Abstract
Genome-wide analysis of cell-free DNA methylation profile is a promising approach for sensitive and specific detection of many cancers. However, scaling such assays for clinical translation is impractical because of the high cost of whole-genome bisulfite sequencing. We show that the small fraction of GC-rich genome is highly enriched in CpG sites and disproportionately harbors most of the cancer-specific methylation signature. Here, we report on the simple and effective heat enrichment of CpG-rich regions for bisulfite sequencing (Heatrich-BS) platform that allows for focused methylation profiling in these highly informative regions. Our novel method and bioinformatics algorithm enable accurate tumor burden estimation and quantitative tracking of colorectal cancer patient’s response to treatment at much reduced sequencing cost suitable for frequent monitoring. We also show tumor epigenetic subtyping using Heatrich-BS, which could enable patient stratification. Heatrich-BS holds great potential for highly scalable screening and monitoring of cancer using liquid biopsy.
Heatrich-BS enables low-cost, genome-wide epigenetic profiling of cell-free DNA for noninvasive cancer detection and monitoring.
INTRODUCTION
Recent studies have demonstrated the promising use of methylation profiling in cell-free DNA (cfDNA) for multicancer detection, leveraging on tissue- and cancer-specific methylation patterns (1, 2). Compared to mutation-based circulating tumor DNA detection methods, where there are a limited number of recurrent mutations available to distinguish between tumor and normal cfDNA, tissue- and cancer-specific DNA methylation patterns are more abundant. A distinct pattern of methylation change—hypermethylation of CpG islands (CGIs) and hypomethylation of the rest of the genome—is widely regarded as a hallmark of cancer (3). Hence, methylation profiling of cfDNA has been shown to outperform mutation assays in cancer detection and tissue of origin localization (2).
At present, cfDNA methylation profiling methods can be categorized into two groups: untargeted and targeted methods. Untargeted methods [e.g., whole-genome bisulfite sequencing (WGBS)] provide unparalleled breadth for interrogating ~30 million CpG sites, enabling broad characterization and discovery of cancer-associated methylation patterns, but it comes at a high cost for sequencing the whole genome (3000 Mb). Targeted approaches, exemplified by hybridization capture methods, reduce sequencing requirement by capturing genomic regions that are informative for specific diseases using synthetic probes, but characterization is restricted to the enriched target. In a recent report (2), multicancer detection is demonstrated from targeted sequencing using a panel of methylation probes capturing 17.2 Mb of the genome and targeting 1.1 million CpG sites. Nonetheless, there is substantial upfront and operational cost for designing, synthesizing, validating, and using these capture probes. Commercial hybridization capture kits of comparable coverage cost >$200 (4), setting a minimum cost for performing these assays. Furthermore, once a panel is designed, it is inflexible to be applied to other types of diseases or subtypes that have a different set of differentially methylated regions (DMRs). To date, there is still a lack of a universal, simple, and cost-effective method to enrich for disease-relevant loci in the liquid biopsy context.
To overcome these problems, we developed heat enrichment of CpG-rich regions for bisulfite sequencing (BS) (Heatrich-BS), which very effectively enriches for CpG-dense regions harboring cancer-associated methylation changes. Together with a bioinformatic approach that enables quantitative tumor fraction measurement from low-depth sequencing, this universal probe-free method enables scalable genome-wide methylation profiling of cfDNA at very low cost (<$30). In a proof-of-principle study, we demonstrate the improved sensitivity of Heatrich-BS for noninvasive longitudinal monitoring of tumor progression in patients with colorectal cancer (CRC). In addition, we showed tumor methylation subtyping from cfDNA enabled by enhanced coverage of Heatrich-BS in epigenetic regulatory regions. Together, these results demonstrate that Heatrich-BS is a cost-effective, scalable platform that can complement existing cancer diagnosis and monitoring tools to enable early intervention and personalized therapy in cancer.
RESULTS
CpG-rich DMRs are highly enriched in regions with high GC content
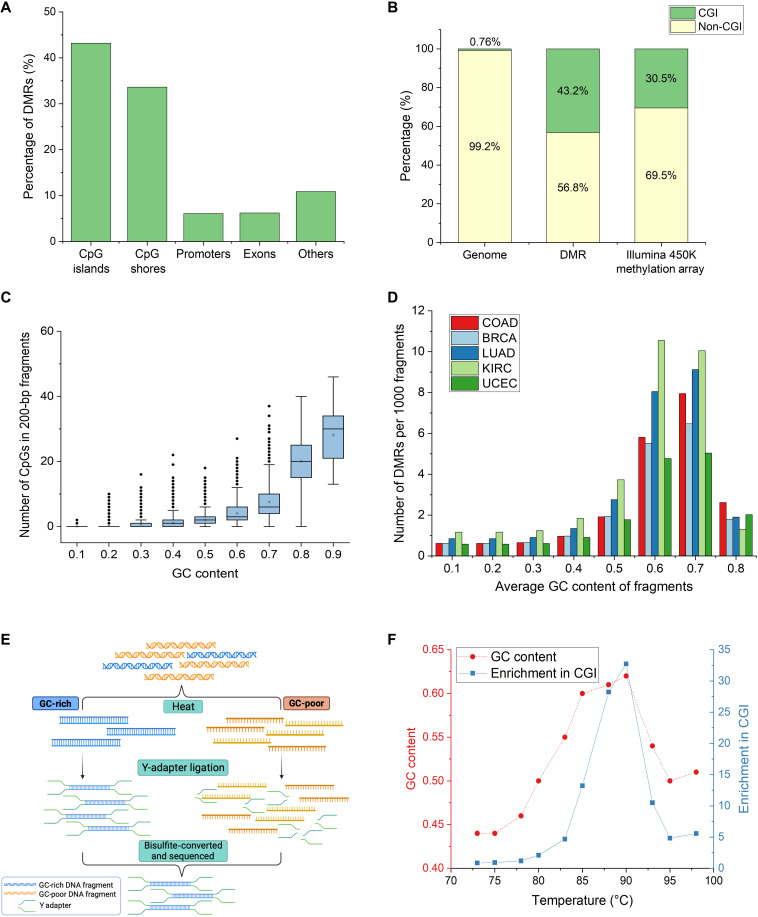
The sequence content in the human genome is highly nonuniform. Long stretches of CpG-poor regions are punctuated by short stretches of CpG-dense regions that coincide with important gene regulatory elements such as promoters. These CpG-dense regions are often differentially methylated between tissues and disease such as cancer (1, 5, 6). Of the DMRs we identified between CRC tissue and healthy plasma (Materials and Methods), nearly 45% of the DMRs lie within CGIs (Fig. 1A), which originates from less than 1% of the genome (Fig. 1B). CGIs are also highly overrepresented in manually curated DNA methylation arrays (e.g., Infinium HumanMethylation450 Bead Chip) (Fig. 1B), reflecting their functional importance and fundamental interest to biology. Hence, it is of value to focus on this small fraction of CpG-rich genome for epigenetic profiling.
Fig. 1. CpG-rich DMRs are highly enriched in regions with high GC content.
(A) Percentage of DMRs between CRC tissue and healthy plasma in different genomic regions. More than 40% of DMRs lie in CpG-rich CGIs. (B) Proportion of DMRs and Illumina 450K Methylation Array probes in CGI with respect to the genomic distribution. Forty-three percent of DMRs and 30% of Illumina 450K methylation array probes lie in 0.76% of the genome. (C) Relationship between GC content and number of CpGs in 0.5 million randomly generated 200-bp fragments from the human genome. Fragments with high GC content also contain more CpG within each fragment. (D) Number of DMRs of different cancers detected per 1000 fragments using different GC-content thresholds. COAD, colorectal adenocarcinoma; BRCA, breast invasive carcinoma; LUAD, lung adenocarcinoma; KIRC, kidney renal clear cell carcinoma; UCEC, uterine corpus endometrial carcinoma. Fragments above 0.6 GC content contain nearly eightfold more DMRs across different cancers. (E) Workflow of Heatrich-BS to select for GC-rich fragments. GC-poor fragments are denatured by heat, and intact GC-rich fragments are selected by Y-adapter ligation. (F) Trend of GC content and read enrichment at CGI over a range of temperatures. Optimal enrichment is achieved at 88°C.
While there is no known means to physically enrich for CpG-dense DNA, it is long known that the G + C content of a double-stranded DNA (dsDNA) fragment is closely related to its thermal stability. It has been shown that the G-C bond in DNA has a binding energy of 25.4 kcal mol−1, which is two times stronger than the A-T bond (7). As the presence of a CpG dinucleotide in a fragment adds two GC bonds to the duplex, we ask whether effective selection of CpG-dense fragments can be achieved by selection of GC-rich fragments. To verify this hypothesis, we calculated the GC content and number of CpGs in 200–base pair (bp) fragments of the human genome (Fig. 1C). Our results showed a strong correlation between GC content and number of CpGs in each fragment. DNA fragments with GC content greater than 0.6 constitutes only 2.5% of the genome but disproportionately includes 85% of CGIs and 58% of identified DMRs between CRC tissues and healthy plasma. We further verified the relationship between different cancer-specific DMRs and GC content (Fig. 1D). Selection of fragments above 0.6 GC content affords nearly eightfold enrichment in proportion of reads in DMRs across different cancers [colorectal adenocarcinoma (COAD), breast invasive carcinoma, lung adenocarcinoma, kidney renal clear cell carcinoma, and uterine corpus endometrial carcinoma]. Therefore, we established that DMRs of various cancers, which are universally overrepresented in CpG-dense regions, can be effectively enriched with selection of high-GC DNA fragments.
Heatrich uses thermal denaturation to select for DNA fragments with high GC content (Fig. 1E). Fragmented DNA was first end-repaired and A-tailed. Following this, the sample was heated to denature the GC-poor fragments, and adapter ligation was immediately performed at low temperature (20°C) to allow any partially denatured fragment to reanneal. The process of adapter ligation allows selection of intact nondenatured GC-rich double-stranded fragments, as T4 DNA ligase has a high selectivity for dsDNA (8). The selected fragments were bisulfite-converted and subsequently sequenced. We found through empirical optimization experiments using sheared genomic DNA (gDNA) (Fig. 1F) that heating DNA to 88°C immediately before adapter ligation yields the best enrichment of reads in CGI (28%) at high GC content (0.63 ± 0.006) compared to the average GC content of the unheated samples (0.42 ± 0.009) (fig. S1A). Additional experiments showed that heating time of 5 min improved upon enrichment of reads in CGIs, whereas longer heating time could denature even some fragments that are found in CGIs (table S1). We also observed that comparable heat enrichment of high GC content fragments can occur in different length DNA fragments commonly found in cfDNA (fig. S1B).
Heatrich enables effective CpG enrichment in fragmented gDNA and cfDNA
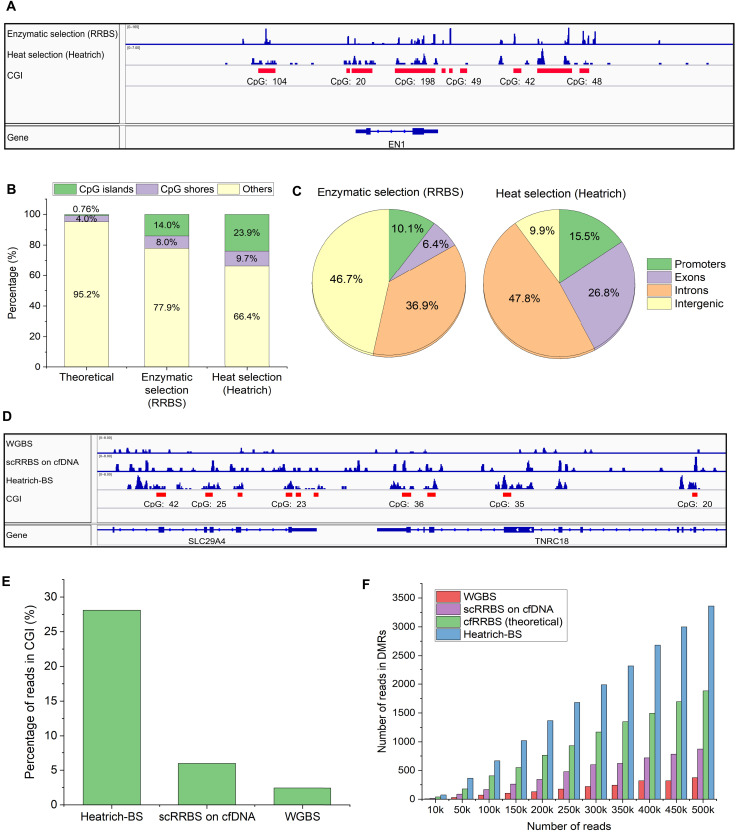
To evaluate the effectiveness of heat denaturation for enrichment of CpG-rich regions, we performed parallel comparisons with reduced representation bisulfite sequencing (RRBS), a conventional enzymatic approach for CpG enrichment. Analysis of the mapping of heat-treated sheared gDNA showed that Heatrich samples displayed a notable accumulation of reads around CGIs, similar to RRBS (SRR222486, Fig. 2A). Quantitatively, heat denaturation is even more effective than the enzymatic approach in enriching for DNA fragments in CGIs and shores (Fig. 2B). Most Heatrich reads are located in promoters, exons, and introns (Fig. 2C), suggesting that this nonenzymatic approach can be an attractive alternative to RRBS for detailed methylation profiling in important genomic regulatory elements. When comparing DNA methylation profiles obtained from Heatrich-BS on sheared K562 gDNA with gold standard WGBS assay (ENCODE), a high Pearson correlation of 0.93 is observed (fig. S1C). We note that unlike deterministic (e.g., RRBS) and targeted (e.g., hybridization capture) approaches, Heatrich may not measure the exact same sites in different samples because of random DNA fragmentation. Nevertheless, we observed that the majority (6143) of DMRs covered in replicate K562 Heatrich-BS libraries (average of 8850 DMRs in each sample) are overlapping (fig. S1D). The degree of overlap between samples would further improve with higher-sequencing depths.
Fig. 2. Enrichment of CpG-rich regions in gDNA and cfDNA using Heatrich.
(A) Localization of Heatrich and RRBS reads to CGI regions. CGIs are marked in red. Heatrich displays substantial piling of reads at CGIs comparable to RRBS. (B) Distribution of Heatrich and RRBS reads in CGIs, shores, and other genomic regions. Heatrich yields more reads in CGIs (24%) compared to RRBS (14%). (C) Distribution of RRBS and Heatrich reads in different genomic regions. Heatrich produces more than twice as many reads in informative promoters and exons (42%) compared to RRBS (16%). (D) Visualization of WGBS, RRBS, and Heatrich-BS reads from cfDNA. CGI regions are marked in red. Heatrich-BS results in substantial piling up of reads and localization to CGI compared to RRBS and WGBS. (E) Percentage of cfDNA reads in CGI using Heatrich-BS, RRBS and WGBS. Heatrich-BS produces nearly 30% reads in CGI compared to 6% in RRBS and 3% in WGBS. (F) Number of cfDNA reads in DMR for different total reads using different methods. Heatrich-BS results in up to 10-fold enrichment of reads in DMRs compared to WGBS and nearly 4-fold enrichment compared to RRBS.
Heatrich is ideally suited for cfDNA methylation profiling because of the low abundance of total DNA and its fragmented nature, where current methods are limited in their CpG enrichment capability. We tested the performance of Heatrich-BS on cfDNA samples obtained from patients with CRC. As both cost and performance are important considerations for routine liquid biopsy, the performance of Heatrich-BS is benchmarked against other sequencing methods at equivalent read counts to establish its cost benefit in the following analysis. We first visualized the reads obtained from Heatrich-BS and compared it with WGBS (no heat treatment) and previously reported single-cell RRBS (scRRBS) on cfDNA (1) (Fig. 2D). Mapped reads from scRRBS on cfDNA (GSM2090507) and WGBS (EGAS00001001219) were distributed almost uniformly across all genomic regions. On the other hand, Heatrich-BS reads were highly concentrated at CGIs and shores. The number of common DMRs profiled between samples is notably higher using Heatrich-BS workflow compared to WGBS and RRBS protocols (table S2). Heatrich-BS samples displayed up to 15-fold enrichment of reads in CGIs compared to WGBS (fig. S1E) and nearly 5-fold enrichment compared to scRRBS on cfDNA (Fig. 2E). This demonstrates that Heatrich-BS outperforms RRBS in terms of CGI enrichment when the input DNA is fragmented. Furthermore, Heatrich-BS had up to 10-fold more reads localizing to DMRs (Fig. 2F) and was able to detect up to 10-fold more DMRs than WGBS using comparable number of sequencing reads (fig. S1F). This would provide higher sensitivity in detecting fragments of tumor origin for the same total sequencing reads.
Our results showed limited DMR coverage with WGBS and RRBS at low sequencing, but enrichment with Heatrich-BS can overcome this important challenge. Hence, Heatrich-BS can be particularly promising to enable sensitive detection of circulating tumor DNA at low cost. We note, however, that Heatrich-BS would not be a substitute for unbiased methods for applications that require comprehensive genome coverage (e.g., de novo DMR discovery). Unbiased approaches such as WGBS can achieve a much broader and uniform coverage across the genome, although at substantially increased cost. We envision that WGBS and Heatrich-BS would complement each other in DMR discovery and routine implementation of liquid biopsy.
Estimating the tumor fraction
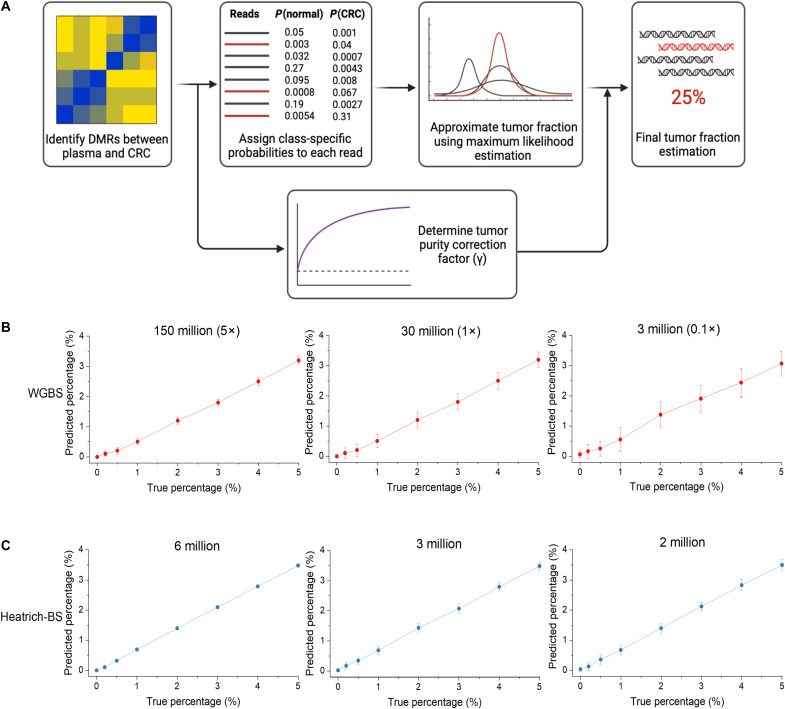
Recent studies have shown that the comethylation patterns within individual DNA fragments can be used to distinguish the origins of cfDNA fragments with higher sensitivity. This has led to the development of methods such as those that make use of methylation haplotype load (1) or α-values (9) for tumor fraction estimation in cfDNA. However, population-averaged measurements at the marker level are still invariably needed, either as a metric for read discordance within a marker (1) or as a requisite for removing confounding markers (9). The need for a minimum read-depth in marker regions, especially for low tumor fraction samples, imposes a bottleneck to further reducing sequencing cost. In this work, we developed a bioinformatics algorithm that estimates global tumor fraction by considering only the tumor probability of individual sequenced fragments without having to estimate population metrics from individual marker regions. The workflow of the algorithm is shown in Fig. 3A and detailed in Materials and Methods. Our developed algorithm allows for accurate estimation of low cfDNA tumor fractions (0.5%) at very low coverage (1×) sequencing data.
Fig. 3. Development and validation of the tumor fraction prediction algorithm.
(A) Workflow of the tumor fraction prediction algorithm. DMRs were identified by comparing healthy volunteer plasma with The Cancer Genome Atlas (TCGA) CRC methylation array data. Class-specific probabilities were assigned to each sequencing fragment, and maximum likelihood estimation was used to infer global tumor fraction. Tumor purity correction was applied to account for normal cell infiltration in TCGA data. (B) True and algorithm-predicted values of simulated plasma WGBS cfDNA samples at different sequencing depths. Confident tumor fraction prediction is achieved beyond 150 million reads (5× sequencing depth). (C) True and algorithm-predicted value of simulated Heatrich-BS samples using different total sequencing reads. Confident tumor fraction prediction is achieved with as few as 3 million reads.
In accordance with previous practices (1, 9–11), we identified DMRs for CRC using WGBS datasets (12) of cfDNA from 23 healthy subjects and Illumina 450K methylation array datasets of 353 COAD samples from The Cancer Genome Atlas (TCGA). The full list of DMRs used in this study is provided in data S1. To validate our algorithm with precisely controlled tumor fractions and sequencing depths, we simulated cfDNA of different tumor fractions by mixing WGBS reads from plasma of three other healthy individuals (12) (that are not included in reference generation) and tumors of patients with CRC (SRR1035745) (13) at different proportions. Tumor estimation from healthy cfDNA samples resulted in a nonzero baseline value that was stable regardless of the sequencing depth (fig. S2A). Similar observations of nonzero baseline of healthy cfDNA have been reported when tissue samples rather than pure cell populations were used as references. This nonzero baseline is attributed to the contribution of non-neoplastic cells from tumor tissue references (14–16) and commonly leads to overestimation of the tumor fraction in methylation-based analysis. To identify the contribution of these non-neoplastic cells to the CRC reference and eliminate its effect on our tumor fraction determination, we determined a tumor purity correction factor by performing receiver operating characteristic (ROC) analysis on healthy and simulated plasma cfDNA WGBS samples at 0.5% tumor fraction. The specific correction factor (γ) that maximized sensitivity and specificity across multiple sequencing depths (table S3) was determined for the generated CRC reference.
Using the determined correction factor, we tested our algorithm on simulated plasma WGBS cfDNA samples from 0 to 5% tumor fraction at different sequencing depths (Fig. 3B). At sequencing depths of 5× and 1×, we obtained a high degree of linearity (Pearson correlation >0.99) between the simulated and predicted tumor fraction values, while the estimated tumor fraction for the healthy individuals was correctly called as zero. Notably, at 1× depth, where each DMR is covered only once on average, our algorithm can accurately detect the presence of small tumor fractions. This is achieved by aggregation of reads from multiple loci, without requiring high depth at individual DMRs. Despite this improvement, excessively low coverage would lead to limited number of DMRs being interrogated, which would in turn affect the specificity and confidence of tumor calling, as evidenced by the larger variations in the predicted tumor fractions, including a higher likelihood of false positives in healthy cfDNA samples at 0.1× sequencing depth. Using the CpG enrichment offered by Heatrich-BS, the sequencing requirement can be kept low without sacrificing coverage of DMRs. To validate this, we approximated the Heatrich-BS assay by selecting only plasma cfDNA fragments with >0.6 GC content (simulated Heatrich-BS samples). We observed that even using very modest number of total sequencing reads (2 million to 6 million reads), high specificity and tumor calling confidence could be achieved (Fig. 3C). Notably, the tumor fraction prediction from simulated Heatrich-BS samples had much higher specificity and lower variance compared to a similar read count WGBS samples (0.1×) at 3 million reads. ROC analysis of WGBS and Heatrich-BS for low tumor burden detection in cfDNA (0.5% tumor fraction) showed that the predictive accuracy of Heatrich-BS samples is much better than conventional WGBS samples [AUC (area under the curve) 0.988 versus 0.547] (fig. S2B). These results demonstrate that Heatrich-BS and the corresponding algorithm enable accurate tumor DNA detection in cfDNA with considerably lesser sequencing requirement compared to existing methods.
Application of Heatrich-BS on patient cfDNA
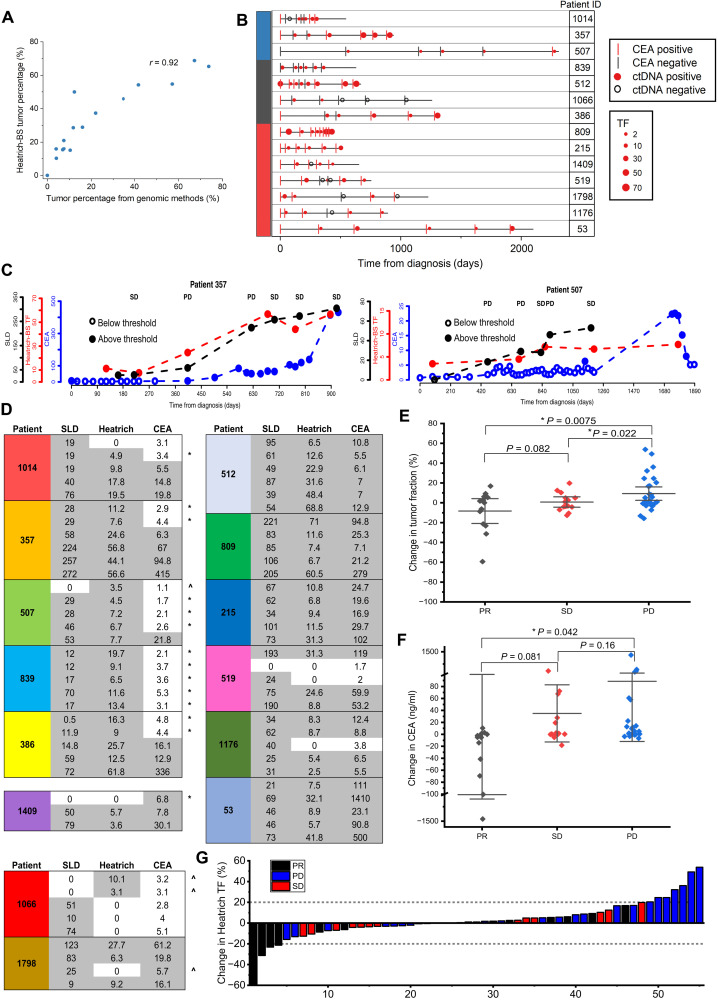
To validate the performance of Heatrich-BS on clinical samples, we first applied the Heatrich-BS assay on 5 healthy volunteers’ and 15 CRC patients’ cfDNA samples (2 million to 8 million sequencing reads each) and compared the tumor fraction obtained from either whole-genome sequencing or deep-targeted sequencing (Fig. 4A) (17). We obtained a Pearson correlation of 0.92 between the tumor fractions predicted by Heatrich-BS and genomic methods (estimates from variant allele frequencies or copy number variations; Materials and Methods), demonstrating that Heatrich-BS can accurately measure tumor fractions from genome-wide methylation profiles with minimal sequencing efforts.
Fig. 4. Application of Heatrich-BS on patient cfDNA samples.
(A) Tumor fractions predicted by genomic methods and Heatrich-BS for patient cfDNA samples. High degree of concordance (Pearson r = 0.92) was achieved between the tumor percentages obtained through the two methods. (B) Longitudinal monitoring of 14 patients with CRC with Heatrich-BS tumor fraction, SLD, and CEA measurements. CEA positive (>5.3 ng/ml) and negative (<5.3 ng/ml) is indicated by red and black lines, respectively. Circulating tumor DNA (ctDNA) positive (>0.5%) and negative (<0.5%) is indicated by filled red circles and open black circles, respectively. Tumor fraction (TF) is represented by the size of the red dots. (C) Heatrich-BS tumor fraction, CEA, and SLD values for patients 357 and 507, which show Heatrich-BS tumor fractions increasing before CEA values, enabling earlier cancer recurrence detection. CEA, Heatrich-BS tumor fractions, and SLD values are represented by blue, red, and black circles, respectively. Open circles indicate values below threshold. (D) Comparison between SLD, Heatrich-BS tumor fraction, and CEA measurements at concurrent time points. “*” indicates that Heatrich-BS outperforms CEA, and “^” indicates that CEA outperforms Heatrich-BS. (E) The relative change in Heatrich-BS tumor fraction is significantly different between patients with PD and patients with SD and PR. (F) The relative change in CEA concentration is significantly different between patients with PD and PR but not between patients with PD and SD. (G) Waterfall plot indicating the relative Heatrich-BS tumor fraction change associated with patient response status (PR, SD, and PD) assessed by CT scans. Changes in tumor fraction exceeding 20% accurately predict patient response.
Current noninvasive surveillance methods for monitoring CRC therapy efficacy and detecting cancer recurrence have their limitations. Radiation exposure and cost limit the frequency at which computed tomography (CT) scans can be performed. Serum protein biomarker such as carcinoembryonic antigen (CEA) can be measured frequently, but it lack sensitivity and specificity (18). We performed a cost analysis of Heatrich-BS (at 3 million reads per sample) and estimated the assay cost to be less than $30 (table S4). Because of its simple workflow and low cost, Heatrich-BS has the potential to be a sensitive assay for frequent monitoring of patients undergoing treatment and those in remission to detect possibility of relapse. To validate the applicability of Heatrich-BS in cancer progression monitoring, we further profiled a cohort of 79 samples from 14 patients with CRC across their course of treatment, with five to seven time points per patient (table S5). Concurrently, we obtained longitudinal CEA measurements and CT scans of these patients for benchmarking tumor fraction predictions by Heatrich-BS (fig. S3). From the aggregated measurements of our cohort, we observed that CEA values do not correlate well with the sum of longest diameter (SLD) of lesions in CT scan [Pearson correlation coefficient (r) = 0.26; fig. S4A], highlighting the limitation of CEA as a quantitative measurement. On the other hand, Heatrich-BS tumor fraction correlate better and more linearly with SLD measurements (Pearson r = 0.62; fig. S4, B and C). Further analyzing the time points for each patient, the Heatrich-BS tumor fractions were compared with CEA status (Fig. 4B) and radiology measurements. To evaluate the potential of using Heatrich-BS for detecting cancer recurrence, we first focus on comparing the detection sensitivity for residual tumor in patients using cfDNA (Heatrich-BS tumor fraction) and CEA (5.3 ng/ml represents cancer detection threshold). Figure 4C lists the tumor detection by radiology (SLD), tumor fractions estimated from Heatrich-BS measurement, and concurrent CEA measurement at the same (or very close) time points for each patient. We observed that Heatrich-BS was able to detect tumor recurrence earlier than CEA measurements in five patients (patients 1014, 357, 507, 839, and 386). Occasionally, CEA was high even when no tumor lesion was detected via radiology (patient 1409), but it was correctly resolved with Heatrich-BS. In contrast, CEA outperformed Heatrich-BS in detection of residual tumors in only two patients (patients 1066 and 1798). Overall, fewer measurements were discordant between detection of tumor via Heatrich-BS (14%) and CT scans as compared to CEA (26%). The detection sensitivity of Heatrich-BS was previously established in silico to be 0.5%, and it has also been showed to be highly specific in a different cohort (Fig. 4A). Our results suggest that Heatrich-BS could be used for frequent monitoring of patients for the risk of recurrence, where a positive cfDNA measurement would be followed up with CT scans for confirmation.
We next investigate whether serial cfDNA monitoring can be used to predict treatment response in patients. To do so, we evaluated whether relative change in tumor fraction detected via Heatrich-BS corresponds to radiographic response in patients and compared how this measure performed relative to CEA changes. We calculated the changes in CEA and Heatrich-BS tumor fraction between successive time points (Fig. 4C) and obtained the response evaluation according to radiology measurement. Relative change in Heatrich-BS tumor fraction is positively correlated with relative change in tumor size (fig. S4D). We observed that patients achieving partial response (PR) or stable disease (SD) had a significantly greater decrease of Heatrich-BS tumor fraction (means, −8.3 and −0.8%, respectively) compared to patients achieving progressive disease (PD) (Fig. 4D; mean, +9.3%; P = 0.0075 for PR versus PD and P = 0.002 for PR versus SD), indicating that Heatrich-BS tumor fraction is predictive of treatment response. On the other hand, patients with PR had a significant reduction of CEA (mean, −125.4 ng/ml) compared to patients with PD (Fig. 4E; mean, +88.6%; P = 0.042 for PR versus PD), but CEA changes between SD and PD did not reach statistical significance, presumably because of their larger nontumor-specific variations. A waterfall plot of the change in Heatrich-BS tumor fraction across different treatment response showed that when the magnitude of change in tumor fraction exceeds 20%, it is completely predictive of tumor response. Therefore, serial cfDNA profiling with Heatrich-BS may provide a good predictor of treatment response to patients with cancer and may outperform standard tumor markers.
Characterization of tumor methylation subtypes in patient cfDNA using Heatrich-BS
Tumorigenesis can be driven by a myriad of genetic or epigenetic factors, resulting in distinct subtypes within a type of cancer. The different driving factors of these subtypes also influence treatment options, prognosis, and survival. One common methylation subtype, known as CpG island methylator phenotype (CIMP), is observed in multiple cancers, such as CRC, breast cancer, gastric cancer, and glioma among others, and is characterized by epigenetic instability, where tumor suppressor genes are inactivated by methylation rather than mutation (19). Studies have shown that patients with CIMP-positive tumors have poorer prognosis and shorter overall survival (20), while CIMP-positive CRCs respond better to irinotecan-based regimen rather than oxaliplatin-based regimen (21). To our knowledge, no existing targeted cfDNA methylation assay is capable of performing cancer methylation subtyping. The untargeted nature of Heatrich-BS provides an opportunity to obtain this additional insight.
Fittingly, most differentially methylated loci in CIMP are found in CGIs that are highly enriched in Heatrich-BS. We found that 41.7% (1121 of 2686) of the loci used to classify and annotate CIMP status in TCGA CRC samples (22) are effectively represented in Heatrich-BS (covered in more than 50 samples). We identified a final set of 635 most informative CpGs that can collectively distinguish the different CIMP subtypes (Fig. 5A). On the other hand, methylation profiles of the DMRs used to predict tumor fraction remained invariant across the different CIMP subtypes (fig. S5). We noted that there is no overlap between CIMP markers and the DMRs used to predict tumor fraction, indicating that Heatrich-BS regions encompass orthogonal sets of markers that are useful for tumor load quantification and methylation subtype prediction.
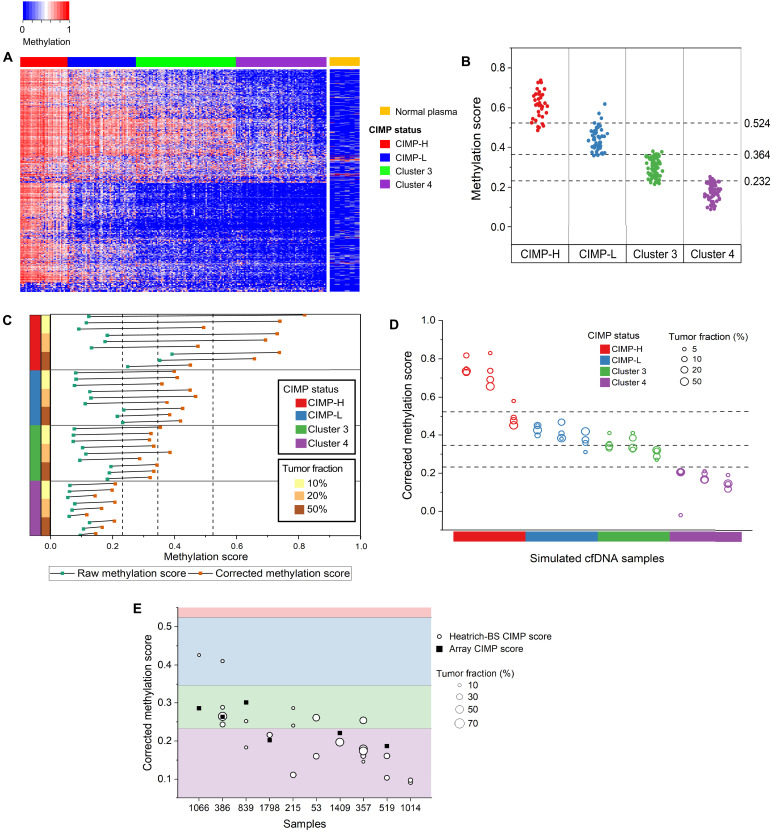
Fig. 5. Characterization of patient cfDNA using Heatrich-BS.
(A) Methylation status of TCGA CRC samples and normal plasma at 635 CpG markers identified for CIMP subtype classification. (B) Methylation score of 233 CRC tissues with CIMP annotation in TCGA. Cutoff thresholds to distinguish different CIMP subtypes were determined by a decision tree classifier. (C) Raw and corrected methylation score of simulated cfDNA with different tumor fractions and methylation subtypes. Deconvolution removes the effect of tumor fraction on raw methylation score calculated from cfDNA. (D) Corrected methylation scores of different CIMP subtypes and tumor fraction simulated samples with decision tree classifier thresholds for CIMP classification. Corrected methylation scores are concordant across tumor fractions more than 10% for different CIMP clusters. (E) Corrected methylation score for patient cfDNA samples with tumor fractions above 10%. CIMP scores from tumors of six patients are measured using DNA methylation array.
We next developed a scoring system that would allow easy classification of tumor methylation subtypes (Materials and Methods). Applying this scoring system to the 233 TCGA CRC samples (22), we observed that CIMP subtypes of CRC tissues are well defined by ranges of methylation scores, and a series of threshold values in methylation scores allow 89% accuracy in classifying CIMP subtypes (Fig. 5B). Nevertheless, raw methylation scores from cfDNA are not expected to reflect the methylation subtype of the underlying tumor because cfDNA is derived from mixture of normal and cancerous cells. To validate the approach used to determine methylation score of the underlying tumor in cfDNA, we simulated cfDNA containing different tumor methylation subtypes at varying tumor fractions by creating mixtures of sequencing reads drawn from TCGA tumor and healthy plasma methylation measurements (Fig. 5C). While the raw methylation score from cfDNA is confounded by its tumor fraction, incorporating tumor fraction estimation from Heatrich-BS assay enables the calculation of corrected methylation score that accurately reflected the underlying tumor methylation subtype (86% accuracy). Nevertheless, at present, tumor subtyping exhibits higher uncertainty at tumor fractions below 10% due to fewer tumor-derived cfDNA fragments (Fig. 5D). Finally, we applied this algorithm to infer tumor methylation subtype of our longitudinal tracking cohort (Fig. 5E). We calculated methylation score for 23 of 79 samples that had tumor fractions >10%. Our results showed that the corrected methylation scores of longitudinal samples from the same patient are often tightly clustered and independent of tumor fraction (15 of 20 or 75% of measurements from same patients conform to consistent CIMP subtypes), while methylation scores between patients could differ greatly, suggesting that the methylation subtype of a patient tumor does not change substantially through disease progression. Our results predicted that there were no CIMP-high patients in the profiled cohort, with most patients falling into CIMP-negative subtypes, cluster 3 or 4. It has been reported that CIMP-high tumors in CRC are strongly associated with microsatellite instability (23). Our tumor methylation subtype prediction from cfDNA is well in line with expectation as all the patient tumors in this longitudinal cohort were profiled to be microsatellite stable during standard clinical evaluation (table S5). Finally, to further validate the accuracy of subtype prediction from cfDNA, we were able to perform DNA methylation profiling (Infinium Methylation EPIC array) of matched tumors from six patients. These patient tumors showed variable methylation at the CpG sites used for CIMP subtyping (fig. S6), and the inferred CIMP subtypes for these patients are plotted in Fig. 5E. Our results showed that the CIMP subtypes from tumor samples largely matches the subtypes inferred from Heatrich-BS. Of the 11 subtype inferences made from Heatrich-BS when tumor subtypes are known, 8 (73%) were attributed to the correct tumor subtypes.
DISCUSSION
In this study, we present the Heatrich-BS assay, which is the first assay that uses the concept of thermal denaturation to achieve CpG enrichment in fragmented DNA. Heatrich selects for DNA fragments with GC content exceeding 60%, and we have demonstrated that nearly 30% of Heatrich-BS reads are in CGIs, which comprise less than 1% of the genome. We also developed a tumor fraction prediction algorithm to augment our assay and validated its application for tumor fractions as low as 0.5% from low-depth sequencing. Compared to existing methods such as CancerDetector (9), which can detect 0.5% tumor from 2× WGBS (~60 million reads), our method and algorithm could detect similar tumor burden with 20-fold less sequencing (~3 million reads). With this two-pronged approach, we realized a universal low-cost ($30) cfDNA methylation assay for quantitative cancer detection.
We showed that Heatrich-BS provides accurate tumor fraction estimates that correspond to cfDNA mutation and copy number measurements. Because of its high sensitivity and low cost, as well as the ability to perform quantitative analysis of tumor from cfDNA, Heatrich-BS is particularly amenable to noninvasive monitoring of cancer progression or recurrence, in which frequent measurements are needed and current methods are inadequate. We showed that Heatrich-BS can provide superior sensitivity for detection of CRC at low tumor fractions compared to conventional CEA protein biomarker assay in longitudinal monitoring of patients with cancer. In addition, we demonstrated the elucidation of tumor methylation subtypes from cfDNA, further confirming the advantage of broad genomic coverage using Heatrich-BS assay.
Heatrich-BS offers many advantages compared to current assays: (i) The workflow of Heatrich-BS is short and easy to perform. The entire assay, from sample collection to sequencing, can be performed in less than 48 hours, resulting in a short turnaround time even for a sequencing assay. (ii) Heat denaturation is independent of DNA sequence biases that can arise from the use of restriction enzymes in assays like RRBS. (iii) Heatrich is based on GC content, which is a physical property of DNA. This enables effective CpG enrichment even in fragmented DNA, such as cfDNA and FFPE samples, where the enrichment capability of current assays such as RRBS is limited (24). (iv) Heatrich-BS requires fewer sequencing reads compared to conventional untargeted assays, which makes it highly cost effective (>10-fold cost saving) to perform.
Finally, the extensive coverage of epigenetically informative regions using Heatrich-BS assay could enable exploration of other important applications. The vast majority (83%) of tissue-specific methylation haplotype blocks (1) identified in previous reports could be detected using Heatrich-BS, suggesting a potential for its use as a universal and affordable multicancer screening and discrimination assay. Because of its high coverage of epigenetic regulatory regions, Heatrich-BS could also be useful for validation of candidate DMRs from tissues of different cancers. There would be need to substantially increase the sequencing reads from the current nonsaturating sequencing throughput to achieve reproducible coverage between samples and to accurately determine the average methylation at each site. Nevertheless, we expect that the cost savings gained by deep methylation sequencing only at CpG-dense regions would be an attractive advantage for this endeavor. We envision that the Heatrich-BS platform would be an important innovation to enable practical and scalable implementation of cfDNA methylation profiling in liquid biopsy for clinical translation.
MATERIALS AND METHODS
Generating sheared DNA
K562 cells (American Type Culture Collection, CCL-243) were cultured in high-glucose Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin-streptomycin (Gibco). gDNA was extracted from cultured K562 cells using the DNeasy Blood and Tissue Kit (Qiagen). The extracted gDNA was fragmented using the LE220 Focused ultrasonicator (Covaris) at the following settings: 450-W peak incidence power, 30% duty factor, and 200 cycles per burst for 420 s. The fragmented DNA was size-selected for 100- to 200-bp fragments using a BluePippin 2% agarose cassette (Sage Sciences).
Tumor DNA methylation profiling
gDNA is extracted from fresh-frozen tumor samples. DNA (1.5 μg) was bisulfite-converted following the recommended protocol of a Zymo EZ DNA Methylation-Gold kit (Zymo Research). Genome-wide DNA methylation profiling of the bisulfite-converted DNA was performed using the Infinium EPIC Beadchip array (Illumina). IDAT files were processed using the minfi package in R using the preprocessIllumina function to yield b values at each locus.
Patient recruitment and extraction of cfDNA from patient blood samples
Patients with CRC were recruited at the National Cancer Centre Singapore under studies 2018/2795 and 2019/2401 approved by the SingHealth Centralised Institutional Review Board. From these patients, blood specimens and tumor specimens were collected where possible and consented for. Blood samples from healthy individuals were collected under study 2012/733/B. Retrospective review of medical records was performed to collect clinicopathological details, such as patient demographics, tumor staging, serum CEA, and mutational status from clinical testing where available (table S5). To assess the sensitivity of Heatrich-BS for tumor monitoring in comparison to CEA measurements, patients included were those whose CEA measurements were informative or uninformative of disease progression. All plasma was separated from whole blood collected in EDTA tubes within 2 hours of venipuncture via centrifugation at 10 min × 300g and 10 min × 9730g and subsequently frozen at −80°C. cfDNA was extracted using the QiaAmp Circulating Nucleic Acids Kit (Qiagen) as per the manufacturer’s protocol.
Heatrich-BS protocol
cfDNA (5 to 10 ng) was used as input for the Heatrich-BS protocol. Library preparation was done using the KAPA HyperPrep Kit (Kapa Biosystems). 1.4 μl of End Repair and A-tailing buffer (Kapa Biosystems) and 0.6 μl of End Repair and A-tailing enzyme mix (Kapa Biosystems) were added to 10 μl of input DNA and incubated at 20°C for 30 min and 65°C for 30 min. Following this, the sample was heated at 88°C for 5 min and immediately placed on ice. The sample was then topped up with 6 μl of ligation buffer (Kapa Biosystems), 2 μl of DNA ligase (Kapa Biosystems), 1 μl of nuclease-free water, and 1 μl of 750 nM methylated TruSeq adapter (Illumina). For no-heat controls, 1 μl of 1.5 μM methylated TruSeq adapter (Illumina) was used instead. After adding these reagents, the sample was incubated at 25°C for 1 hour and then cleaned up by performing two rounds of 1.2× SPRISelect (Beckman Coulter). The sample was then subject to bisulfite conversion following the recommended protocol of Zymo EZ DNA Methylation-Gold kit (Zymo Research). The bisulfite-converted DNA was amplified for 15 cycles using Pfu Polymerase (Agilent) that can overcome uracil stalling, cleaned up using 1.2× SPRISelect (Beckman Coulter), and reamplified using KAPA Hyper Hot-Start Polymerase (Kapa Biosystems) in a real-time polymerase chain reaction machine until plateau was reached. The amplified sample was cleaned up using 1.2× SPRISelect (Beckman Coulter), size-selected for 190- to 400-bp fragments using 2% agarose Bluepippin kits (Sage Sciences), quantified using Kapa Library quantification kits (Kapa Biosystems), and sequenced using MiSeq or NovaSeq (Illumina). Pair-end sequencing of 75 bp each was performed.
Heatrich-BS analysis pipeline
Fastqc (25) was used to check the quality of the pair-end reads generated by MiSeq. After adapter trimming using Cutadapt (26), the reads were aligned to the hg38 human genome using Bismark (27). Aligned fragments with the same start and end positions were deduplicated using Picard tools (28), following which the Bismark methylation extractor was used to obtain per-base methylation status of each fragment.
GC content calculation
To calculate the GC content of each fragment, the forward and reverse reads were aligned separately and then combined to generate a single coordinate range encompassing the entire fragment. The coordinates of the fragment were then used to obtain its sequence from the reference genome. For each fragment, the GC content was defined as the number of Cs and Gs, divided by the total length of the fragment. Percentage of reads in CGI was defined as the proportional of sequenced reads that coincided with hg38 CGI annotation from UCSC Genome Browser.
Tumor fraction determination algorithm
The tumor fraction determination algorithm has three major steps:
Step 1: Identifying the differentially methylated clusters
To identify differentially methylated clusters for tumor-specific cfDNA detection, normal plasma whole-genome methylation data (12) and COAD methylation array from TCGA were used. Twenty-three WGBS datasets for normal plasma (EGAS00001001219) and 353 Illumina 450K methylation array datasets from TCGA were used for cluster generation. The TCGA methylation values were extrapolated to ±100 bp of each probe site. To ensure selection of only consistent sites, only methylation values with an SD less than 0.4 between the various samples in that class were chosen to ensure confidence for the reference. DMRfinder (29) was used to identify differentially methylated clusters. Within these clusters, sites with a 0.5 difference in methylation were selected.
Step 2: Calculating the class-specific probability of each site
Using the generated reference, a normal and tumor class-specific probability must be assigned to each assayed fragment. Because methylation values are binary, the average methylation value observed in the reference is a proportional combination of the unmethylated and methylated reads. For every site in the reference, the contribution from the unmethylated and methylated modes (0 and 1) was calculated. The relative contributions of each mode in the two classes were used to assign class-specific probabilities for the methylation values in the assayed fragment. In this way, a normal or tumor probability value was assigned to each site assayed.
Step 3: Using maximum likelihood estimation to predict the tumor fraction of the sample
After assigning class-specific probabilities to each fragment, the fraction of fragments that come from the tumor must be enumerated. The tumor-derived cfDNA in a sample, also known as tumor fraction, can be denoted as θ, where 0 ≤ θ < 1. To estimate the tumor fraction θ, a maximum likelihood estimation approach and grid search, adapted from CancerDetector (9), was used to calculate the raw tumor fraction for each sample. The determined tumor purity correction factor (γ) of 0.057 is then applied to the raw tumor fraction to generate the final tumor fraction.
GC content analysis in DMRs
The human genome was split into 200-bp tiling windows, and the GC content of each window was calculated. Windows with GC content exceeding 60% were used as a theoretical representation of Heatrich output. To investigate the relationship between GC content and CpG density, we used a random sequence generator to create half a million 200-bp fragments. The number of G + C bases (GC content) and number of CG dinucleotides (CpG content) was calculated as a fraction of the total length. To calculate the number of DMRs per 1000 fragments, we first generated DMRs for each cancer using the earlier mentioned approach (step 1 of tumor fraction determination). We then used random 1000 fragments generated from a plasma cfDNA dataset subject to different GC content thresholds and counted the number of DMRs of each cancer that was detected. To generate the number of DMRs covered by different number of total sequencing reads, a similar approach was used where different datasets were subsamples to the required number of reads, and the number of fragments that contained DMRs was counted. For cell-free RRBS, all fragments with Msp I cut sites between 20 and 160 bp were used as theoretical data.
Tumor burden estimation by whole-genome or targeted sequencing
DNA libraries were prepared using the Kapa Hyper Prep Kit (Kapa Biosystems) and sent for whole-genome sequencing or targeted sequencing. Hybridization capture was done for targeted sequencing using an IDT Xgen custom panel of 101 cancer genes and reagents as per the manufacturer’s instructions. Sequencing was performed on an Illumina HiSeq 4000 (2 × 150-bp paired-end reads). Tumor fraction estimation from whole-genome sequencing data was carried out using the ichorCNA algorithm (17). Variant calling from targeted sequencing data was performed using MuTect (30) with the tumor fraction estimation being the mean variant allele frequency of seven known CRC hotspots (KRAS, NRAS, BRAF, EGFR, APC, TP53, and PIK3CA) present in a particular sample.
Tumor measurements and disease status classification
For each profiled time point, the nearest available CT scan image was retrieved from the patient’s clinical records. Each lesion on the scan was measured in two dimensions (maximum width and maximum length). Indeterminate lesions were not measured. For each time point, the SLD was determined, providing a representation of the total tumor load present at the time point. To ensure consistency, all measurements were carried out by the same clinician. Disease classification for each time point was carried out according to the following criteria: CR, disappearance of all lesions; PR, ≥30% decrease in the SLD of the lesions compared with the SLD of the previous measured time point; PD, ≥20% increase of at least 5 mm in the SLD of the lesion compared with the SLD of the previous measured time point or the appearance of new lesions >10 mm in diameter; and SD, neither PR, PD, nor CR.
Subtype classification
To identify marker sets for CIMP subtype classification, we used the data and annotations from TCGA Network’s publication (22). We selected CpG sites with a minimum SD of 0.25 and showed distinct methylation patterns in the different CIMP subtypes. Because Heatrich-BS disproportionately enriches for CpG-rich regions, we further restricted the marker list to CpGs that were covered by at least 50 Heatrich-BS samples. To determine the threshold values to distinguish CIMP clusters, we used the decision tree classifier from KNIME (31). The TCGA dataset with CIMP classification was split 70 to 30 for training and testing.
To perform CIMP classification of cfDNA samples, a raw methylation score across the marker sites was calculated for each sample. The methylation score, defined as the average of methylation values of the 635 loci, was used to summarize the degree of methylation in CIMP loci. To estimate the methylation score of the underlying tumor in cfDNA, we note that McfDNA = θMtumor + (1 − θ)Mnormal-plasma. Substituting Mnormal-plasma (methylation scores from cfDNA of healthy plasma) and θ (the tumor fraction calculated from Heatrich-BS), the methylation score of the underlying tumor (Mtumor) can be estimated. To calculate Mnormal-plasma, samples with negative methylation scores or fewer than 100 reads falling into marker sites were excluded.
Acknowledgments
We thank O. J. L. Rackham for valuable discussions on the algorithm. We also acknowledge Y. M. D. Lo and his team for sharing normal plasma cfDNA methylation data (12). Patient samples provided were part of the CaLiBRE program, supported by A*STAR under its IAF-PP scheme (grant ID: H1801a0019).
Funding: This work was supported by iHealthtech Precision Medicine and Personalized Therapeutics seed grant (L.F.C.) and National Medical Research Council Singapore MOH-OFIRG18nov-0003 (L.F.C.).
Author contributions: Conceptualization and study design: E.C. and L.F.C. Experimentation: E.C., R.V., and H.J.W. Bioinformatics: E.C. Genomic profiling: Y.L., A.G., P.S.Y.P., A.S., and S.B.N. Clinical sample collection and characterization: P.-M.W., S.H., B.T., A.Y.C., D.Q.C., and I.B.T. Writing manuscript: E.C. and L.F.C. All authors commented on the manuscript and approved the submission. I.B.T. and L.F.C. jointly supervised this work.
Competing interests: L.F.C., E.C., I.B.T., and D.Q.C. are inventors on a patent application related to this work filed by the National University of Singapore and Singapore Health Services (PCT/SG2022/050021, filed on 20 January 2022). The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Heatrich sequencing data have been deposited at the GEO database and are publicly available under the accession codes GSE202606 and GSE208596.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Tables S1 to S5
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.Guo S., Diep D., Plongthongkum N., Fung H. L., Zhang K., Zhang K., Identification of methylation haplotype blocks AIDS in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nat. Genet. 49, 635–642 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu M. C., Oxnard G. R., Klein E. A., Swanton C., Seiden M. V.; CCGA Consortium , Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 31, 745–759 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sproul D., Meehan R. R., Genomic insights into cancer-associated aberrant CpG island hypermethylation. Brief. Funct. Genomics 12, 174–190 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Philibert W., Andersen A. M., Hoffman E. A., Philibert R., Dogan M., The reversion of dna methylation at coronary heart disease risk loci in response to prevention therapy. Processes 9, 699 (2021). [Google Scholar]
- 5.Wang J., Duan Y., Meng Q. H., Gong R., Guo C., Zhao Y., Zhang Y., Integrated analysis of DNA methylation profiling and gene expression profiling identifies novel markers in lung cancer in Xuanwei, China. PLOS ONE 13, 1–19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wen L., Li J., Guo H., Liu X., Zheng S., Zhang D., Zhu W., Qu J., Guo L., Du D., Jin X., Zhang Y., Gao Y., Shen J., Ge H., Tang F., Huang Y., Peng J., Genome-scale detection of hypermethylated CpG islands in circulating cell-free DNA of hepatocellular carcinoma patients. Cell Res. 25, 1250–1264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.M. Y, Probing the nature of hydrogen bonds in DNA base pairs. J. Mol. Model. 12, 665–672 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Doherty A. J., Wigley D. B., Functional domains of an ATP-dependent DNA ligase. J. Mol. Biol. 285, 63–71 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Li W., Li Q., Kang S., Same M., Zhou Y., Sun C., Liu C. C., Matsuoka L., Sher L., Wong W. H., Alber F., Zhou X. J., CancerDetector: Ultrasensitive and non-invasive cancer detection at the resolution of individual reads using cell-free DNA methylation sequencing data. Nucleic Acids Res. 46, e89 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehmann-Werman R., Neiman D., Zemmour H., Moss J., Magenheim J., Vaknin-Dembinsky A., Rubertsson S., Nellgård B., Blennow K., Zetterberg H., Spalding K., Haller M. J., Wasserfall C. H., Schatz D. A., Greenbaum C. J., Dorrell C., Grompe M., Zick A., Hubert A., Maoz M., Fendrich V., Bartsch D. K., Golan T., Sasson S. A. B., Zamir G., Razin A., Cedara H., Shapiroo A. M. J., Glaser B., Shemer R., Dor Y., Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl. Acad. Sci. U.S.A. 113, E1826–E1834 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang S., Li Q., Chen Q., Zhou Y., Park S., Lee G., Grimes B., Krysan K., Yu M., Wang W., Alber F., Sun F., Dubinett S. M., Li W., Zhou X. J., CancerLocator: Non-invasive cancer diagnosis and tissue-of-origin prediction using methylation profiles of cell-free DNA. Genome Biol. 18, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun K., Jiang P., Chan K. C. A., Wong J., Cheng Y. K. Y., Liang R. H. S., Chan W. K., Ma E. S. K., Chan S. L., Cheng S. H., Chan R. W. Y., Tong Y. K., Ng S. S. M., Wong R. S. M., Hui D. S. C., Leung T. N., Leung T. Y., Lai P. B. S., Chiu R. W. K., Lo Y. M. D., Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl. Acad. Sci. U.S.A. 112, E5503–E5512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heyn H., Vidal E., Ferreira H. J., Vizoso M., Sayols S., Gomez A., Moran S., Boque-Sastre R., Guil S., Martinez-Cardus A., Lin C. Y., Royo R., Sanchez-Mut J. V., Martinez R., Gut M., Torrents D., Orozco M., Gut I., Young R. A., Esteller M., Epigenomic analysis detects aberrant super-enhancer DNA methylation in human cancer. Genome Biol. 17, 1–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leary R. J., Lin J. C., Cummins J., Boca S., Wood L. D., Parsons D. W., Jones S., Sjöblom T., Park B.-H., Parsons R., Willis J., Dawson D., Willson J. K. V., Nikolskaya T., Nikolsky Y., Kopelovich L., Papadopoulos N., Pennacchio L. A., Wang T.-L., Markowitz S. D., Parmigiani G., Kinzler K. W., Vogelstein B., Velculescu V. E., Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc. Natl. Acad. Sci. U.S.A. 105, 16224–16229 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter S. L., Cibulskis K., Helman E., McKenna A., Shen H., Zack T., Laird P. W., Onofrio R. C., Winckler W., Weir B. A., Beroukhim R., Pellman D., Levine D. A., Lander E. S., Meyerson M., Getz G., Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moss J., Magenheim J., Neiman D., Zemmour H., Loyfer N., Korach A., Samet Y., Maoz M., Druid H., Arner P., Fu K. Y., Kiss E., Spalding K. L., Landesberg G., Zick A., Grinshpun A., Shapiro A. M. J., Grompe M., Wittenberg A. D., Glaser B., Shemer R., Kaplan T., Dor Y., Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 9, 5068 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adalsteinsson V. A., Ha G., Freeman S. S., Choudhury A. D., Stover D. G., Parsons H. A., Gydush G., Reed S. C., Rotem D., Rhoades J., Loginov D., Livitz D., Rosebrock D., Leshchiner I., Kim J., Stewart C., Rosenberg M., Francis J. M., Zhang C. Z., Cohen O., Oh C., Ding H., Polak P., Lloyd M., Mahmud S., Helvie K., Merrill M. S., Santiago R. A., O’Connor E. P., Jeong S. H., Leeson R., Barry R. M., Kramkowski J. F., Zhang Z., Polacek L., Lohr J. G., Schleicher M., Lipscomb E., Saltzman A., Oliver N. M., Marini L., Waks A. G., Harshman L. C., Tolaney S. M., Van Allen E. M., Winer E. P., Lin N. U., Nakabayashi M., Taplin M. E., Johannessen C. M., Garraway L. A., Golub T. R., Boehm J. S., Wagle N., Getz G., Love J. C., Meyerson M., Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 8, 1324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shinkins B., Nicholson B. D., Primrose J., Perera R., James T., Pugh S., Mant D., The diagnostic accuracy of a single CEA blood test in detecting colorectal cancer recurrence: Results from the FACS trial. PLOS ONE 12, e0171810 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mojarad E. N., Kuppen P. J. K., Aghdaei H. A., Zali M. R., The CpG island methylator phenotype (CIMP) in colorectal cancer. Gastroenterol. Hepatol. from Bed to Bench. 6, 120–128 (2013). [PMC free article] [PubMed] [Google Scholar]
- 20.Juo Y. Y., Johnston F. M., Zhang D. Y., Juo H. H., Wang H., Pappou E. P., Yu T., Easwaran H., Baylin S., van Engeland M., Ahuja N., Prognostic value of CpG island methylator phenotype among colorectal cancer patients: A systematic review and meta-analysis. Ann. Oncol. 25, 2314–2327 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X., Zhang W., Cao P., Advances in CpG island methylator phenotype colorectal cancer therapies. Front. Oncol. 11, 629390 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Cancer Genome Atlas Network , Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weisenberger D. J., Siegmund K. D., Campan M., Young J., Long T. I., Faasse M. A., Kang G. H., Widschwendter M., Weener D., Buchanan D., Koh H., Simms L., Barker M., Leggett B., Levine J., Kim M., French A. J., Thibodeau S. N., Jass J., Haile R., Laird P. W., CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 38, 787–793 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Ludgate J. L., Wright J., Stockwell P. A., Morison I. M., Eccles M. R., Chatterjee A., A streamlined method for analysing genome-wide DNA methylation patterns from low amounts of FFPE DNA. BMC Med. Genomics 10, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.S. Andrews, FastQC: A quality control tool for high throughput sequence data (2010).
- 26.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011). [Google Scholar]
- 27.Krueger F., Andrews S. R., Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Broad Institute, Picard Tools (2018).
- 29.Gaspar J. M., Hart R. P., DMRfinder: Efficiently identifying differentially methylated regions from MethylC-seq data. BMC Bioinformatics 18, 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sougnez C., Gabriel S., Meyerson M., Lander E. S., MuTect. Nat. Biotechnol. 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berthold M. R., Cebron N., Dill F., Gabriel T. R., Kötter T., Meinl T., Ohl P., Thiel K., Wiswedel B., KNIME-the Konstanz information miner: version 2.0 and beyond. AcM SIGKDD Explor. Newsl. 11, 26–31 (2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 to S5
Data S1