Abstract
Genetic polymorphisms are associated with the development of nonalcoholic fatty liver disease (NAFLD). Semaphorin7a (Sema7a) deficiency in mouse peritoneal macrophages reduces fatty acid (FA) oxidation. Here, we identified 17 individuals with SEMA7A heterozygous mutations in 470 patients with biopsy-proven NAFLD. SEMA7A heterozygous mutations increased susceptibility to NAFLD, steatosis severity, and NAFLD activity scores in humans and mice. The Sema7aR145W mutation (equivalent to human SEMA7AR148W) significantly induced small lipid droplet accumulation in mouse livers compared with WT mouse livers. Mechanistically, the Sema7aR145W mutation increased N-glycosylated Sema7a and its receptor integrin β1 proteins in the cell membranes of hepatocytes. Furthermore, Sema7aR145W mutation enhanced its protein interaction with integrin β1 and PKC-α and increased PKC-α phosphorylation, which were both abrogated by integrin β1 silencing. Induction of PKCα_WT, but not PKCα_dominant negative, overexpression induced transcriptional factors Srebp1, Chrebp, and Lxr expression and their downstream Acc1, Fasn, and Cd36 expression in primary mouse hepatocytes. Collectively, our findings demonstrate that the SEMA7AR148W mutation is a potentially new strong genetic determinant of NAFLD and promotes intrahepatic lipid accumulation and NAFLD in mice by enhancing PKC-α–stimulated FA and triglyceride synthesis and FA uptake. The inhibition of hepatic PKC-α signaling may lead to novel NAFLD therapies.
Keywords: Hepatology, Metabolism
Keywords: Genetic variation, Molecular biology, Mouse models
Introduction
Nonalcoholic fatty liver disease (NAFLD) is a common health issue, and its prevalence is rapidly increasing worldwide. NAFLD and its more advanced form, nonalcoholic steatohepatitis (NASH), have the potential to progress into cirrhosis and hepatocellular carcinoma (1–3). NAFLD is characterized by excessive accumulation of lipids in hepatocytes leading to chronic inflammation and liver damage in patients (1–3). The liver is the most important organ for metabolizing fatty acids (FAs) and triglycerides (TGs) as well as maintaining lipid homeostasis. The AMP kinase (AMPK) and protein kinase C (PKC) signaling pathways are mainly responsible for regulating these physiological functions (1). Liver dysregulation can promote the accumulation of excess lipids within, leading to hepatic steatosis and NAFLD (1). It is well known that multiple factors, including genetic and environmental factors, contribute to the development of NAFLD (1–3). However, the mechanisms underlying the pathological process of NAFLD remain to be elucidated. At present, several genetic risk factors for NAFLD have been identified, such as polymorphisms of patatin like phospholipase domain containing 3 (PNPLA3), transmembrane 6 superfamily member 2, and hydroxysteroid 17-beta dehydrogenase 13 (2). The polymorphism (rs738409, c.444C>G) of PNPLA3, encoding an adiponutrin protein associated with lipid droplets, is a strong genetic determinant of NAFLD (2). However, whether additional genetic factors are also important for the development of NAFLD remains to be determined.
Semaphorins are extracellular signaling proteins and can bind to their membrane receptors of plexins and integrins (4). They are essential for the development of many organs and tissues and the maintenance of their functions (4). Semaphorin 7A (SEMA7A), known as the John Milton Hagen antigen or CD108, is a glycosylphosphatidylinositol-anchored membrane protein with chemoattractant and chemorepulsive attributes (5). SEMA7A is expressed in multiple tissues, including the liver (4–7). Functionally, SEMA7A is crucial for axon growth, T cell activation, and other biological processes by binding to its receptors of integrin β1 and plexin C1 (4–9). Mutations in SEMA7A are associated with decreased bone mineral density and Kallmann syndrome (10, 11). Recently, Körner et al. (12) reported that Sema7a deficiency in mouse peritoneal macrophages reduced FA oxidation and oxidative phosphorylation, suggesting that SEMA7A may regulate lipid metabolism. Our preliminary study characterized mice with a missense mutation in Sema7a, and we found that these mice developed hydropic and fatty degeneration in hepatocytes. Therefore, we speculate that SEMA7A mutations may contribute to lipid metabolic disorders and NAFLD development.
In this study, we identified 17 patients with SEMA7A heterozygous mutation in 470 patients with biopsy-proven NAFLD. Remarkably, SEMA7A heterozygous mutations increased susceptibility to and severity of NAFLD in human patients and mouse models. The Sema7aR145W mutation caused intrahepatic accumulation of small lipid droplets in mice by activating PKC-α signaling. In our study, we provide what we believe is new evidence that the SEMA7AR148W mutation is a new genetic determinant of NAFLD and uncover the molecular mechanisms underlying the role of SEMA7AR148W mutation in the development of NAFLD.
Results
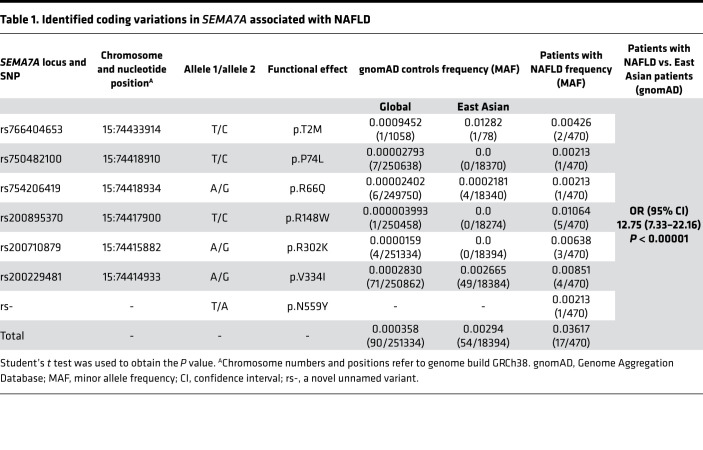
SEMA7A heterozygous mutations are potentially novel risk factors for human NAFLD.
To examine whether SEMA7A mutations contribute to the development of NAFLD in humans, we performed exon sequencing of SEMA7A (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.154113DS1) in 470 patients with biopsy-proven NAFLD (13). We identified 17 NAFLD patients with SEMA7A heterozygous mutations, including p.R148W (n = 5), p.V334I (n = 4), p.R302K (n = 3), p.T2M (n = 2), p.P74L, p.R66Q, and p.N559Y (n = 1 each) (Table 1). There was no patient with in-frame deletion, insertion, or frameshift in exons of SEMA7A. Strikingly, the frequency of candidate variants in SEMA7A in the NAFLD cohort (3.62%; 17 variants in 470 patients with NAFLD) was significantly higher than that of East Asian individuals (0.29%; 54 variants in 18,394 individuals) in the gnomAD, leading to an OR of 12.75 (95% CI: 7.33–22.16; P < 0.00001) (Table 1). These findings indicated that SEMA7A heterozygous mutations were risk factors for the development of NAFLD in humans.
Table 1. Identified coding variations in SEMA7A associated with NAFLD.
SEMA7A heterozygous mutations are significantly associated with human NAFLD severity.
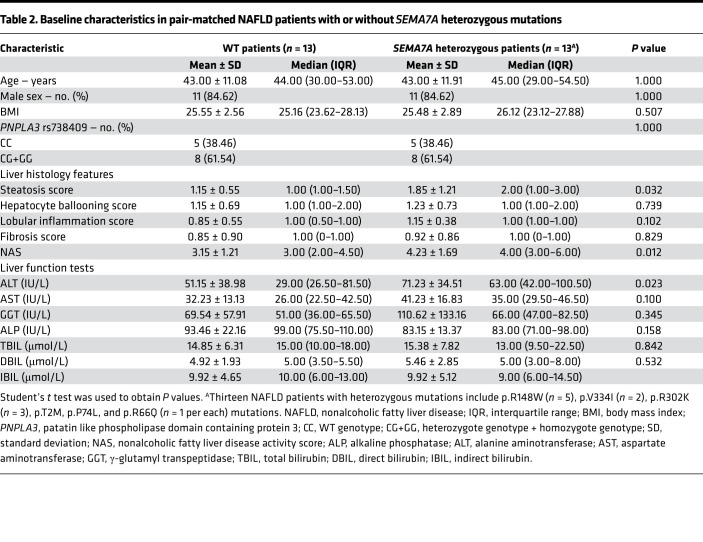
Next, we analyzed livers from 13 out of 17 patients with SEMA7A heterozygous mutations and their pair-matched controls from these 470 NAFLD patients, excluding the 4 patients without pair-matched controls (Table 2). Liver histological analysis showed that the severity of steatosis and NASs were significantly higher in NAFLD patients with SEMA7A heterozygous mutations than those of their matched controls (P = 0.032 and P = 0.012, respectively) (Table 2 and Supplemental Figure 1). Moreover, serum ALT, AST, and GGT levels in NAFLD patients with SEMA7A heterozygous mutations were also higher than in the paired controls (P = 0.023, P = 0.100, and P = 0.345, respectively) (Table 2). However, there were no significant differences in hepatocyte ballooning, lobular inflammation, and fibrosis between these groups (Table 2). Taken together, SEMA7A heterozygous mutations in this population were associated with increased severity of NAFLD.
Table 2. Baseline characteristics in pair-matched NAFLD patients with or without SEMA7A heterozygous mutations.
The Sema7aR145W heterozygous mutation markedly increases NAFLD severity and liver enzyme levels in mice following high-fat diet.
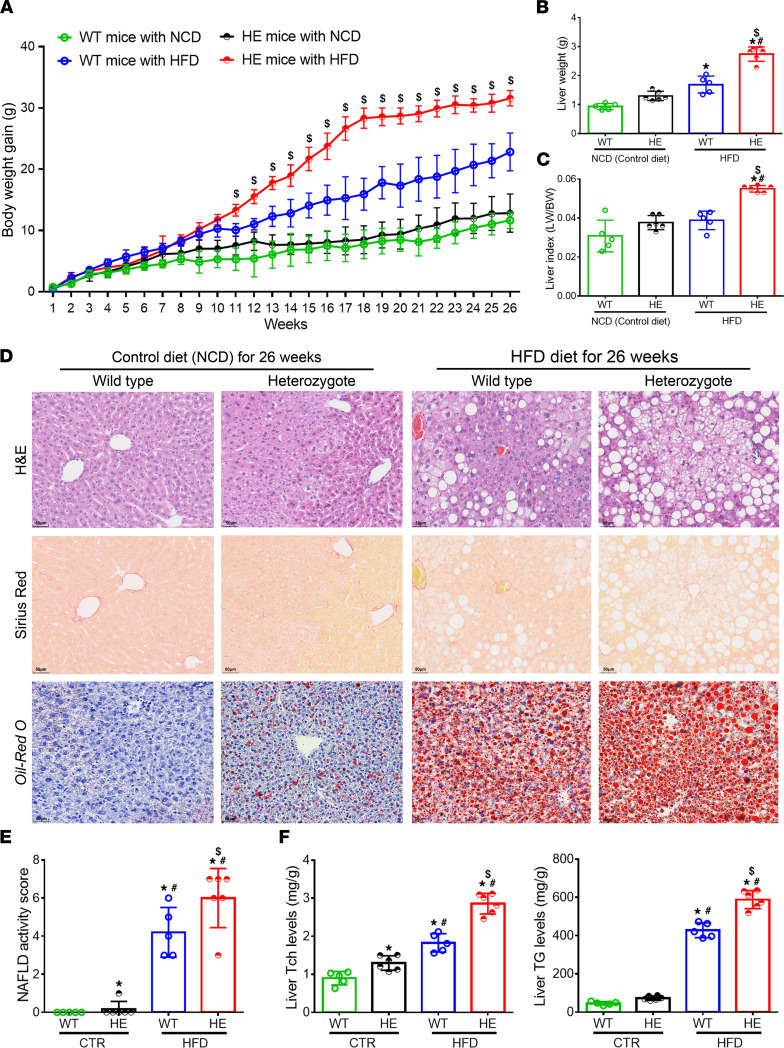
Single-cell RNA sequencing of SEMA7A in human liver cells revealed that SEMA7A was expressed by hepatocytes as well as all other types of liver cells (Supplemental Figure 2). Because 5 out of the 13 NAFLD patients with SEMA7A mutations were heterozygous for SEMA7AR148W, we also characterized Sema7aR145W (equivalent to human SEMA7AR148W) heterozygous mice in order to further investigate the functional role of SEMA7A mutations in the progression of NAFLD. Following feeding with high-fat diet (HFD) for 26 weeks, we found that the body weight gains, liver weights, and liver/body weight ratios in the Sema7aR145W heterozygotes were significantly higher than those of WT controls (Figure 1, A–C). Histological assessments revealed that hepatic lipid droplets and NASs in the HFD-fed Sema7aR145W heterozygous mice were also significantly increased compared with the HFD-fed WT mice (Figure 1, D and E). In comparison with the WT controls, the levels of hepatic TG, total cholesterol (Tch), and serum ALT and AST were also significantly increased (Figure 1F and Supplemental Table 2). Thus, the Sema7aR145W heterozygous mutation promoted the progression of NAFLD in mice.
Figure 1. The Sema7aR145W heterozygous mutation promotes the progression of NAFLD in mice following HFD feeding.
Male WT and Sema7aR145W heterozygous (HE) mice were fed with normal chow diet (NCD) or HFD for 26 weeks. (A) The dynamic changes in the gains of body weights. (B) Liver weights. (C) The liver/body weight ratios. NCD-WT (n = 5), NCD-HE (n = 6), HFD-WT (n = 5), HFD-HE (n = 6). (D) Representative images (original magnification, ×200) of H&E, Oil Red O, and Sirius red staining of liver sections. (E) Quantitative analysis of the NAFLD activity scores (NASs). (F) Hepatic TG and Tch levels. The data were analyzed by 1-way ANOVA with Tukey’s post hoc tests or by Kruskal-Wallis test with Dunn’s post hoc test analysis. *P < 0.05 versus the WT mice with NCD; #P < 0.05 versus the Sema7aR145W heterozygous mice with NCD; $P < 0.05 versus the WT mice with HFD. TG, triglycerides; Tch, total cholesterol.
The Sema7aR145W mutation significantly increases the accumulation of small lipid droplets in mouse livers.
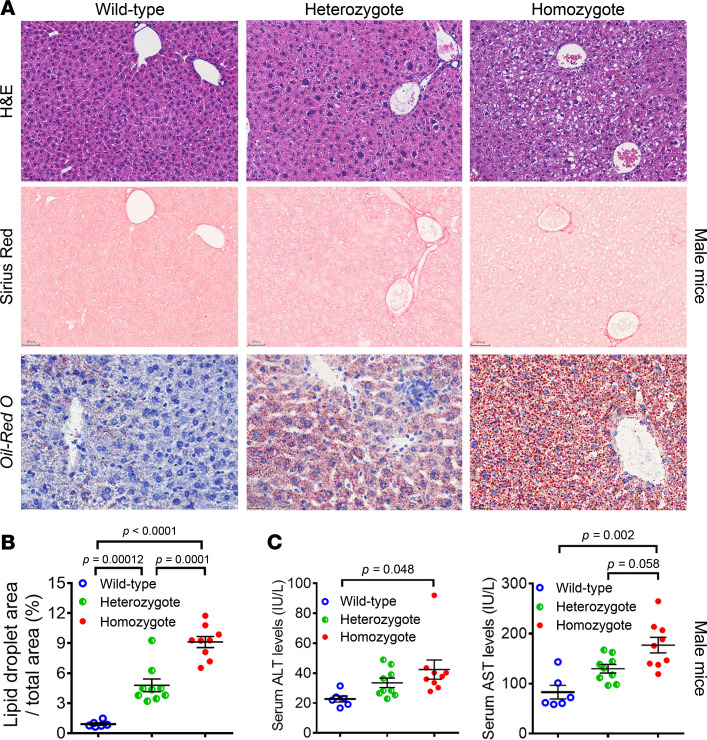
Oil Red O staining displayed the markedly increased accumulation of small lipid droplets in the liver sections of Sema7aR145W homozygous mice at 10 weeks of age, compared with the age-matched WT and heterozygous mice (Figure 2, A and B; and Supplemental Figure 3, A and B). Moreover, the areas of hepatic small lipid droplets in heterozygous mice were also significantly larger than in the WT control mice (Figure 2, A and B; and Supplemental Figure 3, A and B). Furthermore, Sema7aR145W homozygous mice showed levels of serum ALT and AST that were significantly greater than WT and heterozygous mice, although there was no significant difference in these measurements between heterozygotes and WT controls (Figure 2C and Supplemental Figure 3C). Taken together, the Sema7aR145W mutation induced intrahepatic accumulation of small lipid droplets and enhanced liver injury in mice.
Figure 2. The Sema7aR145W mutation causes intrahepatic accumulation of small lipid droplets in male mice at 10 weeks of age.
(A) Representative images (original magnification, ×200) of H&E staining, Sirius red staining, and Oil Red O staining in WT and Sema7aR145W heterozygous and homozygous male mice at the age of 10 weeks. (B) Analysis of lipid droplets in the Oil Red O–stained liver sections of WT (n = 6) and Sema7aR145W heterozygous (n = 9) and homozygous male mice (n = 9). (C) The levels of serum ALT and AST in Sema7aR145W WT (n = 6), heterozygous (n = 9), and homozygous male mice (n = 9). The data were analyzed by 1-way ANOVA with Tukey’s post hoc tests or by Kruskal-Wallis test with Dunn’s post hoc test analysis.
The Sema7aR145W mutation enhances PKC-α activation to stimulate FA and TG synthesis and FA uptake in mouse livers.
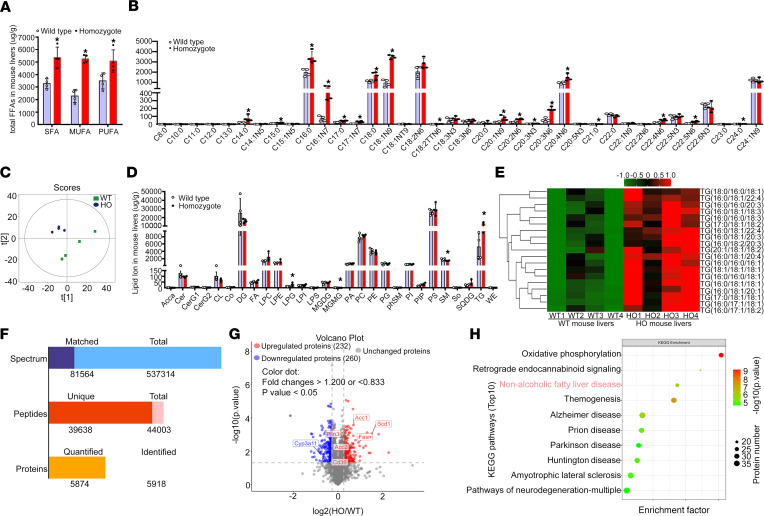
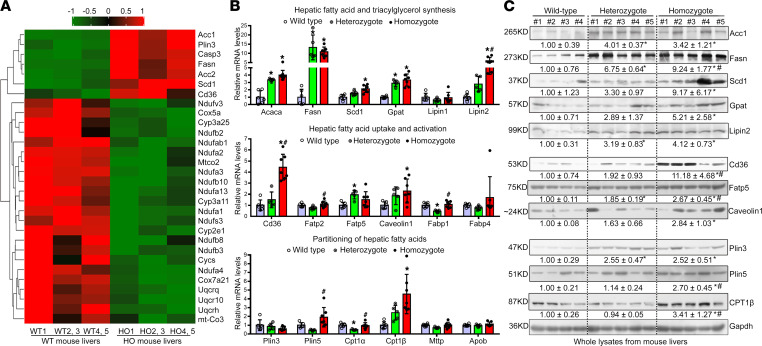
Targeted metabolomics of FA and quantitative lipidomic analyses indicated that the Sema7aR145W homozygous mutation remarkably increased hepatic FA and TG concentrations in mice, relative to the controls (Figure 3, A–E, and Supplemental Tables 3–5). Next, proteomics analysis revealed that, among 5874 quantified proteins, 232 were upregulated and 260 were downregulated (Figure 3, F and G). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis exhibited that the NAFLD pathway (in the top 3) was enriched in Sema7aR145W homozygous mouse livers (Figure 3H and Supplemental Table 6). Proteomics heatmap, real-time quantitative PCR (qPCR), and Western blotting analyses revealed that the Sema7aR145W homozygous mutation increased the expression of key genes for hepatic FA and TG synthesis (Acaca/Acc1, Fasn, Scd1, Gpat, and Lipin2), FA uptake (Cd36, Fatp5, and Caveolin1), and FA partitioning (Plin3, Plin5, and Cpt1β) (Figure 4, A–C). Similar patterns of moderate alterations were detected in heterozygous mice, compared to the WT controls (Figure 4, A–C). These data indicated that the Sema7aR145W mutation stimulated hepatic FA and TG synthesis and FA uptake in mice.
Figure 3. The Sema7aR145W mutation increases hepatic FA and TG concentrations in mouse livers.
Male WT and Sema7aR145W homozygous (HO) mice at 10 weeks old (n = 4 per group) were euthanized and their liver samples were prepared. (A) Gas chromatography tandem mass spectrometry (GC/MS) analysis of total saturated fatty acid (SFA), monounsaturated fatty acid (MUFA), and polyunsaturated fatty acid (PUFA) levels (μg/g of mouse liver) in WT and Sema7aR145W homozygous mouse livers. FFA, free fatty acid. (B) Quantification of hepatic long chain FA (FA μg/g of mouse liver) in WT and Sema7aR145W homozygous mice. (C) Score scatterplot corresponding to a principal component analysis of the lipidomic data in the livers of WT and Sema7aR145W homozygous mice. (D) Quantitative analysis of lipid ion (μg/g of mouse liver) in WT and Sema7aR145W homozygous mouse livers. (E) Heatmap analysis of the TG number of carbons and double bond contents in the livers of WT and Sema7aR145W homozygous mice. (F) Proteomic analysis in the livers of WT and Sema7aR145W heterozygous and homozygous mice (n = 5 per group). (G) Volcano plot of the quantified proteins from WT and Sema7aR145W homozygous mouse livers (n = 5 per group). (H) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of the differentially expressed genes in the pathways between WT and Sema7aR145W homozygous mice. The data were analyzed by independent-sample t test, 2-tailed. *P < 0.05 versus the WT mice.
Figure 4. The Sema7aR145W mutation increases FA and TG synthesis and FA uptake in mouse livers.
(A) Proteomic heatmap analysis of the NAFLD pathway in the livers of WT and Sema7aR145W homozygous mice. The liver extracts from the WT2 and 3, WT4 and 5, HO2 and 3, and HO4 and 5 male mice were combined for proteomics analysis. (B) The relative levels of mRNA transcripts of the genes for FA and TG synthetic enzymes, FA uptake transporters, and FA partitioning in WT mice (n = 6), Sema7aR145W heterozygous mice (n = 5), and Sema7aR145W homozygous mice (n = 7). (C) Western blot analysis of the relative levels of Acc1, Fasn, Scd1, Gpat, Lipin2, Cd36, Fatp5, Caveolin1, Plin3, Plin5, and Cpt1β protein expression in the livers of WT (n = 4), Sema7aR145W heterozygous (n = 5), and Sema7aR145W homozygous (n = 5) mice. The data were analyzed by 1-way ANOVA with Tukey’s post hoc tests or by Kruskal-Wallis test with Dunn’s post hoc test analysis. *P < 0.05 versus the WT mice, #P < 0.05 versus the Sema7aR145W heterozygous mice.
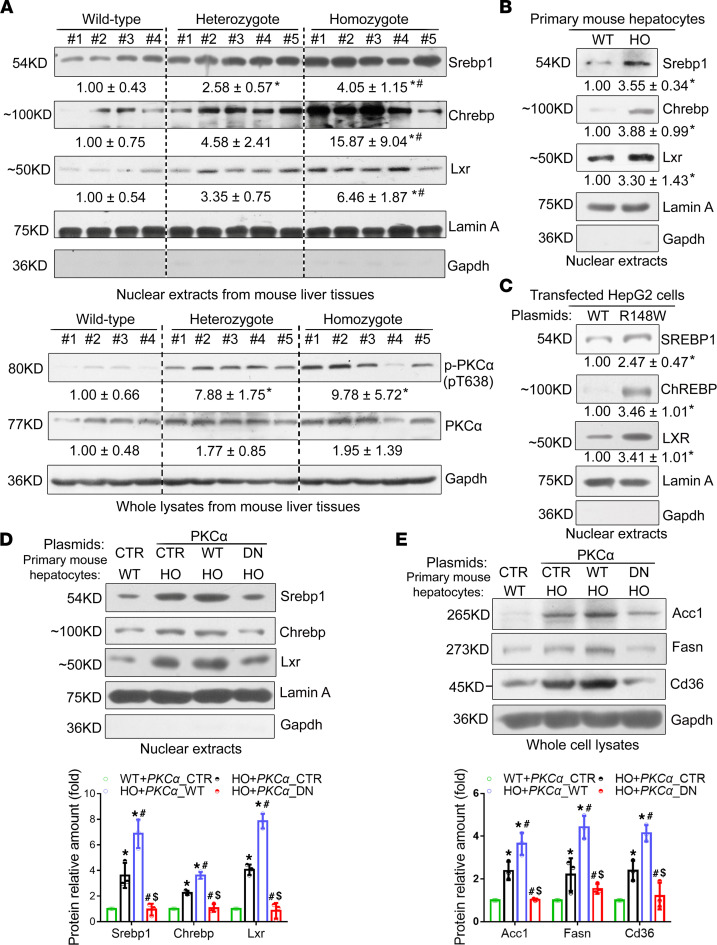
A recent study reported that PKC-α signaling mediates lipid metabolism in diabetic rats (14). Our further analyses of hepatic expression of nuclear Srebp1, Chrebp, Lxr and their targeting genes of Acc1, Fasn, Scd1, Cd36, along with PKC-α phosphorylation, demonstrated that their expression levels were significantly higher in the liver and primary hepatocytes from Sema7aR145W homozygous mice than that from the WT controls (Figure 4, B and C; and Figure 5, A and B). Similar results were obtained in NAFLD patients with SEMA7A mutation, Sema7aR145W heterozygous mouse livers, and HepG2 cells transfected with SEMA7A_R148W construct (Figure 5, A and C, and Supplemental Figure 4). However, there was no significant difference in the relative levels of AMPK phosphorylation among these groups of mice (data not shown). These data indicated that the Sema7aR145W mutation increased PKC-α phosphorylation and nuclear Srebp1, Chrebp, and Lxr expression in the livers of mice. Furthermore, induction of PKCα_WT, but not its dominant negative (PKCα_DN), overexpression markedly increased nuclear Srebp1, Chrebp, and Lxr expression and their downstream Acc1, Fasn, and Cd36 expression in primary Sema7aR145W homozygous mouse hepatocytes (Figure 5, D and E). These findings indicate that the Sema7aR145W mutation enhanced FA and TG synthesis and FA uptake by increasing PKC-α–stimulated nuclear Srebp1, Chrebp, and Lxr expression in hepatocytes of mice.
Figure 5. The Sema7aR145W mutation enhances hepatic FA and TG synthesis and FA uptake by enhancing PKC-α signaling–stimulated expression of transcription factors Srebp1 and Chrebp and nuclear receptor Lxr.
(A) Western blot analysis of the relative levels of Srebp1, Chrebp, Lxr, phosphorylated PKC-α, and PKC-α protein expression in 10-week-old male WT mice (n = 4), Sema7aR145W heterozygous mice (n = 5), and Sema7aR145W homozygous mice (n = 5). Western blot analysis of the relative levels of nuclear Srebp1, Chrebp, and Lxr proteins in primary hepatocytes from WT and Sema7aR145W homozygous mice (B) and in human hepatoma HepG2 cells (C) after transfection with the plasmid for the expression of SEMA7A_WT and SEMA7A_R148W proteins. (D) Representative Western blot of the relative levels of nuclear Srebp1, Chrebp, and Lxr proteins in nuclear extracts and (E) Fasn, Acc1, and Cd36 proteins in whole-cell lysates of primary mouse hepatocytes after transfection with empty vector (CTR) or the plasmid for the expression of PKCα_WT or PKCα_dominant negative (DN) mutant, respectively. All primary mouse hepatocytes were isolated from 12-week-old male WT and Sema7aR145W homozygous mice. Data are representative images or expressed as the mean ± SD of each group from 3 separate experiments. The data were analyzed by 1-way ANOVA with Tukey’s post hoc tests or by Kruskal-Wallis test with Dunn’s post hoc test analysis. *P < 0.05 versus the WT mice, #P < 0.05 versus the Sema7aR145W heterozygous mice, $P < 0.05 versus the primary HO mouse hepatocytes transfected with CTR; n = 3.
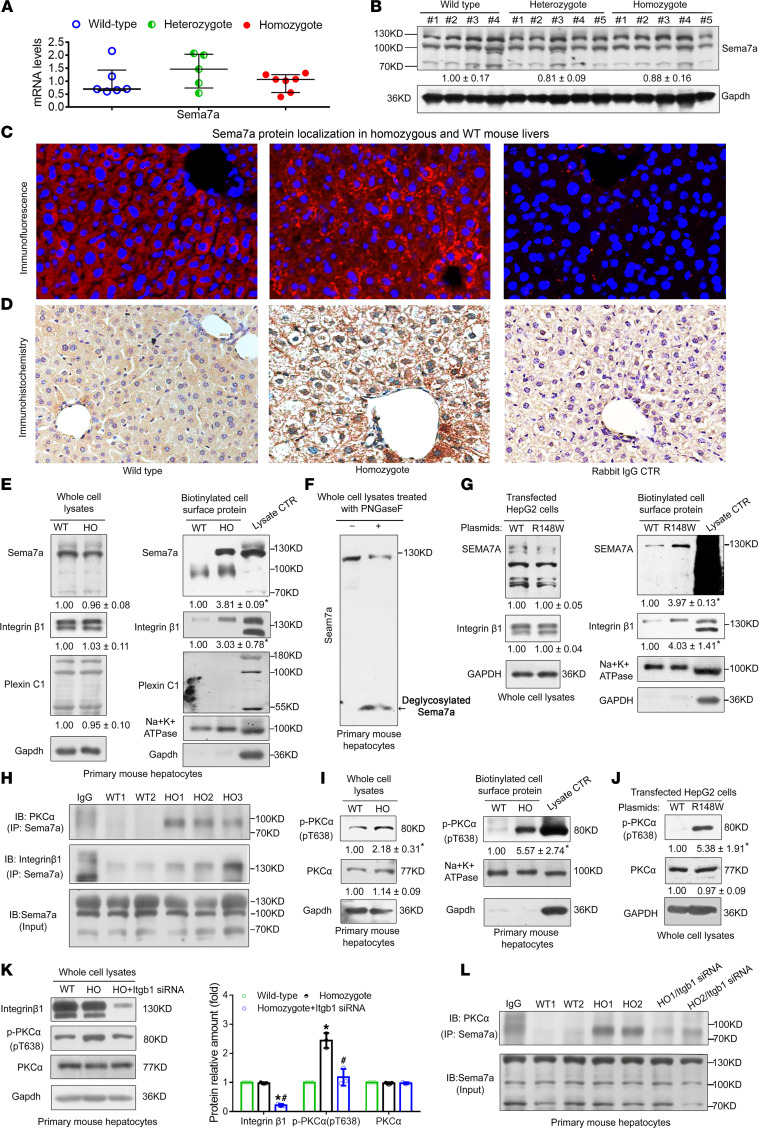
The Sema7aR145W mutation increases its protein in cell surface membranes and activates the PKC-α signaling in hepatocytes.
Real-time qPCR and Western blotting analyses demonstrated that the Sema7aR145W mutation did not result in changes in hepatic Sema7a mRNA and protein expression in mice (Figure 6, A and B). However, immunofluorescence and immunohistochemistry indicated that the Sema7aR145W homozygous mutation markedly increased Sema7a protein on hepatocyte membranes in mouse livers (Figure 6, C and D). Western blotting analysis of cell surface membrane extracts of primary mouse hepatocytes revealed that the Sema7aR145W homozygous mutation dramatically increased the membrane-associated Sema7a and its receptor integrin β1 proteins, but not plexin C1, another Sema7a receptor (Figure 6E). Interestingly, treatment of whole primary mouse hepatocyte lysates with peptide N glycosidase F to remove N-glycans from glycoproteins (15) resulted in a markedly deglycosylated Sema7a protein band (Figure 6F, arrow) with an obviously decreased molecular weight along with a decrease in the ~130 kDa Sema7a protein band (Figure 6F), indicating that the ~130 kDa Sema7a protein was N-glycosylated. A similar pattern of N-glycosylated Sema7a and integrin β1 proteins was observed in HepG2 cells after transfection with the SEMA7A_R148W construct (Figure 6G). However, there was no significant difference in total Sema7a, integrin β1, and plexin C1 protein expression in these mutant mouse hepatocytes and HepG2 cells (Figure 6, E and G). Interestingly, treatment of mouse liver tissues with anti-Sema7a effectively precipitated not only integrin β1, but also PKC-α, a downstream molecule of integrin β1 signaling (16), particularly when using the Sema7aR145W homozygous mouse liver tissues (Figure 6H). Hence, Sema7aR145W mutation markedly increased its interaction with integrin β1 and PKC-α in mouse livers (Figure 6H). Moreover, the Sema7aR145W mutation also increased the relative levels of PKC-α phosphorylation in whole-cell lysates and cell surface membrane fractions of Sema7aR145W homozygous mouse hepatocytes and transfected HepG2 cells, indicating that the Sema7aR145W homozygous mutation enhanced PKC-α activation (16), though it did not change its total protein expression (Figure 6, I and J). Furthermore, integrin β1 silencing dramatically reduced the levels of PKC-α phosphorylation and the interaction between Sema7a and PKC-α in primary Sema7aR145W homozygous mouse hepatocytes, indicating that the increased activation of PKC-α by the Sema7aR145W mutation depended on its receptor integrin β1 (Figure 6, K and L). Together, our data revealed that the Sema7aR148W (human) mutation in hepatocytes increased the cell surface membrane localization of Sema7a and its receptor integrin β1 proteins and activated PKC-α signaling. Subsequently, the enhanced PKC-α activation stimulated FA and TG synthesis and FA uptake and modulated their metabolism in hepatocytes, leading to excessive accumulation of intrahepatic lipids and promoting the progression of NAFLD.
Figure 6. The Sema7aR145W mutation does not alter total Sema7a expression but increases Sema7a and integrin β1 in the cell surface membrane and activates PKC-α signaling in hepatocytes.
Relative levels of Sema7a mRNA transcripts (A) and protein expression (B) in 10-week-old male WT, Sema7aR145W heterozygous, and homozygous mice. One-way ANOVA with post hoc analysis. (C) Immunofluorescence and (D) IHC analyses of Sema7a protein in the livers of WT and Sema7aR145W homozygous mice. Normal rabbit IgG was used as the negative control. Original magnification, ×200. (E) Relative expression levels of Sema7a and its receptors integrin β1 and plexin C1 in whole-cell lysates (left) and membrane fractions (right) extracted from primary mouse hepatocytes. (F) N-glycosylated-Sema7a protein (~130 kDa) was detected in primary mouse hepatocytes. Western blot revealed the deglycosylated Sema7a (black arrow). (G) Relative levels of Sema7a and integrin β1 proteins in whole-cell lysates (left) and membrane fractions (right) from human hepatoma HepG2 cells after transfection with the plasmid for SEMA7A_WT or SEMA7A_R148W. (H) Co-immunoprecipitation analysis of protein interactions among Sema7a, PKCα, and integrin β1 in liver tissues from 10-week-old male WT and Sema7aR145W homozygous mice. (I) Phosphorylated PKC-α (T638) and PKC-α protein levels in whole-cell lysates (left panel) and membrane fractions (right panel) from primary mouse hepatocytes. T638 and PKC-α protein levels in (J) HepG2 cells that were transfected with SEMA7A_WT or SEMA7A_R148W plasmid and (K) primary mouse hepatocytes following integrin β1 silencing. (L) Co-immunoprecipitation analysis of protein interaction between Sema7a and PKC-α in primary mouse hepatocytes after integrin β1 silencing. Data are representative images or expressed as the mean ± SD of each group from 3 separate experiments. The difference among the groups was determined by 1-way ANOVA with Tukey’s post hoc tests or by Kruskal-Wallis test with Dunn’s post hoc test analysis, and the difference between the groups was analyzed by Student’s t test. *P < 0.05 versus WT mice (cells); #P < 0.05 versus Sema7aR145W heterozygous mice (cells).
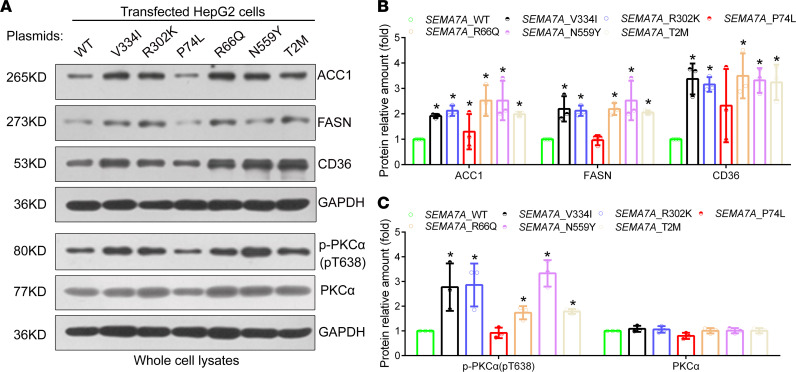
The V334I, R302K, R66Q, N559Y, and T2M mutations in SEMA7A can also induce the activation of the PKC-α signaling and the expression of key genes for FA and TG synthesis and FA uptake in hepatocytes.
To determine the functional significance of other mutations in SEMA7A, we generated serial plasmids for the expression of SEMA7A_V334I, SEMA7A_R302K, SEMA7A_P74L, SEMA7A_R66Q, SEMA7A_N559Y, and SEMA7A_T2M. Next, HepG2 cells were transfected with SEMA7_WT and each type of mutant plasmid (Supplemental Figure 5), and the effects on PKC-α phosphorylation and ACC1, CD36, and FASN expression were analyzed by Western blotting. As shown in Figure 7, A–C, the overexpression of SEMA7A_V334I, _R302K, _R66Q, _N559Y, and _T2M molecules, but not SEMA7A_P74L, significantly increased PKC-α phosphorylation and the expression of CD36, ACC1, and FASN in HepG2 cells. However, there was no single SEMA7A mutant that significantly altered the relative levels of PKC-α expression in HepG2 cells (Figure 7, A and C). Together, these data indicate that SEMA7A_V334I, _R302K, _R66Q, N559Y, and _T2M mutants may act similarly to the R148W mutant in regulating FA and TG synthesis and FA uptake in human hepatocytes.
Figure 7. The SEMA7A mutation activates PKC-α signaling in hepatocytes.
HepG2 cells were transfected with the plasmid for the expression of SEMA7A_WT or SEMA7A_V334I, _P302K, _P74L, _R66Q, _N559Y, or _T2M mutant, and the relative levels of ACC1, FASN, CD36, phosphorylated PKC-α (T638), and PKC-α expression in each group of cells were determined by Western blot. (A) Representative images of Western blot analyses. (B) Quantitative analysis of each mutant protein and (C) the relative levels of T638 and PKC-α in HepG2 cells from 3 separate experiments. The levels of each protein in the SEMA7A_WT–transfected cells were designated as 1. The data were analyzed by 1-way ANOVA with Tukey’s post hoc tests or by Kruskal-Wallis test with Dunn’s post hoc test analysis. *P < 0.05 versus the SEMA7A_WT cells.
Discussion
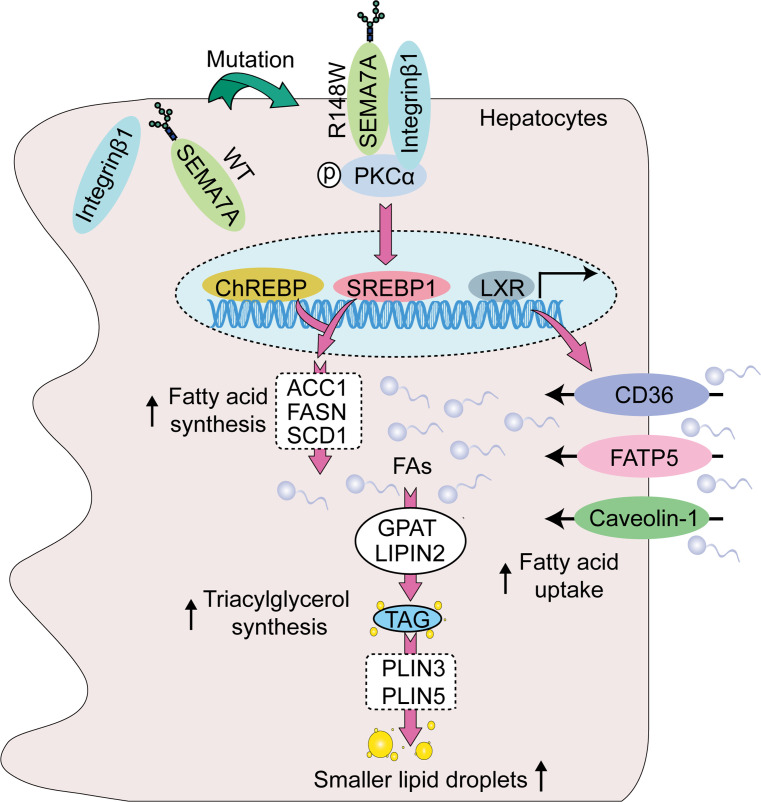
This study reported a potentially novel genetic determinant of NAFLD, the R148W mutation in SEMA7A, and uncovered its pathophysiological mechanisms (Figure 8). Our study highlighted 4 potentially novel findings: (a) SEMA7A mutations were genetic risk factors for human NAFLD and its severity in humans (Tables 1 and 2); (b) the Sema7aR145W mutation significantly increased steatosis severity and NAS in mice (Figure 1); (c) the Sema7aR145W mutation markedly increased N-glycosylated Sema7a and its receptor integrin β1 proteins in hepatocyte surface membranes (Figure 6); and (d) the Sema7aR145W mutation caused intrahepatic lipid accumulation by enhancing PKC-α signaling–stimulated FA and TG synthesis and FA uptake (Figures 2–6).
Figure 8. The potential mechanism by which the SEMA7AR148W mutation causes lipid accumulation in hepatocytes.
First, the mutation increases SEMA7A and its receptor integrin β1 proteins on the surface of cell membranes to promote PKC-α activation in hepatocytes. Second, the activated PKC-α signaling enhances the expression of transcriptional factors SREBP1 and ChREBP and nuclear receptor LXR, increasing FA and TG synthesis and FA uptake in hepatocytes. Finally, these increased the accumulation of small lipid droplets in the liver, leading to the development and progression of NAFLD.
SEMA7A acts on integrin β1 and plexin C1 receptors to regulate multiple physiological and pathological processes (6–11). A recent study has shown that Sema7a deficiency reduces FA oxidation and oxidative phosphorylation in mouse peritoneal macrophages (12). Moreover, mutations in SEMA7A are associated with several disorders, such as Kallmann syndrome (10, 11). Here, we addressed whether mutations in SEMA7A affected lipid metabolism and contributed to the development of NAFLD. First, we identified 17 NAFLD patients with SEMA7A heterozygous mutation in a population of 470 patients with NAFLD, who had been evaluated by histological examination of biopsied liver specimens (Table 1). The Sema7aR145W heterozygous mutation increased susceptibility to NAFLD and NAFLD severity in human patients and mouse models (Figure 1 and Table 2). Mechanistically, the Sema7aR145W mutation markedly increased N-glycosylated SEMA7A and its receptor integrin β1 proteins on the cell membranes of hepatocytes, resulting in intrahepatic accumulation of small lipid droplets by enhancing PKC-α signaling–stimulated FA and TG synthesis and FA uptake (Figure 6). We also provided evidence that the SEMA7AR148W mutation was a gain-of-function mutation, since (a) Sema7a deficiency reduces FA oxidation in mouse peritoneal macrophages and cholestatic liver injury (7), whereas the Sema7aR145W homozygous mutation increased lipid accumulation and elevated levels of serum ALT and AST in mice (Figures 1 and 2); (b) the Sema7aR145W mutation increased Sema7a and its receptor integrin β1 proteins on hepatocyte surface membranes; and (c) PKC-α, a downstream signaling molecule of integrin β1 (16), was detected in the immunoprecipitated Sema7a protein complex, and its phosphorylation was enhanced by the Sema7aR145W mutation (Figure 6). In addition, KEGG analysis of the proteomics data between Sema7aR145W homozygous mice and WT mice demonstrated that the oxidative phosphorylation pathway was the most abundant pathway (Figure 3H). Similarly, a recent study reported that the stability of oxidative phosphorylation subunits was reduced in a diet-induced mouse model of NAFLD (17), suggesting that the oxidative phosphorylation pathway may play a crucial role in NAFLD pathogenesis. Therefore, Sema7aR145W mutation may also influence the oxidative phosphorylation pathway, leading to the progression of NAFLD. Nevertheless, this hypothesis will need to be addressed in the future.
Moreover, SEMA7A is also expressed in multiple tissues, including the brain, lung, intestine, kidney, bone, and immune system (5–14, 16, 18–24). Recent studies have shown that the upregulation of SEMA7A expression is significantly associated with multiple sclerosis (18), systemic sclerosis–related interstitial lung disease (19), rheumatoid arthritis (20), airway inflammation (21), colitis (22), systemic lupus erythematosus (23), and melanoma and other cancers (24). Thus, the SEMA7AR148W mutation may also be the causative genetic factor for these diseases. Notably, SEMA7A is expressed in the cardiovascular system and is crucial for vascularization and angiogenesis (6). Atherosclerosis is characterized by the accumulation of lipids and extracellular matrix on the arterial wall (25). Our present data indicated that the Sema7aR145W mutation caused lipid accumulation in the cell membranes of hepatocytes (Figure 2). Therefore, further studies are needed to ascertain whether the SEMA7AR148W mutation contributes to arterial lipid metabolic dysfunction and atherosclerosis.
In conclusion, our potentially novel data indicated that the SEMA7AR148W mutation was a novel genetic determinant of NAFLD and that PKC-α signaling–induced FA and TG synthesis and FA uptake were enhanced by the SEMA7AR148W mutation. In our study, we discovered a molecular mechanism underlying the pathogenesis of NAFLD caused by the SEMA7AR148W mutation. The inhibition of hepatic PKC-α signaling may lead to novel NAFLD therapies.
Methods
Patients with NAFLD.
Patients with NAFLD were enrolled from a well-characterized Prospective Epidemic Research Specifically of NASH cohort and diagnosed by histological examination of biopsied liver samples from December 2016 to July 2020 (13). Their body weight, height, waist circumference, and hip circumference were measured in light clothing by well-trained nurses in the morning. BMI (kg/m2) was calculated as body weight divided by the height squared. After an 8-hour overnight fast, patients’ blood samples were collected from the antecubital vein by experienced nurses. The levels of serum ALT, AST, GGT, ALP, TBIL, and DBIL in individual patients were analyzed using an automated analyzer (Abbott AxSYM) (13). The detailed methods for liver biopsies have been described in the Supplemental Methods. Genomic DNA was extracted from patients’ peripheral blood mononuclear cells, as described previously (13), and stored at –80°C.
Single gene (SEMA7A) exon sequencing analysis in patients with NAFLD.
The DNA fragments for human SEMA7A exons (exon 1 to exon 14) were amplified by PCR using specific primers (Supplemental Table 1). The PCR products were subjected to Sanger sequencing analysis, which, together with data analysis, were supported technically by Beijing Genomics Institute. The data reported in this paper have been deposited in the OMIX, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (https://ngdc.cncb.ac.cn/omix: accession no. OMIX001290).
OR estimation of SEMA7A heterozygous mutations for NAFLD.
The potential risk of SEMA7A heterozygous missense mutations for the development of NAFLD was estimated by OR and 95% CI through logistic regression analysis using Review Manager version 5.2. Data from East Asian patients (18,394 individuals) were extracted from the gnomAD and used as the controls. A 2-tailed P value of less than 0.05 was considered statistically significant.
Baseline characteristic analysis of NAFLD patients with SEMA7A heterozygous mutations and their pair-matched controls.
We identified 17 NAFLD patients with SEMA7A heterozygous mutations in the 470-patient NAFLD cohort. However, 4 NAFLD patients with SEMA7A heterozygous mutations were excluded from further analyses because of the lack of pair-matched controls. Data from pair-matched controls with WT SEMA7A were extracted from the database for the study. The patients were pair-matched with the controls for age, sex, BMI, and the variation in PNPLA3 (rs738409 C>G encoding PNPLA3 I148M), a risk factor for NAFLD (1, 2). Continuous data are expressed as mean ± SD and median (IQR), whereas categorical data are expressed as frequencies and percentages. The difference between groups was analyzed by χ2 test, paired 2-sample t test, and Wilcoxon’s signed-rank test where applicable.
Generation and characterization of Sema7aR145Wmutant mice and their sample collection.
Sema7aR145W (c.433C>T) mutant C57BL/6J mice were designed and generated by Shanghai Model Organisms Center using the Cas9-targeted single guide RNA of 5′ ATGCCCGGAAGCCCAGCTGCTGG 3′ and a similar protocol described previously (26). The obtained F0 mice were characterized by PCR and sequencing using primer pairs: F1: 5′ GGAGGGAACATGAGTTTGCT 3′; R1: 5′ CCACATGACCACCGGCTACT 3′. Serum and liver samples were collected from 10-week-old WT (n = 10, 6 male/4 female), Sema7aR145W heterozygous (n = 15, 9 male/6 female), and homozygous mice (n = 15, 9 male/6 female) as described previously (26).
Sema7aR145W heterozygous mice with HFD feeding.
Male 8-week-old Sema7aR145W heterozygous and their age-matched WT C57BL/6J mice were randomized and fed with HFD (catalog D12492, Research Diets; WT mice, n = 5; heterozygous mice, n = 6) or NCD (catalog D12450J, Research Diets; WT mice, n = 5; heterozygous mice, n = 6) for 26 weeks. Their body weights were measured weekly. At the end of the HFD feeding, their blood samples were collected for preparing serum samples and the mice were euthanized. Their livers were dissected, imaged (Olympus Corporation), and weighed sequentially. The left lobes of the liver were frozen in liquid nitrogen, and the right lobes of the liver were fixed in formalin for subsequent paraffin-embedding and histological staining.
GC/MS analysis of FA in mouse liver extracts.
Mouse liver samples were prepared from WT, Sema7aR145W heterozygous, and Sema7aR145W homozygous mice and subjected to GC/MS analysis of FA, as described previously (27, 28). The detailed procedures are described in the Supplemental Methods.
Lipidomic analysis.
Hepatic lipids were extracted from WT, Sema7aR145W heterozygous, and Sema7aR145W homozygous mice (n = 4 per group) using the methyl tert-butyl ether (MTBE) method as described previously (29). Briefly, individual samples (30 mg each) were homogenized in 200 μL water and mixed sequentially with 20 μL internal lipid standard mixture, 800 μL of MTBE, and 240 μL of precooled methanol, followed by ultrasonication. The detailed procedures are described in the Supplemental Methods.
Proteomic and bioinformatic analyses.
Liver samples from WT, Sema7aR145W heterozygous, and Sema7aR145W homozygous mice (n = 5 per group) were homogenized in a buffer (4% SDS, 100 mM Tris-HCl, 1 mM DTT, pH 7.6). The obtained proteins (200 μg, each) were digested with 4 μg trypsin (Promega) in 40 μL of 25 mM NH4HCO3 buffer overnight at 37°C, and the obtained peptides were desalted on C18 cartridges (Empore SPE, MilliporeSigma). The detailed procedures are described in the Supplemental Methods.
Preparation and collection of primary mouse hepatocytes.
Primary mouse hepatocytes were isolated from 10- and 12-week-old WT and Sema7aR145W mice using collagenase (Worthington Biochemical Corporation) perfusion as previously described (26, 30). The isolated hepatocytes were cultured in 5% FBS-Williams’ Medium E (Gibco, Thermo Fisher Scientific, catalog 12551) overnight. The cells were harvested and lysed, and their surface membranes were extracted and biotinylated for Western blot analysis. Similarly, their nuclei were extracted for TaqMan qPCR. The details of primer sequences and antibody information are listed in Supplemental Tables 7 and 8.
Cell surface protein biotinylation and extraction.
Cell surface proteins from primary mouse hepatocytes and human HepG2 hepatoma cells were biotinylated using the EZ-Link Sulfo-NHS-SS-Biotin reagent (Thermo Fisher Scientific; catalog 21331), according to the supplier’s protocol (31–33). For biotinylation, cells (1 × 106/well) were cultured in 6-well plates and washed 3 times with chilled phosphate-buffered saline (PBS). The cells were treated with 1.0 mg/mL Sulfo-NHS-SS-Biotin (Thermo Fisher Scientific) in chilled PBS for 60 minutes at 4°C to biotinylate the membrane proteins of the cultured cells. The cells were treated with 100 mM glycine in PBS for 10 minutes to terminate the biotinylation reactions and washed 3 times with ice-cold PBS (pH 8.0) to remove nonreacted biotinylation reagent. Subsequently, the cells were harvested and lysed, followed by centrifugation at 16,000g for 20 minutes at 4°C. The resulting supernatants (250 μg total proteins) were reacted with 200 μL of 10% streptavidin agarose (Thermo Fisher Scientific, 20347) and centrifuged at 500g for 5 minutes at 4°C, followed by washing 5 times with lysis buffer. The biotinylated proteins were eluted in 2× SDS sample buffer supplemented with DTT (0.02 g/mL) and analyzed by SDS-PAGE.
Other information on methods and materials is available in the Supplemental Methods.
Study approval.
The study protocol for patients with NAFLD was approved by the Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University. Written informed consent was obtained from each patient. For single-cell RNA sequencing, the study was carried out in accordance with the Declaration of Helsinki of the World Medical Association. The study protocol was reviewed and approved by the Institutional Ethics Review Board at the Southwest Hospital.
Author contributions
Experiments were conceived and designed by JC. Experiments were performed by NZ, XZ, JD, QP, GL, JQ, ML, LL, and YC. Data analysis was done by JC, MHZ, QP, YP, XZ, JD, QX, and WYL. Special reagents/materials/analysis tools were provided by MHZ, WYL, QL, QW, and LZ. The manuscript was written by JC, SYC, and JLB and critically revised by XO.
Supplementary Material
Acknowledgments
We thank Yong-Feng Yang (Department of Liver Disease, the Second Hospital of Nanjing affiliated to Medical School of South-East University), Xindong Liu, Senlin Xu (Department of Pathology, Southwest Hospital), Xuequan Huang (Department of Radiology, Southwest Hospital), Guohong Deng (Department of Infectious Diseases, Southwest Hospital), Man Li (Liver Center, Yale School of Medicine), and our team members (Department of Gastroenterology, Southwest Hospital of Third Military Medical University) for their technical assistance. This study was supported by grants from the National Natural Science Foundation of China (81922012, 81770583, 32171123), the Outstanding Youth Science Foundation of Chongqing (cstc2021jcyj-jqX0005), the Project of Chongqing Universities Innovation Research Group (2021cqspt01), and Natural Science Foundation of Southwest Hospital (2017YQRC-01 and XZ-2019-505-001).
Version 1. 08/08/2022
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2022, Zhao et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2022;7(15):e154113.https://doi.org/10.1172/jci.insight.154113.
Contributor Information
Nan Zhao, Email: nan.zhao@cldcsw.org.
Xiaoxun Zhang, Email: xiaoxun.zhang@cldcsw.org.
Jingjing Ding, Email: jingjing.ding@cldcsw.org.
Qiong Pan, Email: qiong.pan@cldcsw.org.
Ming-Hua Zheng, Email: zhengmh@wmu.edu.cn.
Wen-Yue Liu, Email: liuwenyue@wmu.edu.cn.
Gang Luo, Email: gang.luo@cldcsw.org.
Jiaquan Qu, Email: jiaquan.qu@cldcsw.org.
Mingqiao Li, Email: mingqiao.li@cldcsw.org.
Ling Li, Email: ling.li@cldcsw.org.
Ying Cheng, Email: ying.cheng@cldcsw.org.
Ying Peng, Email: ying.peng@cldcsw.org.
Qiaoling Xie, Email: qiaoling.xie@cldcsw.org.
Qinglin Wei, Email: qinglin.wei@cldcsw.org.
Qiao Li, Email: qiao.li@cldcsw.org.
Lingyun Zou, Email: lingyun.zou@gmail.com.
Xinshou Ouyang, Email: xinshou.ouyang@yale.edu.
Shi-Ying Cai, Email: shi-ying.cai@yale.edu.
Jin Chai, Email: jin.chai@cldcsw.org.
References
- 1.Hodson L, Gunn PJ. The regulation of hepatic fatty acid synthesis and partitioning: the effect of nutritional state. Nat Rev Endocrinol. 2019;15(12):689–700. doi: 10.1038/s41574-019-0256-9. [DOI] [PubMed] [Google Scholar]
- 2.Friedman SL, et al. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24(7):908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Younossi ZM. Non-alcoholic fatty liver disease — a global public health perspective. J Hepatol. 2019;70(3):531–544. doi: 10.1016/j.jhep.2018.10.033. [DOI] [PubMed] [Google Scholar]
- 4.Alto LT, Terman JR. Semaphorins and their signaling mechanisms. Methods Mol Biol. 2017;1493:1–25. doi: 10.1007/978-1-4939-6448-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamada A, et al. Molecular cloning of a glycosylphosphatidylinositol-anchored molecule CDw108. J Immunol. 1999;162(7):4094–4100. [PubMed] [Google Scholar]
- 6.Song Y, et al. The involvement of semaphorin 7A in tumorigenic and immunoinflammatory regulation. J Cell Physiol. 2021;236(9):6235–6248. doi: 10.1002/jcp.30340. [DOI] [PubMed] [Google Scholar]
- 7.De Minicis S, et al. Semaphorin 7A contributes to TGF-β-mediated liver fibrogenesis. Am J Pathol. 2013;183(3):820–830. doi: 10.1016/j.ajpath.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czopik AK, et al. Semaphorin 7A is a negative regulator of T cell responses. Immunity. 2006;24(5):591–600. doi: 10.1016/j.immuni.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 9.Liu H, et al. Structural basis of semaphorin-plexin recognition and viral mimicry from Sema7A and A39R complexes with PlexinC1. Cell. 2010;142(5):749–761. doi: 10.1016/j.cell.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koh JM, et al. Association study of semaphorin 7a (sema7a) polymorphisms with bone mineral density and fracture risk in postmenopausal Korean women. J Hum Genet. 2006;51(2):112–117. doi: 10.1007/s10038-005-0331-z. [DOI] [PubMed] [Google Scholar]
- 11.Zhao Y, et al. A novel heterozygous intron mutation in SEMA7A causing kallmann syndrome in a female. Gynecol Endocrinol. 2020;36(3):218–221. doi: 10.1080/09513590.2019.1680624. [DOI] [PubMed] [Google Scholar]
- 12.Körner A, et al. Sema7A is crucial for resolution of severe inflammation. Proc Natl Acad Sci U S A. 2021;118(9):e2017527118. doi: 10.1073/pnas.2017527118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng KI, et al. Clinical utility of the MAFLD definition. J Hepatol. 2021;74(4):989–991. doi: 10.1016/j.jhep.2020.12.016. [DOI] [PubMed] [Google Scholar]
- 14.Li X, et al. Regulation of lipid metabolism in diabetic rats by Arctium lappa L. polysaccharide through the PKC/NF-κB pathway. Int J Biol Macromol. 2019;136:115–122. doi: 10.1016/j.ijbiomac.2019.06.057. [DOI] [PubMed] [Google Scholar]
- 15.Bodnar J, et al. Enzymatic removal of N-glycans by PNGase F coated magnetic microparticles. Electrophoresis. 2016;37(10):1264–1269. doi: 10.1002/elps.201500575. [DOI] [PubMed] [Google Scholar]
- 16.Sisková Z, et al. Fibronectin impedes “myelin” sheet-directed flow in oligodendrocytes: a role for a beta 1 integrin-mediated PKC signaling pathway in vesicular trafficking. Mol Cell Neurosci. 2006;33(2):150–159. doi: 10.1016/j.mcn.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Lee K, et al. Hepatic mitochondrial defects in a nonalcoholic fatty liver disease mouse model are associated with increased degradation of oxidative phosphorylation subunits. Mol Cell Proteomics. 2018;17(12):2371–2386. doi: 10.1074/mcp.RA118.000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa C, et al. Expression of semaphorin 3A, semaphorin 7A and their receptors in multiple sclerosis lesions. Mult Scler. 2015;21(13):1632–1643. doi: 10.1177/1352458515599848. [DOI] [PubMed] [Google Scholar]
- 19.Varga J, Pasche B. Transforming growth factor beta as a therapeutic target in systemic sclerosis. Nat Rev Rheumatol. 2009;5(4):200–206. doi: 10.1038/nrrheum.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie J, Wang H. Semaphorin 7A as a potential immune regulator and promising therapeutic target in rheumatoid arthritis. Arthritis Res Ther. 2017;19(1):10. doi: 10.1186/s13075-016-1217-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esnault S, et al. Semaphorin 7A is expressed on airway eosinophils and upregulated by IL-5 family cytokines. Clin Immunol. 2014;150(1):90–100. doi: 10.1016/j.clim.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang S, et al. Intestinal epithelial cell-derived semaphorin 7A negatively regulates development of colitis via αvβ1 integrin. J Immunol. 2012;188(3):1108–1116. doi: 10.4049/jimmunol.1102084. [DOI] [PubMed] [Google Scholar]
- 23.Wang P, et al. Decreased expression of semaphorin 3A and semaphorin 7A levels and its association with systemic lupus erythematosus. Immunol Invest. 2020;49(1–2):69–80. doi: 10.1080/08820139.2019.1649280. [DOI] [PubMed] [Google Scholar]
- 24.Lazova R, et al. The semaphorin 7A receptor plexin C1 is lost during melanoma metastasis. Am J Dermatopathol. 2009;31(2):177–181. doi: 10.1097/DAD.0b013e318196672d. [DOI] [PubMed] [Google Scholar]
- 25. Linton MF, et al. The Role of Lipids and Lipoproteins in Atherosclerosis. In: Feingold KR, et al., eds. Endotext. MDText.com: 2019. [PubMed] [Google Scholar]
- 26.Pan Q, et al. Solute carrier organic anion transporter family member 3A1 is a bile acid efflux transporter in cholestasis. Gastroenterology. 2018;155(5):1578–1592. doi: 10.1053/j.gastro.2018.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lan W, et al. Glycochenodeoxycholic acid impairs transcription factor E3 -dependent autophagy-lysosome machinery by disrupting reactive oxygen species homeostasis in L02 cells. Toxicol Lett. 2020;331:11–21. doi: 10.1016/j.toxlet.2020.05.017. [DOI] [PubMed] [Google Scholar]
- 28.Liu M, et al. KIF5A-dependent axonal transport deficiency disrupts autophagic flux in trimethyltin chloride-induced neurotoxicity. Autophagy. 2021;17(4):903–924. doi: 10.1080/15548627.2020.1739444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, et al. Acyl-CoA thioesterase 1 prevents cardiomyocytes from Doxorubicin-induced ferroptosis via shaping the lipid composition. Cell Death Dis. 2020;11(9):756. doi: 10.1038/s41419-020-02948-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai SY, et al. Hepatic NFAT signaling regulates the expression of inflammatory cytokines in cholestasis. J Hepatol. 2021;74(3):550–559. doi: 10.1016/j.jhep.2020.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park H, et al. IRE1α kinase-mediated unconventional protein secretion rescues misfolded CFTR and pendrin. Sci Adv. 2020;6(8): eaax9914. doi: 10.1126/sciadv.aax9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hui KK, et al. GABARAPs dysfunction by autophagy deficiency in adolescent brain impairs GABAA receptor trafficking and social behavior. Sci Adv. 2019;5(4):eaau8237. doi: 10.1126/sciadv.aau8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ozaki I, et al. Regulation of TGF-β1-induced pro-apoptotic signaling by growth factor receptors and extracellular matrix receptor integrins in the liver. Front Physiol. 2011;2:78. doi: 10.3389/fphys.2011.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.