Abstract
In this study, we have cloned the ankB gene, encoding an ankyrin-like protein in Pseudomonas aeruginosa. The ankB gene is composed of 549 bp encoding a protein of 183 amino acids that possesses four 33-amino-acid ankyrin repeats that are a hallmark of erythrocyte and brain ankyrins. The location of ankB is 57 bp downstream of katB, encoding a hydrogen peroxide-inducible catalase, KatB. Monomeric AnkB is a 19.4-kDa protein with a pI of 5.5 that possesses 22 primarily hydrophobic amino acids at residues 3 to 25, predicting an inner-membrane-spanning motif with the N terminus in the cytoplasm and the C terminus in the periplasm. Such an orientation in the cytoplasmic membrane and, ultimately, periplasmic space was confirmed using AnkB-BlaM and AnkB-PhoA protein fusions. Circular dichroism analysis of recombinant AnkB minus its signal peptide revealed a secondary structure that is ∼65% α-helical. RNase protection and KatB- and AnkB-LacZ translational fusion analyses indicated that katB and ankB are part of a small operon whose transcription is induced dramatically by H2O2, and controlled by the global transactivator OxyR. Interestingly, unlike the spherical nature of ankyrin-deficient erythrocytes, the cellular morphology of an ankB mutant was identical to that of wild-type bacteria, yet the mutant produced more membrane vesicles. The mutant also exhibited a fourfold reduction in KatB activity and increased sensitivity to H2O2, phenotypes that could be complemented in trans by a plasmid constitutively expressing ankB. Our results suggest that AnkB may form an antioxidant scaffolding with KatB in the periplasm at the cytoplasmic membrane, thus providing a protective lattice work for optimal H2O2 detoxification.
Ankyrins are structural proteins in human erythrocytes and brain that bridge the spectrin exoskeleton to the cytoplasmic surface of the plasma membrane (5). They are composed of three domains: (i) an N-terminal membrane-binding domain, (ii) a spectrin-binding domain, and (iii) a C-terminal domain with an apparent regulatory function (5). Integral membrane proteins that associate with ankyrin both in vivo and in vitro include the band III anion exchanger (15), Na+/K+-ATPase (15), and multiple sodium channels (51, 52). The protein-binding N-terminal domain harbors a series of 33-amino-acid tandem repeats (herein termed ank repeats) that extend over 740 residues. The tandem repeat motif is present in 22 contiguous copies with 30 to 35% identity between the repeats (reference 5 and references therein). Closely related repeats (30 to 35% identity to brain ankyrin) were found in seemingly dissimilar proteins of lower and higher eukaryotes that regulate the cell cycle in yeast (e.g., products of cdc10 and SWI6) and are involved in intercellular signaling during development and cell differentiation of Caenorhabditis elegans (products of lin-12, glp-1, and fem-1), Drosophila (Notch), or Xenopus (Xotch) (references 2, 9, 19, and 50 and references therein). Subsequently identified ankyrin-like proteins (ALPs) include transcription factors (e.g., GABP-β and NF-κB), toxins (e.g., black widow spider venom), enzymes (e.g., rat liver-specific glutaminase), and a viral host range factor (Vaccinia hr gene product) (listed in references 5 and 50); a protein-tyrosine kinase in Hydra vulgaris (14); and the Chlorella virus long terminal repeat gene product (GenBank accession no. D14469). Two ALPs were also identified in the higher plant Arabidopsis thaliana (GenBank accession no. M82883), one of which was implicated in membrane transport (GenBank accession no. X62907). So far, more than 150 genes possessing ank repeats have been reported in eukaryotic systems (GenBank search, May 2000). Due to the success in whole genome sequencing, however, genes encoding ankyrin homologs found most recently reside in bacteria.
The first bacterial ALP-encoding gene (phlB), from Serratia liquefaciens, was not recognized as such (21) until Bennett (5) identified an ank repeat consensus sequence (-G-TA/PLM/H-AA--GH---V/A--LL--GAD-N/D--D-). According to various databases, bacterial ALPs have been identified in several actinobacteria (Streptomyces verticillus [13], Streptomyces argillaceus [U43537], Streptomyces coelicolor cosmid 6D7 [AL133213]), two spirochetes (Treponema pallidum [AE001254] and Deinococcus radiodurans [AE002034 and AE001863]), two cyanobacteria (Anabaena sp. strain PCC 7120 [X95645] and Synechocystis sp. strain PCC 6803 [D90900]), and several proteobacteria (S. liquefaciens [21], Chromatium vinosum [17], Rhizobium leguminosarum [AJ243305], Rickettsia prowazekii [AJ235273], Vibrio cholerae [http://www.tigr.org], two Erlichia species [AF047897 and AF153716], and the four species of fluorescent pseudomonads, i.e., Pseudomonas aeruginosa [U59457], Pseudomonas fluorescens [U83328], Pseudomonas putida KT2440 [http://www.tigr.org], and Pseudomonas syringae [32] [AF133262 and AF133263]). Interestingly, unlike eukaryotic ankyrin or ALPs, bacterial ALPs seem to belong to divergent operons: bleomycin and mithramycin antibiotic resistance in S. verticillus (13) and S. argillaceus (U43537), respectively; periplasmic flavocytochrome c and cytoplasmic tetraheme cytochrome c in C. vinosum (17); and a catalase with proposed periplasmic and cytoplasmic locations in P. syringae (32) and P. fluorescens (U83328). The ankyrin gene in V. cholerae (http://www.tigr.org) is also downstream of a gene encoding a type I bacterial catalase. A putative open reading frame (ORF) upstream of a gene encoding a histidinol phosphate aminotransferase, an enzyme required for ethanol tolerance, was found in Acetobacter pasteurianus (DDBJ accession no. D14440) (54). The ALP of S. liquefaciens, whose gene is located downstream of the gene encoding periplasmic phospholipase A1, has a putative regulatory function regarding phospholipase activity (21). Taken together, the bacterial ALP genes are located in close proximity to genes encoding proteins involved in either (i) nutrient acquisition and uptake or (ii) tolerance or resistance to antibiotics, starvation, or oxidative stress.
In this study, we demonstrate the first functional characterization of a bacterial ALP, AnkB, in P. aeruginosa. AnkB was found to be a cytoplasmic membrane-periplasmic protein whose expression is increased upon exposure of bacteria to H2O2. AnkB was also found to be essential for optimal resistance to H2O2, which we believe is in part due to its ability to bind to and stabilize KatB, a type I bacterial catalase.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
All P. aeruginosa and Escherichia coli strains used in this study are listed in Table 1 and were maintained on Luria (L) agar (10 g of tryptone, 5 g of yeast extract, and 10 g of NaCl) or M9 minimal medium (6 g of Na2HPO4, 3 g of KH2PO4, 1 g of NH4Cl, 0.5 g of NaCl, 1 mM MgSO4 · 7H2O, and 0.2% glucose [per liter]) plates, with each medium solidified with 15 g of Bacto agar per liter. All strains were stored indefinitely at −80°C in a 1:1 suspension of overnight-grown culture and either 25% glycerol or 10% skim milk.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristicsa | Source or reference |
|---|---|---|
| E. coli strains | ||
| HB101 | proA2 leuB6 thi-1 lacY1 hsdR hsdM recA13 supE44 rpsL20 | H. Boyer |
| DH5α | F−lacZΔM15 recA1 hsdR17 supE44 Δ(lacZYA argF) | Bethesda Research Laboratories |
| SM10 | Kmr; mobilizer strain | 48 |
| BL21(DE3) | F−dcm ompT hsdS gal λ (DE3); T7 polymerase gene under control of the lacUV5 promoter | 53 |
| P. aeruginosa strains | ||
| PAO1 | Prototrophic, wound isolate | 28 |
| PAO1 ΔankB::Gm | Gmr; deletion of 0.526 kb of the ankB gene | This study |
| PAO1 katA::Tc | Gmr; katA::Tc mutant | 35 |
| PAO1 ΔkatA ΔankB | Gmr Tcr; deletion of 0.526 kb of the ankB gene in a PAO1 ΔkatA background | This study |
| PAO1 katB::Gm | Gmr; katB::Gm mutant | 18 |
| PAO1 ΔkatB ankB::Gm | Gmr; deletion of 1.75 kb of the katB-ankB locus | This study |
| PAO1 radA | Gmr; radA::Gm mutant | This study |
| Plasmids | ||
| pBluescript KS(−/+) | Apr; extended polylinker pUC derivative | Stratagene |
| pKS-T/A | Apr; T/A vector using the EcoRV site for cloning | 35 |
| pKS-ankB | Apr; 174-bp 5′ fragment of ankB in EcoRV site | This study |
| pUCP22 | Apr; broad-host-range extended polylinker pUC derivative | 44 |
| pUCP22-ankB | pUCP22 containing a 0.825-kb EcoRI-HindIII fragment with the ankB gene under lac promoter control | This study |
| pET14b | Expression vector; Apr | Novagen |
| pET14b-ankB | pET14b containing ankB with an amino-terminal His6 tag under T7 promoter control | This study |
| pET23a | Expression vector; Apr | Novagen |
| pET23a-ankB | pET23a containing ankB with a carboxy-terminal His6 tag under T7 promoter control | This study |
| pEX30 | Apr; broad-host-range expression vector | H. P. Schweizer |
| pEX30-ankB | Apr; pEX30 with a 193-bp AflIII-SmaI ankB fragment within the NcoI-SmaI sites | This study |
| pEX30-ankB::phoA | Apr; pEX30-ankB with a 2.6-kb phoA-containing fragment of pPHO7 fused to ankB | This study |
| pEX100T | Apr; oriT mob sacB gene replacement vector | 46 |
| pEX100T-ΔankB::Gm | pEX100T carrying a 2.8-kb ΔankB::Gm gene study replacement construct | This study |
| pEX100T-ΔkatB ankB::Gm | pEX100T carrying a 2.6-kb ΔkatB ankB::Gm gene study replacement construct | This study |
| pKM1 | Apr; broad-host-range blaM β-lactamase fusion plasmid | 10 |
| pPHO7 | Apr; broad-host-range phoA alkaline phosphatase fusion plasmid | 24 |
| pPZ30 | Apr; broad-host-range lacZ-based promoter probe vector | 43 |
| pPZ-ankB-1900 | pPZ30 containing a 1.9-kb fragment of the katB-ankB region including the katB promoter | This study |
| pPZ-ankB-470 | pPZ30 containing a 0.47-kb fragment of the ankB upstream region | This study |
| pPZ-ankB-160 | pPZ30 containing a 0.16-kb fragment of the ankB upstream region | This study |
| pUCGM | Gmr; pUC19 plus 850-bp aaC1 cassette | 45 |
Abbreviations used for genetic markers were as described by Holloway et al. (29). mob, mobilization site (ColE1); Tra+, conjugative phenotype; oriT, origin of transfer (RK2); Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Gmr, gentamicin resistance; Kmr, kanamycin resistance.
Growth conditions.
All bacteria were grown from single-colony isolates or overnight cultures in L broth or M9 minimal medium. Liquid cultures were grown at 37°C with shaking at 300 rpm or on a roller wheel at 70 rpm unless otherwise indicated. Culture volumes were 1/10 of the total Erlenmeyer flask volume to ensure proper aeration.
Cloning and sequence analysis of ankB.
Steps involved in the cloning of the P. aeruginosa PAO1 ankB and radA genes are described in Results. DNA sequencing was performed on both strands using the PRISM Dye Deoxy Terminator Cycle Sequencing Kit and analyzed on an ABI model 373A DNA sequencer. Oligonucleotides for DNA sequencing reactions and PCR analysis were synthesized in the DNA Core Facilities in the Department of Molecular Genetics, Biochemistry and Microbiology at the University of Cincinnati College of Medicine or in the Department of Microbiology at the University of Colorado Health Sciences Center. Sequence analyses were performed using either MacVector 6.5.1 (Eastman Chemical Co., New Haven, Conn.), Sequencher 3.1 (Gene Codes Corp., Ann Arbor, Mich.), or Gene Runner (Hastings Software, Inc.) software. Amino acid alignments were performed using either the BLASTP program provided by the National Center for Biotechnology Information (1) or the Align Plus 3 Global Alignment Program (Sci-Ed Software, Durham, N.C.). Potential membrane-spanning domains (MSDs) of AnkB and other bacterial ankyrin-like proteins were determined using TOP-PRED II (57) or the SOSUI program (http://www.tuat.ac.jp/∼mitaku/adv_sosui/submit.html).
Manipulation of recombinant DNA and RNA.
Plasmid DNA was transformed into E. coli DH5α-MCR (Gibco-BRL, Gaithersburg, Md.) or SM10 (48). 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (40 μg/ml) was routinely added to agar medium to detect the presence of insert DNA. Restriction endonuclease, alkaline phosphatase (AP), Klenow fragment, T4 DNA polymerase, and T4 DNA ligase were used as specified by the vendor (Gibco-BRL). Plasmid DNA was isolated using miniprep kits from Qiagen. DNA fragments used for cloning or in the construction of radiolabeled probes were recovered from agarose gels using SeaPlaque low-melting-point agarose (FMC BioProducts, Rockland, Maine) or with the GeneClean II kit (BIO 101, Inc., La Jolla, Calif.). RNA was isolated by the hot-phenol method and analyzed by RNase protection assays as described in detail elsewhere (3). Radiolabeled riboprobes were generated from cloned DNA fragments (Table 1) using an in vitro runoff transcription system (Promega), and excess probe was hybridized to 20 μg of total RNA.
Overexpression of recombinant AnkB in E. coli.
Overexpression of recombinant AnkB as a His6-tagged protein in E. coli was performed using the T7 promoter-T7 RNA polymerase system (53). A 0.53-kb fragment containing the ankB gene minus the first 19 codons comprising its signal sequence was amplified using primers (XhoI)-ctCGAGGTGCATGGGGTCGAGGT and (BamHI)-ggaTCCAGACTAGCCCAGCAGGC (bases in the XhoI and BamHI restriction sites are underlined, and nonmatching bases near the 5′ end are in lowercase type). This PCR product was cloned into pCRII (Invitrogen), sequenced, and directionally cloned as an XhoI-BamHI fragment into pET14b (Novagen). The resulting plasmid, pET14b-ankB, allowed T7-inducible expression of AnkB containing an in-frame His6 tag at its N terminus spaced by a thrombin cleavage site encoded on pET14b. Similarly, a 0.485-kb fragment containing the ankB gene minus the first 17 codons and lacking the last two codons was amplified using the primers (NheI)-GCTaGCGAGGTGCATGGGGTCGA and (NotI)-GCgGCCGCAGTGCCGTTCAGTTC and subsequently cloned as an NheI-NotI fragment into pET23a linearized with NheI and NotI. In the resulting plasmid, pET23-ankB, the AnkB protein was fused in frame to a carboxy-terminal His6 tag encoded by pET23a. Recombinant plasmids were first selected in E. coli DH5α-MCR and then transformed into E. coli BL21(λDE3), which harbors a single genomic copy of the T7 RNA polymerase gene under control of the lacUV5 promoter. These bacteria were grown in 1 liter of L broth containing ampicillin at 100 mg/ml to an optical density at 600 nm (OD600) of 0.3. At this point, the synthesis of T7 polymerase was induced by the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and the cells were allowed to grow for an additional 3 h at 37°C. Recombinant AnkB proteins were then purified using a nickel-nitrilotriacetic acid column as specified by the manufacturer (Qiagen).
Construction of ankB::blaM, ankB::phoA, and ankB::lacZ fusion plasmids.
For construction of a translational ankB::blaM fusion plasmid, the N-terminal region of ankB was PCR amplified to create a 551-bp fragment harboring the entire ankB gene. This fragment was cut with NaeI, and the 162-bp 5′ fragment containing the predicted MSD and a portion of the predicted periplasmic region of AnkB was ligated into the SmaI site of pKM1 (10). For construction of a translational ankB::phoA fusion plasmid, a 174-bp 5′ fragment of the ankB gene containing an AflIII restriction site was first cloned into a T/A version of pKS(−) (35). The ankB fragment was removed from this plasmid and cloned into the NcoI-SmaI sites of pEX30 (H. P. Schweizer). pPHO7, a broad-host-range phoA-containing plasmid encoding AP (24), was first cut with BamHI, blunted, and then excised with PstI. The resulting phoA gene was then ligated to pEX-ankB that had been previously digested with EcoRI (blunted) and PstI, allowing for directional cloning of ankB in frame with phoA. The resulting construct fuses the first 58 amino acids of AnkB with PhoA. Translational fusions of ankB to the lacZ reporter gene were constructed as follows. PCR products were generated with primer (BamHI)-ggatCCTCGCATCGTCGTCATCTC containing a BamHI site after the third codon of the ankB gene and either primer TACAAGGCTGACAGCGACT (0.16 kb upstream), primer GCAGCGAGGTCAATACCGTC (0.47 kb upstream), or primer CTTGGAACTGCGCCATGCAG (1.9 kb upstream). The PCR products were cloned into pCRII and directionally ligated as EcoRI-BamHI fragments into pPZ30 linearized with EcoRI and BamHI, yielding pPZ-ankB-160, pPZ-ankB-470, and pPZ-ankB-1900, respectively.
Construction of isogenic P. aeruginosa ankB and katB ankB mutants.
To generate a clean ΔankB deletion mutant, the DNA sequences flanking ankB were PCR amplified using primers GCAGCGAGGTCAATACCGTC and (KpnI)-ggtaCCGGTTGCGATCAATCCTGG (0.408-kb ankB upstream fragment) and primers (XbaI)-tctaGATGCGTTGGGCAACAGCGT and (HindIII)-aaGCTTCAACCCCTGCAGCGCCA (0.334-kb ankB downstream fragment). The two PCR products were cloned into pCRII, sequenced, and cloned as an EcoRI-KpnI ankB upstream fragment and an XbaI-HindIII ankB downstream fragment on each side of and in proper orientation to a 1.7-kb aac1 (Gmr) cartridge in pUC-Gm, yielding pUCΔankB::Gm. A 2.8-kb PvuII fragment of pUCΔankB::Gm containing the ΔankB::Gm construct was cloned into the SmaI site of the suicide vector pEX100T (46), and the resulting pEX100T-ΔankB::Gm was transformed into E. coli SM10, which then served as the donor strain in a biparental plate mating (16 h, 37°C) with P. aeruginosa PAO1 or with PAO1 ΔkatA::Tc (35). The mating mixture was plated on brain heart infusion agar containing gentamicin (75 μg ml−1) and irgasan (Ciba-Geigy) (50 μg ml−1) as a counterselective agent. Several colonies were grown to late logarithmic phase in L broth, and serial dilutions were spread onto L agar containing gentamicin (75 μg ml−1) and sucrose (5%). Chromosomal DNA from individual colonies was evaluated for deletion of the ankB gene by PCR and Southern blot analysis (data not shown). A PAO1 ΔkatB ankB deletion mutant was obtained as follows. A 2.7-kb fragment containing the katB-ankB region was PCR amplified using the primers CTTGGAACTGCGCCATGCAG and GCTTCAACCCCTGCAGCGCCA and cloned into pCRII. A 1.75-kb SstII fragment containing most of the katB and ankB genes was excised and replaced by a 1.2-kb Gmr cartridge by blunt-end ligation after filling in the ends with Klenow enzyme. A 2.6-kb PvuII fragment of the resulting plasmid, pCRII-ΔkatB ankB::Gm, was cloned into the SmaI site of pEX100T, yielding the donor plasmid pEX100T-ΔkatB ankB::Gm for a mating as described above.
Cell fractionation: periplasm, cytoplasm, and cytoplasmic membrane.
Bacteria were grown aerobically in 1 liter of L broth at 37°C until the OD600 reached 0.6. At this point, organisms were treated with 350 μM paraquat (Sigma) for 1 h to stimulate katB-ankB transcription prior to harvesting the bacteria by centrifugation at 10,000 × g for 10 min. The pellet was washed twice in ice-cold 10 mM Tris-HCl–30 mM MgCl2 (pH 7.3) (Tris-Mg) and resuspended in 1/25 the volume of the same buffer. Chloroform (15 μl/ml of buffer) was then added, and the cells were incubated on ice for an additional 15 min followed by dilution with an additional 1 ml of buffer. The bacteria were pelleted by centrifugation at 10,000 × g for 10 min at 4°C, and the supernatant was further subjected to centrifugation at 150,000 × g for 2 h at 4°C to remove potential contaminating membranes. Finally, the periplasmic preparation was stored on ice. The bacteria were washed again, resuspended in 5 ml of Tris-Mg, and disrupted by sonication with a Heat Systems-Ultrasonics (Farmingdale, N.Y.) model W-225 sonicator equipped with microtip at output setting 5 at 4°C. Cell debris and membrane fractions were clarified by centrifugation at 35,000 × g for 1 h at 4°C. The supernatant was designated the cytoplasmic fraction.
For preparation of cytoplasmic membrane proteins, the above-described growth conditions were employed. The pellet of paraquat-stimulated bacteria was resuspended in 10 ml of 10 mM Tris-HCl (pH 7.5) (T buffer), containing 20% sucrose, treated with 0.5 mg of DNase I (Gibco) and RNase (Sigma) per ml, and incubated for 15 min at 22°C with periodic agitation. The suspension was placed on ice for 20 min, followed by two passages through a French pressure cell at 1,200 lb/in2 at 4°C. The cell debris was removed by centrifugation at 5,500 × g for 10 min at 4°C. The supernatant containing the membranes was subjected to a two-stage sucrose gradient centrifugation. The first stage involved layering 10 ml of membranes on 14 ml of 50% sucrose and 14 ml of 70% sucrose in T buffer. The membranes were then separated by centrifugation in an SW28 swinging-bucket rotor at 130,000 × g for 17 h at 4°C. Cytoplasmic membranes (the top red band) and outer membranes (the bottom white band) were collected by dropwise collection and diluted to 7 ml in cold T buffer–20% sucrose. The second centrifugation stage involved layering the membrane fractions on 9 ml of 52% sucrose, 9 ml of 58% sucrose, 9 ml of 64% sucrose, and 3 ml of 70% sucrose, each prepared in T buffer. The membranes were again separated by centrifugation in an SW28 swinging bucket rotor at 130,000 × g for 17 h at 4°C. The purified cytoplasmic and outer membrane fractions were concentrated by dropwise collection and dialyzed exhaustively against T buffer for 17 h at 4°C. The purity of each cellular fraction was gauged by measuring enzymatic activities specific for the cytoplasm, periplasm, and inner membrane. Stripping of AP activity from cytoplasmic membrane preparations was accomplished by incubating the membranes in either T buffer (control) or 0.1 M Na2CO3 for 30 min on ice. Thirty milliliters of the membrane suspension was carefully layered on 4 ml of 0.3 M sucrose in T buffer and centrifuged at 150,000 × g for 2 h at 4°C. The pellet was solubilized in 2% Triton X-100 in T buffer and assayed for AP activity as described below. Glucose-6-phosphate dehydrogenase, a cytoplasmic marker, was assayed by monitoring the reduction of NADP+ at 340 nm as previously described (33). β-Lactamase, one periplasmic marker, was assayed by the decomposition of 50 μM nitrocefin (CalBiochem, La Jolla, Calif.) at 486 nm (34). AP, another periplasmic marker, was assayed by monitoring the production of p-nitrophenol through hydrolysis of 1 mM p-nitrophenyl phosphate (Sigma) (20). AP activity in bacterial colonies on L-agar plates containing 10 mM potassium phosphate (pH 7.0) to inhibit endogenous AP activity was monitored using 5-bromo-4-chloro-3-indolyl phosphate (BCIP) (40 μg/ml) in the medium. Succinate dehydrogenase, a cytoplasmic membrane marker, was assayed by monitoring the reduction of dichlorophenol-indophenol as previously described (12).
TEM.
Cells were harvested from exponential-growth-phase cultures, treated with 1 mM H2O2 to induce the katB-ankB operon, and negatively stained for transmission electron microscopy (TEM). For staining, the cells were put on a carbon- and Formvar-coated 200-mesh TEM grid, stained for 15 s with 2% (wt/vol) uranyl acetate, and blotted dry. The negatively stained bacteria were imaged using a Philips EM300 instrument operating at 60 kV under standard conditions with the liquid nitrogen anticontaminator in place.
Circular dichroism spectropolarimetry.
Circular dichroism spectra of purified recombinant AnkB proteins (AnkB-23a and AnkB-14b) were obtained using a JASCO J-710 spectropolarimeter calibrated with d10-camphorsulfonic acid. All samples were dissolved in 10 mM sodium phosphate (pH 7.0) and equilibrated at 22°C for 10 min prior to data collection. The spectra represent the means of four independent scans obtained at a scanning rate of 20 nm min−1. Samples were added to a water-jacketed, cylindrical quartz cuvette with a 0.05-cm path length. Protein content was calculated based upon the equation [protein]μg/ml = 144(A215 − A225), using a 1-cm light path. Percent α-helix was estimated from [θ]222 as described by Greenfield and Fasman (23).
Cell extract preparation and biochemical assays.
Routine cell extracts were prepared from bacteria harvested by centrifugation at 10,000 × g for 10 min at 4°C. Bacteria were washed twice in ice-cold 50 mM potassium phosphate buffer (pH 7.0) and sonicated in an ice-water bath for 10 s as described above. The sonicate was then clarified by centrifugation at 13,000 × g for 10 min at 4°C. Cell extract preparation for native gel electrophoresis was performed as described above except that 50 mM T buffer was used as the diluent. Catalase activity was assayed spectrophotometrically at 240 nm by monitoring the decomposition of 18 mM H2O2 using a Spectronic Genesys 5 spectrophotometer (Spectronic Unicam, Rochester, N.Y.) (4). The concentration of H2O2 was determined using an extinction coefficient of 43.6 mM. One unit of activity is that which degrades 1 μmol of H2O2 per min at 23°C. Nondenaturing polyacrylamide gels (5%) were stained for catalase activity according to the method of Wayne and Diaz (58). β-Galactosidase assays were performed using o-nitrophenyl-β-d-galactopyranoside, and the results were expressed as international units with an extinction coefficient for o-nitrophenyl-β-d-galactopyranoside of 3.1 mM (37). Protein concentrations in cell extracts were estimated by the method of Bradford (8), using bovine serum albumin as a standard.
Nucleotide sequence accession number.
The P. aeruginosa katB, ankB, and radA sequences have been assigned GenBank accession number U89384.
RESULTS
Sequence analysis downstream of the P. aeruginosa katB gene: identification of ankB, encoding an ALP, and radA, a DNA repair protein.
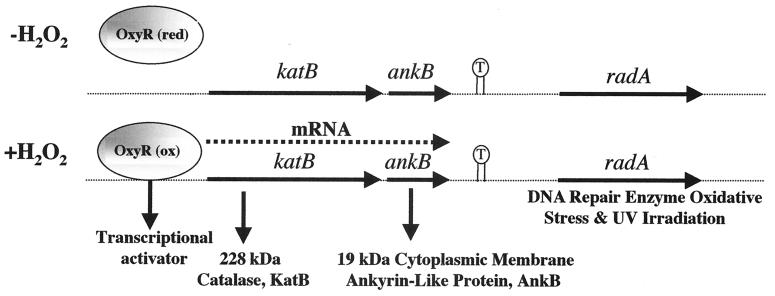
In a previous study, we cloned and characterized the katB gene, encoding a 228-kDa tetrameric catalase (11). The katB gene was recently found to be under the control of the global transactivator OxyR (39), and its transcription is markedly induced upon exposure to H2O2 or the redox-cycling agent paraquat (11). DNA sequence analysis downstream of the katB locus revealed a small, 549-bp ORF (Fig. 1). This ORF, ankB, is predicted to encode a protein of 183 amino acids with a monomeric molecular mass of 19,360 Da and a pI of 5.55. The deduced amino acid sequence demonstrated the highest similarity with genes harboring ank repeats in other bacteria (13, 17, 21, 32) and various ankyrin or ALP genes in eukaryotes (5) (see below). When we sequenced further downstream of the ankB locus, we discovered a large inverted repeat spanning 33 bp (Fig. 1). We then identified a gene downstream of the inverted repeat that was 70% identical to the radA gene of E. coli (49). The radA gene in E. coli encodes a protein that repairs DNA damaged (alkylated) by gamma irradiation. However, because virtually nothing is known of the function of ALPs in bacteria, we chose to focus our efforts on the functional characterization of P. aeruginosa AnkB.
FIG. 1.
Gene map of the ∼5.2-kb insert of pSMB1 containing the katB, ankB, and radA genes. The functions of the gene products are also given. We have previously shown that OxyR, a 34-kDa transactivator, responds to H2O2 by activating katB-ankB (39). T, 33-amino-acid inverted repeat that could represent the transcriptional terminator for the katB-ankB operon.
Amino acid comparison of AnkB with other bacterial ALPs.
Ankyrins are proteins that are characterized by 33-amino-acid ank repeats that are thought to represent an ancient motif that has evolved to allow for functional diversity without compromising specificity (5). Each of the five bacterial ALPs in Fig. 2 possess the 33-amino-acid tandem, nonidentical ank repeats. These repeats are based upon the consensus ank repeat motif put forth by Michaely and Bennett (36). AnkB (PaAnkB), not surprisingly, is similar to AnkF (PsAnkF, in the related plant pathogenic species P. syringae, strain 61 [32]; PsAnkB [in P. syringae B301D; accession no. AF133262]; and PfAnkB [in P. fluorescens; U83328]) in that it possesses four tandem repeats and an 18-residue signal sequence (Fig. 2). However, the Ank signal peptides from P. syringae differ from those of PaAnkB and PfAnkB in that they are followed by a proteolytic proline-threonine-box (hinge [32]). Interestingly, PaAnkB and PfAnkB, but not PsAnkF and PsAnkB, possess an RGD motif, which, in eukaryotes, is responsible for physical interaction with integrins, structural proteins that play a role in the homing and action of immune cells. Other bacterial ALPs (some of which are not listed in Fig. 2) from V. cholerae (http://www.tigr.org/.) (VcAnkB), S. argillaceus (U43537) (SaAnk), C. vinosum (17) (CvAnkA), and S. liquefaciens (21) (SlPhlB, for phospholipase A1-related ALP) possessed longer putative signal sequences (25 to 28 residues), while the ALP from R. leguminosarum (AJ243305) (R1Ank) contained a very short N-terminal MSD of 12 residues. The ALPs of S. verticillus (13) (SvAnkA and SvAnkB) possessed only two 33-amino-acid ank repeats. All other identified bacterial ALPs contain a putative cleavage site following an MSD either in the middle or close to the C terminus, or they do not contain a hydrophobic region long enough to incorporate into the membrane.
FIG. 2.
Alignment of the deduced amino acids from genes coding for bacterial ankyrins PaAnkB (P. aeruginosa; accession no. U59457), PsAnkF (P. syringae; U16026), CvAnkA (C. vinosum; L13419), SlPhlB (S. liquefaciens; P18954), and SvAnkA (S. verticillus; L26954). The 33-amino-acid tandem repeats (underlined) were revealed by using the Ank motif of conserved residues (boldface) as identified by Bennett using the erythrocyte ank repeat consensus sequence (5). Proposed signal sequences are indicated by a double underline. The conserved ank repeat sequences for erythrocyte ankyrin are given below the selected bacterial ALP sequences. RBC, erythrocyte concensus ank repeat sequence.
AnkB is a periplasmic protein: AnkB–β-lactamase and AnkB-alkaline phosphatase protein fusion analysis.
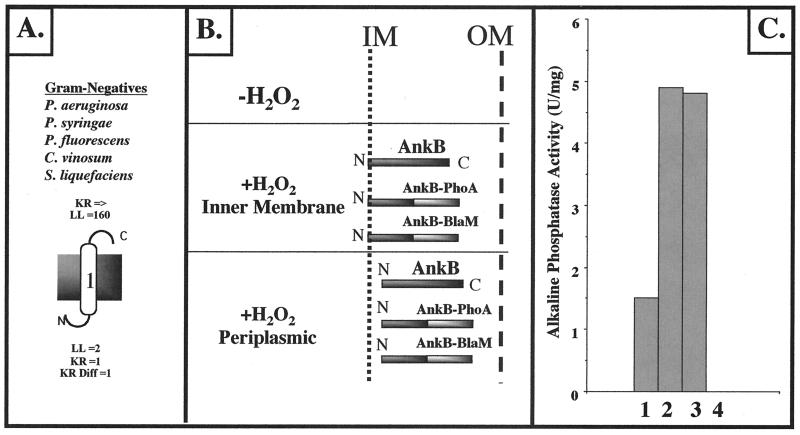
Using the membrane topology program TopPred 3.0, it was predicted that the cytoplasmic N terminus of P. aeruginosa AnkB (3 amino acids) is followed by a 20-amino-acid stretch that is predicted to be the hydrophobic inner-MSD (for a von Heijne schematic, see Fig. 3A) (TMRGWILAGLLLAALAAQAEVH), with the remaining portion of the protein (being highly hydrophilic) predicted to reside in the periplasm. The ALPs of other selected gram-negative bacteria, including C. vinosum, S. liquefaciens, and other Pseudomonas species, are also predicted to span the cytoplasmic membrane (Fig. 3A). To test whether the predicted cytoplasmic membrane location was correct, we constructed AnkB-BlaM and AnkB-PhoA protein fusions linking the C terminus of AnkB with both reporters (Fig. 3B). E. coli and P. aeruginosa harboring an ankB::blaM fusion plasmid were resistant to ampicillin and carbenicillin, respectively. In addition, E. coli and P. aeruginosa harboring pEX30-ankB::phoA were blue on indicator plates containing BCIP, suggesting that the C terminus of AnkB resides in the periplasm and confirming the AnkB–β-lactamase fusion plasmid results. To determine the precise location of AnkB within the cell, membrane fractionation techniques (see Materials and Methods) were employed. Cytoplasm, periplasm, cytoplasmic membrane, and outer membrane fractions were assayed for AP activity (Fig. 3C). The vast majority of AP activity was detected in the periplasmic space and the cytoplasmic membrane. The contaminating AP activity in the cytoplasm is likely due to overexpression and problems associated with transport of a larger-than-normal AnkB-PhoA protein. To show that AnkB traverses the cytoplasmic membrane and is not simply nonspecifically bound to it, cytoplasmic membranes were stripped by treatment with Na2CO3 at high pH (56). Our results showed that AnkB is tightly bound to the cytoplasmic membrane (data not shown). These results suggest that AnkB first exists as a cytoplasmic membrane protein, followed by proteolytic cleavage via a putative LepB signal peptidase cleavage site (GEVHG) to finally reside in the periplasmic space (42).
FIG. 3.
Cellular localization of AnkB in P. aeruginosa. (A) Predicted cytoplasmic membrane organization of P. aeruginosa AnkB bacterial ankyrin-like proteins from P. syringae, P. fluorescens, S. liquefaciens, and C. vinosum based upon the positive-inside-rule algorithm developed by von Heijne (57). For the P. aeruginosa AnkB protein, the large number 1 indicates the predicted single MSD. N, N terminus; C, C terminus; LL, loop length; KR, number of lysine and arginine residues; KR Diff, positive charge difference. (B) Schematic diagram of AnkB–β-lactamase and AnkB-PhoA protein fusions in both E. coli and P. aeruginosa PAO1. In both cases, organisms expressing AnkB–β-lactamase were resistant to ampicillin (E. coli) or carbenicillin (P. aeruginosa). Organisms expressing AnkB-PhoA were found to hydrolyze the alkaline phosphatase substrate BCIP in L-agar plates. IM, inner membrane; OM, outer membrane. (C) AP activity in cellular fractions of P. aeruginosa ankB harboring pEX30-ankB::phoA. Bar 1, cytoplasm; bar 2, periplasm; bar 3, cytoplasmic membrane; bar 4, outer membrane.
Overexpression of AnkB in E. coli: AnkB secondary structure is predominantly α-helical.
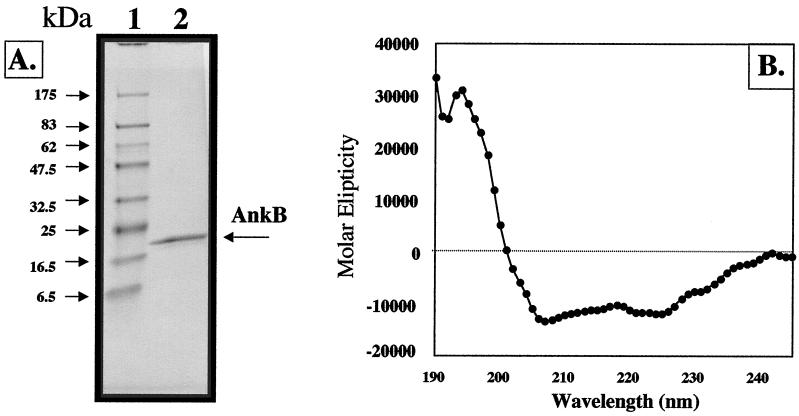
To obtain some preliminary structural analysis of AnkB, we overexpressed and purified two recombinant AnkB proteins with N-terminal (pET23a-AnkB) and C-terminal (pET14b) His6-tagged fusions in E. coli BL21(λDE3) without their predicted MSDs. Figure 4A demonstrates purified pET23a-AnkB. Gorina and Pavletich revealed that the secondary structure of an ank repeat in protein 53BP2, which binds to the p53 tumor suppressor, consists of an L-shaped structure with a β-turn and 2 α-helices (22). Circular dichroism spectropolarimetric analysis of recombinant AnkB-23a suggested that AnkB is ∼60 to 70% α-helical (Fig. 4B). This structure is consistent with the 66% α-helical nature of the ank repeats of the 53BP2 protein (22).
FIG. 4.
Overexpression (A) and circular dichroism analysis (B) of recombinant AnkB proteins. (A) E. coli BL21(λDE3) harboring pET23-ankB-480 was grown aerobically in L broth to mid-logarithmic phase and treated with 1 mM IPTG for 3 h at 37°C. After Ni2+-nitrilotriacetic acid purification, purified protein was separated by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis and the gel was stained with Coomassie blue R-250. Lane 1, molecular mass standard; lane 2, 15 μg of AnkB-480. (B) Circular dichroism spectrum of AnkB-480, using 100 μg ml−1 in 10 mM sodium phosphate (pH 7.0) at 23°C.
Polycistronic nature of katB and ankB: regulation by H2O2.
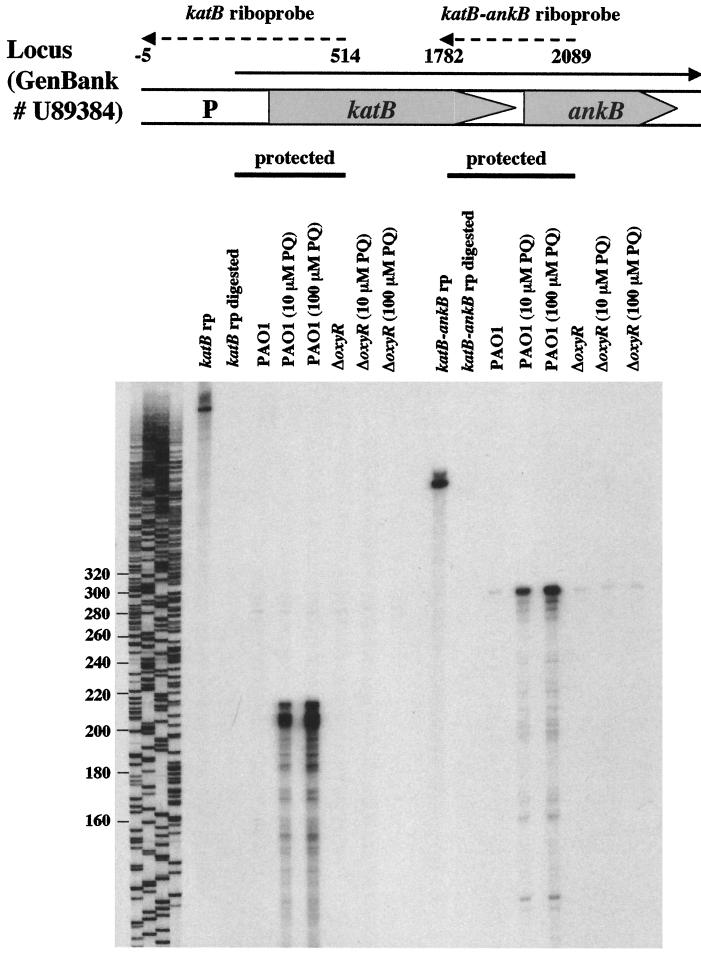
To determine if katB and ankB are part of a small operon, RNase protection assays were performed. Figure 5 demonstrates that transcription of both katB and ankB is stimulated by paraquat in a concentration-dependent fashion. The transcriptional start site was found to be a G 227 bp upstream of the katB start codon. Furthermore, transcription of katB-ankB is dependent upon OxyR, since no katB and very little ankB transcript could be detected in an oxyR mutant. These results were also confirmed using ankB::lacZ reporter fusion studies (data not shown).
FIG. 5.
RNase protection assays indicate that katB and ankB comprise an operon and are regulated by OxyR. Riboprobes specific for the katB promoter (katB rp) and for the katB-ankB overlapping region (katB-ankB rp) were used to detect the corresponding transcripts in P. aeruginosa PAO1 or oxyR mutant total RNA isolated during the exponential growth phase in aerobic M9 minimal medium. Paraquat (PQ) was added to final concentrations of 10 and 100 μM 1 h prior to harvest as indicated. Also shown are the digested probes in the absence of any P. aeruginosa RNA as a control. A DNA sequencing reaction was run in parallel and served as a size marker. Numbers on the left are base pairs.
Phenotypes of a P. aeruginosa ankB mutant. (i) Normal cell size and shape.
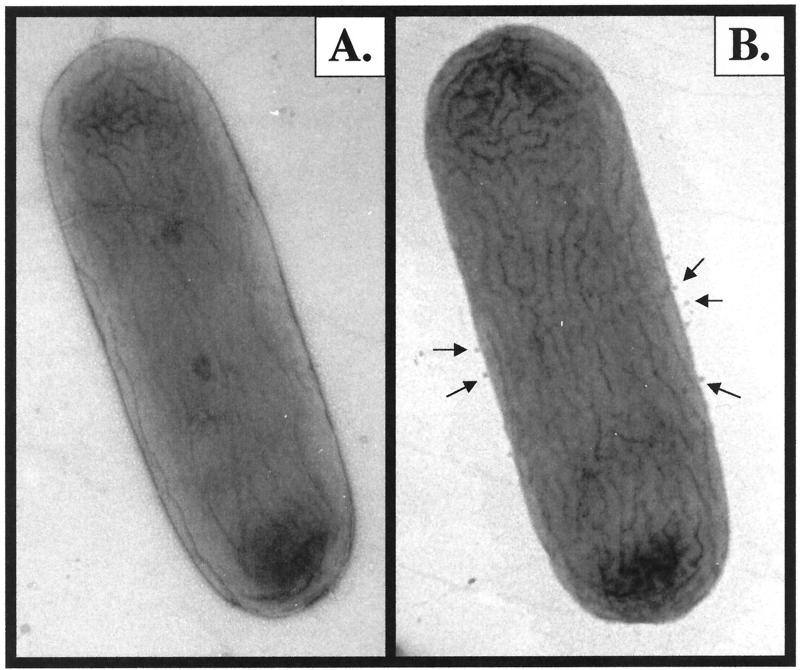
Humans with hereditary spherocytosis (HS) suffer from an ankyrin deficiency. Erythrocytes from individuals with HS lack deformability and stability (40) and are unable to pass through capillaries, resulting in hemolytic anemia and hypersensitivity to osmotic lysis. This disorder has been reproduced in nb/nb (normoblastosis, ankyrin-deficient) mice (7), which have a severe hemolytic anemia throughout life (41). In these settings, it is predicted that a loss of ankyrin from the lipid bilayer causes a reduction in the critical surface area/volume ratio, leading to a shift in the morphology of erythrocytes from discoidal to spherical. Thus, there is a definitive structural role for ankyrins in erythrocytes. In contrast to the case for HS erythrocytes, the ultrastructure of wild-type and ankB mutant bacteria was observed by TEM and no significant differences in overall cell shape were found (Fig. 6). However, the ankB mutant produced more membrane vesicles than wild-type bacteria (Fig. 6B). This implies that a fundamental difference exists between the surfaces of the wild type and the ankB mutant and that their ability to package periplasmic constituents in natural membrane vesicles has changed (i.e., the ankB mutant has more packaging potential). The difference in quantities in membrane vesicles has been confirmed by thin sections (6).
FIG. 6.
Ultrastructural analysis of wild-type PAO (A) and PAO ankB mutant (B) bacteria. Bacteria were grown aerobically in L broth to mid-exponential growth phase and treated with 1 mM H2O2 for 1 h at 37°C to stimulate transcription of katB-ankB. Organisms were then prepared for TEM examination as described in Materials and Methods. The arrows in panel B point to the larger number of membrane vesicles being produced by the ankB mutant. The width of the cells is ∼800 nm.
(ii) Enhanced sensitivity to H2O2.
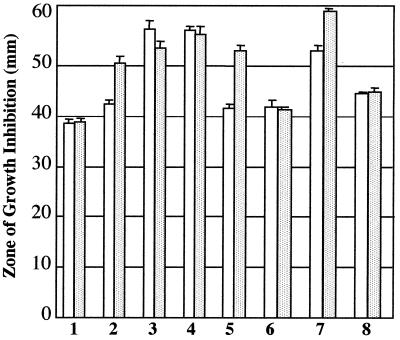
Because we found that ankB is part of a small operon with katB, we postulated that its gene product may play a role in resistance to H2O2. To test this hypothesis, the wild type and ankB, katB, and katB ankB mutants were screened for H2O2 sensitivity. As shown in Fig. 7, an ankB mutant was only slightly more susceptible to H2O2 than wild-type organisms (bars 2). However, when the mutant was pretreated with a sublethal dose of H2O2, which activates the katB-ankB operon, sensitivity was increased dramatically (bars 2 versus bars 1 [shaded bars]). Bars 6 demonstrate that provision of a plasmid that allows for constitutive expression of ankB restored wild-type resistance regardless of H2O2 pretreatment. The katB (bars 3) and katB ankB (bars 4) mutants were equally susceptible to H2O2, and more so than the ankB mutant. Interestingly, provision of ankB alone to the katB ankB mutant dramatically helped these organisms resist H2O2 (bars 8 relative to bars 7).
FIG. 7.
Effect of ankB, katB, and katB ankB on sensitivity to H2O2. All bacteria were grown aerobically overnight in M9F medium at 37°C. Fresh prewarmed medium (1 volume of culture in 10-volume flasks) was inoculated with 1/50 of the final culture volume and allowed to reach an OD600 of 0.6. Some bacteria were pretreated with a sublethal (1 mM) dose of H2O2 for 1 h (shaded bars) relative to control bacteria (open bars). The suspensions were diluted 100-fold in 7 ml of M9F 0.6% top agarose kept at 37°C and poured onto M9F plates. Filter paper disks (7 mm) impregnated with 8.8 M H2O2 were placed on the top agar surface. Zones of growth inhibition were measured after a 24-h aerobic incubation at 37°C. Bars: 1, PAO1; 2, ankB; 3, katB; 4, katB ankB; 5, ankB plus pUCP22; 6, ankB plus pankB; 7, katB ankB plus pUCP22; 8, katB ankB plus pankB. Error bars indicate standard errors of the means.
(iii) Absence of AnkB decreases KatB activity.
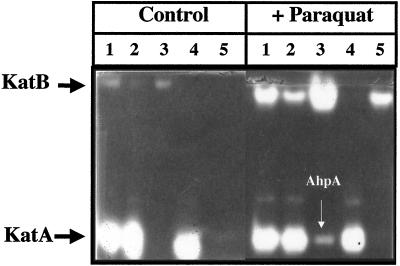
Because of the close proximity of katB and ankB, their organization in a small operon, and the enhanced H2O2 sensitivity of the ankB mutant, we postulated that AnkB could play a role in KatB function. To test this hypothesis, the catalase isozyme profiles of several mutant organisms were examined. Fig. 8 (left panel) shows that the KatB activities of the ankB mutant (lane 2) and a katA ankB (lane 5) mutant are significantly reduced relative to that of wild-type bacteria (lane 1). When transcription of katB was stimulated by the addition of paraquat, there was a robust increase in KatB activity in the wild type (right panel, lane 1) and especially in the katA mutant (right panel, lane 3). The catalase activity band produced in the katA mutant that migrated to the same Rf as KatA could be another, previously undiscovered catalase in P. aeruginosa, although analysis of the recently completed P. aeruginosa genome suggested otherwise (data not shown). We now know that this paraquat-inducible catalase band is one of the alkyl hydroperoxide reductases, AhpA, that possesses weak catalase activity (39). The KatB activities of the ankB mutant (Fig. 8, right panel, lane 2) and a katA ankB mutant (right panel, lane 5) were still reduced relative to that of wild-type bacteria (right panel, lane 1).
FIG. 8.
Absence of AnkB causes a decrease in KatB activity. Bacteria were grown aerobically overnight in L-broth medium at 37°C. Fresh prewarmed medium (1 volume of culture in 10-volume flasks) was inoculated with 1/100 the final culture volume, and the organisms were grown to an OD600 of 0.6. Some bacteria (right panel) were then treated with a sublethal (0.35 mM) (11) dose of paraquat for 1 h, and the others (left panel) served as controls. Cell extracts were prepared, and 10 μg was subjected to nondenaturing polyacrylamide gel electrophoresis (5% polyacrylamide). The gels were then stained for catalase activity (58). Lanes: 1, PAO1; 2, ankB; 3, katA; 4, katB; 5, katA ankB.
(iv) Quantitative effect of AnkB on KatB activity.
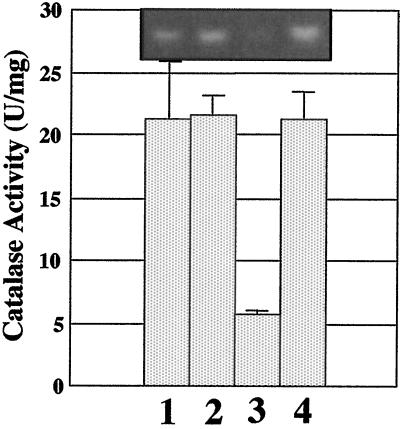
To quantify the effect of AnkB on KatB activity, we examined catalase activity in katA and katA ankB mutants that had been treated with paraquat in stationary-phase culture, where the only catalase activity that can be detected and quantified spectrophotometrically is KatB. As shown in Fig. 9, the KatB activity of a paraquat-treated katA mutant is ∼21.5 U/mg (bar 1). Provision of ankB in trans to the katA mutant had no effect on KatB activity (bar 2). Interestingly, KatB activity in a katA ankB mutant was reduced fourfold (bar 3) relative to that of the katA mutant and was fully complemented by providing ankB in trans (bar 4). There was no observation of the AhpA activity band under these conditions.
FIG. 9.
Quantitative effect of AnkB on KatB activity. Bacteria were grown aerobically overnight in L-broth medium containing 0.35 mM paraquat at 37°C. Catalase activity and activity gel staining were monitored in cell extracts. Bars: 1, katA plus pUCP22; 2, katA plus pankB; 3, katA ankB plus pUCP22; 4, katA ankB plus pankB. Error bars indicate standard errors of the means. The inset photograph is KatB activity staining of cell extracts in a representative native polyacrylamide gel of each strain. It should be noted that no AhpA activity band (see Fig. 8) could be detected under these conditions.
DISCUSSION
The major catalase gene of P. aeruginosa, katA, encoding a constitutive 170-kDa heteromultimer, is positively regulated by iron (35) and maximally expressed in stationary phase, in part through a process of cell-to-cell communication known as quorum sensing (27). Thus, it is not surprising that KatA contributes significant protection against H2O2 in both planktonic and biofilm cultures (26, 27, 35).
In contrast to katA, which is minimally responsive to H2O2, we found in this study that the katB-ankB operon is transcribed dramatically in its presence and requires the global transactivator OxyR (25, 39). When we discovered ankB downstream of katB, we immediately classified its gene product as an ALP because it possessed the characteristic 33-amino-acid ank repeat motifs and showed high similarity to ALPs in the related organisms P. fluorescens and P. syringae (32). Bacterial ALPs differ dramatically from their eukaryotic counterparts in that they contain only the ank repeats, which we believe to be involved in protein-protein interactions (5). Bacterial ALPs should, therefore, lack the structural role in the cell, similar to the function of prototypical erythrocyte spectrin-binding ankyrin. Examination of transmission electron micrographs of the ankB mutant confirmed this assumption (Fig. 6). Due to the polycistronic nature of katB-ankB and the conservation of this operon in the pseudomonads and other proteobacteria such as V. cholerae, we postulated that AnkB might belong to a group of evolutionarily related proteins with a novel, unrecognized function(s), one of which could contribute toward protection against H2O2. Indeed, AnkB appears to play a role in the response of P. aeruginosa to H2O2, because an ankB mutant was more sensitive to it than wild-type organisms (Fig. 7). Furthermore, the enhanced H2O2 sensitivity of an isogenic katB ankB mutant did not change when only katB was provided in trans (39). Although unproven, the nearly fourfold reduction in KatB activity in the ankB mutant suggests that there could be a physical interaction between the two proteins. We found KatB activity in the cytoplasm, periplasm, and cytoplasmic membrane (data not shown). Because AnkB is a cytoplasmic membrane protein whose bitopic integration into the inner membrane ultimately causes its ank repeat domain to reside in the periplasm, we postulated that one function of AnkB may be to bind KatB near inner membrane targets that are sensitive to H2O2 (e.g., F1Fo-ATPase [55]). H2O2 must first enter a protein channel leading to the heme catalytic site of the catalase molecule (47). Without entering this channel, the H2O2 is free to damage cellular components, especially sensitive respiratory chain components and DNA (16). Thus, AnkB may position or anchor KatB so that its H2O2 channel is in the optimal orientation for H2O2 entry. Alternatively, AnkB may serve to stabilize KatB, allowing it to persist longer and function better upon exposure of bacteria to H2O2.
An alternative hypothesis is that AnkB may reinforce the cytoplasmic membrane and prevent crippling of the proton motive force. Microscopic oxygen bubbles could be produced upon H2O2 degradation, thereby increasing cellular turgor pressure. Although unexplained, such cell swelling has been shown in E. coli (38) and in mitochondria treated with H2O2 or agents that generate it (30). Upon H2O2 degradation by catalase, oxygen gas nuclei could be stabilized and even grow in the bacteria at hydrophobic sites. With the production of gas at a rate that saturates the cytoplasm, gas bubbles could readily appear, be stabilized by lipid and/or protein adsorption, and take up considerable volume inside a cell, thereby creating a turgor pressure (31). Thus, AnkB could serve to stabilize the inner membrane against swelling due to the mounting intracellular pressure built by H2O2 degradation. Both hypotheses are being tested experimentally.
ACKNOWLEDGMENTS
The first three authors contributed equally toward completion of this work.
This work was supported by Public Health Service grants AI-40541 (to D.J.H.) and DK-50749 (to K.M.B.) and Cystic Fibrosis grant HASSET98PO (to D.J.H.).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas S, Matsuno K, Fortini M E. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 3.Barton H, Johnson Z, Cox C D, Vasil A I, Vasil M L. Ferric uptake regulator mutants of Pseudomonas aeruginosa with distinct alterations in the iron-dependent repression of exotoxin A and siderophores in aerobic and microaerobic environments. Mol Microbiol. 1996;21:1001–1017. doi: 10.1046/j.1365-2958.1996.381426.x. [DOI] [PubMed] [Google Scholar]
- 4.Beers R F, Jr, Sizer I W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem. 1952;195:133–140. [PubMed] [Google Scholar]
- 5.Bennett V. Ankyrins: adaptors between diverse plasma membrane proteins and the cytoplasm. J Biol Chem. 1992;267:8703–8706. [PubMed] [Google Scholar]
- 6.Beveridge T J. Structures of gram-negative cell walls and their derived membrane vesicles. J Bacteriol. 1999;181:4725–4733. doi: 10.1128/jb.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bodine D M, Birkenmeier C S, Barker J E. Spectrin deficient inherited hemolytic anemias in the mouse: characterization by spectrin synthesis and mRNA activity in reticulocytes. Cell. 1984;37:721–729. doi: 10.1016/0092-8674(84)90408-2. [DOI] [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Breeden L, Nasmyth K. Similarity between cell-cycle genes of budding yeast and fission yeast and the Notch gene of Drosophila. Nature. 1987;329:651–654. doi: 10.1038/329651a0. [DOI] [PubMed] [Google Scholar]
- 10.Broome-Smith J, Spratt B G. A vector for the construction of translational fusions to the TEM β-lactamase and analysis of protein export signals and membrane protein topology. Gene. 1986;49:341–349. doi: 10.1016/0378-1119(86)90370-7. [DOI] [PubMed] [Google Scholar]
- 11.Brown S M, Howell M L, Vasil M L, Anderson A J, Hassett D J. Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J Bacteriol. 1995;177:6536–6544. doi: 10.1128/jb.177.22.6536-6544.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burans J P, Lynn M, Solotorovsky M. Induction of active immunity with membrane fractions from Haemophilus influenzae type b. Infect Immun. 1983;41:285–293. doi: 10.1128/iai.41.1.285-293.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calcutt M J, Schmidt F J. Gene organization in the bleomycin-resistance region of the producer organism Streptomyces verticillus. Gene. 1994;151:17–21. doi: 10.1016/0378-1119(94)90627-0. [DOI] [PubMed] [Google Scholar]
- 14.Chan T A, Chu C A, Rauen K A, Kroiher M, Tatarewicz S M, Steele R E. Identification of a gene encoding a novel protein-tyrosine kinase containing SH2 domains and ankyrin-like repeats. Oncogene. 1994;9:1253–1259. [PubMed] [Google Scholar]
- 15.Davis J, Bennett V. The anion exchanger and Na+K+ ATPase interacts with distinct sites on ankyrin: in vitro assays. J Biol Chem. 1990;265:17252–17256. [PubMed] [Google Scholar]
- 16.Demple B, Linn S. 5,6-saturated lesions in DNA: production by ultraviolet light or hydrogen peroxide. Nucleic Acids Res. 1982;10:3781. doi: 10.1093/nar/10.12.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dolata M M, Van Beeumen J J, Ambler R P, Meyer T E, Cusanovich M A. Nucleotide sequence of the heme subunit of flavocytochrome c from the purple phototrophic bacterium, Chromatium vinosum: a 2.6-kilobase pair DNA fragment contains two multiheme cytochromes, a flavoprotein, and a homolog of human ankyrin. J Biol Chem. 1993;268:14426–14431. [PubMed] [Google Scholar]
- 18.Elkins J G, Hassett D J, Stewart P S, Schweizer H P, McDermott T R. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl Environ Microbiol. 1999;65:4594–4600. doi: 10.1128/aem.65.10.4594-4600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fortini M E, Artavanis-Tsakonas S. Notch: neurogenesis is only part of the picture. Cell. 1993;75:1245–1247. doi: 10.1016/0092-8674(93)90611-s. [DOI] [PubMed] [Google Scholar]
- 20.Garen A, Levinthal C. A fine-structure genetic and chemical study of the enzyme alkaline phosphatase of E. coli I. Purification and characterization of alkaline phosphatase. Biochim Biophys Acta. 1960;38:470. doi: 10.1016/0006-3002(60)91282-8. [DOI] [PubMed] [Google Scholar]
- 21.Givskov M, Olsen L, Molin S. Cloning and expression in Escherichia coli of the gene for extracellular phospholipase A1 from Serratia liquefaciens. J Bacteriol. 1988;170:5855–5862. doi: 10.1128/jb.170.12.5855-5862.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorina S, Pavletich N P. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science. 1996;274:1001–1005. doi: 10.1126/science.274.5289.1001. [DOI] [PubMed] [Google Scholar]
- 23.Greenfield N, Fasman G D. Computed circular dichroism for the evaluation of protein conformation. Biochemistry. 1969;8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 24.Gutierrez C, Devedjian J C. A plasmid facilitating in vitro construction of phoA gene fusions in Escherichia coli. Nucleic Acids Res. 1989;17:3999. doi: 10.1093/nar/17.10.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hassett D J, Alsabbagh E, Parvatiyar K, Howell M L, Wilmott R W, Ochsner U A. A protease-resistant catalase, KatA, released upon cell lysis during stationary phase is essential for aerobic survival of a Pseudomonas aeruginosa oxyR mutant at low cell densities. J Bacteriol. 2000;182:4557–4563. doi: 10.1128/jb.182.16.4557-4563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassett D J, Elkins J G, Ma J-F, McDermott T R. Pseudomonas aeruginosa biofilm sensitivity to biocides: use of hydrogen peroxide as model antimicrobial agent for examining resistance mechanisms. Methods Enzymol. 1999;310:599–608. doi: 10.1016/s0076-6879(99)10046-6. [DOI] [PubMed] [Google Scholar]
- 27.Hassett D J, Ma J-F, Elkins J G, McDermott T R, Ochsner U A, West S E H, Huang C-T, Fredericks J, Burnett S, Stewart P S, McPheters G, Passador L, Iglewski B H. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol Microbiol. 1999;34:1082–1093. doi: 10.1046/j.1365-2958.1999.01672.x. [DOI] [PubMed] [Google Scholar]
- 28.Holloway B W. Genetics of Pseudomonas. Bacteriol Rev. 1969;33:419–443. doi: 10.1128/br.33.3.419-443.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holloway B W, Krishnapillai V, Morgan A F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kakkar P, Mehrotra S, Viswanathan P N. Influence of antioxidants on the peroxidative swelling of mitochrondria in vitro. Cell Biol Toxicol. 1998;14:313–321. doi: 10.1023/a:1007529622874. [DOI] [PubMed] [Google Scholar]
- 31.Kim D. Elastic and viscous properties of fel-phase monolayers measured at the gas-bubble/water interface. Ph.D. thesis. Durham, N.C: Duke University; 1999. [Google Scholar]
- 32.Klotz M G, Anderson A J. Sequence of a gene encoding periplasmic Pseudomonas syringae ankyrin. Gene. 1995;164:187–188. doi: 10.1016/0378-1119(95)00482-l. [DOI] [PubMed] [Google Scholar]
- 33.Klotz M G, Hutcheson S W. Multiple periplasmic catalases in phytopathogenic strains of Pseudomonas syringae. Appl Environ Microbiol. 1992;58:2468–2473. doi: 10.1128/aem.58.8.2468-2473.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kraft A R, Prabhu J, Ursinus A, Holtje J V. Interference with murein turnover has no effect on growth but reduces beta-lactamase induction in Escherichia coli. J Bacteriol. 1999;181:7192–7198. doi: 10.1128/jb.181.23.7192-7198.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma J-F, Ochsner U A, Klotz M G, Nanayakkara V K, Howell M L, Johnson Z, Posey J, Vasil M L, Monaco J J, Hassett D J. Bacterioferritin A modulates catalase A (KatA) activity and resistance to hydrogen peroxide in Pseudomonas aeruginosa. J Bacteriol. 1999;181:3730–3742. doi: 10.1128/jb.181.12.3730-3742.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michaely P, Bennett V. The ANK repeat: a ubiquitous motif involved in macromolecular recognition. Trends Cell Biol. 1992;2:127–130. doi: 10.1016/0962-8924(92)90084-z. [DOI] [PubMed] [Google Scholar]
- 37.Miller J H. A short course in bacterial genetics. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 72–74. [Google Scholar]
- 38.Minakami H, Fridovich I. Effects of paraquat on cultures of Escherichia coli: turbidity versus enumeration. Free Rad Biol Med. 1990;8:387–391. doi: 10.1016/0891-5849(90)90105-r. [DOI] [PubMed] [Google Scholar]
- 39.Ochsner U A, Vasil M L, Alsabbagh E, Parvatiyar K, Hassett D J. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J Bacteriol. 2000;182:4533–4544. doi: 10.1128/jb.182.16.4533-4544.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palek J, Lambert S. Genetics of the red cell membrane. Semin Hematol. 1990;27:290–332. [PubMed] [Google Scholar]
- 41.Peters L L, Turtzo L C, Birkenmeier C S, Barker J E. Distinct fetal Ank-1 and Ank-2 related proteins and mRNAs in normal and nb/nb mice. Blood. 1993;81:2144–2149. [PubMed] [Google Scholar]
- 42.Pugsley A P. The complete secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schweizer H P. Improved broad-host-range lac-based plasmid vectors for the isolation and characterization of protein fusions in Pseudomonas aeruginosa. Gene. 1991;103:87–92. doi: 10.1016/0378-1119(91)90396-s. [DOI] [PubMed] [Google Scholar]
- 44.Schweizer H P. A method for construction of bacterial hosts for lac-based cloning and expression vectors: α-complementation and regulated expression. BioTechniques. 1994;17:452–456. [PubMed] [Google Scholar]
- 45.Schweizer H P. Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–833. [PubMed] [Google Scholar]
- 46.Schweizer H P, Hoang T T. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene. 1995;158:15–22. doi: 10.1016/0378-1119(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 47.Sevinc M S, Mate M J, Switala J, Fita I, Loewen P C. Role of the lateral channel in catalase HPII of Escherichia coli. Protein Sci. 1999;8:490–498. doi: 10.1110/ps.8.3.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon R, Priefer U, Puehler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 49.Song Y, Sargentini N J. Escherichia coli DNA repair gene radA and sms are the same gene. J Bacteriol. 1996;178:5045–5048. doi: 10.1128/jb.178.16.5045-5048.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spence A M, Coulson A, Hodgkin J. The product of fem-1, a nematode sex-determining gene, contains a motif found in cell cycle control proteins and receptors for cell-cell interactions. Cell. 1990;60:981–990. doi: 10.1016/0092-8674(90)90346-g. [DOI] [PubMed] [Google Scholar]
- 51.Srinivasan Y, Elmer L, Davis J, Bennett V, Angelides K. Ankyrin and spectrin associate with voltage-dependent sodium channels in brain. Nature. 1988;333:177–180. doi: 10.1038/333177a0. [DOI] [PubMed] [Google Scholar]
- 52.Srinivasan Y, Lewallen M, Angelides K J. Mapping the binding site of ankyrin for the voltage-dependent sodium channel from brain. J Biol Chem. 1992;267:7483–7489. [PubMed] [Google Scholar]
- 53.Studier F W, Moffatt B H. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 54.Takemura H, Horinouchi S, Beppu T. Suppression of an ethanol-sensitive mutation of Acetobacter pasteurianus by overexpression of the his1 gene encoding histidinol phosphate aminotransferase. J Ferment Bioeng. 1993;76:224–228. [Google Scholar]
- 55.Tamarit J, Cabiscol E, Ros J. Identification of the major oxidatively damaged proteins in Escherichia coli cells exposed to oxidative stress. J Biol Chem. 1998;273:3027–3032. doi: 10.1074/jbc.273.5.3027. [DOI] [PubMed] [Google Scholar]
- 56.van de Goor J, Kelly R B. Association of Drosophila cysteine string proteins with membranes. FEBS Lett. 1996;19:251–256. doi: 10.1016/0014-5793(96)00026-9. [DOI] [PubMed] [Google Scholar]
- 57.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 58.Wayne L G, Diaz G A. A double staining method for differentiating between two classes of mycobacterial catalase in polyacrylamide gels. Anal Biochem. 1986;157:89–92. doi: 10.1016/0003-2697(86)90200-9. [DOI] [PubMed] [Google Scholar]