Abstract
The protein expression profiles of Rhizobium leguminosarum strains in response to specific genetic perturbations in exopolysaccharide (EPS) biosynthesis genes were examined using two-dimensional gel electrophoresis. Lesions in either pssA, pssD, or pssE of R. leguminosarum bv. viciae VF39 or in pssA of R. leguminosarum bv. trifolii ANU794 not only abolished the capacity of these strains to synthesize EPS but also had a pleiotropic effect on protein synthesis levels. A minimum of 22 protein differences were observed for the two pssA mutant strains. The differences identified in the pssD and pssE mutants of strain VF39 were a distinct subset of the same protein synthesis changes that occurred in the pssA mutant. The pssD and pssE mutant strains shared identical alterations in the proteins synthesized, suggesting that they share a common function in the biosynthesis of EPS. In contrast, a pssC mutant that produces 38% of the EPS level of the parental strain showed no differences in its protein synthesis patterns, suggesting that the absence of EPS itself was contributing to the changes in protein synthesis and that there may be a complex interconnection of the EPS biosynthetic pathway with other metabolic pathways. Genetic complementation of pssA can restore wild-type protein synthesis levels, indicating that many of the observed differences in protein synthesis are also a specific response to a dysfunctional PssA. The relevance of these proteins, which are grouped as members of the pssA mutant stimulon, remains unclear, as the majority lacked a homologue in the current sequence databases and therefore possibly represent a novel functional network(s). These findings have illustrated the potential of proteomics to reveal unexpected higher-order processes of protein function and regulation that arise from mutation. In addition, it is evident that enzymatic pathways and regulatory networks are more interconnected and more sensitive to structural changes in the cell than is often appreciated. In these cases, linking the observed phenotype directly to the mutated gene can be misleading, as the phenotype could be attributable to downstream effects of the mutation.
Soil bacteria belonging to the genera Rhizobium, Sinorhizobium, Bradyrhizobium, Mesorhizobium, and Azorhizobium (collectively termed rhizobia) are able to infect the roots of leguminous plants in a host-specific way and stimulate the formation of nodules. Inside these nodules, rhizobia differentiate into bacteroids that reduce atmospheric nitrogen to ammonia, which is used by the plant. One major characteristic of many rhizobia is the production of large amounts of acidic exopolysaccharide (EPS) molecules which serve a variety of roles in free-living rhizobia and in the establishment of symbiosis.
EPS forms a biofilm layer on the cell surface which is thought to contribute to the following processes: cellular protection against environmental stresses, attachment to surfaces, nutrient gathering, and the preferential absorption of plant secreted flavonoids along the membrane surface (5, 20, 45). In addition, EPS biosynthesis by rhizobia is required for the effective nodulation of legumes such as Medicago, Pisum, Trifolium, Leucaena, and Vicia spp., which form indeterminate-type nodules (4, 26). EPS-deficient mutants of Rhizobium leguminosarum bv. viciae, R. leguminosarum bv. trifolii, and Sinorhizobium meliloti induce symbiotically defective phenotypes which include delayed root hair curling, nodules devoid of bacteria due to infection threads that abort within peripheral cells of the developing nodule, and small, partially infected, non-nitrogen-fixing nodules (8, 24, 35, 42, 43). The precise function(s) of the EPS molecules in these associations is still unclear; however, recent studies suggest that the strain-dependent variations in chemical structure of these extracellular polymers may enable EPS to function as a symbiotic signaling molecule which regulates plant responses early in the infection process (8, 10, 14, 28, 43).
In general, EPS synthesised by R. leguminosarum is a polymer of a conserved octasaccharide repeating unit which is assembled by the sequential transfer of sugars to a growing lipid-linked polysaccharide chain. These repeating units can also be decorated by O-acetyl, pyruvyl, and hydroxybutanoyl groups (31, 32). For R. leguminosarum strains, the genetic control of synthesis and regulation of EPS is less well understood than in the case of EPSs (EPS I and EPS II) of S. meliloti (2, 3, 12, 13, 23). Several genes, designated pss genes, have been identified as crucial to the regulation and biosynthesis of EPS in R. leguminosarum bv. viciae, R. leguminosarum bv. trifolii, and R. leguminosarum bv. phaseoli. Functions were assigned to most of the pss genes as a result of genetic complementation or by sequence homology to proteins of known function from other bacteria, including S. meliloti. It should be emphasised that the nucleotide sequences of pss genes from different biovars of R. leguminosarum revealed very high levels of homology, and in most cases they were even identical. Therefore, data obtained with these strains can be considered complementary.
The EPS-deficient mutant strain ANU437, which was used in this study, has been extensively characterized at the phenotypic level. The multiple defects observed suggest pleiotropic effects due to the mutation. In summary, ANU437 is nonmucoid, produces very low levels of acidic EPS (0.3% of that of the parent strain) that lacks an O-acetyl substitution, lacks a capsule, is more sensitive to phytoalexins, and induces delayed formation of small, non-nitrogen-fixing nodules on white and subterranean clovers (35). Cellular growth is more sensitive to addition of glycine and succinate than that of the parental strain, and the symbiotic phenotype observed is condition dependent (35). The phenotypes of VF39 pssA::Tn5, VF39 pssD::Tn5, and VF39 pssE::Tn5, which are also used in this study, are somewhat similar to those of ANU437. These strains are also nonmucoid and induce delayed formation of rare non-nitrogen-fixing nodules on Vicia faba, but they retain their ability to form nitrogen-fixing nodules on Pisum sativum and Vicia sativa (21, 36). In contrast, strains VF39 pssA-Km and VF39 pssC-Km produce EPS at 85 and 38%, respectively, of the wild-type levels. Strain VF39 pssA-Km induces nodules on V. faba with a reduced ability to fix nitrogen, whereas VF39 pssC-Km fails to induce nodules on V. faba (25, 36).
The pssA gene of R. leguminosarum was proposed to encode a isoprenylphosphate (IP) glycosyl transferase responsible for the first glycosyl transferase step in EPS biosynthesis, involving the transfer of a glucose-1-phosphate residue from UDP-glucose to an IP lipid carrier (6, 21, 33, 41). Interestingly, the pssA gene described for R. leguminosarum bv. viciae VF39 has an extension of 63 codons at the 5′ end, since translation was found to start at the first putative GTG start codon rather than the second putative start codon as described for R. leguminosarum bv. phaseoli (6). Work by Ksenzenko et al. (25) indicated that the pssA gene could be expressed and regulated differentially from three different promoters, resulting in two translation variants of PssA, long (263 amino acids [aa]) and short (200 aa). The shorter protein is the glycosyl transferase, whereas the longer protein was postulated to have an additional function(s).
Several other putative glycosyl transferase genes (pssC, -D, -E, -F, -G, -H, and -I) from a new gene cluster of R. leguminosarum that are involved in EPS biosynthesis have recently been isolated and sequenced (24, 36, 41). The pssDE genes are thought to collectively code for a membrane-associated glucuronosyl-(β1→4)-glucosyl transferase that attaches glucuronic acid (GlcA) to Glc-isoprenylpyrophosphate (33). The pssC gene probably encodes a glucuronosyl-(β1→4)-glucuronosyl transferase responsible for the third transferase step, involving the addition of GlcA to GlcA-Glc-1P* (33); however, the pssC mutant used in this study is still capable of making 38% of the amount of EPS made by the wild-type strain.
Transposon mutagenesis is often used in studies on microbes to identify and sequence genes of interest, investigate the phenotypes resulting from the mutation, and possibly assign function via analogy to homologous genes. These studies can be confounded when pleiotropic effects occur as a result of the mutation. In addition, assigning function in the context of molecular interactions and sequence homology does not always offer a full description of protein function. Protein function needs to be described in the context of higher-order processes such as expression levels and turnover, effects of posttranslational modifications on its activity, involvement in metabolic pathways and signal cascades, and effects of gene knockout or overexpression. The application of global gene expression analysis approaches, such as proteomics, can offer a more complete description of protein function (1, 22). In addition, global gene expression studies have increasingly revealed that single mutations can have unexpected regulatory effects beyond the recognized function of the protein (11).
This study utilized two-dimensional (2-D) gel electrophoresis (2-DE) to examine the responses of various R. leguminosarum strains to specific genetic perturbations in the EPS biosynthesis pathway. The proteomes of R. leguminosarum bv. trifolii strain ANU794 and R. leguminosarum bv. viciae strain VF39 carrying lesions in either the pssA, pssD, pssE, or pssC gene, which either abolished or decreased the capacity of these strains to synthesize EPS, were analyzed for pleiotropic effects caused by the mutation. The aims of this study were to examine the interconnection of the EPS biosynthesis pathway with other cellular functions and to investigate the higher-order processes of protein function.
MATERIALS AND METHODS
Bacterial strains and symbiotic properties.
The strains used in this study and their relevant characteristics are listed in Table 1. Since the nucleotide sequences of pssA from R. leguminosarum bv. viciae (GenBank accession number L48804) and R. leguminosarum bv. trifolii (GenBank accession number Y07549) differ only in three positions (substitutions G-2224 for C, T-2226 for G, and T-2228 for A, respectively), all positions are given according to the former sequence. Strain ANU437, a mutant derivative of R. leguminosarum bv. trifolii strain ANU794, contains a transposon Tn5 insertion upstream of the second GTG start codon after position 2185, the mutant R. leguminosarum bv. viciae VF39 pssA-Km contains a Kmr cassette insertion also upstream of the second start codon after position 2060, and the mutant VF39 pssA::Tn5 contains a Tn5 insertion within the central part of the pssA gene, downstream of the second GTG start codon after position 2359. The symbiotic phenotypes of the pssA mutant strains have previously been described (21, 25, 35). The nucleotide sequence of the VF39 pssE, pssD, and pssC genes is presented in GenBank under accession number AF028810. The mutant strains with Tn5 insertions in either pssD or pssE (after positions 6466 and 6833, respectively) fail to produce EPS and to nodulate V. faba (36). The strain VF39 pssC-Km (a Kmr cassette was introduced into the SalI site after position 5063) produces EPS at levels which are 38% of wild-type levels (36). This strain fails to induce nodules on V. faba.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference(s) or source |
|---|---|---|
| Strains | ||
| R. leguminosarum | ||
| ANU794 | Smr derivative of R. leguminosarum bv. trifolii strain TA1; Nod+ Fix+ on white and subterranean clovers | (7, 35) |
| ANU437 | pssA::Tn5 derivative of ANU794; Smr Kmr Exo− Nod+ Fix− on white and subterranean clovers | (7, 35) |
| ANU437/pVF18 | ANU437 complemented with pSP329 carrying the pssA gene from R. leguminosarum bv. viciae VF39; Smr Exo+ Nod+ Fix+ on white and subterranean clovers | This study |
| ANU437/pSPRH4 | ANU437 complemented with pSP329 carrying the pssA gene from R. leguminosarum bv. phaseoli; Smr Exo+ Nod+ Fix+ on white and subterranean clovers | This study |
| VF39 | Wild-type R. leguminosarum bv. viciae; Rifr Nod+ Fix+ on P. sativum, V. sativa, and V. faba | (21) |
| VF39 pssA::Tn5 | pssA::Tn5 derivative of VF39; Rifr Exo− Nod− on V. faba | (21) |
| VF39 pssA-Km | Derivative of VF39 containing an insertion of a Kmr cassette within pssA; Rifr Kmr Exo+/− Nod+ Fix+/− on V. faba | (25) |
| VF39 pssA::Tn5/pVF18 | VF39 pssA::Tn5 complemented with pSP329 carrying the pssA gene from R. leguminosarum bv. viciae VF39; Rifr Exo+ Nod+ Fix+ on V. faba | (21) |
| VF39 pssA::Tn5/pSPRH4 | VF39 pssA::Tn5 complemented with pSP329 carrying the pssA gene from R. leguminosarum bv. phaseoli; Rifr Exo+ Nod+ Fix+ on V. faba | This study |
| VF39 pssD::Tn5 | pssD::Tn5 derivative of VF39; Rifr Exo− Nod− on V. faba | (36) |
| VF39 pssE::Tn5 | pssE::Tn5 derivative of VF39; Rifr Exo− Nod− on V. faba | (36) |
| VF39 pssC-Km | pssC-Km derivative of VF39; Rifr Exo+ Nod− on V. faba | (36) |
| E. coli S17-1 | MM294, RP4-2, Tc::Mu-Km::Tn7 chromosomally intergrated | (39) |
| Plasmids | ||
| pSP329 | RK2ori, Tcr; derivative of pTJS75 containing pUC18 cloning site | Gift from S. Lory (37) |
| pSPRH4 | Constructed by cloning pssA gene from R. leguminosarum bv. phaseoli 8002 into pSP329 | This study |
| pVF18 | Constructed by cloning pssA gene from R. leguminosarum bv. viciae VF39 into pSP329 | (21) |
Nod+, ability to nodulate; Nod−, inability to nodulate; Fix+, ability to fix nitrogen; Fix−, inability to fix nitrogen; Fix+/−, reduced ability to fix nitrogen; Exo+, ability to produce EPS; Exo−, inability to produce EPS; Exo+/−, reduced production of EPS; Rifr, rifampin resistant; Smr, streptomycin resistant; Kmr, kanamycin resistant; Tcr, tetracycline resistant.
Complementation.
Plasmid pVF18, containing the R. leguminosarum bv. viciae VF39 pssA gene, was described earlier (21). It was constructed by cloning of the EcoRI fragment at positions 954 to 4357 into the broad-host-range vector pSP329. Plasmid pSPRH4, containing the R. leguminosarum bv. phaseoli pssA gene, was constructed by cloning of the EcoRV-HindIII fragment at positions 588 to 1659 of the nucleotide sequence under GenBank accession number X12568. The resulting hybrid plasmids were transferred from the mobilizing strain Escherichia coli S17-1 (39) to VF39 pssA::Tn5 or ANU437 strains as described by Simon (38).
Growth conditions and sample preparation.
Rhizobium strains were grown at 28°C, on a shaker at 200 rpm, in 1 liter of BIII medium (9) containing 2 μg of rifampin per liter (where appropriate) and supplemented with (per liter) 0.2 mg of biotin, 2 mg of thiamine-HCl, and trace elements (0.25 mg of CoCl · 6H2O, 0.25 mg of CuSO4 · 5H2O, 0.25 mg of Na2MoO4 · 2H2O, 3 mg of H3BO3, 3 mg of ZnSO4 · 7H2O, and 10 mg of MnSO4 · 4H2O). During mid-exponential-phase growth (optical density at 600 nm of 0.5 to 0.6) the cells were harvested, washed and lysed as previously described (17).
2-DE and N-terminal sequencing.
2-DE and electroblotting to polyvinylidene difluoride (PVDF) membranes were done by previously described methods (17). Isoelectric focusing in the first dimension was carried out on linear pH 4 to 7 and nonlinear pH 3 to 10 18-cm immobilized pH gradient (IPG) strips (Pharmacia-Biotechnology, Uppsala, Sweden) loaded with 100 μg (0.5 to 1 mg for preparative 2-D gels) of total cellular protein and run for 200 kV · h. N-terminal protein sequencing was done on a PROCISE-HT sequencer system or on a PROCISE-CLC (both from Perkin-Elmer Applied Biosystems, Foster City, Calif.) for very-low-abundance proteins. Proteins were excised from Coomassie blue-stained PVDF blots (17). N-terminal protein sequences were used to search the nonredundant protein databases (SWISS-PROT, PIR, TREMBL and GenPept) using the FASTA program at URL http://www.angis.su.oz.au/. The N-terminal sequences were also screened against the shotgun sequence database made from S. meliloti genomic DNA generated by Melanie Barnett and Sharon Long from Stanford University (see http://cmgm.stanford.edu/∼mbarnett/1xgenome.htm or follow the links from http://sequence.toulouse.inra.fr).
Staining and image analysis of 2-D protein arrays.
Proteins on analytical 2-D gels were visualized by silver staining (34) and digitized at 600 dots per inch (dpi). Silver-stained 2-D protein arrays were analyzed using the MELANIE II program (Bio-Rad, Hercules, Calif.) for the qualitative and quantitative analysis of differentially displayed protein spots. Internal protein standards were used as pI and Mr reference points (17, 18). Three independent experiments with each mutant strain were performed for the analysis of the differences in protein synthesis. A spot was classified as being differentially displayed if the relative spot volume ratio varied more than twofold between mutants and parental wild-type strains in at least two out of three independent experiments.
RESULTS
Altered levels of expression of several gene products in an R. leguminosarum bv. trifolii strain in response to Tn5-induced mutation of pssA.
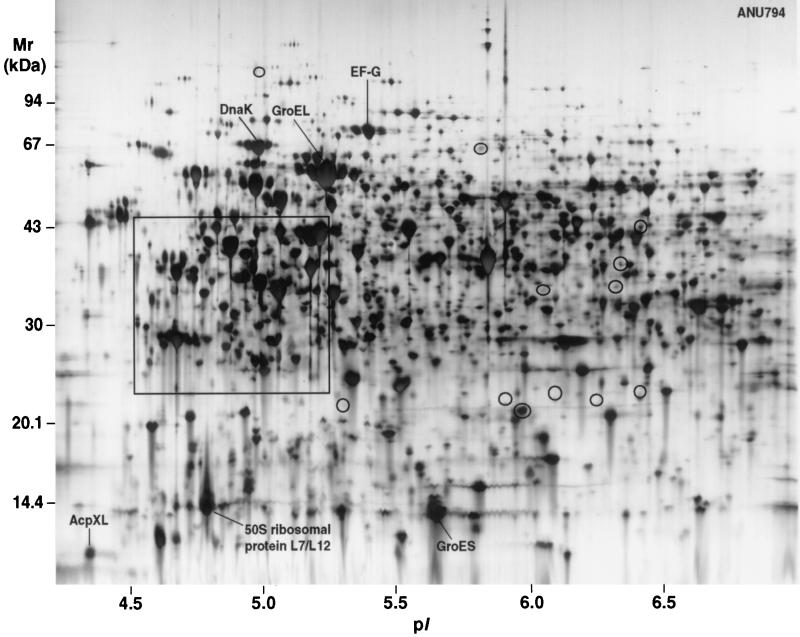
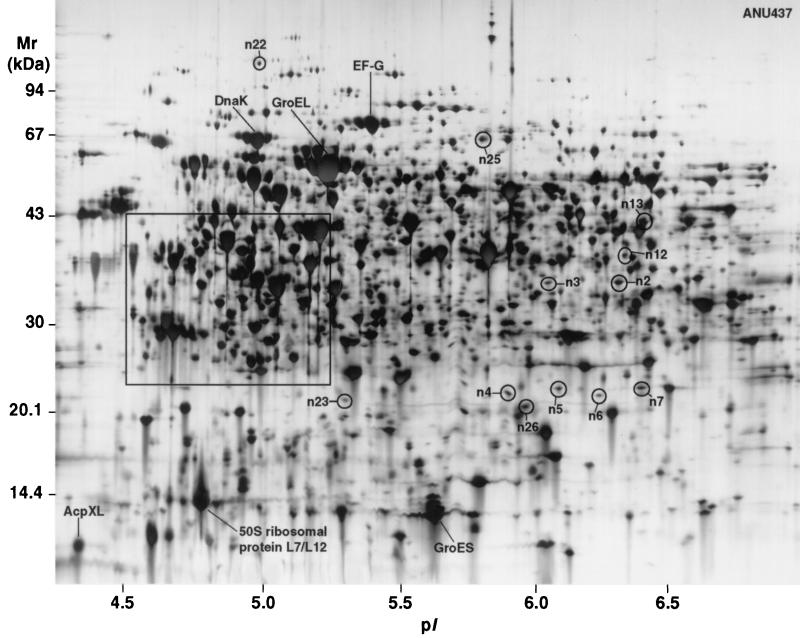
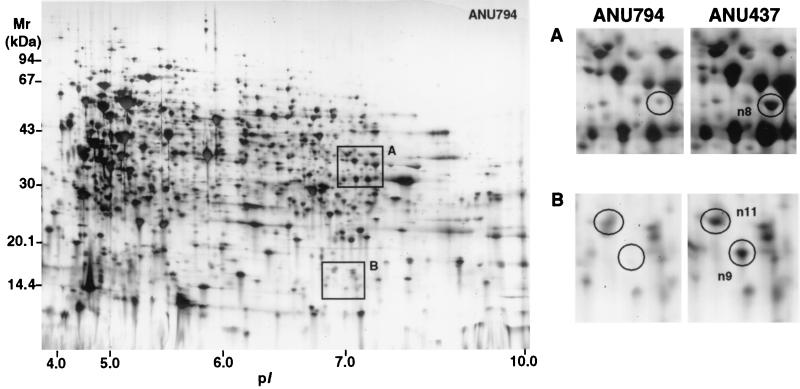
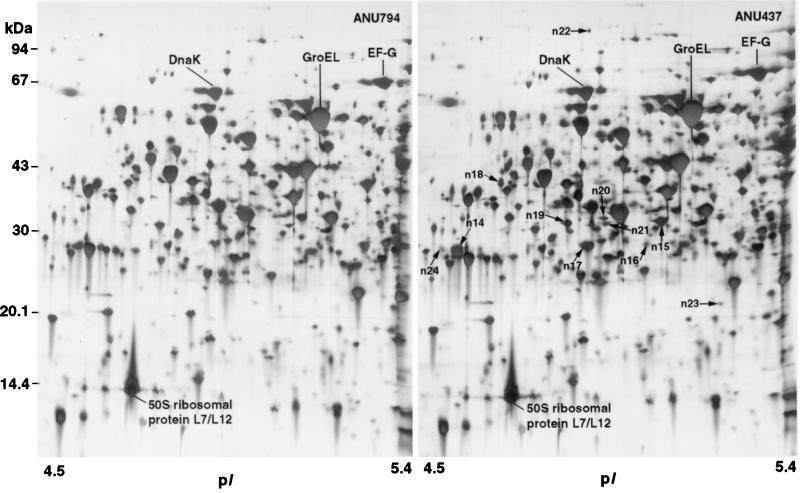
The proteomes of the EPS mutant ANU437 and the wild-type parent ANU794 are shown in Fig. 1, 2, and 3. Image analysis revealed over 2,760 soluble proteins of R. leguminosarum bv. trifolii, resolved as spots in the pI range of 3 to 10 and size range of 10 to 122 kDa. Twenty-three reproducible differences in the levels of protein synthesis were detected when ANU437 was compared with ANU794. Since these proteins are altered in the mutant strain, they have been termed pssA mutant-responsive proteins, and these were grouped as members of the pssA mutant stimulon. The pssA mutant stimulon consists of 15 up-regulated gene products, designated n2, n3, n8, n11 to n13, n15, n17 to n23, and n25 (Fig. 2 and 3), many of which were grossly up-regulated in their levels of abundance. Seven gene products, designated n4 to n7, n9, n16, and n24, were newly synthesized, and protein spot n26 was down-regulated. Narrow-range IPG strips were used for the first dimension of 2-DE to further resolve the proteins occurring in the pI range of 4.5 to 5.4, where many of the spot differences were located, and these are depicted in Fig. 4. The changes in the relative synthesis levels (relative spot volume ratios) of the pssA mutant stimulon proteins are given in Table 2.
FIG. 1.
2-D protein array of R. leguminosarum bv. trifolii strain ANU794. Isoelectric focusing in the first dimension was on IPG strips with a linear gradient ranging from pH 4 to 7 (18 cm) and loaded with 100 μg of total cellular protein. For the second dimension, sodium dodecyl sulfate-polyacrylamide gels (12 to 14% total acrylamide) were used. Proteins were visualized by silver staining. The circled areas correspond to regions on the gel where differences between the wild type and the pssA mutant were identified, and these areas are numbered in Fig. 2. The framed area was further resolved using narrow-range IPG strips, and this is shown in Fig. 4.
FIG. 2.
2-D protein array of R. leguminosarum bv. trifolii strain ANU437 (ANU794 pssA::Tn5). The protein differences identified between ANU794 and ANU437 are circled and assigned arbitrary numbers. The framed area was further resolved using narrow-range IPG strips, and this is shown in Fig. 4.
FIG. 3.
2-D protein array of proteins expressed in ANU794 and resolved in the pI range of 3 to 10. The framed areas are enlarged and represent regions on the map where reproducible spot differences were identified between ANU794 and ANU437. The differentially displayed proteins are circled and assigned arbitrary numbers.
FIG. 4.
2-D gel image representing a protein expression window (proteomic contig) showing proteins from ANU794 and ANU437 in the pI range of 4.5 to 5.4. Narrow-range IPG strips (pH 4.5 to 5.4; 11 cm) loaded with 100 μg of protein were used for the first-dimension separation of 2-DE. The differentially displayed proteins are circled and assigned arbitrary numbers.
TABLE 2.
Relative differential levels of synthesis of proteins in response to pssA mutation in R. leguminosarum bv. trifolii ANU437
| Spota | % Vol in:
|
Change in synthesis levelb
|
Mr/pIc | |||
|---|---|---|---|---|---|---|
| ANU794 | ANU437 | 2-fold | 3-fold | >5-fold | ||
| n2 | 0.031 | 0.080 | + | 37,436/6.31 | ||
| n3 | 0.015 | 0.037 | + | 37,368/6.05 | ||
| n4 | 0 | 0.043 | 22,147/5.90 | |||
| n5 | 0 | 0.035 | 22,880/6.09 | |||
| n6 | 0 | 0.024 | 22,147/6.23 | |||
| n7 | 0 | 0.052 | 22,880/6.38 | |||
| n8 | 0.025 | 0.082 | + | 34,000/7.50 | ||
| n9 | 0 | 0.056 | 16,100/7.00 | |||
| n11 | 0.028 | 0.075 | + | 17,000/6.90 | ||
| n12 | 0.023 | 0.044 | + | 42,570/6.32 | ||
| n13 | 0.034 | 0.064 | + | 50,012/6.39 | ||
| n15 | 0.064 | 0.236 | + | 34,321/5.15 | ||
| n16 | 0 | 0.063 | 30,566/5.11 | |||
| n17 | 0.149 | 0.273 | + | 30,456/4.97 | ||
| n18 | 0.013 | 0.096 | + | 45,109/4.78 | ||
| n19 | 0.048 | 0.154 | + | 34,633/4.94 | ||
| n20 | 0.010 | 0.057 | + | 35,467/5.02 | ||
| n21 | 0.012 | 0.078 | + | 34,197/5.02 | ||
| n22 | 0.004 | 0.025 | + | 114,862/4.99 | ||
| n23 | 0.006 | 0.026 | + | 21,632/5.30 | ||
| n24 | 0 | 0.095 | 31,408/4.64 | |||
| n25 | 0.026 | 0.068 | + | 74,480/5.80 | ||
| n26 | 0.160 | 0.062 | − | 20,939/5.96 | ||
Arbitrary numbers were assigned to proteins which exhibited a change in their relative synthesis levels in response to a Tn5 insertion in the pssA gene of ANU794.
Relative spot volume ratio for a protein from ANU794 and ANU437 of greater (+) or less (−) than two-, three-, or fivefold.
Observed Mr and pI of the protein based on the migration of proteins in the gels.
Identifying members of the R. leguminosarum bv. trifolii pssA mutant stimulon.
The N-terminal amino acid sequences of 15 members of the pssA mutant stimulon were analyzed after they had been electroblotted onto a PVDF membrane and visualized with Coomassie brilliant blue (Table 3). Interestingly, four pairs of newly induced or up-regulated proteins (n4 and n6, n5 and n7, n15 and n21, and n17 and n22) had identical N-terminal sequences. These proteins, except for n17 and n22, most likely represent distinct protein isoforms of similar molecular masses but different pI values. This heterogeneity in charge is likely due to in vivo posttranslational modifications, which can significantly alter the charge on the proteins (46). Interestingly, spots n17 and n22 shared similar pI values, but n22 is approximately four times the size of n17. Spot n22 may represent a protein complex comprised of four n17 subunits, which failed to completely dissociate with the 2-D solubilization and separation procedures used.
TABLE 3.
Proteins in the pssA mutant stimulon from R. leguminosarum bv. trifolii ANU437 analyzed by N-terminal microsequencing and FASTA searching
| ANU437 mutant-responsive proteins
|
Characteristics of matching protein from database
|
|||||
|---|---|---|---|---|---|---|
| Spota | Mr/pI | N-terminal sequenceb | Homologous protein | Organism | Mr/pIc | Swiss-Prot accession no. |
| n2 | 37,436/6.31d | QTDAPLLSKV | MigA | P. aeruginosa | 34,376/8.31 | P95448 |
| :::::.: : | ||||||
| TTDAPLVSVV | ||||||
| 13 22 | ||||||
| n4 | 22,147/5.90e | SNDLIVGFSG | Unknown | |||
| n5 | 22,880/6.09e | SAPKLVGLAGSF | Unknown | |||
| n6 | 22,147/6.23e | SNDLIVGFSGNL | Unknown | |||
| n7 | 22,880/6.38e | SAPKLVGLAGSF | Unknown | |||
| n8 | 34,000/7.50d | SNPVLVNQIIPESRVV | Unknown | |||
| [G/T]LT[A/D]SV | ||||||
| n15 | 34,321/5.15d | ADKKVVVAYQTDALP::::.::: .: .: | Glutamine-binding periplasmic protein | E. coli | 24,963/6.87 | P10344 |
| ADKKLVVATDTAFVP | ||||||
| 22 36 | ||||||
| n16 | 30,566/5.11d | LXXQQKKEFGEFIKQYL | Unknown | |||
| n17 | 30,456/4.97d | EDKSIKVGIMAGEEE | Unknown | |||
| n18 | 45,109/4.78d | DGLSGAPAPFDKGGV | Unknown | |||
| n19 | 34,633/4.94d | TDTVKLRYLASQGNL | Unknown | |||
| n20 | 35,467/5.02d | SILKDDNIDNHADDR | Unknown | |||
| n21 | 34,197/5.02d | ADKKVVVAYQTDALP::::.::: .: .: | Glutamine-binding periplasmic protein | E. coli | 24,963/6.87 | P10344 |
| ADKKLVVATDTAFVP | ||||||
| 22 36 | ||||||
| n22 | 114,862/4.99e | EDKSIKVGIMAGEEEDIXRVVASEAAK | Unknown | |||
| n23 | 21,632/5.30e | WSTFLYT | Unknown | |||
Arbitrary numbers were assigned to proteins which exhibited a change in their level of synthesis in response to a Tn5-induced mutation in the pssA gene of R. leguminosarum bv. trifolii strain ANU794. These numbers correspond to those in Fig. 3, 4, and 5.
N-terminal sequences are shown by using the single-letter for amino acid residues. A comparison of similar sequences is displayed. Colons indicate identical amino acids, and single dots indicate conserved amino acid substitutions.
The predicted molecular weights and isoelectric points of the matched sequences were calculated from the amino acid sequence of the protein using the “compute pI/M.m” tool accessed on the ExPasy server (http://www.expasy.ch).
Sequence alignment searches of the nonredundant protein database revealed that only three protein species exhibited significant sequence homology to previously identified proteins. One differentially displayed protein (n14) was identified as aminoglycoside-3′-O-phosphotransferase, encoded by the Tn5 transposon present in the pssA mutant but not the parental strain (Fig. 4), as expected. Protein spot n2 showed sequence homology to MigA from Pseudomonas aeruginosa. MigA is homologous to glycosyltransferases of other gram-negative bacteria that are involved in the biosynthesis of lipopolysaccharides or EPSs (44). The protein isoforms n15 and n21 matched the glutamine-binding periplasmic protein (GlnH) from E. coli, starting from the residue Ala-23 of the precursor. It has been shown that a signal peptide of the GlnH precursor consists of 22 N-terminal residues (30). Twelve proteins representing nine protein species did not display any significant sequence homology to proteins currently available in the database. All of the open reading frames of Tn5 are known, so none of the nine species are due to Tn5 proteins. This low success in cross-species homology searching is not surprising considering that rhizobia are poorly characterized at the molecular level and that the mutant-responsive proteins of the pssA mutant stimulon are condition specific.
A further attempt to identify proteins with homology to those altered in abundance in strain ANU437 was done by screening the N-terminal sequences against a shotgun database of the entire S. meliloti genome that was translated in all six reading frames (see Materials and Methods). None of the sequenced proteins showed significant homology to entries in this database.
Pleiotropic alterations in the levels of cellular protein synthesis in R. leguminosarum bv. viciae containing a lesion in the pssA gene.
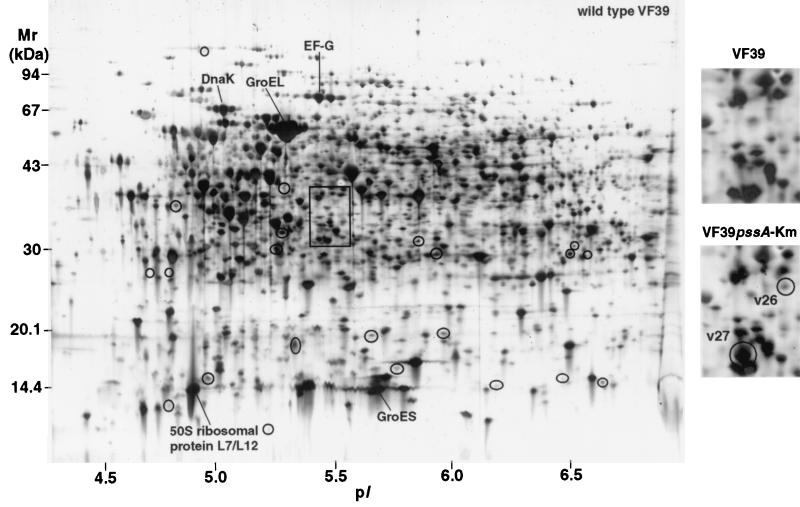
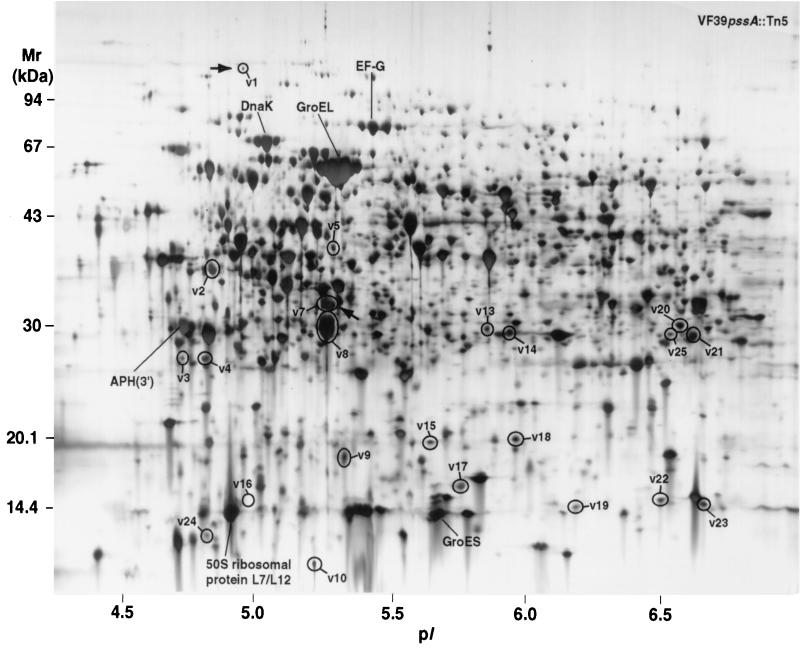
Proteins synthesized in the R. leguminosarum bv. viciae VF39 pssA::Tn5 and VF39 pssA-Km strains were analyzed by 2-DE and compared to those of the wild-type strain (Fig. 5 and 6). Consistent with the results found with ANU437 and ANU794, 22 reproducible spot differences in the level of synthesized proteins were observed between wild-type VF39 and VF39 pssA::Tn5 (Fig. 6). The relative spot volume ratios of these mutant-responsive proteins are given in Table 4. Most of the observed differences involved the up-regulation of gene product expression, while some proteins appeared to be newly synthesized, repressed, or down-regulated in response to the Tn5 mutation. Of the differences observed for VF39 pssA::Tn5, only two mutant-responsive proteins exhibited electrophoretic mobilities similar to those observed for ANU437. These similar spot differences are arbitrarily designated n22 and n15 in the protein profile of ANU437 (Fig. 2) and, respectively, v1 and v7 in the protein profile of VF39 pssA::Tn5 (Fig. 6). Only two protein differences were observed between VF39 pssA-Km and wild-type VF39 (Fig. 5). They involved the induction of two proteins (v26 and v27).
FIG. 5.
2-D protein array of proteins expressed in R. leguminosarum bv. viciae strain VF39 and resolved in the pI range of 4 to 7. The circled areas correspond to regions on the gel where differences between the wild type and the pssA mutant were identified, and these areas are numbered in Fig. 6. Differences between VF39 and VF39 pssA-Km were found only within the framed area, which is enlarged on the right. The spot differences are designated v26 and v27.
FIG. 6.
2-D protein array of R. leguminosarum bv. viciae strain VF39 pssA::Tn5. A total of 22 protein differences between VF39 and VF39 pssA::Tn5 were identified. The differentially displayed proteins are circled and assigned arbitrary numbers. Arrows point to conserved differences found in both VF39 pssA::Tn5 and ANU437 compared to their respective wild-type strains and are based on the spot electrophoretic mobilities. The arrows correspond to spots n22 and n15 in ANU437. APH(3′), aminoglycoside-3′-O-phosphotransferase.
TABLE 4.
Relative differential levels of synthesis of proteins in response to pssA mutation in R. leguminosarum bv. viciae VF39
| Spota | % Vol in:
|
Change in synthesis levelb
|
Mr/pIc | |||
|---|---|---|---|---|---|---|
| VF39 | VF39 pssA::Tn5 | 2-fold | 3-fold | >5-fold | ||
| v1 | 0.005 | 0.011 | + | 114,000/4.98 | ||
| v2 | 0.043 | 0.181 | + | 35,178/4.85 | ||
| v3 | 0 | 0.024 | 26,005/4.74 | |||
| v4 | 0 | 0.100 | 25,620/4.82 | |||
| v5 | 0.013 | 0.068 | + | 38,385/5.30 | ||
| v7 | 0.074 | 0.396 | + | 31,186/5.28 | ||
| v8 | 0.335 | 0.920 | + | 29,294/5.28 | ||
| v9 | 0.086 | 0.175 | + | 17,142/5.34 | ||
| v10 | 0.007 | 0.046 | + | 10,091/5.23 | ||
| v13 | 0.033 | 0.071 | + | 28,990/5.87 | ||
| v14 | 0.057 | 0.118 | + | 28,562/5.98 | ||
| v15 | 0.028 | 0.056 | + | 18,722/5.65 | ||
| v16 | 0.060 | 0 | 14,093/4.99 | |||
| v17 | 0.029 | 0.111 | + | 14,793/5.78 | ||
| v18 | 0.036 | 0.091 | + | 19,047/5.97 | ||
| v19 | 0.012 | 0.042 | + | 13,500/6.19 | ||
| v20 | 0.018 | 0.127 | + | 29,557/6.57 | ||
| v21 | 0 | 0.293 | 28,477/6.62 | |||
| v22 | 0.008 | 0.060 | + | 14,277/6.48 | ||
| v23 | 0.025 | 0.055 | + | 13,764/6.67 | ||
| v24 | 0.018 | 0.049 | + | 11,463/4.83 | ||
| v25 | 0.075 | 0.036 | − | 28,604/6.53 | ||
Arbitrary numbers are assigned to proteins which exhibited a change in their relative synthesis levels in response to a Tn5 insertion in the pssA gene of VF39.
Relative spot volume ratio for a protein from VF39 and VF39 pssA::Tn5 of greater (+) or less (−) than two-, three-, or fivefold.
Observed Mr and pI of the protein based on the migration of the gels.
Proteome analysis of the pssC, pssD, and pssE mutant derivatives of R. leguminosarum bv. viciae.
To examine whether the observed changes in gene expression of the EPS mutants were a result of a dysfunctional pssA gene product or due to a response by the cell to the loss of EPS, we analyzed the proteomes of mutants VF39 pssC-Km, VF39 pssD::Tn5, and VF39 pssE::Tn5, which carry mutations in putative glycosyl transferases involved in EPS biosynthesis. Of the 22 protein changes associated with VF39 pssA::Tn5, 9 were also present in the VF39 pssD::Tn5 and pssE::Tn5 mutants at levels similar to those found in the pssA mutant. These were spots v2 to v5, v9, v13 to v15, and v20. The other 13 pssA::Tn5-associated spot changes were present at wild-type levels in the pssD and pssE mutants. Interestingly, no differences were detected between the mutant VF39 pssC-Km and wild-type VF39, except for one induced spot with an electrophoretic mobility similar to that of v27 in the VF39 pssA-Km proteome which is, therefore, most likely associated with the Kmr cassette.
Complementation analysis of pssA mutants.
The plasmids pVF18 and pSPRH4, containing cloned pssA genes from R. leguminosarum bv. viciae VF39 and R. leguminosarum bv. phaseoli 8002, respectively, were transferred into the pssA mutants ANU437 and VF39 pssA::Tn5. In the case of R. leguminosarum bv. phaseoli, pssA translation starts from the second GTG start codon, leading to a 200-aa-long polypeptide (6). In all four cases, the transconjugants acquired a mucoid morphology comparable to that of the parental strains and were fully complemented for their nodulation-deficient phenotype.
The proteomes of the transconjugants were analyzed to determine the extent to which the wild-type gene can correct the gene expression changes associated with the Tn5 mutation of pssA. 2-D gel comparisons between the ANU437 transconjugants and parental strain ANU794 revealed that all of the protein differences previously observed in ANU437, except for aminoglycoside-3′-O-phosphotransferase encoded by Tn5, were completely corrected. In contrast, the VF39 mutants harboring plasmid pVF18 or pSPRH4 were not fully complemented at the protein synthesis level. Most of the differences observed between VF39 pssA::Tn5 and wild-type VF39 were still present, except for spots v1, v5, v9, v10, v13, v15, and v18, which were corrected to wild-type levels in both VF39 transconjugants. The intensity of spot v22 was reduced to a level closer to that of the wild type.
Grouping the mutant-responsive proteins into EPS- and pssA-dependent responses.
Table 5 shows a comparative analysis of the 22 protein differences identified in the VF39 pssA::Tn5 mutant for the transconjugants and pssD, pssE, and pssC mutants. From the results obtained, the 22 mutant-responsive proteins can be divided into three groups. Group 1 consists of proteins which are responsive to the absence of EPS and include those protein spots whose synthesis levels differed only in strains failing to produce EPS (spots v5, v9, v13, and v15). Group 2 consists of proteins whose wild-type synthesis levels were dependent on the presence of a functional PssA. This group included the protein spots v1, v7, v8, v10, v16 to v19, and v21 to v25. Group 3 consists of proteins which are up-regulated to the same levels in all strains, including pssA transconjugants, but are at wild-type levels in the pssC mutant (v2 to v4, v14, and v20).
TABLE 5.
Comparative analysis of the 22 protein differences associated with VF39 pssA::Tn5 in the pssC, pssD, and pssE mutants and the VF39 pssA::Tn5 transconjugants
| Spot no. | Spot differencea in:
|
|||||
|---|---|---|---|---|---|---|
| VF39 pssA::Tn5b | VF39 pssA::Tn5/pVF18c | VF39 pssA::Tn5/pSPRH4c | VF39 pssD::Tn5c | VF39 pssE::Tn5c | VF39 pssC-Kmc | |
| v1 | Up-reg (+++) | 0 | 0 | 0 | 0 | 0 |
| v2 | Up-reg (+++) | +++ | +++ | +++ | +++ | 0 |
| v3 | Induced (+++) | +++ | +++ | +++ | +++ | 0 |
| v4 | Induced (+++) | +++ | +++ | +++ | +++ | 0 |
| v5 | Up-reg (+++) | 0 | 0 | +++ | +++ | 0 |
| v7 | Up-reg (+++) | +++ | +++ | 0 | 0 | 0 |
| v8 | Up-reg (+++) | +++ | +++ | 0 | 0 | 0 |
| v9 | Up-reg (+++) | 0 | 0 | +++ | +++ | 0 |
| v10 | Induced (+++) | 0 | 0 | 0 | 0 | 0 |
| v13 | Induced (+++) | 0 | 0 | ++ | ++ | 0 |
| v14 | Up-reg (+++) | +++ | +++ | +++ | +++ | 0 |
| v15 | Up-reg (+++) | 0 | 0 | ++ | ++ | 0 |
| v16 | Repressed (−−−) | −−− | −−− | 0 | 0 | 0 |
| v17 | Induced (+++) | +++ | +++ | 0 | 0 | 0 |
| v18 | Up-reg (+++) | 0 | 0 | 0 | 0 | 0 |
| v19 | Up-reg (+++) | +++ | +++ | 0 | 0 | 0 |
| v20 | Up-reg (+++) | +++ | +++ | +++ | +++ | 0 |
| v21 | Induced (+++) | +++ | +++ | 0 | 0 | 0 |
| v22 | Up-reg (+++) | ++ | ++ | 0 | 0 | 0 |
| 23 | Up-reg (+++) | +++ | +++ | 0 | 0 | 0 |
| v24 | Up-reg (+++) | +++ | +++ | 0 | 0 | 0 |
| v25 | Down-reg (−−−) | −−− | −−− | 0 | 0 | 0 |
For comparison, the symbols +++ and −−− are assigned to the spot differences in VF39 pssA::Tn5 and represent the maximum degree of up-regulation (up-reg) or repression, respectively. 0 indicates that the synthesis has been restored to a level similar to that of wild-type VF39.
Protein differences compared with wild-type VF39.
Protein differences compared with VF39 pssA::Tn5.
DISCUSSION
Transposon mutagenesis combined with 2-DE-based proteomics has provided an effective approach to analyzing the higher-order processes of protein function and the complex interconnection of the EPS biosynthetic pathway with other cellular networks. A lesion in either pssA, pssD, or pssE, all of which abolish EPS biosynthesis, resulted in a pleiotropic response at the protein synthesis level in R. leguminosarum bv. trifolii and R. leguminosarum bv. viciae backgrounds. These results demonstrate the complexities of EPS biosynthesis and regulation, which are likely to involve cross talk between regulatory components from different networks, and it is possible that these networks are conserved in these two biovars of R. leguminosarum.
The pleiotropic response to pssA mutation included the up- and down-regulation, repression, and induction in the levels of synthesis of 22 and 23 proteins, respectively, in the R. leguminosarum bv. viciae and R. leguminosarum bv. trifolii mutant strains. A polar effect of the mutation has been ruled out as a reason for these changes in protein synthesis, since (i) a rather strong transcriptional terminator has been identified downstream of pssA, (ii) an open reading frame situated further downstream is transcribed in the opposite direction (21) (GenBank accession number L48804), and (iii) pssD and pssE are not transcribed from the same promoter as pssA. The members of the pssA mutant stimulon represent a functional group, and by further characterizing the members of this group, one can infer more about the function of PssA.
The biological significance of the pssA mutant stimulon proteins remains unclear, as the majority of the N-terminal protein sequences obtained from ANU437 show no homology to sequences available in the current databases. Furthermore, several of these proteins were present as distinct isoforms exhibiting identical N-termini and Mrs but different pIs. Two up-regulated proteins, however, showed significant homology at the N terminus to a putative glycosyl transferase, MigA, from the human pathogen P. aeruginosa, and to a glutamine-binding periplasmic protein GlnH from E. coli. MigA is thought to be involved in the biosynthesis of lipopolysaccharides or EPS in P. aeruginosa, and its expression is induced by respiratory mucus derived from individuals with cystic fibrosis (44). Interestingly, P. aeruginosa is nonmucoid when first recovered from the respiratory tracts of infected individuals, compared to the normal mucoid phenotype (15), suggesting that MigA is induced in response to signals which regulate the levels of lipopolysaccharide and EPS biosynthesis. The expression of the R. leguminosarum bv. trifolii MigA homologue could also be regulated by a similar condition-dependent mechanism, but this requires further investigation. Previous analysis of the extracellular polysaccharides made by ANU437 (35) did not shed light upon a role that this potential MigA homologue might have in the elaboration of an alternative EPS or lipopolysaccharide. GlnH from E. coli forms part of ABC-type solute uptake systems that transport glutamine across the cytoplasmic membrane. The expression of glnD is significantly up-regulated in E. coli cells grown under conditions where glutamine is limiting (29), as would be expected. A possible explanation for the up-regulation of the R. leguminosarum bv. trifolii GlnH homologue in ANU437 is that EPS is required for nutrient gathering and concentrating diffusible molecules from the external milieu along the cell surface. The inability to concentrate nutrients on the cell surface of ANU437 may lead to nutrient deficiency and an up-regulation of this gene and its protein product. We could not find any evidence in the literature to directly link GlnH with EPS synthesis.
The mode of pssA gene expression in the R. leguminosarum biovars trifolii and viciae remains unclear. It is not known whether it is translated in the form of long (263-aa) or short (200-aa) polypeptide. In this study, however, it is clear from the proteomes of the complemented R. leguminosarum bv. trifolii ANU437 transconjugants that the putative 63-aa extension does not play a role in the observed protein differences. We deduced this since the pssA gene from R. leguminosarum bv. phaseoli, which does not have the 63-aa extension, completely restored the proteome of ANU437 to that of the wild type. This is also consistent with the observed proteome for the VF39 pssA-Km mutant. In the latter strain, a Kmr cassette was inserted into the pssA gene just upstream of the −10 sequence of P3 promoter, in contrast to ANU437, where the Tn5 insertion was located downstream of this promoter. Therefore, in the case of VF39 pssA-Km, one can expect the synthesis only of the short version of PssA, although at a reduced level, from the altered P3 promoter or a promoter localized in the Kmr cassette. The mode of pssA gene expression in rhizobia and the functional significance of a 63-aa extension is currently being investigated using antibodies against PssA (V. N. Ksenzenko and T. V. Ivashina unpublished data).
In contrast to the case for ANU437, complementation of VF39 pssA::Tn5 did not completely restore protein synthesis levels to that of the wild type, even though EPS biosynthesis and nodulation efficiency were restored. The transconjugants are merodiploid, since the mutated pssA allele on the genome is genetically complemented rather than genetically replaced and Tn5 is retained in the complemented strain (the strain retains kanamycin resistance, and the protein product of the kanamycin resistance gene is observed on 2-D gels). The only obvious difference between the two pssA mutant strains is the location of the transposon Tn5 within the pssA gene. In the case of VF39 pssA::Tn5, one can hypothesize the synthesis of a truncated PssA protein, since the Tn5 insertion is localized 40 codons downstream of the second GTG codon. In the case of ANU437, a truncated protein cannot be synthesised at all if translation starts from the second GTG codon, or only a short 45-aa polypeptide can be synthesized if translation starts from the first GTG codon. The coexpression of a truncated product with the wild-type protein in the transconjugants may interfere with the function(s) of the latter in a well-recognized process called the dominant-negative inhibition effect (19). Indeed, dominant-negative inhibition has been previously observed in genes involved in EPS synthesis (16). This effect has been used to determine protein function and is common to proteins which form multicomponent complexes but has also been observed for monomeric enzymes (27, 40). In most cases, the mutant variants interfere with the functional assembly of wild-type proteins whose activity depends on oligomerization, interfere with the folding pathway of monomeric wild-type enzymes, or act as competitive inhibitors of the wild-type enzyme when substrate is limiting (19, 27). One could speculate that a truncated pssA gene product may have an inhibitory effect on the activity of unknown functional domains in the wild-type protein, other than the glycosyl transferase domain, because the merodiploid strain produce EPS and nodulates effectively. This hypothetical truncated protein may affect the levels of expression of a number of proteins involved in other regulatory networks. This phenomenon may explain why full complementation is not observed for the VF39 pssA::Tn5 transconjugants. In the case of ANU437, a putative 45-aa polypeptide could be either unstable or insufficient for interaction with wild-type protein compared to the longer truncated protein synthesized in VF39 pssA::Tn5. To determine if a dominant-negative effect is occurring, immunochemical analysis of different forms of the PssA in rhizobium cells needs to be carried out, and this work is currently in progress (Ksenzenko and Ivashina, unpublished data).
By comparing the proteomes from the pssA, pssC, pssD, and pssE mutants of VF39 along with the proteomes from the VF39 pssA::Tn5 transconjugants, the 22 protein differences were grouped into three categories. Mutations in either pssD or pssE led to the loss of EPS synthesis and to the alteration of 9 of the same 22 proteins affected in the corresponding pssA mutant. The 13 other proteins were present at wild-type levels in these two mutants, and these are likely to represent a group of proteins whose synthesis levels change in response to a dysfunctional pssA gene product. Together with the dominant-negative inhibition effect observed, these results further suggest a previously unrecognized multifunctional nature of PssA. Others have proposed earlier that the pssE and pssD genes encode different subunits of the same glycosyl transferase (36). The identical differences in protein synthesis observed for both the pssD and pssE mutants support this hypothesis. Interestingly, no differences in protein synthesis levels were detected in the case of the pssC mutant. From this one can conclude that a complete or nearly complete block in EPS synthesis is needed to induce the observed changes, since reduction of EPS production by more than 60% does not cause any alterations in the VF39 proteome.
A response to the absence of EPS is likely to involve proteins such as the previously mentioned GlnH homologue which are likely to be dependent on the putative functional roles attributed to EPS in nutrient gathering. Alternatively, a block in the EPS biosynthetic pathway, particularly at the pssA step, may lead to an increase in the cellular pool of UDP-glucose, which would likely be filtered through to other pathways, and thus an absence of EPS may indirectly affect the levels of expression of proteins involved in these pathways. In addition, Pollock et al. (33) have suggested that the formation and accumulation of incomplete lipid-linked intermediates in EPS mutants may be detrimental largely because the cells are likely not to release the IP carrier from the incomplete repeat structure for other essential cellular functions such as the synthesis of peptidoglycan. It is possible that this disruption provides a secondary signal to disrupt regulatory networks or other metabolic pathways. However, in the pssD, pssE, and pssC mutants the presence of incomplete lipid-linked intermediates in the cells did not appear to further affect protein synthesis, as no differences apart from to those 22 observed for pssA were seen and a free IP carrier is available in the pssA mutant.
One other group of proteins exhibited elevated synthesis levels in all mutant strains and the transconjugants, except the pssC mutant. These proteins are unlikely to be associated with the Tn5 transposon, as they are not present in ANU437 and have Mrs and pIs that differ from those of Tn5-encoded proteins. Sequence information would need to be obtained to investigate the relevance of these proteins. Since these protein differences are absent from the pssC mutant, one explanation is that the gene products of pssA, pssD, and pssE may form a functional protein complex, as postulated for EPS production in S. meliloti (12), and that the disruption of this complex by mutagenesis or by a truncated pssA gene product may lead to the up-regulation of a unique set of proteins.
Studying the proteomes of EPS mutants has become particularly relevant to functional analysis, especially in terms of higher-order processes such as protein-protein interactions and the description of novel networks of interactions. The results from this study have indicated that the pssA gene product, in addition to being a glycosyl transferase, may also serve other functions by directly or indirectly influencing the expression of a complex series of unknown genes. The more that is known about the proteins exhibiting the response, the more that can be inferred about the function of PssA. In addition, the large number of genes whose expression is altered in response to a perturbation in the pssA gene highlights the challenges of understanding the higher-order processes of protein function and the complex interconnection of functional networks.
ACKNOWLEDGMENTS
This work was partially supported by the Russian Foundation for Basic Research (grant 98-04-48918).
We thank Jill McGovern from the Biomolecular Resource Facility, Australian National University, for performing the N-terminal sequence analysis of proteins. Sharon Long and Melanie Barnett are thanked for providing access to the S. meliloti shotgun database of genomic sequences constructed in their laboratory.
REFERENCES
- 1.Anderson N, Anderson N. Proteome and proteomics: new technologies, new concepts, and new words. Electrophoresis. 1998;19:1853–1861. doi: 10.1002/elps.1150191103. [DOI] [PubMed] [Google Scholar]
- 2.Becker A, Kleickmann A, Keller M, Arnold W, Puhler A. Identification and analysis of the Rhizobium meliloti exoAMONP genes involved in exopolysaccharide biosynthesis and mapping of promoters located on the exoHKLAMONP fragment. Mol Gen Genet. 1993;241:367–379. doi: 10.1007/BF00284690. [DOI] [PubMed] [Google Scholar]
- 3.Becker A, Kleickmann A, Kuster H, Keller M, Arnold W, Puhler A. Analysis of the Rhizobium meliloti genes exoU, exoV, exoW, exoT, and exoI involved in exopolysaccharide biosynthesis and nodule invasion: exoU and exoW probably encode glucosyltransferases. Mol Plant-Microbe Interact. 1993;6:735–744. doi: 10.1094/mpmi-6-735. [DOI] [PubMed] [Google Scholar]
- 4.Becker A, Puhler A. Production of exopolysaccharides. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae: molecular biology of model plant-associated bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 97–118. [Google Scholar]
- 5.Beveridge T J, Graham L L. Surface layers of bacteria. Microbiol Rev. 1991;55:684–705. doi: 10.1128/mr.55.4.684-705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borthakur D, Barker R F, Latchford J W, Rossen L, Johnston A W. Analysis of pss genes of Rhizobium leguminosarum required for exopolysaccharide synthesis and nodulation of peas: their primary structure and their interaction with psi and other nodulation genes. Mol Gen Genet. 1988;213:155–162. doi: 10.1007/BF00333413. [DOI] [PubMed] [Google Scholar]
- 7.Chakravorty A K, Zurkowski W, Shine J, Rolfe B G. Symbiotic nitrogen fixation: molecular cloning of Rhizobium genes involved in exopolysaccharide synthesis and effective nodulation. J Mol Appl Genet. 1982;1:585–596. [PubMed] [Google Scholar]
- 8.Cheng H-P, Walker G C. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J Bacteriol. 1998;180:5183–5191. doi: 10.1128/jb.180.19.5183-5191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dazzo F B. Leguminous root nodules. In: Burns R G, Slater J L, editors. Experimental microbiol ecology. Oxford, United Kingdom: Blackwell Scientific; 1982. pp. 431–446. [Google Scholar]
- 10.Djordjevic S P, Chen H, Batley M, Redmond J W, Rolfe B G. Nitrogen fixation ability of exopolysaccharide synthesis mutants of Rhizobium sp. strain NGR234 and Rhizobium trifolii is restored by the addition of homologous exopolysaccharides. J Bacteriol. 1987;169:53–60. doi: 10.1128/jb.169.1.53-60.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fey S, Nawrocki A, Larsen M, Gorg A, Roepstorff P, Skews G, Williams R, Larson P M. Proteome analysis of Saccharomyces cerevisiae: a methodological outline. Electrophoresis. 1997;18:1361–1372. doi: 10.1002/elps.1150180811. [DOI] [PubMed] [Google Scholar]
- 12.Glucksmann M A, Reuber T L, Walker G C. Family of glycosyl transferases needed for the synthesis of succinoglycan by Rhizobium meliloti. J Bacteriol. 1993;175:7033–7044. doi: 10.1128/jb.175.21.7033-7044.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glucksmann M A, Reuber T L, Walker G C. Genes needed for the modification, polymerization, export, and processing of succinoglycan by Rhizobium meliloti: a model for succinoglycan biosynthesis. J Bacteriol. 1993;175:7045–7055. doi: 10.1128/jb.175.21.7045-7055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzales J E, York G M, Walker G C. Rhizobium meliloti exopolysaccharides: synthesis and symbiotic function. Gene. 1996;179:141–146. doi: 10.1016/s0378-1119(96)00322-8. [DOI] [PubMed] [Google Scholar]
- 15.Govan J R W. Alginate biosynthesis and unusual characteristics associated with the pathogenesis of Pseudomonas aeruginosa in cystic fibrosis. In: Griffiths E, Donachie W, Stephen J, editors. Bacterial infections of respiratory and gastrointestinal mucosae. Oxford, United Kingdom: IRL Press; 1988. pp. 67–96. [Google Scholar]
- 16.Gray J X, Djordjevic M A, Rolfe B G. Two genes that regulate exopolysaccharide production in Rhizobium sp. strain NGR234: DNA sequences and resultant phenotypes. J Bacteriol. 1990;172:193–203. doi: 10.1128/jb.172.1.193-203.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerreiro N, Redmond J W, Rolfe B G, Djordjevic M A. New Rhizobium leguminosarum flavonoid-induced proteins revealed by proteome analysis of differentially displayed proteins. Mol Plant-Microbe Interact. 1997;10:506–516. doi: 10.1094/MPMI.1997.10.4.506. [DOI] [PubMed] [Google Scholar]
- 18.Guerreiro N, Stepkowski T, Rolfe B G, Djordjevic M A. Determination of plasmid-encoded functions in Rhizobium leguminosarum biovar trifolii using proteome analysis of plasmid-cured derivatives. Electrophoresis. 1998;19:1972–1979. doi: 10.1002/elps.1150191115. [DOI] [PubMed] [Google Scholar]
- 19.Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- 20.Hubac C, Ferran J, Guerrier D, Tremolieres A, Kondorosi A. Luteolin absorption in Rhizobium meliloti wild-type and mutant strains. J Gen Microbiol. 1993;139:1571–1578. [Google Scholar]
- 21.Ivashina T V, Khmelnitsky M A, Shlyapnikov M G, Kanapin A A, Ksenzenko V N. The pssA gene from Rhizobium leguminosarum bv. viciae VF39: cloning, sequence and the possible role in polysaccharide production and nodule formation. Gene. 1994;150:111–116. doi: 10.1016/0378-1119(94)90868-0. [DOI] [PubMed] [Google Scholar]
- 22.James P. Protein identification in the post-genome era: the rapid rise of proteomics. Q Rev Biophys. 1997;30:279–331. doi: 10.1017/s0033583597003399. [DOI] [PubMed] [Google Scholar]
- 23.Keller M, Roxlau A, Weng W M, Schmidt M, Quandt J, Niehaus K, Jording D, Arnold W, Puhler A. Molecular analysis of the Rhizobium meliloti mucR gene regulating the biosynthesis of the exopolysaccharides succinoglycan and galactoglucan. Mol Plant-Microbe Interact. 1995;8:267–277. doi: 10.1094/mpmi-8-0267. [DOI] [PubMed] [Google Scholar]
- 24.Krol J, Wielbo J, Mazur A, Kopcinska J, Lotocka B, Golinowski W, Skorupska A. Molecular characterisation of pssCDE genes of Rhizobium leguminosarum bv. trifolii strain TA1: pssD mutant is affected in exopolysaccharide synthesis and endocytosis of bacteria. Mol Plant-Microbe Interact. 1998;11:1142–1148. doi: 10.1094/MPMI.1998.11.11.1142. [DOI] [PubMed] [Google Scholar]
- 25.Ksenzenko V N, Ivashina T V, Sadykhov M R, Chatuev B M, Khmelnitsky M I, Kanapin A A, Shlyapnikov M G. A role of the Rhizobium leguminosarum bv. viciae pss1 and pss4 genes in nitrogen fixing symbiosis. In: Kiss G B, Endre G, editors. First European Nitrogen Fixation Conference and Workshop on Safe Application of Genetically Modified Microorganisms in the Environment. Szeged, Hungary: Officina Press; 1994. pp. 142–146. [Google Scholar]
- 26.Leigh J A, Walker G C. Exopolysaccharides of Rhizobium: synthesis, regulation and symbiotic function. Trends Genet. 1994;10:63–67. doi: 10.1016/0168-9525(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 27.Michaels J E, Schimmel P, Shiba K, Miller W T. Dominant negative inhibition by fragments of a monomeric enzyme. Proc Natl Acad Sci USA. 1996;93:14452–14455. doi: 10.1073/pnas.93.25.14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niehaus K, Kapp D, Puhler A. Plant defence and delayed infection of alfalfa pseudonodules induced by an exopolysaccharide (EPSI)-deficient Rhizobium meliloti mutant. Planta. 1993;190:415–425. [Google Scholar]
- 29.Nohno T, Saito T. Two transcriptional start sites found in the promoter region of Escherichia coli glutamine permease operon, glnHPQ. Nucleic Acids Res. 1987;15:2777. doi: 10.1093/nar/15.6.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nohno T, Saito T, Hong J S. Cloning and complete nucleotide sequence of the Escherichia coli glutamine permease operon (glnHPQ) Mol Gen Genet. 1986;205:260–269. doi: 10.1007/BF00430437. [DOI] [PubMed] [Google Scholar]
- 31.Philip-Hollingsworth S, Hollingsworth R I, Dazzo F B. Host-range related structural features of the acidic extracellular polysaccharides of Rhizobium trifolii and Rhizobium leguminosarum. J Biol Chem. 1989;264:1461–1466. [PubMed] [Google Scholar]
- 32.Philip-Hollingsworth S, Hollingsworth R I, Dazzo F B, Djordjevic M A, Rolfe B G. The effect of interspecies transfer of Rhizobium host-specific nodulation genes on acidic polysaccharide structure and in situ binding by host lectin. J Biol Chem. 1989;264:5710–5714. [PubMed] [Google Scholar]
- 33.Pollock T J, van Workum W A T, Thorne L, Mikolajczak M J, Yamazaki M, Kijne J W, Armentrout R W. Assignment of biochemical functions to glycosyl transferase genes which are essential for biosynthesis of exopolysaccharides in Sphingomonas strain S88 and Rhizobium leguminosarum. J Bacteriol. 1998;180:586–593. doi: 10.1128/jb.180.3.586-593.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rabilloud T, Vuillard L, Gilly C, Lawrence J C. Silver-staining of proteins in polyacrylamide gels: a general overview. Cell Mol Biol. 1994;40:57–75. [PubMed] [Google Scholar]
- 35.Rolfe B G, Carlson R W, Ridge R W, Dazzo F B, Mateos P F, Pankhurst C E. Defective infection and nodulation of clovers by exopolysaccharide mutants of Rhizobium leguminosarum bv. trifolii. Aust J Plant Physiol. 1996;23:285–303. [Google Scholar]
- 36.Sadykov M R, Ivashina T V, Kanapin A A, Shlyapnikov M G, Ksenzenko V N. Structural and functional organisation of the exopolysaccharide biosynthesis genes in Rhizobium leguminosarum bv. viciae VF39. Mol Biol. 1998;32:665–671. [PubMed] [Google Scholar]
- 37.Schmidhauser T J, Helinski D R. Regions of broad-host-range plasmid RK2 involved in replication and stable maintenance in nine species of gram-negative bacteria. J Bacteriol. 1985;164:446–455. doi: 10.1128/jb.164.1.446-455.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon R. High frequency mobilisation of gram-negative bacterial replicons by the in vivo constructed Tn5-Mob transposon. Mol Gen Genet. 1984;196:413–420. doi: 10.1007/BF00436188. [DOI] [PubMed] [Google Scholar]
- 39.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 40.Singh S, Tang H K, Lee J-Y, Saunders G F. Truncation mutations in the transactivation region of PAX6 result in dominant-negative mutants. J Biol Chem. 1998;273:21531–21541. doi: 10.1074/jbc.273.34.21531. [DOI] [PubMed] [Google Scholar]
- 41.van Workum W A T, Canter Cremers H C J, Wijfjes A H M, van der Kolk C, Wijffelman C A, Kijne J W. Cloning and characterisation of four genes of Rhizobium leguminosarum bv. trifolii involved in exopolysaccharide production and nodulation. Mol Plant-Microbe Interact. 1997;10:290–301. doi: 10.1094/MPMI.1997.10.2.290. [DOI] [PubMed] [Google Scholar]
- 42.van Workum W A T, van Brussel A A N, Tak T, Wijffelman C A, Kijne J W. Ethylene prevents nodulation of Vicia sativa sp. Nigra by exopolysaccharide-deficient mutants of Rhizobium leguminosarum bv. viciae. Mol Plant-Microbe Interact. 1995;8:278–285. [Google Scholar]
- 43.van Workum W A T, van Slageren S, van Brussel A A N, Kijne J W. Role of exopolysaccharides of Rhizobium leguminosarum bv. viciae as host plant-specific molecules required for infection thread formation during nodulation of Vicia sativa. Mol Plant-Microbe Interact. 1998;11:1233–1241. [Google Scholar]
- 44.Wang J, Lory S, Ramphal R, Jin S. Isolation and characterization of Pseudomonas aeruginosa genes inducible by respiratory mucus derived from cystic fibrosis patients. Mol Microbiol. 1996;22:1005–1012. doi: 10.1046/j.1365-2958.1996.01533.x. [DOI] [PubMed] [Google Scholar]
- 45.Whitfield C. Bacterial extracellular polysaccharides. Can J Microbiol. 1988;34:415–420. doi: 10.1139/m88-073. [DOI] [PubMed] [Google Scholar]
- 46.Wilkins M R, Sanchez J C, Williams K L, Hochstrasser D F. Current challenges and future applications for protein maps and post-translational vector maps in proteome projects. Electrophoresis. 1996;17:830–838. doi: 10.1002/elps.1150170504. [DOI] [PubMed] [Google Scholar]