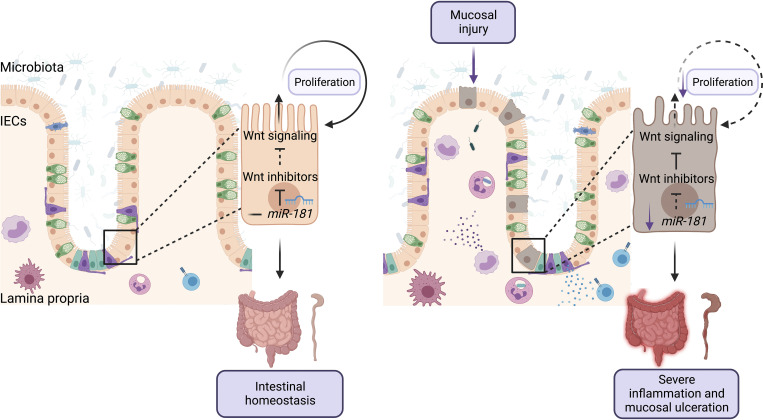
The miR-181 family is downregulated in IECs from human IBD patients and in mice with colitis, a process that is critical for the development of severe colonic inflammation as this microRNA family promotes IEC proliferation and re-epithelialization by potentially enhancing Wnt signaling.
Abstract
The intestinal epithelium is a key physical interface that integrates dietary and microbial signals to regulate nutrient uptake and mucosal immune cell function. The transcriptional programs that regulate intestinal epithelial cell (IEC) quiescence, proliferation, and differentiation have been well characterized. However, how gene expression networks critical for IECs are posttranscriptionally regulated during homeostasis or inflammatory disease remains poorly understood. Herein, we show that a conserved family of microRNAs, miR-181, is significantly downregulated in IECs from patients with inflammatory bowel disease and mice with chemical-induced colitis. Strikingly, we showed that miR-181 expression within IECs, but not the hematopoietic system, is required for protection against severe colonic inflammation in response to epithelial injury in mice. Mechanistically, we showed that miR-181 expression increases the proliferative capacity of IECs, likely through the regulation of Wnt signaling, independently of the gut microbiota composition. As epithelial reconstitution is crucial to restore intestinal homeostasis after injury, the miR-181 family represents a potential therapeutic target against severe intestinal inflammation.
Graphical Abstract
Introduction
The mammalian gastrointestinal (GI) tract houses a complex array of microbes crucial for processing dietary nutrients, thereby facilitating energy consumption by the host (Bäckhed et al., 2004). The intestinal epithelium is a key physical interface that integrates signals from the diet and the microbiota to regulate nutrient uptake and immune cell function by relaying signals from the gut lumen, ultimately promoting intestinal homeostasis or protective immune responses against GI pathogens (Peterson and Artis, 2014). While the gut microbiota plays critical roles in promoting intestinal homeostasis, translocation of bacteria or bacterial products can lead to severe inflammation and fibrosis of the intestinal mucosa (Rieder and Fiocchi, 2009). Indeed, several studies have shown a significant association between microbial translocation and the development of inflammatory bowel disease (IBD) in humans (Mankertz and Schulzke, 2007). As such, it is critical to understand the mechanisms by which the intestinal epithelium promotes host–microbiota interactions during homeostasis and the means by which these interactions are dysregulated during disease.
Intestinal epithelial cells (IECs) are organized in a crypt-villus structure with intestinal epithelial stem cells (IESCs) located at or near the crypt base with differentiated subtypes residing toward the villus (Beumer and Clevers, 2021). Fully differentiated IECs turn over rapidly and are replaced via IESC proliferation in the crypt and subsequent differentiation during transmigration to the villus. Moreover, epithelial cell reconstitution is critical to restore intestinal homeostasis during recovery from mucosal injury (Karin and Clevers, 2016). The transcriptional networks regulating IESC self-renewal, proliferation, and differentiation at steady state and during inflammatory processes have been extensively studied (Beumer and Clevers, 2021). However, how these gene networks are posttranscriptionally regulated to rapidly respond to dynamic host and environmental signals remains unclear. Interestingly, IEC-specific depletion of Dicer, a key enzyme in the processing of microRNAs (miRNAs), a class of well-known translational repressors, results in increased crypt cell death, dysregulation of enterocyte populations, and increased mucosal inflammation (McKenna et al., 2010; Biton et al., 2011). Yet how individual miRNAs directly regulate IEC development and function in response to microbial and dietary cues remain poorly understood.
The miR-181 family is a highly conserved family of miRNAs expressed across many tissues and cell types and regulates inflammatory processes in multiple physiological contexts (Virtue et al., 2019; Henao-Mejia et al., 2013; Williams et al., 2013; Indrieri et al., 2020; Hutchison et al., 2013; Sun et al., 2014). It is now well established that miRNAs function largely by tuning gene expression programs, rather than acting as genetic switches (Bartel and Chen, 2004; Bartel, 2004). Interestingly, this family of miRNAs has been shown to regulate key pathways associated with IEC turnover and function such as the Notch, Wnt, TGFβ, PI3K, and NF-κB pathways (Cichocki et al., 2011; Fragoso et al., 2012; Hori et al., 2017; Zheng et al., 2019; Liu et al., 2018; Henao-Mejia et al., 2013; Zhu and Yang, 2019; Pei et al., 2020; Qin et al., 2010). Moreover, we recently demonstrated that expression of the miR-181 family in adipose tissue is controlled by signals from the gut microbiota to modulate host metabolism and glucose homeostasis (Virtue et al., 2019). However, whether the miR-181 family regulates intestinal homeostasis through its activity in IECs in response to microbial cues remains unknown.
Herein, we show that miR-181 levels are significantly downregulated in colonic IECs from patients with IBD and mice with chemical-induced colitis. Strikingly, we found that IEC-intrinsic miR-181 expression is necessary for protection against severe chemical-induced colitis in mice, potentially by increasing the proliferative capacity of colonic IECs during recovery, in a manner that is largely independent of gut microbiota composition (Karin and Clevers, 2016). As epithelial reconstitution is crucial to restoring intestinal homeostasis after injury, miR-181 thus represents a novel posttranscriptional regulatory factor in IECs critical for protection against severe intestinal inflammation in response to mucosal injury.
Results and discussion
miR-181 expression is dysregulated during colitis
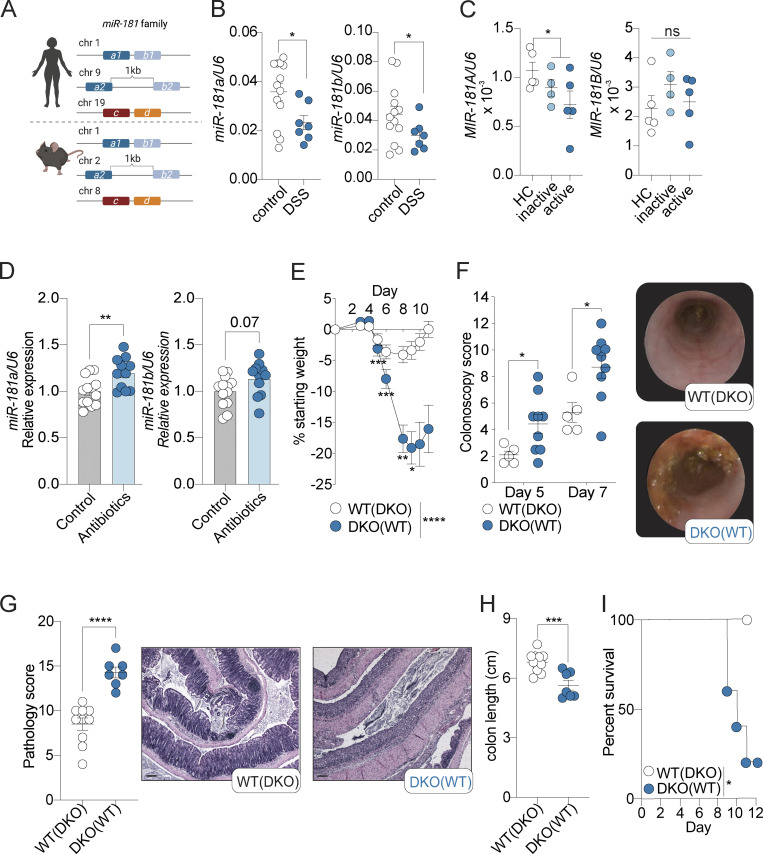
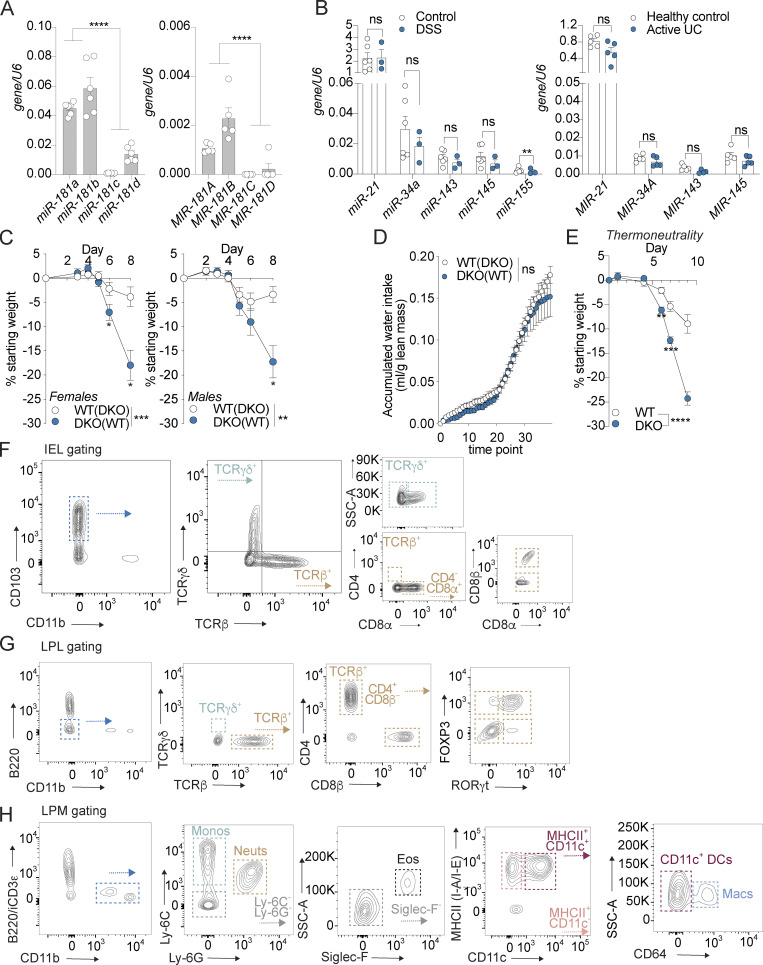
The miR-181 family is composed of six mature family members encoded on three independent loci in mice and humans (Fig. 1 A). Previous studies have shown that signals from the gut microbiota are critical for the regulation of the miR-181 family in distal tissues to promote metabolic homeostasis (Virtue et al., 2019). Moreover, miR-181 levels have been shown to be dysregulated in multiple tissues as a result of inflammatory signals, and alterations in miR-181 expression have been associated with the development of several inflammatory disorders (Cichocki et al., 2011; Hutchison et al., 2013; Virtue et al., 2019; Henao-Mejia et al., 2013; Williams et al., 2013; Hori et al., 2017; Fragoso et al., 2012; Indrieri et al., 2020; Sun et al., 2014; Zhu and Yang, 2019; Carrella et al., 2015; Witkowski et al., 2020). However, whether the miR-181 family regulates intestinal inflammation in response to microbiota-derived signals is unknown. To begin to test this, we first measured the levels of mature miR-181 family members in colonic IECs from adult WT mice at steady state and healthy human individuals. We found that miR-181a and miR-181b were expressed in mouse and human colonic IECs, whereas miR-181c and miR-181d were virtually undetectable (Fig. S1 A). As miR-181a and miR-181b were the most highly expressed members of the miR-181 family in both mouse and human colonic IECs, we then measured the levels of these miR-181 family members in colonic IECs from mice treated with the chemical colitogen dextran sulfate sodium (DSS) and from patients with inactive and active ulcerative colitis (UC; Eichele and Kharbanda, 2017; Chassaing et al., 2014). We found that the levels of mature miR-181a and miR-181b were significantly decreased in mouse IECs during DSS-induced colitis (Fig. 1 B), while only MIR-181A levels were significantly downregulated in patients with UC relative to healthy donor controls (Fig. 1 C). To assess the specificity of miR-181a and miR-181b downregulation in colonic IECs during colitis in mice and humans, we measured the expression of four to five miRNAs previously reported to be critical for regulating intestinal homeostasis and inflammation (miR-21, miR-34a, miR-143, miR-145, and miR-155). Of these miRNAs, only miR-155 was significantly decreased in colonic IECs from mice with DSS-induced colitis (Fig. S1 B). These results suggest that the development of colitis does not cause a global defect in miRNA processing and that expression of the miR-181 family is dysregulated during colonic inflammation in mice and humans.
Figure 1.
miR-181 protects against DSS-induced colitis. (A) Schematic of the relative genomic locations of the three miR-181 clusters (miR-181a1-b1, miR-181a2-b2, and miR-181c-d) in humans (top) and mice (bottom). (B) Expression of mature miR-181a and miR-181b in colonic IECs from WT mice at baseline (n = 13) or after 6–7 d of DSS treatment followed by 4–7 d of recovery (n = 7) starting at 8 wk of age. Each dot represents a single mouse. Values normalized to U6 expression. Data pooled from two independent experiments. (C) Expression of mature MIR-181A and MIR-181B in colonic IECs from human patients with inactive or active UC or from healthy individuals (healthy controls [HC]). Each dot represents a single donor. Values normalized to U6 expression. (D) Expression of mature miR-181a and miR-181b in colonic IECs from WT mice after treatment with broad-spectrum antibiotics for 7 d starting at 8 wk of age (control n = 13, antibiotic-treated n = 11). Each dot represents a single mouse. Values normalized to U6 expression. Data pooled from three independent experiments and normalized to miR-181 levels in control samples. Discrete P value shown for unpaired t test of miR-181b data (P = 0.07). (E) Mean percent change in weight from day 0 of cohoused WT (n = 22) and miR-181 DKO (n = 17) mice treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn from all animals when DKO mice reached 7.5–10% weight loss. Data pooled from four independent experiments. Parentheses indicate cohousing with specified group of mice. (F) Colonoscopy scores and representative colonoscopy images of cohoused WT (n = 5) and DKO (n = 9) mice assessed after 5 and 7 d of treatment with 2% DSS starting at 6–8 wk of age. Each dot represents a single mouse. Parentheses indicate cohousing with specified group of mice. (G) Pathology scores and representative H&E images of longitudinally sectioned colon rolls (rolled distal to proximal) from WT and DKO mice treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn from all animals when DKO mice reached 7.5–10% weight loss and allowed to recover for 3–4 d. Each dot represents a single animal. Data pooled from two independent experiments. Parentheses indicate cohousing with specified group of mice. Scale bars correspond to 0.1 mm. (H) Colon length of WT (n = 11) and DKO (n = 7) mice treated with 2% DSS. DSS was withdrawn from all animals when DKO mice reached 7.5–10% weight loss. Each dot represents a single animal. Data pooled from two independent experiments. Parentheses indicate cohousing with specified group of mice. (I) Percent survival of WT (n = 5) and DKO (n = 5) treated with 2% DSS for 6 d and allowed to recover for 6 d. Data representative of six independent experiments. Parentheses indicate cohousing with specified group of mice. Error bars indicate mean ± SEM. Unpaired Student’s t test (B–D, G, and H), multiple t tests with FDR correction (F), Kruskal–Wallis test with Dunn’s multiple comparisons (E), or Mantel–Cox log-rank test (I); *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Significance values for paired genotype comparisons from multivariate analyses are indicated with a vertical bar and appropriate P- or q-value (represented by asterisk) when appropriate. Significance values for multiple comparisons tests assessing day-to-day changes in weight loss between groups are indicated by P values next to individual data points if <0.05.
Figure S1.
miR-181 is expressed throughout the GI tract and protects against colitis independently of sex and temperature. (A) Expression of mature miR-181a, miR-181b, miR-181c, and miR-181d in colonic IECs from adult WT male mice (left) or human individuals (right) normalized to U6 expression. Each dot represents a single mouse or human. Mouse data representative of two independent experiments. (B) Levels of candidate miRNAs shown to be critical for maintaining intestinal homeostasis (miR-21, miR-34a, miR-143, miR-145, and miR-155) in mice (left) and humans (right) with colitis and healthy controls. Each dot represents a single mouse or human. (C) Mean percent change in weight from day 0 of cohoused WT IHB and DKO female (WT n = 11, DKO n = 9) and male (WT n = 11, DKO n = 8) mice treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn from all animals when DKO mice reached 7.5–10% weight loss. Data pooled from four independent experiments. (D) Evaluation of accumulated water consumption of WT and DKO mice by Comprehensive Laboratory Animal Monitoring System for 48 h (12-h light/dark cycles). Each dot represents grouped averages for the indicated timepoints. (E) Mean percent change in weight from day 0 of WT IHB (n = 14) and DKO (n = 9) male mice acclimated to thermoneutrality for 3 d and then treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn from all animals when DKO mice reached 7.5–10% weight loss. (F) IEL gating strategy. IELs were defined as CD45+ live CD103+ CD11b− cells isolated from the epithelial fraction of the large intestine. Subpopulations of IELs were categorized based on expression of TCRγδ and TCRβ. Further subcategorization of TCRγδ IELs was assessed based on expression of CD8α, while for TCRβ+ IELs, expression of CD4 and CD8α was then assessed. CD8β+ cells were quantified within the TCRβ+ CD8α+ population. (G) LPL gating strategy. LPLs were defined as CD45+ live (viability−) B220− CD11b− cells isolated from the LP of the large intestine. Subpopulations of LPLs were categorized based on expression of TCRγδ and TCRβ. Further subcategorization of TCRβ+ LPLs was assessed based on expression of CD4 and CD8β. CD4+ cells were further categorized based on expression of FOXP3 and RORγt. (H) LPM gating strategy. LPMs were defined as CD45+ live (viability−) B220− intracellular CD3ε− CD11b+ cells isolated from the LP of the large intestine. Subpopulations of LPMs were categorized as follows: monocytes (Ly-6C+ Ly-6G−, monos), neutrophils (Ly-6C− Ly-6G+, neuts), eosinophils (Ly-6C− Ly-6G− Siglec-F+ SSC-Ahi, eos), CD11b+ dendritic cells (Ly-6C− Ly-6G− Siglec-F− MHCII+ CD11c+ CD64−, DCs), and macrophages (Ly-6C− Ly-6G− Siglec-F− MHCII+ CD11c+/− CD64+, macs). Error bars indicate mean ± SEM. Student’s unpaired t test (A), multiple t tests with FDR correction (B), and two-way ANOVA with Sidak’s multiple comparisons test (C–E). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Significance values for paired genotype comparisons from multivariate analyses are indicated with a vertical bar and appropriate P- or q-value (represented by asterisk) when appropriate. Significance values for multiple comparisons tests assessing day-to-day changes in weight loss between groups are indicated by P values next to individual data points if <0.05.
Microbiota-derived signals are key regulators of gene expression programs in the GI tract during health and disease (Muller et al., 2020b; Ohnmacht et al., 2015; Lavelle and Sokol, 2020). As such, we tested if colonic miR-181 levels are regulated by the microbiota. To do so, we treated WT specific pathogen–free mice with broad-spectrum antibiotics and determined mature miR-181a and miR-181b expression in colonic IECs. We found that antibiotic treatment significantly increased mature miR-181 levels in colonic IECs (Fig. 1 D). Altogether, these results suggest that the miR-181 family may play a role in regulating colitis progression through its activity in the GI tract and that colonic miR-181 levels are controlled by inflammatory and microbiota-derived signals.
The miR-181 family protects against the development of colitis
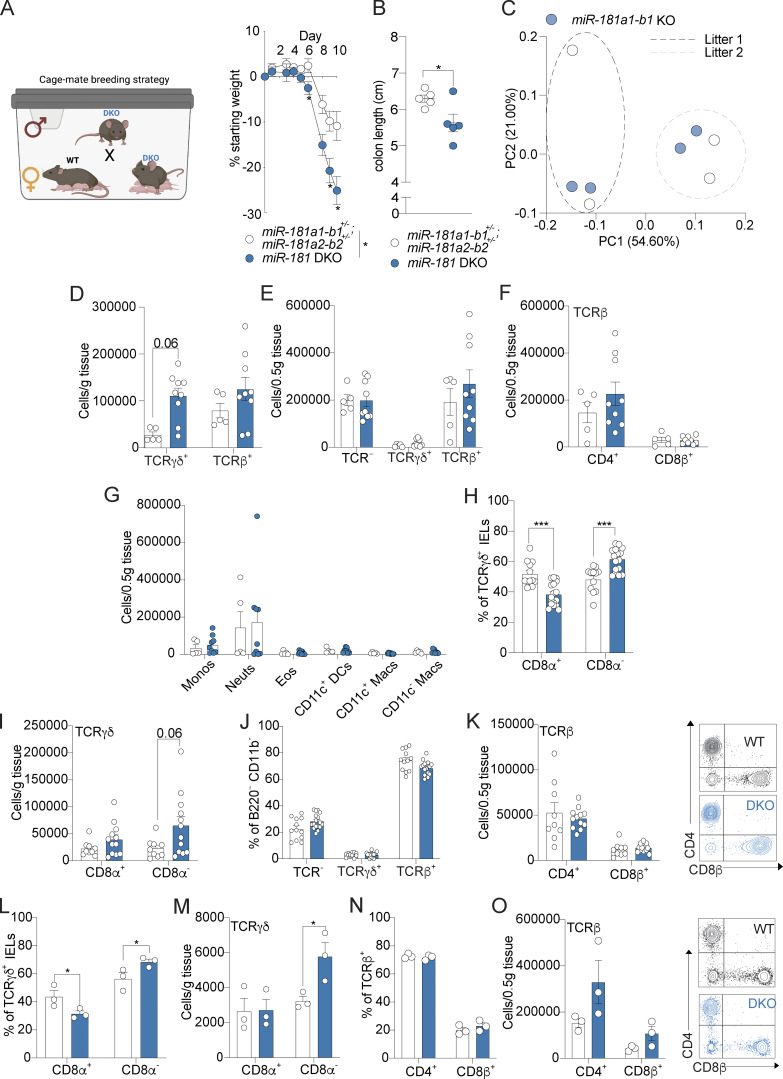
To establish whether the miR-181 family plays a role in regulating mucosal inflammation, we first determined if progression of acute DSS-induced colitis is impacted in the absence of this miRNA family. As triple KO of all three miR-181 clusters is embryonically lethal (Henao-Mejia et al., 2013), we generated mice deficient in the two loci encoding the most highly expressed mature miR-181 species of the miR-181 family in colonic IECs (miR-181a1-b1−/−; miR-181a2-b2−/−; Fig. S1 A) referred to here as double KO (DKO) mice. We then cohoused WT and DKO mice for 3–4 wk, indicated throughout by parentheses. We administered 2% DSS in drinking water for 6 d and assessed colitis severity by measuring weight loss, colonic pathology via colonoscopy and histopathological examination, colon length, and overall survival. Strikingly, and in concordance with our expression data in mice and humans, we found that DKO mice were highly susceptible to colitis development as they lost a significantly greater percentage of their body weight and had more severe colonic pathology and inflammation, ultimately resulting in poorer survival (Fig. 1, E–I; and Tables S1 and S2). Furthermore, this phenotype was not due to sex-dependent differences (Fig. S1 C) and drinking water consumption as determined by a Comprehensive Lab Animal Monitoring System (Fig. S1 D) or altered energy expenditure as DKO mice were still more susceptible to DSS-induced colitis under thermoneutral conditions compared to their WT counterparts (Fig. S1 E). Altogether, our results indicate that the miR-181 family plays a crucial role in protecting against severe colitis in response to mucosal injury.
Multiple miR-181 loci contribute to protection against colitis
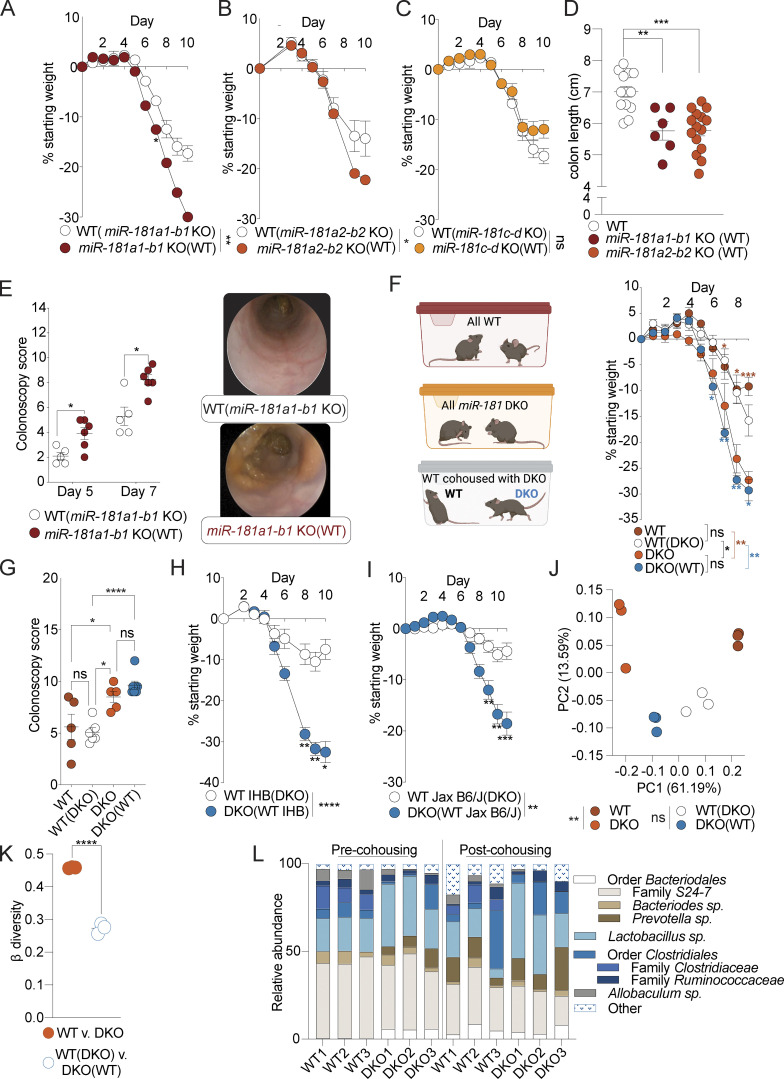
Discrete loci of the miR-181 family have been shown to have nonredundant roles in regulating inflammatory processes and immune cell development (Henao-Mejia et al., 2013; Fragoso et al., 2012). As such, we aimed to determine which miR-181 loci contribute to protection against DSS-induced colitis in mice. We generated single KO mice deficient for one of three loci of the miR-181 family and measured their susceptibility to DSS colitis as described above. We found that loss of the miR-181a1-b1 or miR-181a2-b2 locus, but not the miR-181c-d locus, caused a significant increase in susceptibility to colitis as measured by body weight loss (Fig. 2, A–C), in concordance with our expression data (Fig. S1 A). Moreover, loss of either the miR-181a1-b1 or miR-181a2-b2 locus also led to increased colonic shortening (Fig. 2 D). Finally, colonoscopy revealed significant increases in disease severity over the course of DSS colitis as a result of the loss of miR-181a1-b1 (Fig. 2 E). These data suggest that the expression of microRNAs encoded by the miR-181a1-b1 and miR-181a2-b2 loci have protective effects against the development of severe DSS-induced colitis.
Figure 2.
Multiple miR-181 loci are sufficient to protect against colitis. (A) Mean percent change in weight from day 0 of cohoused WT (n = 3) and miR-181a1-b1 KO mice (n = 6) treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn at day 6, and animals were allowed to recover for 4 d. Data from one experiment. Parentheses indicate cohousing with specified group of mice. (B) Mean percent change in weight from day 0 of cohoused WT (n = 19) and miR-181a2-b2 KO mice (n = 28) treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn at day 7, and animals were allowed to recover for 3 d. Data pooled from two independent experiments. Parentheses indicate cohousing with specified group of mice. (C) Mean percent change in weight from day 0 of cohoused WT (n = 3) and miR-181c-d KO mice (n = 6) treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn at day 6, and animals were allowed to recover for 4 d. Parentheses indicate cohousing with specified group of mice. (D) Colon length of WT (n = 13), miR-181a1-b1 KO (n = 6), and miR-181a2-b2 KO (n = 15) mice treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn at day 7, and animals were allowed to recover for 3 d. Parentheses indicate cohousing with specified group of mice. Each dot represents a single animal. Data from one to two independent experiments. (E) Colonoscopy scores and representative colonoscopy images of cohoused WT B6/J (n = 5) and miR-181a1-b1 KO (n = 6) mice assessed after 5 and 7 d of treatment with 2% DSS starting at 6–8 wk of age. Each dot represents an individual animal. (F and G) Mean percent change in weight from day 0 (F) and terminal colonoscopy scores (G) of cohoused WT (n = 5) and DKO mice (n = 7) and noncohoused WT (n = 5) and DKO mice (n = 5) treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn at day 6, and animals were allowed to recover. Group comparisons are indicated by colored asterisks and brackets (red = WT vs. DKO, blue = WT[DKO] vs. DKO[WT]). Experimental set up is illustrated for clarity (F). For colonoscopy scores, each dot represents an individual animal. Parentheses indicate cohousing with specified group of mice. (H) Mean percent change in weight from day 0 of IHB WT mice cohoused with DKO mice and treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn at day 6, and animals were allowed to recover for 4 d. Data representative of six independent experiments. Parentheses indicate cohousing with specified group of mice. (I) Mean percent change in weight from day 0 of WT mice from Jax cohoused with DKO mice and treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn from all animals when DKO mice reached 7.5–10% weight loss. Data pooled from three independent experiments. Parentheses indicate cohousing with specified group of mice. (J and K) PCA (J) and β diversity (K) analysis of fecal microbial communities as determined by 16s rRNA-seq of stool from WT (n = 3) and DKO mice (n = 3) pre- and post-cohousing. Each dot represents an individual animal at a given condition (J) or discrete grouped comparisons (K). PERMANOVA statistics reported for PCA for grouped comparisons (J). Data representative of two independent experiments. Parentheses indicate cohousing with specified group of mice. (L) Individual mouse fecal microbiota composition at the genus level as determined by 16s rRNA-seq of stool from WT (n = 3) and DKO mice (n = 3) pre- and post-cohousing. The most-resolved taxonomic information is indicated. Nested taxonomic categories indicate related groups. Data representative of two independent experiments. Error bars indicate mean ± SEM. Two-way ANOVA with Sidak’s multiple comparisons test (A–C, F, H, and I), multiple t tests with FDR correction (D, E, and G), PERMANOVA (J), or unpaired Student’s t test (K). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Significance values for paired genotype comparisons from multivariate analyses are indicated with a vertical bar and appropriate P- or q-value (represented by asterisk) when appropriate. Significance values for multiple comparisons tests assessing day-to-day changes in weight loss between groups are indicated by P values next to individual data points if <0.05.
Increased susceptibility of miR-181–deficient mice to colitis is independent of microbiota composition
The development of colitis has been shown to be highly dependent on the composition of the gut microbiota in both mice and humans (Kaiko et al., 2016; Lavelle and Sokol, 2020; Britton et al., 2019). To determine whether the gut microbiota composition was the key determinant of the enhanced susceptibility to DSS-induced colitis in DKO mice, we administered DSS to WT and DKO mice generated from homozygous breeding and housed with littermates of the same genotype or cohoused at weaning with DKO or WT mice, respectively. We found that cohousing DKO mice with WT mice did not rescue the increased severity of DSS colitis driven by the loss of miR-181a and miR-181b and did not significantly increase colitis susceptibility in WT mice as measured by percentage body weight loss and colonoscopy score (Fig. 2, F and G). Moreover, cohousing DKO mice with WT mice derived from in-house breeding (UPenn vivarium) or from Jackson Labs did not rescue the development of severe colitis in miR-181–deficient mice (Fig. 2, H and I). 16s sequencing of feces from WT and DKO mice pre- and post-cohousing showed that cohousing reduced the variability between the gut microbial communities of WT and DKO mice via principal components analysis (PCA) and assessment of β diversity (Fig. 2, J–L). Furthermore, miR-181–deficient offspring from DKO male mice crossed to WT and DKO females in the same cage were more susceptible to DSS colitis compared to their double heterozygous cage-mates as measured by body weight loss and colonic shortening (Fig. S2, A and B). Finally, to formally establish whether miR-181 directly regulates the composition of the gut microbiota, we performed 16s sequencing on feces from miR-181a1-b1+/+ and miR-181a1-b1−/− littermates (Fig. S2 C). Using PCA, we found that most of the variation between miR-181a1-b1+/+ and miR-181-a1-b1−/− gut microbial communities was driven by litter and not by genotype. While cohousing does not eliminate all microbial variation between WT and DKO mice, altogether these results indicate that the composition of the gut microbiota is not the key determinant of increased susceptibility to DSS colitis in miR-181–deficient mice.
Figure S2.
miR-181 regulates the IEL milieu at homeostasis. (A and B) Mean percent change in weight from day 0 (A) and colon length (B) of cage-mate miR-181a1-b1+/−; miR-181a2-b2+/− (n = 5) and DKO mice (n = 12) treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn at day 8, and animals were allowed to recover for 2 d. Breeding strategy is illustrated for clarity (A). For colon length (B), each dot represents a single animal. (C) PCA of fecal microbial communities as determined by 16s rRNA-seq of stool from WT (n = 2 per litter) and miR-181a1-b1 KO littermates (n = 2 per litter) from two separate litters. Each dot represents an individual animal. (D) Total number of TCRγδ+ and TCRβ+ large intestinal IELs from WT and DKO mice at baseline. Each dot represents an individual mouse. Gated on: Singlets CD45+ Live (viability−) CD103+ CD11b− cells. Data pooled from two (WT n = 5, DKO n = 9) independent experiments. (E) Total number of TCR-, TCRγδ+, and TCRβ+ large intestinal LP cells from WT and DKO mice at baseline. Each dot represents an individual mouse. Gated on: Singlets CD45+ Live (viability−) B220− CD11b− cells. Data pooled from two (WT n = 5, DKO n = 9) independent experiments. (F) Total number of TCRβ+ CD4+/− CD8β+/− large intestinal LP cells from WT and DKO mice at baseline. Each dot represents an individual mouse. Gated on: Singlets CD45+ Live (viability−) B220− CD11b− TCRβ+ LP cells. Data pooled from two (WT n = 5, DKO n = 9) independent experiments. (G) Total number of large intestinal LPMs: monocytes (Ly-6C+ Ly-6G−, monos), neutrophils (Ly-6C− Ly-6G+, neuts), eosinophils (Ly-6C− Ly-6G− Siglec-F+ SSC-Ahi, eos), CD11b+ dendritic cells (Ly-6C− Ly-6G− Siglec-F- MHCII+ CD64− CD11c+, DCs), and macrophages (Ly-6C− Ly-6G− Siglec-F− MHCII+ CD11c+/− CD64+, macs) in WT and DKO mice at baseline. Gated on: Singlets CD45+ Live (viability−) B220− intracellular CD3ε− CD11b+ cells. Each dot represents a single mouse. Data pooled from two (WT n = 5, DKO n = 9) independent experiments. (H and I) Percentage (H) and total number (I) of TCRγδ+ large intestinal IELs expressing CD8α from WT (n = 8–9) and DKO (n = 12) mice after 5 d of 2% DSS treatment. Each dot represents an individual mouse. Gated on: Singlets CD45+ Live CD103+ CD11b− TCRγδ+ cells. Data pooled from two independent experiments. (J) Percentage of TCR−, TCRγδ+, and TCRβ+ large intestinal LPLs from WT (n = 8–9) and DKO (n = 12) mice after 5 d of 2% DSS treatment. Each dot represents an individual mouse. Gated on: Singlets CD45+ Live CD103+ CD11b− cells. Data pooled from two independent experiments. (K) Total number of TCRβ+ large intestinal LPLs expressing CD4 and CD8β from WT (n = 8–9) and DKO (n = 12) mice after 5 d of 2% DSS treatment. Each dot represents an individual mouse. Gated on: Singlets CD45+ Live CD103+ CD11b− TCRβ+ cells. Representative FACS plot shown on the right. Data pooled from two independent experiments. (L and M) Percentage (L) and total number (M) of TCRγδ+ large intestinal IELs expressing CD8α from WT (n = 3) and DKO (n = 3) mice after 7 d of 2% DSS treatment and 7 d of recovery. Each dot represents an individual mouse. Gated on: Singlets CD45+ Live CD103+ CD11b− TCRγδ+ cells. (N) Percentage of TCR−, TCRγδ+, and TCRβ+ large intestinal LPLs from WT (n = 3) and DKO (n = 3) mice after 7 d of 2% DSS treatment and 7 d of recovery. Each dot represents an individual mouse. Gated on: Singlets CD45+ Live CD103+ CD11b− cells. (O) Total number of TCRβ+ large intestinal LPLs expressing CD4 and CD8β from WT (n = 3) and DKO (n = 3) mice after 7 d of 2% DSS treatment and 7 d of recovery. Each dot represents an individual mouse. Gated on: Singlets CD45+ Live CD103+ CD11b− TCRβ+ cells. Representative FACS plot shown on the right. Error bars indicate mean ± SEM. Two-way ANOVA with Sidak’s multiple comparisons test (A), Student’s unpaired t test (B), multiple t tests with FDR correction (D–O). *, P < 0.05; ***, P < 0.001. Each dot represents a single mouse (B–O). Significance values for paired genotype comparisons from multivariate analyses are indicated with a vertical bar and appropriate P- or q-value (represented by asterisk) when appropriate. Significance values for multiple comparisons tests assessing day-to-day changes in weight loss between groups are indicated by P values next to individual data points if <0.05.
miR-181 regulates the immune cell milieu in the intestinal epithelium
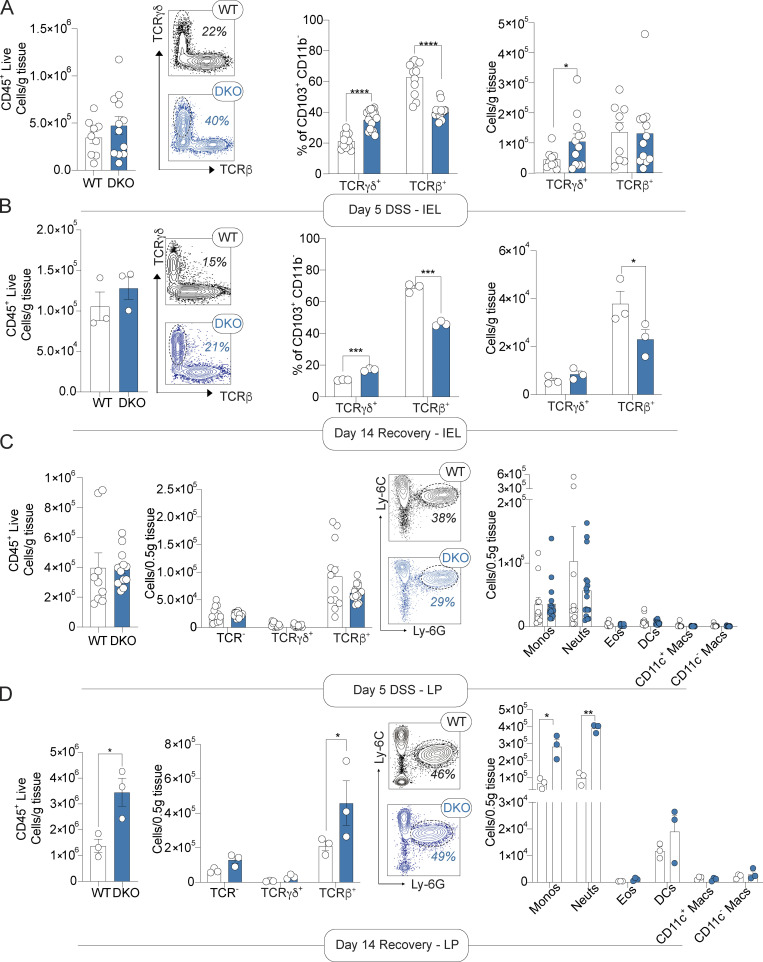
The miR-181 family has been shown to be a critical regulator of the development and function of multiple innate and adaptive immune cell types that contribute to colonic inflammation (Cichocki et al., 2011; Hutchison et al., 2013; Henao-Mejia et al., 2013; Williams et al., 2013; Su et al., 2015; Weng et al., 2015; Sun et al., 2014). Thus, we aimed to determine the composition of the colonic immune cell milieu of miR-181 DKO mice at baseline and during the development of DSS colitis via flow cytometry (Fig. S1, F–H). Distinct populations of immune cells within the GI tract can be found in the lamina propria (LP) and between IECs (intestinal intraepithelial lymphocytes [IELs]). At baseline, we found that the loss of miR-181 caused a dramatic shift in the frequency of IEL populations and an overall increase in the total number of IELs largely driven by an increase in the number of TCRγδ+ IELs (Fig. S2 D). However, we found few differences in the abundance of T cells or myeloid cells in the LP (Fig. S2, E–G). During early DSS colitis, changes observed in the IEL compartment as a result of miR-181 loss and were maintained with an increased abundance of TCRγδ+ IELs, while there were few changes in the LP T cell compartment (Fig. 3 A and Fig. S2, H–K). Although miR-181–dependent changes in the IEL compartment persisted during early colitis, increases in IEL number were diminished during recovery (Fig. 3 B and Fig. S2, L and M; Yang et al., 2016; Muñoz-Rojas and Mathis, 2021). More importantly, we observed a striking elevation in the number of myeloid cells within the LP of DKO mice during recovery from colitis, indicating persistent colonic inflammation (Fig. 3, C and D; and Fig. S2, N and O). These data suggest that the colonic immune cell landscape is dramatically altered by the absence of miR-181 at homeostasis and in the context of mucosal injury.
Figure 3.
miR-181 protects against chronic inflammation in response to mucosal injury. (A and B) Large IELs after 5 d of (A) and recovery from (B) DSS treatment. From left to right, total number of CD45+ live cells, representative FACS plots of WT and DKO mouse IELs showing distribution of TCRγδ and TCRβ populations with percentage of TCRγδ IELs in each, and percentage and total number of TCRγδ+ and TCRβ+ IELs from WT and DKO mice. Each dot represents an individual mouse. (C and D) Large intestine LP immune cells after 5 d of (C) and recovery from (D) DSS treatment. From left to right, total number of CD45+ live cells, total number of TCRγδ+ and TCRβ+ LPLs from WT and DKO mice, representative FACS plots of WT and DKO mouse LPMs showing distribution of Ly-6C and Ly-6G populations with percentage of neutrophils in each, and total number of LP monocytes (Ly-6C+ Ly-6G−, monos), neutrophils (Ly-6C− Ly-6G+, neuts), eosinophils (Ly-6C− Ly-6G− Siglec-F+, SSC-Ahi, eos), CD11b+ dendritic cells (Ly-6C− Ly-6G− Siglec-F− MHCII+ CD11c+ CD64−, DCs), and macrophages (Ly-6C− Ly-6G− Siglec-F− MHCII+ CD11c+/− CD64+, macs) in WT and DKO mice. Each dot represents an individual mouse. For early DSS analysis, data are pooled from two independent experiments with a total of n = 8–9 WT and n = 12 DKO mice. Data collected from mice during recovery from DSS (7 d of DSS treatment, 7 d of recovery) were from one experiment with a total of n = 3 WT and n = 3 DKO animals. Error bars indicate mean ± SEM. Unpaired t test (CD45+ live quantification), multiple t tests with FDR correction (A–D). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
miR-181 does not protect against colitis via its activity in the immune system
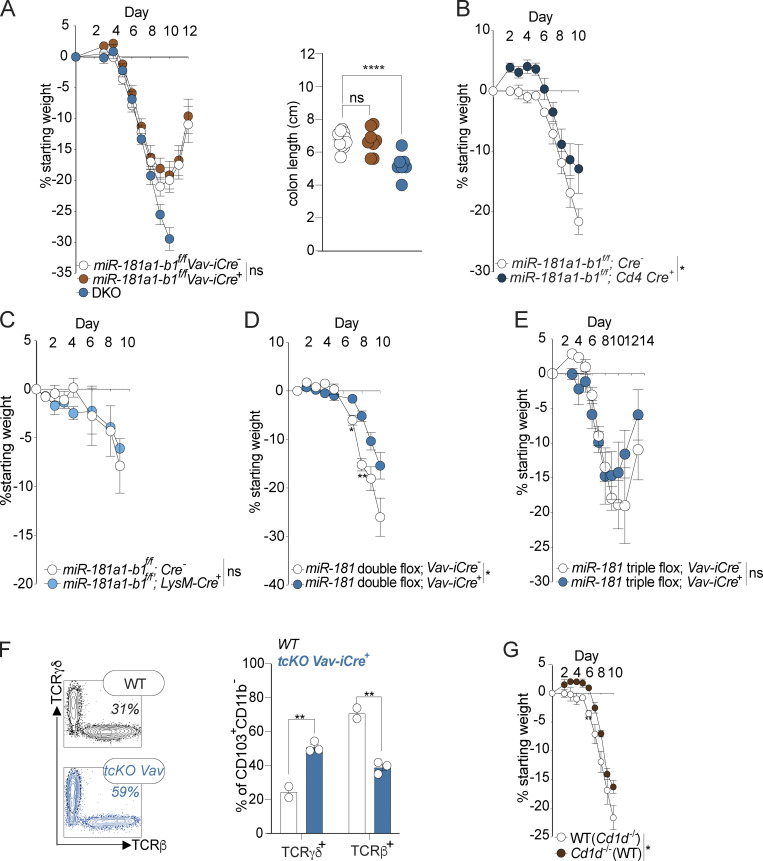
Dysregulation of the miR-181 family, particularly the miR-181a1-b1 locus, has been associated with the development of multiple inflammatory disorders (Liu et al., 2018; Fragoso et al., 2012; Indrieri et al., 2020; Sun et al., 2014; Zhu and Yang, 2019; Witkowski et al., 2020). Moreover, miR-181 been shown to regulate the development and function of immune cells to control a multitude of homeostatic and inflammatory processes. As full body KO of miR-181a1-b1 results in increased susceptibility to DSS-induced colitis (Fig. 2 A), we hypothesized that expression of miR-181a and miR-181b from this locus within the hematopoietic system contributes to the protection against severe DSS-induced colitis. To test this hypothesis, we conditionally depleted miR-181a1-b1 within the entire hematopoietic compartment (Vav-iCre), or specifically in T cells (Cd4-Cre) or myeloid cells (LysM-Cre), and thereafter assessed susceptibility to DSS colitis (Fig. 4, A–C). Interestingly, we did not observe any changes in colitis severity in any of these miR-181a1-b1–deficient mouse strains. Next, we tested whether depletion of multiple protective miR-181 loci within the hematopoietic system is required to cause increased susceptibility to colonic inflammation after mucosal injury. We depleted both miR-181a1-b1 and miR-181a2-b2 loci (double conditional KO [dcKO]) as well as the entire miR-181 family (triple conditional KO [tcKO]) within the hematopoietic compartment and determined susceptibility to DSS-induced colonic inflammation. Concordantly, we did not observe an increase in susceptibility to colitis as determined by weight loss in miR-181 dcKO; Vav-iCre and miR-181 tcKO; and Vav-iCre mice (Fig. 4, D and E). Importantly, we observed an increase in TCRγδ+ IELs in mice deficient for miR-181a and miR-181b within the hematopoietic system, indicating that dysregulated numbers of TCRγδ+ IELs in DKO mice is not the main driver of increased susceptibility to DSS-induced colitis as a result of miR-181 loss (Fig. 4 F). Finally, as miR-181a1-b1 KO mice lack natural killer T (NKT) cells (Henao-Mejia et al., 2013), we assessed the possibility that NKT cell deficiency could be responsible for increased DSS-induced colitis severity in these mice. Importantly, CD1d-deficient mice, which lack NKT cells, did not show increased colonic inflammation or disease severity and in fact were less susceptible to DSS-induced weight loss (Fig. 4 G). Altogether these results suggest that the miR-181 family does not act within the immune system to protect against colitis and that the changes in the IEL milieu driven by miR-181 deficiency do not cause increased susceptibility to colitis.
Figure 4.
miR-181 does not act within the hematopoietic system to protect against severe colitis. (A) Mean percent change in weight from day 0 and colon length of miR-181a1-b1f/f; Vav-iCre− (n = 17), miR-181a1-b1f/f; Vav-iCre+ (n = 11), and DKO mice (n = 10) treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn at day 7 and animals were allowed to recover for 5 d. For colon length data, each dot represents an individual mouse. Data are representative of two independent experiments. (B) Mean percent change in weight from day 0 miR-181a1-b1f/f; Cd4-Cre− (n = 5) and miR-181a1-b1f/f; Cd4-Cre+ (n = 3) mice treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn at day 7, and animals were allowed to recover for 3 d. Data representative of two independent experiments. (C) Mean percent change in weight from day 0 of miR-181a1-b1f/f; LysM-Cre− (n = 3) and miR-181a1-b1f/f; LysM-Cre+ (n = 3) mice treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn at day 8, and animals were allowed to recover. Data representative of two independent experiments. (D) Mean percent change in weight from day 0 of miR-181a1-b1f/f; miR-181a2-b2f/f; Vav-iCre− (n = 4) and miR-181a1-b1f/f; miR-181a2-b2f/f; Vav-iCre+ (n = 6) mice treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn at day 7, and animals were allowed to recover. Data pooled from two independent experiments. (E) Mean percent change in weight from day 0 of miR-181a1-b1f/f; miR-181a2-b2f/f; miR-181c-df/f; Vav-iCre− (n = 3) and miR-181a1-b1f/f; miR-181a2-b2f/f; miR-181c-df/f; Vav-iCre+ (n = 4) mice treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn at day 7, and animals were allowed to recover. (F) Percentage (left) and representative FACS plots (right) of TCRγδ+ and TCRβ+ IELs from 6–8-wk-old miR-181a1-b1f/f; miR-181a2-b2f/f; miR-181c-df/f; Vav-iCre− (WT) and miR-181a1-b1f/f; miR-181a2-b2f/f; miR-181c-df/f; Vav iCre+ mice (tcKO Vav-iCre+) after a 7-d treatment with 2% DSS followed by 1 wk of recovery. Each data point represents an individual mouse. Gated on: Singlets CD45+ Live (viability−) CD103+ CD11b− cells. Data representative of two independent experiments. (G) Mean percent change in weight from day 0 WT (n = 5) and Cd1d−/− (n = 3) mice treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn at day 8, and animals were allowed to recover. Error bars indicate mean ± SEM. Two-way ANOVA with Sidak’s multiple comparisons test (A–E and G), multiple Kruskal–Wallis tests with FDR correction (A and F). *, P < 0.05; **, P < 0.01; ****, P < 0.0001. Significance values for genotype comparisons are indicated with a vertical bar and appropriate P value represented by asterisk. Significance values for paired genotype comparisons from multivariate analyses are indicated with a vertical bar and appropriate P- or q-value (represented by asterisk) when appropriate. Significance values for multiple comparisons tests assessing day-to-day changes in weight loss between groups are indicated by P values next to individual data points if <0.05.
IEC-intrinsic miR-181 protects against colitis
Our earlier results indicate that expression of the miR-181 family in nonhematopoietic cells protects against the development of severe DSS-induced colitis, while miR-181 within the hematopoietic compartment appears to cause increased susceptibility to colonic inflammation (Fig. 4). To formally test this hypothesis, we first generated bone marrow (BM) chimeras by transplanting WT or DKO BM cells into lethally irradiated WT or DKO mice. As expected, the severity of DSS-induced colitis was significantly greater in DKO mice engrafted with DKO BM cells when compared to WT mice engrafted with WT BM cells as determined by body weight loss and colonoscopy. Importantly, and in concordance with our results indicating that expression of hematopoietic miR-181 does not contribute to protection from colitis, DKO mice engrafted with WT BM cells were significantly more susceptible to DSS-induced colitis compared to WT mice engrafted with DKO BM cells (Fig. 5, A and B). Moreover, DKO mice that were transplanted with WT BM were significantly more susceptible to colitis as measured by weight loss compared to WT mice transplanted with WT BM, suggesting that miR-181 expression within the radioresistant compartment, but not the radiosensitive compartment, protects against colitis.
Figure 5.
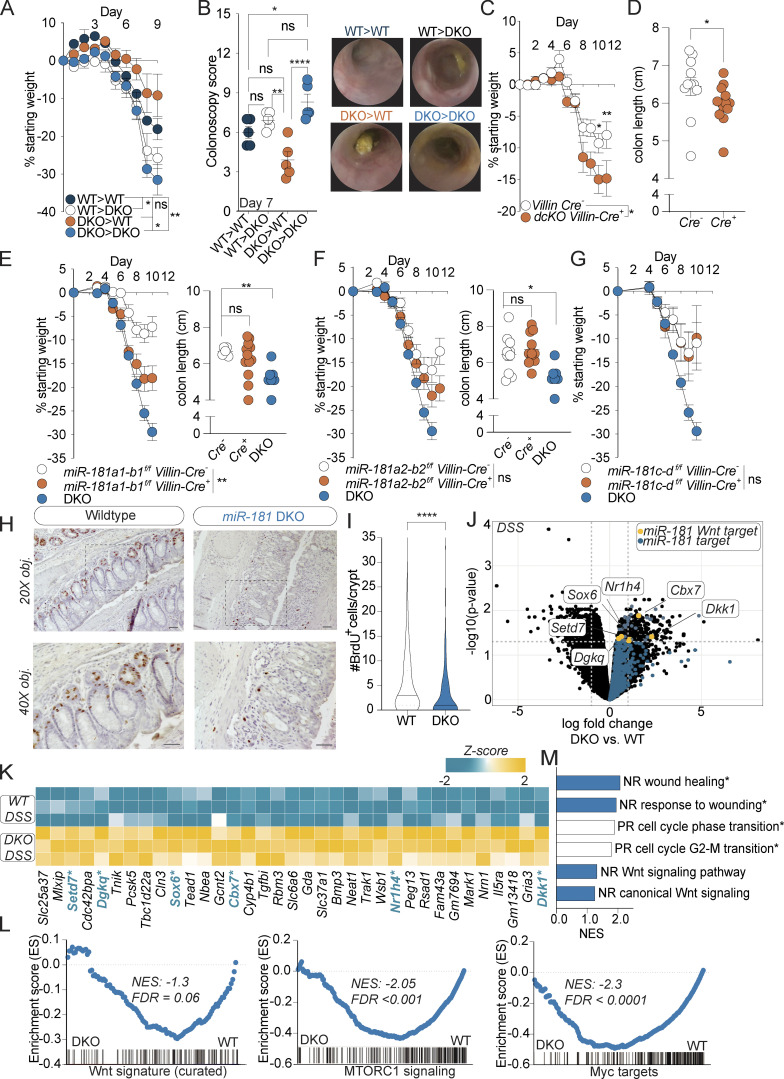
miR-181 acts within IECs to protect against colitis. (A and B) Mean percent change in weight from day 0 (A) and colonoscopy scores (B) of WT and DKO BM chimeric mice treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn at day 7, and colonoscopy was performed. Animals were thereafter allowed to recover. For colonoscopy data, each dot represents an individual mouse. Representative colonoscopy images are shown on the right. (C and D) Mean percent change in weight from day 0 (C) and colon length (D) of miR-181a1-b1f/f; miR-181a2-b2f/f; Villin-Cre− (n = 12) and miR-181a1-b1f/f; miR-181a2-b2f/f; Villin-Cre+ (n = 15) mice treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn from all animals when DKO mice reached 7.5–10% weight loss. For colon length data, each dot represents an individual mouse. Data pooled from three independent experiments. (E) Mean percent change in weight from day 0 and colon length of miR-181a1-b1f/f; Villin-Cre− (n = 6), miR-181a1-b1f/f; Villin-Cre+ (n = 13), and DKO (n = 10) mice treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn at day 7, and animals were allowed to recover. For colon length data, each dot represents an individual mouse. Data representative of two independent experiments. (F) Mean percent change in weight from day 0 and colon length of miR-181a2-b2f/f; Villin-Cre− (n = 16), miR-181a2-b2f/f; Villin-Cre+ (n = 12), and DKO (n = 10) mice treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn at day 7, and animals were allowed to recover. For colon length data, each dot represents an individual mouse. Data representative of two independent experiments. (G) Mean percent change in weight from day 0 of miR-181c-df/f; Villin-Cre− (n = 6), miR-181c-df/f; Villin-Cre+ (n = 5), and DKO (n = 10) mice treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn at day 7, and animals were allowed to recover. Data representative of two independent experiments. (H and I) Representative images and quantification of the number of BrdU+ cells per colonic crypt (50 crypts counted per mouse) in WT and DKO mice after 8 d of 2% DSS administration and 4 d of recovery. Mice were then injected intraperitoneally with 100 mg/g of body weight of BrdU and sacrificed 4 h after injection for histology with representative images shown. Data pooled from two independent experiments. Median values indicated by horizontal bar. Scale bars in H indicate 0.1 mm. (J) Volcano plot of genes expressed in colonic IECs from WT (n = 3) and DKO (n = 3) mice treated with DSS for 7 d and allowed to recover for 2 d. All miR-181 targets increased in DKO mice are highlighted in blue with specific negative regulators of the Wnt pathway highlighted in yellow. (K) Heat map of all significantly enriched miR-181 target genes (n = 35) in IECs from DSS-treated DKO mice relative to WT. Negative regulators of the Wnt pathway (n = 6) are highlighted in blue and indicated by asterisk. (L) GSEA of RNA-seq data (TMM-normalized log2-transformed gene counts per million) from colonic IECs derived from WT and DKO mice treated with DSS for 7 d and allowed to recover for 2 d. Shown are the GSEA plots for a curated Wnt signature from Van der Flier et al. (2007), and hallmark pathways mTORC1 and Myc targets (V1). Normalized enrichment scores (NES) and FDR values are included. (M) Gene ontology pathways as assessed by GSEA that were enriched in colonic IECs from DSS-treated WT (white bars) or DKO (blue bars) mice. Significant pathways (FDR < 0.01) indicated by asterisk. NR, negative regulation; PR, positive regulation. For E–G, representative experiments were performed at the same time and the same noncohoused DKO mice (n = 10) were used as positive controls for these experiments. Error bars indicate mean ± SEM. Two-way ANOVA with Sidak’s multiple comparisons test (A, C, and E–G), multiple t tests with false discovery rate correction (B and D–F), Kolmogorov–Smirnov test (I). *, P < 0.05; **, P < 0.01; ****, P < 0.0001. Significance values for paired genotype comparisons from multivariate analyses are indicated with a vertical bar and appropriate P- or q-value (represented by asterisk) when appropriate. Significance values for multiple comparisons tests assessing day-to-day changes in weight loss between groups are indicated by P values next to individual data points if <0.05.
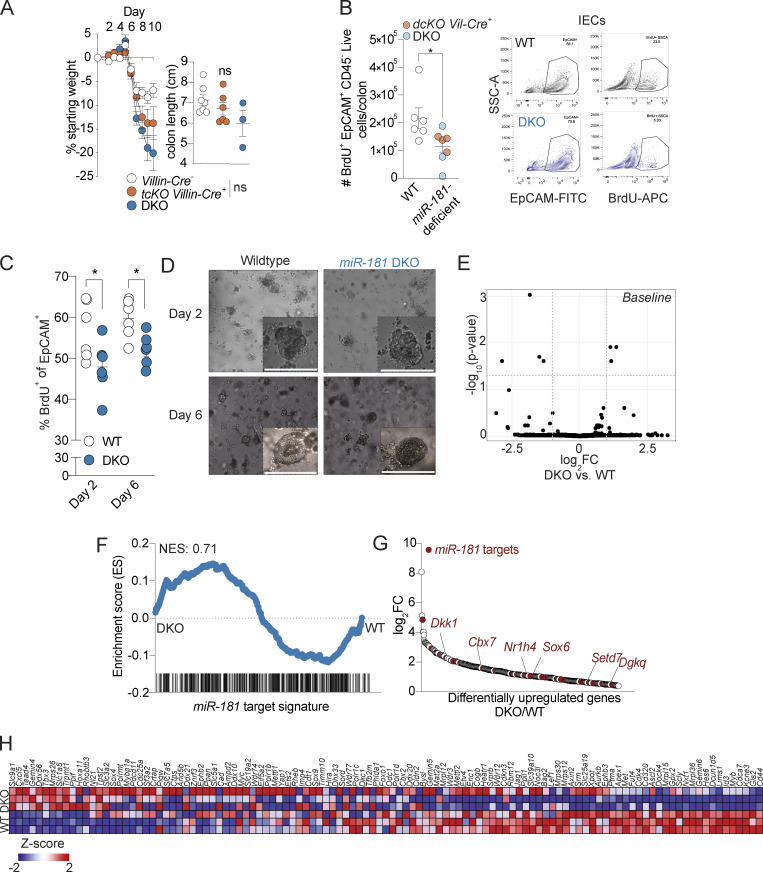
We previously showed that miR-181a and miR-181b are expressed in colonic IECs and that their expression within IECs is significantly decreased in the context of colitis in mice and humans. Thus, we hypothesized that expression of the miR-181 family in IECs prevents the development of severe colonic inflammation. To test this hypothesis, we generated mice deficient in miR-181a1-b1 and miR-181a2-b2 loci in IECs (dcKO; Villin-Cre) in addition to mice with full depletion of the miR-181 family within IECs (tcKO; Villin-Cre). Strikingly, we found that loss of multiple miR-181 loci within IECs leads to a significant increase in colitis severity as measured by weight loss and colon length (Fig. 5, C and D; and Fig. S3 A). We then assessed the contributions of each miR-181 locus within IECs to protect against severe colitis by generating the following mouse strains: miR-181a1-b1f/f; Villin-Cre, miR-181a2-b2f/, Villin-Cre, and miR-181c-df/f; Villin-Cre. We thereafter treated these mice with DSS and measured weight loss and colon length to determine colitis susceptibility (Fig. 5, E–G). Strikingly, miR-181a1-b1 depletion within IECs significantly increased colitis susceptibility, while loss of IEC-specific miR-181c-d did not protect against colonic inflammation. Interestingly, depletion of miR-181a2-b2 in IECs led to a modest nonsignificant increase in body weight loss during DSS treatment (Fig. 5 F). These results indicate that expression of the miR-181a1-b1 locus within IECs contributes to protection against colonic inflammation in response to mucosal injury. Moreover, it suggests that the expression of the miR-181a1-b1 and miR-181a2-b2 loci in other radioresistant cell compartments could also contribute to protection from the development of severe colitis in response to DSS.
Figure S3.
miR-181 regulates the proliferative capacity of IECs. (A) Mean percent change in weight from day 0 (left) and colon length (right) of miR-181a1-b1f/f; miR-181a2-b2f/f; miR-181c-df/f; Villin-Cre− (n = 22), miR-181a1-b1f/f; miR-181a2-b2f/f; miR-181c-df/f; Villin-Cre+ (n = 14), and DKO (n = 12) mice treated with 2% DSS starting at 6–8 wk of age. DSS was withdrawn from all animals when DKO mice reached 7.5–10% weight loss. For colon length data, each dot represents an individual mouse. Data pooled from two independent experiments. (B) Quantification of in vivo BrdU incorporation in colonic IECs from WT (n = 3), miR-181a1-b1f/f; miR-181a2-b2f/f Villin-Cre− (n = 3), miR-181a1-b1f/f; miR-181a2-b2f/f Villin-Cre+ (n = 4), and DKO (n = 3) mice treated with DSS for 7–8 d and allowed to recover for 2 d. Mice were injected intraperitoneally with 100 mg/kg of BrdU and allowed to rest for 4 h prior to sacrifice. Colonic IECs were isolated and assayed for BrdU incorporation via flow cytometry. Each dot represents an individual animal. The gating strategy is shown on the right. Gated on: Singlets Live EpCAM+ CD45− BrdU+ cells. Data pooled from two independent experiments. (C and D) Percentage of in vitro BrdU incorporation in 3D colonoids derived from WT and DKO crypts after two or 6 d of plating. Representative images of day 2 and 6 colonoids are shown with a scale bar indicating 0.1 mm. The percentage of BrdU+ cells was quantified by flow cytometry at the indicated time points. Each dot represents an individual technical replicate derived from crypts pooled from two to three mice per group per experiment. Data pooled from two independent experiments. Gated on: Singlets EpCAM+ BrdU+ cells. (E) Volcano plot of genes expressed in colonic IECs derived from WT (n = 3) and DKO (n = 2) mice at steady-state as measured via RNA-seq. (F) GSEA of miR-181 targets expressed in colonic IECs derived from WT (n = 3) and DKO (n = 3) mice treated with DSS for 7 d and allowed to recover for 2 d as measured via RNA-seq. (G) Differentially upregulated genes in colonic IECs derived from DSS-treated DKO mice relative to WT DSS-treated mice as measured via RNA-seq. Predicted miR-181 targets are indicated by red dots. Within these targets, putative negative regulators of the Wnt pathway (Dkk1, Cbx7, Sox6, Nr1h4, Setd7, and Dgkq) are indicated by gene name in red. (H) Heat map of curated Wnt signature genes from Van der Flier et al. (2007) expressed in colonic IECs derived from WT and DKO mice treated with DSS. GSEA for this gene signature is shown in Fig. 5 L. Error bars indicate mean ± SEM. Unpaired t test (A), multiple t tests with FDR correction (C and D). *, P < 0.05.
Re-epithelialization of the intestinal mucosa is necessary to decrease inflammation after injury (Beumer and Clevers, 2021; Karin and Clevers, 2016; Peterson and Artis, 2014). We and others have shown that the miR-181 family is a critical regulator of cellular proliferation, growth, and metabolism in multiple contexts (Cichocki et al., 2011; Virtue et al., 2019; Henao-Mejia et al., 2013; Williams et al., 2013; Liu et al., 2018; Carrella et al., 2015). Thus, we reasoned that the absence of this miRNA family may lead to impaired proliferation within IECs. To test this, we first injected bromodeoxyuridine (BrdU) into WT and DKO mice treated with DSS and measured BrdU incorporation within colonic IECs by immunohistochemistry (IHC) and flow cytometry. Interestingly, we found that there was a significant reduction in the number of BrdU+ IECs in colons from DSS-treated miR-181–deficient mice when compared to WT mice via IHC and flow cytometry (Fig. 5, H and I; and Fig. S3 B). Concordantly, 3D organoids produced from colonic crypts derived from DKO mice were significantly less proliferative than those derived from WT mouse crypts as measured by in vitro BrdU incorporation (Fig. S3, C and D). These results indicate that miR-181a and miR-181b expression in IECs is required for appropriate cell proliferation after mucosal injury.
In addition to the more recognized role of miRNAs in development and tumorigenesis, emerging data suggest that miRNAs are critical regulators of stress responses. In order to understand the potential regulatory mechanisms of the miR-181 family in IECs during homeostasis and inflammation, we performed RNA sequencing (RNA-seq) on WT and DKO colonic IECs at steady state and during DSS treatment (7 d of DSS, 2 d of recovery). In concordance with the critical role of miRNAs during stress, we observed few significant differentially expressed genes between WT and DKO IECs at steady state (Fig. S3 E). Moreover, the gene signature of miR-181–predicted targets was not enriched in DKOs at baseline (Fig. S3 E). In contrast, the levels of 1,193 genes were significantly different between DKO and WT IECs during DSS-induced colitis, and the miR-181 gene signature was enriched in DKO IECs (Fig. S3 F and Table S3). These results suggest that the miR-181 family is a key posttranscriptional regulator of gene expression programs in IECs during intestinal inflammation.
A key mechanism by which miRNAs repress gene expression is by decreasing mRNA stability. As miR-181 is downregulated during intestinal inflammation in colonic IECs (Fig. 1, B and C), we then asked which predicted miR-181 targets were significantly upregulated during DSS colitis in DKO IECs when compared to IECs from WT mice (Fig. 5, J and K). Interestingly, the fourth most significantly upregulated miR-181 predicted target is the key canonical inhibitor of Wnt, Dickkopf-1 (Dkk1; Fig. 5, J and K; and Fig. S3 G). The Wnt signaling pathway has been shown to be critical for IEC proliferation and renewal of intestinal epithelial stem cells, and mutations in this pathway are strongly associated with colon cancer. Importantly, among the 35 significantly upregulated miR-181–predicted targets in DKO IECs during DSS-induced colitis, we also found five additional putative inhibitors of the Wnt signaling pathway (Cbx7, Sox6, Nr1h4, Setd7, and Dgkq; Kim et al., 2015; Li et al., 2021; Pei et al., 2020; Leow et al., 2016; Yu et al., 2020; Shen et al., 2015; Jacob et al., 2011). While upregulation of some miR-181–predicted targets can be the consequence of enhanced colon inflammation, these results also suggest that the miR-181 family could directly promote the activation of the Wnt signaling pathway during DSS-induced colitis to enhance IEC proliferation and repair. In concordance with this hypothesis, a gene signature associated with Wnt activity, as well as pathways activated by Wnt signaling such as Myc and mTOR, were significantly enriched in IECs from DSS-treated WT mice relative to those from DSS-treated DKO mice (Fig. 5 L and Fig. S3 H). Moreover, gene signatures negatively associated with wound healing were enriched in DKO IECs (Fig. 5 M). Although we have not formally validated these genes as miR-181 targets in IECs, these data suggest that miR-181 promotes intestinal homeostasis by increasing IEC proliferation and re-epithelialization of the GI mucosa after injury, potentially by downregulating inhibitors of the Wnt signaling pathway.
Herein, we show that miR-181 is a key regulator of intestinal inflammation and protects against severe colitis by promoting IEC proliferation in response to mucosal injury in a cell-intrinsic manner. Furthermore, we show that while miR-181 in colonic IECs is regulated by the gut microbiota, changes in the composition of the gut microbiota do not alter the ability of this key miRNA family to protect against the development of severe colitis after intestinal injury. Interestingly, while there is a well-established role for miR-181 in immune cell development and function, miR-181 does not act via the hematopoietic system to protect against injury-induced GI inflammation. Finally, our integrative genetic and transcriptomic analysis revealed that miR-181 promotes intestinal homeostasis by increasing IEC proliferation after colonic injury, potentially by downregulating inhibitors of the Wnt signaling pathway. Altogether, our results suggest that the miR-181 microRNA family is a critical regulator of IEC homeostasis and intestinal inflammation and therefore may be a novel therapeutic target for the treatment of GI inflammatory and neoplastic disorders.
While the remarkable regenerative capacity of the intestinal epithelium is well appreciated, the diversity of IESC populations necessary to restore intestinal homeostasis after discrete types of mucosal injury or environmental challenges is still being elucidated. Although multipotent Lgr5+ crypt-base columnar cells are critical for homeostatic renewal of the intestinal epithelium, studies have suggested that these cells are not principally responsible for recovery from DSS-induced colitogenic injuries (Metcalfe et al., 2014). Moreover, depletion of IESCs expressing the transcription factor Hopx prior to or during DSS treatment does not exacerbate colitis severity (Wang et al., 2019). A recent study reported the existence of an injury-responsive revival stem cell (revSC) population expressing Clusterin, a protein known to regulate protein folding and inhibit apoptosis (Zhang et al., 2005). Clusterin+ cells are exceedingly rare at homeostasis but are necessary for recovery from both irradiation- and DSS-induced epithelial injury (Ayyaz et al., 2019). While loss of these revSCs does not lead to gross defects at homeostasis, DSS treatment induces acute weight loss and increased morbidity in revSC-deficient mice, similar to what we observe in miR-181–deficient animals. Interestingly, expression of Clusterin is dependent on Yes-associated protein 1 (YAP) signaling, and loss of two key YAP repressors involved in Hippo signaling, Lats1 and Lats2, leads to increased homeostatic expression of Clusterin. Strikingly, miR-181 is predicted to target not only Lats1 but also one of its binding partners Mob1b. While we did not observe significant changes in the levels of these targets between WT and DKO mouse IECs in our RNA-seq analysis, both Lats1 and Lats2 levels were increased in DKO IECs during DSS. Therefore, as DKO mice are acutely more susceptible to DSS, it is possible that these changes may be more robust during early disease. More importantly, it is now well appreciated that the Wnt signaling pathway cooperates with YAP/TAZ to promote the proliferation of IESCs and other stem cell populations (Park et al., 2015; Piersma et al., 2015; Azzolin et al., 2014; Li et al., 2019). It is, therefore, possible that the miR-181 family contributes to revSC proliferation by promoting Wnt and YAP signaling by downregulating canonical inhibitors of these potent cell growth pathways.
The miR-181 family has a well-established role in regulating inflammatory processes in multiple tissue types. The earlier studies have shown that the loss of miR-181 results in reduced differentiation of double-negative (DN) to double-positive thymocytes, decreased thymocyte proliferation, and loss of NKT cells (Henao-Mejia et al., 2013). Interestingly, TCRγδ+ IELs are thought to develop not from double-positive thymocytes but instead from DN precursors that migrate to the intestinal epithelium (Jandke et al., 2020; Hoytema van Konijnenburg et al., 2017; Olivares-Villagómez and Van Kaer, 2018). Moreover, these IELs have been shown to be protective against intestinal inflammation and promote IEC homeostasis (Hoytema van Konijnenburg et al., 2017). In this study, we have shown that the loss of miR-181 in the hematopoietic compartment results in protection against DSS colitis. We also observed an increase in TCRγδ+ IELs at steady state in miR-181 DKO mice and mice deficient for miR-181 in the hematopoietic compartment. Therefore, the increase in DN precursors as a result of miR-181 loss may lead to the observed increase in TCRγδ+ IELs and resultant protection against colitis in mice deficient for miR-181 within the hematopoietic compartment. If so, these data indicate that miR-181 regulates intestinal homeostasis by acting in both local and distal tissues to control GI inflammation. We believe that these findings warrant further investigation to elucidate the role of miR-181 in IEL development.
In this study, we showed that mice with IECs deficient in miR-181a1-b1 are more susceptible to severe colitis. Yet, they do not fully recapitulate the striking DSS-induced weight loss and colonic shortening seen in the whole body miR-181 DKO mice. Moreover, mice deficient in miR-181 within the hematopoietic compartment are protected against colitis in both genetic and BM chimeric models, most likely due to increases in TCRγδ+ IELs seen in miR-181 DKO mice at steady state (Hoytema van Konijnenburg et al., 2017). As we have exhaustively ruled out the possibility that loss of the miR-181 family within the hematopoietic system protects from severe colitis, these results indicate that there are likely other nonhematopoietic cell types expressing miR-181 that are required for protection against severe GI inflammation after epithelial injury. Interestingly, miR-181 is highly expressed in the brain and is predicted to target genes critical for neuronal function and neurotransmitter release (Sambandan et al., 2017). It is now clear that both the central and peripheral nervous systems play important roles in the regulation of enteric processes such as peristalsis, sensorimotor function, and immunoregulation to maintain GI homeostasis (Jarret et al., 2020; Yoo and Mazmanian, 2017; Muller et al., 2020b; Muller et al., 2020a; Veiga-Fernandes and Mucida, 2016). Therefore, future work should address the possibility that miR-181 acts within both IECs and neurons to regulate intestinal inflammation after injury.
The severity of colitis induction by DSS has been shown to be heavily dependent on the composition of the gut microbiota, which contributes to considerable variability in disease progression and severity (Elinav et al., 2011). Importantly, we showed that loss of miR-181 leads to increased colitis severity in multiple vivaria, under different cohousing conditions, and when compared to cage-mate control mice, altogether indicating that the composition of the microbiota does not significantly impact the progression of colitis in miR-181–deficient mice. However, microbiota-derived signals regulate miR-181 levels in colonic IECs, as IECs derived from mice treated with antibiotics have increased levels of miR-181 (Fig. 1 D). Interestingly, previous studies have shown that miR-181 levels in distal tissues are modulated by tryptophan-derived metabolites, the levels of which have been shown to be dysregulated during intestinal injury (Virtue et al., 2019; Guo et al., 2020). While IEC proliferation is key to recovery from mucosal damage, aberrant proliferation of IECs can lead to the development of colorectal cancer. As such, titrating the proliferative capacity of IECs based on the needs of the host is critical to maintaining homeostasis. Therefore, there may be both positive and negative microbial signals that tune IEC proliferation via miR-181 to both promote epithelial reconstitution after injury and prevent the development of neoplastic disease. Regardless, these data suggest that miR-181 may be a robust posttranscriptional regulator of IEC proliferation in response to microbiota-derived cues.
An emerging concept in the field of miRNA biology is that miRNAs largely function as redundant systems to maintain organismal homeostasis in the face of environmental or physiological challenges (Biggar and Storey, 2018). Interestingly, a regulatory role for miR-143/miR-145 in IEC regeneration was found by assessing their activity within intestinal mesenchyme (Chivukula et al., 2014). While there were no effects at homeostasis on intestinal pathology in mice deficient in these miRNAs, intestinal injury induced by DSS led to a striking increase in mortality relative to WT controls. These findings are concordant with our results in that miR-181 DKO mice do not display increased inflammation at baseline, as evidenced by no changes myeloid cell abundance, while rapidly succumbing to severe GI inflammation after colitogenic injury.
Altogether, we showed that miR-181 is downregulated in IECs from human IBD patients and in mice with DSS-induced colitis and that this downregulation promotes the development of severe colonic inflammation. Thus, our work proposes that miR-181 is a critical regulator of GI homeostasis potentially through the regulation of IEC proliferation and cell metabolism in response to microbial or inflammatory cues. Therefore, our findings may have clinical implications as they suggest that alleviating the repression of miR-181 levels observed during inflammatory disease may ameliorate colitis severity and promote epithelial reconstitution.
Materials and methods
Mice
C57BL/6J (WT) mice and mice expressing Cre-recombinase (Cd4-Cre [B6.Cg-Tg(Cd4-cre)]1Cwi/BfluJ), LysM-Cre (B6.129P2-Lyz2tm1[cre]Ifo/J), Vav-iCre (B6.Cg-Commd10Tg[Vav1-icre]A2Kio/J), and Villin-Cre B6.Cg-Tg[Vil1-cre]1000Gum/J) were purchased from The Jackson Laboratory (Jax). Mice deficient in miR-181a1-b1, miR-181a2-b2, or miR-181c-d, or with the above loci flanked by loxP sites, were individually generated as previously described (Henao-Mejia et al., 2013). miR-181a1-b1−/− and miR-181a2-b2−/− were crossed to generate miR-181a1-b1−/−; miR-181a2-b2−/− DKO mice. miR-181a1-b1fl/fl and miR-181a2-b2fl/fl mice were crossed to generate miR-181a1-b1fl/fl; miR-181a2-b2fl/fl mice, while miR-181a1-b1fl/fl, miR-181a2-b2fl/fl, and miR-181c-dfl/fl mice were crossed to generate miR-181a1-b1fl/fl; miR-181a2-b2fl/fl; miR-181c-dfl/fl mice. These floxed animals were then bred to mice expressing the Cre-recombinase under the control of the appropriate promoter as described above to generate dcKOs and tcKOs, respectively.
WT mice termed “in-house bred” (IHB) were generated from WT C57BL/6J mice purchased from Jax. They received a bedding exchange upon arrival to promote colonization by facility microbiota. These Jax-born WT animals were then bred in our facility for multiple generations, and their progeny were used to initiate the IHB WT line.
Mice were age-matched in all experiments (age difference between groups <7 d), and cage- and sex-matched littermates were used for experiments when possible. All mice were bred and maintained under specific pathogen–free conditions at American Association for the Accreditation of Laboratory Animal Care–accredited animal facilities at the University of Pennsylvania. Mice were given ad libitum access to food and water and housed in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals under an animal study protocol #805188 approved by an Institutional Animal Care and Use Committee at the University of Pennsylvania. All mice used were between 8 and 20 wk of age at the start of experimentation unless otherwise stated. Sample sizes are indicated in figure legends.
DSS treatment
Starting at 6–8 wk of age, mice were weighed at experimental day 0 and given 2% DSS (mol wt = 36,000–50,000 daltons; MP Biomedicals) diluted in acidified drinking water provided by University Lab Animal Resources. Mice were monitored for body weight loss, and DSS was withdrawn once mice reached 7.5–10% total body weight loss, initiating the recovery period. Animals were closely and continually monitored for weight changes and morbidity throughout the duration of recovery from DSS. Details regarding experimental duration, cohousing, and littermate controls are indicated in figure legends. For experiments in which mice were cohoused, animals were weaned into the same cage at 3–4 wk of age followed by 3–4 wk of cohousing unless otherwise indicated.
Patient intestinal epithelial samples
Human intestinal tissue was obtained from the University of Pennsylvania IBD Immunology Initiative. The study was approved by the Institutional Review Board in Philadelphia, PA (#814428) at the University of Pennsylvania. Colonic tissue biopsies were collected according to a standard collection protocol on consented patients in the IBD-Immunology Initiative during the patient’s routine colonoscopy. Biopsies were given to trained research coordinators, placed in cold PBS, and delivered to the lab within 1 h of collection. The biopsies were submerged in 3 ml epithelial digestion buffer (PBS + 5 mM EDTA + 1 mM dithiothreitol + 5% FBS + 1% penicillin/streptomycin) and vortexed followed by incubation at 37°C for 15 min. After incubation, the samples were vortexed for 20–30 s and 2 ml of supernatant containing IECs were collected. Cells were pelleted for 10 min and frozen at −80°C.
Mouse colonoscopy
Mouse colonoscopy was performed and scored as stated in Elinav et al. (2011). Briefly, colonoscopy was performed using a high-resolution mouse video endoscopic system (“Coloview,” Carl Storz). The severity of colitis was blindly scored using Murine Endoscopic Index of Colitis Severity, which is based on five parameters: granularity of mucosal surface, vascular pattern, translucency of the colon mucosa, visible fibrin, and stool consistency (Becker et al., 2006).
IHC, pathology scoring, and imaging
Colons from WT and miR-181 DKO mice treated with DSS were Swiss rolled distal to proximally and fixed in 10% neutral buffered formalin for 24 h. Tissues were then stored in 70% ethanol followed by paraffin embedding, histological sectioning, and H&E staining all performed by the Molecular Pathology and Imaging Core at the University of Pennsylvania. Brightfield images were acquired with a Nikon 6000 microscope.
Pathological assessment was performed on H&E-stained sections in a blinded fashion without prior knowledge of the study design. Histopathological analysis was focused on assessing and scoring the extension and severity of the subacute/chronic mucosal changes (inflammatory and proliferative) affecting the mid- and distal-colorectal tract present in each sample. Most of the samples displayed multifocal to diffuse inflammatory changes with extensive mucosal ulceration/loss mainly affecting the distal colon and rectum. In some samples, focal crypt dysplasia of the proximal colonic mucosa was observed. This lesion consists of an irregular proliferation of undifferentiated epithelium diffusely expanding the transit amplifying zone and multifocally extending into the crypt-mucosal surface junction. In less affected samples, the pathological picture was very heterogeneous combining small multifocal ulcerative lesions and/or inflammatory cell infiltrate with nearly normal mucosal segments. This finding reflects the typical multifocal nature of the ulcerative and inflammatory lesions induced by DSS administration. The extensive loss of mucosal glands observed in some of the samples from DKO mice precluded in part the evaluation of pathological changes specifically affecting the mucosal crypts including inflammation and hyperplasia.
BrdU IHC and flow cytometry
Mice were intraperitoneally injected with 100 mg/kg of BrdU 4 h prior to sacrifice. Colons were processed and section as above for IHC or processed for IEC isolation as detailed below. BrdU staining was performed with the BrdU In-Situ Detection Kit (BD Pharmigen) or the APC BrdU Flow Kit (BD Pharmigen), according to manufacturer’s instructions, for paraffin-embedded sections or primary colonic IECs, respectively. Brightfield images were acquired with a Nikon 6000 microscope.
Organoids
We employed a 3D colonic organoid system using the intestinal organoid culture protocol from stem cell to assess proliferation in WT and DKO IECs by a flow-based method (adapted from Padmanaban et al., 2020). Briefly, we first isolated the crypts from adult WT and DKO male mice by extensively cleaning the colonic tissue with PBS and then dissociating the crypts from the basement membrane using 10 ml/colon of gentle cell dissociation buffer (stem cell) and rocking at room temperature for 20 min at 20 rpm. Using a serological pipet, we then removed the crypts via gentle pipetting and pooled them by genotype. To plate the crypts, we resuspended 500 crypts in 25 μl intesticult organoid growth media (OGM; stem cell) followed by the addition of 25 μl of growth-factor-reduced Matrigel (Corning), and plated all 50 μl in a 24-well plate prewarmed at 37°C in a tissue culture incubator. Matrigel was allowed to solidify for 10 min at 37°C. Then, 750 μl OGM was added to the wells. Cells were cultured at 5% CO2 37°C in a humidified incubator, and OGM was changed three times per week. To assess proliferation, we added BrdU to a final concentration of 100 μg/ml for 2 h in culture. We then dissolved the Matrigel in 5 U/ml of dispase (stem cell) and dissociated the organoids to single-cell suspension with TrypsinLE. BrdU incorporation was assessed via FACS using the APC BrdU Flow Kit (BD Pharmigen).
Antibiotic treatment
WT mice were treated with broad-spectrum antibiotics to deplete GI microbiota as previously described (Virtue et al., 2019). Briefly, mice were treated with a combination of ampicillin, vancomycin, metronidazole, and kanamycin at a concentration of 1 g/liter of acidified drinking water provided by University Lab Animal Resources. Mice were given antibiotics for 7 d and sacrificed.
Fecal DNA extraction, 16s ribosomal RNA (rRNA)-seq, and microbiota analysis
At 6–14 wk of age, feces from WT and miR-181 DKO female mice were collected prior to cohousing. Mice were then cohoused for 2 wk, and a second fecal sample was collected. For miR-181a1-b1 and WT littermates, fecal samples were collected at 6–7 wk of age from two independent litters derived from breeding miR-181a1-b1+/− mice. Stool samples were collected into 2 ml tubes, snap frozen, and stored at −80°C until DNA extraction. DNA extraction was performed by using DNeasy PowerSoil Kit (Qiagen) according to manufacturer instructions. DNA was amplified by using the KAPA Hifi HotStart ReadyMix (Roche) and specific primers targeting the variable region 1 and 2 (V1/V2) of the 16S rRNA gene. The following barcoded primers were generated: Forward i27- 5′-TATGGTAATTGTAGAGTTTGATCCTGGCTCAG-3′, Reverse 338-3′-XXXXXXXXXXXXTGCTGCCTCCCGTAGGAGT-5′, where X represents a barcode base (Integrated DNA Technologies). After PCR amplification, the library was sequenced using 500 bp paired-end sequencing (Illumina MiSeq) resulting in 180,000–490,000 reads per sample. The paired-end reads were then processed using the QIIME 2 pipeline (Bolyen et al., 2019). As input for analysis, the forward and reverse fastq files and a sample-mapping file with unique barcode sequence were used. First, paired-end reads were split according to the unique barcodes, quality control analysis was performed, and reads that passed were used for taxonomical classification. Taxonomy classification was performed by aligning representative operational taxonomic unit sequences to Greengenes database allowing 97% operational taxonomic unit identity. Core diversity metrics for α and β diversity assessment were analyzed using QIIME diversity core-metrics-phylogenetic plugin.
RNA extraction, cDNA synthesis, and quantitative RT-PCR
Total RNA was extracted from the tissue samples using TRIzol (Life Technologies) and Zymo Research Direct-zol RNA Kit or the Purelink RNA Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Isolated RNA was quantified with a Nanodrop 2000 (Thermo Fisher Scientific). cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The resulting cDNA was analyzed via KAPA Probe, a Taqman-based quantitative PCR (qPCR) system (KAPABiosystems). The following Taqman assays were used: pri-miRNA: ID Mm03307120_pri (pri-miR-181a1b1), Mm03306417_pri (pri-miR181a2b2), Mm03306417_pri (pri-miR-181 cd); mature miRNA: ID 000480 hsa-miR181a, 001098 hsa-miR-181b, 000482 hsa-miR-181c, 000397 hsa-miR-21, 000426 hsa-miR-34a, 002249 hsa-miR-143, 002278 hsa-miR-145, 002571 mmu-miR-155, and 001973 U6 snRNA purchased from Thermo Fisher Scientific. All mature miRNA assays were for the 5p miRNA. qPCR reactions were performed in duplicate using a QuantStudio 6 Flex Real-Time PCR instrument (Thermo Fisher Scientific). To determine the expression of mature miRNA species, total RNA was extracted using the same procedure as stated above. cDNA for miRNA gene expression analysis was synthesized using the Applied Biosystems TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific). Briefly, cDNA synthesis was performed by adding 20 ng of RNA to a 1× reaction containing 0.1 μl 100M dNTP, 1 μl 10× RT buffer, 0.67 μl MultiScribe RT enzyme (50 U/μl), 0.13 μl RNase inhibitor (20 U/μl), 4.29 μl nuclease free water (Invitrogen), and 0.5 μl of 20× stem-loop cDNA primers for the mature miRNA assays indicated above. Gene expression was quantified with TaqMan-based qPCR systems (KAPABiosystems) using indicated primers.
Isolation of IECs, IELs, and LP immune cell populations
Mice were humanely euthanized via CO2 and cervically dislocated. Fur was sprayed with 70% ethanol and the skin incised along the ventral midline using tissue forceps and surgical scissors. The peritoneum was then incised and reflected to open the abdominal cavity. The colon was located by finding the distal end at the rectum and tracing proximally with tissue forceps, ending at the cecum. The distal end of the colon was transected from the rectum and pulled ventrally away from the abdominal cavity and the proximal end of the colon was transected at the cecocolic junction. Once removed, mesenteric fat was pulled from the serosal surface of the colon using curved forceps. The colon was then placed in ice-cold PBS (Corning) in a Petri dish where residual mesenteric fat was removed. Artery scissors and tissue forceps (Roboz) were then used to open the lumen by incising longitudinally. The open colon was then serially shaken in cold PBS in a Petri dish until no large pieces of feces were visible by eye on the luminal surface of the colon. The colon was then placed in a 50 ml conical with ice-cold PBS and stored on ice until all colons were processed. Once all mice were processed, colons were briefly shaken in the 50-ml conical to remove any small particulates and poured over a mesh strainer. A 50-ml conical was then filled with 25 ml of digestion buffer containing 2% FBS (Corning), 20 mM Hepes (Corning), L-glutamine (Corning), penicillin/streptomycin (Corning), 5 mM EDTA (Sigma-Aldrich), and 1 mM dithiothreitol (Sigma-Aldrich) diluted in 1× PBS. Intestines were then cut into 1-cm pieces and placed into the conicals containing digestion buffer. They were then shaken at 225 rpm at 37°C for 15 min. Tissues were then vortexed for 5 s, shaken, and poured over a 70-micron filter into a 50-ml conical on ice. The colonic tissue was then removed from the strainer and placed into a 50-ml conical containing 25 ml of IEC digestion buffer and shaken again at 225 rpm at 37°C for 15 min, vortexed for 5 s, shaken, and poured over the 70-micron filter into a 50-ml conical on ice to pool the two fractions. The IECs were then spun at 450–500 g at 4°C for 5 min, and supernatants were decanted. IEC pellets were then washed in 20 ml of ice-cold PBS and spun at 450–500 g at 4°C for 5 min and decanted. This wash was then repeated. If IECs were to be separated from IELs, a Percoll gradient was performed, wherein IECs were resuspended in 10 ml of 40% Percoll (GE Healthcare; first made isotonic with 10× DPBS, then diluted to 40% in complete media [see below]) in a 15 ml conical and spun at 600 g for 20 min. IECs floated to the top, while IELs and red blood cells were pelleted. IECs were taken from the top, washed in PBS, and, if they were to be isolated for RNA, resuspended in 1 ml of TRIzol in a 2-ml RNAse-free tube, vortexed, and stored at −80°C. To establish the relative contribution of RNA from CD45+ cells to our IEC RNA samples, we determined the expression levels of Ptprc (CD45) and Villin (IEC marker) relative to the housekeeping gene Hprt. If IELs were to be used for flow, the pellet from the Percoll gradient, containing IELs and red blood cells, was washed in PBS and ACK lysed. The reaction was neutralized by the addition of 10–20 ml of complete media containing DMEM with high glucose and L-glutamine (Corning), 1% penicillin/streptomycin, 1× 2-mercaptoethanol, 10% FBS (Corning), and 20 mM Hepes (Corning). Cells were then prepped for downstream processing.
LP lymphocytes (LPLs) and myeloid cells (LPMs) were isolated by first taking the remaining tissue fractions from the epithelial digestion and washing twice with cold PBS. Then, tissues were minced and placed in 6.5 ml of prewarmed LP digestion media containing RPMI 1640 with L-glutamine (Thermo Fisher Scientific), 10% FBS (Corning), 10 mM Hepes (Corning), 1.5 mg/ml Collagenase D (Roche), and 0.5 mg/ml DNase I (Sigma-Aldrich). Tissues were shaken at 225 rpm at 37°C for 1 h. After digestion, the samples were vortexed and decanted over a 70-μm filter into a conical on ice. Tissues were then mashed through the filter with a syringe plunger and washed with PBS repeatedly. Cells were pelleted, washed, ACK lysed, and prepped for downstream processing.
Flow cytometry analysis
Immune cells were isolated from large intestinal tissue as described above. Cells were then stained with the indicated fluorochrome-conjugated antibodies as well as eBioscience Fixable Viability Dye eFluor 506 to exclude nonviable cells. All flow cytometry analysis was done at the University of Pennsylvania Flow Cytometry Facility using BD LSRII cell analyzers. FlowJo software (v. 10 TreeStar) was used for data analysis and graphic rendering. The fluorochrome-conjugated antibodies used were Anti-mouse B220 (RA3-6B2; BioLegend), Anti-mouse CD3ε (145-2C11; Thermo Fisher Scientific), Anti-mouse CD4 (GK1.5; BioLegend), Anti-mouse CD8α (53-6.7; BioLegend), Anti-mouse CD8β (YTS156.7.7; BioLegend), Anti-mouse/human CD11b (M1/70; BioLegend), Anti-mouse CD45 (30-F11; BioLegend), Anti-mouse CD103 (2E7; BioLegend), Anti-mouse TCRβ (H57-597; BioLegend), Anti-mouse TCRγδ (GL3; BioLegend), Anti-mouse Ly6G (1A8; BioLegend), Anti-mouse NK1.1 (PK136; BioLegend), Anti-mouse RORγt (B2D; eBioscience), Anti-mouse Siglec-F (E50-2440; BD Biosciences), and Anti-mouse FOXP3 (FJK-16s; BioLegend). Anti-BrdU antibodies were provided with the APC Flow Kit (BD Pharmigen). Gating strategies are: CD45+ Live cells: gated on singlets CD45+ Live (viability−) cells; IELs: gated on singlets CD45+ Live (viability−) CD103+ CD11b− cells from epithelium; LP non-B nonmyeloid cells: gated on Singlets CD45+ Live (viability−) B220− CD11b− cells; LPMs: gated on Singlets CD45+ Live (viability−) B220− intracellular CD3ε− CD11b+ cells.
BM chimeras
BM chimeras were performed as previously described (Henao-Mejia et al., 2013). Briefly, 0.5 × 106 WT and 0.5 × 106 miR-181–deficient BM cells were injected into lethally irradiated hosts. 6 wk later, engraftment was assessed by percentages of CD45.1 and CD45.2 cells in the spleen. DSS treatment was then performed as described above.
RNA-seq and analysis
Percolled colonic IECs were isolated from WT and DKO mice at baseline and during DSS (7 d of DSS, 2 d of recovery) as detailed above. RNA was isolated using a Purelink RNA kit (Thermo Fisher Scientific). Quality control, library preparation, and library sequencing were performed by Azenta. Briefly, quality control was performed via TapeStation with RNA integrity score values for all samples >6. Libraries were prepped and then sequenced using Illumina HiSeq configured for 150 bp paired-end reads. Raw reads were aligned to the GRCm38/mm10 genome using STAR and transcript counts were generated (Dobin et al., 2013). Data processing and differential gene expression were performed using the R packages DESeq2, edgeR, and limma (voom function used for modeling mean–variance relationships and for deriving Bayesian statistics for differentially expressed genes; Love et al., 2014; McCarthy et al., 2012; Robinson et al., 2009; Ritchie et al., 2015). miR-181 targets were found via TargetScan, microT-CDS (DIANA Tools), miRDB, and mirBase to find putative miR-181 targets in IECs (Agarwal et al., 2015; Chen and Wang, 2019; Kozomara et al., 2018). For mirDB hits, high confidence predicted targets (score >80) were analyzed. Gene set enrichment analysis (GSEA) was performed with a curated Wnt signature generated from a previous study (Van der Flier et al., 2007) as well as Hallmark Pathway and Gene Ontology biological process signatures. RNA-seq data were deposited at the Gene Expression Omnibus under GSE208410.
Quantification and statistical analysis
Statistical analyses were performed in Prism 9. Data were tested for normality using either a Kolmogorov–Smirnov test or a Shapiro–Wilk test and analysis performed using two-tailed Student’s t test or one- or two-way ANOVA with Sidak’s multiple comparisons test for parametric data. False discovery rate (FDR) corrections were performed when indicated. For nonparametric data, a Mann–Whitney test or a Kruskal–Wallis with Dunn’s multiple comparisons test was performed as indicated in the figure legends. In all cases, P < 0.05 was considered statistically significant. All error bars represent SEM.
Online supplemental material
Fig. S1 shows the quantification of five microRNAs in mouse and human IECs, and the impact of miR-181 deficiency on DSS colitis progression in males, females, and under thermoneutral conditions. Moreover, it shows the gating strategy for quantifying intraepithelial lymphocytes and immune cells in the LP. Fig. S2 shows results from miR-181–sufficient and –deficient cage-mate DSS and 16s sequencing results from miR-181a1-b1 KO and WT littermate feces. It also shows flow cytometric characterization of IEL and LP immune cell populations from WT and DKO mice at baseline, during acute DSS treatment, and during recovery from DSS colitis. Fig. S3 shows the analysis of IEC proliferation in WT and DKO organoids and mice. In addition, it shows the RNA-seq analysis of WT and DKO IEC at steady state and under DSS-induced colitis. Tables S1 and S2 show the histopathological assessment of mucosal damage in colons from WT and DKO mice during DSS-induced colitis. Table S3 shows the gene counts from STAR alignment of our RNA-seq studies.
Supplementary Material
shows pathological assessment of mucosal damage in colons from WT and DKO mice during DSS-induced colitis.
shows pathological assessment of neoplastic and inflammatory changes in colons from WT and DKO mice during DSS-induced colitis.
shows gene counts of RNA-seq data from WT and DKO mouse colonic IECs at baseline and during recovery from DSS (7 d of 2% DSS treatment, 2 d of recovery).
Acknowledgments
This work was supported by funds from the Children’s Hospital of Philadelphia, University of Pennsylvania Institute for Immunology, the National Institutes of Health (grants R21AI128060, R21DK111755, and R01HL136572), the PEW Charitable Trusts, the Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases Award, and the Chan Zuckerberg Initiative (to J. Henao-Mejia); National Institutes of Health grants T32AI070077 and F31DK122677 (to M.T. Jimenez); the PEW Charitable Trusts (to J. Tello-Cajiao); and F. Hoffmann-La Roche (to N. Gagliani).
Author contributions: M.T. Jimenez and J. Henao-Mejia designed and performed experiments and wrote the manuscript. J. Henao-Mejia supervised the project. M.L. Clark, M.F. Michieletto, and S. Liu assisted with in vivo experiments, protocol development, and experimental design. J.M. Wright and I. Erickson maintained mouse lines and assisted with in vivo experiments. L. Dohnalova processed, sequenced, and analyzed all metagenomic data. G.T. Uhr performed in vivo studies. J. Tello-Cajiao provided conceptual guidance related to RNA-sequencing experiments. L. Joannas, A. Williams, and N. Gagliani assisted with in vivo studies. M. Bewtra and V.T. Tomov provided human samples from IBD-Immunology Initiative. C.A. Thaiss provided conceptual guidance and assisted with manuscript development.
Data availability
RNA-seq datasets have been uploaded to the Gene Expression Omnibus repository (accession number GSE208410).
References
- Agarwal, V., Bell G.W., Nam J.-W., and Bartel D.P.. 2015. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 4:e05005. 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyaz, A., Kumar S., Sangiorgi B., Ghoshal B., Gosio J., Ouladan S., Fink M., Barutcu S., Trcka D., Shen J., et al. 2019. Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature. 569:121–125. 10.1038/s41586-019-1154-y [DOI] [PubMed] [Google Scholar]
- Azzolin, L., Panciera T., Soligo S., Enzo E., Bicciato S., Dupont S., Bresolin S., Frasson C., Basso G., Guzzardo V., et al. 2014. YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell. 158:157–170. 10.1016/j.cell.2014.06.013 [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 116:281–297. 10.1016/s0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bartel, D.P., and Chen C.-Z.. 2004. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 5:396–400. 10.1038/nrg1328 [DOI] [PubMed] [Google Scholar]
- Becker, C., Fantini M.C., and Neurath M.F.. 2006. High resolution colonoscopy in live mice. Nat. Protoc. 1:2900–2904. 10.1038/nprot.2006.446 [DOI] [PubMed] [Google Scholar]
- Beumer, J., and Clevers H.. 2021. Cell fate specification and differentiation in the adult mammalian intestine. Nat. Rev. Mol. Cell Biol. 22:39–53. 10.1038/s41580-020-0278-0 [DOI] [PubMed] [Google Scholar]
- Biggar, K.K., and Storey K.B.. 2018. Functional impact of microRNA regulation in models of extreme stress adaptation. J. Mol. Cell Biol. 10:93–101. 10.1093/jmcb/mjx053 [DOI] [PubMed] [Google Scholar]
- Biton, M., Levin A., Slyper M., Alkalay I., Horwitz E., Mor H., Kredo-Russo S., Avnit-Sagi T., Cojocaru G., Zreik F., et al. 2011. Epithelial microRNAs regulate gut mucosal immunity via epithelium–T cell crosstalk. Nat. Immunol. 12:239–246. 10.1038/ni.1994 [DOI] [PubMed] [Google Scholar]
- Bolyen, E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37:852–857. 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton, G.J., Contijoch E.J., Mogno I., Vennaro O.H., Llewellyn S.R., Ng R., Li Z., Mortha A., Merad M., Das A., et al. 2019. Microbiotas from humans with inflammatory bowel disease alter the balance of gut Th17 and RORγt+ regulatory T cells and exacerbate colitis in mice. Immunity. 50:212–224.e4. 10.1016/j.immuni.2018.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed, F., Ding H., Wang T., Hooper L.V., Koh G.Y., Nagy A., Semenkovich C.F., and Gordon J.I.. 2004. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA. 101:15718–15723. 10.1073/pnas.0407076101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrella, S., Barbato S., D’Agostino Y., Salierno F.G., Manfredi A., Banfi S., and Conte I.. 2015. TGF-β controls miR-181/ERK regulatory network during retinal axon specification and growth. PloS One. 10:e0144129. 10.1371/journal.pone.0144129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing, B., Aitken J.D., Malleshappa M., and Vijay-Kumar M.. 2014. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr. Protoc. Immunol. 104:15.25. 1–15.25. 14. 10.1002/0471142735.im1525s104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., and Wang X.. 2020. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 48:D127–D131. 10.1093/nar/gkz757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivukula, R.R., Shi G., Acharya A., Mills E.W., Zeitels L.R., Anandam J.L., Abdelnaby A.A., Balch G.C., Mansour J.C., Yopp A.C., et al. 2014. An essential mesenchymal function for miR-143/145 in intestinal epithelial regeneration. Cell. 157:1104–1116. 10.1016/j.cell.2014.03.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichocki, F., Felices M., McCullar V., Presnell S.R., Al-Attar A., Lutz C.T., and Miller J.S.. 2011. Cutting edge: MicroRNA-181 promotes human NK cell development by regulating Notch signaling. J. Immunol. 187:6171–6175. 10.4049/jimmunol.1100835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichele, D.D., and Kharbanda K.K.. 2017. Dextran sodium sulfate colitis murine model: An indispensable tool for advancing our understanding of inflammatory bowel diseases pathogenesis. World J. Gastroenterol. 23:6016–6029. 10.3748/wjg.v23.i33.6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav, E., Strowig T., Kau A.L., Henao-Mejia J., Thaiss C.A., Booth C.J., Peaper D.R., Bertin J., Eisenbarth S.C., Gordon J.I., and Flavell R.A.. 2011. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 145:745–757. 10.1016/j.cell.2011.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso, R., Mao T., Wang S., Schaffert S., Gong X., Yue S., Luong R., Min H., Yashiro-Ohtani Y., and Davis M.. 2012. Modulating the strength and threshold of NOTCH oncogenic signals by mir-181a-1/b-1. PLos Genet. 8:e1002855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Chou W.-C., Lai Y., Liang K., Tam J.W., Brickey W.J., Chen L., Montgomery N.D., Li X., Bohannon L.M., et al. 2020. Multi-omics analyses of radiation survivors identify radioprotective microbes and metabolites. Science. 370:eaay9097. 10.1126/science.aay9097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao-Mejia, J., Williams A., Goff L.A., Staron M., Licona-Limón P., Kaech S.M., Nakayama M., Rinn J.L., and Flavell R.A.. 2013. The microRNA miR-181 is a critical cellular metabolic rheostat essential for NKT cell ontogenesis and lymphocyte development and homeostasis. Immunity. 38:984–997. 10.1016/j.immuni.2013.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori, D., Dunkerly-Eyring B., Nomura Y., Biswas D., Steppan J., Henao-Mejia J., Adachi H., Santhanam L., Berkowitz D.E., Steenbergen C., et al. 2017. miR-181b regulates vascular stiffness age dependently in part by regulating TGF-β signaling. PloS One. 12:e0174108. 10.1371/journal.pone.0174108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoytema van Konijnenburg, D.P., Reis B.S., Pedicord V.A., Farache J., Victora G.D., and Mucida D.. 2017. Intestinal epithelial and intraepithelial T cell crosstalk mediates a dynamic response to infection. Cell. 171:783–794.e13. 10.1016/j.cell.2017.08.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison, E.R., Kawamoto E.M., Taub D.D., Lal A., Abdelmohsen K., Zhang Y., Wood W.H. III, Lehrmann E., Camandola S., Becker K.G., et al. 2013. Evidence for miR‐181 involvement in neuroinflammatory responses of astrocytes. Glia. 61:1018–1028. 10.1002/glia.22483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indrieri, A., Carrella S., Carotenuto P., Banfi S., and Franco B.. 2020. The pervasive role of the miR-181 family in development, neurodegeneration, and cancer. Int. J. Mol. Sci. 21:2092. 10.3390/ijms21062092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob, L.S., Wu X., Dodge M.E., Fan C.-W., Kulak O., Chen B., Tang W., Wang B., Amatruda J.F., and Lum L.. 2011. Genome-wide RNAi screen reveals disease-associated genes that are common to hedgehog and Wnt signaling. Sci. Signal. 4:ra4. 10.1126/scisignal.2001225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandke, A., Melandri D., Monin L., Ushakov D.S., Laing A.G., Vantourout P., East P., Nitta T., Narita T., Takayanagi H., et al. 2020. Butyrophilin-like proteins display combinatorial diversity in selecting and maintaining signature intraepithelial γδ T cell compartments. Nat. Commun. 11:3769. 10.1038/s41467-020-17557-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarret, A., Jackson R., Duizer C., Healy M.E., Zhao J., Rone J.M., Bielecki P., Sefik E., Roulis M., Rice T., et al. 2020. Enteric nervous system-derived IL-18 orchestrates mucosal barrier immunity. Cell. 180:813–814. 10.1016/j.cell.2020.02.004 [DOI] [PubMed] [Google Scholar]
- Kaiko, G.E., Ryu S.H., Koues O.I., Collins P.L., Solnica-Krezel L., Pearce E.J., Pearce E.L., Oltz E.M., and Stappenbeck T.S.. 2016. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell. 167:1137–1720. 10.1016/j.cell.2016.10.034 [DOI] [PubMed] [Google Scholar]
- Karin, M., and Clevers H.. 2016. Reparative inflammation takes charge of tissue regeneration. Nature. 529:307–315. 10.1038/nature17039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.Y., Park J.H., Won H.Y., Lee J.Y., and Kong G.. 2015. CBX7 inhibits breast tumorigenicity through DKK-1-mediated suppression of the Wnt/β-catenin pathway. FASEB J. 29:300–313. 10.1096/fj.14-253997 [DOI] [PubMed] [Google Scholar]
- Kozomara, A., Birgaoanu M., and Griffiths-Jones S.. 2018. miRBase: From microRNA sequences to function. Nucleic Acids Res. 47:D155–D162. 10.1093/nar/gky1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavelle, A., and Sokol H.. 2020. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 17:223–237. 10.1038/s41575-019-0258-z [DOI] [PubMed] [Google Scholar]
- Leow, S.C., Poschmann J., Too P.G., Yin J., Joseph R., McFarlane C., Dogra S., Shabbir A., Ingham P.W., Prabhakar S., et al. 2016. The transcription factor SOX6 contributes to the developmental origins of obesity by promoting adipogenesis. Development. 143:950–961. 10.1242/dev.131573 [DOI] [PubMed] [Google Scholar]
- Li, J., Ouyang T., Li M., Hong T., Alriashy M., Meng W., and Zhang N.. 2021. CBX7 is dualistic in cancer progression based on its function and molecular interactions. Front. Genet. 12:740794. 10.3389/fgene.2021.740794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N., Lu N., and Xie C.. 2019. The Hippo and Wnt signalling pathways: Crosstalk during neoplastic progression in gastrointestinal tissue. FEBS J. 286:3745–3756. 10.1111/febs.15017 [DOI] [PubMed] [Google Scholar]
- Liu, J., Xing Y., and Rong L.. 2018. miR-181 regulates cisplatin-resistant non-small cell lung cancer via downregulation of autophagy through the PTEN/PI3K/AKT pathway. Oncol. Rep. 39:1631–1639. 10.3892/or.2018.6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankertz, J., and Schulzke J.-D.. 2007. Altered permeability in inflammatory bowel disease: Pathophysiology and clinical implications. Curr. Opin. Gastroenterol. 23:379–383. 10.1097/MOG.0b013e32816aa392 [DOI] [PubMed] [Google Scholar]
- McCarthy, D.J., Chen Y., and Smyth G.K.. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40:4288–4297. 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, L.B., Schug J., Vourekas A., McKenna J.B., Bramswig N.C., Friedman J.R., and Kaestner K.H.. 2010. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology. 139:1654–1664.e1. 10.1053/j.gastro.2010.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe, C., Kljavin N.M., Ybarra R., de Sauvage F.J., and Frederic J.. 2014. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell. 14:149–159. 10.1016/j.stem.2013.11.008 [DOI] [PubMed] [Google Scholar]
- Muller, P.A., Matheis F., Schneeberger M., Kerner Z., Jové V., and Mucida D.. 2020a. Microbiota-modulated CART+ enteric neurons autonomously regulate blood glucose. Science. 370:314–321. 10.1126/science.abd6176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, P.A., Schneeberger M., Matheis F., Wang P., Kerner Z., Ilanges A., Pellegrino K., del Mármol J., Castro T.B.R., Furuichi M., et al. 2020b. Microbiota modulate sympathetic neurons via a gut–brain circuit. Nature. 583:441–446. 10.1038/s41586-020-2474-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Rojas, A.R., and Mathis D.. 2021. Tissue regulatory T cells: Regulatory chameleons. Nat. Rev. Immunol. 21:597–611. 10.1038/s41577-021-00519-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnmacht, C., Park J.-H., Cording S., Wing J.B., Atarashi K., Obata Y., Gaboriau-Routhiau V., Marques R., Dulauroy S., Fedoseeva M., et al. 2015. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORγt⁺ T cells. Science. 349:989–993. 10.1126/science.aac4263 [DOI] [PubMed] [Google Scholar]
- Olivares-Villagómez, D., and Van Kaer L.. 2018. Intestinal intraepithelial lymphocytes: Sentinels of the mucosal barrier. Trends Immunol. 39:264–275. 10.1016/j.it.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanaban, V., Grasset E.M., Neumann N.M., Fraser A.K., Henriet E., Matsui W., Tran P.T., Cheung K.J., Georgess D., and Ewald A.J.. 2020. Organotypic culture assays for murine and human primary and metastatic-site tumors. Nat. Protoc. 15:2413–2442. 10.1038/s41596-020-0335-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H.W., Kim Y.C., Yu B., Moroishi T., Mo J.-S., Plouffe S.W., Meng Z., Lin K.C., Yu F.-X., Alexander C.M., et al. 2015. Alternative Wnt signaling activates YAP/TAZ. Cell. 162:780–794. 10.1016/j.cell.2015.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, Y.-f., He Y., Hu L.-z., Zhou B., Xu H.-y., and Liu X.-q.. 2020. The crosstalk between lncRNA-SNHG7/miRNA-181/cbx7 modulates malignant character in lung adenocarcinoma. Am. J. Pathol. 190:1343–1354. 10.1016/j.ajpath.2020.02.011 [DOI] [PubMed] [Google Scholar]
- Peterson, L.W., and Artis D.. 2014. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 14:141–153. 10.1038/nri3608 [DOI] [PubMed] [Google Scholar]
- Piersma, B., Bank R.A., and Boersema M.. 2015. Signaling in fibrosis: TGF-β, WNT, and YAP/TAZ converge. Front. Med. 2:59. 10.3389/fmed.2015.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, L., Chen Y., Niu Y., Chen W., Wang Q., Xiao S., Li A., Xie Y., Li J., and Zhao X.. 2010. A deep investigation into the adipogenesis mechanism: Profile of microRNAs regulating adipogenesis by modulating the canonical Wnt/β-catenin signaling pathway. BMC Genom. 11:1–10. 10.1186/1471-2164-11-320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, F., and Fiocchi C.. 2009. Intestinal fibrosis in IBD—a dynamic, multifactorial process. Nat. Rev. Gastroenterol. Hepatol. 6:228–235. 10.1038/nrgastro.2009.31 [DOI] [PubMed] [Google Scholar]
- Ritchie, M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., and Smyth G.K.. 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandan, S., Akbalik G., Kochen L., Rinne J., Kahlstatt J., Glock C., Tushev G., Alvarez-Castelao B., Heckel A., and Schuman E.M.. 2017. Activity-dependent spatially localized miRNA maturation in neuronal dendrites. Science. 355:634–637. 10.1126/science.aaf8995 [DOI] [PubMed] [Google Scholar]
- Shen, C., Wang D., Liu X., Gu B., Du Y., Wei F.Z., Cao L.L., Song B., Lu X., Yang Q., et al. 2015. SET7/9 regulates cancer cell proliferation by influencing β-catenin stability. FASEB J. 29:4313–4323. 10.1096/fj.15-273540 [DOI] [PubMed] [Google Scholar]
- Su, R., Lin H.S., Zhang X.H., Yin X.L., Ning H.M., Liu B., Zhai P.F., Gong J.N., Shen C., Song L., et al. 2015. MiR-181 family: Regulators of myeloid differentiation and acute myeloid leukemia as well as potential therapeutic targets. Oncogene. 34:3226–3239. 10.1038/onc.2014.274 [DOI] [PubMed] [Google Scholar]
- Sun, X., Sit A., and Feinberg M.W.. 2014. Role of miR-181 family in regulating vascular inflammation and immunity. Trends Cardiovasc. Med. 24:105–112. 10.1016/j.tcm.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Flier, L.G., Sabates–Bellver J., Oving I., Haegebarth A., De Palo M., Anti M., Van Gijn M.E., Suijkerbuijk S., Van de Wetering M., Marra G., and Clevers H.. 2007. The intestinal Wnt/TCF signature. Gastroenterology. 132:628–632. 10.1053/j.gastro.2006.08.039 [DOI] [PubMed] [Google Scholar]
- Veiga-Fernandes, H., and Mucida D.. 2016. Neuro-immune interactions at barrier surfaces. Cell. 165:801–811. 10.1016/j.cell.2016.04.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtue, A.T., McCright S.J., Wright J.M., Jimenez M.T., Mowel W.K., Kotzin J.J., Joannas L., Basavappa M.G., Spencer S.P., Clark M.L., et al. 2019. The gut microbiota regulates white adipose tissue inflammation and obesity via a family of microRNAs. Sci. Transl. Med.. 11:eaav1892. 10.1126/scitranslmed.aav1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Chiang I.L., Ohara T.E., Fujii S., Cheng J., Muegge B.D., Ver Heul A., Han N.D., Lu Q., Xiong S., et al. 2019. Long-term culture captures injury-repair cycles of colonic stem cells. Cell. 179:1144–1159.e15. 10.1016/j.cell.2019.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, H., Lal K., Yang F.F., and Chen J.. 2015. The pathological role and prognostic impact of miR-181 in acute myeloid leukemia. Cancer Gen. 208:225–229. 10.1016/j.cancergen.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, A., Henao-Mejia J., Harman C.C.D., and Flavell R.A.. 2013. miR-181 and metabolic regulation in the immune system. Cold Spring Harbor Symp. Quant. Biol. 78:223–230. 10.1101/sqb.2013.78.020024 [DOI] [PubMed] [Google Scholar]
- Witkowski, M., Witkowski M., Saffarzadeh M., Friebel J., Tabaraie T., Ta Bao L., Chakraborty A., Dörner A., Stratmann B., Tschoepe D., et al. 2020. Vascular miR-181b controls tissue factor-dependent thrombogenicity and inflammation in type 2 diabetes. Cardiovasc. Diabetol. 19:20. 10.1186/s12933-020-0993-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, B.H., Hagemann S., Mamareli P., Lauer U., Hoffmann U., Beckstette M., Föhse L., Prinz I., Pezoldt J., Suerbaum S., et al. 2016. Foxp3+ T cells expressing RORγt represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol. 9:444–457. 10.1038/mi.2015.74 [DOI] [PubMed] [Google Scholar]
- Yoo, B.B., and Mazmanian S.K.. 2017. The enteric network: Interactions between the immune and nervous systems of the gut. Immunity. 46:910–926. 10.1016/j.immuni.2017.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., Li S., Guo J., Xu Z., Zheng J., and Sun X.. 2020. Farnesoid X receptor antagonizes Wnt/β-catenin signaling in colorectal tumorigenesis. Cell Death Dis. 11:640. 10.1038/s41419-020-02819-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Kim J.K., Edwards C.A., Xu Z., Taichman R., and Wang C.Y.. 2005. Clusterin inhibits apoptosis by interacting with activated Bax. Nat. Cell Biol. 7:909–915. 10.1038/ncb1291 [DOI] [PubMed] [Google Scholar]
- Zheng, H., Liu J., Tycksen E., Nunley R., and McAlinden A.. 2019. MicroRNA-181a/b-1 over-expression enhances osteogenesis by modulating PTEN/PI3K/AKT signaling and mitochondrial metabolism. Bone. 123:92–102. 10.1016/j.bone.2019.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, L.M., and Yang M.. 2019. The suppression of miR-181 inhibits inflammatory responses of osteoarthritis through NF-κB signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 23:5567–5574. 10.26355/eurrev_201907_18290 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows pathological assessment of mucosal damage in colons from WT and DKO mice during DSS-induced colitis.
shows pathological assessment of neoplastic and inflammatory changes in colons from WT and DKO mice during DSS-induced colitis.
shows gene counts of RNA-seq data from WT and DKO mouse colonic IECs at baseline and during recovery from DSS (7 d of 2% DSS treatment, 2 d of recovery).
Data Availability Statement
RNA-seq datasets have been uploaded to the Gene Expression Omnibus repository (accession number GSE208410).