Abstract
Mitogen-activated protein kinase (MAPK) cascades are conserved and universal signal transduction modules that play important roles in regulating stress responses in plants. Although MAP3K, MP2K, and MPK family in tea plant (Camellia sinensis) have been investigated, little is known about MPK family genes responding to various abiotic stresses in tea plant. In this study, we performed a comprehensive genome-wide analysis of the tea plant MAPKs (CsMPKs) family gene based on the genomic data of tea plants by bioinformatics-based methods. Here, 21 putative CsMPK genes were identified in the tea plant and divided into 4 subfamilies according to the homologous to Arabidopsis and their phylogenetic relationships. The gene structure and conserved motifs of these CsMPKs in the same group showed high similarity, suggesting that they were highly conserved and might have a similar function. The expression profiles of the CsMPK genes were further investigated by quantitative real-time reverse transcription PCR, indicating that many CsMPK genes were involved in response to cold, drought, heat, or heat combined with drought treatment, suggesting their potential roles in abiotic stress responses in tea plant. These results would provide valuable information for further exploring the functional characterization of CsMPK genes in tea plants.
Keywords: tea plant, mitogen-activated protein kinase, gene expression, abiotic stresses
1. Introduction
As sessile organisms, plants often encounter various adverse conditions, such as high and low temperatures, drought, salt stresses, and pathogen invasion, during their growth and development. To coordinate the biotic and abiotic stresses, plants have evolved complex signaling networks to perceive and transmit environmental stimuli. Mitogen-activated protein kinase (MAPK) cascades play vital roles in controlling intracellular response to extracellular signals [1].
MAPK cascades classically are composed of three subsequently protein kinases, namely MAP kinase kinase kinase (MAPKKK/MAP3K/MEKK), MAP kinase kinase (MAPKK/MKK/MEK), and MAP kinase (MAPK/MPK), but sometimes contain an upstream MAP4K [2]. In general, MAPKs are phosphorylated by MAPK kinase at their conserved threonine and tyrosine residues in the activation loop (T-loop), and in turn, MAPKK is phosphorylated in the S/T-X3–5-S/T motif of their activation loop by MAPKKKs when development or environmental signals incurred [3]. Compared with MAPKKKs and MAPKKs, MAPKs act at the bottom of MAPK cascades showing more complexity and sequence diversity. Plant MAPKs can be divided into four groups, including A, B, C, and D subfamilies based on the phosphorylation motif and phylogenetic relationships of amino acid sequence. Members of the A, B, and C subfamilies have a TEY motif in their active sites, while members in the D subfamily possess a TDY motif in their phosphorylation site [1].
In plants, numerous researches demonstrated that MPKs play indispensable regulatory roles in response to abiotic stresses [4]. Such as MPK3 and MPK6 were reported to play a critical role in response to drought, cold, salt, and heat stress in Arabidopsis [4,5], Jammes et al. demonstrated that AtMPK9 and AtMPK12 were preferentially and highly expressed in guard cells and positively regulated ROS-mediated ABA signaling [6]. A role for the MAPK module consisting of MEKK1-MKK1/2-MPK4 has been confirmed in response to drought, cold, and salt stresses in Arabidopsis [7,8]. In rice, OsMPK5 was shown to be a double-faced player in the stress response network: a positive regulator of abiotic stress tolerance and ABA-mediated defense responses to brown spots but a negative regulator of ET-controlled resistance to Magnaporthe oryzae [9]. OsMPK33 was demonstrated to play a negative role in salt tolerance through unfavorable ion homeostasis [10]. The MKK1–MPK4 module was reported to mediate salt signaling in rice [11]. OsMKK6 and OsMPK3 constitute a moderately low-temperature signaling pathway and regulate the tolerance to cold stress in rice [12]. In addition, in maize, MPK4, MPK5, MPK6, and MPK7 were active by diverse stresses, such as drought, low temperature, and salt [13].
In light of the importance of MAPK genes, the identification and characterization of MAPK members have been conducted in various plant species. Such as Arabidopsis thaliana genome contains 20 MAPKs [3], there are 17 MAPKs in rice (Oryza sativa) genome [14], 54 MAPK genes were identified in wheat (Triticum aestivum) [15], 28 MAPKs in cotton (Gossypium raimondii) [16], and 28 MAPKs in sunflower (Helianthus annuus) [17]. Tea plant (Camellia sinensis), an evergreen woody plant, is an important economic crop that is widely distributed in subtropical to tropical climate regions [18]. Due to the local climate changes, tea plant frequently experiences various environmental stresses during its lifecycle, and heat, drought, and low-temperature stresses are the main factors that affect the yield and quality of tea products [19,20]. Recently, high-quality genome sequencing data for tea plants were presented, which provides convenience for further understanding the tea plant CsMPK gene family [21,22,23]. In the present study, 21 CsMPK genes were identified using the tea plant genome by exploring the genomic data. The analysis of identified CsMPK genes in the sequence features, phylogenetic relationships, cis-elements in promoters, and dynamic expression patterns in response to various abiotic stresses was conducted. The results of this study provided valuable information for further investigation of tea plant MPK gene family.
2. Materials and methods
2.1. Identification and characterization of CsMPK gene family
The identification of the CsMPK gene family of tea plant was conducted according to the method described by Wang et al. with some modifications [24]. The MPK protein sequences of A. thaliana, O. sativa, Vitis vinifera, and Populus trichocarpa were downloaded from TAIR (http://www.Arabidopsis.org), the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/index.shtml), the V. vinifera proteome 12 × database (http://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis/), and Ensembl database (http://plants.ensembl.org), respectively. Whole genome sequences of tea plant were downloaded from the Tea Plant Genome Database (http://tpia.teaplant.org/index.html). The MPK sequences of A. thaliana, O. sativa, V. vinifera, and P. trichocarpa were used as queries to search against tea plant proteins using BLASTP program with an e-value of 1 × 10−5 as the threshold. All the acquired MPK protein sequences from the four plants mentioned above were used to build the local Hidden Markov Model-based searches (HMMER) that were used to identify the CsMPKs in tea plant with the threshold of E < 1 × 10−20. Then, the BLASTP results were integrated with the HMMER hits and parsed by manual editing to remove redundant. Furthermore, these identified CsMPK members were confirmed by CDD (https://www.ncbi.nlm.nih.gov/cdd/), PFAM (http://pfam.xfam.org/), InterProScan database (http://www.ebi.ac.uk/interpro/), and SMARAT (http://smart.embl-heidelberg.de/). The molecular weight (MW) and isoelectric point (pI) of the CsMPKs were predicted using ProtParam (http://web.expasy.org/protparam/).
2.2. Sequence alignments and phylogenetic tree construction of CsMPK genes
Multiple alignments of amino acid sequences of MPKs were performed using ClustalW program with the default parameters [25]. Phylogenetic tree of MPKs from C. sinensis, A. thaliana, O. sativa, V. vinifera, and P. trichocarpa was generated by MEGA 6.0 software using a neighbor-joining (NJ) method with 1,000 bootstrap replicates [26], which was visualized in Figtree (http://tree.bio.ed.ac.uk/software/figtree/).
2.3. Gene structure and conserved motif analysis
The exon/intron structure of the CsMPK genes was retrieved from a gene annotation file (http://www.plantkingdomgdb.com/tea_tree/data/gff3/), and the diagrams were displayed by comparing CDSs and their corresponding gene sequences from genome using the online program GSDS 2.0 (http://gsds.cbi.pku.edu.cn/) [27]. The conserved motifs of CsMAPKs were analyzed by the program of MEME with the following parameters: any number of repetitions, maximum of 10 motifs, and an optimum motif width of 6–50 amino acid residues [28].
2.4. cis-element analysis of putative promoter regions
The upstream sequences (1.5 kb) of CsMPK genes were submitted to the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to predict various cis-regulatory elements according to consensus sequences, positional matrices, and individual sites on particular promoter sequences [29].
2.5. Plant materials and treatments
Two-year-old cutting seedlings of the tea cultivar “Longjing43” were grown in a controlled environment chamber maintained for 12 h day (25 ± 3°C) and 12 h night (20 ± 3°C), and 75% relative humidity for 2 weeks. Then, cold, heat, drought, and heat combined with drought stress treatments were performed. Untreated tea plants were used as control. For cold treatment, the tea plants were kept in a growth chamber at 4°C. The first and second tender leaves were harvested 24 and 48 h after the treatment from control and cold-treated plants. The treatments of heat, drought, and heat combined with drought stress were conducted simultaneously. For heat treatment, the tea plants were transferred to chambers maintained at 38°C. For drought treatment, the tea plants were irrigated with 20% (w/v) polyethylene glycol (PEG) 6000. For heat combined with drought stress, the tea plants were irrigated PEG 6000 and then quickly put in chambers at 38°C. The first and second tender leaves treated with drought, heat, heat combined with drought stresses, and control leaves were collected 24 h post-treatment, respectively. The collected leaves were immediately frozen in liquid nitrogen and then stored at −80°C for gene expression analysis. For each treatment, three independent biological replicates were performed.
2.6. Total RNA isolation and quantitative real-time reverse transcription PCR (qRT-PCR) expression analysis
Total RNA was extracted from young leaves of tea plants using the “TaKaRa MiniBEST Plant RNA Extraction” Kit (TaKaRa, Dalian, China) in accordance with the manufacturer’s protocol. RNase-free DNaseI was added to each sample to digest the genomic DNA. Then, cDNA was synthesized following the kit instruction (Promega Corp., Madison, WI, USA) and stored at −20°C. The primer sequences used in this experiment were designed based on gene sequences and the Beacon designer software (NJ, USA). CsGAPDH was used as the reference gene [30]. The primer sequences are shown in Table S1. The qRT-PCR was performed on a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.) with three independent biological replicates. The reaction conditions were: denaturation at 95°C for 30 s, 40 cycles of 95°C for 5 s, and 60°C extensions for 30 s. Relative gene expression levels were calculated using the 2−∆∆CT method [31].
3. Results
3.1. Identification and characterization of CsMPK family genes in tea plant
To identify MPK encoding genes from tea plant genome, both BLASTP and Hidden Markov Model (HMM) searches were conducted using A. thaliana, O. sativa, V. vinifera, and P. trichocarpa MPK proteins as query sequences and then used these search results to identify all the CsMPK family members by CDD, PFAM, and SMART programs. Finally, a total of 21 CsMPK members were identified from tea plant genome. To distinguish from these identified CsMPKs, we provisionally named them according to the homologous gene in A. thaliana (Table 1). The sequence data of all the above CsMPKs are shown in Table S2. The information analysis of CsMPKs showed that the open reading frame (ORF) length for CsMPK genes ranged from 850 to 6,765 bp, and they encoded proteins ranging from 284 to 1,482 amino acids in size. The deduced protein MWs ranged from 32.65 to 169.32 kDa, and the protein isoelectric points (pIs) were between 4.9 and 9.37 (Table 1).
Table 1.
The information of MAPK gene family in tea plant
| Gene | Homologous gene | Sequence ID | Group | ORF length (bp) | Protein length (aa) | MW (kDa) | pI |
|---|---|---|---|---|---|---|---|
| CsMPK1-1 | AtMPK1 | TEA031435 | C | 1,262 | 421 | 47.94 | 5.97 |
| CsMPK1-2 | AtMPK1 | TEA016315 | C | 1,152 | 384 | 44.23 | 6.70 |
| CsMPK3-1 | AtMPK3 | TEA026040 | A | 1,113 | 372 | 42.75 | 5.25 |
| CsMPK3-2 | AtMPK3 | TEA020852 | A | 1,092 | 365 | 41.68 | 5.89 |
| CsMPK3-3 | AtMPK3 | TEA020851 | A | 850 | 284 | 32.65 | 5.62 |
| CsMPK3-4 | AtMPK3 | TEA017807 | A | 2,461 | 822 | 91.59 | 8.64 |
| CsMPK3-5 | AtMPK3 | TEA017811 | A | 1,110 | 371 | 41.43 | 6.27 |
| CsMPK4-1 | AtMPK4 | TEA006273 | B | 6,765 | 1,482 | 169.32 | 4.90 |
| CsMPK4-2 | AtMPK4 | TEA021759 | B | 988 | 330 | 38.21 | 5.41 |
| CsMPK4-3 | AtMPK4 | TEA006436 | B | 1,098 | 367 | 42.24 | 6.28 |
| CsMPK4-4 | AtMPK4 | TEA007103 | B | 2,379 | 798 | 90.21 | 6.18 |
| CsMPK6 | AtMPK6 | TEA024415 | A | 1,516 | 507 | 57.24 | 6.06 |
| CsMPK7 | AtMPK7 | TEA032724 | C | 1,105 | 368 | 42.22 | 7.60 |
| CsMPK9-1 | AtMPK9 | TEA022268 | D | 1,554 | 517 | 59.34 | 6.75 |
| CsMPK9-2 | AtMPK9 | TEA015676 | D | 1,698 | 569 | 64.24 | 6.43 |
| CsMPK15 | AtMPK15 | TEA012905 | D | 1,753 | 587 | 66.87 | 6.99 |
| CsMPK16-1 | AtMPK16 | TEA026883 | D | 2,916 | 975 | 110.3 | 9.22 |
| CsMPK16-2 | AtMPK16 | TEA031053 | D | 1,752 | 587 | 66.62 | 9.09 |
| CsMPK19-1 | AtMPK19 | TEA018880 | D | 1,791 | 599 | 67.28 | 9.22 |
| CsMPK19-2 | AtMPK19 | TEA022253 | D | 2,363 | 792 | 90.12 | 9.25 |
| CsMPK20 | AtMPK20 | TEA004137 | D | 855 | 286 | 33.27 | 9.37 |
3.2. Phylogenetic relationship analysis of CsMPK genes
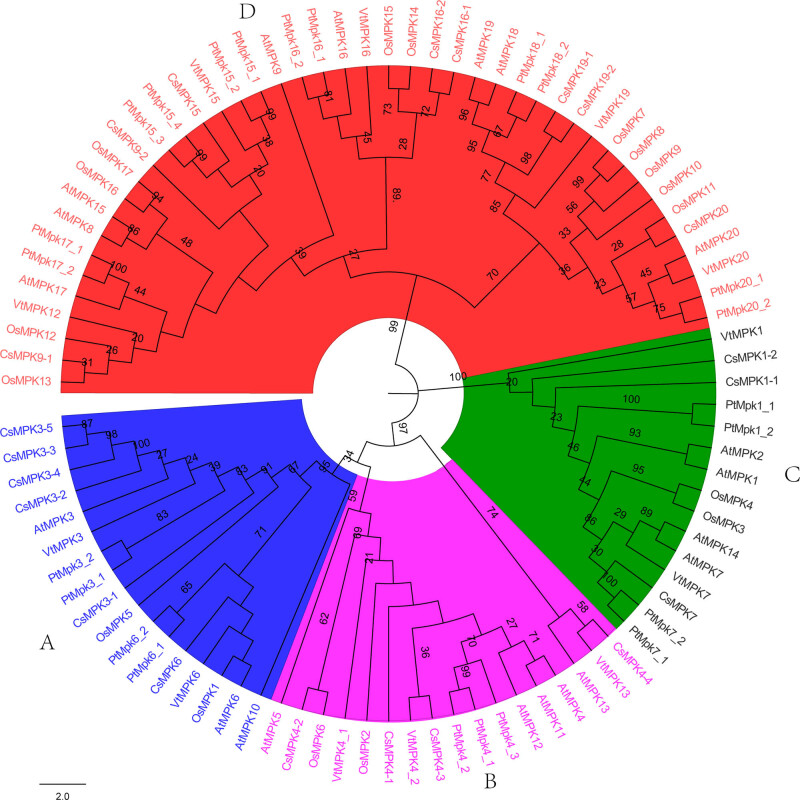
To analyze the phylogenetic relationships of the MPKs, an NJ phylogenetic tree was constructed with the amino acid sequences of 20 AtMPKs from Arabidopsis, 17 OsMPKs from O. sativa, 12 VvMPKs from grapevine, 23 PtMPKs from poplar, and 21 putative CsMPKs from tea plant (Figure 1). Through phylogenetic analyses, these MPKs were classified into four groups, designated as A, B, C, and D. Group D was the first largest clade, including eight CsMPKs, followed by group A (six members). Group C (three members) had the fewest number of CsMPKs; the rest belonged to group B, containing four CsMPKs. CsMPKs belonging to groups A, B, and C possess a TEY motif, and the D group has a TDY motif at the activation site (Figure S1).
Figure 1.
Phylogenetic relationship of putative MPK genes from A. thaliana (AtMPK), O. sativa (OsMPK), V. vinifera (VvMPK), P. trichocarpa (PtMPK), and C. sinensis (CsMPK). The phylogenetic tree with 100 bootstrap replicates was constructed by MEGA6.0 program with the NJ method. Letters A–D named as different groups of MPKs.
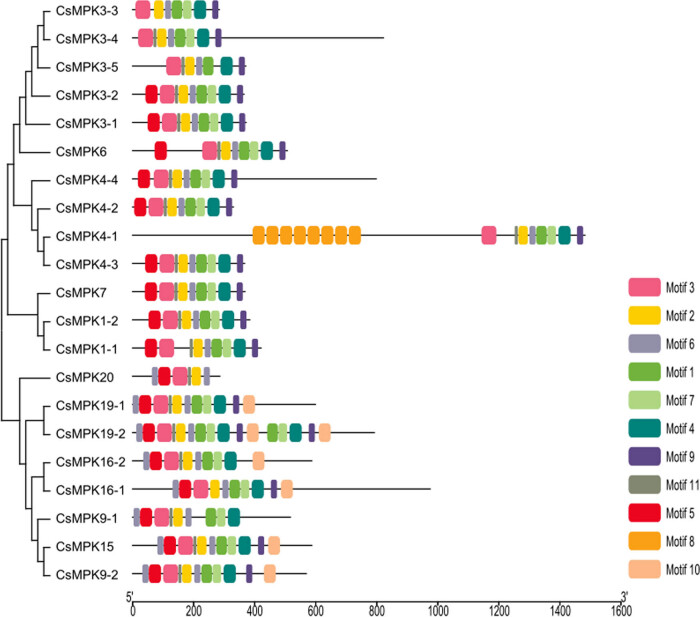
3.3. Conserved motifs analysis of CsMPKs
The conserved protein domains in CsMPKs were identified and compared by the online MEME program. A total of 11 conserved motifs were detected (Figure 2). The lengths of the identified motifs varied from 11 to 50 amino acids (motif logos shown in Figure S2). Two CsMPKs (CsMPK4-1 and CsMPK19-2) contained the maximum number of motifs (16), while the minimum number of motifs (6) occurred in CsMPK20. Furthermore, all of CsMPKs contained motifs 2, 3, and 6 (contained the TXY signature motif), but motif 8 was found only in CsMPK4-1. The alignment analysis and sequence logo revealed that the sequences of CsMPKs in the motif regions were highly conserved (Figures S1 and S2). In addition, the members in the same subfamily were found to share similar conserved motifs. For example, the majority of CsMPKs in groups A, B, and C possessed 9 motifs, whereas most members in group D had at least 10 motifs. Interestingly, motif 6 in group D members occurred two times. These CsMPKs in the same subgroup showed similar motif distribution indicating that these genes might have relatively high conservation.
Figure 2.
The distribution of conserved motifs of CsMPKs from tea plant according to the phylogenetic relationship. The motifs were identified by MEME program with the complete amino acid sequences of the 21 CsMPKs. Each motif is represented by one color box.
3.4. Gene structure analysis of CsMPK genes
Gene structures of the identified 21 CsMPK genes were analyzed by aligning tea plant genomic DNA sequences. As shown in Figure 3, the exon/intron structures of CsMPK genes could be divided into four groups on the basis of their phylogenetic relationship, and those CsMPK genes belonged to the same clade and shared similar intron/exon organization structures. In group A, the number of introns in CsMPKs varied from 4 to 7. Intron numbers in group B varied widely, from 4 (CsMPK4-2) to 17 (CsMPK4-4). Group C had no more than 3 introns, while group D had at least 10 introns except CsMPK20 and CsMPK19-1, containing 5 and 8 introns, respectively.
Figure 3.
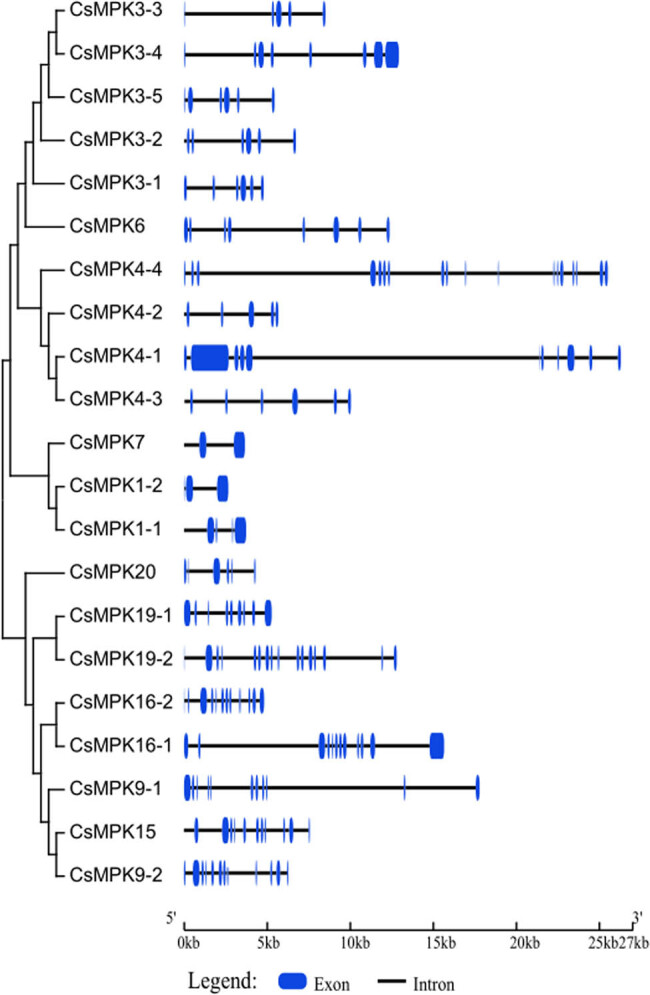
Phylogenetic relationship and gene structure of CsMPK genes. The phylogenetic tree was created using MEGA6.0 program with the NJ method. Exons and introns are indicated by blue bars and thin lines, respectively. Gene models were drawn to scale as indicated on bottom.
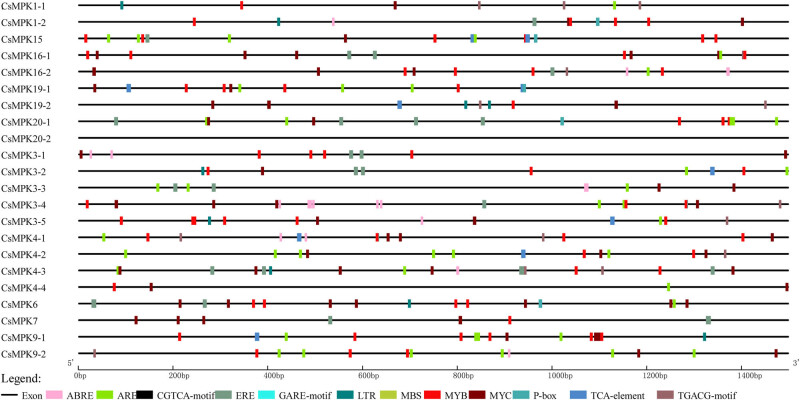
3.5. Stress-related cis-elements analysis of CsMPK genes
To further explore the regulatory mechanisms and potential functions of CsMPK genes under stress conditions, the 1.5 kb promoter sequences of the CsMPK genes were used to analyze the cis-regulatory elements. Two categories of cis-elements were found in the promoter region of CsMPK genes (Figure 4 and Table S3). The first class was constituted by hormone-responsive elements, such as ABRE, ERE, and P-box. TGACG-motif and CGTCA-motif are involved in methyl jasmonate responsiveness, which existed in 10 CsMPKs. ERE is related to an ethylene-responsive element found in 12 CsMPKs. The ABRE element existed in nine CsMPKs. The results suggested that some phytohormones (e.g., ABA and JA) are involved in regulating the expression of some CsMPK genes. The other category was constituted by abiotic stress response-related cis-elements, including LTR, MBS, MYB, MYC, and ARE. MYC responds to abiotic stress signals, which existed in all of CsMPK genes. MYB responds to dehydration and ABA signals and were also found in all CsMPK genes except CsMPK3-3. Eight CsMPKs possessed LTRE, which responds to cold, drought, and ABA signals. MBS is involved in drought response and detected in 8 CsMPKs.
Figure 4.
Predicted cis-elements in the 1.5 kb upstream promoter regions of CsMPK genes. Different cis-elements are represented by different colors.
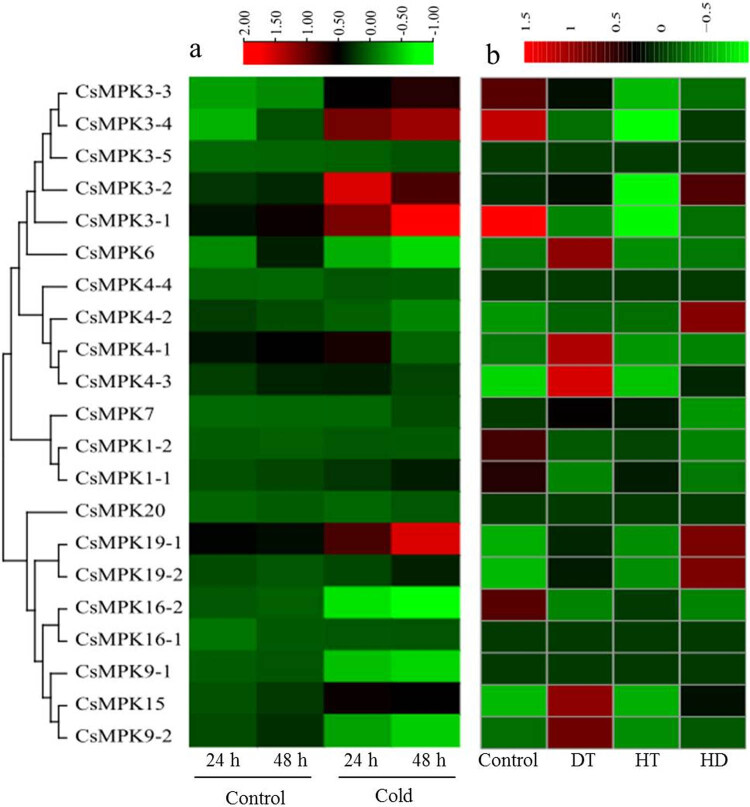
3.6. Expression patterns of CsMPK genes under abiotic stresses
To examine the potential role of CsMPK genes in various abiotic stresses, the expression patterns of all of these genes were investigated under heat, drought, or cold stress conditions (Figure 5a and b). For cold treatment (Figure 5a), the expressions of six CsMPK genes (CsMPK3-1, CsMPK3-2, CsMPK3-3, CsMPK3-4, CsMPK4-3, CsMPK15, and CsMPK19-1) were induced to present a significant upregulation, while CsMPK9-1, CsMPK9-2, and CsMPK16-2 were downregulated under cold stress condition. CsMPK4-1 was upregulated at 24 h of treatment and then downregulated at 48 h. Among the upregulated CsMPK genes, four genes belonged to group A, one in group B, and another two in group D. In response to drought treatment (Figure 5b), most group A and group C members were downregulated, whereas CsMPK genes in groups B and D showed a significantly upregulated expression. For heat treatment, only six CsMPK genes (CsMPK3-3, CsMPK3-4, CsMPK3-2, CsMPK3-1, CsMPK1-2, and CsMPK16-2) displayed downregulation, and no gene was found to show significant up-regulation (Figure 5). Under heat combined with drought treatments (Figure 5b), six CsMPK genes (CsMPK3-2, CsMPK4-2, CsMPK4-3, CsMPK19-1, CsMPK19-2, and CsMPK15) were significantly upregulated, and another six genes (CsMPK3-3, CsMPK3-4, CsMPK3-1, CsMPK1-1, CsMPK1-2, and CsMPK16-2) displayed a downregulated expression trend (Figure 5b). In addition, no changes in the expression of three CsMPK genes (CsMPK3-5, CsMPK4-4, and CsMPK20) were observed under cold, heat, drought, and heat combined with drought treatments. Notably, CsMPK3-2 showed differential expression under three stress conditions (cold, heat, and heat combined drought), indicating that it might play an important role in response to several stresses.
Figure 5.
Hierarchical clustering of the expression patterns of CsMPK genes in tea plant leaves under cold, drought, heat, and heat combined with drought stresses. The red and green colors indicate higher or lower expression levels, respectively. (a) Heatmap of the CsMPK genes expression under cold treatment. (b) Heatmap of the CsMPK genes expression under drought, heat, and heat combined with drought treatments. DT, HT, and HD were denoted as drought, heat, and heat combined with drought treatments, respectively.
4. Discussion
Due to the importance of plant MPKs in multifaceted biological processes such as growth, development, and stress responses [32,33], lots of MPK family members were identified in many plants through whole genome sequencing [34,35]. Recently, the tea plant genome database has been published [21,22,23], which provides the basis for the comprehensive analysis of MAPK cascade members. As the upstream components of the MAPK cascade, MAPKKK family members have been described in detail [36]. Chatterjee et al. analyzed the expression profiles and regulatory network of the MAPK cascade (MAPKK–MAPK) gene family of the tea plant [37]. Tea is the main industrial crop of the Xinyang district, which is often challenged by cold, heat, drought, and heat combined with drought stresses. To implore the tolerance mechanism of tea plant to above-mentioned stresses, we performed a detailed analysis of the CsMPK family genes and the expression characteristics of these genes under abiotic stresses. In this study, 21 CsMPK genes were identified in the tea plant genome. The number of CsMPKs is close to the MPK gene number in Arabidopsis (20 MPKs) and poplar (23 MPKs) (Nicole et al. identified 21 PtMPK genes from P. trichocarpa genome [38]. The variation in PtMPKs amount within the same species in the two studies might come as a result of different parameters employed during HMM profiling and motif analysis), but the genome size of tea plant (∼2.85 Gb) is approximately 23 times that of the Arabidopsis genome (∼125 Mb) and 6 times than poplar genome (∼485 Mb) [39]. The number of CsMPK members in tea plant was more than in grapevine (12 members), coffee (12 members), and kiwifruit (18 members) and slightly lower than that in apple (26 members). The genome sizes of these plants (grapevine is ∼490 Mb, coffee is ∼710 Mb, apple is ∼742 Mb, and kiwifruit is ∼616 Mb) are significantly smaller than the tea plant genome [40,41,42,43]. The phenomenon indicated that the number of MPK genes is irrelevant to the size of the plant genome.
The sequence alignment and phylogenetic tree analysis revealed four distinct groups, which were consistent with the MPKs previously identified in Arabidopsis [3], rice [14], poplar [38], and sunflower [17]. In group A, tea plants had four extra copies of MPK3 that might have been because of gene duplications. Such extra gene copies have also been observed in sunflowers [17] and apples [44]. Four copies of CsMPK4 were also found in group B. In plants, gene duplications play vital roles in the genomic expansion and diversification of gene function [45]. Two rounds of whole genome duplication events have occurred in the tea tree genome, which resulted in the expansion of several gene classes [22,23]. More copies of CsMPKs might suggest that their capabilities were broken down into more detailed functions.
The exon/intron structure plays an important role in organismal evolution [46]. In the study, we observed that the positions, sizes, and sequences of the introns differed between CsMPK genes (Figure 3). Members in group C had the fewest introns, while group D possessed the most introns. Similar exon/intron structures were found in apples [44]. Generally, genes with fewer introns could be rapidly activated in response to environmental stresses [47]. MPK genes are one of the rapidly induced genes under different environmental challenges [3]. The less of introns fit in with the needs for the rapid activation of MPK genes. In this study, we found that group A members with fewer introns were induced by various stress treatments, whereas genes with many introns, such as CsMPK4-4, CsMPK9-1, and CsMPK16-1, did not show any significant expression changes under cold, heat, and drought stresses (Figure 5). In addition, CsMPK members with similar conserved motifs and exon/intron structure showed a tendency to cluster into one subgroup (Figures 2 and 3), which implied functional consistency among these CsMPKs in the same subfamily. This correlation between motif arrangement and intron numbers supported the previous classification of the CsMPK genes. These results might provide an excellent reference to explore the functions of the CsMPKs.
Numerous reports demonstrated that plant MPK genes play important roles in determining response to abiotic stresses [48]. For example, AtMAPK3, AtMAPK4, ZmMAPK1, and SiMAPK7 were reported to regulate abiotic stress responses [49,50,51]. In this study, the expression levels of the CsMPK3-1, CsMPK3-3, and CsMPK3-4, which were the orthologue of the AtMAPK3 gene, were induced by cold (Figure 5). Furthermore, these genes possessed at least four types of cis-elements in the promoter region (Figure 4), suggesting that they were related to the transcriptional control of stress responses. Notably, CsMPK3-2 was significantly upregulated under cold and heat combined with drought, implying that CsMPK3-2 might be an important regulator in response to abiotic stresses. The expressions of CsMPK3-1, CsMPK3-2, CsMPK3-3, and CsMPK3-4 were downregulated under heat stress. These results are similar to the previous report in which the SlMPK3 was proposed to negatively regulate heat stress in tomato plants [52]. However, CsMPK3-5 had nine types of cis-elements and fewer introns, and no change was found in the expression level. The results suggested that the cis-elements of gene were not all of decisive factor in responding to stresses, or they could be induced by other stresses. CsMPK4-3, a member of group B, was also induced by drought stress, consisting of the AtMPK4 [53]. The CsMPK genes belonging to group D have not been as well studied as those of groups A and B. In the present study, the expression of CsMPK19-1 and CsMPK19-2 was increased in cold and heat combined with drought stress, and CsMPK15 and CsMPK9-2 were upregulated under drought stress, suggesting the possible roles of these CsMPK genes in abiotic stress responses. In addition, we found that most CsMPK genes possessed at least one hormone-responsive cis-element, including ABA, JA, and ethylene-responsive elements (Table S3). The relationship between MAPK signaling pathways and ABA, JA, SA, and ethylene in plant abiotic stress responses was characterized [54,55]. However, little is known about how the CsMPK genes respond to phytohormones in tea plant. These questions need further research. The regulatory mechanism of CsMPK genes under abiotic stress is complex. More research is needed to explore the specific functions of the CsMPK family genes in tea plants through additional experiments.
5. Conclusions
In this study, a genome-wide analysis of CsMPK family genes in tea plant was performed, and 21 putative CsMPK genes were identified. The identified CsMPK genes were classified into four subfamilies (groups A, B, C, and D). The gene structure and conserved motifs of the CsMPK genes supported the classification results. Numerous cis-elements associated with abiotic stresses were found in the promoter regions of CsMPK genes. Further expression profile analysis using qRT-PCR showed that most CsMPK genes have a positive or negative response to cold, drought, or heat stresses, indicating that CsMPK genes play important roles in abiotic stress response. This work provided useful information on the CsMPK family genes of tea plant and will contribute to investigating the function and regulatory mechanism of CsMPK genes.
Supplementary Material
Footnotes
Funding information: This work was supported by the Youth Foundation of Xinyang Agriculture and Forestry University (2019LG007), the Foundation of Central Laboratory of Xinyang Agriculture and Forestry University (FCL202009 and FCL202015), Natural Science Foundation of Henan Province of China (202300410025), and Key Scientific Research Projects of Colleges and Universities of Henan Province of China (20B210019).
Author contributions: Conceptualization, L.X.H. and L.J.; methodology, investigation, and formal analysis, L.X.H., Z.M., G.C.H., and J.H.D.; writing original draft, L.J.; writing review and editing, S.J.Y. and L.J.; visualization, supervision, and project administration, L.X.H., S.J.Y., and L.J.; funding acquisition, L.X.H. and L.J. All authors have read and agreed to the published version of the manuscript.
Conflict of interest: Authors state no conflict of interest.
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- [1].Cristina M, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol. 2010;61:621–49. [DOI] [PubMed]
- [2].Champion A, Picaud A, Henry Y. Reassessing the MAP3K and MAP4K relationships. Trends Plant Sci. 2004;9:123–9. [DOI] [PubMed]
- [3].MAPK Group. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002;7:301–8. [DOI] [PubMed]
- [4].Raja V, Majeed U, Kang H, Andrabi KI, John R. Abiotic stress: Interplay between ROS, hormones and MAPKs. Env Exp Bot. 2017;137:142–57.
- [5].de Zelicourt A, Colcombet J, Hirt H. The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci. 2016;21:677–85. [DOI] [PubMed]
- [6].Jammes F, Song C, Shin D, Munemasa S, Kwak JM. MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. PNAS. 2009;106:20520–5. [DOI] [PMC free article] [PubMed]
- [7].Furuya T, Matsuoka D, Nanmori T. Membrane rigidification functions upstream of the MEKK1-MKK2-MPK4 cascade during cold acclimation in Arabidopsis thaliana. FEBS Lett. 2014;588:2025–30. [DOI] [PubMed]
- [8].Qiu JL, Zhou L, Yun BW, Nielsen HB, Fiil BK, Petersen K, et al. Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS11. Plant Physiol. 2008;148:212–22. [DOI] [PMC free article] [PubMed]
- [9].Sharma R, Vleesschauwer D, Sharma MK, Ronald PC. Recent advances in dissecting stress-regulatory crosstalk in rice. Mol plant. 2013;6:250–60. [DOI] [PubMed]
- [10].Lee SK, Kim BG, Kwon TR, Jeong MJ, Park SR, Lee JW, et al. Overexpression of the mitogen-activated protein kinase gene OsMAPK33 enhances sensitivity to salt stress in rice (Oryza sativa L.). J Biosci. 2011;36:139–51. [DOI] [PubMed]
- [11].Wang F, Wen J, Zhang W. The mitogen-activated protein kinase cascade MKK1-MPK4 mediates salt signaling in rice. Plant Sci. 2014;227:181–9. [DOI] [PubMed]
- [12].Xie G, Kato H, Imai R. Biochemical identification of the OsMKK6-OsMPK3 signalling pathway for chilling stress tolerance in rice. Biochem J. 2012;443:95–102. [DOI] [PubMed]
- [13].Wu T, Kong X, Zong X, Li D, Li D. Expression analysis of five maize MAP kinase genes in response to various abiotic stresses and signal molecules. Mol Biol Rep. 2011;38:3967–75. [DOI] [PubMed]
- [14].Reyna NS, Yang Y. Molecular analysis of the rice MAP kinase gene family in relation to Magnaporthe grisea infection. MPMI. 2006;19:530–40. [DOI] [PubMed]
- [15].Zhan H, Yue H, Zhao X, Wang M, Song W, Nie X. Genome-wide identification and analysis of MAPK and MAPKK gene families in bread wheat (Triticum aestivum L.). Genes. 2017;8:284. [DOI] [PMC free article] [PubMed]
- [16].Zhang X, Xu X, Yu Y, Chen C, Wang J, Cai C, et al. Integration analysis of MKK and MAPK family members highlights potential MAPK signaling modules in cotton. Sci Rep. 2016;6:29781. [DOI] [PMC free article] [PubMed]
- [17].Neupane S, Schweitzer SE, Neupane A. Identification and characterization of mitogen-activated protein kinase (MAPK) genes in sunflower (Helianthus annuus L.). Plants (Basel). 2019;8:28. [DOI] [PMC free article] [PubMed]
- [18].Liang C, Zhou Z, Yang Y. Genetic improvement and breeding of tea plant (Camellia sinensis) in China: from individual selection to hybridization and molecular breeding. Euphytica. 2007;154:239–48.
- [19].Zheng C, Wang Y, Ding Z, Zhao L. Global transcriptional analysis reveals the complex relationship between tea quality, leaf senescence and the responses to cold-drought combined stress in Camellia sinensis. Front Plant Sci. 2016;7:1858. [DOI] [PMC free article] [PubMed]
- [20].Zhu X, Liao J, Xia X, Xiong F, Li Y, Shen J, et al. Physiological and iTRAQ-based proteomic analyses reveal the function of exogenous γ-aminobutyric acid (GABA) in improving tea plant (Camellia sinensis L.) tolerance at cold temperature. BMC plant biol. 2019;19:43. [DOI] [PMC free article] [PubMed]
- [21].Wei C, Yang H, Wang S, Zhao J, Wan X. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. PNAS. 2018;115:4151–8. [DOI] [PMC free article] [PubMed]
- [22].Xia E, Zhang H, Sheng J, Li K, Zhang Q, Kim C, et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol Plant. 2017;10:866–77. [DOI] [PubMed]
- [23].Wang X, Feng H, Chang Y, Ma C, Wang L, Hao X, et al. Population sequencing enhances understanding of tea plant evolution. Nat Commun. 2020;11:4447. [DOI] [PMC free article] [PubMed]
- [24].Wang G, Wang T, Jia ZH, Xuan JP, Pan DL, Guo ZR, et al. Genome-wide bioinformatics analysis of MAPK gene family in Kiwifruit (Actinidia Chinensis). Int J Mol Sci. 2018;19:2510–6. [DOI] [PMC free article] [PubMed]
- [25].Hung JH, Weng Z. Sequence alignment and homology search with BLAST and ClustalW. Cold Spring Harb Protoc. 2016;11:1–6. [DOI] [PubMed]
- [26].Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis Version 6.0. Mol Biol Evol. 2013;30:2725–9. [DOI] [PMC free article] [PubMed]
- [27].Hu B, Jin J, Guo AY, Zhang H, Gao G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 2014;31:1296–7. [DOI] [PMC free article] [PubMed]
- [28].Bailey TL, Mikael B, Buske FA, Martin F, Grant CE, Luca C, et al. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:202–8. [DOI] [PMC free article] [PubMed]
- [29].Lescot M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–7. [DOI] [PMC free article] [PubMed]
- [30].Wu Z, Tian C, Jiang Q, Li X, Zhuang J. Selection of suitable reference genes for qRT-PCR normalization during leaf development and hormonal stimuli in tea plant (Camellia sinensis). Sci Rep. 2016;6:19748. [DOI] [PMC free article] [PubMed]
- [31].Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods. 2001;25:402–8. [DOI] [PubMed]
- [32].Andrasi N, Rigo G, Zsigmond L, Perz-Salamo I, Papdi C, Klement E, et al. The mitogen-activated protein kinase 4-phosphorylated heat shock factor A4A regulates responses to combined salt and heat stresses. J Exp Bot. 2019;70:4903–18. [DOI] [PMC free article] [PubMed]
- [33].Xu J, Zhang S. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 2015;20:56–64. [DOI] [PubMed]
- [34].McNeece BT, Sharma K, Lawrence GW, Lawrence KS, Klink VP. The mitogen activated protein kinase (MAPK) gene family functions as a cohort during the Glycine max defense response to Heterodera glycines. Plant Physiol Biochem. 2019;137:25–41. [DOI] [PubMed]
- [35].Yan Y, Wang LZ, Ding Z, Tie W, Ding X, Zeng C, et al. Genome-wide identification and expression analysis of the mitogen-activated protein kinase gene family in Cassava. Front Plant Sci. 2016;7:1294. [DOI] [PMC free article] [PubMed]
- [36].Paul A, Srivastava AP, Subrahmanya S, Shen GX, Mishra N. In-silico genome wide analysis of Mitogen activated protein kinase kinase kinase gene family in C. sinensis. PLoS One. 2021;16(11):e0258657. [DOI] [PMC free article] [PubMed]
- [37].Chatterjee A, Paul A, Unnati GM, Rajput R, Biswas T, Kar T, et al. MAPK cascade gene family in Camellia sinensis: In-silico identification, expression profiles and regulatory network analysis. BMC Genomics. 2020;21:613. [DOI] [PMC free article] [PubMed]
- [38].Nicole MC, Hamel LP, Morency MJ, Beaudoin N, Ellis BE, Seguin A. MAP-ping genomic organization and organ-specific expression profiles of poplar MAP kinases and MAP kinase kinases. BMC Genomics. 2006;7:223. [DOI] [PMC free article] [PubMed]
- [39].Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science. 2006;313:1596–604. [DOI] [PubMed]
- [40].Daccord N, Celton JM, Linsmith G, Becker C, Choisne N, Schijlen E, et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Genet. 2017;49:1099–106. [DOI] [PubMed]
- [41].Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–7. [DOI] [PubMed]
- [42].Denoeud F, Carretero-Paulet L, Dereeper A, Droc G, Guyot R, Pietrella M, et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science. 2014;345:1181–4. [DOI] [PubMed]
- [43].Huang SX, Ding J, Deng DJ, Tang W, Sun HH, Liu DY, et al. Draft genome of the kiwifruit Actinidia chinensis. Nat commun. 2013;4:2640. [DOI] [PMC free article] [PubMed]
- [44].Zhang S, Xu R, Luo X, Jiang Z, Shu H. Genome-wide identification and expression analysis of MAPK and MAPKK gene family in Malus domestica. Gene. 2013;531:377–87. [DOI] [PubMed]
- [45].Panchy N, Lehti-Shiu M, Shiu SH. Evolution of gene duplication in plants. Plant Physiol. 2016;171(4):2294–316. [DOI] [PMC free article] [PubMed]
- [46].Xu G, Guo C, Shan H, Kong H. Divergence of duplicate genes in exon-intron structure. PNAS. 2012;109:1187–92. [DOI] [PMC free article] [PubMed]
- [47].Jeffares DC, Penkett CJ, Bahler J. Rapidly regulated genes are intron poor. Trends Genet. 2008;24:375–8. [DOI] [PubMed]
- [48].Sinha AK, Jaggi M, Raghuram B, Tuteja N. Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signa Behav. 2011;6:196–203. [DOI] [PMC free article] [PubMed]
- [49].Ortiz-Masia D, Perez-Amador MA, Carbonell J, Marcote MJ. Diverse stress signals activate the C1 subgroup MAP kinases of Arabidopsis. Febs Lett. 2007;581:1834–40. [DOI] [PubMed]
- [50].Yu L, Yan J, Yang Y, He L, Zhu W. Enhanced tolerance to chilling stress in tomato by overexpression of a mitogen-activated protein kinase, SlMPK7. Plant Mol Biol Rep. 2006;34:76–88.
- [51].Wu L, Zu X, Zhang H, Wu L, Xi Z, Chen Y. Overexpression of ZmMAPK1 enhances drought and heat stress in transgenic Arabidopsis thaliana. Plant Mol Biol. 2015;88:429–43. [DOI] [PubMed]
- [52].Yu W, Wang L, Zhao R, Sheng J, Zhang S, Li R, et al. Knockout of SlMAPK3 enhances tolerance to heat stress involving ROS homeostasis in tomato plants. BMC Plant Biol. 2019;19:354. [DOI] [PMC free article] [PubMed]
- [53].Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K. Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J. 2000;24:655–65. [DOI] [PubMed]
- [54].Danquah A, Zelicourt A, Colcombet J, Hirt H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol Adv. 2014;32:40–52. [DOI] [PubMed]
- [55].Meldau S, Ullman-Zeunert L, Govind G, Bartram S, Baldwin IT. MAPK-dependent JA and SA signalling in Nicotiana attenuata affects plant growth and fitness during competition with conspecifics. BMC Plant Biol. 2012;12:213. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.