The central PdII ion of the complex cation has an N3O square-planar coordination sphere defined by the three N atoms of the tridentate 2,2′:6′,2′′-terpyridine ligand and one O atom from the NO3 − anion.
Keywords: crystal structure; palladium(II) complex; square-planar structure; 2,2′:6′,2′′-terpyridine; tridentate ligand; nitrate salt
Abstract
The title complex, [Pd(NO3)(C15H11N3)]NO3, comprises a cationic PdII complex and a nitrate anion. In the complex, the PdII cation is four-coordinated in a distorted square-planar coordination geometry defined by the three N atoms of the tridentate 2,2′:6′,2′′-terpyridine ligand and one O atom from the NO3
− anion. In the crystal, the complex molecules are stacked in columns along the a axis being connected by π–π stacking [closest inter-centroid separation between pyridyl rings = 3.878 (3) Å]. The connections between columns and anions to sustain a three-dimensional architecture are C—H⋯O hydrogen bonds.
Structure description
With reference to the title complex, [Pd(terpy)(NO3)](NO3) (terpy = 2,2′:6′,26′6′-terpyridine), the crystal structures of related PdII complexes [Pd(terpy)(pyridine)](ClO4)2 (Bugarčić et al., 2004 ▸), [Pd(terpy)(NO3)](NTf2) [NTf2 = bis(trifluoromethylsulfonyl)amide anion; Illner et al., 2009 ▸) and [Pd2(terpy)2(NO3)]2(PF6)6·CH3CN (Mei et al., 2007 ▸) have been determined previously.
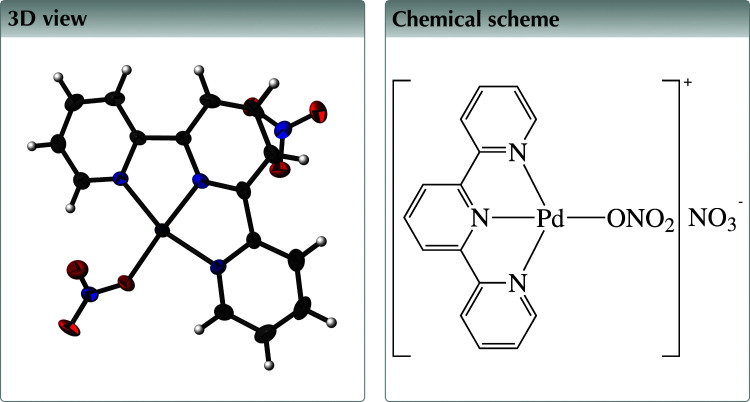
The title complex comprises a cationic PdII complex [Pd(terpy)(NO3)]+ and an NO3 − anion (Fig. 1 ▸). In the complex, the central PdII cation is four-coordinated in a distorted square-planar coordination geometry defined by the pyridyl N1, N2 and N3 atoms derived from the tridentate terpy ligand and the O1 atom from the nitrato ligand. The tight N—Pd—N chelating angles of <N1—Pd1—N2 = 81.26 (17)° and <N2—Pd1—N8 = 81.03 (16)° contribute to the distortion of the square-plane. The Pd—N [1.917 (4) to 2.030 (4) Å] and Pd—O [2.028 (3) Å] bond lengths are close. The pyridine rings of the terpy ligand are located approximately parallel to the least-squares plane of the PdN3O unit [maximum deviation = 0.023 (2) Å], with dihedral angles of 1.4 (2)° (ring N1/C1–C5), 3.1 (2)° (ring N2/C6–C10) and 3.0 (2)° (ring N3/C11–C15). In the crystal (Fig. 2 ▸), the complex molecules are stacked in columns along the a axis. Within the columns, numerous intermolecular π–π interactions between adjacent pyridine rings are present. For Cg1 (the centroid of ring N2/C6–C10) and Cg2i [the centroid of ring N3/C11–C15; symmetry code: (i) x + 1, y, z], the centroid-centroid distance is 3.878 (3) Å and the dihedral angle between the ring planes is 3.2 (3)° (Spek, 2020 ▸). The complex cations and anions form intermolecular C—H⋯O hydrogen bonds (Table 1 ▸) to stabilize the three-dimensional packing.
Figure 1.
The molecular structure of the title complex showing the atom labelling and displacement ellipsoids drawn at the 50% probability level for non-H atoms.
Figure 2.
A view of the packing in the crystal of the title complex, viewed approximately along the a axis. Hydrogen-bonding interactions are drawn as dashed lines.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C3—H3⋯O2i | 0.94 | 2.55 | 3.419 (7) | 153 |
| C4—H4⋯O6ii | 0.94 | 2.37 | 3.303 (7) | 172 |
| C7—H7⋯O6ii | 0.94 | 2.30 | 3.231 (6) | 171 |
| C8—H8⋯O5iii | 0.94 | 2.43 | 3.088 (6) | 127 |
| C9—H9⋯O6iv | 0.94 | 2.35 | 3.254 (6) | 160 |
| C13—H13⋯O5v | 0.94 | 2.46 | 3.402 (7) | 176 |
| C15—H15⋯O3vi | 0.94 | 2.38 | 3.280 (7) | 161 |
Symmetry codes: (i)
 ; (ii)
; (ii)
 ; (iii)
; (iii)
 ; (iv)
; (iv)
 ; (v)
; (v)
 ; (vi)
; (vi)
 .
.
Synthesis and crystallization
To a solution of Pd(NO3)2·2H2O (0.1320 g, 0.495 mmol) in acetone (30 ml) was added 2,2′:6′,2′′-terpyridine (0.1179 g, 0.505 mmol) followed by stirring for 3 h at room temperature. The formed precipitate was separated by filtration, washed with acetone and dried at 323 K to give a light-yellow powder (0.2123 g). Yellow crystals of the product suitable for X-ray analysis were obtained by slow evaporation of its CH3NO2 solution at room temperature.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | [Pd(NO3)(C15H11N3)]NO3 |
| M r | 463.69 |
| Crystal system, space group | Orthorhombic, P n a21 |
| Temperature (K) | 223 |
| a, b, c (Å) | 6.2190 (2), 33.9728 (15), 7.4819 (3) |
| V (Å3) | 1580.75 (11) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 1.22 |
| Crystal size (mm) | 0.21 × 0.14 × 0.06 |
| Data collection | |
| Diffractometer | PHOTON 100 CMOS detector |
| Absorption correction | Multi-scan (SADABS; Bruker, 2016 ▸) |
| T min, T max | 0.688, 0.745 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 41749, 3116, 2745 |
| R int | 0.084 |
| (sin θ/λ)max (Å−1) | 0.618 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.027, 0.048, 1.09 |
| No. of reflections | 3116 |
| No. of parameters | 244 |
| No. of restraints | 1 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.34, −0.43 |
| Absolute structure | Flack x determined using 1141 quotients [(I +)−(I −)]/[(I +)+(I −)] Parsons et al. (2013 ▸). |
| Absolute structure parameter | 0.006 (16) |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2414314621000857/tk4067sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314621000857/tk4067Isup2.hkl
CCDC reference: 2058389
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The author thanks the KBSI, Seoul Center, for the X-ray data collection.
full crystallographic data
Crystal data
| [Pd(C15H11N3)(NO3)]NO3 | Dx = 1.948 Mg m−3 |
| Mr = 463.69 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pna21 | Cell parameters from 9942 reflections |
| a = 6.2190 (2) Å | θ = 2.4–27.7° |
| b = 33.9728 (15) Å | µ = 1.22 mm−1 |
| c = 7.4819 (3) Å | T = 223 K |
| V = 1580.75 (11) Å3 | Plate, yellow |
| Z = 4 | 0.21 × 0.14 × 0.06 mm |
| F(000) = 920 |
Data collection
| PHOTON 100 CMOS detector diffractometer | 2745 reflections with I > 2σ(I) |
| Radiation source: sealed tube | Rint = 0.084 |
| φ and ω scans | θmax = 26.1°, θmin = 2.4° |
| Absorption correction: multi-scan (SADABS; Bruker, 2016) | h = −7→7 |
| Tmin = 0.688, Tmax = 0.745 | k = −41→42 |
| 41749 measured reflections | l = −9→9 |
| 3116 independent reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.027 | H-atom parameters constrained |
| wR(F2) = 0.048 | w = 1/[σ2(Fo2) + (0.0152P)2 + 0.8349P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.09 | (Δ/σ)max = 0.001 |
| 3116 reflections | Δρmax = 0.34 e Å−3 |
| 244 parameters | Δρmin = −0.43 e Å−3 |
| 1 restraint | Absolute structure: Flack x determined using 1141 quotients [(I+)-(I-)]/[(I+)+(I-)] Parsons et al. (2013). |
| Primary atom site location: structure-invariant direct methods | Absolute structure parameter: 0.006 (16) |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Hydrogen atoms on C atoms were positioned geometrically and allowed to ride on their respective parent atoms: C—H = 0.94 Å and Uiso(H) = 1.2Ueq(C). The Flack parameter = 0.006 (16) after the final cycles of refinement. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Pd1 | 0.22807 (4) | 0.15784 (2) | 0.48682 (9) | 0.02233 (10) | |
| O1 | 0.1702 (6) | 0.21529 (10) | 0.4313 (4) | 0.0343 (10) | |
| O2 | 0.2291 (7) | 0.19950 (12) | 0.1521 (6) | 0.0463 (10) | |
| O3 | 0.1758 (7) | 0.26017 (11) | 0.2256 (6) | 0.0540 (13) | |
| N1 | 0.4977 (6) | 0.16769 (12) | 0.6321 (6) | 0.0261 (10) | |
| N2 | 0.2920 (6) | 0.10452 (11) | 0.5520 (5) | 0.0220 (9) | |
| N3 | −0.0259 (6) | 0.13077 (12) | 0.3685 (5) | 0.0214 (9) | |
| N4 | 0.1927 (7) | 0.22538 (14) | 0.2626 (7) | 0.0364 (12) | |
| C1 | 0.5913 (9) | 0.20210 (16) | 0.6668 (8) | 0.0344 (13) | |
| H1 | 0.5322 | 0.2252 | 0.6180 | 0.041* | |
| C2 | 0.7726 (9) | 0.2048 (2) | 0.7722 (8) | 0.0450 (16) | |
| H2 | 0.8307 | 0.2295 | 0.8015 | 0.054* | |
| C3 | 0.8672 (9) | 0.17072 (19) | 0.8340 (8) | 0.0415 (15) | |
| H3 | 0.9939 | 0.1719 | 0.9023 | 0.050* | |
| C4 | 0.7740 (9) | 0.13467 (19) | 0.7944 (8) | 0.0323 (13) | |
| H4 | 0.8378 | 0.1112 | 0.8343 | 0.039* | |
| C5 | 0.5864 (9) | 0.13370 (17) | 0.6959 (7) | 0.0232 (12) | |
| C6 | 0.4710 (8) | 0.09759 (15) | 0.6481 (6) | 0.0232 (11) | |
| C7 | 0.5267 (8) | 0.05923 (15) | 0.6877 (7) | 0.0293 (13) | |
| H7 | 0.6493 | 0.0536 | 0.7566 | 0.035* | |
| C8 | 0.3979 (8) | 0.02930 (15) | 0.6235 (7) | 0.0308 (13) | |
| H8 | 0.4354 | 0.0031 | 0.6481 | 0.037* | |
| C9 | 0.2163 (7) | 0.03682 (14) | 0.5247 (7) | 0.0282 (16) | |
| H9 | 0.1295 | 0.0161 | 0.4828 | 0.034* | |
| C10 | 0.1642 (6) | 0.07567 (11) | 0.4881 (12) | 0.0219 (8) | |
| C11 | −0.0190 (8) | 0.09089 (14) | 0.3876 (7) | 0.0222 (11) | |
| C12 | −0.1803 (8) | 0.06726 (16) | 0.3173 (7) | 0.0285 (12) | |
| H12 | −0.1745 | 0.0398 | 0.3300 | 0.034* | |
| C13 | −0.3493 (8) | 0.08484 (17) | 0.2285 (7) | 0.0302 (13) | |
| H13 | −0.4597 | 0.0694 | 0.1793 | 0.036* | |
| C14 | −0.3552 (9) | 0.12527 (18) | 0.2122 (8) | 0.0266 (14) | |
| H14 | −0.4706 | 0.1376 | 0.1535 | 0.032* | |
| C15 | −0.1895 (8) | 0.14736 (16) | 0.2831 (8) | 0.0269 (12) | |
| H15 | −0.1927 | 0.1749 | 0.2705 | 0.032* | |
| O4 | 0.1900 (5) | 0.09192 (10) | 1.0047 (10) | 0.0396 (10) | |
| O5 | 0.2336 (6) | 0.03043 (12) | 1.0689 (6) | 0.0505 (11) | |
| O6 | −0.0235 (6) | 0.04770 (11) | 0.8974 (5) | 0.0425 (10) | |
| N5 | 0.1345 (5) | 0.05686 (11) | 0.9909 (9) | 0.0287 (8) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Pd1 | 0.02599 (16) | 0.01582 (15) | 0.02517 (16) | 0.00131 (14) | −0.0033 (4) | −0.0007 (3) |
| O1 | 0.049 (2) | 0.0166 (18) | 0.037 (3) | 0.0048 (15) | −0.0099 (16) | 0.0013 (15) |
| O2 | 0.064 (3) | 0.036 (2) | 0.040 (2) | −0.010 (2) | −0.010 (2) | 0.000 (2) |
| O3 | 0.059 (3) | 0.020 (2) | 0.083 (3) | −0.003 (2) | −0.020 (2) | 0.021 (2) |
| N1 | 0.030 (2) | 0.025 (3) | 0.023 (2) | −0.0059 (18) | −0.0040 (19) | −0.0003 (19) |
| N2 | 0.0229 (19) | 0.019 (2) | 0.024 (2) | 0.0028 (17) | 0.0051 (17) | 0.0003 (16) |
| N3 | 0.025 (2) | 0.019 (2) | 0.020 (2) | −0.0004 (17) | 0.0018 (18) | −0.0021 (19) |
| N4 | 0.030 (2) | 0.026 (3) | 0.054 (3) | −0.007 (2) | −0.016 (2) | 0.011 (3) |
| C1 | 0.042 (3) | 0.024 (3) | 0.037 (3) | −0.002 (3) | −0.004 (3) | 0.000 (3) |
| C2 | 0.051 (4) | 0.047 (4) | 0.038 (3) | −0.021 (3) | −0.008 (3) | −0.002 (3) |
| C3 | 0.035 (3) | 0.063 (4) | 0.027 (3) | −0.014 (3) | −0.009 (3) | 0.003 (3) |
| C4 | 0.029 (3) | 0.044 (4) | 0.025 (3) | 0.003 (3) | 0.001 (3) | 0.006 (3) |
| C5 | 0.026 (3) | 0.026 (3) | 0.018 (3) | 0.000 (2) | 0.000 (2) | 0.003 (2) |
| C6 | 0.025 (3) | 0.027 (3) | 0.018 (3) | 0.003 (2) | 0.003 (2) | 0.005 (2) |
| C7 | 0.027 (3) | 0.029 (3) | 0.031 (3) | 0.003 (2) | 0.002 (2) | 0.009 (3) |
| C8 | 0.034 (3) | 0.020 (3) | 0.039 (3) | 0.008 (2) | 0.007 (3) | 0.012 (2) |
| C9 | 0.029 (2) | 0.019 (2) | 0.037 (5) | −0.0034 (19) | 0.008 (2) | 0.000 (2) |
| C10 | 0.0234 (19) | 0.018 (2) | 0.024 (2) | 0.0004 (15) | 0.000 (5) | −0.001 (4) |
| C11 | 0.026 (3) | 0.018 (3) | 0.022 (3) | 0.002 (2) | 0.006 (2) | 0.000 (2) |
| C12 | 0.028 (3) | 0.024 (3) | 0.033 (3) | −0.006 (2) | 0.007 (2) | −0.004 (2) |
| C13 | 0.024 (3) | 0.037 (4) | 0.030 (3) | −0.004 (2) | 0.003 (2) | −0.009 (3) |
| C14 | 0.021 (3) | 0.034 (4) | 0.024 (4) | 0.006 (3) | 0.004 (3) | 0.001 (3) |
| C15 | 0.029 (3) | 0.024 (3) | 0.027 (3) | 0.007 (2) | 0.003 (3) | −0.002 (2) |
| O4 | 0.0415 (17) | 0.0291 (18) | 0.048 (3) | −0.0045 (14) | −0.002 (3) | −0.003 (3) |
| O5 | 0.041 (2) | 0.042 (3) | 0.068 (3) | 0.011 (2) | −0.020 (2) | 0.013 (2) |
| O6 | 0.039 (2) | 0.039 (2) | 0.050 (2) | −0.0026 (18) | −0.0192 (19) | 0.002 (2) |
| N5 | 0.0249 (17) | 0.035 (2) | 0.027 (2) | 0.0011 (16) | 0.004 (4) | −0.007 (4) |
Geometric parameters (Å, º)
| Pd1—N2 | 1.917 (4) | C5—C6 | 1.466 (8) |
| Pd1—N1 | 2.026 (4) | C6—C7 | 1.381 (7) |
| Pd1—O1 | 2.028 (3) | C7—C8 | 1.380 (7) |
| Pd1—N3 | 2.030 (4) | C7—H7 | 0.9400 |
| O1—N4 | 1.315 (6) | C8—C9 | 1.374 (7) |
| O2—N4 | 1.228 (6) | C8—H8 | 0.9400 |
| O3—N4 | 1.218 (6) | C9—C10 | 1.386 (6) |
| N1—C1 | 1.332 (6) | C9—H9 | 0.9400 |
| N1—C5 | 1.366 (7) | C10—C11 | 1.460 (7) |
| N2—C6 | 1.346 (6) | C11—C12 | 1.388 (7) |
| N2—C10 | 1.349 (6) | C12—C13 | 1.379 (7) |
| N3—C15 | 1.328 (6) | C12—H12 | 0.9400 |
| N3—C11 | 1.363 (6) | C13—C14 | 1.379 (8) |
| C1—C2 | 1.379 (7) | C13—H13 | 0.9400 |
| C1—H1 | 0.9400 | C14—C15 | 1.381 (8) |
| C2—C3 | 1.379 (9) | C14—H14 | 0.9400 |
| C2—H2 | 0.9400 | C15—H15 | 0.9400 |
| C3—C4 | 1.387 (8) | O4—N5 | 1.245 (5) |
| C3—H3 | 0.9400 | O5—N5 | 1.236 (6) |
| C4—C5 | 1.380 (7) | O6—N5 | 1.245 (5) |
| C4—H4 | 0.9400 | ||
| N2—Pd1—N1 | 81.26 (17) | N2—C6—C7 | 119.1 (5) |
| N2—Pd1—O1 | 176.54 (15) | N2—C6—C5 | 112.9 (4) |
| N1—Pd1—O1 | 95.62 (15) | C7—C6—C5 | 128.0 (5) |
| N2—Pd1—N3 | 81.03 (16) | C8—C7—C6 | 118.4 (5) |
| N1—Pd1—N3 | 162.23 (16) | C8—C7—H7 | 120.8 |
| O1—Pd1—N3 | 102.04 (15) | C6—C7—H7 | 120.8 |
| N4—O1—Pd1 | 115.4 (3) | C9—C8—C7 | 121.8 (5) |
| C1—N1—C5 | 119.8 (5) | C9—C8—H8 | 119.1 |
| C1—N1—Pd1 | 127.7 (4) | C7—C8—H8 | 119.1 |
| C5—N1—Pd1 | 112.5 (4) | C8—C9—C10 | 118.4 (5) |
| C6—N2—C10 | 123.3 (4) | C8—C9—H9 | 120.8 |
| C6—N2—Pd1 | 118.2 (3) | C10—C9—H9 | 120.8 |
| C10—N2—Pd1 | 118.3 (3) | N2—C10—C9 | 118.9 (5) |
| C15—N3—C11 | 119.8 (4) | N2—C10—C11 | 112.6 (4) |
| C15—N3—Pd1 | 127.9 (3) | C9—C10—C11 | 128.4 (4) |
| C11—N3—Pd1 | 112.4 (3) | N3—C11—C12 | 120.8 (5) |
| O3—N4—O2 | 123.9 (5) | N3—C11—C10 | 115.5 (4) |
| O3—N4—O1 | 117.5 (5) | C12—C11—C10 | 123.7 (4) |
| O2—N4—O1 | 118.6 (4) | C13—C12—C11 | 118.9 (5) |
| N1—C1—C2 | 121.9 (6) | C13—C12—H12 | 120.6 |
| N1—C1—H1 | 119.1 | C11—C12—H12 | 120.6 |
| C2—C1—H1 | 119.1 | C12—C13—C14 | 119.6 (5) |
| C1—C2—C3 | 118.9 (6) | C12—C13—H13 | 120.2 |
| C1—C2—H2 | 120.5 | C14—C13—H13 | 120.2 |
| C3—C2—H2 | 120.5 | C13—C14—C15 | 119.1 (5) |
| C2—C3—C4 | 119.5 (5) | C13—C14—H14 | 120.4 |
| C2—C3—H3 | 120.3 | C15—C14—H14 | 120.4 |
| C4—C3—H3 | 120.3 | N3—C15—C14 | 121.8 (5) |
| C5—C4—C3 | 119.2 (6) | N3—C15—H15 | 119.1 |
| C5—C4—H4 | 120.4 | C14—C15—H15 | 119.1 |
| C3—C4—H4 | 120.4 | O5—N5—O4 | 121.2 (5) |
| N1—C5—C4 | 120.5 (5) | O5—N5—O6 | 118.5 (4) |
| N1—C5—C6 | 115.1 (5) | O4—N5—O6 | 120.3 (5) |
| C4—C5—C6 | 124.3 (5) | ||
| Pd1—O1—N4—O3 | −174.4 (3) | C6—C7—C8—C9 | −0.9 (8) |
| Pd1—O1—N4—O2 | 5.6 (6) | C7—C8—C9—C10 | 0.6 (8) |
| C5—N1—C1—C2 | −2.6 (8) | C6—N2—C10—C9 | 1.0 (9) |
| Pd1—N1—C1—C2 | 178.2 (4) | Pd1—N2—C10—C9 | 175.9 (5) |
| N1—C1—C2—C3 | 4.2 (9) | C6—N2—C10—C11 | −179.7 (4) |
| C1—C2—C3—C4 | −2.4 (9) | Pd1—N2—C10—C11 | −4.8 (7) |
| C2—C3—C4—C5 | −0.9 (8) | C8—C9—C10—N2 | −0.6 (9) |
| C1—N1—C5—C4 | −0.8 (8) | C8—C9—C10—C11 | −179.7 (6) |
| Pd1—N1—C5—C4 | 178.5 (4) | C15—N3—C11—C12 | 0.5 (7) |
| C1—N1—C5—C6 | −179.4 (5) | Pd1—N3—C11—C12 | 179.3 (4) |
| Pd1—N1—C5—C6 | −0.2 (5) | C15—N3—C11—C10 | −178.3 (5) |
| C3—C4—C5—N1 | 2.5 (8) | Pd1—N3—C11—C10 | 0.5 (6) |
| C3—C4—C5—C6 | −179.0 (5) | N2—C10—C11—N3 | 2.6 (8) |
| C10—N2—C6—C7 | −1.4 (8) | C9—C10—C11—N3 | −178.2 (6) |
| Pd1—N2—C6—C7 | −176.3 (4) | N2—C10—C11—C12 | −176.1 (5) |
| C10—N2—C6—C5 | 177.7 (5) | C9—C10—C11—C12 | 3.0 (11) |
| Pd1—N2—C6—C5 | 2.8 (5) | N3—C11—C12—C13 | −0.4 (8) |
| N1—C5—C6—N2 | −1.6 (6) | C10—C11—C12—C13 | 178.3 (5) |
| C4—C5—C6—N2 | 179.8 (5) | C11—C12—C13—C14 | −0.3 (8) |
| N1—C5—C6—C7 | 177.4 (5) | C12—C13—C14—C15 | 1.0 (8) |
| C4—C5—C6—C7 | −1.2 (8) | C11—N3—C15—C14 | 0.1 (8) |
| N2—C6—C7—C8 | 1.3 (7) | Pd1—N3—C15—C14 | −178.4 (4) |
| C5—C6—C7—C8 | −177.7 (5) | C13—C14—C15—N3 | −0.9 (9) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C3—H3···O2i | 0.94 | 2.55 | 3.419 (7) | 153 |
| C4—H4···O6ii | 0.94 | 2.37 | 3.303 (7) | 172 |
| C7—H7···O6ii | 0.94 | 2.30 | 3.231 (6) | 171 |
| C8—H8···O5iii | 0.94 | 2.43 | 3.088 (6) | 127 |
| C9—H9···O6iv | 0.94 | 2.35 | 3.254 (6) | 160 |
| C13—H13···O5v | 0.94 | 2.46 | 3.402 (7) | 176 |
| C15—H15···O3vi | 0.94 | 2.38 | 3.280 (7) | 161 |
Symmetry codes: (i) x+1, y, z+1; (ii) x+1, y, z; (iii) −x+1, −y, z−1/2; (iv) −x, −y, z−1/2; (v) x−1, y, z−1; (vi) x−1/2, −y+1/2, z.
Funding Statement
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (grant No. 2018R1D1A1B07050550).
References
- Bruker (2016). APEX2, SAINT and SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Bugarčić, D., Petrović, B. & Zangrando, E. (2004). Inorg. Chim. Acta, 357, 2650–2656.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Illner, P., Puchta, R., Heinemann, F. W. & van Eldik, R. (2009). Dalton Trans. pp. 2795–2801. [DOI] [PubMed]
- Mei, G.-Q., Huang, K.-L. & Huang, H.-P. (2007). Acta Cryst. E63, m2510–m2511.
- Parsons, S., Flack, H. D. & Wagner, T. (2013). Acta Cryst. B69, 249–259. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Spek, A. L. (2020). Acta Cryst. E76, 1–11. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2414314621000857/tk4067sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314621000857/tk4067Isup2.hkl
CCDC reference: 2058389
Additional supporting information: crystallographic information; 3D view; checkCIF report




