PURPOSE
The utility of circulating tumor DNA (ctDNA) analyses has not been established in the risk stratification of Wilms tumor (WT). We evaluated the detection of ctDNA and selected risk markers in the serum and urine of patients with WT and compared findings with those of matched diagnostic tumor samples.
PATIENTS AND METHODS
Fifty of 395 children with stage III or IV WT enrolled on Children's Oncology Group trial AREN0533 had banked pretreatment serum, urine, and tumor available. Next-generation sequencing was used to detect ctDNA. Copy-number changes in 1q, 16q, and 1p, and single-nucleotide variants in serum and urine were compared with tumor biopsy data. Event-free survival (EFS) was compared between patients with and without ctDNA detection.
RESULTS
ctDNA was detected in the serum of 41/50 (82%) and in the urine in 13/50 (26%) patients. Agreement between serum ctDNA detection and tumor sequencing results was as follows: 77% for 1q gain, 88% for 16q deletions, and 70% for 1p deletions, with ĸ-coefficients of 0.56, 0.74, and 0.29, respectively. Sequencing also demonstrated that single-nucleotide variants detected in tumors could be identified in the ctDNA. There was a trend toward worse EFS in patients with ctDNA detected in the serum (4-year EFS 80% v 100%, P = .14).
CONCLUSION
ctDNA demonstrates promise as an easily accessible prognostic biomarker with potential to detect tumor heterogeneity. The observed trend toward more favorable outcome in patients with undetectable ctDNA requires validation. ctDNA profiling should be further explored as a noninvasive diagnostic and prognostic tool in the risk-adapted treatment of patients with WT.
INTRODUCTION
Wilms tumor (WT) is the most common malignant kidney tumor in the pediatric population,1 with 500 new cases being diagnosed annually in the United States. Although outcomes of children diagnosed with WT have significantly improved over the past few decades, there remain significant challenges to optimizing therapy such that all patients are cured while side effects of therapy are minimized. A subset of patients experience relapse after therapy, and survival after relapse is unacceptably low.2-4 Recent clinical research efforts have focused on identifying patients who are at increased risk of relapse, demonstrating the prognostic relevance of specific tumor genomic features at diagnosis, including loss of heterozygosity (LOH) of 16q and 1p and, more recently, gain of 1q.5,6 On the basis of these observations, the Children's Oncology Group (COG) recently implemented treatment intensification for patients with LOH of 1p and 16q tumor genetics, resulting in marked improvements in outcome.7
CONTEXT
Key Objective
Is circulating tumor DNA (ctDNA) detectable in the blood and/or urine of patients with stage III and IV Wilms tumor with next-generation sequencing? If ctDNA is detectable, can prognostically relevant somatic variants be identified? Is detection associated with outcome?
Knowledge Generated
In this retrospective cohort, ctDNA was detected in the majority of serum but less than half of urine samples from the same patients. Identification of deletions of chromosomal arms 1p and 16q and gains of 1q were detectable in ctDNA. In most but not all patients, ctDNA findings correlated with genomic alterations in corresponding tumors. All recurrences and deaths occurred in patients who had detectable levels of ctDNA at the time of diagnosis.
Relevance
This study demonstrates that ctDNA analysis may improve prognostication and risk-stratified treatment selection for patients with Wilms tumor with advantages of using minimally invasive technology and potential for overcoming challenges of tumor heterogeneity.
Liquid biopsy is a novel technology that is recently being applied to adult and pediatric malignancies to detect, quantify, and profile ultrarare strands of circulating tumor DNA (ctDNA) as a screening method.8-10 These technologies are already used clinically to select patients for targeted therapies in the setting of colorectal cancer, NSCLC, and breast cancer.11-17 Previous studies indicate that ctDNA is detectable in a range of pediatric solid tumors; however, those studies exploring ctDNA in patients with WT have been limited by sample size and cohorts of patients with a mix of risk-defining clinical features.9,18-26 Whether recently identified stratifying genomic features, namely 16q and 1p LOH and 1q copy gains, can be identified in the blood of patients with WT using liquid biopsy has yet to be explored.
In this study, we used a next-generation sequencing approach to profile cell-free DNA extracted from banked serum and urine samples collected from patients with stage III or IV WT. We leveraged genome-wide assessment of copy-number alterations (CNAs) to determine which patients had detectable levels of ctDNA, explore whether the presence of ctDNA in the blood or urine of these patients correlates with outcome, and evaluate whether ctDNA can serve as an alternative source of tumor material for detecting the presence of 1q copy gains and 16q and 1p LOH.
PATIENTS AND METHODS
Patient Selection
Our cohort consisted of patients who had previously been enrolled and treated on the COG prospective AREN0533 trial with a diagnosis of stage III or IV WT for which matched serum, urine, and tumor samples were banked and available at the COG biopathology center.7,27 All patients provided written informed consent. Eligible patients were those younger than 15 years diagnosed on the basis of imaging with a stage III or IV WT. Patients with stage III WT were eligible for AREN0533 only if tumors were found to have LOH in 1p and 16q. All participating institutions from which data and samples are derived obtained institutional review board approval before study conduct.
Sample Preparation, DNA Extraction, and Sequencing
Specimen collection and processing procedures as well as the details of standard DNA extraction techniques and sequencing library preparation are detailed in the Data Supplement (online only). All samples were profiled for the detection of CNAs with ultra-low-passage whole-genome sequencing (ULP-WGS) and analyzed with the ichorCNA software as previously described.22,28
A targeted next-generation sequencing assay, OncoPanel, was performed to detect mutations in formalin-fixed paraffin-embedded tumor tissue, serum, and urine ctDNA (additional details on sequencing and analysis are presented in the Data Supplement).29,30
Annotation of Recurrent CNAs
CNAs of chromosomal arms 1p, 1q, and 16q were identified from ULP-WGS data by extracting the log2 fold-change for data from the segmental files generated from the ichorCNA algorithm. The low coverage of this technique required that we rely on a large segment of the chromosome to confidently identify the copy-number variants. In this study, copy-number losses and gains were called if they involved at least half of the chromosome arms of interest as indicated by the start and stop genomic coordinates in the segmental files.
Statistical Methods
Fifty patients were eligible for inclusion in this study. Descriptive statistics were computed as relative frequencies for categorical variables and means, standard deviations, and ranges for continuous variables. Previously analyzed individual patient data for LOH of 1p, LOH 16q, combined LOH 1p AND 16q, and 1q gain from AREN03B2 were included for comparison.
To evaluate ctDNA detection rates overall, the distribution of % ctDNA detected in urine and serum were separately plotted as box/violin plots. Rates of marker positivity were calculated separately for each marker, specimen type, and source (current study v AREN03B2). Raw rates of agreement between specimen sources and associated 95% CIs were computed. As these rates can be skewed by underlying prevalence of a marker as well as chance sampling variability, kappa coefficients for inter-rater reliability in detecting each marker were also computed.31
Event-free survival (EFS; defined as the time from enrollment on AREN0533 to the earliest of disease relapse, progression, secondary malignancy, or death due to any cause) and overall survival (OS; defined as the time from enrollment to death due to any cause) were compared by ctDNA detection (yes v no) using Kaplan-Meier estimates and log-rank tests, where patients not experiencing an event of interest were right-censored at their last known disease assessment (EFS) or last known vital status (OS).
At the request of reviewers, potential associations between ctDNA detection in serum or urine and disease burden variables (metastatic disease, tumor diameter, lymph node status, local stage, and renal sinus involvement) were tested with Fisher exact tests. All analyses were performed using the R program for statistical computing.
RESULTS
ctDNA Is Detectable in the Serum and Urine of Patients With WT
Descriptive statistics for the included 50 patients are given in Table 1. Eighty-six percent of patients presented with stage III disease at the primary tumor site, defined by an upfront biopsy, positive lymph nodes, positive margins, tumor rupture, or peritoneal implants. Ninety-two percent of included patients presented with stage IV disease. All patients were initially considered to have favorable-histology WT, but five patients (10%) were found to have focal or diffuse anaplasia at delayed nephrectomy.
TABLE 1.
Baseline Characteristics of Patients
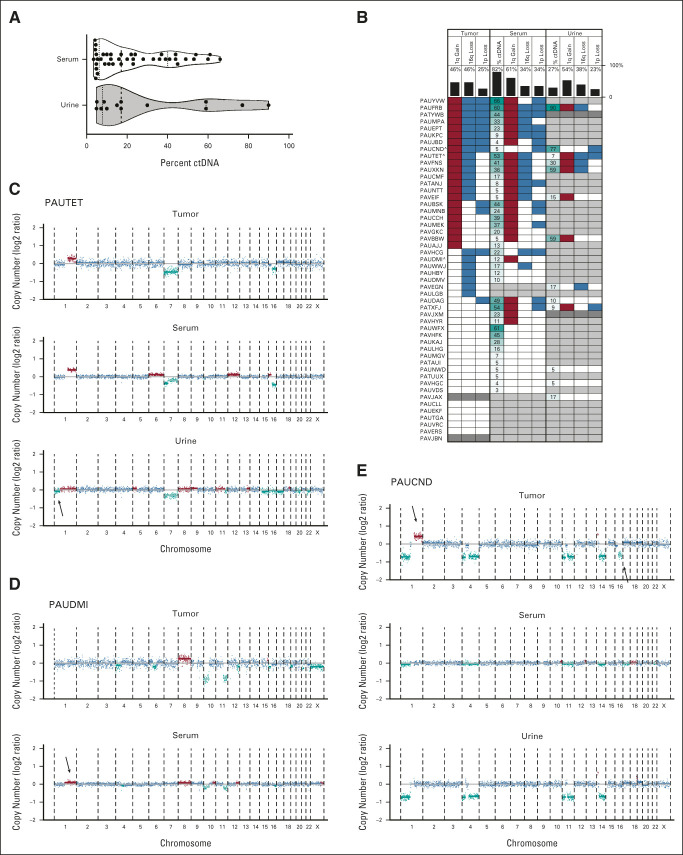
ctDNA was detected in the serum of 82% of patients, whereas ctDNA was only detected in the urine of 27% of patients. The lower limit of detection for ctDNA by ULP-WGS with ichorCNA analysis is 3% of the total cell-free DNA extracted from a sample.28 The maximum amount of ctDNA detection in the serum and urine was 66% and 90%, respectively (Fig 1A). All five patients found to have anaplasia at delayed resection had detectable levels of ctDNA in the serum, and 2/5 anaplastic patients additionally had detectable ctDNA in the urine. In reviewer-requested ad hoc analyses, ctDNA detection in serum was more likely with increased local stage (P = .051 among all patients; P = .036 in the subset with upfront nephrectomies wherein stage was not due to biopsy). Other disease burden variables explored were not associated with ctDNA detection in serum. No disease burden variables were associated with ctDNA detection in urine (Data Supplement).
FIG 1.
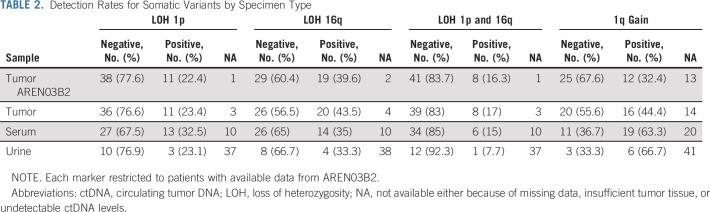
Detection of ctDNA by identification of copy-number variants in serum and urine of patients with WT. (A) Violin plot of ctDNA levels in serum and urine samples restricted to cases with ctDNA above the limit of detection by ULP-WGS. Dashed vertical line within each violin plot indicates the median and the dotted lines indicate the boundaries between the first and second quartiles and the third and fourth quartiles. Probability densities are reflected by the upper and lower borders of the violin plot. Individual ctDNA data are indicated by black dots. (B) Summary of CNAs in chromosomes 1p, 1q, and 16q by case across sample types. Tissue is indicated at the top of the plot, data type indicated in the column label, and vertical black bars indicate the percent of total evaluable samples containing the variant (or with detectable ctDNA). Red blocks indicate copy gains, blue indicates copy losses, white indicates that a variant was not detected, dark gray indicates an unevaluable sample (no tissue or insufficient DNA for sequencing), and light gray indicates that no ctDNA was detectable in the sample. Percent ctDNA content indicated by number with higher ctDNA content shaded in dark teal and lower ctDNA by light teal. Cases plotted in (C-E) include “a” in the patient identifier. (C-E) Genome-wide plots represent the log2 ratio for each data point with blue data equivalent to two copies of the genomic location, teal equal to copy-number loss, and red equal to copy-number gains. Chromosomal segmental medians are also plotted as horizontal lines with light teal segments representing likely subclonal events. Arrows indicate CNAs that are not present in all matched samples from the patient. Case identifiers are present at the top of the panels. CNA, copy-number alteration; ctDNA, circulating tumor DNA; WT, Wilms tumor.
Somatic Variants Identified From Tumor Biopsies Can Be Detected in ctDNA
Chromosomal CNAs of 1p, 16q, and 1q are associated with outcome in patients with WT.5,6 We next studied whether these variants could be detected in the tumors or liquid biopsy samples by ULP-WGS. Rates of detection of LOH 1p, LOH 16q, LOH 1p AND 16q were fairly stable across specimen sources (Fig 1B and Data Supplement), whereas rates of detection of 1q gain were substantially higher in serum ctDNA (63.3%) and urine ctDNA (66.7%) than in matched diagnostic tumor analyzed contemporaneously as part of this project (44.4%) or as reported previously on AREN03B2 (32.4%; Table 2).7,32 This enrichment could be attributed, at least in part, to the significantly higher rate of ctDNA detection in the serum among patients whose tumors demonstrated gain in 1q, which was noted in ad hoc analyses (P = .01; Fig 1B and Data Supplement). In fact, all patients that were found to have a 1q gain in the tumor by ULP-WGS had detectable levels of ctDNA in the serum. Rates of agreement in biomarker classification among pairs of specimen types ranged from 67% to 90% for LOH 1p, 75% to 88% for LOH 16q, 80% to 90% for combined LOH 1p AND 16q, and 77% to 89% for 1q gain (Table 3 and Figs 1C-1E). In general, across the markers evaluated, agreement was lowest between urine and tumor and highest between serum and tumor, where tumor was presently analyzed or reported from AREN03B2.
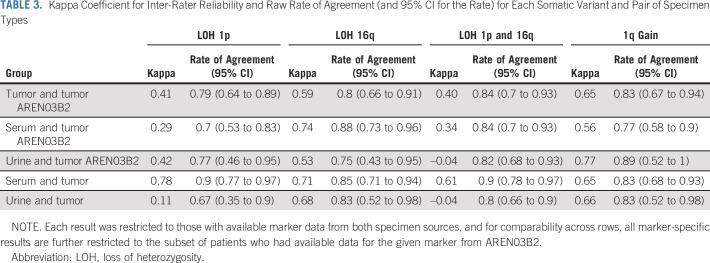
TABLE 2.
Detection Rates for Somatic Variants by Specimen Type
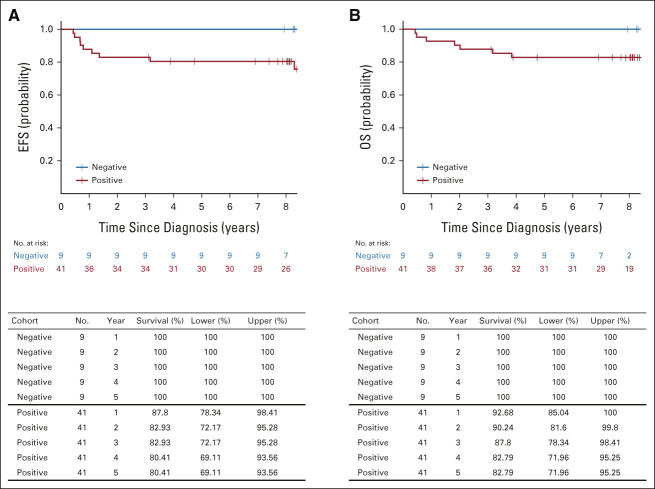
TABLE 3.
Kappa Coefficient for Inter-Rater Reliability and Raw Rate of Agreement (and 95% CI for the Rate) for Each Somatic Variant and Pair of Specimen Types
Kappa coefficients for agreement between analytic sources ranged from 0.11 to 0.78 (slight to substantial) for LOH 1p; 0.53-0.74 (moderate to substantial) for LOH 16q; –0.04 to 0.61 (none to moderate) for combined LOH 1p AND 16q; and 0.56-0.77 (moderate to substantial) for 1q gain. For all but 1q gain, the strongest association was between serum ctDNA and tumor; for 1q gain, the strongest association (kappa = 0.77) was between urine ctDNA and tumor as analyzed on AREN03B2 (Table 3).
Panel sequencing of tumor, serum, and urine samples with detectable levels of tumor DNA content also revealed that single-nucleotide variants (SNVs) detected in one tissue type were often detected in other tissue types. We focused our analysis of somatic SNVs on genes known to be recurrently mutated in WT samples and targeted in our existing OncoPanel hybrid capture assay (WT1, CTNNB1, DICER1, MYCN, TP53, BCOR, BCORL1, ASXL1, MAP3K4, and ARID1A).33 In patients with available tumor sequencing and sufficient ctDNA levels in serum for sequencing, we found that the most commonly mutated genes were CTNNB1 and WT1, consistent with previous reports (Fig 2).33 We also found that the majority of SNVs detected in tumors were also detected at lower allelic fraction in ctDNA but detection of some SNVs also suggested subclonal heterogeneity within the patient's disease (Fig 2).
FIG 2.
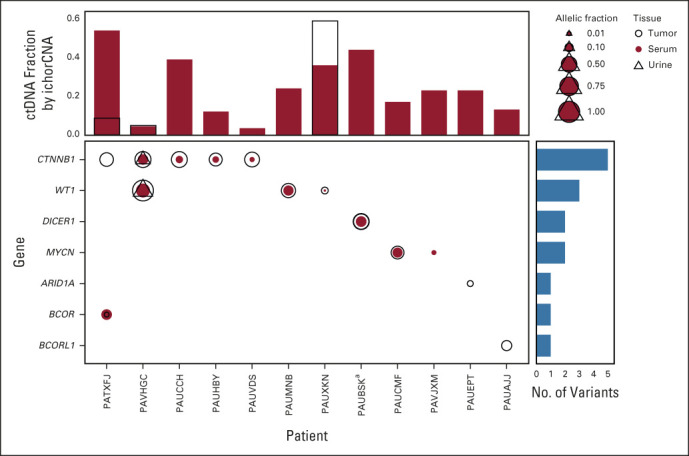
SNVs identified in tumors and matched ctDNA samples. Plotted are somatic variants identified in genes frequently mutated in WTs. Open circles indicate variants identified in tumors, red circles indicate variants identified in serum, and open triangles identified from urine. The size of the symbols reflects the relative allelic fraction of the identified events. Vertical bar plots reflect the fraction of ctDNA identified in the serum (red bars) and urine (open bars with black outline). Horizontal blue bars summarize the number of variants identified in the indicated gene. aA case with two variants in the DICER1 gene identified in both the tumor and the serum. ctDNA, circulating tumor DNA; SNV, single-nucleotide variant; WT, Wilms tumor.
Rare Events Occur in Patients With Detectable ctDNA
Within our cohort of 50 patients, those with ctDNA detected in their serum trended toward worse long-term EFS (4-year EFS = 80.41%) than those without (4-year EFS = 100%; log-rank P = .14; Fig 3A). Patients with ctDNA detected in serum had a 4-year OS of 82.79%, whereas those without ctDNA detected in serum had a 4-year OS of 100% (log-rank P = .20; Fig 3B). Although the overall sample size and few EFS and OS events in this 50-patient cohort precluded statistical significance, the nearly 20% difference in long-term EFS and 17% difference in long-term OS may be clinically significant and warrants further validation in a prospective cohort. Removing patients from this analysis whose diagnoses were updated to anaplastic WT at postchemo delayed resection had minimal impact on these comparisons (Data Supplement).
FIG 3.
EFS and OS in patients with and without detectable ctDNA in the serum: (A) EFS and (B) OS by ctDNA detection in the serum of patients with stage III and IV WT. ctDNA, circulating tumor DNA; EFS, event-free survival; OS, overall survival; WT, Wilms tumor.
The trends toward worse EFS and OS observed among patients with ctDNA detected in serum were also observed for patients with ctDNA detected in urine, but with a lower level of discrimination. Patients with ctDNA detected in urine had a 4-year EFS of 76.92% and 4-year OS of 76.92%, whereas those without had a 4-year EFS of 88.57% and 4-year OS of 91.43% (EFS log-rank P = .39 and hazard ratio = 1.85; OS log-rank P = .13 and hazard ratio = 3.20; Data Supplement). Excluding patients with anaplasia at delayed resection had a more substantial impact on relative EFS and OS (Data Supplement).
DISCUSSION
The past several decades of clinical trials for patients with WTs has resulted in a dramatic improvement in outcomes, with the majority of patients now cured. However, patients who experience relapses have a markedly worse outcome even when treated with intensified regimens. Furthermore, emerging evidence demonstrates that the use of clinical measures of disease burden alone does not risk stratify as precisely as integration of histopathologic and molecular features of a patient's disease. It also remains unclear how to include molecular prognostic features present in only subclonal populations of tumor cells. Ongoing efforts to implement these additional prognostic biomarkers of outcome suggest that improved precision of risk stratification could further reduce the rate of disease progression or relapse after initial therapy and mitigate risks of long-term toxicities by facilitating further intensity reduction for some patients without jeopardizing outcomes.
Liquid biopsies are emerging as a new tool for prognostication and precision medicine in cancer.11-18,24 Previous studies have demonstrated that the detection of ctDNA in the blood of patients with WT was feasible, but these studies were too small to demonstrate associations with other prognostic features or outcomes.19,21-23,25 In this study, we profiled samples collected from a cohort of 50 patients with matched tumor, serum, and urine who were treated on the COG study AREN0533 for patients with newly diagnosed stage III and IV disease. We found that the majority of these patients shed sufficient levels of ctDNA into their serum to be reliably detectable by a low-sensitivity next-generation sequencing approach that identifies segmental and chromosomal copy-number variants. By contrast, only a minority of patients had detectable levels of ctDNA in their urine, suggesting that more sensitive assays or larger sample volumes may be needed to make urine-based liquid biopsies useful in this disease.
ULP-WGS can be used to detect ctDNA when it comprises at least 3% of the cell-free DNA content isolated from a liquid biopsy sample.28 In previous studies of other pediatric solid tumors, we have found that this limit of detection is a convenient and clinically relevant cutoff for distinguishing patients with high ctDNA (≥ 3%) from patients with low ctDNA (< 3%).24 Consistent with this previous work, we found that none of the patients enrolled on this study with low (undetectable) ctDNA in their serum experienced a relapse and all survived. Although this difference in PFS and OS was not significant, these findings justify further validation in prospective cohorts and suggest that ctDNA levels could further refine risk stratification for patients with stage III-IV WT. We also found that all patients later found to have localized or diffuse anaplasia in their tumors at the time of surgery, shed ctDNA into the serum and urine, further supporting the concept that detection of ctDNA by ULP-WGS may be associated with more aggressive disease in patients with WT, an important finding to also validate in an expanded cohort.
In this study, we demonstrated that somatic variants present in the tumor could be detected in the serum and urine of patients with detectable levels of ctDNA. These findings demonstrate the practice changing potential of liquid biopsies to detect the presence of validated prognostic molecular events in the context of up-front chemotherapy without biopsy. Although the level of correlation between tumor profiling and ctDNA samples was high, we also observed discrepancies between samples that suggest the presence of subclonal heterogeneity in these patients, most of whom had metastatic disease. In other cancers, ctDNA has been shown to better capture the presence of subclonal variants than solitary biopsies, especially in patients with metastatic disease.9,16,20,34-37 In five cases from our study, we detected a 1q gain in the serum but not in the matched tumor, and in two cases, 1q gain was detected in the tumor but not the serum, even when samples were profiled by using the same technology (ULP-WGS), suggesting the possible utility of a more comprehensive profiling strategy for risk stratification at diagnosis. However, it is still unclear how to interpret the detection of these somatic events when present in subclonal fractions. Interestingly, we did not identify TP53 mutations in either the tumors or ctDNA from the five patients found to have anaplasia at delayed resection. It is possible these cases were representative of TP53 wild-type anaplasia31 or that our sequencing techniques were not sensitive enough to detect rare subclones containing these variants. We anticipate that more comprehensive detection of clonal and subclonal CNVs and SNVs will be possible by cell-free DNA profiling as more sensitive sequencing approaches are implemented.38 Additional studies will be needed to determine whether ctDNA can better identify clinically relevant tumor genetic biomarkers than tumor biopsies in WT and to determine how to incorporate subclonal variants into risk-stratification strategies.
One limitation of this study is that our cohort was enriched for patients with stage IV disease because only a subset of patients with stage III disease, those with LOH of 1p and 16q, were eligible for AREN0533. Levels of detectable ctDNA could be lower in patients with biologically less aggressive stage III disease or patients with localized WT, which will need to be further studied. This patient cohort was only a subset of the patients enrolled on AREN0533, which could have introduced unexpected selection bias. Another limitation of this study is that only serum was available from these patients, and samples demonstrated a high degree of contamination with cellular DNA.39-41 For this reason, accurate ctDNA levels could not be confidently calculated (beyond detect or not detect), and this precluded studying whether the quantity of ctDNA was directly associated with an increased risk of relapse or death. Furthermore, only small volumes of urine were available for our study. Given the potential to collect much larger volumes of urine from patients, this analysis does not rule out the possibility that urine could be a promising source of material for ctDNA analysis. Moreover, as only pretreatment samples were available for this cohort, we were not able to evaluate whether changes in ctDNA detection on and after therapy were associated with prognosis or relapse, but we recognize this as an important area of future study.17,20,42-44 Each of these limitations can be overcome through routine collection of serial blood and urine samples using validated ctDNA-optimized sample handling protocols, such as the use of cell-stabilizer tubes, in future clinical trials.39-41 Finally, leveraging ULP-WGS for detection of segmental CNAs could have resulted in some false-negative results for focal CNVs picked up by array profiling for the AREN03B2 study, potentially accounting for some of the rare discrepancies seen in our analysis. Improvements in computational analysis would be expected to shorten the minimum length of CNVs detectable by ULP-WGS to one megabase, equivalent to the length needed to call an alteration in the AREN03B2 protocol.
In summary, to our knowledge, our study is the first to define the feasibility of detecting and quantifying ctDNA from liquid biopsy samples collected from patients with WT enrolled on a multi-institutional prospective clinical trial, with available correlative treatment and outcome data. The findings highlight the potential advantage of these assays in refining risk-stratified treatment strategies that have already improved outcome for patients with this pediatric malignancy. We believe prospective validation of these results is warranted, and studies to determine whether ctDNA studies in patients with higher-risk and lower-risk disease would also be informative.
ACKNOWLEDGMENT
The authors thank the Children's Oncology Group (COG) protocol coordinators, research coordinators, clinical research assistants, and other health professionals who contributed to acquiring samples used in this study. The authors wish to thank the extended AREN0533, AREN0532, and AREN03B2 teams who supported the clinical trial and biobanking data and samples used in this study. Finally, the authors are very grateful to the patients and their families for consenting to sample deposition through AREN03B2.
Jeffrey S. Dome
Patents, Royalties, Other Intellectual Property: Rockland Immunochemicals
Lisa R. Diller
Stock and Other Ownership Interests: Novartis, Amgen, Roche, CRISPR Therapeutics, Baxter, LabCorp, Apellis Pharmaceuticals, AstraZeneca, Intellia Therapeutics, Novo Nordisk
Patents, Royalties, Other Intellectual Property: My husband holds a patent with Massachusetts General Hospital for a drug currently in clinical trials. The drug is an inhibitor of BMP-1 (Alk2) receptor and is being studied in anemia of myelodysplastic syndrome and high-hepcidin level–associated anemias
Brian D. Crompton
Employment: Acceleron Pharma
Stock and Other Ownership Interests: Acceleron Pharma
Research Funding: Gradalis
No other potential conflicts of interest were reported.
See accompanying editorial on page 3006
DISCLAIMER
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
PRIOR PRESENTATION
Presented in part at the International Society of Paediatric Oncology Virtual Congress, October 21-24, 2021.
SUPPORT
Supported by a Children's Oncology Group Translational Pilot Studies Program for Solid Malignancies (B.D.C.) and the Solder True Life Foundation (E.A.M.). The project was also supported by the COG Chair's Grant (U10CA098543), NCTN Network Group Operations Center Grant (U10CA180886), Statistics & Data Center Grant (U10CA098413), NCTN Statistics & Data Center (U10CA180899), Human Specimen Banking in NCI-Sponsored Clinical Trials (U24CA114766), Human Specimen Banking in NCI-Sponsored Clinical Trials (1U24-CA196173), and St Baldrick's Foundation.
E.A.M. and B.D.C. jointly and equally supervised this work.
DATA SHARING STATEMENT
Upon publication, sequencing data will be available through the dbGAP repository (https://www.ncbi.nlm.nih.gov/gap/) at accession number phs002847. Clinical data for the study cohort (study ID: AREN0533, NCTN Trial Number: NCT00379340) are available through the NCTN/NCORP Data Archive (https://nctn-data-archive.nci.nih.gov/). Sequencing and clinical data from both databases are linkable using universal patient-specific research identifiers.
AUTHOR CONTRIBUTIONS
Conception and design: Laura M. Madanat-Harjuoja, Kelly Klega, Brett Tornwall, Jeffrey S. Dome, Lisa R. Diller, Conrad V. Fernandez, Elizabeth A. Mullen, Brian D. Crompton
Financial support: Lisa R. Diller, Elizabeth A. Mullen, Brian D. Crompton
Collection and assembly of data: Laura M. Madanat-Harjuoja, Lindsay A. Renfro, Kelly Klega, Brett Tornwall, Aaron R. Thorner, Anwesha Nag, David Dix, Elizabeth A. Mullen, Brian D. Crompton
Data analysis and interpretation: Laura M. Madanat-Harjuoja, Lindsay A. Renfro, Kelly Klega, Brett Tornwall, Anwesha Nag, David Dix, Jeffrey S. Dome, Lisa R. Diller, Conrad V. Fernandez, Elizabeth A. Mullen, Brian D. Crompton
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Circulating Tumor DNA as a Biomarker in Patients With Stage III and IV Wilms Tumor: Analysis From a Children's Oncology Group Trial, AREN0533
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Jeffrey S. Dome
Patents, Royalties, Other Intellectual Property: Rockland Immunochemicals
Lisa R. Diller
Stock and Other Ownership Interests: Novartis, Amgen, Roche, CRISPR Therapeutics, Baxter, LabCorp, Apellis Pharmaceuticals, AstraZeneca, Intellia Therapeutics, Novo Nordisk
Patents, Royalties, Other Intellectual Property: My husband holds a patent with Massachusetts General Hospital for a drug currently in clinical trials. The drug is an inhibitor of BMP-1 (Alk2) receptor and is being studied in anemia of myelodysplastic syndrome and high-hepcidin level–associated anemias
Brian D. Crompton
Employment: Acceleron Pharma
Stock and Other Ownership Interests: Acceleron Pharma
Research Funding: Gradalis
No other potential conflicts of interest were reported.
REFERENCES
- 1.Murphy WM, Grignon DJ, Perlman EJ: Kidney Tumors in Children, Atlas of Tumor Pathology. Washington DC, Armed Forces Institute of Pathology, 2004, pp 57-64 [Google Scholar]
- 2.Malogolowkin M, Cotton CA, Green DM, et al. : Treatment of Wilms tumor relapsing after initial treatment with vincristine, actinomycin D, and doxorubicin. A report from the National Wilms Tumor Study Group. Pediatr Blood Cancer 50:236-241, 2008 [DOI] [PubMed] [Google Scholar]
- 3.Spreafico F, Pritchard Jones K, Malogolowkin MH, et al. : Treatment of relapsed Wilms tumors: Lessons learned. Expert Rev Anticancer Ther 9:1807-1815, 2009 [DOI] [PubMed] [Google Scholar]
- 4.Mullen EA, Chi YY, Hibbitts E, et al. : Impact of surveillance imaging modality on survival after recurrence in patients with favorable-histology Wilms tumor: A report from the Children's Oncology Group. J Clin Oncol 36:3396-3403, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gratias EJ, Dome JS, Jennings LJ, et al. : Association of chromosome 1q gain with inferior survival in favorable-histology Wilms tumor: A report from the Children's Oncology Group. J Clin Oncol 34:3189-3194, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grundy PE, Breslow NE, Li S, et al. : Loss of heterozygosity for chromosomes 1p and 16q is an adverse prognostic factor in favorable-histology Wilms tumor: A report from the National Wilms Tumor Study Group. J Clin Oncol 23:7312-7321, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Dix DB, Fernandez CV, Chi YY, et al. : Augmentation of therapy for combined loss of heterozygosity 1p and 16q in favorable histology Wilms tumor: A Children's Oncology Group AREN0532 and AREN0533 study report. J Clin Oncol 37:2769-2777, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bettegowda C, Sausen M, Leary RJ, et al. : Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6:224ra24, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abbou SD, Shulman DS, DuBois SG, et al. : Assessment of circulating tumor DNA in pediatric solid tumors: The promise of liquid biopsies. Pediatr Blood Cancer 66:e27595, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiser DA, West-Szymanski DC, Fraint E, et al. : Progress toward liquid biopsies in pediatric solid tumors. Cancer Metastasis Rev 38:553-571, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcuello M, Vymetalkova V, Neves RPL, et al. : Circulating biomarkers for early detection and clinical management of colorectal cancer. Mol Aspects Med 69:107-122, 2019 [DOI] [PubMed] [Google Scholar]
- 12.Chabon JJ, Hamilton EG, Kurtz DM, et al. : Integrating genomic features for non-invasive early lung cancer detection. Nature 580:245-251, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alimirzaie S, Bagherzadeh M, Akbari MR: Liquid biopsy in breast cancer: A comprehensive review. Clin Genet 95:643-660, 2019 [DOI] [PubMed] [Google Scholar]
- 14.Haber DA, Velculescu VE: Blood-based analyses of cancer: Circulating tumor cells and circulating tumor DNA. Cancer Discov 4:650-661, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chabon JJ, Simmons AD, Lovejoy AF, et al. : Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 7:11815, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corcoran RB, Chabner BA: Application of cell-free DNA analysis to cancer treatment. N Engl J Med 379:1754-1765, 2018 [DOI] [PubMed] [Google Scholar]
- 17.Dawson S, Tsui D, Murtaza M, et al. : Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 368:1199-1209, 2013 [DOI] [PubMed] [Google Scholar]
- 18.Bosse KR, Buongervino S, Lane M, Hyman A, et al. : Serial profiling of ctDNA identifies clinically actionable genomic evolution in high-risk neuroblastoma. Cancer Res, 79, 2019 (suppl 13; abstr 3105) [DOI] [PMC free article] [PubMed]
- 19.Charlton J, Williams RD, Weeks M, et al. : Methylome analysis identifies a Wilms tumor epigenetic biomarker detectable in blood. Genome Biol 15:434, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chicard M, Colmet-Daage L, Clement N, et al. : Whole-exome sequencing of cell-free DNA reveals temporo-spatial heterogeneity and identifies treatment-resistant clones in neuroblastoma. Clin Cancer Res 24:939-949, 2018 [DOI] [PubMed] [Google Scholar]
- 21.Jimenez I, Chicard M, Colmet-Daage L, et al. : Circulating tumor DNA analysis enables molecular characterization of pediatric renal tumors at diagnosis. Int J Cancer 144:68-79, 2019 [DOI] [PubMed] [Google Scholar]
- 22.Klega K, Imamovic-Tuco A, Ha G, et al. : Detection of somatic structural variants enables quantification and characterization of circulating tumor DNA in children with solid tumors. JCO Precis Oncol 10.1200/PO.17.00285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miguez ACK, Barros BDF, de Souza JES, et al. : Assessment of somatic mutations in urine and plasma of Wilms tumor patients. Cancer Med 9:5948-5959, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shulman DS, Klega K, Imamovic-Tuco A, et al. : Detection of circulating tumour DNA is associated with inferior outcomes in Ewing sarcoma and osteosarcoma: A report from the Children's Oncology Group. Br J Cancer 119:615-621, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Treger TD, Chagtai T, Butcher R, et al. : Somatic TP53 mutations are detectable in circulating tumor DNA from children with anaplastic Wilms tumors. Transl Oncol 11:1301-1306, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Roy N, Van Der Linden M, Menten B, et al. : Shallow whole genome sequencing on circulating cell-free DNA allows reliable noninvasive copy-number profiling in neuroblastoma patients. Clin Cancer Res 23:6305-6314, 2017 [DOI] [PubMed] [Google Scholar]
- 27.Dix DB, Seibel NL, Chi YY, et al. : Treatment of stage IV favorable histology Wilms tumor with lung metastases: A report from the Children's Oncology Group AREN0533 study. J Clin Oncol 36:1564-1570, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adalsteinsson VA, Ha G, Freeman SS, et al. : Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun 8:1324, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia EP, Minkovsky A, Jia Y, et al. : Validation of OncoPanel: A targeted next-generation sequencing assay for the detection of somatic variants in cancer. Arch Pathol Lab Med 141:751-758, 2017 [DOI] [PubMed] [Google Scholar]
- 30.Sholl LM, Do K, Shivdasani P, et al. : Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight 1:e87062, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landis JR, Koch GG: The measurement of observer agreement for categorical data. Biometrics 33:159-174, 1977 [PubMed] [Google Scholar]
- 32.Cajaiba MM, Dyer LM, Geller JI, et al. : The classification of pediatric and young adult renal cell carcinomas registered on the Children's Oncology Group (COG) protocol AREN03B2 after focused genetic testing. Cancer 124:3381-3389, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gadd S, Huff V, Walz AL, et al. : A Children's Oncology Group and TARGET initiative exploring the genetic landscape of Wilms tumor. Nat Genet 49:1487-1494, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbosh C, Birkbak NJ, Wilson GA, et al. : Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545:446-451, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spina V, Bruscaggin A, Cuccaro A, et al. : Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood 131:2413-2425, 2018 [DOI] [PubMed] [Google Scholar]
- 36.Alix-Panabieres C, Pantel K: Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov 6:479-491, 2016 [DOI] [PubMed] [Google Scholar]
- 37.Bardelli A, Pantel K: Liquid biopsies, what we do not know (yet). Cancer Cell 31:172-179, 2017 [DOI] [PubMed] [Google Scholar]
- 38.Casbon JA, Osborne RJ, Brenner S, et al. : A method for counting PCR template molecules with application to next-generation sequencing. Nucleic Acids Res 39:e81, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang Q, Henry NL, Paoletti C, et al. : Comparative analysis of circulating tumor DNA stability in K3EDTA, Streck, and CellSave blood collection tubes. Clin Biochem 49:1354-1360, 2016 [DOI] [PubMed] [Google Scholar]
- 40.van Dessel LF, Beije N, Helmijr JC, et al. : Application of circulating tumor DNA in prospective clinical oncology trials—Standardization of preanalytical conditions. Mol Oncol 11:295-304, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merker JD, Oxnard GR, Compton C, et al. : Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. Arch Pathol Lab Med 142:1242-1253, 2018 [DOI] [PubMed] [Google Scholar]
- 42.Garcia-Murillas I, Schiavon G, Weigelt B, et al. : Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 7:302ra133, 2015 [DOI] [PubMed] [Google Scholar]
- 43.Krumbholz M, Eiblwieser J, Ranft A, et al. : Quantification of translocation-specific ctDNA provides an integrating parameter for early assessment of treatment response and risk stratification in Ewing sarcoma. Clin Cancer Res 27:5922-5930, 2021 [DOI] [PubMed] [Google Scholar]
- 44.Stankunaite R, George SL, Gallagher L, et al. : Circulating tumour DNA sequencing to determine therapeutic response and identify tumour heterogeneity in patients with paediatric solid tumours. Eur J Cancer 162:209-220, 2022 [DOI] [PubMed] [Google Scholar]