Abstract
A bacterium, Sphingomonas sp. strain A1, can incorporate alginate into cells through a novel ABC (ATP-binding cassette) transporter system specific to the macromolecule. The transported alginate is depolymerized to di- and trisaccharides by three kinds of cytoplasmic alginate lyases (A1-I [66 kDa], A1-II [25 kDa], and A1-III [40 kDa]) generated from a single precursor through posttranslational autoprocessing. The resultant alginate oligosaccharides were degraded to monosaccharides by cytoplasmic oligoalginate lyase. The enzyme and its gene were isolated from the bacterial cells grown in the presence of alginate. The purified enzyme was a monomer with a molecular mass of 85 kDa and cleaved glycosidic bonds not only in oligosaccharides produced from alginate by alginate lyases but also in polysaccharides (alginate, polymannuronate, and polyguluronate) most efficiently at pH 8.0 and 37°C. The reaction catalyzed by the oligoalginate lyase was exolytic and thought to play an important role in the complete depolymerization of alginate in Sphingomonas sp. strain A1. The gene for this novel enzyme consisted of an open reading frame of 2,286 bp encoding a polypeptide with a molecular weight of 86,543 and was located downstream of the genes coding for the precursor of alginate lyases (aly) and the ABC transporter (algS, algM1, and algM2). This result indicates that the genes for proteins required for the transport and complete depolymerization of alginate are assembled to form a cluster.
Alginate is a linear polysaccharide composed of β-d-mannuronate and the C5 epimer α-l-guluronate, arranged in three different ways: poly-β-d-mannuronate (polyM), poly-α-l-guluronate (polyG), and heteropolymeric (polyMG) region in which there is a random arrangement of the monomers (4). Alginate produced by brown seaweeds is not acetylated and is widely used in food and pharmaceutical industries because of the ability of the polymer to chelate metal ions and form a highly viscous solution (18). The acetylated form of alginate is synthesized by certain bacteria, such as mucoid cells of Pseudomonas aeruginosa and Azotobacter vinelandii. P. aeruginosa causes serious chronic pulmonary infections in the lungs of patients with cystic fibrosis (CF) (2), and alginate produced by the bacterial cells seems to play a crucial role in the adherence of the bacterium to target cells (21). The alginate functions as a biofilm and decreases the effect of antimicrobial agents by repressing the penetration of the agent into the biofilm, thus making it difficult to treat biofilm-dependent bacterial infectious diseases (5).
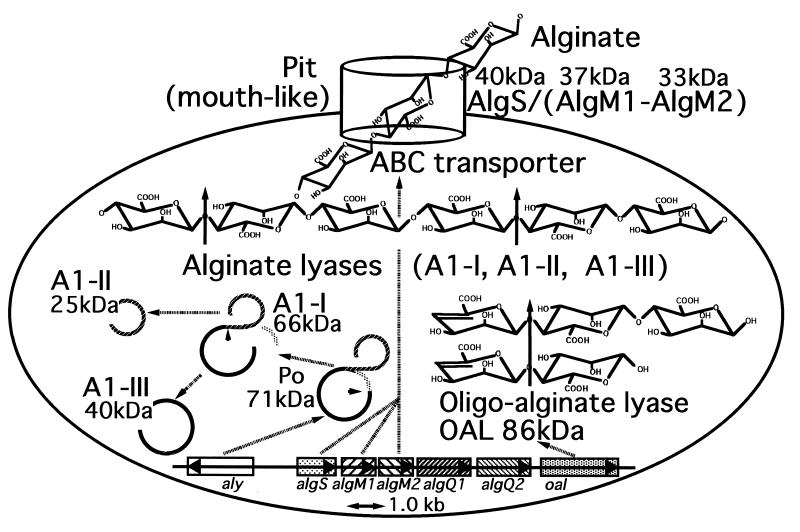
A bacterium, Sphingomonas sp. strain A1, incorporates alginate into the cells through a pit formed on the cell surface (8), and an ABC (ATP-binding cassette) transporter specific to the macromolecule is responsible for transport of alginate (16) (Fig. 1). The incorporated alginate is depolymerized by three types of cytoplasmic alginate-depolymerizing enzymes (alginate lyases A1-I [66 kDa], A1-II [25 kDa], and A1-III [40 kDa]) formed from a common precursor protein through the consecutive processes of posttranslational modification (Fig. 1) (17). A1-I is autoprocessed to give rise A1-II and A1-III (9), and these alginate lyases cleave glycosidic bonds endolytically in the alginate molecule by β-elimination reaction. Briefly, A1-I is active on acetylated and nonacetylated alginates. A1-II prefers polyG and nonacetylated alginate produced by brown seaweeds. A1-III efficiently liquefies polyM and acetylated alginates produced by mucoid cells of P. aeruginosa derived from the lungs of CF patients (17, 32); this property may be useful as a clinical agent for the therapy of CF and other infectious diseases caused by P. aeruginosa.
FIG. 1.
Overall pathway for alginate metabolism in Sphingomonas sp. strain A1. Alginate is first concentrated in a mouth-like pit on the cell surface and then incorporated into cells through the ABC transporter system consisting of AlgS, AlgM1, and AlgM2. The incorporated alginate is depolymerized by three kinds of alginate lyase (A1-I, A1-II, and A1-III) to give rise to di- and trisaccharides with an unsaturated uronyl residue at the nonreducing terminus. Alginate lyases are first synthesized as a precursor protein (Po), followed by the transformation to A1-I by excising the N-terminal peptide. A1-I is autocatalytically processed to A1-II and A1-III. The resultant oligosaccharides by the actions of A1-I, A1-II, and A1-III are degraded by oligoalginate lyase to an unsaturated monosaccharide, which is nonenzymatically converted into α-keto acid. Genes encoding proteins responsible for alginate transport and assimilation form a cluster. aly, alginate lyase (Po) gene; algS, algM1, and algM2, alginate ABC transporter genes; algQ1 and algQ2, genes with unknown functions; oal, oligoalginate lyase gene.
Three alginate lyases (A1-I, A1-II, and A1-III) can produce di- and trisaccharides from alginate as major final products (6, 32), thus implying that the cells of Sphingomonas sp. strain A1 have an additional enzyme responsible for the degradation of alginate oligosaccharides to the constituent monosaccharides. To construct the complete metabolic pathway for alginate in Sphingomonas sp. strain A1, we have purified and characterized the enzyme (oligoalginate lyase) catalyzing the degradation of alginate oligosaccharides and determined its primary structure by cloning the gene.
MATERIALS AND METHODS
Materials.
Sodium alginate (average molecular weight, 25,700; polymerization degree, more than 100, viscosity, 1,000 cp) from Eisenia bicyclis and DEAE-cellulose were purchased from Nacalai Tesque Co. Ltd., Kyoto, Japan. PolyM and polyG (polymerization degree, ∼100) were kind gifts from T. Muramatsu, Nagasaki University, Nagasaki, Japan. Silica gel 60/Kieselguhr F254 thin-layer chromatography (TLC) plates were obtained from E. Merck, Darmstadt, Germany. Butyl-Toyopearl 650M and QAE-Toyopearl 650C were purchased from Tosoh Co. (Tokyo, Japan), Sephacryl S-200HR was from Pharmacia Biotech (Uppsala, Sweden), and Bio-Gel P2 was from Bio-Rad Laboratories (Hercules, Calif.). Restriction endonucleases, DNA-modifying enzymes, a vector (pUC118), and Escherichia coli DH5α competent cells were from Takara Shuzo Co. (Kyoto, Japan) and Toyobo Co. (Tokyo, Japan). A broad-host-range cosmid vector of pKS13 (11) was a kind donation from M. Takagi, University of Tokyo, Tokyo, Japan.
Microorganism and culture conditions.
For the purification of oligoalginate lyase, cells of Sphingomonas sp. strain A1 were aerobically cultured at 30°C and 100 rpm for 48 h in a liquid alginate medium consisting of 0.1% (NH4)2SO4, 0.1% KH2PO4, 0.1% Na2HPO4, 0.01% MgSO4 · 7H2O, 0.01% yeast extract, and 0.5% alginate (pH 7.2). To investigate the effect of a carbon source on the enzyme activity, alginate in the medium (20 ml) was replaced by pectin or glucose (0.5%).
Preparation of substrates for oligoalginate lyase.
Alginate depolymerization products (di- and trisaccharides) produced from alginate by alginate lyase (A1-III) were purified by a gel filtration of Bio-Gel P2 column (0.9 by 122 cm) previously equilibrated with distilled water (6).
TLC.
Degradation of alginate oligosaccharides by oligoalginate lyase was analyzed by TLC with a solvent system of 1-butanol–acetic acid–water (2:1:1, vol/vol). The reaction products were visualized by heating the TLC plate at 110°C for 5 min after spraying with 10% (vol/vol) sulfuric acid in ethanol. Unsaturated saccharides and α-keto acids on TLC plates were detected by thiobarbituric acid (TBA) and o-phenylenediamine staining methods, respectively (13, 30).
Assay for enzymes and protein.
Oligoalginate lyase was assayed using tri- and/or disaccharides as substrates. One unit of enzyme activity was defined as the amount required to degrade 1 μg of alginate trisaccharide per min. The amount of degradation products on TLC plates was densitometrically measured using the NIH1.52 program. Protein was determined by the method of Bradford (3), with bovine serum albumin as a standard, or by measuring absorbance at 280 nm, assuming that E280 = 1.0 corresponds to 1 mg/ml.
Cell fractionation.
Cells grown at 30°C for 48 h in alginate medium were harvested by centrifugation at 13,000 × g and 4°C for 10 min, and the resulting culture fluid was used as an extracellular enzyme source. Collected cells were washed in 0.2 M Tris acetate buffer, pH 7.8, resuspended in the same buffer, and then subjected to preparation of periplasmic, cytoplasmic, and membrane fractions as described previously (10).
Purification of oligoalginate lyase.
Unless otherwise specified, all procedures were performed at 0 to 4°C. Cells of Sphingomonas sp. strain A1 were cultured at 30°C for 48 h in 15 liters of alginate medium (1.5 liters/flask). After cultivation, the cells were collected by centrifugation at 13,000 × g and 4°C for 10 min, washed with 20 mM Tris-HCl buffer (pH 7.2) (buffer A), and resuspended in the same buffer to prepare cell fractions. Spheroplasts were ultrasonically disrupted (Insonator, model 201M; Kubota, Tokyo, Japan) at 0°C and 9 kHz for 40 min, and the clear solution (cytoplasmic fraction) obtained after centrifugation at 150,000 × g and 4°C for 2 h was dialyzed against buffer A overnight. The dialysate was applied to a DEAE-cellulose column (4.7 by 41 cm) previously equilibrated with buffer A. The enzyme was eluted with a linear gradient of NaCl (0 to 2.0 M) in buffer A (2 liters), and 17-ml fractions were collected every 9 min. The active fractions, which were eluted with 0.7 M NaCl, were saturated with ammonium sulfate (30%), and then the enzyme solution was applied to a Butyl-Toyopearl 650M column (2.7 by 17 cm) previously equilibrated with buffer A saturated with ammonium sulfate (30%). The enzyme was eluted with a linear gradient of ammonium sulfate (30 to 0%) in buffer A (500 ml), and 4-ml fractions were collected every 4 min. The active fractions, which were eluted with buffer A saturated with ammonium sulfate (less than 3%), were combined, concentrated by ultrafiltration with an Amicon model 8200 (Amicon Co., Beverly, Mass.) to about 3 ml, and applied to a Sephacryl S-200HR column (2.7 by 64 cm) previously equilibrated with 20 mM potassium phosphate buffer (KPB), pH 7.0, containing 0.15 M NaCl. The enzyme was eluted with the same buffer, and 3-ml fractions were collected every 6 min. The enzyme eluted between fraction 44 and 47. These fractions were combined and dialyzed against buffer A overnight. The dialysate was applied to QAE-Toyopearl 650C column (0.8 by 2.8 cm) previously equilibrated with buffer A. The enzyme was eluted with a linear gradient of NaCl (0 to 0.5 M) in buffer A (100 ml), and 1-ml fractions were collected every 0.5 min. The active fractions, which were eluted with 0.2 M NaCl, were used as a purified enzyme source.
Electrophoresis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and native gradient PAGE were done as described previously (12; Pharmacia LKB manual, Pharmacia LKB, Uppsala, Sweden).
N-terminal amino acid sequence of oligoalginate lyase.
The N-terminal amino acid sequence of the enzyme was determined by Edman degradation using the Procise 492 protein sequencing system (Applied Biosystems Division of Perkin-Elmer, Foster City, Calif.).
Molecular cloning of oligoalginate lyase gene.
A genomic DNA library (16) of Sphingomonas sp. strain A1 previously constructed in E. coli using a pKS13 cloning vector was screened by colony hybridization method (1) with 32P-labeled probe (ATGAARAARYTNGARCARCC) corresponding to the N-terminal amino acid sequence of the enzyme. Several positive clones were obtained and cultivated in Luria-Bertani (LB) medium (24) supplemented with tetracycline at 50 μg/ml. A plasmid vector was extracted from one of these clones and then subjected to subcloning and DNA sequence.
DNA sequence and DNA manipulations.
The nucleotide sequence of the oligoalginate lyase gene was determined by the dideoxy-chain termination method using automated DNA sequencer model 377 (Applied Biosystems Division, Perkin-Elmer) (25). Subcloning, transformation, and gel electrophoresis were performed as described previously (24).
Transformation of Sphingomonas sp.
A plasmid constructed with pKS13 was transferred to cells of Sphingomonas species by the triparental mating method (23).
Nucleotide sequence accession number.
The nucleotide sequence for the oligoalginate lyase gene reported in this study has been deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. AB011415.
RESULTS AND DISCUSSION
Oligoalginate lyase activity in Sphingomonas sp. strain A1.
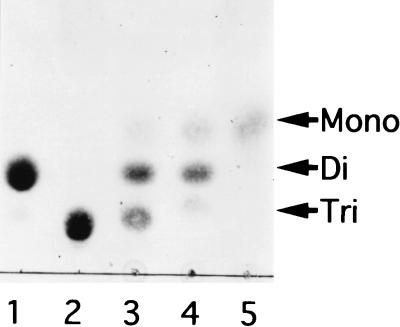
Alginate oligosaccharides (di- and trisaccharides) with an unsaturated saccharide at the nonreducing terminus are released as final products from alginate by cytoplasmic alginate lyases (A1-I, A1-II, and A1-III) in cells of Sphingomonas sp. strain A1 (6, 32). The alginate trisaccharide was found to be degraded via alginate disaccharide to monosaccharide by the cytoplasmic enzyme fraction prepared from the cells of Sphingomonas sp. strain A1 (Fig. 2), although extracellular, periplasmic, and membrane enzyme fractions failed to degrade the substrate at the same protein level as the cytoplasmic fraction. The alginate disaccharide produced from alginate trisaccharide by the enzyme fraction was confirmed to be unsaturated by the TLC TBA staining method (data not shown), indicating the presence of an enzyme, designated here as an oligoalginate lyase, catalyzing the release of unsaturated saccharide from alginate oligosaccharides by β-elimination reaction. Cells of Sphingomonas sp. strain A1 can grow in the medium containing alginate, pectin, or glucose as the sole carbon source (15). The level of expression of oligoalginate lyase in the bacterium grown on alginate was 71 mU/mg of protein, although it was significantly lower (less than 2 mU/mg of protein) when grown in the medium containing pectin or glucose.
FIG. 2.
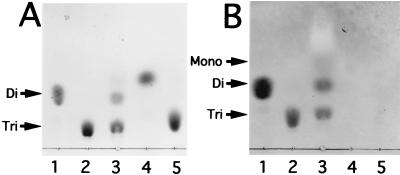
Degradation of alginate trisaccharide by cytoplasmic enzyme fraction. Alginate trisaccharide (10 μg) was incubated with the enzyme fraction (5.6 μg of protein) for 15 min (lane 3), 1 h (lane 4), and 24 h (lane 5), and the products were analyzed by TLC. Tri, Di, and Mono indicate the tri-, di-, and monosaccharides generated from alginate, respectively. Lane 1, alginate disaccharide (10 μg); lane 2, alginate trisaccharide (10 μg).
Purification and characterization of oligoalginate lyase.
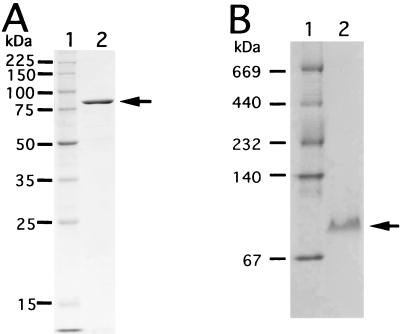
Oligoalginate lyase was purified 40.8-fold with a recovery yield of 1.20% from the cytoplasmic fraction of Sphingomonas sp. strain A1 cells grown on alginate (Table 1) and was homogeneous on SDS- and native gradient polyacrylamide gels (Fig. 3).
TABLE 1.
Purification of oligoalginate lyase
| Stepa | Total amt (mg) of protein | Total activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Cytoplasmic fraction | 3,530 | 250 | 0.071 | 100 | 1.00 |
| DEAE-cellulose | 1,470 | 128 | 0.087 | 51.1 | 1.23 |
| Butyl-Toyopearl 650M | 99.8 | 59.2 | 0.593 | 23.7 | 8.39 |
| Sephacryl S-200HR | 8.06 | 14.4 | 1.79 | 5.77 | 25.3 |
| QAE-Toyopearl 650C | 1.04 | 3.00 | 2.89 | 1.20 | 40.8 |
Purification procedures are described in Materials and Methods.
FIG. 3.
Electrophoretic profiles of oligoalginate lyase. (A) SDS-PAGE. Lane 1, molecular mass standards (synthetic polypeptides with molecular masses of 225, 150, 100, 75, 50, 35, 25, and 15 kDa); lane 2, purified enzyme. The arrow indicates the position of purified oligoalginate lyase. (B) Native gradient PAGE. Lane 1, molecular mass standards thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), lactate dehydrogenase (140 kDa), and bovine serum albumin (67 kDa); lane 2, purified enzyme. The arrow indicates the position of purified oligoalginate lyase.
(i) Molecular mass.
The molecular mass of the enzyme was determined to be 85 kDa by SDS-PAGE (Fig. 3A) and 83 kDa by native gradient PAGE (Fig. 3B), thus indicating that the enzyme is monomeric.
(ii) pH and temperature.
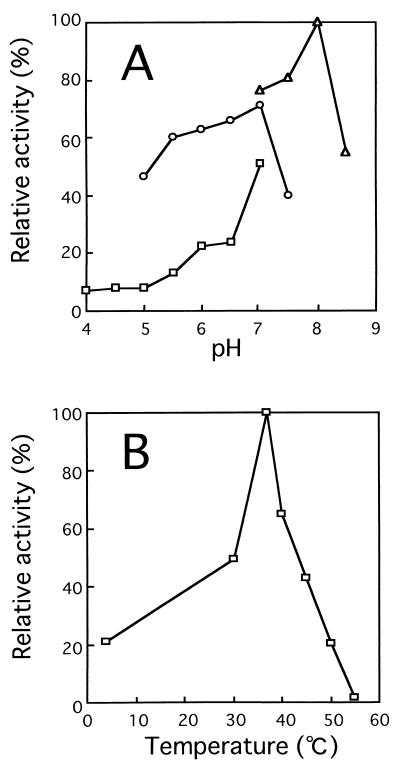
The enzyme was most active at pH 8.0 in 50 mM Tris-HCl buffer and at 37°C (Fig. 4) and was stable below 40°C when treated at various temperatures (30 to ∼80°C) for 10 min in 20 mM Tris-HCl buffer, pH 7.2.
FIG. 4.
Effects of pH (A) and temperature (B) on activity of oligoalginate lyase. Experiments were carried out at 30°C using alginate trisaccharide (10 μg) and purified enzyme (1.8 mU). (A) To determine the effect of pH, reactions were performed at 30°C for 30 min in the following 50 mM buffers; sodium acetate (□), potassium phosphate (○), and Tris-HCl (▵). Activity at pH 8.0 in Tris-HCl buffer was set at 100%. (B) To determine the optimal temperature, reactions were performed for 30 min at various temperatures in 50 mM Tris-HCl buffer, pH 8.0. Activity at 37°C was set at 100%.
(iii) Metal ions and other compounds.
The activity of the enzyme was determined in the presence and absence of various compounds. The activity of the enzyme was enhanced in the presence of Ca2+, Mn2+, Fe2+, and Mg2+ (1.4- to approximately twofold at 1 mM) or dithiothreitol (sevenfold at 1 mM). Other compounds tested (Co2+, Cu2+, Hg2+, glutathione, 2-mercaptoethanol, iodoacetic acid, N-ethylmaleimide, and EDTA tested at 1 mM; l-fucose, d-galactose, d-glucose, d-glucuronic acid, d-mannose, l-rhamnose, and d-xylose tested at 5 mM) showed no meaningful effect on activity of the enzyme.
(iv) Substrate specificity and mode of action.
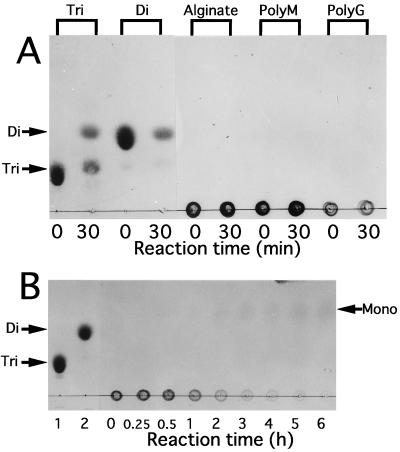
To examine the substrate specificity of oligoalginate lyase, the enzyme was incubated at 30°C for 30 min in a mixture containing 50 mM Tris-HCl buffer (pH 8.0), 0.1 mM dithiothreitol, and various substrates (Fig. 5A). The enzyme degraded alginate oligosaccharides (di- and trisaccharides), while polymers (alginate, polyM, and polyG) seemed not to be depolymerized. Therefore, the main function of oligoalginate lyase is to degrade the products (alginate oligosaccharides) formed by alginate lyase reactions in cells of Sphingomonas sp. strain A1. However, after prolonged incubation of these polymers with the enzyme, they were completely depolymerized. In the case of alginate, the enzyme released monosaccharide without producing any intermediates (Fig. 5B) and did not change an absorbance at 235 nm in the reaction mixture though alginate lyases with a endolytical manner generally increase the absorbance at 235 nm derived from the reaction products. The activity on alginate was also observed when the enzyme expressed in E. coli by the use of a plasmid containing only the oligoalginate lyase gene was used (data not shown). Furthermore, TLC TBA staining showed that the enzyme converted unsaturated alginate trisaccharide to unsaturated alginate disaccharide (Fig. 6B). These results indicate that the enzyme acts on alginate exolytically and catalyzes the β-elimination reaction. The resultant monosaccharide formed from alginate by oligoalginate lyase is also thought to be unsaturated and nonenzymatically transformed to α-keto acid, since the product (α-keto acid) was detected on TLC plates by staining with TBA (Fig. 6B) or o-phenylenediamine (data not shown) to detect ketohexonate. This is the reason why the product on TLC plates was scarcely detected by the method with sulfuric acid (Fig. 5 and 6A). The nonenzymatic conversion of unsaturated monosaccharide to α-keto acid has also been reported in the study of bacterial alginate metabolism (20).
FIG. 5.
Substrate specificity of oligoalginate lyase. (A) Various substrates (0.5%) indicated on the top were incubated with oligoalginate lyase (1.8 mU) at 30°C, and products formed after 0- and 30-min incubations were analyzed by TLC. Tri, alginate trisaccharide; di, alginate disaccharide. (B) Alginate (0.5%) was incubated with oligoalginate lyase (1.8 mU) at 30°C for several hours, followed by TLC analysis. Lane 1, alginate trisaccharide (10 μg); lane 2, alginate disaccharide (10 μg). Tri, di, and mono indicate the tri-, di-, and monosaccharides from alginate, respectively.
FIG. 6.
Mode of action of oligoalginate lyase. Alginate trisaccharide (10 μg) was incubated with oligoalginate lyase (1.8 mU) for 30 min at 30°C, and the products were analyzed on TLC plates (lanes 3 in panels A and B) by staining with sulfuric acid (A) or TBA (B). Lane 1, alginate disaccharide (10 μg); lane 2, alginate trisaccharide (10 μg); lane 4, alginate disaccharide (10 μg) prepared by acid hydrolysis; lane 5, alginate trisaccharide (10 μg) prepared by acid hydrolysis. Tri, Di, and Mono indicate the unsaturated tri-, di-, and monosaccharides from alginate, respectively.
(v) N-Terminal amino acid sequence.
The N-terminal amino acid sequence of oligoalginate lyase was determined to be 1MKKLEQPQSADMTIRYFP18.
Molecular cloning and sequence analysis of oligoalginate lyase gene.
The gene for oligoalginate lyase was screened in a genomic DNA library of Sphingomonas sp. strain A1, which was constructed in E. coli DH5α with no oligoalginate lyase activity. Several positive clones hybridized with the probe corresponding to N-terminal amino acid sequence were obtained. One clone harbored the plasmid (designated pOAL) having a 21-kb fragment of genomic DNA in pKS13 cloning vector and showed an apparent oligoalginate lyase activity. The EcoRI-digested fragment (about 9 kb) of the genomic DNA inserted in pOAL was ligated with EcoRI-digested pUC118 and the construct was designated pOAL-1. The nucleotide sequence of the EcoRI fragment of the plasmid pOAL-1 was determined from the first EcoRI site of the inserted genomic DNA. The fragment was found to contain a gene coding for N-terminal amino acid sequence (MKKLEQPQSADMTIRYFP) (1Met to 18Pro) of oligoalginate lyase. The gene (2,286 bp) began with an ATG start codon at nucleotide 6600 from the first EcoRI site of inserted genomic DNA and ended with a TAA stop codon at nucleotide 8885 downstream from the same EcoRI site. The gene encoded a polypeptide composed of 761 amino acid residues with a molecular weight of 86,543, thus confirming that the predicted amino acid sequence represents the primary structure of the enzyme. Signal sequence was not found in the N terminus of the polypeptide, thus suggesting that the enzyme is localized in cells as has actually been purified from the cytoplasmic fraction. A predicted ribosome-binding site (Shine-Dalgarno sequence, GAAGGA) (28) was found just before the start codon of the gene. An apparent promoter with homology to E. coli consensus promoter (7) and terminator were not found in the 5′ and 3′ regions of the gene, respectively.
The oligoalginate lyase of Sphingomonas sp. strain A1 was used to search for similarity in the protein databases by FASTA program (19). The enzyme exhibited no significant identity (more than 25%) with other proteins including alginate lyases on the databases.
Genetic mapping of oligoalginate lyase gene.
Since Sphingomonas sp. strain A1 has a cluster of genes involved in alginate transport (ABC transporter; algS, algM1, and algM2) and assimilation (alginate lyase as a precursor, aly) (16) (Fig. 1), the location of oligoalginate lyase gene was investigated. A transductant of strain AL-L, which is a alginate transport-deficient mutant derived from Sphingomonas sp. strain A1 (16), with pOAL exhibited growth in alginate medium, though the mutant cells did not. This result shows that the inserted genomic DNA in pOAL contains the cluster of alginate-ABC transporter genes (algS, algM1, and algM2). In fact, the nucleotide sequence of the genomic fragment in pOAL-1 completely matched the sequence of alginate-ABC transporter genes, and the first EcoRI site of inserted fragment in pOAL-1 existed in the middle of algS. Therefore, the oligoalginate lyase gene was confirmed to be localized in about 4 kb downstream of algM2 (Fig. 1).
Thus, we have purified and characterized oligoalginate lyase responsible for the complete depolymerization of alginate in Sphingomonas sp. strain A1 and elucidated the gene structure for the enzyme. In combination with results published previously (6, 16, 17, 32), an overall depolymerization route of alginate in Sphingomonas sp. strain A1 was established (Fig. 1). The high-molecular-weight-alginate in medium is first concentrated in the pit formed on the cell surface of the bacterium, directly incorporated into cells by the ABC transporter consisting of AlgS, AlgM1, and AlgM2, and then depolymerized to di- and trisaccharides by the action of three kinds of cytoplasmic alginate lyases (A1-I, A1-II, and A1-III). The alginate oligosaccharides thus formed are finally degraded by the cytoplasmic oligoalginate lyase to monosaccharides, which are nonenzymatically converted into α-keto acid. A disruptant with a mutation in the gene for endo-type alginate lyases (A1-I, A1-II, and A1-III) can grow sufficiently on a medium containing alginate trisaccharide as a carbon source but grows very poorly on medium containing alginate (K. Momma, W. Hashimoto, and K. Murata, unpublished results). The growth of the disruptant in both media suggests the significance of oligoalginate lyase in cells of Sphingomonas sp. strain A1, that is, the enzyme is prerequisite for the complete depolymerization of alginate, especially for degradation of alginate oligosaccharides produced by alginate lyases. As far as we know, this is the first report on the identification of molecular machineries equipped for the transport, depolymerization, and assimilation of alginate in bacteria, and oligoalginate lyase is the first cytoplasmic enzyme responsible for the exolytic depolymerization of alginate oligosaccharides in addition to alginate, polyM, and polyG.
Marine bacteria of Alteromonos sp. have been proposed to depolymerize alginate to oligosaccharides by extracellular endo-type alginate lyase and further degraded by intracellular exo-type enzyme (22, 27). However, in the case of Sphingomonas sp. strain A1, alginate is directly incorporated into cells without depolymerization. Therefore, the assimilation route of alginate in Sphingomonas sp. strain A1 is quite different from that found in marine bacteria. Most alginate lyases so far examined are specific to either polyM or polyG (29) with the exception of extracellular alginate lyase from Bacillus circulans 1351 (14) or Alteromonas sp. strain H-4 (26), which is able to depolymerize both polyM and polyG. Alginate lyases from Bacillus and Alteromonas species are thought to be of the endo type because they depolymerize alginate with a formation of oligoalginates with different molecular sizes. Therefore, the exo-type oligoalginate lyase we found in Sphingomonas sp. strain A1 is a novel cytoplasmic enzyme with a broad specificity for alginate and its depolymerization products.
In order to clarify the relationship between structure and function of alginate lyase, we have already determined the three-dimensional structure of A1-III specific to polyM (33). Crystals of A1-II (31) specific to polyG and A1-I (B. Mikami, H.-J. Yoon, W. Hashimoto, and K. Murata, unpublished results) active on both polyM and polyG were also prepared and have now been subjected to X-ray crystallographic studies. Since these alginate lyases with different substrate specificities function in an endolytic manner (6, 17, 32), the results on structural analysis of oligoalginate lyase with a broad substrate specificity may add to our understanding of the difference in action mode and substrate specificity of these lyases. The determination of the crystal structure of oligoalginate lyase is also in progress.
ACKNOWLEDGMENTS
We thank Tsuyoshi Muramatsu, Nagasaki University, for his kind gifts of polyM and polyG, and Masamichi Takagi, University of Tokyo, for his kind donation of pKS13.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience, John Wiley and Sons; 1987. [Google Scholar]
- 2.Boat T F, Beadet A L, Welsh M J. The metabolic basis of inherited disease. In: Seriver C R, editor. Cystic fibrosis. New York, N.Y: McGraw-Hill; 1989. pp. 2649–2680. [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Gacesa P. Alginates. Carbohydr Polym. 1988;8:161–182. [Google Scholar]
- 5.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hashimoto W, Okamoto M, Hisano T, Momma K, Murata K. Sphingomonas sp. A1 lyase active on both poly-β-d-mannuronate and heteropolymeric regions in alginate. J Ferment Bioeng. 1998;86:236–238. [Google Scholar]
- 7.Hawley D K, McClure W R. Comparison and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hisano T, Kimura N, Hashimoto W, Murata K. Pit structure on bacterial cell surface. Biochem Biophys Res Commun. 1996;220:979–982. doi: 10.1006/bbrc.1996.0526. [DOI] [PubMed] [Google Scholar]
- 9.Hisano T, Nishimura M, Yamashita T, Sakaguchi K, Murata K. On the self-processing of bacterial alginate lyase. J Ferment Bioeng. 1994;78:109–110. [Google Scholar]
- 10.Kawasaki S, Moriguchi R, Sekiya K, Nakai T, Ono E, Kume K, Kawahara K. The cell envelope structure of the lipopolysaccharide-lacking gram-negative bacterium Sphingomonas paucimobilis. J Bacteriol. 1994;176:284–290. doi: 10.1128/jb.176.2.284-290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimbara K, Hashimoto T, Fukuda M, Koana T, Takagi M, Oishi M, Yano K. Cloning of two tandem genes involved in degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. J Bacteriol. 1989;171:2740–2747. doi: 10.1128/jb.171.5.2740-2747.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Lanning M C, Cohen S S. The detection and estimation of 2-ketohexonic acids. J Biol Chem. 1951;189:109–114. [PubMed] [Google Scholar]
- 14.Larsen B, Hoøen K, Østgaard K. Kinetics and specificity of alginate lyases. Hydrobiologia. 1993;260/261:557–561. [Google Scholar]
- 15.Momma K, Hashimoto W, Miyake O, Yoon H-J, Kawai S, Mishima Y, Mikami B, Murata K. Special cell surface structure, and novel macromolecule transport/depolymerization system of Sphingomonas sp. A1. J Ind Microbiol Biotechnol. 1999;23:425–435. doi: 10.1038/sj.jim.2900719. [DOI] [PubMed] [Google Scholar]
- 16.Momma K, Okamoto M, Mishima Y, Mori S, Hashimoto W, Murata K. A novel bacterial ATP-binding cassette (ABC) transporter system that allows uptake of macromolecules. J Bacteriol. 2000;182:3998–4004. doi: 10.1128/jb.182.14.3998-4004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murata K, Inose T, Hisano T, Abe S, Yonemoto Y, Yamashita T, Takagi M, Sakaguchi K, Kimura A, Imanaka T. Bacterial alginate lyase: enzymology, genetics and application. J Ferment Bioeng. 1993;76:427–437. [Google Scholar]
- 18.Onsøyen E. Commercial applications of alginates. Carbohydr Eur. 1996;14:26–31. [Google Scholar]
- 19.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Preiss J, Ashwell G. Alginic acid metabolism in bacteria. J Biol Chem. 1962;237:309–316. [PubMed] [Google Scholar]
- 21.Ramphal R, Pier G B. Role of Pseudomonas aeruginosa mucoid exopolysaccharide in adherence to tracheal cells. Infect Immun. 1985;47:1–4. doi: 10.1128/iai.47.1.1-4.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romeo T, Bromley J C, Preston J F. Alginate lyase of varying substrate specificities from marine bacteria. In: Smith W H, editor. Biomass energy development. New York, N.Y: Plenum Press; 1986. pp. 303–320. [Google Scholar]
- 23.Ruvkun G B, Ausubel F M. A general method for site-directed mutagenesis in prokaryotes. Nature. 1981;289:85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawabe T, Ohtsuka M, Ezura Y. Novel alginate lyases from marine bacterium Alteromonas sp. strain H-4. Carbohydr Res. 1997;304:69–76. doi: 10.1016/s0008-6215(97)00194-8. [DOI] [PubMed] [Google Scholar]
- 27.Sawabe T, Sawada C, Suzuki E, Ezura Y. Intracellular alginate-oligosaccharide degrading enzyme activity that is incapable of degrading intact sodium alginate from a marine bacterium Alteromonas sp. Fisheries Sci. 1998;64:320–324. [Google Scholar]
- 28.Shine J, Dalgarno L. The 3′ terminal sequence of E. coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosomal binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutherland I W. Polysaccharide lyases. FEMS Microbiol Rev. 1995;16:323–347. doi: 10.1111/j.1574-6976.1995.tb00179.x. [DOI] [PubMed] [Google Scholar]
- 30.Warren L. Thiobarbituric acid spray reagent for deoxy sugars and sialic acids. Nature. 1960;186:237. doi: 10.1038/186237a0. [DOI] [PubMed] [Google Scholar]
- 31.Yoon H-J, Hashimoto W, Katsuya Y, Mezaki Y, Murata K, Mikami B. Crystallization and preliminary X-ray crystallographic analysis of alginate lyase A1-II from Sphingomonas species A1. Biochim Biophys Acta. 2000;1476:382–385. doi: 10.1016/s0167-4838(99)00244-7. [DOI] [PubMed] [Google Scholar]
- 32.Yoon H-J, Hashimoto W, Miyake O, Okamoto M, Mikami B, Murata K. Overexpression in Escherichia coli, purification, and characterization of Sphingomonas sp. A1 alginate lyases. Protein Expr Purif. 2000;19:84–90. doi: 10.1006/prep.2000.1226. [DOI] [PubMed] [Google Scholar]
- 33.Yoon H-J, Mikami B, Hashimoto W, Murata K. Crystal structure of alginate lyase A1-III from Sphingomonas species A1 at 1.78 Å resolution. J Mol Biol. 1999;290:505–514. doi: 10.1006/jmbi.1999.2883. [DOI] [PubMed] [Google Scholar]