Abstract
Pseudomonas aeruginosa possesses an extensive armament of genes involved in oxidative stress defense, including katB-ankB, ahpB, and ahpC-ahpF. Transcription of these genes was regulated in response to H2O2, paraquat, or organic peroxides. Expression of katB-lacZ and the observed KatB catalase levels in P. aeruginosa PAO1 were induced up to 250-fold after exposure to oxidative stress-generating compounds. Also, ahpB-lacZ and ahpC-lacZ expression was 90- and 3-fold higher, respectively, upon exposure to paraquat. The dose- and time-response curves revealed that 1 μM paraquat was sufficient for half-maximal activation of each reporter fusion within 5 min of exposure. Expression of these genes was not observed in a ΔoxyR mutant, indicating that OxyR was essential for this response. The transcriptional start sites of katB-ankB, ahpB, and ahpC-ahpF were mapped, putative OxyR-binding sites were identified upstream of the −35 promoter elements, and direct binding of purified OxyR protein to these target promoters was demonstrated. The oxyR mutant was hypersusceptible to oxidative stress-generating agents, including H2O2 and paraquat, in spite of total KatA catalase activity being comparable to that of the wild type. The oxyR phenotype was fully complemented by a plasmid containing the oxyR gene, while any of the katB, ahpB, or ahpCF genes alone resulted in only marginal complementation. Increased katB-lacZ expression and higher KatB catalase levels were detected in a ΔahpCF background compared to wild-type bacteria, suggesting a compensatory function for KatB in the absence of AhpCF. In P. aeruginosa, oxyR is located upstream of recG, encoding a putative DNA repair enzyme. oxyR-lacZ and recG-lacZ reporter activities and oxyR-recG mRNA analysis showed that oxyR and recG are organized in an operon and expressed constitutively with regard to oxidative stress from a single promoter upstream of oxyR. Mutants affected in recG but not oxyR were dramatically impaired in DNA damage repair as measured by sensitivity to UV irradiation. In conclusion, we present evidence that the oxyR-recG locus is essential for oxidative stress defense and for DNA repair.
Pseudomonas aeruginosa generates metabolic energy primarily through aerobic respiration. This process, involving a four-electron reduction of molecular oxygen (O2) to water, can be potentially dangerous to the cell. Specifically, aberrant electron flow from the electron transport chain or cellular redox enzymes to O2 can lead to the production of reactive oxygen intermediates (ROIs). These include superoxide (O2−), hydrogen peroxide (H2O2), and hydroxyl radical (HO·). Furthermore, bacteria can be exposed to exogenous ROIs, especially during infection of humans, where phagocytes (e.g., neutrophils) mount a dramatic oxygen-dependent antimicrobial response (16, 38). The unchecked production or accumulation of these species can lead to cell damage, mutations, or death. The generation of HO·, the most destructive of the above-mentioned compounds, is in part dependent upon the presence of a transition metal, such as iron or copper, and H2O2. Defense against ROIs is provided by antioxidant enzymes (superoxide dismutase [SOD], catalase, and peroxidase), iron sequestration, free-radical-scavenging agents, DNA-binding proteins, and DNA repair enzymes (4, 25, 26, 32, 35, 62). P. aeruginosa possesses an impressive antioxidant armament for defense against ROIs, including two SODs (cofactored by either iron [Fe-SOD] or manganese [Mn-SOD] [19, 20] to disproportionate O2− to H2O2 and O2), three catalases (KatA, KatB, and KatC) (6, 32), and four alkyl hydroperoxide reductases (AhpA, AhpB, AhpCF, and Ohr) (U. A. Ochsner, D. J. Hassett, and M. L. Vasil, unpublished data).
We have now investigated the roles of individual oxidative stress defense genes by phenotypic assessment of specific mutants and have monitored the responses of these genes to oxidative stress. It appears that redundancy of oxidative stress defense systems allows P. aeruginosa to optimally cope with ROIs generated by its own vigorous aerobic metabolism and to respond rapidly to exogenous ROIs. Some of the genes involved in oxidative stress defense, including katA (encoding the major catalase in P. aeruginosa [32]), and ahpA (encoding a ferredoxin-dependent alkyl hydroperoxide reductase [U. Ochsner and D. Hassett, unpublished data]), are expressed at high levels during aerobic growth. Their activities are maintained at such high levels that even significant oxidative stress causes only a twofold increase in expression, suggesting that high KatA and AhpA activities are critical for detoxification of ROIs produced endogenously during normal aerobic growth. On the other hand, several oxidative stress defense genes, including katB-ankB (24), ahpB, and ahpC-ahpF, are dramatically induced by ROI-generating agents, suggesting a specific and tightly regulated response.
Key regulators modulating the oxidative stress response in bacteria are SoxR and OxyR, both of which are activated at the posttranslational level. O2− activates SoxR through oxidation of its [2Fe-2S] cluster (11, 13), and oxidized SoxR induces the expression of the second transcription factor SoxS, which directly activates transcription of several genes, including sodA in Escherichia coli (27, 30, 60). H2O2 induces at least 30 genes in E. coli, and the response of a subset of these genes depends on OxyR, a 34-kDa LysR-type transcriptional activator (8, 53). E. coli oxyR mutants are hypersensitive to H2O2 and have increased rates of spontaneous mutagenesis during aerobic growth (52). OxyR-regulated genes in E. coli include katG (encoding hydroperoxidase I), gorA (encoding glutathione reductase), ahpCF (encoding alkyl hydroperoxide reductase) (52), and fur (for ferric uptake regulator) (66). Furthermore, E. coli OxyR also controls the formation of a small RNA, designated oxyS, that can act as a positive or negative regulator in response to oxidative stress. The abundant and relatively stable 109-nucleotide oxyS RNA is transcribed immediately upstream and divergently of oxyR in E. coli. Several oxyS-regulated genes were identified in E. coli, including dps (DNA-binding protein of stationary phase) and rpoS (ςS) (1, 14). Recent biochemical studies have shed light on the molecular mechanism of OxyR activation in E. coli. OxyR is redox sensitive and can switch rapidly between oxidized and reduced states, but only the oxidized form of OxyR acts as a transcriptional activator (54). In the presence of H2O2, OxyR forms an intramolecular disulfide bond which can be deactivated by enzymatic reduction upon relief of oxidative stress (2, 65). Both the oxidized and the reduced forms of the E. coli OxyR protein have been shown to possess DNA binding activity (55). Oxidized OxyR recognizes a motif comprised of four ATAG elements spaced at 10-bp intervals (56).
A better understanding of the oxidative stress response in P. aeruginosa, a ubiquitous gram-negative opportunist, is of great industrial and clinical importance. In this work, we provide evidence for the existence of an OxyR homolog in P. aeruginosa and characterize three OxyR-regulated genes essential for the optimal defense against oxidative stress. We describe significant differences in the OxyR response between P. aeruginosa and E. coli, including a link to DNA repair since the P. aeruginosa oxyR gene is located in an operon with recG, and OxyR regulation of a novel type of alkyl hydroperoxide reductase (AhpB) not found in E. coli that is very important for resistance to H2O2.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and chemicals.
All P. aeruginosa and E. coli strains and plasmids used in this study are listed in Table 1. Luria broth (LB) was used for strain maintenance and contained 1.5% agar (Difco) in solid media. M9 minimal medium (45) was used for cultivating P. aeruginosa in the presence of oxidative stress-generating agents. Liquid cultures were grown aerobically at 37°C in shake flasks or, for smaller volumes up to 2 ml, in 14-ml plastic tubes (Fisher Scientific) shaken at 250 rpm. Antibiotics were added as follows: for E. coli, ampicillin (Sigma Chemical Co., St. Louis, Mo.) (100 μg ml−1), gentamicin (Abbott Laboratories) (15 μg ml−1), kanamycin (Sigma) (100 μg ml−1), and tetracycline (Sigma) (15 μg ml−1); for P. aeruginosa, carbenicillin (Research Products International) (750 μg ml−1), gentamicin (75 μg ml−1), and tetracycline (150 μg ml−1). Paraquat (methyl viologen), H2O2 (30%), cumene hydroperoxide (CHP) (80% stock solution diluted with ethanol), t-butyl hydroperoxide (tBHP) (70% solution), and o-nitrophenyl-β-d-galactopyranoside (ONPG) were from Sigma Chemical Co.). Bovine liver catalase was from Boehringer Mannheim, and concentrated protein dye (Bradford reagent) was from Bio-Rad. X-Gal (5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside) was from Research Products International and was used at 40 μg ml−1.
TABLE 1.
Strains and plasmids used in this study
| Strains or plasmids | Genotype or characteristicsa | Source or reference |
|---|---|---|
| E. coli strains | ||
| BL21(DE3) | High-stringency T7 expression host; hsdS DE3 | Novagen |
| DH5α-MCR | F−lacZΔM15 recA1 hsdR17 supE44 Δ(lacZYA argF) | Bethesda Research Laboratories |
| SM10 | Kmr; mobilizer strain | 49 |
| P. aeruginosa strains | ||
| PAO1 | Prototrophic, wound isolate | 22 |
| ΔahpB::Gm | Gmr; ΔahpB::Gm mutant of PAO1 | This study |
| ΔahpCF::Gm | Gmr; ΔahpCF::Gm mutant of PAO1 | This study |
| ΔkatA::Gm | Gmr; ΔkatA::Gm mutant of PAO1 | 32 |
| ΔkatB::Gm | Gmr; ΔkatB::Gm mutant of PAO1 | 24 |
| ΔoxyR::Gm | Gmr; ΔoxyR::Gm mutant of PAO1 | This study |
| ΔoxyR | Unmarked ΔoxyR mutant of PAO1 | This study |
| ΔrecC::Tc | Tcr; ΔrecC::Tc mutant of PAO1 | This study |
| recG::Gm | Gmr; recG::Gm insertion mutant of PAO1 | This study |
| recG::Gm ΔrecC::Tc | Gmr Tcr; recG::Gm ΔrecC::Tc double mutant | This study |
| Plasmids | ||
| pCRII-2.1 | Apr Kmr TA cloning vector for PCR fragments | Invitrogen |
| pCRII-ahpB-606 | pCRII-2.1 containing the ahpB promoter on a 606-bp PCR fragment generated with primers GCTTCAACTCGAAGTCCAG and GATGGGTGAACTGCGAGTC; source of T7-expressed ahpB riboprobe and of DNA fragment for mobility shift | This study |
| pCRII-ahpC-355 | pCRII-2.1 containing the ahpC promoter on a 355-bp PCR fragment generated with primers GACCATCCTGGTGCTGGTC and TGCCCTTCAGGGATTCCTC; source of T7-expressed ahpB riboprobe and of DNA fragment for mobility shift | This study |
| pCRII-omlA-444b | pCRII-2.1 containing a 444-bp omlA promoter fragment; source of T7-expressed omlA riboprobe | 39 |
| pET-OxyR | pET14b (Novagen) carrying the oxyR gene on a 954-bp NdeI-BamHI fragment for the production of His6-tagged OxyR | This study |
| pEX100T | AproriT mob sacB | 47 |
| pFLP2 | AprsacB; broad-host-range recombination system | 21 |
| pPS856 | Apr; source of FRT-Gmr-FRT cassette | 21 |
| pPZ30 | Apr; broad-host-range lacZ fusion vector | 46 |
| pPZ-oxyR-126 | Apr; pPZ30 containing an oxyR::lacZ fusion | This study |
| pPZ-recG-1133 | Apr; pPZ30 containing an oxyR::recG::lacZ fusion | This study |
| pPZ-recG-360 | Apr; pPZ30 containing an recG::lacZ fusion | This study |
| pPZ-katB-480 | Apr; pPZ30 containing a katB::lacZ fusion | This study |
| pPZ-ahpB-377 | Apr; pPZ30 containing an ahpB::lacZ fusion | This study |
| pPZ-ahpC-270 | Apr; pPZ30 containing an ahpC::lacZ fusion | This study |
| pPZ-katA | Apr; pPZ30 containing a katA::lacZ fusion | 32 |
| pBluescript SK+ | Apr; lacZ′; cloning vector | Stratagene |
| pSK-ahpC-ahpF-484 | pBluescript SK(+) containing the ahpC-ahpF intergenic region on a 484-bp PCR fragment generated with primers GATCAAGACCGTCGAGATC and GTCGGTCTTCAGGGTGATC; source of T7-expressed ahpC-ahpF riboprobe | This study |
| pSK-oxyR-384 | pBluescript SK(+) containing the oxyR promoter on a 384-bp BamHI-XhoI fragment; source of T7-expressed oxyR riboprobe and of DNA fragment for mobility shift | This study |
| pSK-oxyR-recG-365 | pBluescript SK(+) containing the oxyR-recG overlapping region on a 365-bp PCR fragment generated with primers TGGAGTCCTCGTCGCTGGA and TGTCCTGCAGGGTTTCCAG; source of T7-expressed oxyR-recG riboprobe | This study |
| pSK-katB-731 | pBluescript SK(+) containing the katB promoter on a 731-bp PCR fragment generated with primers GCTTTGAATTCACTCAGAAG and TCCTGCAGCAGCACCGAAC; source of T7-expressed katB riboprobe and of DNA fragment for mobility shift | This study |
| pUC19 | Apr, ColE1; E. coli cloning vector | 61 |
| pUCP19, pUCP22 | Apr; broad-host-range expression vectors | 59 |
| pUCP-oxyR | Apr; pUCP19 containing oxyR under Plac control | This study |
| pUCP-katB | Apr; pUCP19 containing katB under Plac control | This study |
| pUCP-ahpB | Apr; pUCP19 containing ahpB under Plac control | This study |
| pUCP-ahpCF | Apr; pUCP19 containing ahpCF under Plac control | This study |
| pUCP-recG | Apr; pUCP19 containing recG under Plac control | This study |
mob, mobilization site; oriT, origin of transfer (RK2); Apr, ampicillin resistance; Kmr, kanamycin resistance; Gmr, gentamicin resistance; Tcr, tetracycline resistance; Plac, lac promoter; FRT, Flp recombinase target.
Oxidative stress and UV irradiation sensitivity assays.
To test susceptibility of P. aeruginosa strains to oxidative stress agents, 100 μl of cells grown overnight in LB were inoculated into 100 ml of M9 medium, grown to mid-exponential phase (optical density at 600 nm [OD600 = 0.5), and split into 2-ml aliquots. Paraquat, H2O2, CHP, or tBHP was added at appropriate concentrations, and the subcultures were shaken aerobically. For disk inhibition assays, a culture volume representing 0.2 OD600 unit of cells was mixed with 3 ml of 0.7% low-melting-point M9 agarose at 37°C and poured onto M9 agar plates. Sterile filter disks containing 10 μl of either 2% H2O2 or 20% CHP were placed in triplicate on the top agar, the plates were incubated overnight at 37°C, and the zones of growth inhibition were recorded. For UV sensitivity assays, 100 μl of cells grown overnight in LB was diluted with 10 ml of M9 medium in an uncovered glass petri dish and shaken at 60 rpm at room temperature. Irradiation was performed with a UV lamp (Fotodyne model 3-6000) placed 5 cm above the cells. Samples were removed at 0, 10, 20, 30, 40, and 60 s of UV irradiation and serially diluted with LB in a microtiter dish. Catalase (1,300 U ml−1) was added for the dilution of oxyR mutant strains. Appropriate dilutions (50 μl) were spotted on LB agar, and the colonies were enumerated after overnight incubation at 37°C in the dark.
General genetic procedures.
PCR was performed using Taq polymerase and custom-made primers (Bethesda Research Laboratories, Gaithersburg, Md.) in a Perkin-Elmer Cetus thermal cycler, with 30 cycles of denaturing (1 min, 94°C), annealing (1 min, 54°C), and extending (1 min per kb of DNA, 72°C). The PCR products were purified in low-melting-point agarose gels, routinely cloned into pCRII-2.1 (Invitrogen), and sequenced with Sequenase 2.0 (United States Biochemical) and M13 primers or custom-made 18-mer oligonucleotides. Published procedures were followed for Southern blot analysis, colony hybridization, end labeling of DNA fragments, and other recombinant DNA methods (45), using DNA modifying enzymes from Bethesda Research Laboratories. Standard protocols were used for the isolation of plasmid DNA (23) and chromosomal DNA (9). Plasmids were maintained in E. coli DH5α-MCR (Bethesda Research Laboratories) and transformed into P. aeruginosa strains using the magnesium chloride method (42). RNA was isolated by the hot-phenol method and analyzed by RNase protection assays as described in detail elsewhere (3). Radiolabeled riboprobes were generated from cloned DNA fragments (Table 1), using an in vitro runoff transcription system (Promega), and excess probe was hybridized to 20 μg of total RNA.
Construction of isogenic mutant strains.
Mutant strains affected in oxyR were constructed as follows. A 1.36-kb PCR product containing the oxyR region was generated with primers oxyR-226 (5′TGTACACCAGGTAGTCGAG) and oxyR-1585 (5′-GTTTCCAGGCCTACCCGAG), cloned into pCRII-2.1, sequenced, excised with EcoRI, and cloned into the EcoRI site of pUC19. A 0.62-kb XhoI-SstII internal fragment of the oxyR gene was removed, and the ends were blunted with Klenow enzyme and ligated to a 1.3-kb FRT-Gmr-FRT cassette (Gmr) excised from pPS856 (21) with BamHI and followed by end polishing. The resulting plasmid, pUCΔoxyR::Gm, was digested with PvuII, yielding a 2.5-kb ΔoxyR::Gm construct which was ligated into the SmaI site of the gene replacement vector pEX100T (47). E. coli SM10 containing pEX100T-ΔoxyR::Gm was used as the donor strain in a biparental mating with P. aeruginosa PAO1. Transconjugants were selected on brain heart infusion agar containing gentamicin (75 μg ml−1) and irgasan (50 μg ml−1) and subsequently plated on LB agar containing gentamicin (75 μg ml−1) and 5% sucrose. Successful double-crossover events leading to the replacement of the oxyR gene with the Gmr cassette in the putative ΔoxyR::Gm mutant strain were verified by the loss of pEX100T-encoded Cbr and by PCR across the oxyR gene using the primers oxyR-226 and oxyR-1585. To obtain an unmarked ΔoxyR mutant strain, E. coli SM10 harboring pFLP2 (21) was mated into ΔoxyR::Gm, and ΔoxyR::Gm/pFLP2 was grown overnight in LB to allow excision of the Gmr cartridge via the adjacent FRT sequences (21). Single colonies on LB-carbenicillin were then checked for the loss of Gmr. Finally, pFLP2 was cured from ΔoxyR by selection for sucrose resistance, indicating the loss of the pFLP2-borne sacB gene, and the resulting unmarked ΔoxyR mutant was also checked for loss of plasmid-encoded Cbr.
The other mutant strains used in this study were constructed by essentially the same method as described above. In brief, a 1.2-kb DNA fragment containing the ahpB region was PCR amplified using primers ahpB-1321 (5′GATGGCGCTT CAACTCGAAG) and ahpB-2537 (5′TGCATGCCGGTGATCAGCAG). A 0.63-kb HincII-SmaI fragment containing the entire ahpB coding sequence minus the four first codons was then replaced by a Gmr cartridge, resulting in a ΔahpB::Gm mutant. To obtain a ΔahpCF::Gm mutant, a 2.1-kb region containing the ahpC-ahpF locus was isolated by PCR using primers ahpC-621 (5′GACCATCCTGGTGCTGGTC) and ahpF-2741 (5′TTCCAGCAGGGTCACATGG). A 1.5-kb HincII fragment containing most of the ahpC gene and a 5′ portion of ahpF was replaced by a Gmr cartridge. A recG::Gm mutant was constructed by insertion of a Gmr cartridge into the unique BglII site within the recG gene that had been PCR amplified with primers recG-1557 (5′GAGAAGCTCGCTCGGGTAG) and recG-3107 (5′GAAGGCTTCCATCACCACG).
Construction of lacZ reporter fusions.
DNA fragments containing the relevant promoter regions, including the translational start sites, were PCR amplified, cloned into pCRII-2.1, sequenced, and ligated into pPZ30. To achieve an in-frame translational fusion to the promoterless lacZ gene, a PstI site was incorporated in the primer sequence at an appropriate 3′-end position, when necessary. Specifically, plasmid pPZ-oxyR-126 contained a 126-bp EcoRI-PstI fragment harboring the oxyR promoter region plus the first six codons of the oxyR gene. In pPZ-recG-1133 and pPZ-recG-360 the first 32 codons of the recG gene were fused to lacZ, and these plasmids contained increasing upstream sequence as depicted in Fig. 1. Plasmid pPZ-katB-480 contained the katB promoter plus 59 codons of the katB gene on a 480-bp EcoRI-PstI fragment that had been generated by PCR with primers katB-38 (5′CTTGGAACTGCGCCATGCAG) and katB-514 (5′TCCTGCAGCAGCACCGAAC [the PstI site is underlined]). Construct pPZ-ahpB-377 harbored the first four codons of the ahpB gene and the ahpB promoter on a 377-bp EcoRI-PstI fragment obtained with primers ahpB-1321 (5′GATGGCGCTTCAACTCGAAG) and ahpB-1697 (5′ctgCAGTACGCTCATCGCGAGG [nonmatching nucleotides in lowercase type]). A 270-bp EcoRI-PstI fragment containing the ahpC promoter plus the first three codons of ahpC was PCR amplified with primers ahpC621 (5′GACCATCCTGGTGCTGGTC) and ahpC-890 (5′ctgCAGGGACATCAGTCGTTCCT) and cloned into pPZ30 linearized with EcoRI and PstI, yielding pPZ-ahpC-270. Besides those mentioned above, a minilibrary of several additional genes were also tested for their dependence on OxyR, and they included katA, katC, ahpA, ohr, fur, omlA, sodA, sodB, bfrA, bfrB, phuR, plcH, toxA, pvdS, rpoS, dps and ptxR.
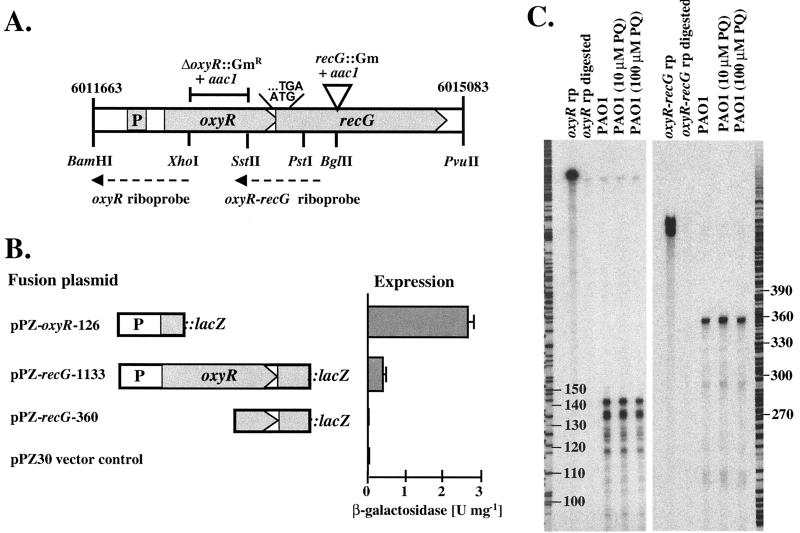
FIG. 1.
Characterization of the oxyR-recG operon. (A) Genetic map showing the putative promoter, the overlapping TGA stop codon for oxyR and ATG start codon for recG, the locations of the riboprobes, relevant restriction sites, and the deletions and insertions made in the ΔoxyR and recG mutants. The numbers flanking the maps indicate the coordinates of these loci in the PAO1 genome (Pathogenesis Corp., 12-15-99 release). (B) Expression of oxyR and recG as fusions to lacZ. The portions of the oxyR-recG DNA sequence contained in the lacZ fusion plasmids are indicated, together with the corresponding β-galactosidase activities expressed from these constructs. Error bars indicate standard deviations. (C) RNase protection assays. Riboprobes specific for the oxyR promoter (oxyR-rp) and for the oxyR-recG overlapping region (oxyR-recG rp) were used to detect the corresponding transcripts in P. aeruginosa PAO1 total RNA isolated during the exponential growth phase in M9 medium. Paraquat (PQ) was added to final concentrations of 10 and 100 μM 1 h prior to harvest as indicated. Also shown are the digested probes in the absence of any P. aeruginosa RNA as a control. A DNA sequencing reaction was run in parallel and served as a size marker. Numbers indicate nucleotides.
Construction of complementing plasmids.
The P. aeruginosa-E. coli multicopy shuttle vectors pUCP19 and pUCP22 (59) were used for the construction of recombinant plasmids containing the oxyR, katB, ahpB, ahpC-ahpF, and recG genes under the control of the plasmid-borne lac promoter that drives constitutive expression in P. aeruginosa. The complete oxyR gene was PCR amplified with primers oxyR-460 (5′GCAGTGTAGGCGTCGAATC) and oxyR-1585 (5′GTTTCCAGGCCTACCCGAG), and the PCR product was cloned into pCRII-2.1 and transferred as a 1.13-kb EcoRI fragment into pUCP19, resulting in pUCP-oxyR. Plasmid pUCP-katB was constructed similarly using a 1.9-kb PCR product obtained with primers katB-38 (see above) and katB-1887 (5′CCAGGATTGATCGCAACCGG). The ahpB gene was amplified by PCR with primers ahpB-1321 and ahpB-2537 (see above) and was directionally cloned as a 1.2-kb HindIII-XbaI fragment from pCRII-2.1 into pUCP22, yielding pUCP-ahpB. The ahpC-ahpF region was located on a 3.5-kb SphI fragment as predicted from the P. aeruginosa genome sequence. Accordingly, chromosomal DNA of P. aeruginosa PAO1 was cut with SphI, and fragments of the size range of 3 to 4 kb were cloned into pUCP19. A pUCP-ahpCF plasmid harboring the ahpC-ahpF genes under lac promoter control was subsequently isolated by colony hybridization using a 270-bp ahpC promoter fragment (see above) as a radiolabeled probe. For the construction of pUCP-recG, it had to be considered that the native recG gene lacked a Shine-Dalgarno sequence due to the overlap of its ATG start codon with the oxyR TGA stop codon (Fig. 1). Therefore, a Shine-Dalgarno motif (underlined, see below) was incorporated 7 bp upstream of the recG ATG. A 2.1-kb PCR product containing recG was obtained with primers recG-1490 (5′aggagAAATAGCATGACCGAGCTGTC) and recG-3616 (5′GCTTCAAGACTGAGACCTACG), cloned into pCRII-2.1, and directionally cloned as a HindIII-XbaI fragment into pUCP22, resulting in pUCP-recG.
Purification of OxyR and DNA mobility shift assays.
The oxyR gene was PCR amplified as an NdeI-BamHI fragment using primers (NdeI)-catATGACCCTCACCGAACTGC and (BamHI)-ggatCCTGGACAGCTCGGTCATG and cloned into pCRII-2.1. After verification of its sequence, the oxyR gene was cloned into the NdeI-BamHI sites of pET14b (Novagen), generating an in frame fusion with the vector-encoded His tag sequence. OxyR protein with an amino-terminal His6 tag was overexpressed from pET-OxyR in the BL21(DE3) T7 expression strain and purified through metal affinity chromatography on Ni-nitrilotriacetic acid (Qiagen). End-labeled DNA fragments (1 to 2 ng) harboring the relevant promoter sequences (Table 1) were incubated for 15 min with increasing amounts (up to 1 μM) of freshly purified His6-OxyR protein in 20 μl of binding buffer [20 mM bis-Tris borate (pH 7.5), 40 mM KCl, 5 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, 100 μg of bovine serum albumin ml−1, 50 μg of poly(dI-dC) ml−1, 10% glycerol], and 10 μl of the mixture was loaded on a 6% polyacrylamide gel in running buffer (20 mM bis-Tris borate, pH 7.5). After electrophoresis for 3 to 4 h at 250 V, the gel was dried and autoradiographed.
Biochemical procedures.
β-Galactosidase activities were determined as follows. Bacterial cell extracts from 2-ml cultures were prepared by centrifugation (10,000 × g, 10 min, 4°C), resuspension of the cells in 0.5 ml of 50 mM potassium phosphate buffer (pH 7.0), and sonication for 5 s (Branson Sonifier; output level 5). The insoluble fraction was removed by centrifugation (13,000 × g, 10 min, 4°C), and protein concentrations were estimated by the Bradford assay using bovine serum albumin as a standard (5). β-Galactosidase assays were performed using ONPG as the substrate and expressed as international units with a millimolar extinction coefficient for ONPG of 3.1 (37). Catalase activity of normalized soluble protein samples was detected in stained 5% nondenaturing polyacrylamide gels (57).
RESULTS
Characterization of the oxyR-recG operon in P. aeruginosa PAO1.
A putative OxyR homolog was identified using the E. coli OxyR amino acid sequence to search the P. aeruginosa genome sequence (www.pseudomonas.com). This P. aeruginosa OxyR homolog is predicted to be a 34-kDa protein with 40% amino acid sequence identity to E. coli OxyR (8). OxyR is a positive regulator of H2O2-inducible genes in E. coli and Salmonella enterica serovar Typhimurium and belongs to the LysR family of bacterial regulatory proteins (8). Immediately downstream of oxyR was an open reading frame encoding a 76-kDa protein with 59% amino acid sequence identity to the E. coli RecG protein, which functions as an ATP-dependent DNA helicase involved in replication and repair of DNA (29, 31). The stop codon of P. aeruginosa oxyR overlapped the start codon of recG, suggesting that oxyR and recG are organized in an operon (Fig. 1A). Expression of oxyR and recG was monitored by translational fusions to the lacZ reporter gene, as depicted in Fig. 1B. oxyR::lacZ activity was detected from pPZ-oxyR-126 containing a promoter immediately upstream of the oxyR gene. recG::lacZ expression was absent in pPZ-recG-360, harboring roughly 300 bp of recG upstream sequence, but was detected in pPZ-recG-1133, which contained, in addition, the entire oxyR gene including the oxyR promoter region, indicating that recG is coexpressed from the oxyR promoter in an operon with oxyR. This finding was supported by RNase protection assays using an intergenic 365-nucleotide oxyR-recG riboprobe (Fig. 1A), which was entirely protected (Fig. 1C). A single transcriptional start site for the oxyR-recG operon was found 21 nucleotides upstream of the oxyR translational start, as determined by RNase protection with a 384-nucleotide probe of which 141 nucleotides were protected. The oxyR-recG transcription did not respond to oxidative stress (Fig. 1C) or to other stimuli, such as growth phase or iron concentration (data not shown).
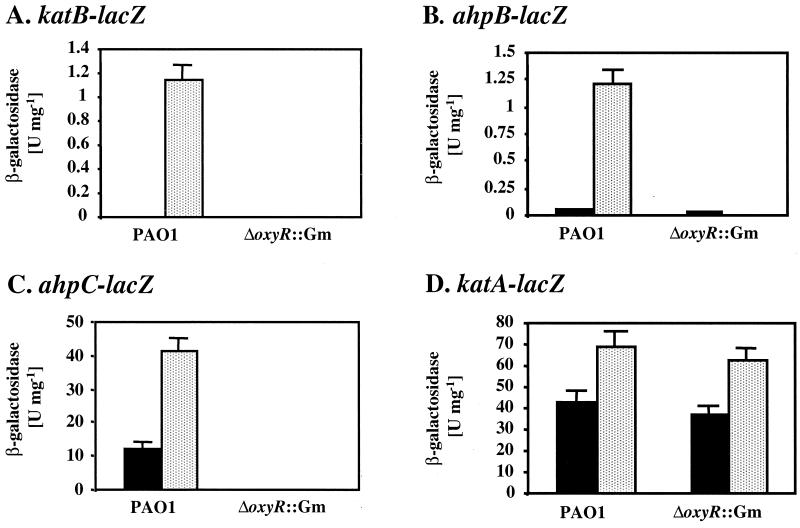
OxyR-dependent activation of katB, ahpB, and ahpC expression.
To identify OxyR-regulated genes in P. aeruginosa, we screened a plasmid minilibrary of lacZ reporter fusions of about 20 candidate genes potentially involved in oxidative stress defense, iron uptake and storage, and DNA repair, as listed in Materials and Methods. The reporter activities in the wild type and a ΔoxyR mutant were compared at mid-exponential growth phase in the presence and absence of 100 μM paraquat. Applying a fivefold difference in the expression levels of the candidate genes between wild-type and oxyR mutant cells as the cutoff, we found three fusions (pPZ-katB-480, pPZ-ahpB-377, and pPZ-ahpC-270) that were OxyR dependent (Fig. 2). Expression of katB-lacZ was not detected in unstimulated wild-type organisms and was induced 250-fold upon exposure to paraquat, while no activity was detected in the ΔoxyR mutant (Fig. 2A). The ahpB-lacZ fusion was expressed at very low levels in both wild-type and ΔoxyR bacteria, and a 90-fold induction by paraquat was observed in the wild type but not in the ΔoxyR mutant (Fig. 2B). Expression of ahpC-lacZ was substantial in untreated wild-type cells and increased threefold in the presence of paraquat. In contrast, ahpC-lacZ was not expressed in the ΔoxyR mutant (Fig. 2C). All other tested fusions did not depend on OxyR; e.g., katA-lacZ in plasmid pPZ-katA was expressed at similar levels in either wild-type or ΔoxyR cells, although a roughly twofold response to paraquat was observed (Fig. 2D). This regulation of katA has been reported previously (32), and it appears that it involves a mechanism different from OxyR activation, which is, in part, controlled by quorum sensing and iron levels (18). Among other genes that were expressed independently of OxyR were sodA and sodB (encoding Fe-SOD and Mn-SOD, respectively), dps (encoding DNA-binding protein of stationary phase), ahpA (encoding alkyl hydroperoxide reductase A), bfrA and bfrB (encoding bacterioferritins A and B, respectively), fur, oxyR itself, and all additional genes of the minilibrary listed in Materials and Methods (data not shown).
FIG. 2.
OxyR-dependent gene expression in response to oxidative stress. Wild-type and ΔoxyR bacteria containing plasmid-borne katB-lacZ (A), ahpB-lacZ (B), ahpC-lacZ (C), and katA-lacZ (D) fusions were grown in M9 medium to mid-exponential phase. The reporter activities were measured without paraquat treatment (black bars) or after treatment with 100 μM paraquat for 1 h (stippled bars). The β-galactosidase activities are presented as international units, and the error bars represent the standard deviations from four independent experiments.
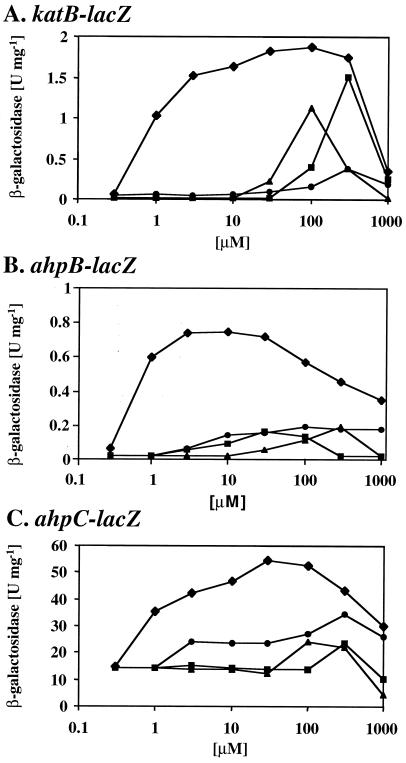
Dose-response effect of OxyR-dependent expression of katB, ahpB, and ahpC.
To obtain dose-response curves for OxyR-mediated gene activation, mid-exponential-phase cultures of wild-type bacteria containing katB-lacZ, ahpB-lacZ, and ahpC-lacZ were exposed for a given time to oxidative stress compounds, including paraquat, CHP, tBHP, and H2O2, at concentrations ranging from 0.3 μM to 1 mM (Fig. 3). The strongest induction was evoked by paraquat, which caused a half-maximal response at a concentration of 1 μM and resulted in a sustained response to concentrations of up to 300 μM, above which it became lethal. Interestingly, all three fusions were responsive to organic hydroperoxides. Approximately 100 to 300 μM CHP or tBHP was typically required to elicit a significant response, and at higher concentrations (1 mM), the cells were killed. Repeated addition of H2O2 caused activation of ahpB-lacZ and ahpC-lacZ at a concentration of 3 μM or higher; in contrast, at least 100 μM was required for activation of katB-lacZ. Generally, H2O2 had a less pronounced effect on OxyR-dependent gene activation than any of the other tested compounds, presumably due to rapid detoxification by endogenous catalase. Interestingly, the ahpB-lacZ fusion responded somewhat more strongly to lower concentrations of all of the oxidative stress compounds (e.g., 1 to 10 μM paraquat) compared to katB-lacZ and ahpC-lacZ (Fig. 3).
FIG. 3.
Dose-response curves for OxyR-dependent gene expression. Wild-type cells containing plasmid-borne katB-lacZ (A), ahpB-lacZ (B), and ahpC-lacZ (C) fusions were grown in M9 medium to mid-exponential phase and treated with increasing concentrations of paraquat for 1 h (diamonds), of CHP for 30 min (squares), of tBHP for 30 min (triangles), or of H2O2 for 1 h (circles). The β-galactosidase activities are shown as a function of the indicated concentrations of the oxidative stress-generating compounds and are the mean values from triplicate assays.
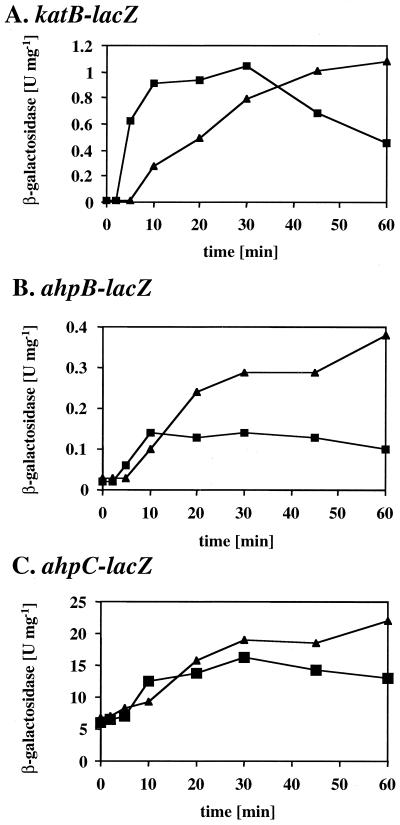
Time course of OxyR-mediated gene activation.
The efficiency of the OxyR-mediated response was further evaluated by monitoring the timing of target gene expression. Mid-exponential-phase cultures of wild-type bacteria containing katB-lacZ, ahpB-lacZ, and ahpC-lacZ were treated with a fixed concentration of paraquat (100 μM) or CHP (300 μM), and the β-galactosidase reporter activities in samples taken at several time points postinduction were determined (Fig. 4). Paraquat elicited a response within 10 min of exposure, and the katB-lacZ, ahpB-lacZ, and ahpC-lacZ activities increased for at least 1 h. This response was expected because paraquat is not degraded and is capable of continuous redox cycling in viable aerobic bacteria. CHP caused activation within 5 min of exposure, but the response reached a plateau after 30 min and declined somewhat after that. This result was also expected, since CHP can be detoxified (e.g., by Ahp activities) and thus elicits only a transient oxidative stress response.
FIG. 4.
Time-response curves for OxyR-dependent gene expression. Wild-type cells containing katB-lacZ (A), ahpB-lacZ (B), and ahpC-lacZ (C) were grown in M9 medium to mid-exponential phase and treated with 100 μM paraquat (triangles) or 300 μM CHP (squares). Samples were removed before treatment and at 2, 5, 10, 20, 30, 45, and 60 min postexposure, and their β-galactosidase activities were determined. The values are the means from triplicate experiments.
Genetic analysis of the OxyR-regulated genes katB-ankB, ahpB, and ahpC-ahpF.
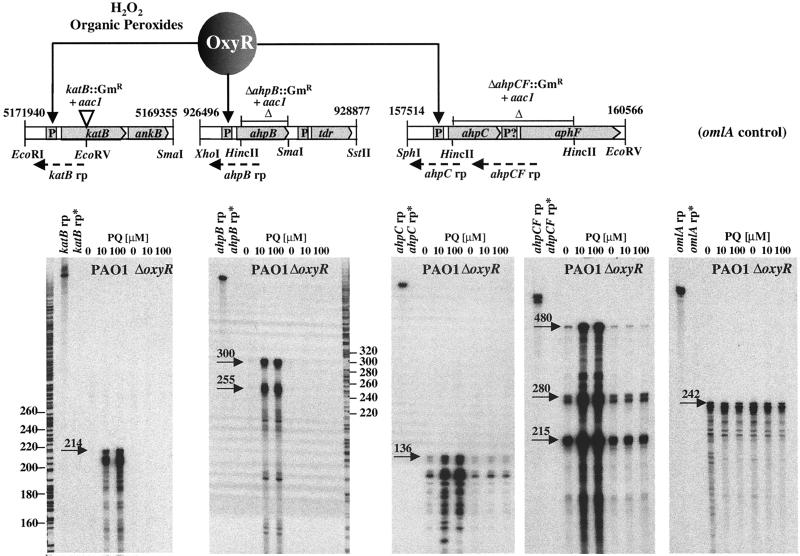
Genetic maps and the introduced mutations in katB, ahpB, and ahpCF are depicted in Fig. 5. Also shown are the results of RNase protection assays using specific riboprobes to map the individual transcriptional start sites. The katB, ahpB, and ahpCF transcripts were strongly induced in the presence of the oxidative stress-generating agent paraquat, and this response was dependent on OxyR. A single katB transcriptional start site was detected 42 nucleotides upstream of the katB translational initiation site. The katB::Gm mutation has a polar negative effect on ankB, which is in an operon with katB and encodes an ankyrin-like protein required for optimal catalase B activity (24). Transcription of ahpB started 60 nucleotides upstream of its start codon. The ahpB gene encodes a 22-kDa protein harboring a motif typical for antioxidant reductases as determined by the e-motif search (Department of Biochemistry, Stanford University) and contains a candidate membrane-spanning helix suggesting a localization in either the cytoplasmic membrane or periplasm (data not shown). The putative AhpB protein is 60% identical at the amino acid sequence level to the product of the Legionella pneumophila alkyl hydroperoxide reductase (tsaA) gene (GenBank accession number L46863) and 51% identical to a mouse thiol-specific antioxidant (GenBank accession number X82067). This type of peroxidase reduces hydroperoxides with reducing power from thioredoxin (28). Interestingly, the ahpB gene is located immediately upstream of an open reading frame (tdr) encoding a putative thioredoxin reductase (Fig. 5). The tdr gene, however, was not in an operon with ahpB but was expressed from its own promoter and independent of OxyR (data not shown). The ahpC mRNA start site was mapped to 43 nucleotides upstream of the ahpC translational start. Low levels of this transcript were also detectable in unstimulated wild-type cells and in oxyR mutant cells, suggesting that ahpC is expressed at low basal levels in an OxyR-independent way. The ahpC and ahpF coding sequences were spaced apart by a 144-bp intergenic sequence. RNase protection assays using a 484-nucleotide ahpC-ahpF riboprobe over this region were performed to address the question of whether ahpC and ahpF form an operon. Clearly, a fraction of the probe was protected over its entire length, suggesting an organization of ahpC and ahpF in an operon. However, additional protected RNA species of 280 and 215 nucleotides were detected, indicating that some ahpC transcripts may terminate within the ahpC-ahpF intergenic region and that ahpF may be transcribed from a separate promoter and independently of ahpC. In either case, both ahpC and ahpF transcription appeared to be OxyR responsive. The P. aeruginosa ahpC and ahpF genes encode a 21- and a 56-kDa proteins, respectively, with 59 and 66% amino acid sequence identities, respectively, to the E. coli AhpC and AhpF alkyl hydroperoxide reductase subunits. This type of Ahp is widely found in most bacterial species (51) and requires NADH or NADPH for activity (41).
FIG. 5.
Genetic maps and transcripts of OxyR-regulated genes. The katB, ahpB, and ahpC-ahpF loci are shown with their coordinates in the PAO1 genome (Pathogenesis Corp., 12-15-99 release), the sites of insertions or deletions in the corresponding mutants, the locations of the riboprobes (rp), and relevant restriction sites. The katB-ankB operon encodes a previously characterized inducible catalase (6) and an ankyrin-like factor required for optimal catalase activity (24). The ahpB gene encodes a thiol-specific peroxidase and is located upstream of a thioredoxin reductase (tdr). The ahpC-ahpF operon encodes the two subunits of the classic alkyl hydroperoxide reductase. The RNase protection assays were done with total RNA isolated from PAO1 wild-type or oxyR mutant cells in the absence or presence of paraquat (PQ) as indicated. A probe specific for the constitutively expressed omlA gene (39) was used as a control. Also loaded were diluted probes (rp) and the digested probes (rp*) as controls. Arrows point to the relevant protected riboprobe bands, and their approximate sizes (in nucleotides) are given.
Characterization of the katB, ahpB, and ahpC promoters.
The mapping of the transcriptional start sites for katB-ankB, ahpB, ahpC-ahpF, and oxyR allowed the localization of the corresponding −10 and −35 elements (Fig. 6). Putative OxyR-binding sites were identified upstream of the katB, ahpB, and ahpCF promoters. Four ATAG elements spaced at 10-bp intervals comprise the binding sites for oxidized E. coli OxyR (56), and such elements were found in proper spacing and distance within the katB, ahpB, and ahpCF promoters. The number of bases matching the OxyR consensus binding sequence were 9 of 16 (katB), 13 of 16 (ahpB), and 12 of 16 (ahpCF), and in all cases, the OxyR binding motif was located exactly adjacent to the −35 promoter elements (Fig. 6). Purified His6-tagged OxyR protein at a concentration of at least 100 μM caused a mobility shift of DNA fragments containing these target promoters, indicating direct binding of OxyR. A DNA fragment harboring the oxyR promoter was not shifted by OxyR (Fig. 6), which is in agreement with the finding that oxyR expression did not respond to oxidative stress.
FIG. 6.
Binding of OxyR to the katB, ahpB, and ahpC promoters. The alignment of the OxyR-regulated promoters indicates four putative OxyR-binding tetranucleotide sequences (underlined), the residues matching the consensus sequence derived from E. coli OxyR-regulated promoters (asterisks), and the −35 promoter elements in proper distance of the mapped transcriptional start sites. The gel mobility shift assays of radiolabeled DNA fragments containing the corresponding promoter regions were performed with purified His6-tagged OxyR protein at the given concentrations.
Phenotypic comparison of mutants affected in oxyR, katB, ahpB, ahpCF, and recG.
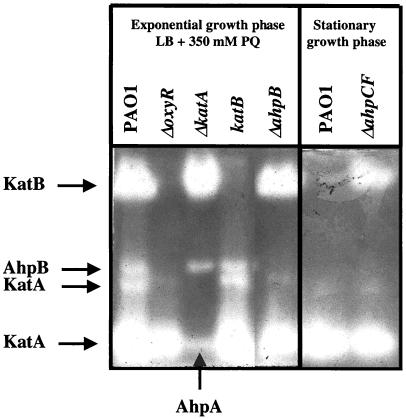
The susceptibility of a ΔoxyR::Gm mutant to oxidative stress compounds was compared to the phenotypes of mutants affected in single OxyR-regulated genes in order to dissect their specific roles in the oxidative stress response. Wild-type, ΔoxyR::Gm, recG::Gm, ΔkatB::Gm, ΔahpB::Gm, and ΔahpCF::Gm bacteria were tested for their susceptibilities to H2O2 and CHP using standardized disk inhibition assays (Table 2). The ΔoxyR::Gm mutant containing the control plasmid pUCP19 was dramatically susceptible to both compounds. Plasmid pUCP-oxyR fully complemented this phenotype, while plasmid-borne copies of the recG gene, which is located downstream and in an operon with oxyR (see above), resulted in minimal complementation. These findings strongly suggest that the oxyR phenotype was caused by the lack of OxyR-mediated oxidative stress defense. Still, recG appeared to be essential for optimal resistance to H2O2 and CHP, and the recG mutant could be complemented by the recG gene in trans. Multiple copies of single OxyR-regulated genes expressed from the constitutive lac promoter on pUCP resulted in only marginal complementation. The katB::Gm mutant showed increased susceptibility to H2O2, and this phenotype could not be complemented by providing only katB in trans, presumably due to the polar negative effect on ankB (24). The ΔahpB::Gm mutant was hypersusceptible to H2O2 but not to CHP and was successfully complemented by ahpB in trans. The ΔahpCF::Gm mutant exhibited a somewhat intriguing phenotype. While ΔahpCF::Gm mutant cells were hypersusceptible to CHP, they were more resistant to H2O2 than the wild type, and plasmid pUCP-ahpCF reversed that trend. Elevated KatB catalase levels were measured in the ΔahpCF::Gm mutant, and the KatB catalase activity was detectable even in the absence of paraquat as an inducer (Fig. 7). In agreement with that observation was the finding that a katB-lacZ fusion was expressed at severalfold higher levels in a ΔahpCF::Gm background than in wild-type cells (data not shown). This compensatory mechanism between ahpCF and katB expression indicates that the absence of AhpCF leads to internal oxidative stress. During experiments to measure catalase levels in various catalase-deficient and ahp mutant strains using activity staining, we unexpectedly observed extra bands that we suspected might reflect the ability of some alkyl hydroperoxidases to also use H2O2 as a substrate (Fig. 7). Wild-type organisms produced KatA and KatB activities, and two additional smaller activity bands migrated between KatA and KatB. We determined that the lower band was an electrophoretic variant of KatA, since this band was absent in a katA mutant. The upper band most likely represented AhpB, since it was absent in the oxyR and ahpB mutants. Both middle bands were retained in a katB mutant. Interestingly, the katA mutant possessed a catalase activity band that migrated with KatA. We determined that this band is AhpA, because it was absent in an ahpA mutant (data not shown). Furthermore, preliminary catalase assays indicated that both AhpB and AhpA possessed some catalase activity (data not shown), while it remained uncertain whether AhpCF had such activity.
TABLE 2.
Oxidative stress susceptibility of mutant strainsa
| Strain/plasmidb | Zone of growth inhibition (mm)c with:
|
|
|---|---|---|
| 2% H2O2 | 20% CHP | |
| PAO1/pUCP19 | 20 ± 1 | 19 ± 1 |
| ΔoxyR::Gm/pUCP19 | 46 ± 4 | 31 ± 3 |
| ΔoxyR::Gm/pUCP-oxyR | 19 ± 1 | 19 ± 1 |
| ΔoxyR::Gm/pUCP-recG | 29 ± 3 | 25 ± 2 |
| ΔoxyR::Gm/pUCP-katB | 38 ± 3 | 26 ± 3 |
| ΔoxyR::Gm/pUCP-ahpB | 33 ± 2 | 25 ± 2 |
| ΔoxyR::Gm/pUCP-ahpCF | 37 ± 1 | 25 ± 1 |
| ΔrecG::Gm/pUCP19 | 25 ± 1 | 27 ± 2 |
| ΔrecG::Gm/pUCP-recG | 21 ± 1 | 22 ± 1 |
| ΔkatB::Gm/pUCP19 | 24 ± 1 | 18 ± 1 |
| ΔkatB::Gm/pUCP-katB | 24 ± 1 | 19 ± 1 |
| ΔahpB::Gm/pUCP19 | 28 ± 3 | 19 ± 2 |
| ΔahpB::Gm/pUCP-ahpB | 21 ± 1 | 19 ± 1 |
| ΔahpCF::Gm/pUCP19 | 18 ± 1 | 24 ± 2 |
| ΔahpCF::Gm/pUCP-ahpCF | 24 ± 3 | 20 ± 1 |
The strains were grown overnight in M9 medium, and 0.2 OD600 unit of culture was mixed with 3 ml of 0.8% low-melting-point agarose in M9 medium containing carbenicillin and poured onto M9 agar. Triplicate filter disks containing 10 μl of 2% H2O2 or 20% CHP were immediately placed on the plates, and zones of growth inhibitions were measured after overnight incubation at 37°C.
Genes were cloned into pUCP19 in the orientation of the plasmid Plac promoter, which is constitutively expressed in P. aeruginosa.
Results are means and standard deviations.
FIG. 7.
Catalase activity gel of soluble cell extracts. Bacterial cultures of PAO1 wild-type and oxyR, katA, katB, and ahpB mutant cells were grown under aerobic conditions in LB to mid-exponential phase and then exposed to 350 μM paraquat (PQ) for 1 h. PAO1 and the ahpCF mutant were grown to stationary phase (16 h) as indicated. Normalized amounts (15 μg) of the soluble protein fractions were separated on a nondenaturing gel and stained for catalase activity. The arrows indicate the positions of KatA, KatB, AhpA, and AhpB activities.
Role of the oxyR-recG operon in DNA repair.
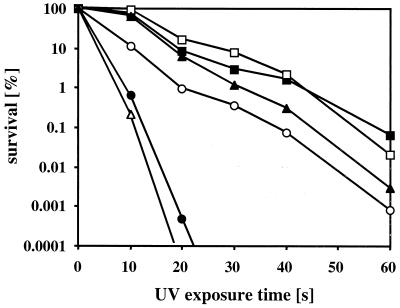
Since P. aeruginosa oxyR was located in an operon with the recG gene, encoding a putative DNA helicase, a possible function of the oxyR-recG locus in DNA repair was investigated. Wild-type bacteria, an unmarked ΔoxyR mutant harboring an in-frame deletion of oxyR with an unlikely polar effect on recG, a recG::Gm mutant, and a ΔrecC::Tc mutant were compared for their sensitivity to UV irradiation-induced DNA damage (Fig. 8). Wild-type cells and the ΔoxyR mutant showed similar killing patterns, characterized by roughly 3 to 4 log units of killing over 60 s of UV irradiation. The recG::Gm mutant was hypersensitive to UV and was killed by more than 5 log units within 20 s of irradiation. The UV sensitivity of recG::Gm was more dramatic than that of the ΔrecC::Tc mutant. Expression of the recG gene in trans partially restored UV tolerance in the recG::Gm mutant. A recG::Gm ΔrecC::Tc double mutant was slightly more sensitive than the recG::Gm single mutant, suggesting an additive effect. Clearly, our data show that recG plays an important role in DNA damage repair.
FIG. 8.
Kill curves upon exposure to UV irradiation. Wild-type PAO1 (■), an unmarked nonpolar ΔoxyR mutant (□), a recG::Gm mutant (●), a recG::Gm mutant complemented with pUCP-recG (○), a ΔrecC::Tc mutant (▴), and a recG::Gm ΔrecC::Tc double mutant (▵) were grown overnight in LB. The cells were UV irradiated while shaking, and aliquots were removed at 10-s intervals. Serial dilutions were plated on L agar to determine the viable cell counts. The UV killing assays were performed five times with independent cultures, and the outcome of one representative experiment is shown.
DISCUSSION
H2O2 is a powerful antimicrobial agent commonly used in health care as a topical anti-infective, as well as in industry for the treatment of problematic bacterial biofilms. Also, human phagocytes produce H2O2 as a natural weapon during the respiratory burst to combat microbial infections (38). Thus, it is not surprising that microbes have evolved several strategies to cope with oxidative stress. In P. aeruginosa, the primary defense against H2O2 involves a constitutively expressed catalase, KatA (12, 17, 32), but little is known about a specific response of P. aeruginosa to oxidative stress agents. While the existence of a second, H2O2- or paraquat-inducible, catalase (KatB) has been described (6), the regulatory mechanism governing this response remained unknown. In this report, we characterize a regulatory gene, oxyR, and present evidence that the OxyR protein is involved in transcriptional activation of at least three genes encoding antioxidants. The OxyR-mediated stress response has been well studied in E. coli by both genetic and biochemical means (52–56). Also, a possible role of OxyR to combat host defense systems has been investigated in numerous pathogenic bacteria, including Enterococcus faecalis (44), Haemophilus influenzae (33), and Mycobacterium tuberculosis (10, 50). In M. tuberculosis, the OxyR-regulated genes katG and ahpC play crucial roles in isoniazid resistance, since isoniazid requires activation by KatG to exert lethal effects, while AhpC could play a detoxifying role (63, 64). Interestingly, the oxyR gene in members of the M. tuberculosis complex is located next to ahpC; however, oxyR is nonfunctional, due to numerous deletions and point mutations (10). M. tuberculosis katG mutant strains were found to have acquired a compensatory mutation resulting in an upregulation of AhpC, and it has been shown that this protein confers resistance to isoniazid and protection against H2O2, even in the absence of adequate catalase and peroxidase activities (48). We found a compensatory cross-regulation of OxyR-dependent katB-ankB and ahpC-ahpF expression in P. aeruginosa. A ΔahpCF::Gm mutant strain was more resistant to H2O2, and this phenotype correlated with higher levels of KatB observed in a catalase activity gel. Such an increased resistance due to elevated expression from all peroxide regulon promoters has been reported for a Bacillus subtilis ahpC mutant (7). Similarly, the lack of AhpC-AhpF peroxidase expression in E. coli has been shown to lead to constitutive OxyR activation due to the accumulation of endogenous oxidants (43). Somewhat surprising was the hypersusceptibility to H2O2 of strains harboring the ahpCF genes on a multicopy plasmid. However, a similar phenomenon has been observed upon overexpression of ahpCF in S. enterica serovar Typhimurium (51). The reasons for this are unclear, but possible explanations are that multiple copies of the ahpCF promoter titrate out OxyR or that increased AhpC-AhpF hydroperoxide reductase activity could somehow interfere with the proper sensing of oxidative stress, e.g., by maintaining the oxidized state of the OxyR protein.
In the course of this study, we learned that P. aeruginosa possesses multiple lines of OxyR-dependent, inducible oxidative stress defense systems with potentially overlapping functions. The expression of the three identified OxyR targets, katB-ankB, ahpB, and ahpC-ahpF, responded to any of the exogenously added oxidative stress compounds, including H2O2, paraquat, and organic hydroperoxides, suggesting that all of these agents or products derived from their action cause oxidation of the OxyR protein, which then indiscriminately activates the target promoters. However, one of the OxyR target genes, ahpB, responded to significantly lower concentrations of oxidative stress agents than the other targets. The reason for this dose-response shift is unknown, but interestingly, the putative OxyR-binding site in the ahpB promoter had a higher identity to the consensus “OxyR box” than the OxyR boxes in the katB and ahpCF promoters. The strength of an OxyR-binding site could possibly determine the affinity of OxyR to a target promoter and could allow the sequential activation of antioxidant genes with regard to the extent of oxidative stress encountered. Furthermore, we found that not only the KatB catalase, but also AhpB, which belongs to the alkyl hydroperoxide reductase family, possesses catalase activity. In fact, a ΔahpB::Gm mutant strain exhibited a more pronounced hypersusceptibility to H2O2 than to organic hydroperoxides. These findings suggest overlapping functions of KatB, AhpB, and AhpC-AhpF in the detoxification processes. Clearly, detailed biochemical studies on purified KatB, AhpB, and AhpC-AhpF are needed to investigate their potential broad substrate specificities. Multiple enzymatic activities have been demonstrated for mycobacterial KatG, which can act both as a catalase-peroxidase (34) and as a peroxynitritase (58). The observed redundancy of overlapping oxidative stress defense systems in P. aeruginosa may also be explained, in part, by the localization of the antioxidant enzymes in different cellular compartments. While KatA is found in the cytoplasm and in the extracellular milieu (see the accompanying paper by Hassett et al. [15]), KatB is found in the cytoplasm, cytoplasmic membrane, and periplasm (24). AhpB possesses a single cytoplasmic membrane-spanning domain, suggesting a function in the protection of membrane-bound respiratory chain components from H2O2.
We present evidence that expression of katB-ankB, ahpB, and ahpC-ahpF depends on OxyR. Putative OxyR boxes were found at the proper location within the ahpB and ahpC promoters, and binding of OxyR to these target promoters was demonstrated, indicating a direct activation of these genes by OxyR.
The phenotypes of a ΔoxyR mutant included a dramatic susceptibility to oxidative stress agents and a low plating efficiency (see also the accompanying paper by Hassett et al. [15]). The oxidative stress susceptibility of P. aeruginosa was significantly increased in a low-iron environment compared to iron-rich conditions (data not shown). Although the presence of iron is known to trigger the formation of HO·, which has deleterious effects on the cells, iron is required for the function of the heme-containing antioxidant enzymes. The hypersusceptibility to oxidative stress in low-iron media was even more drastic in a ΔoxyR mutant, suggesting a potential role of OxyR in iron metabolism. Moreover, none of the OxyR-regulated factors characterized in this study was capable of fully complementing the ΔoxyR mutant phenotype, suggesting the existence of additional members of the OxyR regulon. Some genes, including fur and dps of E. coli, are expressed in both OxyR-dependent and OxyR-independent ways, and the situation in P. aeruginosa may be similar. In our screening of a minilibrary of translational fusions to the lacZ gene, we pulled out those genes that were expressed at at least a fivefold higher level in wild-type compared to oxyR mutant cells under oxidative stress conditions. As a consequence, we did not pick up those genes that are expressed OxyR independently but can be further upregulated by OxyR. Also, the possibility of indirect OxyR regulation exists and could involve the small RNA oxyS, which has been shown to regulate several genes in E. coli (1). However, we have not found an oxyS-like gene in a search of the P. aeruginosa genome (www.pseudomonas.com). A future goal is the isolation of other OxyR-regulated genes, through an in vitro cycle selection procedure that has been successful in the past to identify Fur-regulated genes of P. aeruginosa (40).
Antioxidant enzymes represent the first line of defense in the battle against oxidative stress. A second strategy to survive these harsh conditions is to maintain an efficient DNA repair system. Interestingly, the P. aeruginosa oxyR gene was found in an operon with recG, encoding a homolog of the E. coli RecG DNA helicase, which is an ATP-dependent DNA recombinase implicated in DNA replication, recombination, and repair (29, 31). To our knowledge, P. aeruginosa is the first microorganism for which such a genetic link between an oxidative stress gene and a DNA repair gene has been identified. In some other organisms, including mycobacteria, the oxyR gene is located in a cluster with genes encoding antioxidant enzymes, but we did not find any alkyl hydroperoxide reductases or catalases encoded near P. aeruginosa oxyR (data not shown). The precise role of RecG is somewhat elusive, but it has been postulated that the DNA binding and unwinding activities of RecG are involved in promoting branch migration by catalyzing the formation of four-strand Holliday junctions from three-strand junctions (36). Clearly, we demonstrated that a recG::Gm mutant of P. aeruginosa was hypersensitive to UV irradiation-induced DNA damage, indicating that recG is essential for optimal DNA repair. Also, the P. aeruginosa recG::Gm mutant was hypersusceptible to oxidative stress agents, thus directly demonstrating the DNA-damaging effects of ROIs. Taken together, the two coordinately expressed factors encoded by the oxyR-recG operon play a crucial role the survival in response to environmental challenges.
ACKNOWLEDGMENTS
This work was supported by grant AI-15490 (to M.L.V.) from the National Institutes of Health and by Public Health Service grant AI-40541 (to D.J.H.) and a Pilot Grant from the Cystic Fibrosis Foundation (to D.J.H.).
REFERENCES
- 1.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 2.Aslund F, Zheng M, Beckwith J, Storz G. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc Natl Acad Sci USA. 1999;96:6161–6165. doi: 10.1073/pnas.96.11.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton H A, Johnson Z, Cox C D, Vasil A I, Vasil M L. Ferric uptake regulator mutants of Pseudomonas aeruginosa with distinct alterations in the iron-dependent repression of exotoxin A and siderophores in aerobic and microaerobic environments. Mol Microbiol. 1996;21:1001–1017. doi: 10.1046/j.1365-2958.1996.381426.x. [DOI] [PubMed] [Google Scholar]
- 4.Beyer W, Imlay J, Fridovich I. Superoxide dismutases. Prog Nucleic Acid Res Mol Biol. 1991;40:221–253. doi: 10.1016/s0079-6603(08)60843-0. [DOI] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Brown S M, Howell M L, Vasil M L, Anderson A J, Hassett D J. Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J Bacteriol. 1995;177:6536–6544. doi: 10.1128/jb.177.22.6536-6544.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bsat N, Chen L, Helmann J D. Mutation of the Bacillus subtilis alkyl hydroperoxide reductase (ahpCF) operon reveals compensatory interactions among hydrogen peroxide stress genes. J Bacteriol. 1996;178:6579–6586. doi: 10.1128/jb.178.22.6579-6586.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christman M F, Storz G, Ames B N. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc Natl Acad Sci USA. 1989;86:3484–3488. doi: 10.1073/pnas.86.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics, a manual for genetic engineering. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 10.Deretic V, Philipp W, Dhandayuthapani S, Mudd M H, Curcic R, Garbe T, Heym B, Via L E, Cole S T. Mycobacterium tuberculosis is a natural mutant with an inactivated oxidative-stress regulatory gene: implications for sensitivity to isoniazid. Mol Microbiol. 1995;17:889–900. doi: 10.1111/j.1365-2958.1995.mmi_17050889.x. [DOI] [PubMed] [Google Scholar]
- 11.Ding H, Demple B. Thiol-mediated disassembly and reassembly of [2Fe-2S] clusters in the redox-regulated transcription factor SoxR. Biochemistry. 1998;37:17280–17286. doi: 10.1021/bi980532g. [DOI] [PubMed] [Google Scholar]
- 12.Elkins J G, Hassett D J, Stewart P S, Schweizer H P, McDermott T R. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl Environ Microbiol. 1999;65:4594–4600. doi: 10.1128/aem.65.10.4594-4600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaudu P, Weiss B. SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc Natl Acad Sci USA. 1996;93:10094–10098. doi: 10.1073/pnas.93.19.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez-Flecha B, Demple B. Role for the oxyS gene in regulation of intracellular hydrogen peroxide in Escherichia coli. J Bacteriol. 1999;181:3833–3836. doi: 10.1128/jb.181.12.3833-3836.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassett D J, Alsabbagh E, Parvatiyar K, Howell M L, Wilmott R W, Ochsner U A. A protease-resistant catalase, KatA, released upon cell lysis during stationary phase is essential for aerobic survival of a Pseudomonas aeruginosa oxyR mutant at low cell densities. J Bacteriol. 2000;182:4557–4563. doi: 10.1128/jb.182.16.4557-4563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hassett D J, Cohen M S. Bacterial adaptation to oxidative stress: implications for pathogenesis and interaction with phagocytic cells. FASEB J. 1989;3:2574–2582. doi: 10.1096/fasebj.3.14.2556311. [DOI] [PubMed] [Google Scholar]
- 17.Hassett D J, Elkins J G, Ma J F, McDermott T R. Pseudomonas aeruginosa biofilm sensitivity to biocides: use of hydrogen peroxide as model antimicrobial agent for examining resistance mechanisms. Methods Enzymol. 1999;310:599–608. doi: 10.1016/s0076-6879(99)10046-6. [DOI] [PubMed] [Google Scholar]
- 18.Hassett D J, Ma J-F, Elkins J G, McDermott T R, Ochsner U A, West S E H, Huang C-T, Fredericks J, Burnett S, Stewart P S, McPheters G, Passador L, Iglewski B H. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol Microbiol. 1999;34:1082–1093. doi: 10.1046/j.1365-2958.1999.01672.x. [DOI] [PubMed] [Google Scholar]
- 19.Hassett D J, Schweizer H P, Ohman D E. Pseudomonas aeruginosa sodA and sodB mutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. J Bacteriol. 1995;177:6330–6337. doi: 10.1128/jb.177.22.6330-6337.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassett D J, Woodruff W A, Wozniak D J, Vasil M L, Cohen M S, Ohman D E. Cloning and characterization of the Pseudomonas aeruginosa sodA and sodB genes encoding manganese- and iron-cofactored superoxide dismutase: demonstration of increased manganese superoxide dismutase activity in alginate-producing bacteria. J Bacteriol. 1993;175:7658–7665. doi: 10.1128/jb.175.23.7658-7665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoang T T, Karkhoff-Schweizer R R, Kutchma A J, Schweizer H P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 22.Holloway B W. Genetics of Pseudomonas. Bacteriol Rev. 1969;33:419–443. doi: 10.1128/br.33.3.419-443.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmes D S, Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 24.Howell M L, Alsabbagh E, Ma J-F, Ochsner U A, Klotz M G, Beveridge T J, Blumenthal K M, Niederhoffer E C, Morris R E, Needham D, Dean G E, Wani M A, Hassett D J. AnkB, a periplasmic ankyrin-like protein in Pseudomonas aeruginosa, is required for optimal catalase B (KatB) activity and resistance to hydrogen peroxide. J Bacteriol. 2000;182:4545–4556. doi: 10.1128/jb.182.16.4545-4556.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imlay J A, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 26.Imlay J A, Linn S. Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide. J Bacteriol. 1987;169:2967–2976. doi: 10.1128/jb.169.7.2967-2976.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jair K W, Fawcett W P, Fujita N, Ishihama A, Wolf R E., Jr Ambidextrous transcriptional activation by SoxS: requirement for the C-terminal domain of the RNA polymerase alpha subunit in a subset of Escherichia coli superoxide-inducible genes. Mol Microbiol. 1996;19:307–317. doi: 10.1046/j.1365-2958.1996.368893.x. [DOI] [PubMed] [Google Scholar]
- 28.Jeong W, Cha M K, Kim I H. Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol-specific antioxidant protein (TSA)/alkyl hydroperoxide peroxidase C (AhpC) family. J Biol Chem. 2000;275:2924–2930. doi: 10.1074/jbc.275.4.2924. [DOI] [PubMed] [Google Scholar]
- 29.Kalman M, Murphy H, Cashel M. The nucleotide sequence of recG, the distal spo operon gene in Escherichia coli K-12. Gene. 1992;110:95–99. doi: 10.1016/0378-1119(92)90449-y. [DOI] [PubMed] [Google Scholar]
- 30.Li Z, Demple B. SoxS, an activator of superoxide stress genes in Escherichia coli. Purification and interaction with DNA. J Biol Chem. 1994;269:18371–18377. [PubMed] [Google Scholar]
- 31.Lloyd R G, Buckman C. Genetic analysis of the recG locus of Escherichia coli K-12 and of its role in recombination and DNA repair. J Bacteriol. 1991;173:1004–1011. doi: 10.1128/jb.173.3.1004-1011.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma J-F, Ochsner U A, Klotz M G, Nanayakkara V K, Howell M L, Johnson Z, Posey J, Vasil M L, Monaco J J, Hassett D J. Bacterioferritin A modulates catalase A (KatA) activity and resistance to hydrogen peroxide in Pseudomonas aeruginosa. J Bacteriol. 1999;181:3730–3742. doi: 10.1128/jb.181.12.3730-3742.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maciver I, Hansen E J. Lack of expression of the global regulator OxyR in Haemophilus influenzae has a profound effect on growth phenotype. Infect Immun. 1996;64:4618–4629. doi: 10.1128/iai.64.11.4618-4629.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcinkeviciene J A, Magliozzo R S, Blanchard J S. Purification and characterization of the Mycobacterium smegmatis catalase-peroxidase involved in isoniazid activation. J Biol Chem. 1995;270:22290–22295. doi: 10.1074/jbc.270.38.22290. [DOI] [PubMed] [Google Scholar]
- 35.Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol. 1997;179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGlynn P, Lloyd R G. RecG helicase activity at three- and four-strand DNA structures. Nucleic Acids Res. 1999;27:3049–3056. doi: 10.1093/nar/27.15.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller J H. A short course in bacterial genetics. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 72–74. [Google Scholar]
- 38.Miller R A, Britigan B E. Role of oxidants in microbial pathophysiology. Clin Microbiol Rev. 1997;10:1–18. doi: 10.1128/cmr.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ochsner U A, Vasil A I, Johnson Z, Vasil M L. Pseudomonas aeruginosa fur overlaps with a gene encoding a novel outer membrane lipoprotein, OmlA. J Bacteriol. 1999;181:1099–1109. doi: 10.1128/jb.181.4.1099-1109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ochsner U A, Vasil M L. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc Natl Acad Sci USA. 1996;93:4409–4414. doi: 10.1073/pnas.93.9.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poole L B, Ellis H R. Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium. 1. Purification and enzymatic activities of overexpressed AhpF and AhpC proteins. Biochemistry. 1996;35:56–64. doi: 10.1021/bi951887s. [DOI] [PubMed] [Google Scholar]
- 42.Potter A A. Gene cloning in Pseudomonas aeruginosa. In: Dillon J A, Nasim A, Nestman E R, editors. Recombinant DNA methodology. New York, N.Y: John Wiley and Sons; 1985. pp. 147–156. [Google Scholar]
- 43.Rosner J L, Storz G. Effects of peroxides on susceptibilities of Escherichia coli and Mycobacterium smegmatis to isoniazid. Antimicrob Agents Chemother. 1994;38:1829–1833. doi: 10.1128/aac.38.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross R P, Claiborne A. Evidence for regulation of the NADH peroxidase gene (npr) from Enterococcus faecalis by OxyR. FEMS Microbiol Lett. 1997;151:177–183. doi: 10.1111/j.1574-6968.1997.tb12567.x. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Schweizer H P. Improved broad-host-range lac-based plasmid vectors for the isolation and characterization of protein fusions in Pseudomonas aeruginosa. Gene. 1991;103:87–92. doi: 10.1016/0378-1119(91)90396-s. [DOI] [PubMed] [Google Scholar]
- 47.Schweizer H P, Hoang T T. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene. 1995;158:15–22. doi: 10.1016/0378-1119(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 48.Sherman D R, Mdluli K, Hickey M J, Arain T M, Morris S L, Barry C E, 3rd, Stover C K. Compensatory ahpC gene expression in isoniazid-resistant Mycobacterium tuberculosis. Science. 1996;272:1641–1643. doi: 10.1126/science.272.5268.1641. [DOI] [PubMed] [Google Scholar]
- 49.Simon R, Priefer U, Puehler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 50.Sreevatsan S, Pan X, Zhang Y, Deretic V, Musser J M. Analysis of the oxyR-ahpC region in isoniazid-resistant and -susceptible Mycobacterium tuberculosis complex organisms recovered from diseased humans and animals in diverse localities. Antimicrob Agents Chemother. 1997;41:600–606. doi: 10.1128/aac.41.3.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Storz G, Jacobson F S, Tartaglia L A, Morgan R W, Silveira L A, Ames B N. An alkyl hydroperoxide reductase induced by oxidative stress in Salmonella typhimurium and Escherichia coli: genetic characterization and cloning of ahp. J Bacteriol. 1989;171:2049–2055. doi: 10.1128/jb.171.4.2049-2055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Storz G, Tartaglia L A. OxyR: a regulator of antioxidant genes. J Nutr. 1992;122:627–630. doi: 10.1093/jn/122.suppl_3.627. [DOI] [PubMed] [Google Scholar]
- 53.Storz G, Tartaglia L A, Ames B N. The OxyR regulon. Antonie Leeuwenhoek. 1990;58:157–161. doi: 10.1007/BF00548927. [DOI] [PubMed] [Google Scholar]
- 54.Storz G, Tartaglia L A, Ames B N. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science. 1990;248:189–194. doi: 10.1126/science.2183352. [DOI] [PubMed] [Google Scholar]
- 55.Tartaglia L A, Gimeno C J, Storz G, Ames B N. Multidegenerate DNA recognition by the OxyR transcriptional regulator. J Biol Chem. 1992;267:2038–2045. [PubMed] [Google Scholar]
- 56.Toledano M B, Kullik I, Trinh F, Baird P T, Schneider T D, Storz G. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell. 1994;78:897–909. doi: 10.1016/s0092-8674(94)90702-1. [DOI] [PubMed] [Google Scholar]
- 57.Wayne L G, Diaz G A. A double staining method for differentiating between two classes of mycobacterial catalase in polyacrylamide gels. Anal Biochem. 1986;157:89–92. doi: 10.1016/0003-2697(86)90200-9. [DOI] [PubMed] [Google Scholar]
- 58.Wengenack N L, Jensen M P, Rusnak F, Stern M K. Mycobacterium tuberculosis KatG is a peroxynitritase. Biochem Biophys Res Commun. 1999;256:485–487. doi: 10.1006/bbrc.1999.0358. [DOI] [PubMed] [Google Scholar]
- 59.West S E, Schweizer H P, Dall C, Sample A K, Runyen-Janecky L J. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 60.Wu J, Weiss B. Two divergently transcribed genes, soxR and soxS, control a superoxide response regulon of Escherichia coli. J Bacteriol. 1991;173:2864–2871. doi: 10.1128/jb.173.9.2864-2871.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Q M, Takemoto T, Mito S, Yonei S. Induction of repair capacity for oxidatively damaged DNA as a component of peroxide stress response in Escherichia coli. J Radiat Res (Tokyo) 1996;37:171–176. doi: 10.1269/jrr.37.171. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, Dhandayuthapani S, Deretic V. Molecular basis for the exquisite sensitivity of Mycobacterium tuberculosis to isoniazid. Proc Natl Acad Sci USA. 1996;93:13212–13216. doi: 10.1073/pnas.93.23.13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 65.Zheng M, Åslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 66.Zheng M, Doan B, Schneider T D, Storz G. OxyR and SoxRS regulation of fur. J Bacteriol. 1999;181:4639–4643. doi: 10.1128/jb.181.15.4639-4643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]