0.328896 | 0.036* | |
| C11 | 0.3362 (2) | 0.4177 (3) | 0.2862 (2) | 0.0312 (6) | |
| C12 | 0.3794 (2) | 0.4120 (3) | 0.2043 (2) | 0.0329 (6) | |
| H12A | 0.354823 | 0.324715 | 0.167285 | 0.040* | |
| H12B | 0.347003 | 0.489267 | 0.158080 | 0.040* | |
| C13 | 0.5040 (2) | 0.4207 (3) | 0.2430 (2) | 0.0302 (5) | |
| C14 | 0.5668 (2) | 0.3511 (3) | 0.33598 (19) | 0.0276 (5) | |
| C15 | 0.7354 (2) | 0.4908 (3) | 0.4252 (2) | 0.0307 (6) | |
| O16 | 0.68175 (18) | 0.5800 (2) | 0.4402 (2) | 0.0579 (7) | |
| O17 | 0.84313 (17) | 0.5003 (2) | 0.4521 (2) | 0.0606 (8) | |
| C18 | 0.8958 (3) | 0.6248 (3) | 0.5039 (3) | 0.0611 (11) | |
| H18A | 0.976272 | 0.616654 | 0.525273 | 0.092* | |
| H18B | 0.877556 | 0.637293 | 0.561397 | 0.092* | |
| H18C | 0.869276 | 0.704400 | 0.460044 | 0.092* | |
| O19 | 0.89116 (15) | 0.19842 (19) | 0.41368 (14) | 0.0334 (4) | |
| C20 | 0.8901 (2) | 0.1983 (3) | 0.6859 (2) | 0.0369 (6) | |
| H20A | 0.971000 | 0.190656 | 0.712677 | 0.055* | |
| H20B | 0.865283 | 0.204928 | 0.740014 | 0.055* | |
| H20C | 0.867042 | 0.281133 | 0.644830 | 0.055* | |
| C21 | 0.8723 (2) | −0.0569 (3) | 0.6899 (2) | 0.0407 (7) | |
| H21A | 0.841283 | −0.139699 | 0.650705 | 0.061* | |
| H21B | 0.844160 | −0.048635 | 0.741779 | 0.061* | |
| H21C | 0.953273 | −0.063927 | 0.719617 | 0.061* | |
| C22 | 0.3647 (2) | 0.5576 (3) | 0.3387 (2) | 0.0386 (7) | |
| H22A | 0.332418 | 0.631853 | 0.290953 | 0.058* | |
| H22B | 0.445306 | 0.568687 | 0.369190 | 0.058* | |
| H22C | 0.334574 | 0.561723 | 0.389470 | 0.058* | |
| C23 | 0.2108 (2) | 0.3990 (3) | 0.2434 (2) | 0.0386 (7) | |
| H23A | 0.175703 | 0.475736 | 0.198843 | 0.058* | |
| H23B | 0.184611 | 0.397602 | 0.296837 | 0.058* | |
| H23C | 0.191455 | 0.311806 | 0.207127 | 0.058* | |
| O24 | 0.55164 (16) | 0.4811 (2) | 0.19806 (15) | 0.0394 (5) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0238 (11) | 0.0255 (12) | 0.0370 (14) | −0.0006 (10) | 0.0111 (10) | 0.0008 (10) |
| C2 | 0.0257 (12) | 0.0230 (12) | 0.0356 (14) | −0.0012 (10) | 0.0110 (11) | −0.0004 (10) |
| C3 | 0.0251 (12) | 0.0255 (12) | 0.0373 (15) | −0.0023 (10) | 0.0125 (11) | −0.0049 (11) |
| C4 | 0.0301 (13) | 0.0271 (13) | 0.0409 (16) | 0.0045 (11) | 0.0142 (12) | 0.0013 (11) |
| C5 | 0.0253 (12) | 0.0290 (13) | 0.0340 (14) | 0.0027 (10) | 0.0095 (11) | 0.0048 (11) |
| C6 | 0.0294 (12) | 0.0268 (12) | 0.0375 (15) | −0.0010 (11) | 0.0131 (11) | 0.0041 (11) |
| C7 | 0.0209 (11) | 0.0234 (11) | 0.0349 (14) | −0.0004 (9) | 0.0091 (10) | −0.0030 (10) |
| O8 | 0.0231 (8) | 0.0295 (9) | 0.0348 (10) | 0.0014 (7) | 0.0109 (7) | 0.0041 (8) |
| C9 | 0.0268 (12) | 0.0237 (12) | 0.0326 (14) | 0.0025 (10) | 0.0100 (10) | 0.0026 (10) |
| C10 | 0.0240 (12) | 0.0266 (13) | 0.0385 (15) | 0.0018 (10) | 0.0110 (11) | 0.0034 (11) |
| C11 | 0.0251 (12) | 0.0267 (13) | 0.0402 (15) | 0.0024 (10) | 0.0113 (11) | 0.0050 (11) |
| C12 | 0.0279 (12) | 0.0315 (13) | 0.0380 (15) | 0.0011 (11) | 0.0115 (11) | 0.0021 (11) |
| C13 | 0.0297 (12) | 0.0256 (12) | 0.0351 (14) | 0.0012 (10) | 0.0125 (11) | 0.0017 (11) |
| C14 | 0.0244 (11) | 0.0229 (12) | 0.0344 (14) | 0.0005 (10) | 0.0103 (10) | 0.0008 (10) |
| C15 | 0.0262 (12) | 0.0258 (12) | 0.0396 (15) | −0.0022 (10) | 0.0124 (11) | 0.0033 (11) |
| O16 | 0.0392 (12) | 0.0427 (13) | 0.094 (2) | −0.0050 (10) | 0.0282 (13) | −0.0255 (13) |
| O17 | 0.0286 (11) | 0.0317 (11) | 0.116 (2) | −0.0079 (9) | 0.0229 (13) | −0.0240 (13) |
| C18 | 0.0410 (17) | 0.0309 (16) | 0.104 (3) | −0.0120 (14) | 0.0204 (19) | −0.0201 (18) |
| O19 | 0.0305 (9) | 0.0317 (10) | 0.0425 (11) | −0.0013 (8) | 0.0193 (9) | −0.0026 (8) |
| C20 | 0.0311 (13) | 0.0402 (16) | 0.0355 (15) | 0.0013 (12) | 0.0089 (12) | −0.0025 (12) |
| C21 | 0.0368 (15) | 0.0381 (16) | 0.0460 (18) | 0.0077 (13) | 0.0151 (13) | 0.0103 (13) |
| C22 | 0.0377 (14) | 0.0295 (14) | 0.0512 (18) | 0.0048 (12) | 0.0204 (14) | −0.0010 (13) |
| C23 | 0.0289 (13) | 0.0376 (15) | 0.0461 (17) | 0.0024 (12) | 0.0112 (12) | 0.0106 (13) |
| O24 | 0.0367 (10) | 0.0391 (11) | 0.0470 (12) | 0.0054 (9) | 0.0214 (10) | 0.0129 (9) |
Geometric parameters (Å, º)
| C1—C14 | 1.501 (3) | C11—C12 | 1.532 (4) |
| C1—C2 | 1.510 (4) | C11—C22 | 1.534 (4) |
| C1—C15 | 1.530 (4) | C12—C13 | 1.508 (3) |
| C1—H1 | 1.0000 | C12—H12A | 0.9900 |
| C2—C7 | 1.337 (3) | C12—H12B | 0.9900 |
| C2—C3 | 1.468 (3) | C13—O24 | 1.229 (3) |
| C3—O19 | 1.228 (3) | C13—C14 | 1.461 (4) |
| C3—C4 | 1.513 (4) | C15—O16 | 1.191 (3) |
| C4—C5 | 1.526 (4) | C15—O17 | 1.314 (3) |
| C4—H4A | 0.9900 | O17—C18 | 1.450 (4) |
| C4—H4B | 0.9900 | C18—H18A | 0.9800 |
| C5—C21 | 1.529 (4) | C18—H18B | 0.9800 |
| C5—C20 | 1.533 (4) | C18—H18C | 0.9800 |
| C5—C6 | 1.541 (3) | C20—H20A | 0.9800 |
| C6—C7 | 1.490 (4) | C20—H20B | 0.9800 |
| C6—H6A | 0.9900 | C20—H20C | 0.9800 |
| C6—H6B | 0.9900 | C21—H21A | 0.9800 |
| C7—O8 | 1.374 (3) | C21—H21B | 0.9800 |
| O8—C9 | 1.381 (3) | C21—H21C | 0.9800 |
| C9—C14 | 1.338 (3) | C22—H22A | 0.9800 |
| C9—C10 | 1.486 (3) | C22—H22B | 0.9800 |
| C10—C11 | 1.538 (4) | C22—H22C | 0.9800 |
| C10—H10A | 0.9900 | C23—H23A | 0.9800 |
| C10—H10B | 0.9900 | C23—H23B | 0.9800 |
| C11—C23 | 1.526 (3) | C23—H23C | 0.9800 |
| C14—C1—C2 | 108.7 (2) | C22—C11—C10 | 110.2 (2) |
| C14—C1—C15 | 110.2 (2) | C13—C12—C11 | 112.6 (2) |
| C2—C1—C15 | 110.4 (2) | C13—C12—H12A | 109.1 |
| C14—C1—H1 | 109.2 | C11—C12—H12A | 109.1 |
| C2—C1—H1 | 109.2 | C13—C12—H12B | 109.1 |
| C15—C1—H1 | 109.2 | C11—C12—H12B | 109.1 |
| C7—C2—C3 | 118.7 (2) | H12A—C12—H12B | 107.8 |
| C7—C2—C1 | 120.7 (2) | O24—C13—C14 | 120.7 (2) |
| C3—C2—C1 | 120.5 (2) | O24—C13—C12 | 122.0 (2) |
| O19—C3—C2 | 121.0 (2) | C14—C13—C12 | 117.2 (2) |
| O19—C3—C4 | 121.2 (2) | C9—C14—C13 | 119.2 (2) |
| C2—C3—C4 | 117.7 (2) | C9—C14—C1 | 121.3 (2) |
| C3—C4—C5 | 114.0 (2) | C13—C14—C1 | 119.4 (2) |
| C3—C4—H4A | 108.8 | O16—C15—O17 | 122.7 (3) |
| C5—C4—H4A | 108.8 | O16—C15—C1 | 125.7 (2) |
| C3—C4—H4B | 108.8 | O17—C15—C1 | 111.6 (2) |
| C5—C4—H4B | 108.8 | C15—O17—C18 | 116.8 (2) |
| H4A—C4—H4B | 107.7 | O17—C18—H18A | 109.5 |
| C4—C5—C21 | 109.6 (2) | O17—C18—H18B | 109.5 |
| C4—C5—C20 | 109.8 (2) | H18A—C18—H18B | 109.5 |
| C21—C5—C20 | 108.6 (2) | O17—C18—H18C | 109.5 |
| C4—C5—C6 | 107.8 (2) | H18A—C18—H18C | 109.5 |
| C21—C5—C6 | 109.6 (2) | H18B—C18—H18C | 109.5 |
| C20—C5—C6 | 111.5 (2) | C5—C20—H20A | 109.5 |
| C7—C6—C5 | 111.5 (2) | C5—C20—H20B | 109.5 |
| C7—C6—H6A | 109.3 | H20A—C20—H20B | 109.5 |
| C5—C6—H6A | 109.3 | C5—C20—H20C | 109.5 |
| C7—C6—H6B | 109.3 | H20A—C20—H20C | 109.5 |
| C5—C6—H6B | 109.3 | H20B—C20—H20C | 109.5 |
| H6A—C6—H6B | 108.0 | C5—C21—H21A | 109.5 |
| C2—C7—O8 | 123.0 (2) | C5—C21—H21B | 109.5 |
| C2—C7—C6 | 125.6 (2) | H21A—C21—H21B | 109.5 |
| O8—C7—C6 | 111.4 (2) | C5—C21—H21C | 109.5 |
| C7—O8—C9 | 117.1 (2) | H21A—C21—H21C | 109.5 |
| C14—C9—O8 | 122.5 (2) | H21B—C21—H21C | 109.5 |
| C14—C9—C10 | 125.8 (2) | C11—C22—H22A | 109.5 |
| O8—C9—C10 | 111.7 (2) | C11—C22—H22B | 109.5 |
| C9—C10—C11 | 111.8 (2) | H22A—C22—H22B | 109.5 |
| C9—C10—H10A | 109.2 | C11—C22—H22C | 109.5 |
| C11—C10—H10A | 109.2 | H22A—C22—H22C | 109.5 |
| C9—C10—H10B | 109.2 | H22B—C22—H22C | 109.5 |
| C11—C10—H10B | 109.2 | C11—C23—H23A | 109.5 |
| H10A—C10—H10B | 107.9 | C11—C23—H23B | 109.5 |
| C23—C11—C12 | 110.2 (2) | H23A—C23—H23B | 109.5 |
| C23—C11—C22 | 108.8 (2) | C11—C23—H23C | 109.5 |
| C12—C11—C22 | 109.9 (2) | H23A—C23—H23C | 109.5 |
| C23—C11—C10 | 109.5 (2) | H23B—C23—H23C | 109.5 |
| C12—C11—C10 | 108.1 (2) | ||
| C14—C1—C2—C7 | −25.2 (3) | O8—C9—C10—C11 | 159.3 (2) |
| C15—C1—C2—C7 | 95.7 (3) | C9—C10—C11—C23 | 167.9 (2) |
| C14—C1—C2—C3 | 151.0 (2) | C9—C10—C11—C12 | 47.7 (3) |
| C15—C1—C2—C3 | −88.1 (3) | C9—C10—C11—C22 | −72.5 (3) |
| C7—C2—C3—O19 | 170.0 (2) | C23—C11—C12—C13 | −176.3 (2) |
| C1—C2—C3—O19 | −6.3 (4) | C22—C11—C12—C13 | 63.7 (3) |
| C7—C2—C3—C4 | −5.4 (4) | C10—C11—C12—C13 | −56.7 (3) |
| C1—C2—C3—C4 | 178.2 (2) | C11—C12—C13—O24 | −144.6 (3) |
| O19—C3—C4—C5 | 156.8 (2) | C11—C12—C13—C14 | 36.9 (3) |
| C2—C3—C4—C5 | −27.7 (3) | O8—C9—C14—C13 | 178.5 (2) |
| C3—C4—C5—C21 | 173.5 (2) | C10—C9—C14—C13 | −1.7 (4) |
| C3—C4—C5—C20 | −67.3 (3) | O8—C9—C14—C1 | −5.9 (4) |
| C3—C4—C5—C6 | 54.3 (3) | C10—C9—C14—C1 | 173.9 (2) |
| C4—C5—C6—C7 | −49.8 (3) | O24—C13—C14—C9 | 174.9 (3) |
| C21—C5—C6—C7 | −168.9 (2) | C12—C13—C14—C9 | −6.5 (4) |
| C20—C5—C6—C7 | 70.8 (3) | O24—C13—C14—C1 | −0.8 (4) |
| C3—C2—C7—O8 | −168.6 (2) | C12—C13—C14—C1 | 177.8 (2) |
| C1—C2—C7—O8 | 7.7 (4) | C2—C1—C14—C9 | 24.4 (3) |
| C3—C2—C7—C6 | 9.2 (4) | C15—C1—C14—C9 | −96.7 (3) |
| C1—C2—C7—C6 | −174.5 (2) | C2—C1—C14—C13 | −160.0 (2) |
| C5—C6—C7—C2 | 20.1 (4) | C15—C1—C14—C13 | 78.9 (3) |
| C5—C6—C7—O8 | −161.9 (2) | C14—C1—C15—O16 | 7.8 (4) |
| C2—C7—O8—C9 | 13.7 (3) | C2—C1—C15—O16 | −112.2 (3) |
| C6—C7—O8—C9 | −164.4 (2) | C14—C1—C15—O17 | −173.5 (2) |
| C7—O8—C9—C14 | −14.7 (4) | C2—C1—C15—O17 | 66.4 (3) |
| C7—O8—C9—C10 | 165.5 (2) | O16—C15—O17—C18 | 1.4 (5) |
| C14—C9—C10—C11 | −20.5 (4) | C1—C15—O17—C18 | −177.2 (3) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C6—H6B···O24i | 0.99 | 2.50 | 3.324 (4) | 140 |
| C18—H18A···O17ii | 0.98 | 2.53 | 3.451 (5) | 156 |
| C18—H18A···O19ii | 0.98 | 2.41 | 3.093 (4) | 126 |
Symmetry codes: (i) x, −y+1/2, z+1/2; (ii) −x+2, −y+1, −z+1.
References
- Burla, M. C., Caliandro, R., Camalli, M., Carrozzini, B., Cascarano, G. L., De Caro, L., Giacovazzo, C., Polidori, G. & Spagna, R. (2005). J. Appl. Cryst. 38, 381–388.
- Gross, H. & Freiberg, J. (1966). Chem. Ber. 99, 3260–3267.
- Gustafsson, C. (1948). Suomen Kemistilehti B, 21, 3–5.
- Rohr, K. & Mahrwald, R. (2009). Bioorg. Med. Chem. Lett. 19, 3949–3951. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Spek, A. L. (2020). Acta Cryst. E76, 1–11. [DOI] [PMC free article] [PubMed]
- Steiner, T. (1996). Crystallogr. Rev. 6, 1–51.
- Stoe & Cie (2019). X-RED and X-AREA. Stoe & Cie, Darmstadt, Germany.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2414314620010184/gg4005sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314620010184/gg4005Isup2.hkl
schematic diagram of the reaction. DOI: 10.1107/S2414314620010184/gg4005sup3.tif
Supporting information file. DOI: 10.1107/S2414314620010184/gg4005Isup4.cml
CCDC reference: 2018468
Additional supporting information: crystallographic information; 3D view; checkCIF report




