Abstract
Proprotein convertase subtilisin/kexin type-9 (PCSK9) is a posttranslational regulator of the LDL receptor (LDLR). Recent studies have proposed a role for PCSK9 in regulating immune responses. Using RNA-Seq–based variant discovery, we identified a possible psoriasis-susceptibility locus at 1p32.3, located within PCSK9 (rs662145 C > T). This finding was verified in independently acquired genomic and RNA-Seq data sets. Single-cell RNA-Seq (scRNA-Seq) identified keratinocytes as the primary source of PCSK9 in human skin. PCSK9 expression, however, was not uniform across keratinocyte subpopulations. scRNA-Seq and IHC demonstrated an epidermal gradient of PCSK9, with expression being highest in basal and early spinous layer keratinocytes and lowest in granular layer keratinocytes. IL36G expression followed the opposite pattern, with expression highest in granular layer keratinocytes. PCSK9 siRNA knockdown experiments confirmed this inverse relationship between PCSK9 and IL36G expression. Other immune genes were also linked to PCSK9 expression, including IL27RA, IL1RL1, ISG20, and STX3. In both cultured keratinocytes and nonlesional human skin, homozygosity for PCSK9 SNP rs662145 C > T was associated with lower PCSK9 expression and higher IL36G expression, when compared with heterozygous skin or cell lines. Together, these results support PCSK9 as a psoriasis-susceptibility locus and establish a putative link between PCSK9 and inflammatory cytokine expression.
Keywords: Autoimmunity, Dermatology
Keywords: Clinical practice, Skin
Introduction
Over the past decade, GWAS have identified over 60 psoriasis-susceptibility loci (1). However, psoriasis has an estimated heritability of 80%; thus, association studies have likely only captured a fraction of this heritability. Contrariwise, there are thousands of differentially expressed genes (DEGs) in psoriatic skin (2), but only a small fraction are known to contribute to psoriasis pathophysiology. The vast majority of DEGs remain unexplored in psoriasis. Several of these unexplored psoriasis DEGs are being actively pursued as therapeutic targets for other diseases, making the list of ready-to-administer drugs with potential efficacy in psoriasis expansive. Thus, increasing our knowledge of genetic predisposition to psoriasis may identify additional therapeutic targets.
Recently, expression of PCSK9, a posttranslational regulator of the LDL receptor (LDLR) that leads to LDLR internalization and degradation (3), has been shown to be altered in animal models of psoriasis and in the serum and skin of patients with psoriasis (4, 5). Furthermore, the PCSK9 inhibitor evolocumab, which is approved for the treatment of hypercholesterolemia, has been associated with the development of various inflammatory skin conditions including psoriasis (6, 7). Studies have also demonstrated a role of PCSK9 in the regulation of apoptosis and proinflammatory cytokine secretion from macrophages (8). Although PCSK9 SNPs (e.g., rs662145 C > T) have been linked to cardiovascular disease and Alzheimer’s disease (9, 10), their association with psoriasis remains unknown. Similarly, the expression patterns and role of PCSK9 in human keratinocytes in healthy individuals and patients with psoriasis are uninvestigated.
Here, we explore PCSK9 as a possible psoriasis-susceptibility locus. We show that an SNP at the 3′ untranslated region of PCSK9 (rs662145 C > T) predisposed individuals to psoriasis, a finding that was verified in independently acquired psoriasis RNA-Seq and genomic data sets. Using single-cell RNA-Seq (scRNA-Seq), keratinocytes were identified as the predominant cellular source of PCSK9 in human skin. Analysis of PCSK9 in cultured primary keratinocytes revealed a strong negative correlation between PSCK9 and IL36G and IL36B, a relationship further supported by IHC data. Specifically, an epidermal gradient of PCSK9 was detected with expression highest in basal and early spinous layer keratinocytes and lowest in granular layer keratinocytes. In contrast, the gradient was reversed for IL-36, which was predominantly expressed in granular layer keratinocytes. PCSK9 siRNA knockdown in immortalized keratinocytes confirmed the inverse relationship between PCSK9 and IL36. Finally, we found that IL36 expression was increased in individuals homozygous for the PCSK9 (rs662145 C > T) variant when compared with their heterozygous counterparts. Together these results link PCSK9 to psoriasis and inflammatory cytokine production in human skin.
Results
PCSK9 is a possible psoriasis-susceptibility locus.
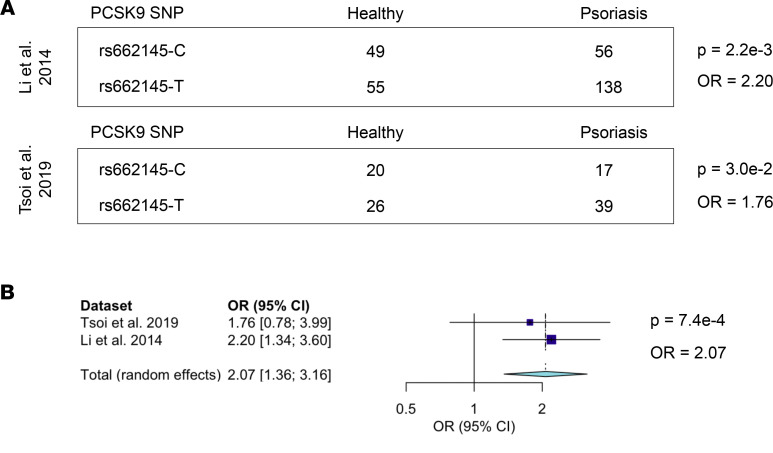
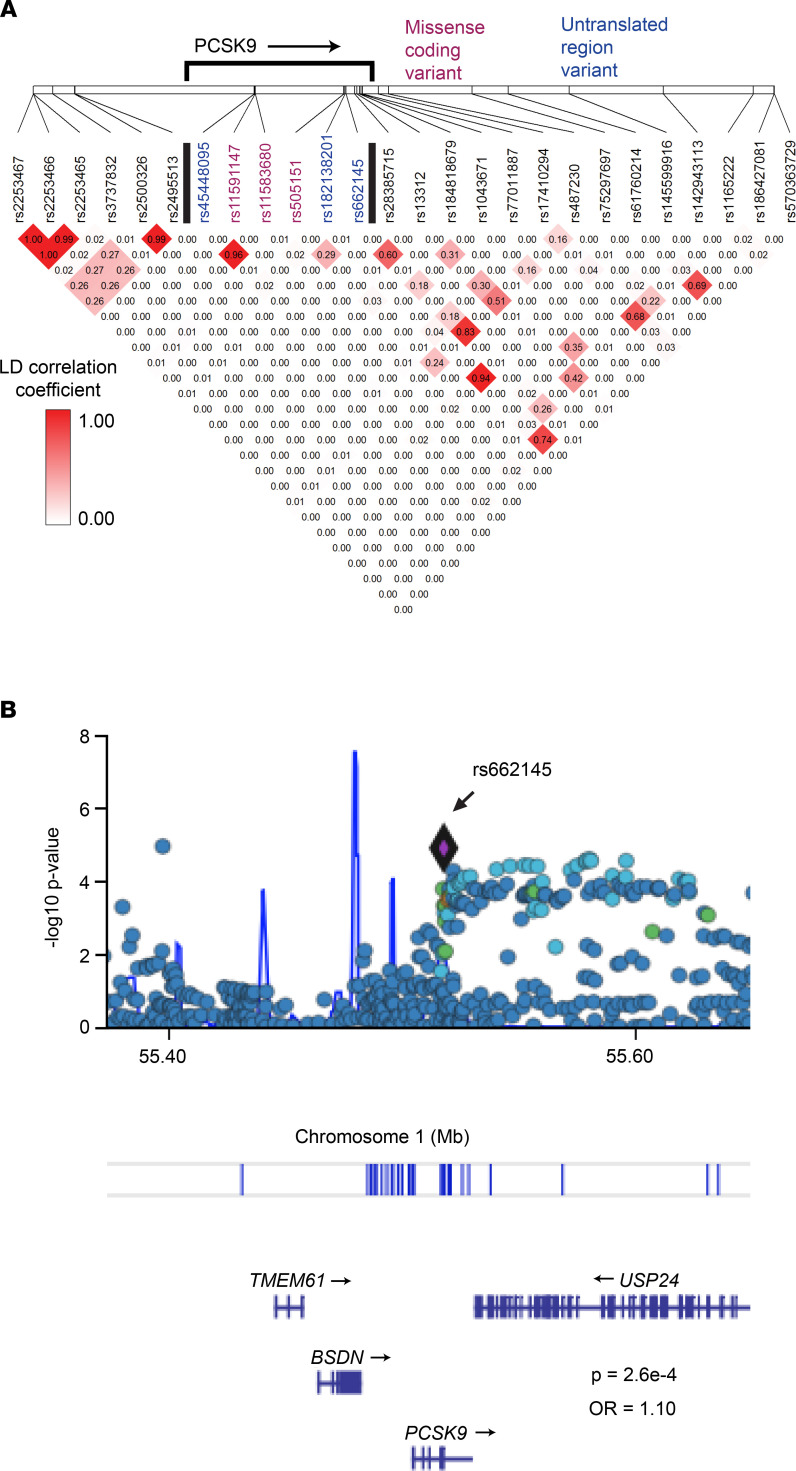
Given that patients on PCSK9 inhibitors have been reported to develop various inflammatory skin eruptions (6, 7), we used RNA-Seq–based genetic variant discovery to mine the largest published psoriasis transcriptome data set (11) for SNPs of PCSK9. This analysis identified 13 PCSK9 SNPs. When compared with healthy controls, SNP rs662145 C > T was more prevalent among patients with psoriasis, a finding that remained significant after adjusting for multiple testing (OR = 2.20, P = 2.2 × 10–3). This finding was then confirmed in a second published independent psoriasis data set (OR = 1.76, P = 3.0 × 10–2) (Figure 1, A and B). A meta-analysis of these 2 data sets yielded a final combined OR of 2.07 (P = 6.5 × 10–3) (Figure 1B, and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.141193DS1). The RNA-Seq–based genetic variant discovery method used to establish this linkage was then validated by calculating the linkage disequilibrium of rs662145 C > T with other nearby alleles (12), comparing the RNA-based results to those obtained from the 1000 Genomes Project data set as the gold standard. This analysis revealed a similar pattern of linkage disequilibrium for rs662145 C > T regardless of the data type used (Figure 2A, and Supplemental Figure 1). To further confirm the association between PCSK9 SNP rs662145 C > T and psoriasis, we next examined the 1p32.3 genomic region in 2590 patients with psoriasis and 1720 controls, using data from previously published and recently acquired data sets (13). Similar to the results obtained from our variant discovery method, the genomic data revealed a significant linkage between PCSK9 rs662145 C > T and psoriasis (OR = 1.10, P = 2.6 × 10–4; Figure 2B, purple diamond). Thus, in multiple independent data sets, PCSK9 rs662145 C > T was associated with an increased risk of psoriasis.
Figure 1. RNA-Seq variant calling identified a psoriasis-associated SNP in 3′ UTR of the PCSK9 gene.
(A) SNP calling was performed on 2 separate psoriasis RNA-Seq data sets (12, 14), and ORs were calculated using the allele counting method. Fisher’s exact test was performed to calculate P values. Names of analyzed data sets are shown on the left with OR and P values on the right. The presence of SNP rs662145 C > T was associated with increased psoriasis risk. SNP rs662145-C constitutes the reference and minor allele, and SNP rs662145-T constitutes the alternative and major allele. (B) Meta-analysis of both RNA-Seq data sets using a random-effects model.
Figure 2. Genomic data confirmed a significant linkage between PCSK9 SNP rs662145 C > T and psoriasis.
(A) PCSK9 SNP linkage disequilibrium estimated from a psoriasis RNA-Seq data set (12) (see data set details in Supplemental Table 1). (B) Analysis of GWAS data from 2590 cases of psoriasis and 1720 controls (13) revealed a significant linkage between PCSK9 rs662145 C > T and psoriasis (P = 2.6 × 10–4).
Psoriasis-linked PCSK9 variant rs662145 C > T is associated with altered PCSK9 and IL36 expression.
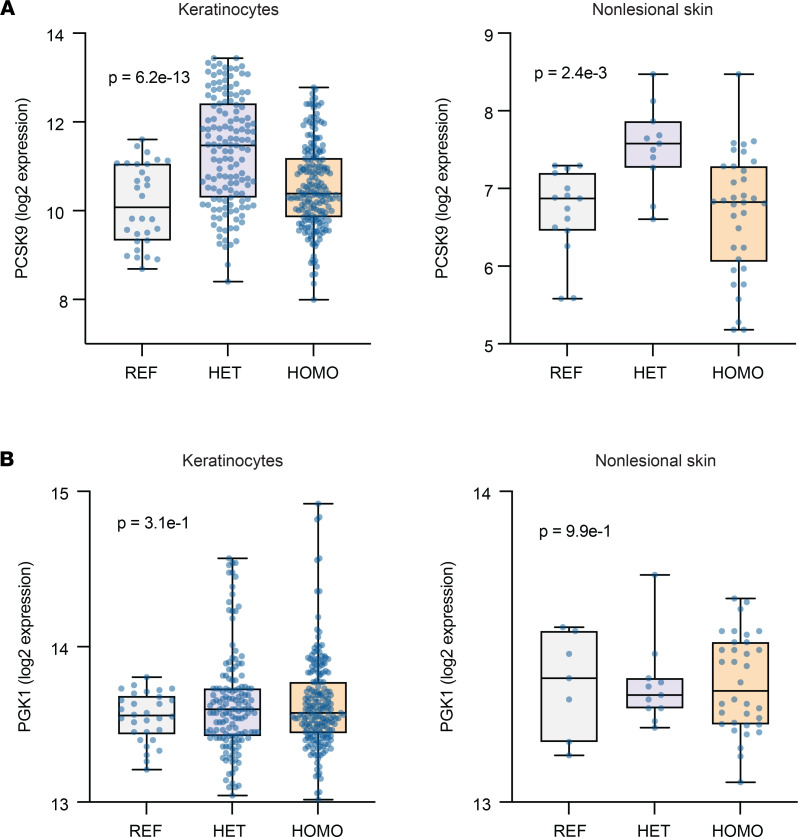
After identifying PCSK9 as a possible psoriasis-susceptibility locus, we next sought to determine how the psoriasis-associated PCSK9 variant, rs662145 C > T, may predispose individuals to psoriasis. We first parsed 2 RNA-Seq data sets — a newly generated data set derived from primary human keratinocytes isolated from healthy individuals without psoriasis and a data set derived from nonlesional skin that we previously generated (14) — to characterize the effect of SNP rs662145 on PCSK9 expression. Box-and-whisker plots of PCSK9 expression were constructed for each data set (Figure 3, A and B). Within each data set, samples were parsed into 3 groups based on the PCSK9 allele they expressed — i.e., homozygous for the reference allele (SNP rs662145-C), heterozygous for the alternative allele (SNP rs662145-T), and homozygous for the alternative allele. This analysis revealed that when compared with their homozygous counterparts, primary human healthy keratinocytes heterozygous for PCSK9 SNP rs662145 C > T exhibited increased expression of PCSK9 (P = 6.2 × 10–13; Figure 3A). The same pattern of PCSK9 expression was also observed in nonlesional human skin (P = 2.4 × 10–3; Figure 3A).
Figure 3. PCSK9 SNP rs662145 C > T is associated with altered expression of PCSK9 in cultured keratinocytes and nonlesional skin.
(A) In cultured keratinocyte cell lines and nonlesional skin, PCSK9 SNP rs662145 C > T HOMO phenotypes expressed lower levels of PCSK9 compared with PCSK9 SNP rs662145 C > T HET phenotypes. Box-and-whisker plots show normalized PCSK9 expression (log transformed reads on y axis). Each dot represents 1 keratinocyte line or skin sample. Differential gene expression was calculated using DESeq2. FDR-adjusted P values are displayed on each plot. (B) In cultured keratinocyte cell lines and nonlesional skin, there was no difference in the expression of PGK1, a housekeeping gene, between the reference allele and PCSK9 SNP rs662145 C > T HET and HOMO phenotypes. HOMO, homozygous; HET, heterozygous.
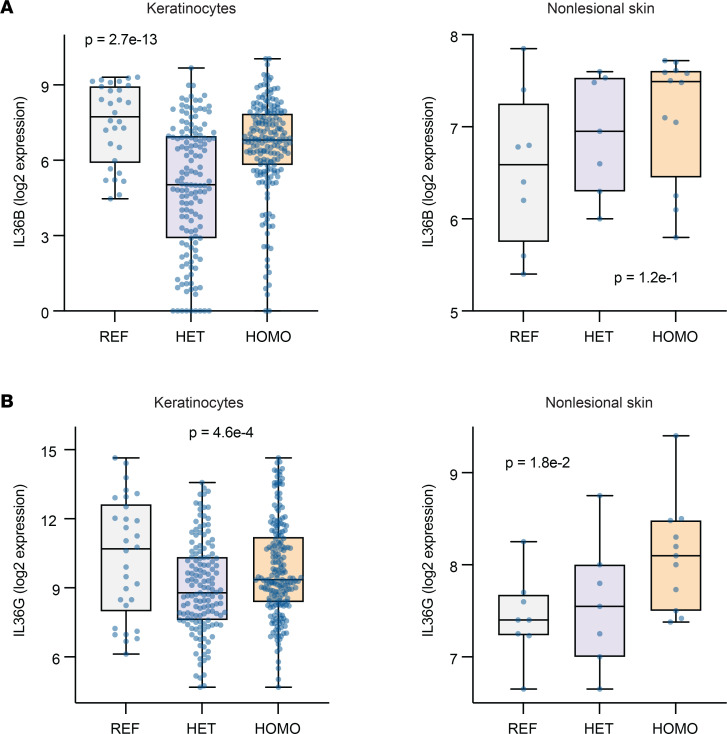
We next aimed to establish if PCSK9 expression was linked to any well-known mediator of psoriasis. Examining PCSK9 expression in cultured keratinocytes revealed that SNP rs662145 C > T was associated with differential expression of IL36, an IL-1 family member and psoriasis-defining cytokine (15, 16). Comparing IL36 expression in primary human keratinocytes homozygous and heterozygous for PCSK9 SNP rs662145 C > T revealed that homozygosity for rs662145 C > T was associated with lower expression levels of PCSK9 (Figure 3A) and higher levels of IL36B and IL36G (Figure 4, A and B). This same expression pattern was also observed in nonlesional human skin — i.e., homozygosity for PCSK9 SNP rs662145 C > T was associated with lower PCSK9 expression (Figure 3A) and higher IL36G expression (Figure 4B) when compared with heterozygosity for PCSK9 SNP rs662145 C > T.
Figure 4. PCSK9 SNP rs662145 C > T is associated with altered expression of IL36 in cultured keratinocytes and nonlesional skin.
(A) In cultured keratinocyte cell lines and nonlesional skin, PCSK9 SNP rs662145 C > T HOMO phenotypes expressed higher levels of IL36B compared with PCSK9 SNP rs662145 C > T HET phenotypes. Box-and-whisker plots show normalized PCSK9 expression (log transformed reads on y axis). Each dot represents 1 keratinocyte line or skin sample. Differential gene expression was calculated using DESeq2. FDR-adjusted P values are displayed on each plot. (B) In cultured keratinocyte cell lines and nonlesional skin, PCSK9 SNP rs662145 C > T HOMO phenotypes expressed higher levels of IL36G compared with PCSK9 SNP rs662145 C > T HET phenotypes. HOMO, homozygous; HET, heterozygous.
However, the relationship between homozygosity for the PCSK9 reference allele and IL36 expression diverged between keratinocyte cultures and nonlesional skin. In human keratinocyte cultures, homozygosity for the PCSK9 reference allele was associated with lower expression of PCSK9 (Figure 3A) and higher expression of IL36B and IL36G (Figure 4, A and B). In contrast, in nonlesional human skin, IL36G expression was lower in individuals homozygous for the PCSK9 reference allele, with an apparent dose-response relationship between the PCSK9 rs662145 C > T variant and baseline IL36G expression (Figure 4B). Thus, while homozygosity for the psoriasis-linked PCSK9 variant rs662145 C > T was consistently associated with lower PCSK9 expression and higher IL36G expression, homozygosity for the PCSK9 reference allele appeared to yield different results in vitro and in vivo.
PCSK9 is negatively related to keratinocyte-derived IL36.
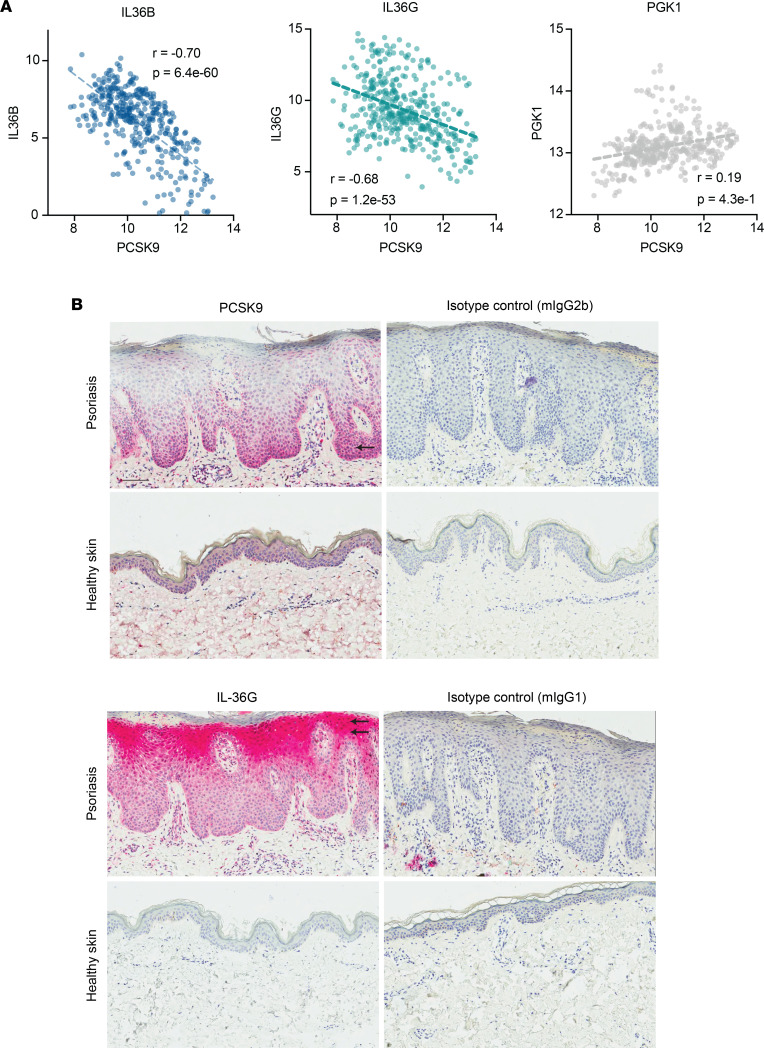
Having established that expression of PCSK9 rs662145 C > T variant was associated with increased expression of keratinocyte-derived IL36, we next aimed to elucidate the relationship between PCSK9 and IL36. We cultured healthy primary keratinocytes and found that PCSK9 negatively correlated with IL36B and IL36G expression (r = –0.70, P = 6.4 × 10–60; and r = –0.68, P = 1.2 × 10–53, respectively; Figure 5A). No significant correlation was noted between PCSK9 and PGK1, a housekeeping gene (Figure 5A). We then performed IHC staining, which also highlighted this negative relationship between PCSK9 and IL-36γ expression in psoriatic skin — i.e., where epidermal expression of PCSK9 was high, IL-36γ expression was low (Figure 5B). PCSK9 expression was most prominent in the basal and early spinous layers of the epidermis, whereas IL-36γ expression was highest in the granular layer (Figure 5B).
Figure 5. PCSK9 expression negatively correlates with IL36B and IL36G expression in keratinocytes and skin.
(A) In cultured keratinocyte cell lines, PCSK9 expression negatively correlated with IL36B and IL36G expression. PCSK9 expression did not correlate with PGK1 expression, a housekeeping gene. In these plots, each dot represents an in vitro cultured keratinocyte cell line under a different culture condition (control, IL-4, IL-13, IL-17A, IFN-α, IFN-γ, TNF-α, IL-4 and IL-13, IL-17A and IFN-γ, IL-17A, and TNF-α). Normalized log2 transformed reads for each gene are plotted on the x axis and y axis. Pearson’s correlation coefficients and P values are displayed on each plot. (B) Epidermal expression of PCSK9 and IL36G. Representative pictures of an IHC staining for PCSK9 and IL36G as well as the matching isotype controls in lesional skin of a patient with psoriasis (upper row) and healthy control nonlesional skin (lower row). The single arrow points to the basal layer. Double arrows point to the granular layer. Scale bar: 100 μm.
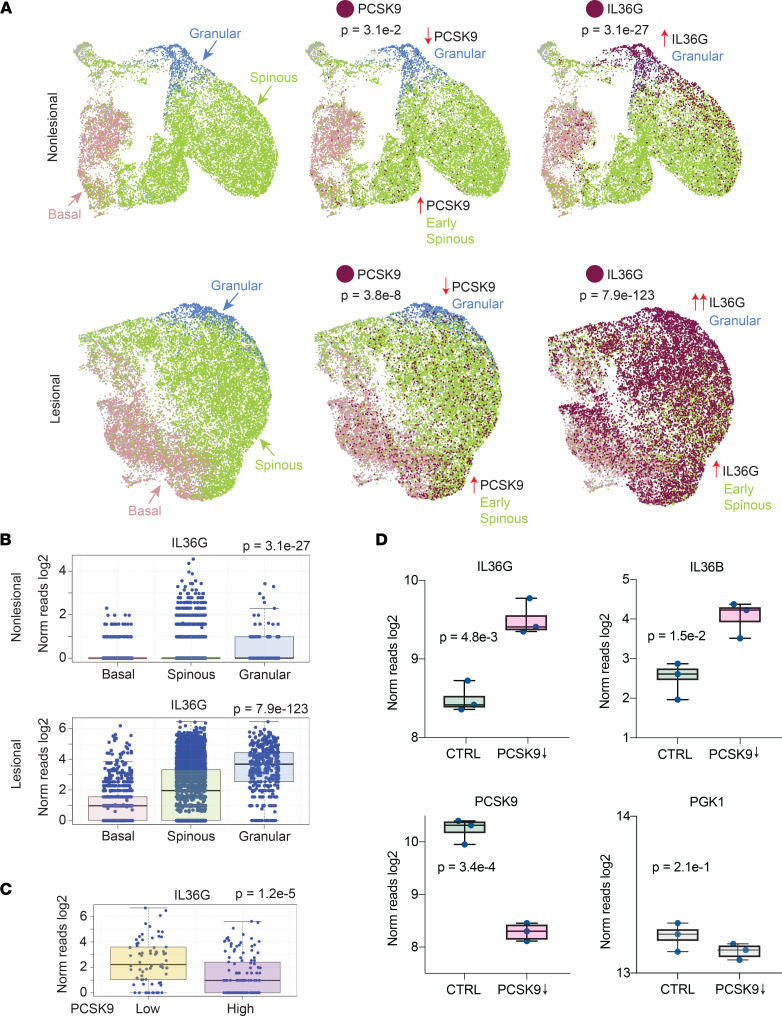
The same expression pattern was also noted in our scRNA-Seq data of psoriatic nonlesional and lesional skin (n = 9) (Figure 6, A–C). The Uniform Manifold Approximation and Projection (UMAP) method was used to create 2-dimensional representations of the resulting data. Basal, spinous, and granular layer keratinocyte populations were identified using validated keratinocyte-expressed genes. scRNA-Seq also revealed that, in human skin, the vast majority of PCSK9 expression could be attributed to keratinocytes (Supplemental Figure 2). Analysis of gene expression within each keratinocyte subpopulation demonstrated a gradient of PCSK9 expression with expression being highest in basal and early spinous layer keratinocytes and lowest in granular layer keratinocytes (psoriatic nonlesional skin, P = 3.1 × 10–2; psoriatic lesional skin, P = 3.8 × 10–8; Figure 6A). In contrast, IL36G expression was highest in granular layer keratinocytes (psoriatic nonlesional skin, P = 3.1 × 10–27; psoriatic lesional skin, P = 7.9 × 10–123; Figure 6, A and B). Furthermore, when keratinocytes were parsed into PCSK9 high and low expressors, IL36G expression was significantly higher in the PCSK9 low-expressing keratinocytes (P = 1.2 × 10–5; Figure 6C).
Figure 6. PCSK9 expression is negatively and directly related to IL36B and IL36G expression.
(A) Single-cell sequencing of psoriatic nonlesional and lesional skin (n = 9). The UMAP method was used to create 2-dimensional representation of the resulting data. Keratinocyte populations were identified by the expression levels of established keratinocyte markers (red, basal layer keratinocytes, DST high; green, spinous layer keratinocytes, KRT5 low and KLK7 low; and blue, granular layer keratinocytes, KLK7 high). PCSK9- and IL36G-expressing cells are depicted in maroon. (B) Expression of IL36G in individual basal, spinous, and granular layer keratinocytes shown as box-and-whisker plots of log2 transformed gene expression. P values were calculated for each data set using 1-way ANOVA. (C) PCSK9-positive keratinocytes were parsed into 2 groups, PCSK9-high and PCSK9-low (x axis). Box-and-whisker plots of indicated intracellularly expressed genes are plotted on the y axis (log2 reads). P values were calculated using Student’s t test. (D) Box plots showing the effects of in vitro siRNA knockdown of PCSK9 in keratinocyte cell lines on IL36B and IL36G. PCSK9 (positive control) and PGK1 (negative control) expression is also shown for scrambled siRNA transfected and PCSK9 siRNA transfected cultures. Each dot represents an independently cultured and independently transfected HaCaT keratinocyte cell line (n = 3). P values were calculated with Student’s t test.
To further verify this negative relationship between PCSK9 and IL36, we used PCSK9 siRNA to knock down PCSK9 expression in an IL36-expressing HaCaT human keratinocyte cell line. The siRNA knockdown experiment was performed in 3 independently cultured HaCaT cell lines, and gene expression was assessed by RNA-Seq. In comparison to control siRNA knockdown cell cultures, PCSK9 knockdown cultures expressed significantly more IL36B and IL36G (P = 1.5 × 10–2 and P = 4.8 × 10–3, respectively; Figure 6D), which confirmed the inverse relationship between PCSK9 and IL36. No significant change in gene expression was noted for PGK1, a housekeeping gene.
PCSK9 is linked to other potential mediators of psoriasis.
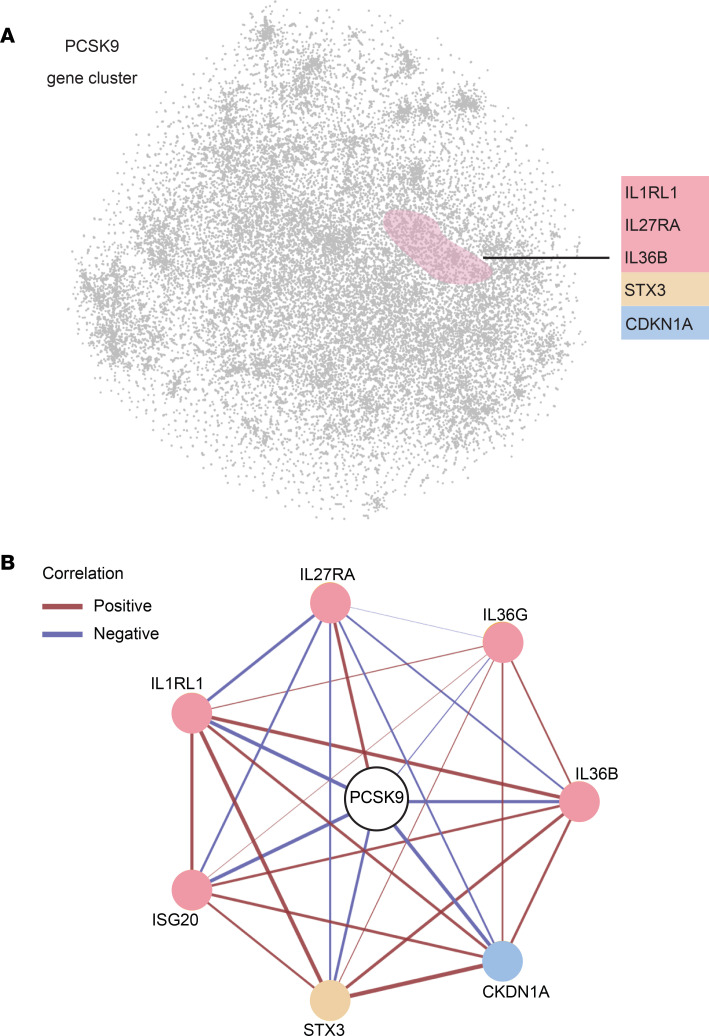
We then investigated whether PCSK9 is linked to other potential inflammatory mediators of psoriasis in addition to IL36B and IL36G. To further assess how PCSK9 expression was associated with other keratinocyte-expressed psoriasis-relevant genes, we created a 2-dimensional plot of the keratinocyte transcriptome using the t-Distributed Stochastic Neighbor Embedding (t-SNE) dimensionality reduction method. In this plot, each data point represents a different gene expressed in keratinocytes and the distance between any 2 points represents how strongly the genes correlate with each other in their expression. Within this representation of the keratinocyte transcriptome, PCSK9 clustered with a variety of genes related to cytokines (IL1RL1, IL27RA, and IL36B), cytokine secretion (STX3), and keratinocyte proliferation (CDKN1A) (Figure 7A). To visualize these relationships, PCSK9 gene coexpression network analysis was performed, which highlighted the different relationships between PCSK9 and the various keratinocyte-expressed mediators of inflammation (Figure 7B).
Figure 7. PCSK9 expression clusters with inflammatory mediators of psoriasis.
(A) A 2-dimensional plot of the keratinocyte transcriptome was constructed using a nonlinear dimensionality reduction strategy, the t-SNE method. Each point represents a gene and the distance between the points is inversely related to how well the genes correlate with one another. Within this plot, PCSK9 clusters with various genes of interest, shown on the right of the plot. (B) PCSK9 coexpression network with each circle representing a different gene and the lines connecting each circle representing the strength (thickness of line) and direction (red = positive, blue = negative) of each correlation.
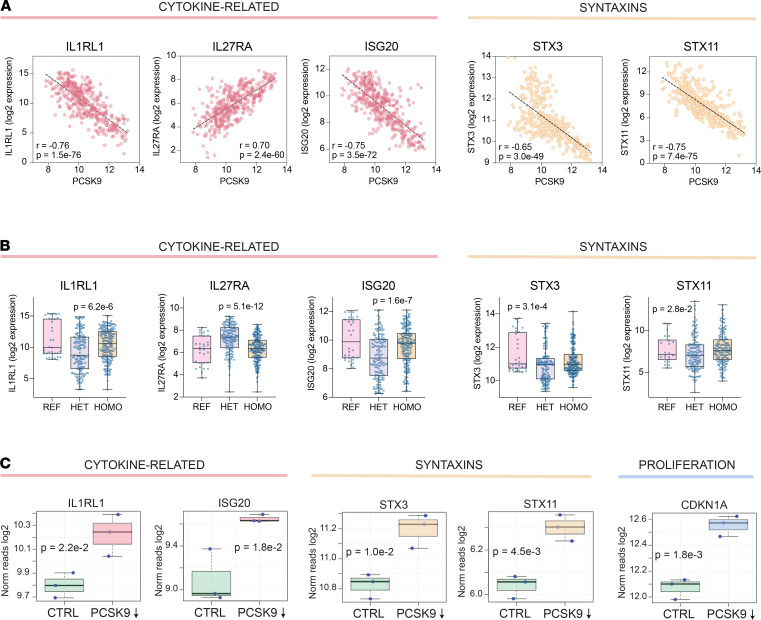
To determine how well PCSK9 correlates with these and other keratinocyte-expressed genes, we investigated patterns of gene expression in our cultured keratinocyte cell lines. We found that the expression of IL1RL1, IL27RA, ISG20, STX3, and STX11 appeared to be linked to PCSK9 in keratinocytes. Specifically, scatter plots of gene expression revealed that the expression of PCSK9 positively correlated with IL27RA (Figure 8A; r = 0.70, P = 2.4 × 10–60) and negatively correlated with IL1RL1, ISG20, STX3, and STX11 (r = –0.76, P = 1.5 × 10–76; r = –0.75, P = 3.5 × 10–72; r = –0.65, P = 3.0 × 10–49; and r = –0.75, P = 7.4 × 10–75, respectively) (Figure 8A). IL1RL1, IL27RA, ISG20, STX3, and STX11 were also differentially expressed in primary human keratinocyte cell lines heterozygous and homozygous for the PCSK9 variant rs662145 C > T (P = 6.2 × 10–6, P = 5.1 × 10–12, P = 1.6 × 10–7, P = 3.1 × 10–4, and P = 2.8 × 10–2, respectively) (Figure 8B). Similar to the pattern noted for PCSK9, primary keratinocyte cell lines heterozygous for PCSK9 SNP rs662145 C > T had increased expression of IL27RA (Figure 8B). In contrast, the opposite pattern was associated with IL1RL1, ISG20, STX3, and STX11, where expression was decreased in lines heterozygous for PCSK9 SNP rs662145 C > T (Figure 8B). Finally, PCSK9 siRNA knockdown resulted in increased expression of IL1RL1, ISG20, STX3, STX11, and CDKN1A (Figure 8C), confirming a direct negative correlative relationship between PCSK9 and these genes.
Figure 8. PCSK9 expression directly correlates with inflammatory mediators of psoriasis.
(A) Individual scatter plots showing correlations between PCSK9 and IL1RL1, IL27RA, ISG20, STX3, and STX11 in cultured keratinocytes. In these plots, each dot represents an in vitro cultured keratinocyte cell line under a different culture condition (control, IL-4, IL-13, IL-17A, IFN-α, IFN-γ, TNF-α, IL-4 and IL-13, IL-17A and IFN-γ, IL-17A, and TNF-α). PCSK9 expression is shown on the x axis with each plot depicting a different gene on the y axis. (B) Box-and-whisker plots showing expression of genes of interest in cultured keratinocytes displaying the REF allele or rs662145 C > T variant PCSK9 allele. Differential gene expression was calculated using DESeq2. REF, reference; HET, heterozygous; HOMO, homozygous. (C) Box plots showing effects of in vitro siRNA knockdown of PCSK9 in keratinocyte cell lines on expression of genes of interest. Each dot represents an independently cultured and independently transfected HaCaT keratinocyte cell line (n = 3). The y axis depicts log2 transformed normalized reads and the x axis compares control keratinocyte cell lines with PCSK9 knockdown cell lines, with P values calculated via Student’s t test.
Discussion
The major finding of our study is the identification of PCSK9 as a psoriasis-susceptibility locus (Figures 1 and 2). This was established by mining psoriasis and healthy control RNA-Seq data sets for differential occurrences of PCSK9 variants. A meta-analysis across 2 independently acquired RNA data sets yielded a highly statistically significant final model for this association (P = 7.4 × 10–4; Figure 1B). Furthermore, PCSK9 genotyping data from 2590 psoriasis cases and 1720 controls also associated the exact same SNP, rs662145 C > T, to the psoriasis phenotype (P = 2.6 × 10–4; Figure 2B). Thus, across multiple data sets (RNA and DNA), SNP rs662145 C > T was consistently associated with psoriasis. However, there are major differences between traditional genotyping and the RNA-Seq–based variant discovery method used here. First, the RNA-Seq–based variant discovery method examined only a fraction of the total PCSK9 variants, those which resided in the transcribed region of the gene. Also, the relatively small sample size of the RNA-Seq data sets makes it difficult to estimate the OR of the association. In fact, the OR estimated via RNA-Seq–based variant discovery was higher than the OR identified by genotyping the PCSK9 locus. However, as these are different techniques, we expected them to yield different ORs. For example, unlike traditional genotyping methods, the variant discovery method used allele counting to calculate the OR. Thus, further investigation is needed to accurately estimate the OR for this association, especially since heterozygosity and homozygosity for PCSK9 SNP rs662145 C > T may display unexpected effects, as described below.
It is becoming increasingly apparent that many causal variants alter disease risk through regulatory effects on gene expression (17). GWAS have uncovered hundreds of genetic variants linked to human diseases (18), yet functional biomechanistic studies aimed at understanding these linkages have lagged far behind. In addition to identifying PCSK9 as a psoriasis-susceptibility locus, we performed the initial studies to understand how the psoriasis-linked PCSK9 variant, SNP rs662145 C > T, might predispose individuals to psoriasis. Here, we establish that SNP rs662145 C > T is associated with altered PCSK9 expression in keratinocytes. This was determined by parsing the primary human keratinocyte RNA-Seq data sets into 3 groups (homozygous for the PCSK9 reference allele, heterozygous for PCSK9 SNP rs662145 C > T, and homozygous for PCSK9 SNP rs662145 C > T). When compared with heterozygous keratinocyte cell lines, PCSK9 SNP rs662145 C > T homozygous cell lines expressed low levels of PCSK9 and high levels of IL36 cytokines, a family of cytokines induced by IL-17A in autoimmunity and inflammatory skin conditions (19). IL-36β and IL-36γ are known to promote epidermal hyperplasia, hyperkeratosis, and cutaneous inflammation in psoriasis, and IL-36γ is thought to be a key driver of psoriasis pathogenesis (14, 20), especially pustular psoriasis (21). The negative correlative relationship between PCSK9 and IL36 expression is an important finding of our study. It was consistently observed across multiple experimental approaches, including IHC, RNA-Seq, scRNA-Seq, and PCSK9 siRNA knockdown (Figures 5 and 6). This inverse relationship provides a plausible explanation as to how PCSK9 SNP rs662145 C > T may predispose individuals to psoriasis — i.e., lowering PCSK9 expression in keratinocytes increases baseline cutaneous expression of IL-36, a psoriasis-driving cytokine. Although not explored here, other investigators have linked loss-of-function and gain-of-function PCSK9 variants to disease (22).
The fact that keratinocyte cell lines homozygous for the PCSK9 reference allele also had low PCSK9 expression and high IL36 expression was difficult to reconcile. If PCSK9 SNP rs662145 C > T predisposes individuals to psoriasis by increasing their baseline cutaneous expression of IL-36, one would expect that keratinocytes homozygosity for the PCSK9 reference allele should have low IL36 expression, as individuals homozygous for the PCSK9 reference allele are not predisposed to psoriasis. Indeed, these in vitro results differed from those we observed in nonlesional human skin, where homozygosity for the PCSK9 reference allele was associated with low IL36G expression. In fact, in nonlesional skin RNA-Seq data demonstrated a linear relationship between the dose of PCSK9 SNP rs662145 C > T and the amount of baseline IL36G detected (Figure 4B). This finding is highly relevant because polymorphisms in IL36 have been implicated in plaque psoriasis (23), and this cytokine has increasingly been identified as an important driver of other immune-mediated skin diseases, including pyoderma gangrenosum and hidradenitis suppurativa (24–29). Although the differences between cultured keratinocytes and human skin were not further explored here, it is important to note that PCSK9 can exist as a monomer, dimer, and trimer (30, 31). This may be a reason why our in vitro keratinocyte findings did not perfectly translate to in vivo settings. A major finding that was consistent across the in vitro and in vivo settings was the observation that PCSK9 SNP rs662145 C > T homozygous individuals and cell lines express less PCSK9 and more IL36G when compared with their heterozygous counterparts.
To date, reports regarding the effect of PCSK9 inhibition on skin inflammation have been conflicting. In an imiquimod mouse model of psoriasis-like dermatitis, suppressing PCSK9 with siRNA resulted in improvement of skin lesions (5). However, the effects of PCSK9 suppression in human skin remain poorly characterized. Two PCSK9 inhibitors, alirocumab and evolocumab, are now FDA-approved to reduce circulating LDL levels and lower all-cause mortality in patients with coronary artery disease (32). In patients on these inhibitors, case reports have documented various inflammatory skin eruptions, including psoriasis and atopic dermatitis (6, 7). The development of atopic dermatitis while on PCSK9 inhibition was accompanied by a high serum IgE (6). Although most commonly studied in atopic dermatitis, IgE is also elevated in patients with psoriasis (33), which may be due to the increased expression of IL-36 in this disease (14). Specifically, IL-36 has recently been linked to Ig isotype switching to IgE following cutaneous antigen exposure (34).
Thus, in addition to its well-characterized ability to bind to LDLR, PCSK9 appears to be strongly linked to mediators of innate and adaptive immunity. Although the mechanisms by which PCSK9 variants may alter immune responses remain to be investigated, cholesterol-sulfate is a well-known and potent transcriptional regulator in keratinocytes (35). It is possible that the altered intracellular abundance of cholesterol-sulfate and phospholipids in PCSK9 rs662145 C > T variant individuals results in the differential expression of proinflammatory cytokines (Supplemental Figure 3). Another possibility is that PCSK9 regulates removal of inflammatory lipids such as LPS via its effects on LDLR (36).
Here, we have focused on using alternative methods, including RNA-Seq–based genetic variant discovery and expression quantitative trait loci–like analysis strategies, to characterize immune responses from whole tissue RNA-Seq data sets and integrated scRNA-Seq to validate our findings (37, 38).
In summary, our study establishes PCSK9 as a possible psoriasis-susceptibility locus and demonstrates its negative relationship with IL36G at the gene expression and protein levels. We also firmly establish an association between PCSK9 SNP rs662145 C > T and PCSK9 gene expression. Although gaps in our understanding of these relationships remain, our working hypothesis is that PCSK9 SNP rs662145 C > T may predispose individuals to psoriasis by increasing baseline cutaneous expression of IL36G. Future mechanistic studies are needed to better elucidate these findings.
Methods
Human skin RNA-Seq data sets.
RNA-Seq FASTQ files of human normal and psoriatic lesional skin were downloaded from the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra). A total of 4 data sets were used: 3 independent data sets (accession numbers: SRP165679, SRP026042, and SRP057087) and 1 data set comprised of 2 combined experimental data sets published by the same research group (accession numbers: SRP035988 and SRP050971) (11, 12, 14). The latter included 99 lesional psoriatic and 90 normal skin biopsies from people of European descent enrolled in the Southeast Michigan area. Patients with psoriasis who volunteered for a skin biopsy underwent a washout period of all immunosuppressive medications prior to the procedure.
RNA-Seq.
Total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen, catalog 74134). RNA concentrations were then quantified using a Qubit Fluorometer (Invitrogen), and RNA integrity was assessed using the Agilent TapeStation. Samples with an RNA integrity number greater than or equal to 8 were used for this study. Indexed libraries were constructed from 500 ng of total RNA using the TruSeq Stranded mRNA Sample Prep Kit (Illumina, catalog 20020594) following the manufacturer’s instruction. The quantity and quality of the libraries were also assessed by Qubit and Agilent 2100 Bioanalyzer, respectively. The average library size was 400 bp. Libraries’ molar concentrations were validated by quantitative PCR (qPCR) for library pooling. Sequencing was performed on the Illumina HiSeq 4000 platform using PE150 chemistry.
Alignment and variant calling.
Raw sequencing data were received in FASTQ format. Alignment of sequencing reads to the reference genome (hg38) was performed using STAR v2.5.2 (39). Haplotype calling was performed using the Genome Analysis Toolkit (GATK v3.8) (40) and best practice guidelines to call variants from the RNA-Seq data. For the analysis, only known variants were analyzed. The National Center for Biotechnology Information (NCBI) database of genetic variation (dbSNP) release 146 was used as the reference for known SNPs. Gene annotation downloaded from the Ensembl website (http://www.ensembl.org/) was used for STAR mapping and the following read count evaluation. Gene expression level normalization and differential expression analysis was carried out using DESeq2 Bioconductor R package (version 1.6.3). This package provides statistics for determination of differential expression using a model based on the negative binomial distribution.
Differential gene expression, correlation, and linkage.
Gene expression level normalization and differential expression analysis was carried out using DESeq2 Bioconductor R package (version 1.6.3) (41). This package provides statistics for determination of differential expression using a model based on the negative binomial distribution. Differential expression P values were corrected for multiple testing using the FDR method. Genes with FDR-adjusted P values less than 0.05 and fold change greater than 2 or less than 0.5 were considered differentially expressed. Correlation analyses of gene expressions were performed on read counts of each identified gene normalized with the DESeq2 package. Values were subsequently log transformed and winsorized when necessary. Pearson’s correlation coefficient (r) was calculated using the cor.test function in R (42). Spearman’s correlation coefficient (rs) was estimated by algorithm AS 89. When Pearson’s correlation coefficients were used, Cook’s distances were also calculated to ensure that there were no overly influential data points. The igraph software R package was used for the expression network visualization (43). Linkage disequilibrium among genes was calculated and visualized using R package gaston (44).
2D visualization of the keratinocyte transcriptome.
We computed gene pairwise distances using the formula, 1-r2, where r represents Pearson’s correlation coefficient. A visual representation of the gene coexpression network was then created using a dimensionality reduction technique, t-SNE, calculated with R package Rtsne (45).
IHC.
FFPE skin blocks of 3 patients with psoriasis and 3 healthy controls were cut into 3 μm sections. The sections were first incubated at 70°C in an oven for 40 minutes to remove excess paraffin. With the help of Leica BOND RXm, they were deparaffinized and treated with EDTA buffer-based, pH 9.0 Epitope Retrieval Solution (ER2) for 30 minutes at 100°C. Sections were stained with an anti-PCSK9 (Thermo Fisher Scientific, catalog MA5-32843, mouse IgG2b, clone 2F1, 1:2000) and an anti–IL-36G (Abcam, catalog ab156783, mouse IgG1, clone OTI2F4, 1:500) using AP chromogen (BOND Polymer Refine Red Detection). The incubation time for both Abs was 30 minutes at room temperature. Sections were counterstained with hematoxylin.
Meta-analysis.
Meta-analyses were completed using the R package metafor (46). A weighted random-effects model was used to estimate a summary effect size. To estimate between-study variance, a restricted maximum-likelihood estimator was applied. A weighted estimation with inverse-variance weights was used to fit the model.
Single-cell sequencing.
Punch biopsies of 4 mm from patients with psoriasis and healthy controls were obtained (n = 9). Single cell suspensions were then prepared by mincing the biopsy tissue and digesting it in 0.2% Collagenase II (Life Technologies) and 0.2% Collagenase V (Sigma) in plain medium for 1 hour at 37°C and then strained through a 70 μM mesh. Single-cell sequencing libraries were then prepared using a 10X Genomics Chromium Controller. Libraries were sequenced on the Illumina NovaSeq 6000 sequencer to generate 151 bp paired-end reads. Data processing including quality control, read alignment, and gene quantification were conducted using 10X Genomics Cell Ranger v3.1 using their default parameters. Empty droplets and cells with unique molecular identifier (UMI) counts less than a calculated threshold based on the distribution of UMI counts per cell were removed. Data normalization and cell library size correction were performed using the median of ratios method implemented in the R package DESeq2. Clustered cells were mapped to corresponding cell types using the singleR R package and the Human Primary Cell Atlas as a reference data. Keratinocytes were then further subdivided into basal, spinous, and granular layer keratinocytes by matching their cell cluster gene signatures with cell type–specific markers (DST, KRT5, KRT10, and KLK7). Data have been deposited in the NCBI Gene Expression Omnibus (GEO) and are accessible through GEO Series accession numbers GSE121212 and GSE173706.
Keratinocyte cell cultures.
The human immortalized keratinocyte cell line HaCaT (47) was cultured in DMEM (ATCC, catalog 30-2002) containing 1% Penicillin-Streptomycin Solution (ATCC, catalog 30-2300; penicillin G [10,000 U/mL] and streptomycin [10,000 μg/mL]) and 10% of Gibco FBS. Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2 until they reached the exponential phase (2–3 weeks). Three passages were achieved before the experiments. Then, 3 × 105 HaCaT cells were seeded in a 6-well flat-bottom tissue culture plate (Corning-Costar) in 2 mL of DMEM-supplemented per well. HaCaT cultures were treated with or without IL-4, IL-13, IL-17A, IFN-α, IFN-γ, TNF-α, IL-4 and IL-13, IL-17A and IFN-γ, IL-17A, and TNF-α). Expression analysis was performed using RNA-Seq as described above.
siRNA-mediated knockdown of PCSK9.
For siRNA-mediated knockdown experiments, 3 parallel transfections were conducted using the SMARTpool siRNAs (mixture of 4 siRNAs), siRNA targeting PCSK9, and negative control siRNA, which were obtained from Dharmacon-Horizon Discovery. After overnight incubation, HaCaT cells (47) were transfected with 20 pmol of SMARTpool siRNAs using Lipofectamine RNAiMax Reagent (Thermo Fisher Scientific) as per the manufacturer’s instructions. After 48 hours, cells were washed with PBS and were harvested for RNA extraction using a mixture of Gibco Trypsin-EDTA (0.5%) and no phenol red (Thermo Fisher Scientific). Trypsin was neutralized with FBS and the cells were collected by centrifugation and resuspended in 100 μL of RNAlater. qPCR analysis was performed to test the efficiency of the transfection. Further expression analysis was performed using RNA-Seq as described above.
Statistics.
All statistical analyses were performed using R (version 3.1.2) (42). In RNA-Seq analysis, fold changes were compared between groups using the Wald test implemented within the DESeq2 package (41). Correlation analysis was performed using the Pearson’s correlation coefficient, calculated in R (42) using cor.test function, which was also used to estimate P values for Pearson’s correlation coefficient using a 2-tailed Student’s t distribution test. Meta-analysis performed using R and P values were calculated using a Wald-type test for the coefficient of the meta-analysis model. Fisher’s exact test was used for testing association of SNPs with psoriasis. A 1-way ANOVA was used to test for association between SNP genotypes and gene expression levels. P values were adjusted for multiple testing using the Benjamini-Hochberg method, and adjusted P values less than 0.05 were considered statistically significant in all tests.
Study approval.
Skin biopsies were obtained from volunteer patients following protocols approved by the University of Michigan Institutional Review Board. Written informed consent was obtained from all subjects.
Author contributions
AM, AJX, AT, LCT, JEG, IEA, and EM wrote the manuscript. AM, AT, MCB, WL, QL, STL, LCT, GL, XX, RW, JTE, YI, ARL, NYK, AIM, AK, JEG, and EM acquired data. AM, AJX, NYK, LCT, JEG, and EM analyzed the data. All authors read, revised, and approved the final manuscript. The method used to assign the authorship order of the 2 co–first authors is as follows: AM was primarily responsible for data analysis, and AJX was primarily responsible for editing, review, revisions, and figures.
Supplementary Material
Acknowledgments
The research was supported by grant P30AR075043 from the NIH/National Institute for Arthritis and Musculoskeletal and Skin Diseases (NIAMS) (to LCT and JEG) and by NIH/NIAMS grant 1K24AR077313 (to EM).
Version 1. 07/21/2022
In-Press Preview
Version 2. 08/22/2022
Electronic publication
Funding Statement
LCT and JEG were supported by NIAMS, P30AR075043. EM was supported by NIAMS, 1K24AR077313.
Footnotes
Conflicts of interest: The authors have declared that no conflict of interest exists.
Authorship note: AM and AJX are co–first authors.
Copyright: © 2022, Merleev et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2022;7(16):e141193.https://doi.org/10.1172/jci.insight.141193.
Contributor Information
Alexander Merleev, Email: amerleev@ucdavis.edu.
Antonio Ji-Xu, Email: antoniojixu@gmail.com.
Atrin Toussi, Email: amtoussi@ucdavis.edu.
Lam C. Tsoi, Email: alextsoi@umich.edu.
Stephanie T. Le, Email: stvle@ucdavis.edu.
Guillaume Luxardi, Email: gluxardi@ucdavis.edu.
Xianying Xing, Email: xingx@med.umich.edu.
Rachael Wasikowski, Email: wasikowr@med.umich.edu.
William Liakos, Email: william.liakos@gmail.com.
Marie-Charlotte Brüggen, Email: Marie-Charlotte.Brueggen@usz.ch.
James T. Elder, Email: jelder@umich.edu.
Iannis E. Adamopoulos, Email: iannis@outlook.com.
Yoshihiro Izumiya, Email: yizumiya@ucdavis.edu.
Qinyuan Li, Email: li.qinyuan@gmail.com.
Amanda Kirane, Email: akirane@stanford.edu.
Alina I. Marusina, Email: alina.marusina@yahoo.com.
Johann E. Gudjonsson, Email: johanng@med.umich.edu.
Emanual Maverakis, Email: emaverakis@ucdavis.edu.
References
- 1.Tsoi LC, et al. Large scale meta-analysis characterizes genetic architecture for common psoriasis associated variants. Nat Commun. 2017;8:15382. doi: 10.1038/ncomms15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swindell WR, et al. RNA-seq identifies a diminished differentiation gene signature in primary monolayer keratinocytes grown from lesional and uninvolved psoriatic skin. Sci Rep. 2017;7(1):18045. doi: 10.1038/s41598-017-18404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yokoji-Takeuchi M, et al. Indirect regulation of PCSK9 gene in inflammatory response by Porphyromonas gingivalis infection. Heliyon. 2019;5(1):e01111. doi: 10.1016/j.heliyon.2018.e01111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garshick MS, et al. Characterization of PCSK9 in the blood and skin of psoriasis. J Invest Dermatol. 2021;141(2):308–315. doi: 10.1016/j.jid.2020.05.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luan C, et al. Potentiation of psoriasis-like inflammation by PCSK9. J Invest Dermatol. 2019;139(4):859–867. doi: 10.1016/j.jid.2018.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanda N, Okajima F. Atopic dermatitis-like rash during evolocumab treatment of familial hypercholesterolemia. J Nippon Med Sch. 2019;86(3):187–190. doi: 10.1272/jnms.JNMS.2019_86-309. [DOI] [PubMed] [Google Scholar]
- 7.Ragavan A, et al. Evolocumab therapy and a possible association with skin reactions. Atherosclerosis Supplements. 2019;10(38):e7 [Google Scholar]
- 8.Ding Z, et al. PCSK9 and inflammation: role of shear stress, pro-inflammatory cytokines, and LOX-1. CardiovascRes. 2020;116(5):908–915. doi: 10.1093/cvr/cvz313. [DOI] [PubMed] [Google Scholar]
- 9.Gai MT, et al. Polymorphisms of rs2483205 and rs562556 in the PCSK9 gene are associated with coronary artery disease and cardiovascular risk factors. Sci Rep. 2021;11(1):11450. doi: 10.1038/s41598-021-90975-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Picard C, et al. Proprotein convertase subtilisin/kexin type 9 (PCSK9) in Alzheimer’s disease: a genetic and proteomic multi-cohort study. PLoS One. 2019;14(8):e0220254. doi: 10.1371/journal.pone.0220254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsoi LC, et al. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 2015;16:24. doi: 10.1186/s13059-014-0570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li B, et al. Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J Invest Dermatol. 2014;134(7):1828–1838. doi: 10.1038/jid.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stuart PE, et al. Transethnic analysis of psoriasis susceptibility in South Asians and Europeans enhances fine-mapping in the MHC and genomewide. HGG Adv. 2022;3(1):100069. doi: 10.1016/j.xhgg.2021.100069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsoi LC, et al. Atopic dermatitis is an IL-13-dominant disease with greater molecular heterogeneity compared with psoriasis. J Invest Dermatol. 2019;139(7):1480–1489. doi: 10.1016/j.jid.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Towne JE, Sims JE. IL-36 in psoriasis. Curr Opin Pharmacol. 2012;12(4):486–490. doi: 10.1016/j.coph.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Van de Kerkhof PC. From empirical to pathogenesis-based treatments for psoriasis. J Invest Dermatol. 2022;142(7):1778–1785. doi: 10.1016/j.jid.2022.01.014. [DOI] [PubMed] [Google Scholar]
- 17.Gallagher MD, Chen-Plotkin AS. The post-GWAS era: from association to function. Am J Hum Genet. 2018;102(5):717–730. doi: 10.1016/j.ajhg.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6(2):95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 19.Germán B, et al. Disrupting the IL-36 and IL-23/IL-17 loop underlies the efficacy of calcipotriol and corticosteroid therapy for psoriasis. JCI Insight. doi: 10.1172/jci.insight.123390. doi: 10.1172/jci.insight.123390. [published online January 24, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Queen D, et al. Function and regulation of IL-36 signaling in inflammatory diseases and cancer development. Front Cell Dev Biol. 2019;7:317. doi: 10.3389/fcell.2019.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swindell WR, et al. RNA-seq analysis of IL-1B and IL-36 responses in epidermal keratinocytes identifies a shared MyD88-dependent gene signature. Front Immunol. 2018;9:80. doi: 10.3389/fimmu.2018.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seidah NG, et al. PCSK9: a key modulator of cardiovascular health. Circ Res. 2014;114(6):1022–1036. doi: 10.1161/CIRCRESAHA.114.301621. [DOI] [PubMed] [Google Scholar]
- 23.Traks T, et al. Polymorphisms in IL36G gene are associated with plaque psoriasis. BMC Med Genet. 2019;20(1):10. doi: 10.1186/s12881-018-0742-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Y, et al. Cytokinocytes: the diverse contribution of keratinocytes to immune responses in skin. JCI Insight. 2020;5(20):142067. doi: 10.1172/jci.insight.142067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gudjonsson JE, et al. Contribution of plasma cells and B cells to hidradenitis suppurativa pathogenesis. JCI Insight. 2020;5(19):139930. doi: 10.1172/jci.insight.139930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maverakis E, et al. Pyoderma gangrenosum. Nat Rev Dis Primers. 2020;6(1):81. doi: 10.1038/s41572-020-0213-x. [DOI] [PubMed] [Google Scholar]
- 27.Uppala R, et al. “Autoinflammatory psoriasis”-genetics and biology of pustular psoriasis. Cell Mol Immunol. 2021;18(2):307–317. doi: 10.1038/s41423-020-0519-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsoi LC, et al. Progression of acute-to-chronic atopic dermatitis is associated with quantitative rather than qualitative changes in cytokine responses. J Allergy Clin Immunol. 2020;145(5):1406–1415. doi: 10.1016/j.jaci.2019.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang EA, et al. Classic ulcerative pyoderma gangrenosum is a T cell-mediated disease targeting follicular adnexal structures: a hypothesis based on molecular and clinicopathologic studies. Front Immunol. 2017;8:1980. doi: 10.3389/fimmu.2017.01980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan D, et al. Self-association of human PCSK9 correlates with its LDLR-degrading activity. Biochemistry. 2008;47(6):1631–1639. doi: 10.1021/bi7016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen H, et al. Correlation between plasma proprotein convertase subtilisin/kexin type 9 and blood lipids in patients with newly diagnosed primary nephrotic syndrome. Ren Fail. 2020;42(1):405–412. doi: 10.1080/0886022X.2020.1756846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonagh M, et al. A systematic review of PCSK9 inhibitors alirocumab and evolocumab. J Manag Care Spec Pharm. 2016;22(6):641–653q. doi: 10.18553/jmcp.2016.22.6.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kasumagic-Halilovic E. Total serum immunoglobulin E levels in patients with psoriasis. Mater Sociomed. 2020;32(2):105–107. doi: 10.5455/msm.2020.32.105-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patrick GJ, et al. Epicutaneous Staphylococcus aureus induces IL-36 to enhance IgE production and ensuing allergic disease. J Clin Invest. 2021;131(5):143334. doi: 10.1172/JCI143334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elias PM, et al. Role of cholesterol sulfate in epidermal structure and function: lessons from X-linked ichthyosis. Biochim Biophys Acta. 2014;1841(3):353–361. doi: 10.1016/j.bbalip.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walley KR, et al. PCSK9 is a critical regulator of the innate immune response and septic shock outcome. Sci Transl Med. 2014;6(258):258ra143. doi: 10.1126/scitranslmed.3008782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merleev AA, et al. Meta-analysis of RNA sequencing data sets reveals an association between TRAJ23, psoriasis, and IL-17A. JCI Insight. 2018;3(13):120682. doi: 10.1172/jci.insight.120682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dillen CA, et al. Clonally expanded γδ T cells protect against Staphylococcus aureus skin reinfection. J Clin Invest. 2018;128(3):1026–1042. doi: 10.1172/JCI96481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2012;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKenna A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love MI, et al. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. R-Core-Team. R: A language and environment for statistical computing. http://www.R-project.org/ Accessed July 19, 2022.
- 43. Csardi G, Nepusz T. The igraph software package for complex network research. http://static1.squarespace.com/static/5b68a4e4a2772c2a206180a1/t/5cd1e3cbb208fc26c99de080/1557259212150/c1602a3c126ba822d0bc4293371c.pdf Accessed July 19, 2022.
- 44. Perdry HD, et al. gaston: Genetic Data Handling (QC, GRM, LD, PCA) & Linear Mixed Models. R package version 1.5.7. https://CRAN.R-project.org/package=gaston Accessed July 19, 2022.
- 45. Krijthe JH. Rtsne: T-Distributed Stochastic Neighbor Embedding using a Barnes-Hut Implementation. https://cran.r-project.org/web/packages/Rtsne/Rtsne.pdf Accessed July 19, 2022.
- 46.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw. 2010;36(3):1–48. doi: 10.18637/jss.v036.i03. [DOI] [Google Scholar]
- 47.Boukamp P, et al. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106(3):761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.