Abstract
Enterococcus faecalis induces the synthesis of at least 42 proteins during 24 h of glucose starvation. Because of its induction during carbohydrate and complete starvation (incubation in tap water) and CdCl2 and bile salts stresses, one of these proteins (Gls24) was qualified as a “general stress protein” and was analyzed at the molecular level. Its corresponding gene, gls24, seems to be the penultimate gene of an operon composed, altogether, of six open reading frames (ORFs). The ORF preceding gls24 (orf4) showed very strong identity with gls24. The deduced polypeptides of these two genes showed similarity with a 20-kDa hypothetical protein from Lactococcus lactis and an alkaline stress protein from Staphylococcus aureus with no previously known biological significance. Data from the operon sequence and Northern analysis led to the conclusions that (i) gls24 possesses its own promoter which is especially induced at the onset of starvation and (ii) the operon promoter is stress inducible in exponential-phase cells. A mutation in the gls24 gene led to a severe reduction of growth rate and reduction of survival against 0.3% bile salts in the 24-h-starved cells compared to the wild-type strain. Moreover, the chain length of the mutant is significantly reduced during growth. These results argue strongly for a role of the protein Gls24 and/or GlsB in morphological changes and in stress tolerance in E. faecalis. Comparison of two-dimensional protein gels from wild-type cells with those from gls24 mutant cells revealed a pleiotropic effect of the mutation on gene expression. At least nine proteins were present in larger amounts in the mutant. For six of them, the corresponding N-terminal microsequence has been obtained. Three of these sequences map in genes coding for l-lactate dehydrogenase, lipoamide dehydrogenase, and pyruvate decarboxylase, all involved in pyruvate metabolism.
In their natural environment, microbial cells have to sense and to cope with different growth-restricting conditions, like chemical stresses and nutrient deprivation. Therefore, cells develop strategies for survival and resistance against multiple stresses. Sophisticated control mechanisms ensure that selected genes are expressed under the right conditions and at the right time. This expression is regulated through control of transcriptional initiation by alternative sigma factors in some gram-positive and -negative bacteria (20, 26, 37, 39). As described in several reviews, this process triggers dramatic changes in cellular physiology and even in morphology (43). Some bacteria, like Bacillus species, form endospores to survive nutrient-poor conditions. In gram-negative bacteria, some starvation-induced genes are known to be involved in the acquisition of a multiresistant state (i.e., katE, treA, xthA, and dps) (2, 9, 47, 49), in morphological changes (i.e., bolA) (1), or in glycogen synthesis (i.e., glgA and glgS) (31, 55). The situation seems comparable for Bacillus subtilis in which many stress-implicated genes are under the control of the alternative sigma factor, ςB. Furthermore, some genes regulated by ςS in Escherichia coli are under ςB control of in B. subtilis (i.e., katE, dps, and opuE) (29).
A ςB homologous gene has been also identified in other nonsporulating gram-positive bacteria (56, 57). Surprisingly, in the almost-finished Enterococcus faecalis chromosome sequence (The Institute for Genomic Research) as well as in the closely related species Lactococcus lactis, whose genome has been entirely sequenced, no ςS- or ςB-like sigma factors have been found (8). This observation raises the question of what is/are the mechanism(s) which is/are involved in gene regulation and induction during stress responses in these bacteria. Derré et al. recently showed that clpP and clpC, which are well-known stress- and starvation-inducible genes in B. subtilis, are under the control of the novel regulator protein CtsR in E. faecalis (13). However, little information is available about the starvation response in gram-positive, non-spore-forming bacteria such E. faecalis. This last is a resident of the intestinal tract of humans and animals. This bacterium can cause serious diseases and is one of the main causes of hospital-based infections. This hardy organism resists many kinds of stresses (17–19) and is used as a major indicator of the hygienic quality of food, milk, and drinking water. We have previously identified 42 glucose starvation proteins in E. faecalis. Four temporal classes of proteins were defined with respect to their enhanced synthesis after glucose exhaustion (23). Proteins from the two early classes seem to be the most important for long-term survival and acquisition of multiresistance towards several lethal treatments in E. faecalis (22, 23). Comparison of two-dimensional (2D) protein gels led us to discover that, in E. faecalis strain JH2-2 and ATCC 19433, the intensity of the spot corresponding to protein Gls24 increased during glucose and complete starvation and during different stress treatments. Indeed, compared to its level during growth at 37°C, its abundance increased 3- and 2.1-fold after 12 h of glucose starvation and 2 weeks of total starvation (tap water), respectively (28). Moreover, CdCl2 and bile salt stresses induced its level between two- and sixfold (38). Thus, Gls24 can be considered as a general stress protein. Based on the N-terminal sequence of this a priori important glucose starvation protein, we have identified the corresponding gene, gls24. We show here that gls24 is the penultimate gene of a six-gene operon of hitherto unknown function. In this study, we report the sequence and transcriptional analysis of this operon under stress and starvation conditions. The phenotype of the mutant and its 2D protein pattern are compared with those of wild-type cells.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The E. faecalis strain used in this study for chromosomal DNA and RNA preparation, survival, and protein analysis was JH2-2 (32, 58). E. coli XL1Blue (Stratagene, La Jolla, Calif.) was used as a host for the construction of subgenomic libraries. Plasmids pBluescript KS(+) (Stratagene) and pUCB300 (21) were used as cloning and integrational vectors, respectively. Cultures of E. faecalis were grown at 37°C without shaking in 20-ml glass tubes containing 10 ml of semisynthetic medium (Bacto Folic AOAC Medium; Difco, Detroit, Mich.) supplemented with glucose. Preliminary growth yield studies using different concentrations of glucose have led to the choice of 0.15% (wt/vol) glucose to ensure that exhaustion of glucose triggered transition to the stationary phase (22). For plate count, a sample was taken, immediately diluted in 0.9% NaCl, and poured in M17 (51) agar (1.5% [wt/vol]) (Difco) supplemented with 0.5% (wt/vol) glucose. Plates were incubated at 37°C for 48 h. E. coli strains were cultivated under vigorous agitation at 37°C in 2TY medium (48) with ampicillin (100 μg/ml) when required.
Challenge conditions.
After centrifugation, control cells (exponential-growth-phase cells) and 24-h-starved cells were resuspended in fresh semisynthetic medium. Ten milliliters of each culture received one of the following treatments: 62°C, 20 mM H2O2, pH 3.7 (adjusted with lactic acid), pH 11.9 (adjusted with NaOH), 17% (vol/vol) ethanol, 0.3% bile salts, and 50 mg of CdCl2 per ml. After 0, 15, and 30 min, a sample was taken for plate count. Survival at any given time point was determined as the ratio of CFU after treatment to the number of CFU at the zero time point.
Analysis of mRNA transcription by Northern blotting.
Total RNA of E. faecalis was isolated from exponentially growing, stationary-phase, or stressed cells by using the Rneasy Midi Kit (QIAGEN, Santa Clarica, Calif.). Northern blots of exactly 10 μg of electrophoresed RNA were prepared by using Hybond-N+ membranes and standard procedures (48). Sizes of transcripts were estimated by comparing band mobility of standards in an RNA ladder (0.56 to 9.4 kb) (Amersham International, Little Chalfont, United Kingdom). Membrane-bound nucleic acids were hybridized at a temperature 5°C below the melting temperature with 32P-labeled probes that were prepared by using terminal deoxynucleotidyl transferase (Amersham International). Membranes were then exposed to a storage phosphor screen (Packard Instrument Company, Canberra, Australia) for 5 h.
Mapping the transcriptional start sites.
Primers complementary to the 5′ coding region of orf1 (5′-GATCTTGTTGCGGAAATCCATCACTAT-3′) and gls24 (5′-CTGGTGTGTGTGGTCCGTTTCCTG-3′) were labeled with 10 U of polynucleotide kinase (Boehringer Mannheim) and 2 μCi of [γ32P]ATP (10 mCi/ml) (Amersham International). Total RNA was isolated from E. faecalis JH2-2 at the onset of starvation or after 30 min of CdCl2 (50 μg/ml) stress. The labeled primers were mixed to 10 μg of RNA in 14 μl of the reverse transcriptase buffer containing 40 U of RNase inhibitor (Boehringer Mannheim). After heating at 65°C for 5 min, annealing was obtained by a slow decrease of the temperature until 25°C. The extension reaction was then performed in a 20-μl final volume with 50 U of avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim) and 0.5 mM of each deoxynucleoside triphosphate at 42°C for 1 h. After heat denaturation, 2-μl samples were loaded onto a 6% polyacrylamide-urea sequencing gel for electrophoresis.
General molecular methods.
Restriction endonucleases, alkaline phosphatase, and T4 DNA ligase were obtained from Boehringer Mannheim and used according to the manufacturers instructions. PCR was carried out in a reaction volume of 25 μl with 5 μg of chromosomal DNA of E. faecalis by using Ready To Go PCR beads (Pharmacia Biotech). The annealing temperature was 5°C below the melting temperature of primers; 30 cycles were performed, and PCR products were purified by using the QIAquick Kit (QIAGEN) before being cloned into the SmaI site of the vector. E. coli and E. faecalis were transformed by using Gene Pulser Apparatus (Bio-Rad Laboratories, Richmond, Calif.) as described by Dower et al. (16). Plasmids were purified by using QIAprep Miniprep (QIAGEN). DNA and amino acid sequences were analyzed by using the Mac Vector program (Kodak Scientific Imaging Systems), and databases searches were performed with the BLAST program (3). Other standard techniques were carried out as described by Sambrook et al. (48).
Construction of the gls24 insertional mutant.
To construct an insertional mutant with a disruption in the E. faecalis gls24 gene, a 450-bp AcsI/AcsI fragment of the gene gls24 was ligated with the insertional vector pUCB300 which had been digested with EcoRI. The resulting plasmid obtained after transformation of E. coli XL1Blue was used to transform competent cells of E. faecalis JH2-2. Erythromycin-resistant colonies were selected on agar plates containing 15 μg of erythromycin per ml. Integrations were verified by PCR and Southern blot analysis, and the disappearance of protein Gls24 was confirmed by 2D gel electrophoresis.
Electron microscopy.
E. faecalis cells were fixed by the addition of glutaraldehyde to a final concentration of 2% (wt/vol) in 0.1 M sodium cacodylate buffer (SCB) (pH 6.8). After this first fixation, cells were rinsed with 0.1 M SCB and fixed for 1 h in 1% (wt/vol) osmium tetroxide in 0.1 M SCB. The samples were then washed twice with 0.1 M SCB, were dehydrated with acetone, were critical-point dried by the CO2 method of Anderson (4), and were coated with gold. Cells were examined and photographed with a JEOL-JSM 6400F field emission scanning electron microscope operating at 5 kV.
2D protein gel electrophoresis.
Culture conditions were as described before. Culture aliquots of 5 ml were pulse labeled with 250 μCi [35S]methionine-cysteine protein labeling mix (1,000 Ci/mmol) (New England Nuclear Co.). Starved cells were labeled during 24 h of starvation. Bacteria were harvested by centrifugation and were washed twice in cold 0.9% NaCl. Cells, resuspended in 500 μl of buffer I (0.3% sodium dodecyl sulfate, 200 mM dithiothreitol, 28 mM Tris HCl, and 22 mM Tris), were broken by the addition of glass beads (0.1- and 0.25-mm diameter) and by vortexing for 4 min. Unbroken cells were removed by centrifugation, and the supernatant was transferred to another tube. After 5 min at 100°C, samples were chilled on ice and 24 μl of buffer II (24 mM Tris, 476 mM Tris HCl, 50 mM MgCl2, 1 mg of DNase I [Gibco BRL] per ml, and 0.25 mg of RNase A [Sigma Chemical Co.] per ml) were added. The reaction was stopped after 15 min at 4°C by the addition of 4 volumes of ice-cold acetone, and precipitation of proteins was allowed to occur for 20 min on ice. Proteins were collected by centrifugation at 11,000 × g for 10 min and were suspended in 15 μl of buffer at pH 4 to 8 (540 mg of urea per ml, 10 mg of dithiothreitol, 2% [vol/vol] Ampholyte 4-8 [Millipore, Bedford, Mass.], 0.52% [vol/vol] Triton X-100). High-resolution 2D electrophoresis was performed according to the method of O'Farrell (44) with modifications as described by Lopez et al. (41). The first and second dimension were performed by using the Millipore Investigator 2-D electrophoresis system (Millipore). Polyacrylamide gels (14%) without stacking gels were used. Dried gels were exposed to a storage phosphor screen (Packard Instrument Company) for 48 h, and the intensity of synthesis of proteins was determined by the quantification of the corresponding spot using OptiQuant Image Analysis Software (Packard Instrument Company). For the preparative electrophoresis, 50 ml of bacterial culture was used. Protein extraction and the 2D electrophoresis were achieved as above with the following modifications: cells were broken by addition of glass beads and 3 ml of buffer I and by vortexing for 10 min. After 5 min at 100°C, samples were chilled on ice and 145 μl of buffer II was added. At the end, proteins were suspended in 30 μl of buffer, pH 4 to 8. After separation, the gel was transferred onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore) by electroblotting (MilliBlot-Graphite Electroblotter; Millipore) according to the manufacturer's instructions. After Coomassie blue staining of the membrane, interesting spots were cut off, and proteins were sequenced by the Institut für Biochemie (Wien, Austria).
Nucleotide sequence accession number.
The GenBank accession number for the sequence of gls24 and glsB is AJ000042. For the other sequences reported here, preliminary sequence data was obtained from The Institute for Genomic Research website (http://www.tigr.org).
RESULTS
Nucleotide sequence of the E. faecalis gls24 and surrounding genes.
First, the gls24 gene was cloned by the reverse genetic approach. The Gls24 protein was purified by preparative 2D electrophoresis, and the sequence of the first 28 amino acids from the N terminus was determined (23). A 20-bp oligonucleotide probe (5′-AAYGARAARTTYAAYAAYGT-3′) was designed on the basis of the sequence of the first amino acids (XXNEKFNNV) and was hybridized with E. faecalis chromosomal DNA predigested with different restriction enzymes. The entire sequence of the genes gls24 and glsB was obtained by subcloning a 375-bp Sau3AI-Sau3AI and a 1,724-bp AcsI-SspI fragment.
Analysis of the nucleotide sequence revealed an open reading frame (ORF) starting with an ATG codon and encoding a protein of 180 amino acids with a calculated molecular mass of 20.2 kDa and a pI of 5. These data are consistent with the location of Gls24 on the 2D gels (23). The start codon was preceded by a potential ribosome binding site (RBS) sequence GGAGG which was complementary of the 3′ end of the 16S rRNA of E. faecalis CACCUCCAAA (27). The RBS sequence and the start codon were separated by eight nucleotides, which is the optimal spacing (seven to nine nucleotides) determined by Vellanoweth and Rabinowitz (54). It may be noted that an AT-rich inverted repeat (IR) of 18 bp was present 20 bp upstream of the RBS. gls24 was separated from another downstream ORF (named glsB) by 28 bp. An IR located 53 bp downstream of glsB (ΔG = −19 kcal/mol [52]) followed by a stretch of Ts may function as a Rho-independent transcription terminator (12).
By using the E. faecalis genome sequence provided by The Institute for Genomic Research, four ORFs (named orf1, -2, -3, and -4) located upstream of gls24 were identified. No putative transcriptional terminator was present between these four ORFs or between orf4 and gls24. These data suggested that orf1-, -2, -3, -4, gls24, and glsB constitute an operon that was later confirmed (see below). Each ORF is preceded by a GGAGG RBS sequence separated from the start codon by seven or eight nucleotides. The start codon of orf2 seems to be GTG.
Deduced amino acid sequence analysis of E. faecalis gls24 and surrounding genes.
The characteristics and alignment results of the six deduced amino acid sequences with those present in databases are summarized in Table 1. Orf2 and GlsB did not reveal any significant homology with known proteins. All the other genes show homology with hypothetical proteins or polypeptides of unknown functions from gram-positive bacteria. The proteins Orf3 and Orf4 exhibited 38 and 55% identity to likewise adjacent hypothetical 6- and 20-kDa proteins from L. lactis (15) (Table 1). Surprisingly, Orf4 and Gls24 are 71% identical over 122 residues from the central part of the sequences. Otherwise, both genes are 39 and 37% identical to an alkaline shock protein ASP23 identified in Staphylococcus aureus which belong to the SigB regulon (34, 36). Furthermore, the recently completed or nearly completed genome sequences of Streptococcus pyogenes (http://www.genome.ou.edu) and Streptococcus pneumoniae (http://www.tigr.org) revealed proteins with approximately 50% identity to Gls24.
TABLE 1.
Characteristics and homology searches results of the six genes that constitute the operon
| Protein | Size (amino acids [aa]) | Homology searchesa
|
||
|---|---|---|---|---|
| Name (organism, accession number) | % of identity for sequence length | Reference | ||
| Orf1 | 298 | YhxD hypothetical oxidoreductase (B. subtilis, P40398) | 61 over 289 aa | 53 |
| Orf2 | 191 | NHb | ||
| Orf3 | 64 | Putative 6-kDa protein (L. lactis, U23376) | 38 over 62 aa | 15 |
| Orf4 | 171 | Gls24 (E. faecalis, AJ000042) | 71 over 122 aa | This study |
| Putative 20-kDa protein (L. lactis, U23376) | 55 over 152 aa | 15 | ||
| Alkaline shock protein ASP23 (S. aureus, JC2527) | 39 over 117 aa | 36 | ||
| Gls24 | 180 | Orf4 (E. faecalis) | 71 over 122 aa | This study |
| Putative 20-kDa protein (L. lactis, U23376) | 54 over 144 aa | 15 | ||
| Alkaline shock protein ASP23 (S. aureus, JC2527) | 37 over 96 aa | 36 | ||
| GlsB | 64 | NHb | ||
Homology searches were performed with BLAST program (3).
NH, no homology found.
Transcription analysis after stress exposure.
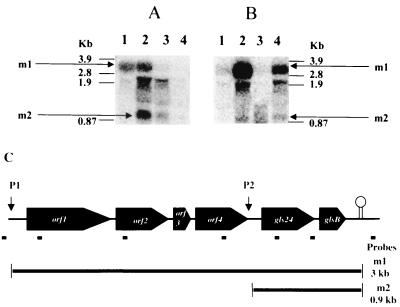
Northern blots prepared with RNA isolated from E. faecalis JH2-2 were hybridized with several different probes indicated in Fig. 1C. Figure 1A shows the results obtained with RNA extracted from cells in exponential growth phase (lane 1) at the onset (lane 2) and after 3 and 12 h of glucose starvation (lanes 3 and 4) hybridized with an oligonucleotide complementary to a sequence between gls24 and glsB. A band named m1, whose size corresponds to the entire operon size, is present in lanes 1 and 2. This approximately 3-kb transcript was no longer detectable after 3 or more hours of starvation (lanes 3 and 4). Unusually shaped bands were present in the region of the blot containing the front edge of the 23S rRNA band. A similar phenomenon, possibly an artifact resulting from the extremely high concentrations of the rRNA species in the gel and the degradation products of the larger transcript, has been observed by other investigators (14, 40). A second high-intensity band of approximately 0.9 kb (Fig. 1A, m2) was specifically present at the onset and in a lesser extent after 3 h of starvation. Since hybridization with probes complementary to orf4, orf2, and orf1 sequences hybridized with the m1 but not with the m2 band (data not shown), the latter corresponds to the gls24-glsB transcript (Fig. 1C). The 375-bp Sau3AI/Sau3AI fragment containing the region upstream of gls24 was cloned in front of the reporter gene gusA (encoding the β-glucuronidase) giving plasmid pNUM24 (M. Uguen, unpublished results). By transformation of the plasmid in E. faecalis, we showed that this construction induced the expression of gusA. At the onset of starvation, this expression is two- to threefold higher than without the cloned promoter fragment, correlating nicely with the observed amplifications on the protein level (data not shown). These results proved that the transcript m2 did not arise from a processing of the m1 mRNA but from the induction of the promoter upstream of gls24. Moreover, no signals were obtained after hybridization with probes based on the sequences downstream of the terminator and upstream of the putative promoter region of the operon. Taken together, these observations suggest a complex transcriptional regulation and that the six-gene operon including gls24 have two promoter regions, one upstream of the first gene orf1 and another between orf4 and gls24.
FIG. 1.
Northern analyses of E. faecalis RNA. (A) Samples of total RNA were prepared from cells in exponential growth phase (lane 1), at the onset of starvation (lane 2), and after 3 and 12 h of glucose starvation (lanes 3 and 4, respectively). (B) Samples of total RNA were prepared from cells in exponential growth phase (lanes 1 and 3) and after 30 min in the presence of 50 μg of CdCl2 per ml (lane 2) or 0.08% bile salts (lane 4). The hybridizations were achieved with the 32P-labelled probe deduced from the sequence between gls24 and glsB (5′-CCATGATTGTTTCCTCCC-3′). The numbers on both sides show the RNA markers (3.9 to 0.87 kb). Positions of the entire operon mRNA (transcripts m1) and the gls24-glsB mRNA (transcripts m2) are indicated by arrows. (C) Schematic representation of the E. faecalis operon containing gls24 encoding the general stress protein Gls24. Promoter regions P1 and P2 are indicated by arrows. The transcripts m1 and m2 derived from this operon observed in the Northern analyses are presented, and their deduced sizes are indicated. Small black bars under the operon indicate positions of the different oligonucleotides used as probes for the Northern analyses.
Since synthesis of Gls24 protein is induced after exposure to several stresses, Northern blots were carried out with RNA extracted from E. faecalis exposed to CdCl2 (50 μg/ml) and bile salts (0.08%) stresses as described by Laplace et al. (38) and Flahaut et al. (19), respectively. Interestingly, the entire operon transcript was strongly induced after CdCl2 and bile salts treatments (Fig. 1B). Otherwise, the small transcript m2 was also detectable in these experiments, though in lesser abundance than in cells entering stationary phase.
Mapping the transcriptional start sites.
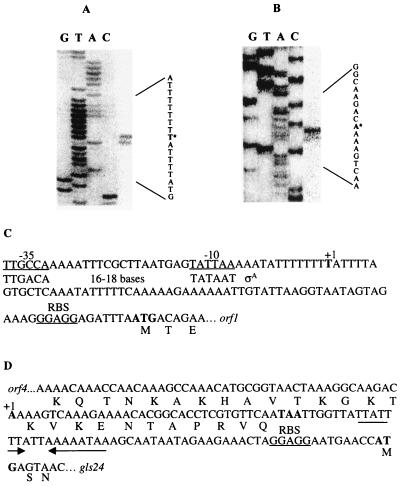
Two potential transcriptional start sites were mapped, one for the entire operon, one starting upstream of the two last genes, gls24 and glsB (Fig. 2A and B). We concluded that the T 72 bp upstream of the ATG codon of orf1 and the A 93 bp upstream of the ATG codon of gls24 correspond to the +1 positions of the m1 and m2 transcripts, respectively. Although no ςA consensus sequences are located at an appropriate distance from the m1 transcriptional start site, sequences sharing similarity to −35 (TTGCCA) and −10 (TATTAA) boxes (deviations underlined) separated by an optimal space of 18 nucleotides are located further upstream (Fig. 2C). No obvious promoter consensus sequence was observed upstream of the m2 start site (Fig. 2D). Note that the m2 transcriptional start site was located inside the terminal part of the orf4 gene.
FIG. 2.
(A and B) Mapping of the 5′ ends of the entire operon mRNA (A) and gls24-glsB mRNA (B) by primer extension analysis. RNA was isolated from E. faecalis JH2-2 after 30 min of CdCl2 stress exposure (A) and at the onset of starvation (B). The potential transcription start sites are marked with asterisks. Lanes G, T, A, and C show the sequencing ladder obtained by using the same primer as was used for the primer extension. (C and D) Sequences of the P1 (C) and P2 (D) promoter regions. Potential −35 and −10 regions and the RBS sequences are underlined. The transcriptional start sites (+1) and translational start and stop codons are indicated in boldface letters. The consensus sequence for ςA-dependent promoter with its appropriate spacer is shown (C). The AT-rich inverted repeat sequence observed between the P2 promoter region is marked by arrows (D).
Phenotypic studies of a gls24 mutant.
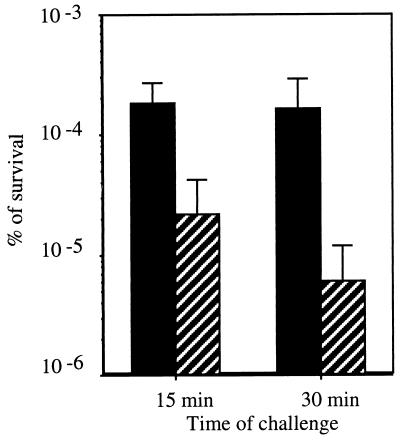
Because of the induction of gls24 mRNA under stationary phase and stress conditions, we examined whether a knockout mutation in this gene affected stress resistance and long term survival. The gls24 mutant displayed an extended generation time (58 min) compared to the wild type (45 min) at 37°C in the semisynthetic medium (data not shown). No changes in survival under glucose starvation and resistance to thermal (62°C), hydrogen peroxide (20 mM), CdCl2 (50 mg/ml), acid pH (3.2), basic pH (11.9), and ethanol (17%) stresses were observed in the mutant cells (data not shown). Nevertheless, gls24 mutants starved for 24 h were more sensitive to a 0.3% bile salts challenge; the percentage of survival was 26-fold lower than in the wild type after 30 min of challenge (Fig. 3). Interestingly, this phenotype was not observed with an orf4 insertional mutant (data not shown). Moreover, the introduction of a plasmid caring the gls24-glsB operon into the wild-type strain increased fourfold the survival against bile salts challenge, which was another proof of the role of this operon in the resistance towards this stress condition (data not shown).
FIG. 3.
Percentage of survival for 24-h-starved cells of E. faecalis JH2-2 (black bars) and gls24 mutant (hatched bars) cells after 15 and 30 min of challenge with 0.3% bile salts. These data are the average of four separate experiments, and standard deviations are indicated at the top of each bar.
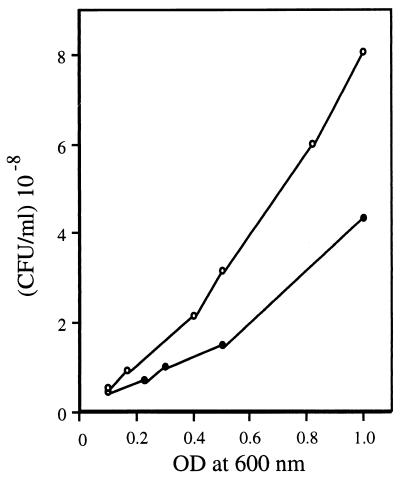
Morphological features of gls24 mutant.
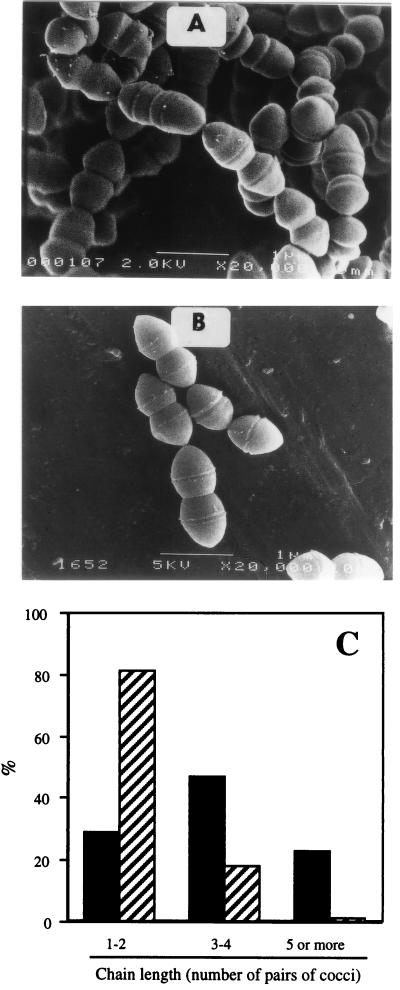
We found that the gls24 mutant and the wild-type strain did not show the same correlation between the optical density at 600 nm (OD600) and the numeration by plate counts. Whereas the size of cells was approximately the same, the number of CFU of gls24 mutants was two- to threefold higher at an equivalent OD600. For example, at an OD600 of 0.5, 1.4 × 108 and 3 × 108 CFU/ml were counted for E. faecalis JH2-2 and the mutant strain, respectively (Fig. 4). Microscopic observations revealed that these differences could be explained by differences in chain length (Fig. 5A and B). During exponential growth phase, the E. faecalis JH2-2 culture comprised 50 and 20% of three or four and five or more pairs of coccal chains, respectively. In contrast, 80% of gls24 mutant cells were organized in pairs or in very short chains (Fig. 5C). Moreover, wild-type cells were able to form some very long chains (10 to 15 pairs of cocci) which were never observed in gls24 mutants. Again, this morphological anomaly was not observed with the orf4 insertional mutant. The wild-type phenotype and morphology were restored when complementing the mutant strain with the entire gls24-glsB operon cloned into the low-copy-number plasmid pNZ273 (46), whereas the plasmid containing only gls24 was inefficient (data not shown). The proper complementation was confirmed by the presence of the corresponding Gls24 spot on 2D gel electrophoresis.
FIG. 4.
Correlation between OD600 and plate count of E. faecalis JH2-2 (closed circle) and gls24 mutant strain (open circle).
FIG. 5.
Morphology of E. faecalis cells. Strains JH2-2 (A) and gls24 mutant (B) were grown at 37°C in semisynthetic medium. Electron micrographs of rapidly growing cells are shown. (C) Ratio of different chain lengths of cells observed in E. faecalis JH2-2 (black bars) and gls24 mutant strain (hatched bars). 100% corresponds to at least 200 chains.
2D polyacrylamide gel electrophoresis of E. faecalis JH2-2 and gls24 mutant strains.
We were interested in whether the mutation in gls24 affected the expression of other genes or operons. Analysis of 2D gels revealed that, in addition to the logical absence of the protein Gls24, several differences in the protein pattern existed between the mutant and wild-type cells (Fig. 6). Nine polypeptides were significantly overexpressed in the gls24 mutant (Fig. 6). Five of these proteins (numbers 1, 5, 7, 8, and 9) were under the limit of detection in the wild-type strain (Table 2). The synthesis of protein 1 appeared independent of the growth phase, whereas the four others were induced during glucose starvation (Fig. 6C and D and Table 2).
FIG. 6.
2D separation of 35S-labelled proteins from growing (A and C) and 24-h starved cells (B and D) of E. faecalis JH2-2 (A and B) and gls24 mutant (C and D) strains. Arrows indicate the positions of polypeptides that are synthesized in higher amounts in the gls24 mutant than in the wild-type cells. The position of the protein Gls24 is also indicated.
TABLE 2.
Rate of synthesis and partial identification of the proteins observed in larger amount in gls24 mutant cells compared to the wild type
| Protein no. | RISa
|
N-terminal sequence | Corresponding gene productb | |||
|---|---|---|---|---|---|---|
|
E. faecalis JH2-2
|
E. faecalis gls24 mutant
|
|||||
| Growing phase | 24 h of starvation | Growing phase | 24 h of starvation | |||
| 1 | UDc | UD | 0.84 | 0.9 (1d) | XXXDFAIE | Lipoamide dehydrogenase |
| 2 | 0.3 | 0.1 (0.3) | 0.96 | 0.37 (0.3) | AAGNKDHQ | l-Lactate dehydrogenase |
| 3 | 0.08 | 0.25 (3) | 0.7 | 1.3 (1.8) | AQKTMIQA | Pyruvate decarboxylase |
| 4 | 0.1 | 0.05 (0.5) | 0.4 | 0.27 (0.6) | XEFLDSLK | Unknown |
| 5 | UD | UD | 0.04 | 0.31 (7.7) | ||
| 6 | 0.4 | 0.13 (0.3) | 0.38 | 0.37 (1) | ||
| 7 | UD | UD | 0.05 | 0.13 (2.2) | XEFQLEKN | Unknown |
| 8 | UD | UD | 0.02 | 0.07 (2.7) | ||
| 9 | UD | UD | 0.03 | 0.52 (14) | XNEGFNNVP | Unknown |
Relative intensity of synthesis (RIS) is the ratio of the intensity of each spot to the total intensity on the autoradiograms × 100.
Gene product was obtained by comparing the ORF from the sequenced genome of E. faecalis containing the N-terminal sequence and databases. See text for more details.
UD, undetectable.
RIS after 24 h of starvation/RIS in exponential phase.
The synthesis of protein 3 increased and that of proteins 2 and 4 decreased to a comparable extent in stationary phase in both mutant and wild-type cells. The reason why they were observed in larger amount in the mutant cells was that their synthesis was already enhanced during the growth phase (Fig. 6 and Table 2). In exponential phase, the level of synthesis of protein 6 appeared similar in the JH2-2 and gls24 mutant strains. Because its synthesis was not repressed in stationary phase as is the case in the wild-type strain, the corresponding spot was more intense in the starved mutant bacteria (Table 2).
Prior to this study, the N-terminal sequences of the six proteins have been determined, and the corresponding genes of four of these microsequences have been found in the E. faecalis genome database (Table 2). Proteins 1, 2, and 3 correspond to the lipoamide dehydrogenase (E3 component of the pyruvate dehydrogenase system), the l-lactate dehydrogenase, and the pyruvate decarboxylase (E1 component of the pyruvate dehydrogenase system), respectively. These enzymes showed at least 60 to 70% identity to the homologous proteins from S. aureus, B. subtilis, and Lactobacillus casei. Interrogation of different databases showed that the gene encoding protein 7 as well as the microsequences of the other polypeptides (proteins 4 and 9) did not reveal any significant homology to known proteins.
The obvious induction of one of these proteins in the mutant strain has been verified at the transcriptional level. Northern blot analysis was performed with RNA from growing and starved cells and a probe based on the sequence of the gene encoding the l-lactate dehydrogenase. The corresponding transcript of about 1.2 kb was strongly induced (10-fold) in the mutant cells during the growth phase compared to the JH2-2 strain (Fig. 7). Moreover, unlike in wild-type cells, the mRNA was still detectable after 3 h of glucose starvation (Fig. 7).
FIG. 7.
Northern blot analysis of total cellular RNA isolated from strain JH2-2 (lanes 1 to 3) and gls24 mutant (lanes 4 to 6). Samples were prepared from cells in exponential growth phase (lanes 1 and 4), at the onset of starvation (lanes 2 and 5), and after 3 h of glucose starvation (lanes 3 and 6). A 32P-labelled 20-bp oligonucleotide deduced from the sequence of the gene encoding l-lactate dehydrogenase (5′-CGTCAGGATTATTTTTCACC-3′) was used as a hybridization probe. Position of the 1.2-kb mRNA is indicated.
DISCUSSION
Whereas considerable knowledge about E. coli and other gram-negative bacteria as well as B. subtilis has been gained in recent years, little is known about the stress and starvation responses in gram-positive non-spore-forming bacteria. In this report, we analyzed the regulation and the consequence of a knockout mutation of a gene encoding a general stress protein of E. faecalis named Gls24. Indeed, this protein has been found to be induced by different starvation conditions as well as by several chemical stresses (18, 19, 23, 28, 38). Moreover, Gls24 is a member of the class A of E. faecalis glucose starvation proteins that comprises polypeptides synthesized in both growing and resting cells and which are overproduced at different times of starvation (23). An extensive analysis of the E. faecalis genome sequence allowed us to identify gls24 as the penultimate gene of a hitherto unknown six-gene operon. Surprisingly, gls24 exhibited strong homology with orf4 located just upstream.
The operon is under complex regulation. The transcriptional start sites of the two promoters P1 (governing the expression of the entire operon) and P2 (controlling the two last genes including gls24) were mapped. Starvation induction of gls24 is due to the activation of this internal promoter, and basal expression and the chemical stress induction is initiated at P1. This difference is reflected by the significant sequence divergences in the promoter areas. Whereas −10 and −35 boxes close to the consensus of ςA-dependent promoters have been found nearby the transcriptional start site in P1, P2 is characterized by less-conserved sequences in these regions. In L. lactis, a promoter upregulated during transition into stationary phase (P170) has recently been described (42). P170 controls the expression of a monocistronic gene orfX (encoding a protein of unknown function) and may be induced by a hitherto-unknown transcriptional factor. In contrast to P2, transcription from P170 was only induced if the pH was between 6.5 and 6.0. Moreover, comparison of promoters P2 and P170 shows that they are very different. Like P2, P170 lacked the consensus −35 region but contained an extended −10 box (42). This motif occurs frequently in promoters from gram-positive bacteria and is strictly required for activity in E. coli promoters that lack a consensus −35 region (6, 24, 33, 35).
The 18-bp IR located in the promoter region upstream of the start codon of gls24 consists exclusively of A and T nucleotides. This characteristic is similar to the Per box, a sequence flanking promoters of genes responding to oxidative stress in B. subtilis (mrgA, katA1, hemAXCDBL operon, and ahpC) (5, 7, 11, 45), Listeria seeligeri (kat) (25), and E. faecalis (ahpC) (10). It has been proposed as the target site for the recently discovered peroxide regulon repressor PerR in B. subtilis (10, 11). However, it is premature to speculate if the IR of the gls24-glsB promoter is also involved in the control of transcription initiation upon entrance into stationary phase.
Disruption of the gls24 gene provoked obvious growth and morphological defect, sensitivity toward a bile salts challenge, and modifications of expression of several genes. Owing to its lifestyle, E. faecalis has to cope with an environment containing bile salts. Therefore, the sensitivity to this agent and the reduced growth rate of the gls24 insertional mutant suggest that gls24-glsB operon gives a selective advantage to survive under these conditions. The altered phenotype of the gls24 mutant, despite the presence of a gene showing important homology to it, revealed that Gls24 and Orf4 seem to have different physiological roles. This conclusion is strengthened by the result that a knock out mutation in orf4 does not lead to the modifications observed in gls24 mutant cultures. However, it cannot be omitted that these paralogues can complement each other under environmental conditions and that GlsB may play a role in gls24 mutant phenotype. Moreover, the presence of these two genes in the same operon structure (probably having evolved by gene duplication) may indicate that their products have important function in cellular metabolism.
To our knowledge, such a mutant reduced-chain-length phenotype has so far not been described for chain-forming bacteria. Because the wild-type phenotype is restored by complementation with a fragment containing gls24 and glsB, but not gls24 alone, it is highly probable that the insertional mutation has a polar effect on glsB expression. So our phenotype and complementation analysis suggest that Gls24-GlsB or, more likely, GlsB alone may be involved in cell attachment. Alternatively, it is possible that the role of these proteins in this phenomenon is indirect, and one or several of the polypeptides showing modified expression in the gls24 mutant strain may be responsible for this effect.
Three of these proteins had been identified to correspond to the l-lactate dehydrogenase, lipoamide dehydrogenase, and pyruvate decarboxylase. The latter two enzymes constitute, with a third polypeptide (dihydrolipoyl transacetylase), the pyruvate dehydrogenase complex (30). Interestingly, these proteins are involved in the pyruvate crossroad, leading either to the formation of lactate or acetyl coenzyme A. The reason why the two branches were activated in the mutant cells is not clear at present. The identification of the remaining six proteins induced in the gls24 mutant strain may contribute to understanding this phenomenon. One possible hypothesis is that induction of these activities is aimed to compensate the mutation of gls24-glsB, which may mask an even more severe impact on morphology and cell physiology.
Shankar and coworkers have recently identified a new cell wall-associated protein (Esp) in E. faecalis (50). A statistical association with infection-derived E. faecalis isolates compared to isolates from healthy individuals provided indirect evidence for a contributory role to virulence for this 202-kDa protein. Surprisingly, instead of gls24-glsB genes, the esp gene is located just downstream of orf4 in the MMH594 E. faecalis strain (V. Shankar, Abstr. Am. Soc. Microbiol. Conf. Streptococcal Genet. 1998, abstr. 15, p. 22–23, 1998). Because the esp gene is not constantly present in E. faecalis isolates, this may also be the case for both gls24 and glsB genes. From these observations and because the JH2-2 strain is also a clinically derived isolate (32, 58), it will be interesting to determine whether gls24 as well as the entire operon are involved in virulence.
In conclusion, this report gives the first insight into the complex regulation and function of a stress- and starvation-inducible operon of E. faecalis. Inactivation in the corresponding gene has a pleiotropic effect on cell morphology, stress sensitivity, and expression of several genes. Work is in progress to understand the molecular basis responsible for these modifications, to analyze the function of the other genes of the operon, and to identify the regulators of this stress- and starvation-inducible genomic island of E. faecalis.
ACKNOWLEDGMENTS
The expert technical assistance of Annick Blandin and Béatrice Gillot was greatly appreciated. We thank A. Benachour, J.-M. Laplace, and V. Pichereau for helpful discussions; M. Uguen and A. Dufour (Laboratoire de Biologie et Chimie Moléculaire, Université de Vannes, France) for providing plasmid pNUM24; and I. L. van Alen-Boerrigter for the pNZ273 plasmid (NIZO food research, The Netherlands).
REFERENCES
- 1.Aldea M, Garrido T, Hernandez-Chico C, Vicente M, Kushner S R. Induction of a growth-phase-dependent promoter trigger transcription of bolA, an Escherichia coli morphogene. EMBO J. 1989;8:3923–3931. doi: 10.1002/j.1460-2075.1989.tb08573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almiron M, Link A J, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 3.Altshul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Anderson T F. Techniques for the preservation of three-dimensional structure in preparing specimens for the electron microscope. Trans N Y Acad Sci. 1951;13:130–134. [Google Scholar]
- 5.Antelmann H, Engelmann S, Schmid R, Hecker M. General and oxidative stress responses in Bacillus subtilis: cloning, expression, and mutation of the alkyl hydroperoxide reductase operon. J Bacteriol. 1996;178:6571–6578. doi: 10.1128/jb.178.22.6571-6578.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bashyam M D, Tyagi A K. Identification and analysis of ‘extended-10’ promoters from mycobacteria. J Bacteriol. 1998;180:2568–2573. doi: 10.1128/jb.180.9.2568-2573.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bol D K, Yasbin R E. The isolation, cloning and identification of a vegetative catalase gene from Bacillus subtilis. Gene. 1991;109:31–37. doi: 10.1016/0378-1119(91)90585-y. [DOI] [PubMed] [Google Scholar]
- 8.Bolotin A, Mauger S, Malarme K, Ehrlich S D, Sorokin A. Low-redundancy sequencing of the entire Lactococcus lactis IL1403 genome. Antonie Leeuwenhoek. 1999;76:27–76. [PubMed] [Google Scholar]
- 9.Boos W, Ehmann U, Bremer E, Middendorf A, Postma P. Trehalase of Escherichia coli. J Biol Chem. 1987;262:13212–13218. [PubMed] [Google Scholar]
- 10.Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann J D. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake Fur and peroxide regulon PerR repressors. Mol Microbiol. 1998;29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, Keramati L, Helman J D. Coordinate regulation of Bacillus subtilis peroxide and metal ions. Proc Natl Acad Sci USA. 1995;92:8190–8194. doi: 10.1073/pnas.92.18.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Auberton Carafa Y, Brody E, Thermes C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stemloop structures. J Mol Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 13.Derré I, Rapoport G, Msadek T. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol Microbiol. 1999;31:117–131. doi: 10.1046/j.1365-2958.1999.01152.x. [DOI] [PubMed] [Google Scholar]
- 14.de Vos W M, Boerrigter I, van Rooijen R J, Reicheand B, Hengstenberg W. Characterization of the lactose-specific enzymes of the phosphotransferase system in Lactococcus lactis. J Biol Chem. 1990;265:22554–22560. [PubMed] [Google Scholar]
- 15.Donkersloot J A, Thompson J. Cloning, expression, sequence analysis, and site-directed mutagenesis of the Tn5306-encoded N5-carboxyethyl ornithine synthase from Lactococcus lactis K1. J Biol Chem. 1995;270:12226–12234. doi: 10.1074/jbc.270.20.12226. [DOI] [PubMed] [Google Scholar]
- 16.Dower W J, Miller J F, Ragsdal C W. High efficiency transformation of Escherichia coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flahaut S. Incidences physiologiques and biochimiques de différents stress chez Enterococcus faecalis ATCC 19433. Ph.D. thesis. Caen, France: University of Caen; 1996. [Google Scholar]
- 18.Flahaut S, Benachour A, Giard J-C, Boutibonnes P, Auffray Y. Defense against lethal treatments and de novo protein synthesis induced by NaCl in Enterococcus faecalis ATCC19433. Arch Microbiol. 1996;165:317–324. doi: 10.1007/s002030050333. [DOI] [PubMed] [Google Scholar]
- 19.Flahaut S, Hartke A, Giard J-C, Benachour A, Boutibonnes P, Auffray Y. Relationship between stress response towards bile salts, acid and heat treatment in Enterococcus faecalis. FEMS Microbiol Lett. 1996;138:49–54. doi: 10.1111/j.1574-6968.1996.tb08133.x. [DOI] [PubMed] [Google Scholar]
- 20.Foster J W. Low pH adaptation and the acid tolerance response of Salmonella typhimurium. Crit Rev Microbiol. 1995;21:215–237. doi: 10.3109/10408419509113541. [DOI] [PubMed] [Google Scholar]
- 21.Frère J, Benachour A, Novel M, Novel G. Identification of the theta-type minimal replicon of the Lactococcus lactis ssp. lactis CNRZ270 lactose protease plasmid pUCL22. Curr Microbiol. 1993;27:97–102. [Google Scholar]
- 22.Giard J-C, Hartke A, Flahaut S, Benachour A, Boutibonnes P, Auffray Y. Starvation-induced multiresistance in Enterococcus faecalis JH2-2. Curr Microbiol. 1996;32:264–271. doi: 10.1007/s002849900048. [DOI] [PubMed] [Google Scholar]
- 23.Giard J-C, Hartke A, Flahaut S, Boutibonnes P, Auffray Y. Glucose starvation response in Enterococcus faecalis JH2-2: survival and protein analysis. Res Microbiol. 1997;148:27–35. doi: 10.1016/S0923-2508(97)81897-9. [DOI] [PubMed] [Google Scholar]
- 24.Graves M C, Rabonowitz J C. In vivo and in vitro transcription of the Clostridium pasteurianum ferrodoxin gene. J Biol Chem. 1986;261:11409–11415. [PubMed] [Google Scholar]
- 25.Haas A, Brehm K, Kreft J, Goebel W. Cloning, characterization, and expression in Escherichia coli of a gene encoding Listeria seeligeri catalase, a bacterial enzyme highly homologous to mammalian catalases. J Bacteriol. 1991;173:5159–5167. doi: 10.1128/jb.173.16.5159-5167.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haldenwang W G. The sigma factors of Bacillus subtilis. Microbiol Rev. 1995;59:1–30. doi: 10.1128/mr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall M C. Are point mutations or DNA rearrangements responsible for the restriction fragment length polymorphisms that are used to type bacteria? Microbiology. 1994;140:197–204. doi: 10.1099/13500872-140-1-197. [DOI] [PubMed] [Google Scholar]
- 28.Hartke A, Giard J-C, Laplace J-M, Auffray Y. Survival of Enterococcus faecalis in an oligotrophic microcosm: changes in morphology, development of general stress resistance, and analysis of protein synthesis. Appl Environ Microbiol. 1998;64:4238–4245. doi: 10.1128/aem.64.11.4238-4245.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hecker M, Völker U. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the ςB regulon. Mol Microbiol. 1998;29:1129–1136. doi: 10.1046/j.1365-2958.1998.00977.x. [DOI] [PubMed] [Google Scholar]
- 30.Hemilä H, Palva A, Paulin L, Arvidson S, Palva I. Secretory S complex of Bacillus subtilis: sequence analysis and identity to pyruvate dehydrogenase. J Bacteriol. 1990;172:5052–5063. doi: 10.1128/jb.172.9.5052-5063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hengge-Aronis R, Fischer D. Identification and molecular analysis of glgS, a novel growth phase-regulated and rpoS-dependent gene involved in glycogen synthesis in Escherichia coli. Mol Microbiol. 1992;6:1877–1886. doi: 10.1111/j.1365-2958.1992.tb01360.x. [DOI] [PubMed] [Google Scholar]
- 32.Jacob A E, Hobbs S J. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol. 1974;117:360–372. doi: 10.1128/jb.117.2.360-372.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keilty S, Rosenberg M. Constitutive function of a positively regulated promoter reveals new sequences essential for promoter activity. J Biol Chem. 1987;262:6389–6395. [PubMed] [Google Scholar]
- 34.Kullik I, Giachino P, Fuchs T. Deletion of the alternative sigma factor sigmaB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J Bacteriol. 1998;180:4814–4820. doi: 10.1128/jb.180.18.4814-4820.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar A, Malloch R A, Fujita N, Smilie D A, Ishihama A, Hayward R S. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an ‘extended minus 10’ promoter. J Mol Biol. 1993;232:406–418. doi: 10.1006/jmbi.1993.1400. [DOI] [PubMed] [Google Scholar]
- 36.Kuroda M, Ohta T, Hayashi H. Isolation and the gene cloning of an alkaline shock protein in methicillin resistant Staphylococcus aureus. Biochem Biophys Res Commun. 1995;207:978–984. doi: 10.1006/bbrc.1995.1281. [DOI] [PubMed] [Google Scholar]
- 37.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 38.Laplace J-M, Boutibonnes P, Auffray Y. Unusual resistance and acquired tolerance to cadmium chloride in Enterococcus faecalis. J Basic Microbiol. 1996;36:311–317. doi: 10.1002/jobm.3620360504. [DOI] [PubMed] [Google Scholar]
- 39.Linquist S. The heat-shock response. Ann Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- 40.Llanos R M, Hillier A J, Davidson B E. Cloning, nucleotide sequence, expression, and chromosomal location of ldh, the gene encoding l-(+)-lactate dehydrogenase, from Lactococcus lactis. J Bacteriol. 1992;174:6956–6964. doi: 10.1128/jb.174.21.6956-6964.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopez M F, Patton W F, Utterback B L, Chung-Welch N, Barry P, Skea W M, Cambria R P. Effect of various detergents on protein migration in the second dimension of two-dimensional gels. Anal Biochem. 1991;199:35–44. doi: 10.1016/0003-2697(91)90266-v. [DOI] [PubMed] [Google Scholar]
- 42.Madsen S M, Arnau J, Vrang A, Givskov M, Israelsen H. Molecular characterization of the pH-inducible and growth phase-dependent promoter P170 of Lactococcus lactis. Mol Microbiol. 1999;32:75–87. doi: 10.1046/j.1365-2958.1999.01326.x. [DOI] [PubMed] [Google Scholar]
- 43.Morita R Y. Bioavailability of energy and the starvation state. In: Kjelleberg S, editor. Starvation in bacteria. New York, N.Y: Academic press; 1993. pp. 8–16. [Google Scholar]
- 44.O'Farrell P H. High two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 45.Petricek M, Rutberg L, Schröder I, Hederstedt L. Cloning and characterization of the hemA region of the Bacillus subtilis chromosome. J Bacteriol. 1990;172:2250–2258. doi: 10.1128/jb.172.5.2250-2258.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Platteeuw C, Simons G, de Vos W M. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl Environ Microbiol. 1994;60:587–593. doi: 10.1128/aem.60.2.587-593.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rockabrand D, Arthur T, Korinek G, Livers K, Blum P. An essential role for the Escherichia coli DnaK protein in starvation-induced thermotolerance, H2O2 resistance, and reductive division. J Bacteriol. 1995;177:3695–3703. doi: 10.1128/jb.177.13.3695-3703.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 49.Schellhorn H E, Hassan H M. Transcriptional regulation of katE in Escherichia coli K-12. J Bacteriol. 1988;170:4286–4292. doi: 10.1128/jb.170.9.4286-4292.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shankar V, Baghdayan A S, Huycke M M, Lindahl G, Gilmore M S. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect Immun. 1999;67:193–200. doi: 10.1128/iai.67.1.193-200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terzaghi B E, Sandine W. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;2:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tinoco I, Borer P N, Dengler B, Levine M D. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973;246:40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- 53.van Sinderen D, ten Berge A, Hayema B J, Hamoen L, Venema G. Molecular cloning and sequence of comK, a gene required for genetic competence in Bacillus subtilis. Mol Microbiol. 1994;11:695–703. doi: 10.1111/j.1365-2958.1994.tb00347.x. [DOI] [PubMed] [Google Scholar]
- 54.Vellanoweth R L, Rabinowitz J C. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol Microbiol. 1992;6:1105–1114. doi: 10.1111/j.1365-2958.1992.tb01548.x. [DOI] [PubMed] [Google Scholar]
- 55.Weichart D, Lange R, Henneberg N, Hengge-Aronis R. Identification and characterization of stationary phase-inducible genes in Escherichia coli. Mol Microbiol. 1993;10:407–420. [PubMed] [Google Scholar]
- 56.Wiedmann M, Arvik T J, Boor K J. General stress transcription factor B and its role in acid tolerance and virulence of Listeria monocytogenes. J Bacteriol. 1998;180:3650–3656. doi: 10.1128/jb.180.14.3650-3656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu S, de Lencastre H, Tomasz A. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J Bacteriol. 1996;178:6036–6042. doi: 10.1128/jb.178.20.6036-6042.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yagi Y, Clewell D B. Recombination-deficient mutant of Streptococcus faecalis. J Bacteriol. 1980;143:966–970. doi: 10.1128/jb.143.2.966-970.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]