Abstract
Circulating tumor cells (CTCs) play a crucial role in tumor recurrence and metastasis, and their early detection has shown remarkable benefits in clinical theranostics. However, CTCs are extremely rare, thus detecting them in the blood is very challenging. New CTC detection techniques are continuously being developed, enabling deeper analysis of CTC biology and potential clinical application. This article reviews current CTC detection techniques and their clinical application. CTCs have provided, and will continue to provide, important insights into the process of metastasis, which could lead to development of new therapies for different cancers.
1. Introduction
Circulating tumor cells (CTCs) were first described by Ashworth in 1869 as a group of tumor cells in the peripheral bloodstream originating from spontaneous solid tumor tissues (primary or metastatic) and a biomarker for cancer diagnosis and progression [1–5]. The mechanism of tumor metastasis caused by circulating tumor cells is shown in Figure 1. Tumor cells shed into the blood in situ cause blood-borne metastases [6]. CTCs with an epithelial-mesenchymal transition (EMT) phenotype are invasive enough to pass through the extracellular matrix (ECM), dissociate from the marginal front, and invade the tumor vasculature. CTCs can evade anoikis cell death in circulation. Disseminated tumor cells (DTCs) exhibiting the EMT phenotype undergo intravascular stasis and develop cell protrusions to promote transendothelial migration (TEM) of cancer cells into the metastatic site, where they may stay dormant for some time before colonizing. This allows cancer cells to evade immune surveillance and successfully colonize distant organs. DTCs then acquire the mesenchymal-epithelial transition phenotype to proliferate and form secondary tumors. Cancer cells promote self-growth and colonization of the metastatic site by secreting exosomes that promote their dynamic interaction with the tumor microenvironment [7]. Therefore, CTCs provide cellular evidence for metastasis and are useful biomarkers for cancer progression in most cancer patients [8]. Several biological characteristics contribute to the shedding of CTCs by the primary tumor and their role in metastasis. Generally, EMT promotes the formation and metastasis of CTCs. Metastasis is driven by cytokines, proteins, and transforming growth factor (TGF)-β-Smad signaling. TGF-β promotes metastasis by reducing the expression of epithelial cadherin (E-cadherin). On the other hand, the resistance of A-kinase anchoring protein 8 (AKAP8) to EMT can inhibit breast cancer metastasis. The infiltration of CTCs into the metastatic site is a complicated process. In addition to producing EMT and proteases, endothelial cells (ECs) secrete CXC chemokine ligand 12 (CXCL12) to promote infiltration and perivascular tumor-associated macrophages (TAMs) to upregulate epidermal growth factor and matrix metalloproteinase-9 [9]. A hardened ECM induces the formation of invasive pseudopodia in cancer cells, enabling them to penetrate the ECM to invade blood vessels. The activity of cancer-associated fibroblasts in the recombinant ECM has been shown to promote drilling and subsequent invasion of tumor cells [7].
Figure 1.
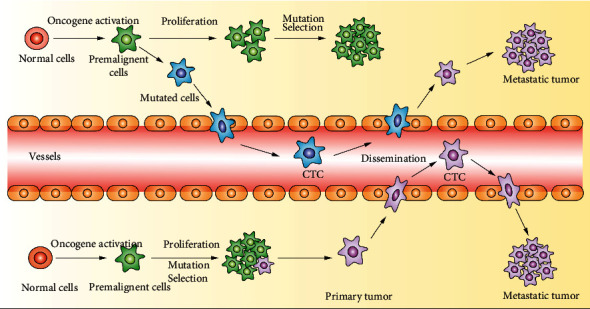
The mechanism of circulating tumor cells (CTCs) driving tumor metastasis. CTCs refer to all kinds of tumor cells in the peripheral blood. Due to their spontaneous or clinic operation, most of the CTCs undergo apoptosis or are swallowed after entering the peripheral blood. A few can escape and are anchored to become metastatic.
CTCs are rare in healthy individuals, and even in patients with malignancies, less than one CTC per 105 to 107 peripheral blood mononuclear cells (PBMCs) could be observed. Thus, isolation and enrichment are often the first steps in CTC detection in laboratories and hospitals [10]. There are two classical approaches to separating CTCs from blood samples: physical separation that exploits unique physical properties of CTCs (such as density and size) and immune adhesion, which depends on the high binding affinity of receptors on CTCs to specific antibodies or aptamers [11]. Compared with immune adhesion, physical isolation is a simpler method as it obviates the need for cell labeling. However, the immune adhesion method achieves higher purity in CTC isolation.
Due to technical limitations, few studies have investigated the precision of CTC detection methods. Keller and Pantel discussed how CTC analysis at single-cell resolution provides unique insights into tumor heterogeneity [12]. Martin et al. reviewed preclinical and clinical data on cancer treatment, CTC mobilization, and other factors that may promote metastasis, establishing that advanced therapeutic strategies could benefit patients with locally advanced cancer [13]. However, a systematic review of the occurrence, development, and outcome of CTCs in metastatic cancer is largely lacking. In this review, we present an overview of the biological characteristics of CTCs, current CTC detection techniques, and principles and methods of CTC isolation. Finally, potential applications of CTCs in the treatment of metastatic cancer are proposed.
2. Biological Characteristics of CTCs
2.1. Cellular Size
Due to high heterogeneity of tumor cells, the pore size in the CellSearch system is typically slightly larger than leukocytes [14]. Other researchers have successfully separated CTCs using size-based platforms that exploit difference in cell sizes. CTCs in prostate cancer are divided into three categories based on size (diameter): very small nuclear CTCs (<8.54 μm), small nuclear CTCs (8.54–14.99 μm), and large nuclear CTCs (>14.99 μm) [3].
Jiang et al. selectively enlarged the size of tumor cells covered with polystyrene microspheres and the modified cells were clearly distinguishable from white blood cells. The modification method had no significant effect on cell survival and proliferation. Using this method, 15 CTC subtypes were detected in 18 cases of colorectal cancer at a concentration of 4–72 CTCs/mL. Thus, this method has great potential in the early diagnosis and individualized treatment of cancer [15]. Zavridou et al. directly compared two different methods of isolating CTCs from head and neck squamous cell carcinoma: a size-dependent microfluidic system and epithelial cell adhesion molecule (EpCAM)-dependent positive selection. The results showed that, in the same blood sample, the label-freesize-dependent CTC separation system had higher sensitivity than the EpCAM-dependent CTC enrichment system [16].
2.2. Cellular Density
Density is the physical property exploited in traditional separation and enrichment methods for CTCs [17]. In Ficoll density gradient centrifugation, CTCs, plasma, and monocytes remain in the upper layer, whereas erythrocytes and polymorphonuclear leukocytes settle in the bottom layer. CTCs may occur in both plasma and separation fluid. Thus, some liquids above the red blood cell layer should be collected for enrichment to prevent the loss of CTCs [5]. Huang et al. developed a new density gradient centrifugation method that uses biodegradable gelatin nanoparticles wrapped on silica beads for isolation, release, and downstream analysis of CTCs from colorectal and breast cancer patients. This method has remarkable CTC capture efficiency (>80%), purity (>85%), high CTC-release efficiency (94%), and viability (92.5%) [18]. Thus, this approach provides new opportunities for personalized cancer diagnosis and treatment and may also be useful in developing drug treatment guidelines for cancer.
2.3. Heteromorphy in CTCs
Marrinucci et al. conducted cellular morphological evaluation of circulating components of highly metastatic breast cancer. They found highly polymorphic CTCs in breast cancer patients, including CTCs with high and low nuclear-to-cytoplasmic ratios and early and late apoptotic changes. In addition, compared with tumor cells in other sites, the complete morphologic spectrum of cancer cells in primary and metastatic tumors was also present in peripheral blood circulation [19]. Several studies have found various forms of CTCs in peripheral blood existing either independently or in clusters, with some CTCs even interacting with platelets to form a shell around them [20–23]. Aceto et al. found that CTC clusters in breast cancer-bearing mice were shed as whole oligomeric colonies rather than as simply a group of CTCs aggregating in the bloodstream [24, 25].
2.4. Proliferation and Apoptosis of CTCs
For tumor cells in circulation, only a few CTCs with high viability and potent metastatic potential survive and colonize distant organs to develop into metastatic foci. CTCs entering the circulatory system have very short survival times, typically less than 24 h, and vary in their indices of proliferation [26]. Driemel et al. found that the high expression of EpCAM was common in cancer cells in the proliferation stage, while the low expression of EpCAM inhibited the proliferation of CTCs [27]. Studies have reported low levels of expression of proliferating nuclear antigen Ki-67 in CTCs, suggesting that most CTCs may remain in a dormant state without entering the cell division cycle [28, 29]. It has also been found that, several years after primary tumorigenesis, dispersed CTCs and micrometastasis niche can remain dormant for a long time during resection of primary tumors [30]. These results suggest that dormant CTCs can be attached to tissues or cell clusters until their activation or that of a certain factor in the isolation procedures. The specific mechanism may be related to the body's immune response.
2.5. The Metastatic Portent of Circulating Tumor Cells
EMT is a biological process by which epithelial cells acquire a mesenchymal phenotype through a series of biochemical changes [31]. In recent years, accumulating evidence suggests EMT phenomena in the process of cancer cell metastasis [32–34]. In this process, cancer cells lose polarity and their connection with ECM, transforming into fusiform mesenchymal cells, which are easily detached from the tumor cell population. Several ECM-degrading proteases are upregulated in cancer cells with EMT, increasing their invasiveness [32–35]. As shown in Figure 2, the occurrence of EMT in CTCs could result in the loss of specific molecular markers in epithelial cells such as EpCAM and cytokeratin and overexpression of specific molecular markers in interstitial cells such as vimentin and cadherin. These cells have a strong survival advantage and high metastasis and potential for transfer in the blood circulation [17, 36–38]. Based on the EMT stage, CTCs are divided into E-type, M-type, E/M-type, and N (null)-type. Several studies have shown that E/M-type CTCs have enhanced epithelial cell adhesion and extravasation capacity, representing more aggressive subtype of cancer cells with the highest metastatic capacity [39–41]. Additionally, M-type CTCs exhibit enhanced resistance to clinically relevant chemotherapeutics.
Figure 2.
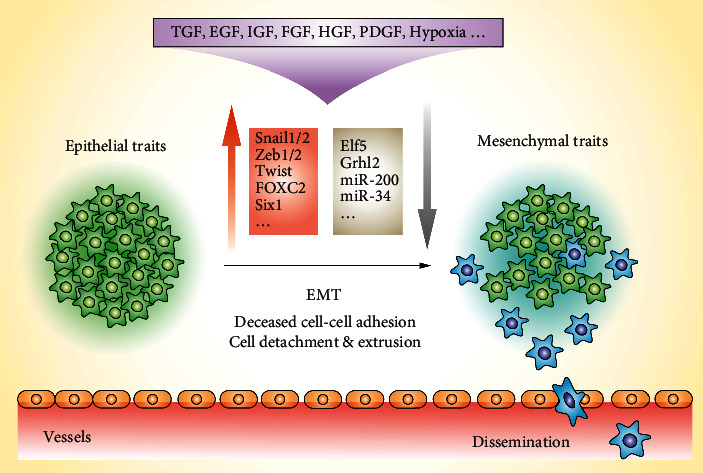
Induction of epithelial-mesenchymal transition (EMT) by various cytokines. It is generally assumed that the metastatic spread of epithelial tumors depends on EMT, a process in which cancer cells lose their polarity and cell-cell adhesion to acquire fibroblast-like features such as migration and invasion.
3. Separation and Enrichment of CTCs
There are two major approaches based on the principle of CTC separation and enrichment: physical property separation and affinity-based identification [42, 43]. For the physical property separation method, tumor cells are separated from other cells based on differences in size [44, 45], density [46], deformability, and adhesion between tumor cells and normal blood cells [18, 47]. The affinity-based identification method involves identification of the specific antigen on the surface of cancer cells using antibodies [44, 48], aptamer [49, 50], or E-selectin [51, 52]. Additional details are shown in Figure 3.
Figure 3.
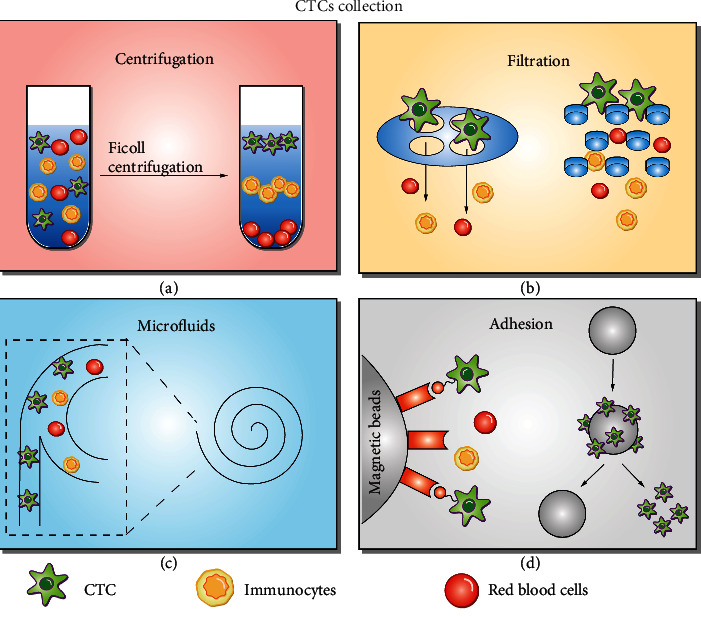
Circulating tumor cell (CTC) enrichment technologies. (a) Density gradient centrifugation. (b) Microfluidic-based separation technology. (c) Different filtration systems depending on the size of blood cells used for separation and enrichment of CTC. (d) Adhesion is dependent on the affinity and specific binding of antibodies or aptamers (e.g., immune adhesion) to the CTC receptor.
3.1. Gradient Density Centrifugation
Two centrifugation-based systems are available in the market today: OncoQuick and AccuCyte [43, 53]. After isolation and enrichment with Ficoll-Paque separation fluid, 24 CTCs were detected in fifty-eight 1 mL blood samples from colorectal cancer patients using real-time reverse transcription-polymerase chain reaction (RT-PCR) [53]. Rosenberg et al. reported that using a new OncoQuick system to isolate cancer cells had a 632-fold enrichment effect compared with less than 4-fold enrichment effect using Ficoll-Paque [53]. In addition, 11 CTCs were detected in 37 samples of gastric cancer patients using a combination of OncoQuick and RT-PCR [53]. In another study, 5 and 25 CTCs were detected in 60 cases of early breast cancer patients after immunofluorescence and 63 samples of patients with advanced breast cancer, respectively [2]. Although density-gradient centrifugation is uncomplicated, it lacks specificity and can easily lead to loss of tumor cells without corresponding density.
Therefore, density gradient centrifugation is often used as the first step to separate CTCs and then combined with other methods to specifically bind and separate CTCs. For example, Hu et al. used density gradient centrifugation and magnetic separation based on CD45 antibody to separate CTCs [46]. Different from the traditional negative enrichment, Hu et al. applied the subtraction enrichment and immunostaining fluorescence in situ hybridization (SE-iFISH) strategy to detect CTCs, which effectively removed red blood cells by centrifugation rather than using hypotonic injury [54].
3.2. Method for Separation and Capture Based on Cell Size
The method takes advantage of the larger size of CTCs compared with erythrocytes [55]. Isolation by size of epithelial tumor cells (ISET) and ScreenCell systems have been used for clinical trials in melanoma, breast, lung, and pancreatic cancers [56, 57]. For the first time, Zheng et al. used parylene-C to make circular and oval microporous filters, achieving a CTC capture efficiency of 90% [58]. A model for gene analysis and analysis of cells after chip electrolysis developed by Birkhahn et al. [59] was subsequently applied to the detection of exfoliated cells from urinary bladder cancer. Hosokawa et al. integrated nickel microporous sieves made from micro-electroforming into a microfluidic chip. The team also applied the improved nickel microporous sieves to the detection of CTCs in the blood of patients with small-cell lung cancer [60].
To improve the capture efficiency of CTCs, Coumans et al. studied factors affecting the trapping of filter cells [61]. In microfluidic chips, the precise fabrication of shapes and microstructures in microchannels makes it possible to separate and enrich tumor cells on a size-by-size basis [62]. Erythrocytes have stronger deformability and smaller volume, thus can easily cross various microstructures [63]. Niciński et al. proposed a new tool that uses microfluidic devices, photovoltaic (PV)-based SERS activity platform, and shell separation nanoparticles (shins) for simultaneous separation and unlabeled analysis of circulating tumor cells in blood samples. The results demonstrated the potential of SERS-based tools for isolating tumor cells from whole blood samples in a simple and minimally invasive way in a scaled-up detection and molecular identification pipeline [64]. Ohnaga et al. used a microchannel to capture circulating tumor cells in esophageal and breast cancers [65]. Zeinali et al. used a sensitive microfluidic CTC capture device to analyze circulating epithelium and EMT-like CTCs in pancreatic cancer [66].
To capture CTCs larger than the maximum pore size regardless of cell surface expression, blood is filtered through pores (usually 8 μm in diameter). However, the success of this process depends on many factors, including blood flow rate, pore size uniformity, and membrane stiffness. High flow velocity will cause CTC to “squeeze” through pores, causing membrane distortion. A very slow flow rate will lead to excessive accumulation of white blood cells, blood coagulation, and prolonged processing time [67]. Moreover, tumor cells undergoing epithelial-mesenchymal transition (EMT) were smaller than those without EMT characteristics [68]. Therefore, CTCs receiving EMT may not be detected using these technologies. Due to the inherent heterogeneity and dynamic expression of EpCAM and the degradation of cytokeratin during the transformation of epithelial cells into mesenchymal cells, the detection of circulating tumor cells in hepatocellular carcinoma with conventional methods is significantly limited, leading to false-negative detection of such CTCs. Wang et al. reported for the first time the existence of small-sized CTCs (<5 μm WBC) with cytogenetic abnormalities in aneuploid chromosome 8, which is predominantly detected in hepatocellular carcinoma (HCC) patients [69].
3.3. Immunomagnetic Beads
Almost all cells in the blood are diamagnetic or weakly magnetic [70]. Therefore, tumor cells are usually labeled with antibody-conjugated magnetic beads or nanoparticles. These antibodies bind primarily to tumor-cellsurface-specific antigens, including some intracellular antigens [71]. The number of CTCs counted using cell search has been used for prognosis of some cancers after metastasis [72–74]. Wu et al. developed a magnetic cell centrifugation platform (MCCP) combining the separation mechanism of magnetically labeled cells with the size-based method and obtained target cells with 97% purity, high throughput of 2 μL/s, and a sample enrichment factor of 66 times [75]. Overall, the performance of the immunomagnetic particle separation method mainly includes the following factors: (1) the expression level and specificity of the target antigen and the binding ability of the corresponding antibody and (2) the efficiency of immunomagnetic particle labeling. Immunomagnetic particles used for cell separation have high recovery and purity and even detect CTCs in one step [76–78].
3.4. Chip Technology
In the 1990s, Manz et al. proposed a microfluidic chip technology [79]. Commonly used CTC antibodies include human EpCAM and leukocyte common antigen CD45 [80]. Affinity sorting includes two types of capture methods. The first type is the positive sorting method, which directly targets and specifically captures target cells. The second type is the negative sorting method, which involves the capture nontarget cells, which are then discarded. A schematic diagram of the working principle is shown in Figure 4.
Figure 4.
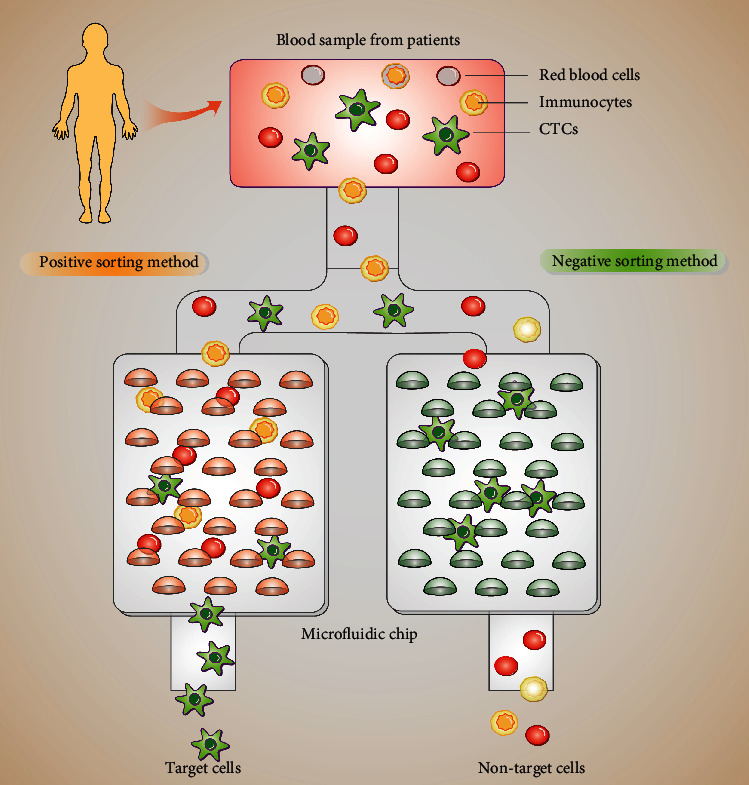
Peripheral blood samples from patients with non-small-cell lung cancer were obtained before any treatment and immediately processed in the circulating tumor cell (CTC) herringbone (HB)-chip that captures anti-EP-CAM-coated column epithelial cell adhesion molecules (left). Negative consumption of untargeted cells by a negative consumption method, including red blood cells and immune cells. Targeted cells such as CTCs were left in the chip for further analysis (right).
3.4.1. Positive Sorting Methods
The traditional affinity sorting method involves direct binding of the antibody to the microfluidic chip channel [81]. Sequist et al. developed the second-generation CTC chip called herringbone (HB)-chip [82]. Compared with first-generation CTC chips, the second-generationHB-chip is easy to use and more efficient, providing comprehensive and easy access to data. Hughes et al. integrated halloysite nanotubes into this chip [83] by immobilizing E-selectin and anti-EpCAM on nanotubes. In this design, E-selectin captures rapidly moving CTCs, whereas anti-EpCAM specifically captures CTCs [84, 85], increasing the purity of the captured CTCs. To simplify the experimental procedures, Stott et al. designed a fishbone-based affinity sorting chip for direct analysis of whole blood samples with a sorting speed of up to 1 mL/h. Captured circulating tumor cells could also be used for other assays or cell culture [86]. Sheng et al. optimized the fishbone structure to achieve a CTC capture efficiency and sorting purity higher than 90% and 84%, respectively [87]. These microfluidic chip technologies have shown good CTC capture capability. However, releasing CTCs from microfluidic chips for subsequent analysis is challenging. Therefore, researchers have introduced magnetic materials into microfluidic chips for CTC sorting [88].
3.4.2. Negative Sorting Methods
EpCAM-based affinity separation cannot be applied to CTCs with weakly expressed or nonexpressed EpCAM in the process of tumor cell metastasis, which leads to the significant decrease or even loss of EpCAM expression. For example, Lee designed a chip called “μ-MixMACS”, which greatly increased the number of CTCs detected [89]. Sajay et al. designed a two-step negative CTC sorting platform where the recovered cells remain bioactive and can be further analyzed for protein or nucleic acid content [90]. A CTC-negative enrichment scheme, which utilized the RosetteSep™ CTC Enrichment Cocktail Containing Anti-CD56 to collect CTCs in peripheral blood, was used to monitor the occurrence and disease response to treatment at different time points [91].
Unlike traditional negative enrichment, researchers utilize subtraction enrichment (SE), independent of cell size, cluster, or surface anchor protein expression. The immunostained proteins were proved to be free from the restriction of antigen epitopes inside and outside cells and membrane-related tumor biomarkers. With the clinical application of SE-iFISH, in addition to the traditional tumor cell types, there are more and more accidental discoveries of various phenotypes of CTCs [54]. Zhang et al. have shown that aneuploidy CD31− CTC and CD31+ CTEC may be used as a pair of biomarkers for circulating cell tumors to predict patients with non-small-cell lung cancer receiving antiangiogenesis combined therapy [40]. Based on the SE-iFISH strategy, Yang et al. demonstrated that patients with early bladder cancer had more triploid CTCs, tetraploid CTCs, and total circulation endothelial cells (CECs). Various CTC/CEC subtypes may have different potential function to guide the diagnosis, prognosis prediction, and treatment decision of bladder cancer [92]. Li et al. found that the presence of circulating tumor-cell-associated white blood cell (CTC-WBC) clusters is an independent prognostic factor for advanced non-small-cell lung cancer [93].
4. Detection of CTCs
4.1. Immunocytochemistry
Immunochemistry is a modern technology that binds specific monoclonal antibodies with CTCs, followed by conjugation of a chromogenic reagent to visualize CTCs. The most commonly used monoclonal antibody are anti-CK antibodies, such as epithelial-specific markers (CK) [94], interstitial cell surface markers (Snail1, E47, and Twist) [95], E-cadherin antagonist [96], stem cell markers (CD133+, CD44+, and CD24−), aldehyde dehydrogenase 1 (ALDH1) [97–100], special marker Survivin [101, 102], estrogen receptor (ER) [103], and progesterone receptor (PR) [104]. EpCAM and CK may be lost during epithelial-mesenchymal transition (EMT), leading to the failure of EpCAM- and CK-dependent strategies to detect CTCs. Therefore, Li et al. used SET-iFISH technology to enrich and characterize CTCs in advanced gastric cancer (AGC) and obtained a higher positive detection rate than that obtained using EpCAM-dependent detection strategies (CellSearch) [105]. In addition, Li et al. captured CTCs in AGC through SE-iFISH and found that cHER2 phenotype is useful to understanding the treatment resistance of AGC patients [106]. Subsequently, scientists used this method to characterize the markers of CTC, such as EpCAM [41], PD-L1 [107], vimentin [40], and CD44 [108].
4.2. RT-PCR
Currently, RT-PCR is considered the gold standard in the detection of some viruses because of its high sensitivity [109, 110]. RT-PCR is also widely used in tumor detection [111, 112]. However, selecting optimal RNA markers can be challenging, limiting its efficacy. An ideal RNA marker should have the following characteristics: all types of tumor cells are expressed in peripheral blood leukocytes, nontumor epithelial cells are not expressed, and no illegitimate transcription events [113]. Using qualitative RT-PCR, Wang et al. found that the expression of androgen receptor variant 7 (AR-V7) in CTCs from patients with prostate cancer was associated with drug resistance. The upregulation of AR-V7 led to the enhancement of cancer cell proliferation, suggesting poor patient prognosis [114]. Wei et al. recruited 78 patients with stage IA2–IIA1 cervical cancer who had undergone radical hysterectomy by laparotomy or laparoscopy and selected 34 uterine fibroid patients and 32 healthy subjects as the positive control group and negative control group, respectively. RT-PCR was used to amplify peripheral blood CK19, CK20, and SCC-Ag from total RNA. The results showed that CTCs were highly expressed in both the open surgery group and the laparoscopic radical mastectomy group, with no significant difference between the two groups [115]. Using Survivin, hTERT and hMAM markers to detect CTC in breast cancer patients, Shen et al. found that these markers improved the sensitivity of parallel tests and the specificity of series tests [116]. The molecular spectrum of four genes, including CK20, CK19, CEA, and GCC, identified 87.7% of tumor metastases with a false-positive rate of only 2.2% [117].
5. Clinical Applications of CTCs
CTCs are a promising biomarker for early disease diagnosis, treatment response and disease progression evaluation, recurrence monitoring, and therapeutic target identification for drug development [118]. Detection of CTCs has been widely used in the diagnosis of early and metastatic cancers (Table 1).
Table 1.
Clinical applications of CTCs
| Cancer type | Patient number | Detection methods | Marker | Significance | Clinical trial no. | Reference |
|---|---|---|---|---|---|---|
| Breast cancer | 549 | CellSearch | EpCAM, CK | In breast cancer patients with first-line chemotherapy, CTC counts were associated with mortality. | NCT00382018 | Paoletti et al. [119] |
| Prostate cancer | 147 mCRPC | VERSA | EpCAM | A transcriptional profile detectable in CTCs can serve as an independent prognostic marker in mCRPC. | NCT01942837, NCT01942837 | Sperger et al. [120] |
| Pancreatic cancer | 209 patients | CellSearch | EpCAM | CTC-positive preoperatively (≥1 CTC/7.5 mL) showed a detrimental outcome despite successful tumor resections. | NCT01919151 | Hugenschmidt et al. [121] |
| Colorectal cancer | 153 | CellSearch | EpCAM, CK | Baseline CTCs ≥ 3 were detected in 19% of the patients. CTC ≥ 3 at baseline and 4 weeks after therapy showed shorter overall survival. | NCT01442935 | Bidard et al. [122] |
5.1. Early Diagnosis and Staging of Cancer
Traditional imaging methods cannot effectively detect early tumor lesions. CTC detection approaches can detect tumor earlier than imaging or clinical manifestations when the lesion is <1 cm, hence facilitate early diagnosis. Besides its role in early tumor diagnosis, CTC is also correlated with tumor grade and TNM stage. Santos et al. found that CTCs have great potential in the early diagnosis of colorectal cancer since they can be detected in the peripheral blood of patients with early-stage colorectal cancer. Therefore, the CTC test may be applied to the diagnosis of colorectal cancer [123]. Clinical staging of colorectal cancer is often based on anatomical alterations of the intestine; however, it is difficult to accurately identify micrometastasis during the prognosis and treatment of patients [124]. Detection of CTCs in the blood does not necessarily indicate the occurrence of metastasis. However, several studies have shown the value of detection of CTCs in the staging of colorectal cancer in clinical practice [125]. Using an advanced CanPatrol CTC enrichment technique and in situ hybridization to sort and classify CTCs in blood samples, 90.18% of hepatocellular carcinoma (HCC) patients were found to be CTC positive, even at the early stage of HCC [126]. CTCs were also detected in 2 of 12 patients with hepatitis B virus (HBV) infection, with both patients developing small HCC tumors in less than five months. Another study by Wang et al. implicated CTCs in tumor staging [127]. Recent studies have shown that CTCs also put into good use in hematologic malignancies. Primary plasma cell leukemia (pPCL) is clinically distinguishable from newly diagnosed multiple myeloma (NDMM) based on the proportion of circulating tumor cells of 20% [128]. Zhang et al. also used a technology based on oncolytic herpes-simplex-virus-1 to detect CTCs in non-Hodgkin's lymphoma [129].
5.2. Treatment Evaluation and Recurrence Monitoring
Treatment evaluation and recurrence monitoring of CTCs has been extensively studied. Lin et al. measured the number of peripheral blood CTCs before and after NK cell immunotherapy in stage IV non-small-cell lung cancer (NSCLC) patients, providing a useful reference for monitoring any change in NK cell therapeutic effect [130]. Nagrath et al. detected and monitored CTCs using a CTC chip and found that the CTC count of patients with lung and prostate cancer decreased significantly before and after chemotherapy and endocrine therapy. Although the changes in the CTC count due to treatment are affected by the differences between individual patients, they can still be used as a reference for evaluating the efficacy of tumor treatment [131–133]. In some cases, CTCs are more sensitive than imaging, thus they are included in efficacy evaluations [134]. In recent years, several detection techniques have been developed for CTC genotyping as well as detection of crucial gene mutations, such as ER [135], HER2 [136], and TP53 [137]. Thus, these techniques can help clinicians in treatment evaluation and monitoring tumor recurrence. Zhou et al. used PCR and fluorescence-activatedsingle-cell sorting (FACS) to detect levels of EpCAM mRNA+ CTCs and CD4+CD25+Foxp3+ Treg cells in 49 HCC patients before surgery. The data showed that CTC/Treg levels were positively correlated with the risk of postoperative recurrence [138].
Due to differences in tumor type and stage among cancer patients and occult, it is difficult to detect metastatic tumor relapse within five years of primary tumor resection. Cancer that persists despite treatment and cannot be detected by current medical imaging modalities is defined as minimal residual disease (MRD), which is in the occult stage of cancer progression. Liquid biopsy methods based on detection of small amounts of circulating tumor cells (CTCs) or trace amounts of circulating cell-free tumor DNA (ctDNA) are now available for MRD detection in patients with various malignant tumors. Monitoring CTCs and ctDNA during postoperative follow-up assessments can detect disease recurrence months earlier than other medical imaging methods. Further characterization of CTCs and ctDNA could provide insights into the molecular evolution of MRD during tumor progression, which has important implications for treatments that delay or even prevent metastatic recurrence. Therefore, the detection of CTCs has become the main method for the assessment of minimal residual disease (MRD) [139].
6. Conclusion
Despite the initial promise of CTCs in clinical application, several challenges must be addressed before CTC analysis gains widespread application in clinical practice. At present, CTC cell count and molecular phenotype analysis are applied in practice. More comprehensive characterization of CTC-based genomes, transcriptomes, and proteomes from high-throughput sequencing projects will further benefit clinical applications but also increase the complexity and difficulty of data analysis.
The survival of CTCs in the peripheral blood is a complex process involving multiple factors and mechanisms. It has been reported that hypoxia, autophagy, and secretion of exosomes may affect the prognosis of CTCs. Whether CTCs can be differentiated based on their phenotypes and karyotypes or not and how to formulate individualized treatment regimens for CTCs to resist apoptosis-induced drug resistance are urgent problems that need to be solved. Given the increasing popularity of molecular diagnosis in clinical practice and continuing decline in the cost of the technology, the detection of CTCs will become a powerful and indispensable tool for the diagnosis of circulating tumor cells (tumor DNA) with its advantages of repeatability, mutation detection at the molecular level, noninvasive diagnosis and broad application potential in targeted therapy, efficacy testing, postoperative prognosis, radiotherapy and chemotherapy strategy guidance, as well as in differential diagnosis.
Acknowledgments
The work was supported by the National Natural Science Foundation of China (Grant Nos. 82172335, 81971994, and 91846103), Zhejiang Provincial Key Research and Development Program (Grant No. 2020C03032), and Technology Project of Zhejiang Provincial Health Commission (Grant No. 2022KY155).
Contributor Information
Dawei Cui, Email: daweicui@zju.edu.cn.
Jue Xie, Email: zyyyxj2011@zju.edu.cn.
Data Availability
The data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
JX and DC conceived the research topic. SW and SZ conducted literature review, drafted the manuscript, and designed the figures and tables. JX, DC, and SW revised the manuscript. All the authors approved the submitted version. Siwen Wu and Shubi Zhao contributed equally to this work.
References
- 1.Hofman V. J., Ilie M. I., Bonnetaud C., et al. Cytopathologic detection of circulating tumor cells using the isolation by size of epithelial tumor cell method. American Journal Of Clinical Pathology . 2011;135(1):146–156. doi: 10.1309/ajcp9x8ozbeiqvvi. [DOI] [PubMed] [Google Scholar]
- 2.Vona G., Sabile A., Louha M., et al. Isolation by size of epithelial tumor cells: a new method for the immunomorphological and molecular characterization of circulatingtumor cells. The American Journal of Pathology . 2000;156(1):57–63. doi: 10.1016/s0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J. F., Ho H., Lichterman J., et al. Subclassification of prostate cancer circulating tumor cells by nuclear size reveals very small nuclear circulating tumor cells in patients with visceral metastases. Cancer . 2015;121(18):3240–3251. doi: 10.1002/cncr.29455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein C. A. The Metastasis Cascade. Science . 2008;321(5897):1785–1787. doi: 10.1126/science.1164853. [DOI] [PubMed] [Google Scholar]
- 5.Mazard T., Cayrefourcq L., Perriard F., et al. Clinical relevance of viable circulating tumor cells in patients with metastatic colorectal cancer: the COLOSPOT prospective study. Cancers (Basel) . 2021;13(12):p. 2966. doi: 10.3390/cancers13122966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klotz R., Thomas A., Teng T., et al. Circulating tumor cells exhibit metastatic tropism and reveal brain metastasis drivers. Cancer Discovery . 2020;10(1):86–103. doi: 10.1158/2159-8290.cd-19-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majidpoor J., Mortezaee K. Steps in metastasis: an updated review. Medical Oncology . 2021;38(1):p. 3. doi: 10.1007/s12032-020-01447-w. [DOI] [PubMed] [Google Scholar]
- 8.Pan R., Yu C., Shao Y., et al. Identification of key genes and pathways involved in circulating tumor cells in colorectal cancer. Analytical Cellular Pathology . 2022;2022:11. doi: 10.1155/2022/9943571.9943571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egeblad M., Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nature Reviews Cancer . 2002;2(3):161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 10.Hazra S., Jayaprakash K. S., Pandian K., Raj A., Mitra S. K., Sen A. K. Non-inertial lift induced migration for label-free sorting of cells in a co-flowing aqueous two-phase system. Analyst . 2019;144(8):2574–2583. doi: 10.1039/c8an02267d. [DOI] [PubMed] [Google Scholar]
- 11.Ignatiadis M., Sledge G. W., Jeffrey S. S. Liquid biopsy enters the clinic—implementation issues and future challenges. Nature Reviews Clinical Oncology . 2021;18(5):297–312. doi: 10.1038/s41571-020-00457-x. [DOI] [PubMed] [Google Scholar]
- 12.Keller L., Pantel K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nature Reviews Cancer . 2019;19(10):553–567. doi: 10.1038/s41568-019-0180-2. [DOI] [PubMed] [Google Scholar]
- 13.Martin O. A., Anderson R. L., Narayan K., MacManus M. P. Does the mobilization of circulating tumour cells during cancer therapy cause metastasis? Nature Reviews Clinical Oncology . 2017;14(1):32–44. doi: 10.1038/nrclinonc.2016.128. [DOI] [PubMed] [Google Scholar]
- 14.Nanou A., Crespo M., Flohr P., De Bono J., Terstappen L. Scanning electron microscopy of circulating tumor cells and tumor-derived extracellular vesicles. Cancers (Basel) . 2018;10(11):p. 416. doi: 10.3390/cancers10110416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin J., Deng J., Wang L., Du C., Zhang W., Jiang X. Detection of circulating tumor cells by fluorescence microspheres-mediated amplification. Analytical Chemistry . 2020;92(10):6968–6976. doi: 10.1021/acs.analchem.9b05844. [DOI] [PubMed] [Google Scholar]
- 16.Zavridou M., Mastoraki S., Strati A., et al. Direct comparison of size-dependent versus EpCAM-dependent CTC enrichment at the gene expression and DNA methylation level in head and neck squamous cell carcinoma. Scientific Reports . 2020;10(1):p. 6551. doi: 10.1038/s41598-020-63055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kallergi G., Papadaki M. A., Politaki E., Mavroudis D., Georgoulias V., Agelaki S. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Research . 2011;13(3):p. R59. doi: 10.1186/bcr2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Q., Wang F. B., Yuan C. H., et al. Gelatin nanoparticle-coated silicon beads for density-selective capture and release of heterogeneous circulating tumor cells with high purity. Theranostics . 2018;8(6):1624–1635. doi: 10.7150/thno.23531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marrinucci D., Bethel K., Bruce R. H., et al. Case study of the morphologic variation of circulating tumor cells. Human Pathology . 2007;38(3):514–519. doi: 10.1016/j.humpath.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 20.Hou J. M., Krebs M. G., Lancashire L., et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. Journal of Clinical Oncology . 2012;30(5):525–532. doi: 10.1200/jco.2010.33.3716. [DOI] [PubMed] [Google Scholar]
- 21.Duda D. G., Duyverman A. M. M. J., Kohno M., et al. Malignant cells facilitate lung metastasis by bringing their own soil. Proceedings of the National Academy of Sciences of the USA . 2010;107(50):21677–21682. doi: 10.1073/pnas.1016234107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho E. H., Wendel M., Luttgen M., et al. Characterization of circulating tumor cell aggregates identified in patients with epithelial tumors. Physical Biology . 2012;9(1) doi: 10.1088/1478-3975/9/1/016001.016001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao J., Pohlmann P. R., Isaacs C., et al. Circulating tumor cells: technologies and their clinical potential in cancer metastasis. Biomedicines . 2021;9(9):p. 1111. doi: 10.3390/biomedicines9091111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aceto N., Bardia A., Miyamoto D. T., et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell . 2014;158(5):1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reduzzi C., Di Cosimo S., Gerratana L., et al. Circulating tumor cell clusters are frequently detected in women with early-stage breast cancer. Cancers (Basel) . 2021;13(10):p. 2356. doi: 10.3390/cancers13102356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theil G., Lindner C., Bialek J., Fornara P. Association of circulating tumor cells with inflammatory and biomarkers in the blood of patients with metastatic castration-resistant prostate cancer. Life (Basel) . 2021;11(7):p. 664. doi: 10.3390/life11070664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Driemel C., Kremling H., Schumacher S., et al. Context-dependent adaption of EpCAM expression in early systemic esophageal cancer. Oncogene . 2014;33(41):4904–4915. doi: 10.1038/onc.2013.441. [DOI] [PubMed] [Google Scholar]
- 28.Flores L. M., Kindelberger D. W., Ligon A. H., et al. Improving the yield of circulating tumour cells facilitates molecular characterisation and recognition of discordant HER2 amplification in breast cancer. British Journal of Cancer . 2010;102(10):1495–1502. doi: 10.1038/sj.bjc.6605676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song J., Yu Z., Dong B., et al. Clinical significance of circulating tumour cells and Ki-67 in renal cell carcinoma. World Journal of Surgical Oncology . 2021;19(1):p. 156. doi: 10.1186/s12957-021-02268-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Werner S., Heidrich I., Pantel K. Clinical management and biology of tumor dormancy in breast cancer. Seminars in Cancer Biology . 2022;78:49–62. doi: 10.1016/j.semcancer.2021.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Dongre A., Rashidian M., Eaton E. N., et al. Direct and indirect regulators of epithelial–mesenchymal transition–mediated immunosuppression in breast carcinomas. Cancer Discovery . 2021;11(5):1286–1305. doi: 10.1158/2159-8290.cd-20-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thiery J. P., Lim C. T. Tumor Dissemination: An EMT Affair. Cancer Cell . 2013;23(3):272–273. doi: 10.1016/j.ccr.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Barrière G., Tartary M., Rigaud M. Epithelial mesenchymal transition: a new insight into the detection of circulating tumor cells. International Scholarly Research Notices . 2012;2012:6. doi: 10.5402/2012/382010.382010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheng G. Defining epithelial-mesenchymal transitions in animal development. Development . 2021;148(8) doi: 10.1242/dev.198036.dev198036 [DOI] [PubMed] [Google Scholar]
- 35.Lin D., Shen L., Luo M., et al. Circulating tumor cells: biology and clinical significance. Signal Transduction and Targeted Therapy . 2021;6(1):p. 404. doi: 10.1038/s41392-021-00817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agnoletto C., Corrà F., Minotti L., et al. Heterogeneity in circulating tumor cells: the relevance of the stem-cell subset. Cancers (Basel) . 2019;11(4):p. 483. doi: 10.3390/cancers11040483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li W., Kang Y. Probing the fifty shades of EMT in metastasis. Trends in Cancer . 2016;2(2):65–67. doi: 10.1016/j.trecan.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao N., Powell R. T., Yuan X., et al. Morphological screening of mesenchymal mammary tumor organoids to identify drugs that reverse epithelial-mesenchymal transition. Nature Communications . 2021;12(1):p. 4262. doi: 10.1038/s41467-021-24545-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J., Antin P., Berx G., et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nature Reviews Molecular Cell Biology . 2020;21(6):341–352. doi: 10.1038/s41580-020-0237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang T., Zhang L., Gao Y., et al. Role of aneuploid circulating tumor cells and CD31+ circulating tumor endothelial cells in predicting and monitoring anti-angiogenic therapy efficacy in advanced NSCLC. Molecular Oncology . 2021;15(11):2891–2909. doi: 10.1002/1878-0261.13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu X., Li J., Cadilha B. L., et al. Epithelial-type systemic breast carcinoma cells with a restricted mesenchymal transition are a major source of metastasis. Science Advances . 2019;5(6) doi: 10.1126/sciadv.aav4275.eaav4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arya S. K., Lim B., Rahman A. R. A. Enrichment, detection and clinical significance of circulating tumor cells. Lab on a Chip . 2013;13(11):1995–2027. doi: 10.1039/c3lc00009e. [DOI] [PubMed] [Google Scholar]
- 43.Li X., Li Y., Shao W., Li Z., Zhao R., Ye Z. Strategies for enrichment of circulating tumor cells. Translational Cancer Research . 2020;9(3):2012–2025. doi: 10.21037/tcr.2020.01.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H. Y., Koch C., Haller A., et al. Evaluation of microfluidic ceiling designs for the capture of circulating tumor cells on a microarray platform. Advanced Biosystems . 2020;4(2) doi: 10.1002/adbi.201900162.e1900162 [DOI] [PubMed] [Google Scholar]
- 45.Nam K. H., Yong W., Harvat T., et al. Size-based separation and collection of mouse pancreatic islets for functional analysis. Biomed Microdevices . 2010;12(5):865–874. doi: 10.1007/s10544-010-9441-2. [DOI] [PubMed] [Google Scholar]
- 46.Hu C. L., Zhang Y. J., Zhang X. F., et al. 3D culture of circulating tumor cells for evaluating early recurrence and metastasis in patients with hepatocellular carcinoma. OncoTargets and Therapy . 2021;14:2673–2688. doi: 10.2147/ott.s298427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sundling K. E., Lowe A. C. Circulating tumor cells: overview and opportunities in cytology. Advances in Anatomic Pathology . 2019;26(1):56–63. doi: 10.1097/pap.0000000000000217. [DOI] [PubMed] [Google Scholar]
- 48.Wang S., Liu K., Liu J., et al. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angewandte Chemie International Edition . 2011;50(13):3084–3088. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu J., Raba K., Guglielmi R., et al. Magnetic-based enrichment of rare cells from high concentrated blood samples. Cancers . 2020;12(4):p. 933. doi: 10.3390/cancers12040933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bamrungsap S., Chen T., Shukoor M. I., et al. Pattern recognition of cancer cells using aptamer-conjugated magnetic nanoparticles. ACS Nano . 2012;6(5):3974–3981. doi: 10.1021/nn3002328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu X. L., Zhu M. L., Liu D., et al. Highly integrated nanoplatform based on an e-selectin-targeting strategy for metastatic breast cancer treatment. Molecular Pharmaceutics . 2019;16(8):3694–3702. doi: 10.1021/acs.molpharmaceut.9b00616. [DOI] [PubMed] [Google Scholar]
- 52.Jyotsana N., Zhang Z., Himmel L. E., Yu F., King M. R. Minimal dosing of leukocyte targeting TRAIL decreases triple-negative breast cancer metastasis following tumor resection. Science Advances . 2019;5(7) doi: 10.1126/sciadv.aaw4197.eaaw4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenberg R., Gertler R., Friederichs J., et al. Comparison of two density gradient centrifugation systems for the enrichment of disseminated tumor cells in blood. Cytometry . 2002;49(4):150–158. doi: 10.1002/cyto.10161. [DOI] [PubMed] [Google Scholar]
- 54.Hu B., Gong Y., Wang Y., Xie J., Cheng J., Huang Q. Comprehensive atlas of circulating rare cells detected by se-ifish and image scanning platform in patients with various diseases. Frontiers in Oncology . 2022;12 doi: 10.3389/fonc.2022.821454.821454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boral D., Marchetti D. Liquid biopsy in prostate cancer: a case for comprehensive genomic characterization of circulating tumor cells. Clinical Chemistry . 2018;64(2):251–253. doi: 10.1373/clinchem.2017.283440. [DOI] [PubMed] [Google Scholar]
- 56.Monterisi S., Castello A., Toschi L., et al. Preliminary data on circulating tumor cells in metastatic NSCLC patients candidate to immunotherapy. American Journal of Nuclear Medicine and Molecular Imaging . 2019;9(6):282–295. [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao Y., Zhao S., Chen Y., et al. Isolation of circulating tumor cells in patients undergoing surgery for esophageal cancer and a specific confirmation method. Oncology Letters . 2019;17(4):3817–3825. doi: 10.3892/ol.2019.10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mu Z., Benali-Furet N., Uzan G., et al. Abstract P2-02-14: detection and characterization of CTCs isolated by screencell®-filtration in metastatic breast cancer. Cancer Research . 2016;76(4):P2–14. doi: 10.1158/1538-7445.sabcs15-p2-02-14. [DOI] [Google Scholar]
- 59.Birkhahn M., Mitra A. P., Williams A. J., et al. A novel precision-engineered microfiltration device for capture and characterisation of bladder cancer cells in urine. European Journal of Cancer . 2013;49(15):3159–3168. doi: 10.1016/j.ejca.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hosokawa M., Yoshikawa T., Negishi R., et al. Microcavity array system for size-based enrichment of circulating tumor cells from the blood of patients with small-cell lung cancer. Analytical Chemistry . 2013;85(12):5692–5698. doi: 10.1021/ac400167x. [DOI] [PubMed] [Google Scholar]
- 61.Coumans F. A. W., van Dalum G., Beck M., Terstappen L. W. M. M. Filter characteristics influencing circulating tumor cell enrichment from whole blood. PLoS One . 2013;8(4) doi: 10.1371/journal.pone.0061770.e61770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baharfar M., Yamini Y., Seidi S., Karami M. Quantitative analysis of clonidine and ephedrine by a microfluidic system: on-chip electromembrane extraction followed by high performance liquid chromatography. Journal of Chromatography B . 2017;1068-1069:313–321. doi: 10.1016/j.jchromb.2017.10.062. [DOI] [PubMed] [Google Scholar]
- 63.Yao Q. Q., Hu J., Zheng P. F., et al. In vitroevaluation of marrow clot enrichment on microstructure decoration, cell delivery and proliferation of porous titanium scaffolds by selective laser melting three-dimensional printing. Journal of Biomedical Materials Research Part B: Applied Biomaterials . 2018;106(6):2245–2253. doi: 10.1002/jbm.b.34032. [DOI] [PubMed] [Google Scholar]
- 64.Niciński K., Krajczewski J., Kudelski A., et al. Detection of circulating tumor cells in blood by shell-isolated nanoparticle—enhanced Raman spectroscopy (SHINERS) in microfluidic device. Scientific Reports . 2019;9(1):p. 9267. doi: 10.1038/s41598-019-45629-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohnaga T., Shimada Y., Takata K., et al. Capture of esophageal and breast cancer cells with polymeric microfluidic devices for CTC isolation. Molecular and Clinical Oncology . 2016;4(4):599–602. doi: 10.3892/mco.2016.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeinali M., Murlidhar V., Fouladdel S., et al. Abstract 1595: analysis of circulating epithelial and EMT-like CTCs in pancreatic cancer using a sensitive microfluidic CTC capture device. Cancer Research . 2015;75(15):p. 1595. doi: 10.1158/1538-7445.am2015-1595. [DOI] [Google Scholar]
- 67.Attard G., de Bono J. S. Utilizing circulating tumor cells: challenges and pitfalls. Current Opinion in Genetics & Development . 2011;21(1):50–58. doi: 10.1016/j.gde.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 68.Ito H., Inoue H., Kimura S., et al. Prognostic impact of the number of viable circulating cells with high telomerase activity in gastric cancer patients: a prospective study. International Journal of Oncology . 2014;45(1):227–234. doi: 10.3892/ijo.2014.2409. [DOI] [PubMed] [Google Scholar]
- 69.Wang L., Li Y., Xu J., et al. Quantified postsurgical small cell size CTCs and EpCAM+ circulating tumor stem cells with cytogenetic abnormalities in hepatocellular carcinoma patients determine cancer relapse. Cancer Letters . 2018;412:99–107. doi: 10.1016/j.canlet.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 70.Yamato M., Kimura T. Magnetic processing of diamagnetic materials. Polymers (Basel) . 2020;12(7):p. 1491. doi: 10.3390/polym12071491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pamme N. On-chip bioanalysis with magnetic particles. Current Opinion in Chemical Biology . 2012;16(3-4):436–443. doi: 10.1016/j.cbpa.2012.05.181. [DOI] [PubMed] [Google Scholar]
- 72.Salvianti F., Gelmini S., Mancini I., et al. Circulating tumour cells and cell-free DNA as a prognostic factor in metastatic colorectal cancer: the OMITERC prospective study. British Journal of Cancer . 2021;125(1):94–100. doi: 10.1038/s41416-021-01399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scher H. I., Armstrong A. J., Schonhoft J. D., et al. Development and validation of circulating tumour cell enumeration (Epic Sciences) as a prognostic biomarker in men with metastatic castration-resistant prostate cancer. European Journal of Cancer . 2021;150:83–94. doi: 10.1016/j.ejca.2021.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kang B. J., Ra S. W., Lee K., et al. Circulating tumor cell number is associated with primary tumor volume in patients with lung adenocarcinoma. Tuberculosis and Respiratory Diseases . 2020;83(1):61–70. doi: 10.4046/trd.2019.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu X., Bai Z., Wang L., et al. Magnetic cell centrifuge platform performance study with different microsieve pore geometries. Sensors (Basel) . 2019;20(1):p. 48. doi: 10.3390/s20010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang Y. Y., Chen P., Wu C. H., et al. Screening and molecular analysis of single circulating tumor cells using micromagnet array. Scientific Reports . 2015;5(1) doi: 10.1038/srep16047.16047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cierna Z., Mego M., Janega P., et al. Matrix metalloproteinase 1 and circulating tumor cells in early breast cancer. BMC Cancer . 2014;14:p. 472. doi: 10.1186/1471-2407-14-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y., Wang W., Guo H., Liu M., Zhu H., Sun H. Hyaluronic acid-functionalized redox responsive immunomagnetic nanocarrier for circulating tumor cell capture and release. Nanotechnology . 2021;32(47) doi: 10.1088/1361-6528/abdf8c.475102 [DOI] [PubMed] [Google Scholar]
- 79.Manz A., Graber N., Widmer H. Miniaturized total chemical analysis systems: A novel concept for chemical sensing. Sensors and Actuators B: Chemical . 1990;1(1-6):244–248. doi: 10.1016/0925-4005(90)80209-i. [DOI] [Google Scholar]
- 80.Kim D. D., Yang C. S., Chae H. D., Kwak S. G., Jeon C. H. Melanoma antigen-encoding gene family member A1-6 and hTERT in the detection of circulating tumor cells following CD45− depletion and RNA extraction. Oncology Letters . 2017;14(1):837–843. doi: 10.3892/ol.2017.6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kotz K. T., Xiao W., Miller-Graziano C., et al. Clinical microfluidics for neutrophil genomics and proteomics. Nature Medicine . 2010;16(9):1042–1047. doi: 10.1038/nm.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sequist L. V., Nagrath S., Toner M., Haber D. A., Lynch T. J. The CTC-chip: an exciting new tool to detect circulating tumor cells in lung cancer patients. Journal of Thoracic Oncology . 2009;4(3):281–283. doi: 10.1097/jto.0b013e3181989565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hughes A. D., Mattison J., Powderly J. D., Greene B. T., King M. R. Rapid isolation of viable circulating tumor cells from patient blood samples. Journal of Visualized Experiments . 2012;64 doi: 10.3791/4248.e4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hughes A. D., King M. R. Use of naturally occurring halloysite nanotubes for enhanced capture of flowing cells. Langmuir . 2010;26(14):12155–12164. doi: 10.1021/la101179y. [DOI] [PubMed] [Google Scholar]
- 85.Hughes A. D., Mattison J., Western L. T., Powderly J. D., Greene B. T., King M. R. Microtube device for selectin-mediated capture of viable circulating tumor cells from blood. Clinical Chemistry . 2012;58(5):846–853. doi: 10.1373/clinchem.2011.176669. [DOI] [PubMed] [Google Scholar]
- 86.Stott S. L., Hsu C. H., Tsukrov D. I., et al. Isolation of circulating tumor cells using a microvortex-generatingherringbone-chip. Proceedings of the National Academy of Sciences of the USA . 2010;107(43):18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sheng W., Ogunwobi O. O., Chen T., et al. Capture, release and culture of circulating tumor cells from pancreatic cancer patients using an enhanced mixing chip. Lab Chip . 2014;14(1):89–98. doi: 10.1039/c3lc51017d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Poudineh M., Aldridge P. M., Ahmed S., et al. Tracking the dynamics of circulating tumour cell phenotypes using nanoparticle-mediated magnetic ranking. Nature Nanotechnology . 2017;12(3):274–281. doi: 10.1038/nnano.2016.239. [DOI] [PubMed] [Google Scholar]
- 89.Lee T. Y., Hyun K. A., Kim S. I., Jung H. I. An integrated microfluidic chip for one-step isolation of circulating tumor cells. Sensors and Actuators B: Chemical . 2017;238(jan):1144–1150. doi: 10.1016/j.snb.2016.05.163. [DOI] [Google Scholar]
- 90.Sajay B. N. G., Chang C. P., Ahmad H., et al. Microfluidic platform for negative enrichment of circulating tumor cells. Biomedical Microdevices . 2014;16(4):537–548. doi: 10.1007/s10544-014-9856-2. [DOI] [PubMed] [Google Scholar]
- 91.Pereira-Veiga T., Martínez-Fernández M., Abuin C., et al. CTCs expression profiling for advanced breast cancer monitoring. Cancers (Basel) . 2019;11(12):p. 1941. doi: 10.3390/cancers11121941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang X., Lv J., Zhou Z., et al. Clinical application of circulating tumor cells and circulating endothelial cells in predicting bladder cancer prognosis and neoadjuvant chemosensitivity. Frontiers in Oncology . 2021;11 doi: 10.3389/fonc.2021.802188.802188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li Z., Fan L., Wu Y., et al. Analysis of the prognostic role and biological characteristics of circulating tumor cell-associated white blood cell clusters in non-small cell lung cancer. Journal of Thoracic Disease . 2022;14(5):1544–1555. doi: 10.21037/jtd-22-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ikeda M., Koh Y., Teraoka S., et al. Detection of AXL expression in circulating tumor cells of lung cancer patients using an automated microcavity array system. Cancer Medicine . 2020;9(6):2122–2133. doi: 10.1002/cam4.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smolkova B., Mego M., Horvathova Kajabova V., et al. Expression of SOCS1 and CXCL12 proteins in primary breast cancer are associated with presence of circulating tumor cells in peripheral blood. Translational Oncology . 2016;9(3):184–190. doi: 10.1016/j.tranon.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hapach L. A., Carey S. P., Schwager S. C., et al. Phenotypic heterogeneity and metastasis of breast cancer cells. Cancer Research . 2021;81(13):3649–3663. doi: 10.1158/0008-5472.can-20-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lisyaniy N. I., Stanetskaya D. N., Lisyaniy A. N., Belskaya L. N. Content of stem tumor CD133 (+) cells in brain neoplasms of different histological type. Experimental Oncology . 2017;39(3):219–223. doi: 10.31768/2312-8852.2017.39(3):219-223. [DOI] [PubMed] [Google Scholar]
- 98.Ahn K. S., Hwang J. Y., Han H. S., Kim S. T., Hwang I., Chun Y. O. The impact of acute inflammation on progression and metastasis in pancreatic cancer animal model. Surgical Oncology . 2018;27(1):61–69. doi: 10.1016/j.suronc.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 99.Savelieva O. E., Tashireva L. A., Kaigorodova E. V., et al. Heterogeneity of stemlike circulating tumor cells in invasive breast cancer. International Journal of Molecular Sciences . 2020;21(8):p. 2780. doi: 10.3390/ijms21082780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sun Y. B., Sun G. H., Xu S., Xu J. J. Tumor-suppressive activity of sTRAIL on circulating CD44+ cells in patients with non-small cell lung cancer. Biological Chemistry . 2020;401(3):417–422. doi: 10.1515/hsz-2019-0339. [DOI] [PubMed] [Google Scholar]
- 101.Alotaibi A. A. A., Najafzadeh M., Davies J. D., Baumgartner A., Anderson D. Inhibition of survivin expression after using oxaliplatin and vinflunine to induce cytogenetic damage in vitro in lymphocytes from colon cancer patients and healthy individuals. Mutagenesis . 2017;32(5):517–524. doi: 10.1093/mutage/gex022. [DOI] [PubMed] [Google Scholar]
- 102.Chen H., Fan X., Zhao Y., et al. Stimuli-responsive polysaccharide enveloped liposome for targeting and penetrating delivery of survivin-shRNA into breast tumor. ACS Applied Materials & Interfaces . 2020;12(19):22074–22087. doi: 10.1021/acsami.9b22440. [DOI] [PubMed] [Google Scholar]
- 103.Kwan T. T., Bardia A., Spring L. M., et al. A digital RNA signature of circulating tumor cells predicting early therapeutic response in localized and metastatic breast cancer. Cancer Discovery . 2018;8(10):1286–1299. doi: 10.1158/2159-8290.cd-18-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Leehy K. A., Truong T. H., Mauro L. J., Lange C. A. Progesterone receptors (PR) mediate STAT actions: PR and prolactin receptor signaling crosstalk in breast cancer models. The Journal of Steroid Biochemistry and Molecular Biology . 2018;176:88–93. doi: 10.1016/j.jsbmb.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li Y., Zhang X., Ge S., et al. Clinical significance of phenotyping and karyotyping of circulating tumor cells in patients with advanced gastric cancer. Oncotarget . 2014;5(16):6594–6602. doi: 10.18632/oncotarget.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li Y., Zhang X., Liu D., et al. Evolutionary expression of HER2 conferred by chromosome aneuploidy on circulating gastric cancer cells contributes to developing targeted and chemotherapeutic resistance. Clinical Cancer Research . 2018;24(21):5261–5271. doi: 10.1158/1078-0432.ccr-18-1205. [DOI] [PubMed] [Google Scholar]
- 107.Zhang L., Zhang X., Liu Y., et al. PD-L1+ aneuploid circulating tumor endothelial cells (CTECs) exhibit resistance to the checkpoint blockade immunotherapy in advanced NSCLC patients. Cancer Letters . 2020;469:355–366. doi: 10.1016/j.canlet.2019.10.041. [DOI] [PubMed] [Google Scholar]
- 108.Xing C., Li Y., Ding C., et al. CD44+ circulating tumor endothelial cells indicate poor prognosis in pancreatic ductal adenocarcinoma after radical surgery: a pilot study. Cancer Management and Research . 2021;13:4417–4431. doi: 10.2147/cmar.s309115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Baek Y. H., Um J., Antigua K. J. C., et al. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerging Microbes & Infections . 2020;9(1):998–1007. doi: 10.1080/22221751.2020.1756698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chung H. Y., Jian M. J., Chang C. K., et al. Novel dual multiplex real-time RT-PCR assays for the rapid detection of SARS-CoV-2, influenza A/B, and respiratory syncytial virus using the BD MAX open system. Emerging Microbes & Infections . 2021;10(1):161–166. doi: 10.1080/22221751.2021.1873073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Buglyó G., Magyar Z., Romicsné Görbe É., et al. miRNA profiling of hungarian regressive wilms’ tumor formalin-fixedparaffin-embedded (FFPE) samples by quantitative real-time polymerase chain reaction (RT-PCR) Med Sci Monit . 2021;27 doi: 10.12659/msm.932731.e932731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kuang Y., Xu P., Wang J., et al. Detecting ALK rearrangement with RT-PCR: a reliable approach compared with next-generation sequencing in patients with NSCLC. Molecular Diagnosis and Therapy . 2021;25(4):487–494. doi: 10.1007/s40291-021-00532-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chou H. C., Sheu J. C., Huang G. T., Wang J. T., Chen D. S. Albumin messenger RNA is not specific for circulating hepatoma cells. Gastroenterology . 1994;107(2):630–631. doi: 10.1016/0016-5085(94)90205-4. [DOI] [PubMed] [Google Scholar]
- 114.Wang S., Yang S., Nan C., Wang Y., He Y., Mu H. Expression of androgen receptor variant 7 (AR-V7) in circulated tumor cells and correlation with drug resistance of prostate cancer cells. Medical Science Monitor . 2018;24:7051–7056. doi: 10.12659/msm.909669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wei X. Q., Ma Y., Chen Y., Liu X., Zhao M., Zhou L. W. Laparoscopic surgery for early cervical squamous cell carcinoma and its effect on the micrometastasis of cancer cells. Medicine (Baltimore) . 2018;97(34) doi: 10.1097/md.0000000000011921.e11921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shen C., Hu L., Xia L., Li Y. The detection of circulating tumor cells of breast cancer patients by using multimarker (Survivin, hTERT and hMAM) quantitative real-time PCR. Clinical Biochemistry . 2009;42(3):194–200. doi: 10.1016/j.clinbiochem.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 117.Gervasoni A., Monasterio Muñoz R. M., Wengler G. S., Rizzi A., Zaniboni A., Parolini O. Molecular signature detection of circulating tumor cells using a panel of selected genes. Cancer Letters . 2008;263(2):267–279. doi: 10.1016/j.canlet.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 118.Jiang M., Jin S., Han J., et al. Detection and clinical significance of circulating tumor cells in colorectal cancer. Biomarker Research . 2021;9(1):p. 85. doi: 10.1186/s40364-021-00326-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Paoletti C., Miao J., Dolce E. M., et al. Circulating tumor cell clusters in patients with metastatic breast cancer: a SWOG S0500 translational medicine study. Clinical Cancer Research . 2019;25(20):6089–6097. doi: 10.1158/1078-0432.ccr-19-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sperger J. M., Emamekhoo H., McKay R. R., et al. Prospective evaluation of clinical outcomes using a multiplex liquid biopsy targeting diverse resistance mechanisms in metastatic prostate cancer. Journal of Clinical Oncology . 2021;39(26):2926–2937. doi: 10.1200/jco.21.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hugenschmidt H., Labori K. J., Brunborg C., et al. Circulating tumor cells are an independent predictor of shorter survival in patients undergoing resection for pancreatic and periampullary adenocarcinoma. Annals of Surgery . 2020;271(3):549–558. doi: 10.1097/sla.0000000000003035. [DOI] [PubMed] [Google Scholar]
- 122.Bidard F. C., Kiavue N., Ychou M., et al. Circulating tumor cells and circulating tumor DNA detection in potentially resectable metastatic colorectal cancer: a prospective ancillary study to the unicancer prodige-14 trial. Cells . 2019;8(6):p. 516. doi: 10.3390/cells8060516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sastre J., Maestro M. L., Puente J., et al. Circulating tumor cells in colorectal cancer: correlation with clinical and pathological variables. Annals of Oncology . 2008;19(5):935–938. doi: 10.1093/annonc/mdm583. [DOI] [PubMed] [Google Scholar]
- 124.Yang M., Rehman A. U., Zuo C., et al. A novel histologic grading scheme based on poorly differentiated clusters is applicable to treated rectal cancer and is associated with established histopathological prognosticators. Cancer Medicine . 2016;5(7):1510–1518. doi: 10.1002/cam4.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hou J. M., Greystoke A., Lancashire L., et al. Evaluation of circulating tumor cells and serological cell death biomarkers in small cell lung cancer patients undergoing chemotherapy. The American Journal of Pathology . 2009;175(2):808–816. doi: 10.2353/ajpath.2009.090078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Qi L. N., Xiang B. D., Wu F. X., et al. Circulating tumor cells undergoing emt provide a metric for diagnosis and prognosis of patients with hepatocellular carcinoma. Cancer Research . 2018;78(16):4731–4744. doi: 10.1158/0008-5472.can-17-2459. [DOI] [PubMed] [Google Scholar]
- 127.Wang S., Zhang C., Wang G., et al. Aptamer-mediatedtransparent-biocompatible nanostructured surfaces for hepatocellular circulating tumor cells enrichment. Theranostics . 2016;6(11):1877–1886. doi: 10.7150/thno.15284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hofste Op Bruinink D., Kuiper R., van Duin M., et al. Identification of high-risk multiple myeloma with a plasma cell leukemia-like transcriptomic profile. Journal of Clinical Oncology . 2022 doi: 10.1200/JCO.21.01217.JCO2101217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang W., Bao L., Yang S., et al. Tumor-selective replication herpes simplex virus-based technology significantly improves clinical detection and prognostication of viable circulating tumor cells. Oncotarget . 2016;7(26):39768–39783. doi: 10.18632/oncotarget.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lin M., Liang S. Z., Shi J., et al. Circulating tumor cell as a biomarker for evaluating allogenic NK cell immunotherapy on stage IV non-small cell lung cancer. Immunology Letters . 2017;191:10–15. doi: 10.1016/j.imlet.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 131.Nagrath S., Sequist L. V., Maheswaran S., et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature . 2007;450(7173):1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Maheswaran S., Sequist L. V., Nagrath S., et al. Detection of mutations inEGFRin circulating lung-cancer cells. The New England Journal of Medicine . 2008;359(4):366–377. doi: 10.1056/nejmoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Stott S. L., Lee R. J., Nagrath S., et al. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Science Translational Medicine . 2010;2(25) doi: 10.1126/scitranslmed.3000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yu M., Bardia A., Wittner B. S., et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science . 2013;339(6119):580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Beije N., Onstenk W., Kraan J., et al. Prognostic impact of HER2 and ER status of circulating tumor cells in metastatic breast cancer patients with a HER2-Negative primary tumor. Neoplasia . 2016;18(11):647–653. doi: 10.1016/j.neo.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Somlo G., Lau S. K., Frankel P., et al. Multiple biomarker expression on circulating tumor cells in comparison to tumor tissues from primary and metastatic sites in patients with locally advanced/inflammatory, and stage IV breast cancer, using a novel detection technology. Breast Cancer Research and Treatment . 2011;128(1):155–163. doi: 10.1007/s10549-011-1508-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Garrido-Navas M. C., García-Díaz A., Molina-Vallejo M. P., et al. The polemic diagnostic role of TP53 mutations in liquid biopsies from breast, colon and lung cancers. Cancers (Basel) . 2020;12(11):p. 3343. doi: 10.3390/cancers12113343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhou Y., Wang B., Wu J., et al. Association of preoperative EpCAM circulating tumor cells and peripheral Treg cell levels with early recurrence of hepatocellular carcinoma following radical hepatic resection. BMC Cancer . 2016;16(1):p. 506. doi: 10.1186/s12885-016-2526-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pantel K., Alix-Panabières C. Liquid biopsy and minimal residual disease—latest advances and implications for cure. Nature Reviews Clinical Oncology . 2019;16(7):409–424. doi: 10.1038/s41571-019-0187-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are included within the article.


