Abstract
Bacteria orchestrate collective behaviors and accomplish feats that would be unsuccessful if carried out by a lone bacterium. Processes undertaken by groups of bacteria include bioluminescence, biofilm formation, virulence factor production, and release of public goods that are shared by the community. Collective behaviors are controlled by signal transduction networks that integrate sensory information and transduce the information internally. Here, we discuss network features and mechanisms that, even in the face of dramatically changing environments, drive precise execution of bacterial group behaviors. We focus on representative quorum-sensing and second-messenger cyclic dimeric GMP signal relays. We highlight ligand specificity versus sensitivity, how small molecule ligands drive discrimination of kin versus non-kin, signal integration mechanisms, single-input sensory systems versus coincidence detectors, and tuning of input-output dynamics via feedback regulation. We summarize how different features of signal transduction systems allow groups of bacteria to successfully interpret and collectively react to dynamically changing environments.
Keywords: Bacterial group behavior, signal transduction, quorum sensing, c-di-GMP, specificity, sensitivity, feedback
COORDINATION OF GROUP BEHAVIORS IN BACTERIA
Much like a cell in a eukaryotic tissue, an insect colony, or a primate troop composed of individuals, bacterial cells participate as members of coordinated groups, allowing them to overcome challenges and carry out tasks that they could not accomplish alone. Bacterial group behaviors include the formation of organized multicellular structures, the production of light on a scale observable from space, and the ability to sicken healthy plants, animals, and humans (1, 21, 53). In the context of infection, bacteria often employ group behaviors that allow them to colonize a host, launch virulence mechanisms, and overcome immune system defenses (1). Specifically, during colonization, bacteria commonly form matrix-encapsulated groups of cells called biofilms (21). Biofilm formation allows the resident bacterial cells to evade host anti-bacterial processes and clinical antibiotics, making the bacteria within these communities notoriously difficult to eradicate (67). Moreover, pathogenic bacteria commonly coordinate the production of virulence factors across the population and profoundly sicken the host (1). Finally, to elude the immune system, bacteria often also work together to suppress host inflammatory responses (66).
To coordinate their actions, bacteria are equipped with signal transduction pathways that allow them to detect and respond to one another and to changing environmental conditions. The information contained in the detected signals is integrated by cells to accurately inform them about their surroundings and enable them to appropriately modulate their output behaviors. Commonly, bacteria use quorum sensing, a cell-to-cell chemical communication process in which bacteria release and detect signal molecules called autoinducers to regulate collective behaviors (56). Autoinducer levels increase in step with bacterial cell density and when a threshold autoinducer concentration is achieved, cells in the population synchronously launch high cell density (HCD) group behaviors (56). Quorum sensing allows bacteria to track and react to both the cell density and the species composition of the vicinal community. It is increasingly appreciated that quorum-sensing circuits can be used to foster inter-domain social interactions between bacteria, eukaryotes, and viruses (15, 94). Bacteria often integrate quorum-sensing information with pathways that rely on the ubiquitous second-messenger cyclic dimeric GMP (c-di-GMP) molecule (79). For example, across the bacterial domain, low levels of c-di-GMP are associated with free-swimming, individualistic behaviors, whereas high c-di-GMP concentrations in the cytoplasm correlate with sessile multicellular community formation (27, 41).
In the current review, we highlight the features of quorum-sensing and c-di-GMP signaling pathways that allow bacteria to make fate-changing decisions in the face of complex and fluctuating environmental conditions. Rather than focusing on one signaling pathway, we emphasize features of different signaling circuits, including receptor properties, sensitivity versus specificity, coincidence detection, and feedback regulation that endow bacteria with the ability to undertake group decisions with high fidelity.
PREVALENT RECEPTOR TYPES THAT CONTROL BACTERIAL COMMUNITY BEHAVIORS
Small-molecule-binding transcription factors
Perhaps the most elementary mechanism that bacteria use to coordinate social behaviors involves the release of autoinducers that diffuse back into cells and are subsequently detected by cytoplasmic transcription factors whose activities are controlled by ligand binding (Fig. 1A). This scenario is typified by the first quorum-sensing circuit experimentally defined, involving the LuxI autoinducer synthase and the LuxR autoinducer receptor/transcription factor in the bioluminescent bacterium Vibrio fischeri (Fig. 1A) (18). Since the discovery of LuxR in V. fischeri, many LuxR family members have been studied, and in some cases, e.g., TraR from Agrobacterium tumefaciens and LasR from Pseudomonas aeruginosa, using structural approaches (6, 89, 97). In brief, LuxI-type autoinducer synthases produce species-specific acyl homoserine lactone autoinducers (AHLs) that diffuse into the extracellular environment (16). At HCD, and thus, high autoinducer concentration, cognate LuxR-type receptors bind AHLs via an N-terminal ligand-binding domain (LBD) which mediates receptor stabilization, folding, and dimerization (Fig. 1A) (86). Liganded LuxR receptors, in turn, bind to so-called “lux boxes” in promoter regions of target genes via their C-terminal helix-turn-helix DNA-binding domains (DBDs) (86). These complexes interact with RNA polymerase and activate the expression of genes that underpin group behaviors (81).
Fig. 1. Classes of receptors regulating bacterial group behaviors.
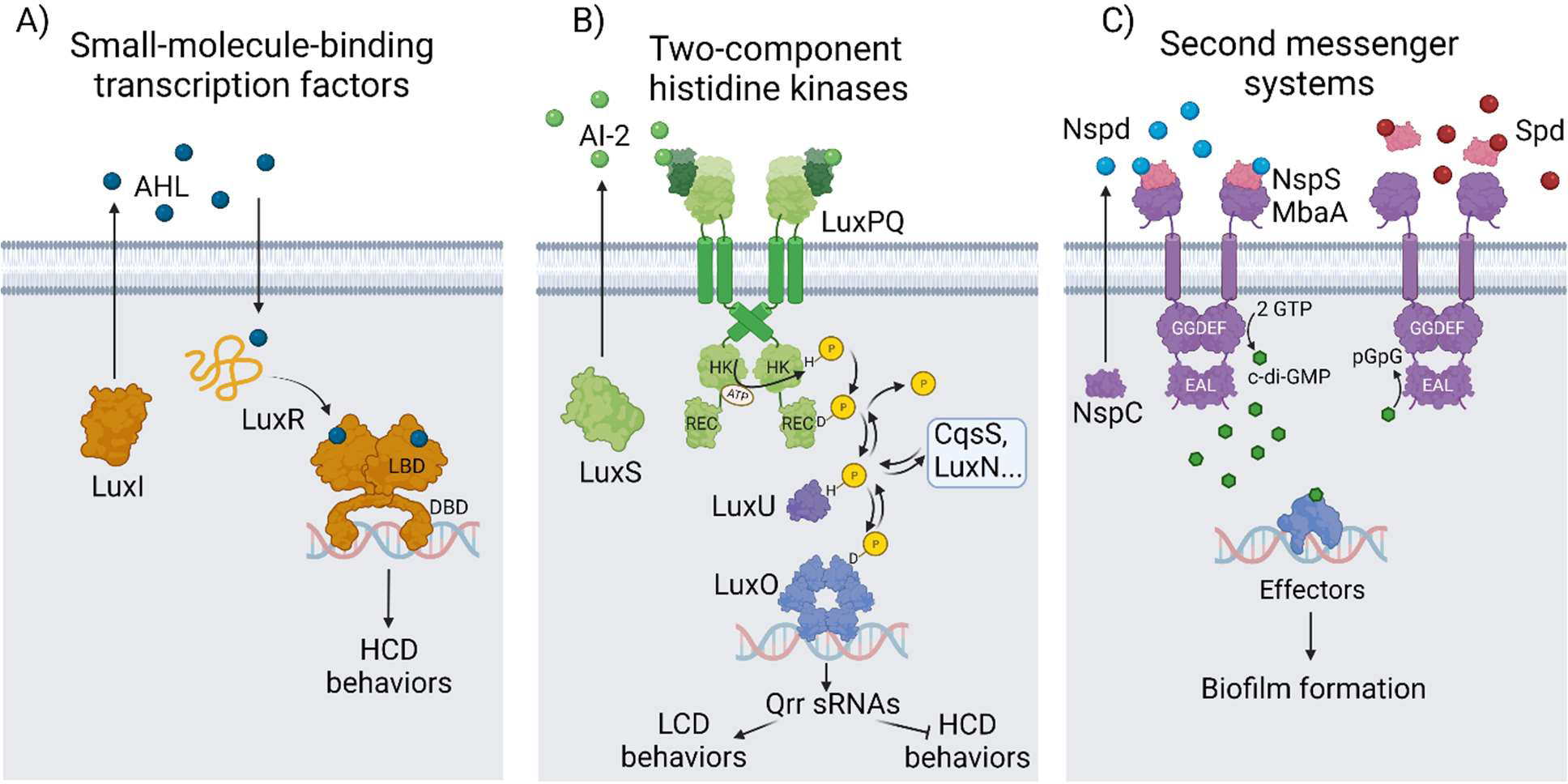
A) Small molecule binding transcription factors of the LuxR family function by detecting acyl homoserine lactones (AHLs). Liganded LuxR-type proteins control genes specifying group behaviors. LBD = ligand binding domain; DBD = DNA binding domain. B) Two-component systems function by phosphorylation/dephosphorylation cascades. The vibrio AI-2 SHK LuxPQ and its downstream signal-relay components are shown. LuxQ is a kinase at LCD and a phosphatase at HCD. HK = histidine kinase; REC = receiver domain. C) The second messenger c-di-GMP controls biofilm formation in many bacteria. The vibrio c-di-GMP regulatory polyamine NspS/MbaA receptor complex detects norspermidine (Nspd) and spermidine (Spd). When norspermidine is detected, MbaA produces c-di-GMP via its GGDEF domain. When spermidine is detected, MbaA degrades c-di-GMP via its EAL domain. MbaA-driven changes in c-di-GMP levels alter activity of downstream effectors which are transcription factors. See text for descriptions of all three types of systems. Figure created with BioRender.com.
To date, thousands of LuxR-type receptors have been identified using bioinformatic approaches across proteobacteria, suggesting that these receptors are fundamental to the ability of bacteria to obtain information regarding their cell densities (37, 82). Interestingly, as many as 75% of sequenced bacterial LuxR-type receptors exist in genomes that lack a gene specifying a cognate LuxI-type autoinducer synthase (termed “LuxR-solos”) (37). LuxR-solos are thought to detect AHLs produced by cohabitating species, for example, SdiA in enteric bacteria (52, 90). Furthermore, some LuxR-solos, which commonly exhibit low similarity to V. fischeri LuxR, particularly in their LBDs, recognize ligands that are structurally unrelated to AHLs (64). For example, in V. cholerae, the LuxR-solo VqmA binds to and is regulated by the autoinducer 3,5-dimethylpyrazin-2-ol (DPO), a non-AHL whose biosynthesis involves threonine, alanine, and the highly conserved enzyme threonine dehydrogenase (Tdh) (62). DPO controls VqmA activity in a cell-density-dependent manner and regulates group behaviors including biofilm formation and virulence factor production (62). As evidenced by VqmA and several other well-studied LuxR-solos, despite significant sequence divergence from canonical LuxR receptors and the lack of cognate LuxI autoinducer synthases, LuxR-solo receptors appear to have largely retained their roles in perceiving chemical communication signals and controlling group behaviors.
Two-component histidine kinases
Bacteria commonly use two-component histidine-kinase sensory systems to dynamically respond to changing environments (Fig. 1B) and quorum-sensing circuits often rely on this type of signal relay (28, 98). In brief, two-component systems are composed of a sensor histidine kinase (SHK) that controls the phosphorylation state of a downstream receiver domain (40, 98). SHKs can function both as kinases and phosphatases (40, 98). SHK detection of environmental stimuli controls the balance between kinase and phosphatase activities and, as a result, dictates the downstream phosphorylation state of the receiver domain (40). In many two-component systems, the receiver domain exists as a separate protein referred to as a response regulator. The phosphorylation state of the response regulator likewise controls its activity, which is typically DNA binding or protein-protein interaction (40). In other two-component systems, the receiver domain is attached to the SHK and functions as an intermediate between the receptor and a downstream target of phosphorylation (40).
Here, we discuss the well-studied autoinducer-2 (AI-2) SHK, LuxPQ from vibrio species. The AI-2 autoinducer is produced by diverse bacterial species via the conserved autoinducer synthase, LuxS (12, 72). AI-2 production, detection, and response analyses suggest that AI-2 functions across bacterial species as a universal indicator of global bacterial cell density (61). In the case of vibrios, at low cell densities (LCD), when LuxPQ exists in its unliganded state, LuxQ dimers function as kinases (57, 58). In the kinase-activate state, LuxQ proteins autophosphorylate (Fig. 1B) (57, 58). The phosphoryl group is channeled to the attached receiver domain, and subsequently, to a phosphotransfer protein called LuxU (Fig. 1B) (22). In the final step of the phosphorylation cascade, the phosphoryl group is relayed from LuxU to the LuxO response regulator (23). Phospho-LuxO (LuxO~P) activates the expression of genes encoding small RNAs (Qrr sRNAs) that promote the LCD gene expression program and repress HCD behaviors (Fig. 1B) (48). Detection of accumulated AI-2, indicative of HCD, occurs via the periplasmic protein LuxP, which exists in a complex with LuxQ. Ligand binding drives a conformational change in the periplasmic domain of LuxQ (57, 58). This alteration is transmitted internally to inhibit LuxQ kinase activity. Because LuxQ harbors intrinsic phosphatase activity in its receiver domain, the flow of phosphate in the cascade reverses, leading to LuxO dephosphorylation, inactivation, and the transition to the HCD gene expression program. Notably, in the vibrio quorum-sensing pathway, additional SHKs, namely CqsS, LuxN, CqsR, and VpsS, also contribute to LuxU/LuxO phosphorylation/dephosphorylation (Fig. 1B) (43, 61). This arrangement, in which several receptors feed information into the same effector allows vibrios to integrate multiple cell density cues into a shared set of output behaviors. We note that such an arrangement is apparently rare among SHK signaling cascades.
Second messenger signaling
The final category of receptors discussed in this review are those that mediate the production or degradation of small-molecule second messengers that regulate bacterial group behaviors. Across the bacterial domain, second messenger signals, commonly linear or cyclized nucleotides, control bacterial responses to changing environmental conditions (31). Here, we focus on regulation of biofilm communities by c-di-GMP. In most bacterial species, low levels of cytoplasmic c-di-GMP are associated with cell motility whereas high c-di-GMP levels drive a sessile, biofilm lifestyle (68). c-di-GMP is produced by proteins called diguanylate cyclases which harbor conserved enzymatic sites composed of a GGDEF amino acid sequence motif (68). Degradation of c-di-GMP is catalyzed by phosphodiesterase enzymes that contain either EAL or HD-GYP catalytic motifs (68). Changes in c-di-GMP levels are detected by downstream effectors including riboswitches, transcription factors, and other regulatory proteins to drive motility (low c-di-GMP) or biofilm formation (high c-di-GMP) (27). An ongoing topic of discussion in the field concerns why bacterial genomes often encode dozens of c-di-GMP biosynthetic/catabolic enzymes (41). Moreover, the enzymes that produce and degrade c-di-GMP exhibit great diversity: some contain ligand-binding domains and some do not, some are membrane bound and some are cytoplasmic, some contain a single catalytic activity and some contain both diguanylate cyclase and phosphodiesterase domains, some appear to signal locally to specific downstream effectors, whereas others alter the shared “global” cytoplasmic c-di-GMP pool (30). It has been proposed that sensor-domain-containing diguanylate cyclases and phosphodiesterases enable cells to respond to environmental cues, whereas diguanylate cyclases and phosphodiesterases that lack these domains maintain c-di-GMP homeostasis (30).
The NspS/MbaA receptor in V. cholerae illustrates how cell-to-cell communication can regulate group behaviors via c-di-GMP signaling (Fig. 1C). NspS/MbaA activity is controlled by two polyamines: norspermidine, a rare polyamine in the biosphere that is produced by vibrios (via the carboxynorspermidine decarboxylase NspC) and by select other organisms, and spermidine, a polyamine produced widely by organisms spanning all domains (10, 11, 44, 78). Both polyamines are detected by the periplasmic protein NspS, and polyamine binding controls the NspS-MbaA interaction and the resulting enzymatic activity of MbaA, a membrane-spanning bifunctional receptor (4, 5, 95). When NspS detects norspermidine it associates with MbaA and this step drives c-di-GMP production via the MbaA cytoplasmic GGDEF domain (11). In contrast, spermidine binding to NspS leads to its dissociation from MbaA, and as a consequence, MbaA exhibits phosphodiesterase activity (11). c-di-GMP is degraded via the MbaA cytoplasmic EAL domain. In response to norspermidine, the c-di-GMP produced by NspS/MbaA promotes biofilm formation by activating the transcription of genes required for matrix production and surface adhesion (11, 78). Degradation of c-di-GMP by apo-MbaA reduces the expression of those same genes (11). The hypothesis is that the norspermidine signal, since it is primarily made by vibrios, represents a high level of relatedness in the community, and when this situation is encountered V. cholerae commits to the biofilm state. In contrast, it is presumed that high levels of spermidine, that as mentioned is broadly produced, indicates a mixed species community, and in this circumstance, V. cholerae disperses from the niche, fleeing competition.
SIGNALING SPECIFICITY AND INTER-DOMAIN COMMUNICATION
Bacteria in natural settings often exist in dynamic multispecies consortia. Quorum sensing is proposed to provide bacteria the means to track cell density and to garner information about the species composition of the vicinal community (56). Quorum-sensing bacteria face daunting tasks in multispecies communities, including accurately and simultaneously tracking changing numbers of kin and non-kin bacteria, robustly executing quorum-sensing-driven behaviors when mixtures of autoinducers are present, and limiting access of competitors and cheaters to quorum-sensing outputs, which are often public goods (56). Accomplishing these feats is likely critical for bacteria to thrive in multispecies communities. The desire to learn how bacteria overcome these fascinating challenges has driven a surge in research to define mechanisms underlying preservation of specificity in quorum-sensing systems. We now have the first inklings of how some quorum-sensing signals are kept private, allowing only select bacteria to tune in and participate in group behaviors and, in the opposite vein, how some bacterial quorum-sensing communication channels are broadcast widely, promoting detection and response by a variety of species in the community. Finally, there are instances of “eavesdropping” on quorum-sensing-mediated conversations. Here we provide a few examples.
Signaling specificity
One mechanism by which quorum-sensing bacteria accurately enact group behaviors in multispecies communities is via reliance on distinct, and context-specific, ligand-binding preferences of their quorum-sensing receptors. For example, LuxR-type receptors exhibit a spectrum of ligand binding stringencies: some LuxR proteins respond only to a specific, usually endogenously-produced, signal, whereas other LuxR-type receptors are promiscuous, and respond to a range of ligands, both endogenously-produced and produced by other bacterial species (25, 45, 93). An example of a LuxR-type receptor with exquisite specificity is AbaR from Acinetobacter baumannii which detects the autoinducer, (R)-N-(3-hydroxydodecanoyl) L-homoserine lactone (Fig. 2A) (25). Screening of AbaR against a panel of AHLs showed that only AHLs with the highest similarity to the native autoinducer drove even modest AbaR activation (25). In contrast, the Pseudomonas aeruginosa LasR receptor, which is activated by N-(3-oxododecanoyl) L-homoserine lactone, is promiscuous and indeed could be agonized by many structurally diverse AHLs from the same panel of ligands (Fig. 2B) (25). The hypothesis is that highly specific LuxR-type receptors regulate gene expression when the population density of kin reaches a quorum, whereas promiscuous LuxR-type receptors drive group behaviors even when other AHL-producing bacteria are in the majority. A recent mutagenesis and structural study identified LasR variants exhibiting enhanced ligand specificity, revealing that a flexible loop near the ligand-binding site governs ligand selectivity (51).
Fig. 2. Specificity in AHL quorum sensing.
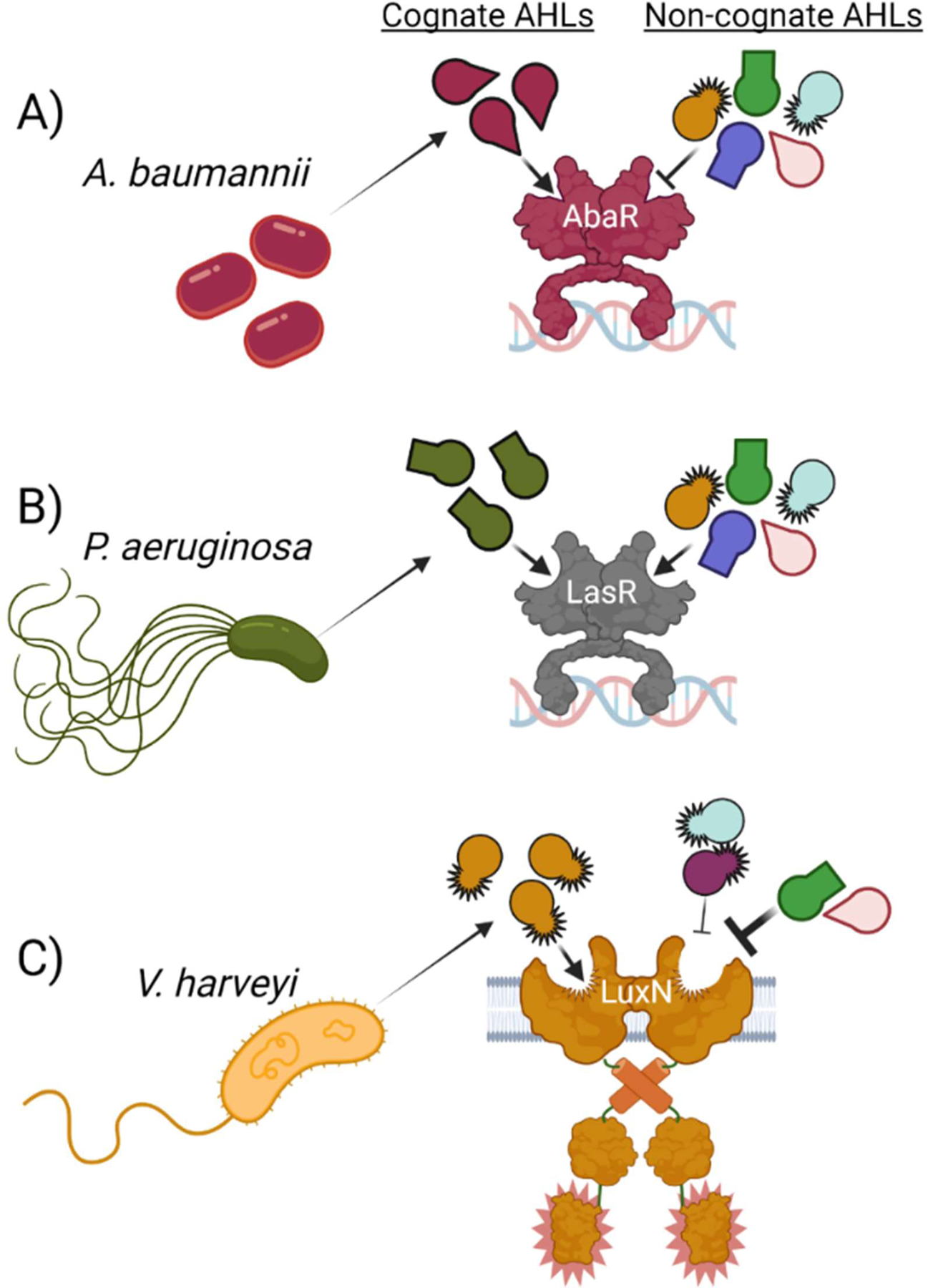
(A) The A. baumannii AbaR receptor is activated by its cognate autoinducer with high specificity. Non-cognate AHLs produced by other bacteria commonly antagonize AbaR, restricting cells to the LCD gene expression mode even when the cognate ligand is abundant. (B) The P. aeruginosa LasR receptor exhibits relaxed specificity and activates the HCD quorum-sensing gene expression program when bound to its cognate ligand or to many AHLs produced by other bacteria. Only select AHLs antagonize LasR. (C) Although unrelated to LuxR-type receptors, the V. harveyi LuxN two-component SHK receptor recognizes an AHL ligand. LuxN is highly specific for its cognate autoinducer and is antagonized by other AHLs. AHLs that are similar in structure to the cognate ligand only partially antagonize LuxN, whereas highly divergent AHLs strongly antagonize its activity. Figure created with BioRender.com.
A fascinating feature of some AHL receptors, including Vibrio harveyi LuxN, a two-component quorum-sensing receptor that detects the AHL ligand N-((R)-3-hydroxybutanoyl)-L-homoserine lactone with remarkable specificity, is that, while they cannot be activated by non-cognate AHL autoinducer ligands, they can be potently antagonized by them (Fig. 2C) (25, 45). In the case of LuxN, AHLs with close structural similarity to the natural AHL ligand do not agonize LuxN, yet do show modest antagonist activity, whereas AHLs with more structural variation are stronger antagonists (Fig. 2C) (45). The hypothesis is that the gradation in ligand antagonism activity tracks with species divergence from V. harveyi, i.e., the more different the signal molecule the more evolutionarily distant is the producer. The consequence is that when species that are not closely related to V. harveyi are present in a community, they produce AHLs that potently antagonize LuxN, inhibiting V. harveyi from launching its quorum-sensing program, which in turn, limits release of public goods that could benefit competitors. Antagonism by non-cognate AHLs is not unique to LuxN. Indeed, the AbaR-LasR screen described above showed that many of the AHLs that were incapable of agonizing AbaR or LasR instead acted as competitive inhibitors of the cognate ligands (25). Antagonism specificity followed the opposite pattern than that for agonism: AbaR was antagonized by many AHLs while LasR was antagonized only by select AHLs. The interpretation was that competitive inhibition prevents quorum-sensing-directed group behaviors when many, in the case of AbaR, or select, in the case of LasR, competitors are present.
Inter-domain communication
Uncovering how hosts and microbes interact in health and disease is of tremendous interest. The focus on these interactions stems first from our increasing understanding of the stunning roles endogenous microbiomes play in host organisms. Second, the topic is of major interest because improved understanding of mechanisms facilitating these interactions is driving the development of strategies to enhance beneficial bacteria and combat pathogens. Here, we discuss recently discovered examples of how quorum sensing is used for interactions across domains.
As discussed above, inter-species bacterial communication can be mediated by AHLs and by AI-2 and related molecules produced by the highly conserved LuxS synthase. Recently, it has been shown that bacteria in the Vibrio and Enterobacteriaceae families detect AI-2-like molecules produced by eukaryotic cells, which do not harbor LuxS enzymes. Specifically, mammalian epithelial cells produce an “AI-2 mimic” upon incubation with bacteria or following tight-junction disruption (39). The structure of the mammalian AI-2 mimic remains unknown. The bacterial response to the AI-2 mimic required the LuxP receptor suggesting that the mimic binds at the same site as AI-2 and likely shares structural features with AI-2 (39). It remains to be determined whether AI-2 mimic release by the host controls bacterial quorum sensing for the benefit of the host, if the bacteria have evolved to eavesdrop on host stress and use that vulnerability to their advantage, or if this mechanism benefits both the host and the bacteria. A subsequent study revealed that AI-2 mimic signaling is not unique to mammalian epithelial cells. Indeed, examination of inter-domain signaling between yeast and bacteria showed that, under stress, Saccharomyces cerevisiae also produces an AI-2 mimic the identity of which is 4-hydroxy-5-methylfuran-3(2H)-one (MHF) (88). All evidence to date indicates that the mammalian AI-2 mimic is not MHF (88). The yeast MHF biosynthetic pathway relies on Cff1p, an enzyme of previously unknown function annotated as a sugar isomerase/epimerase. Notably, cff1 exists in all domains of life, including in pathogenic fungi and bacteria, suggesting that MHF is a widespread molecule possibly used for quorum sensing in diverse species (88). Collectively, these recent discoveries suggest that AI-2 signaling pathways are widespread and could be central to communication across domains. Of note, a newly revealed category of AI-2 receptors, harboring dCACHE domains, exist in prokaryotic organisms including in bacteria that do not possess the established AI-2 receptors LuxP or LsrB (96). As of now it is not known whether cross-domain AI-2 mimic signaling influences the output activity of this new family of receptors.
A nascent area of research explores the roles quorum sensing plays when, rather than being the “guests” (whether beneficial or harmful), bacteria are the hosts. Bacteriophages, which outnumber all other organisms on Earth, are thought to have significantly influenced bacterial evolution (46). Moreover, quorum-sensing-mediated social interactions between bacteria and the phages that prey on them are increasingly being uncovered. New research shows that, during these interactions, the useful information encoded in quorum-sensing autoinducers can dictate output behaviors in both the bacterial hosts and the phage invaders.
On the bacterial side, quorum sensing activates the genes encoding the CRISPR-Cas immune system in P. aeruginosa, presumably protecting the bacteria from phage attack when the bacteria are at their most vulnerable (i.e., at HCD) (36). In an apparent counter strategy, the protein Aqs1 from the Pseudomonas DMS3 phage inhibits LasR, the Pseudomonas master quorum-sensing receptor (73). Aqs1 restricts Pseudomonas to execution of its LCD gene expression program even when cell density is high (73). Thus, group behaviors are not undertaken and that presumably assists phage DMS3 in overpowering host defenses.
On the phage side, remarkably, some phages can “eavesdrop” on bacterial quorum sensing to drive their lysogeny-lysis lifecycle transitions. Specifically, vibriophage VP882, encodes a quorum-sensing receptor, VqmAPhage, that has high similarity to the receptor and transcription factor discussed above called VqmA in vibrios (75). Vibrios make the autoinducer 3,5-dimethylpyrazin-2-ol (DPO), the ligand for VqmA (62). The VqmA-DPO complex launches the downstream quorum-sensing gene expression program. During phage VP882 infection, however, VqmAPhage also detects accumulated DPO, produced by the host. Liganded VqmAPhage drives expression of the gene encoding an antirepressor called Qtip, which in turn, sequesters and inactivates the cI repressor of lysis (75, 77). The consequence is that host-cell lysis occurs. Thus, phage VP882 enacts its lysis program only when host cell density is high, presumably maximizing spread to other bacteria. Notably, phages related to VP882 contain genes encoding analogous transcription factors and Qtip-like antirepressors in the same genomic locations as vqmAPhage and qtip, respectively (75). These findings suggest that it may be common for phages to tune into host sensory information and tailor their lysis-lysogeny lifestyle transitions to particular host-produced cues. Furthermore, early results suggest that phage surveillance of quorum-sensing autoinducers is not restricted to DPO, as bacteriophages have been discovered that encode LuxR-type receptors responsive to the AHL autoinducers produced by their host bacteria (76). The roles that these LuxR-type receptors play in phage lifecycles remain undefined. These initial findings promise an upcoming vibrant period of discovery of additional bacterial-phage quorum-sensing-mediated interactions that will deliver surprising new biological principles.
SIGNAL INTEGRATION
Information flow in quorum sensing
To decipher which bacterial species are in the vicinal community and their abundance, bacteria must extract the information contained in blends of extracellular communication signal molecules and transduce that information internally to drive changes in group behaviors. In this section, we discuss how the information encoded in multiple of autoinducers is integrated by two-component quorum-sensing circuits. In contrast to typical phosphorylation cascades in eukaryotes, in which individual kinases commonly phosphorylate hundreds of downstream targets, exquisite signaling specificity exists between bacterial SHKs and their partner cognate response regulators, a hallmark of information flow in bacterial two-component systems (47, 70). Indeed, each SHK typically displays a strong kinetic preference for a cognate response regulator, ensuring that phosphorylation of non-cognate response regulators, known as “crosstalk,” is limited (20, 26). For example, when activated by autoinducer binding, the Staphylococcus aureus quorum-sensing SHK, AgrC, rapidly transfers a phosphoryl group to its cognate response regulator, AgrA, whereas phosphotransfer to non-cognate response regulators does not occur on biologically relevant time scales (80, 91). Phosphorylated AgrA, in turn, activates HCD gene expression.
Notable exceptions to the single SHK-single response regulator paradigm exist in the well-studied quorum-sensing circuits of V. harveyi and V. cholerae, which we introduced briefly in our discussion of two-component quorum-sensing systems, and which are the focus of this section (Fig. 1B, 3A). As noted, in the V. harveyi and V. cholerae quorum-sensing pathways, multiple SHKs converge to control the phosphorylation state of a common target – the downstream phosphotransfer protein, LuxU – which in turn, shuttles phosphate to the LuxO response regulator/transcription factor (61). Specifically, V. harveyi and V. cholerae both possess the quorum-sensing SHK receptors CqsS, which detects a genus-specific autoinducer CAI-1, and LuxPQ, which detects the widespread autoinducer, AI-2 as described above (Fig. 3A) (12, 33). V. harveyi additionally harbors the LuxN SHK, which responds to the species-specific autoinducer AI-1 (24). Two other SHKs, CqsR and VpsS, also contribute to LuxU phosphorylation/ dephosphorylation (35, 43, 74, 92). Testing of diverse compounds revealed that CqsR responds to ethanolamine and VpsS activity is controlled indirectly by nitric oxide, providing important tools for studies of these two receptors’ mechanisms (35, 92). The authors presume that other native ligands exist because, in response to the addition of cell-free culture fluids, both receptors exhibit increased phosphatase activity (43). The identities of the putative endogenously-made ligands remain unknown.
Fig. 3. Features of the vibrio quorum-sensing circuit.
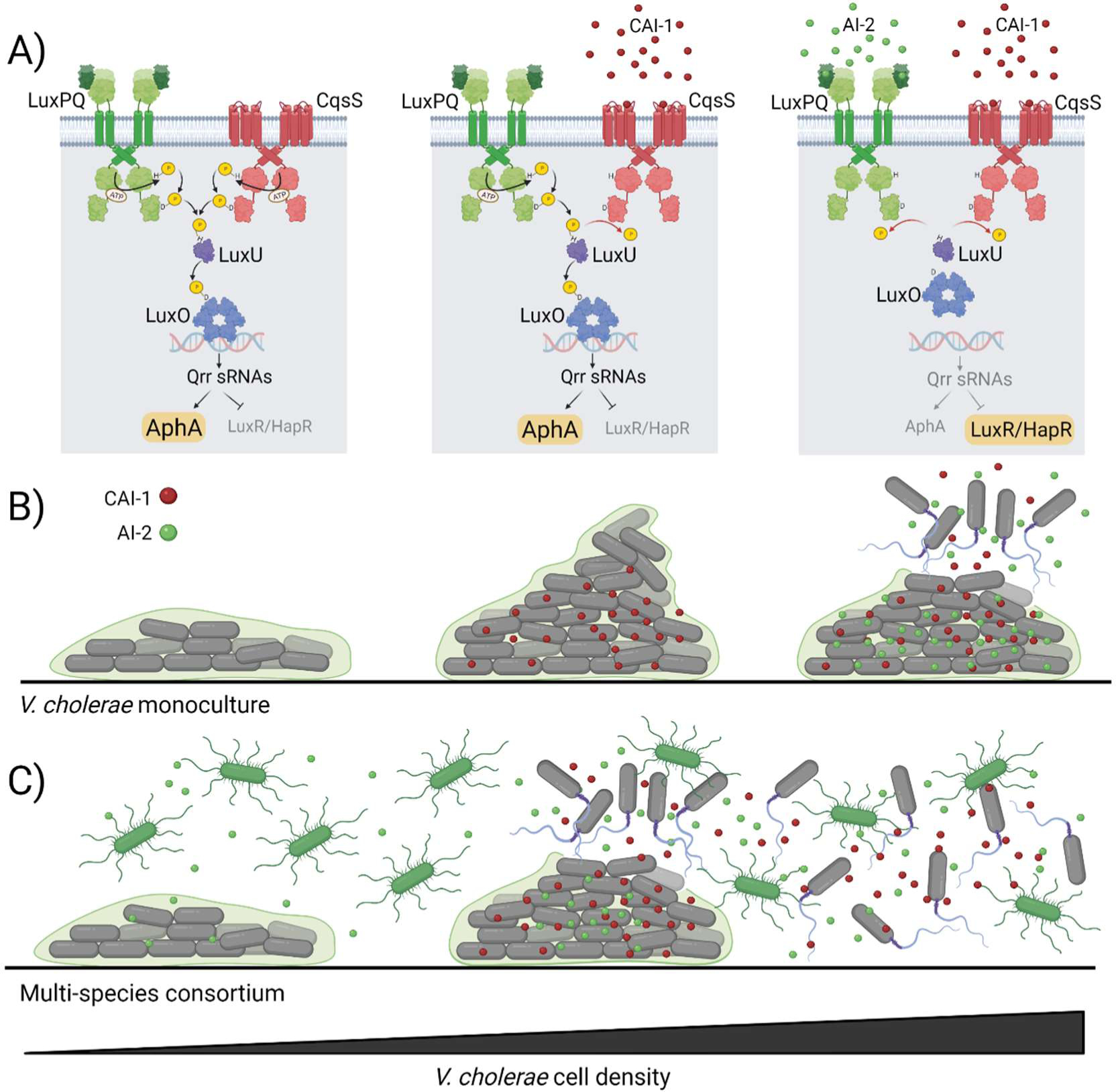
(A) Diagram of coincidence detection for a simplified quorum-sensing circuit containing only two receptors, LuxPQ and CqsS. (Left panel) When both receptors are unliganded, kinase activity drives LuxO phosphorylation and LCD behaviors are enacted. (Middle panel) When one receptor is unliganded, it functions as a kinase (in the scheme that is LuxPQ). The ligand-bound receptor functions as a phosphatase (in the scheme that is CqsS). Kinase activity overpowers phosphatase activity, and the cells remain in the LCD state. (Right panel) HCD behavior occurs only when both autoinducers are simultaneously detected, and thus the coincidence detection requirement is satisfied. (B) The V. cholerae biofilm lifecycle, when cells are grown in monoculture, over increasing cell densities. The genus specific CAI-1 autoinducer accumulates first, but biofilm dispersal does not occur until AI-2, which accumulates later in growth, stimulates the coincidence detector. (C) The V. cholerae biofilm lifecycle in a multi-species consortium containing other AI-2 producing bacteria. Endogenous CAI-1 combined with exogenous sources of AI-2 activate the coincidence detector at an early stage of biofilm formation leading to premature biofilm dispersal. Figure created with BioRender.com.
At LCD, when the V. harveyi and V. cholerae quorum-sensing SHKs are unliganded and they exhibit kinase activity, they collectively funnel phosphate to LuxO, leading to activation of expression of the genes encoding the Qrr sRNAs (61) (Fig. 3A). The Qrr sRNAs, in turn, post-transcriptionally activate production of the LCD master regulator AphA and repress production of the master HCD regulator LuxR (V. harveyi)/HapR (V. cholerae) (48). Under this condition, the bacteria enact LCD behaviors. At HCD, autoinducer binding inhibits the SHK kinase activities resulting in net phosphatase activity (61). LuxO is dephosphorylated and inactivated. The Qrr sRNAs are no longer produced, and as a consequence, AphA production is not activated and LuxR/HapR production is de-repressed. LuxR/HapR drives expression of genes required for HCD behaviors (61) (Fig. 3A).
A major focus of studies of the two vibrio quorum-sensing circuits has been to understand how multiple autoinducers function individually and together to control LuxO~P levels, and in turn, the operation of the cascade. The conundrum is that each autoinducer apparently conveys distinct information about the composition of the community, with AI-1 specifying kin, CAI-1 specifying “closely related”, and AI-2 specifying “other”, yet all autoinducer information flows to LuxO, making it difficult to understand if and how the circuit architecture could promote distinct gene expression patterns depending on which autoinducer(s) are present and at what relative levels. The answer, based on several studies, is that the system functions as a coincidence detector such that multiple autoinducers must be present simultaneously to trigger the HCD state (Fig. 3A, simplified diagram showing only two receptors) (9, 54). Specifically, any unliganded SHK functions as a kinase, funneling phosphate to LuxO, and as a result, imposes the LCD quorum-sensing state. Thus, only when the full suite of autoinducers is present is kinase activity eliminated, allowing phosphatase activity to dominate, permitting the switch to the HCD mode (Fig 3B) (9, 54). The interpretation of these findings is that vibrios only commit to launching expression of the HCD quorum-sensing regulon when two conditions are met: cell density is confirmed to be high and the kin autoinducer is present. The former condition presumably ensures that tasks undertaken will be successful because many cells are participating, and the latter condition ensures that leakage of public goods to competitors is minimized because significant numbers of kin cells are present. Other biological signaling networks, particularly neuronal circuits, also operate as coincidence detectors and commit to state changes only when multiple conditions are satisfied concurrently (42, 69).
The vibrio quorum-sensing coincidence detection arrangement, while possibly providing benefits to kin, would seem to necessarily preclude the ability to fine-tune specific gene expression patterns based on the species composition of the community. Not so! Recent studies demonstrate that the circuit architecture is configured to enable fine adjustments. Specifically, the receptors exhibit an offset in the cell densities at which they detect autoinducers produced by kin (AI-1 and CAI-1) versus the widely produced ligand (AI-2), allowing them to respond uniquely to blends of autoinducers (Fig. 3B) (9). Briefly, in monoculture, the vibrio-specific autoinducers CAI-1 and AI-1 (in V. harveyi), are detected at low- and mid-cell densities of growth, respectively, whereas AI-2 is only detected late in growth at quite HCD (Fig. 3B) (9, 32). Thus, AI-2 acts as the rate limiting step for the coincidence detection conditions to be met. The three receptors possess remarkably similar EC50 values (~10 nM) for their respective ligands in vivo, so presumably, it is the rate that each autoinducer accumulates that determines the detection offset (38, 58, 84). A manifestation of this arrangement has been described for the quorum-sensing-regulated biofilm lifecycle of V. cholerae (Fig. 3B) (9). At the launch of the V. cholerae biofilm lifecycle, at LCD, individual cells attach to a substate. Biofilms begin to form as a consequence of cell division and exopolysaccharide matrix secretion. Early in biofilm development, the endogenously-produced CAI-1 autoinducer is detected by CqsS, and CqsS converts from kinase mode to phosphatase mode. Nonetheless, LCD genes continue to be expressed because LuxPQ remains unliganded and therefore contributes kinase activity to the quorum-sensing cascade. At later stages in biofilm development, endogenously-produced AI-2 accumulates beyond the detection threshold, at which point, both quorum-sensing receptors are liganded and the coincidence detector is satisfied (Fig. 3B). Thus, the cells switch from the LCD to the HCD gene expression pattern, and they disperse from biofilms and return to a planktonic lifestyle (9). Critically, because of the delay in detection of any endogenously-produced AI-2 relative to CAI-1, exogenous supply of AI-2 can fulfill the coincidence detector requirement. Thus, if additional AI-2 is introduced, premature dispersal from biofilms occurs (Fig. 3C) (9). In contrast, introduction of excess CAI-1 does not drive premature biofilm dispersal because CAI-1 is not the limiting ligand required to satisfy the coincidence detector. The biological logic underlying this setup is thought to reflect detection of kin versus detection of non-kin. Delayed detection of any endogenously-produced AI-2 may allow vibrios to “tune into” AI-2 made by other bacterial species in the community (Fig. 3C). Thus, prior to committing to HCD behaviors, a quorum of vibrios is required to be present, and once this condition is satisfied, non-endogenous-sources of AI-2 are sufficient to drive HCD gene expression. Given that, in vibrios, HCD conditions promote biofilm dispersal, the further interpretation of these findings is that when vibrios are in biofilms that are surrounded by kin, they stay put, but when they are surrounded by non-kin, they flee the present locale presumably so seek out more hospitable/less competitive niches.
To enact finely tuned changes in hundreds of group behaviors, vibrios convert the LuxO phosphorylation state, as driven by the coincidence detector, into the production of the set of versatile Hfq-dependent Qrr sRNAs that mediate the outputs (Figs. 1B and 3) (19). The Qrr sRNAs regulate the translation and stability of dozens of target mRNAs, and notably, they control the levels of the LCD and HCD quorum-sensing master regulators AphA and LuxR/HapR, respectively. They also feedback regulate several of the quorum-sensing components including LuxO and LuxN (discussed below). The Qrr sRNAs function by four regulatory mechanisms (Fig. 4) (19). In the case of positive regulation, Qrr sRNAs bind to the target mRNA upstream of a stem loop that occludes the ribosome binding site. Binding exposes the site enabling translation (e.g. aphA) (19). The other three mechanisms are repressive: The Qrr sRNAs can bind to and destabilize mRNA transcripts by catalytic degradation in which the mRNA but not the sRNA is degraded, (e.g. luxR/hapR). They can drive coupled degradation in which both the target mRNA and the Qrr are destroyed (e.g. luxMN). Finally, the Qrr sRNAs can sequester target mRNAs and prevent their translation (e.g. luxO) (19). These different regulatory mechanisms function with distinct dynamics and potencies. For example, catalytic degradation is the most rapid, and is superior in suppressing translation of target mRNAs compared to sequestration (19). Additionally, regulation by Qrr catalytic degradation enables larger-scale changes in expression of the gene encoded on the mRNA than does regulation by Qrr sequestration. Thus, even though all of the extracellular information is channeled through a single regulator, LuxO, the regulatory architecture delivers precise, distinct, and complex patterns of gene expression.
Fig. 4. The Qrr sRNAs control quorum-sensing target genes.
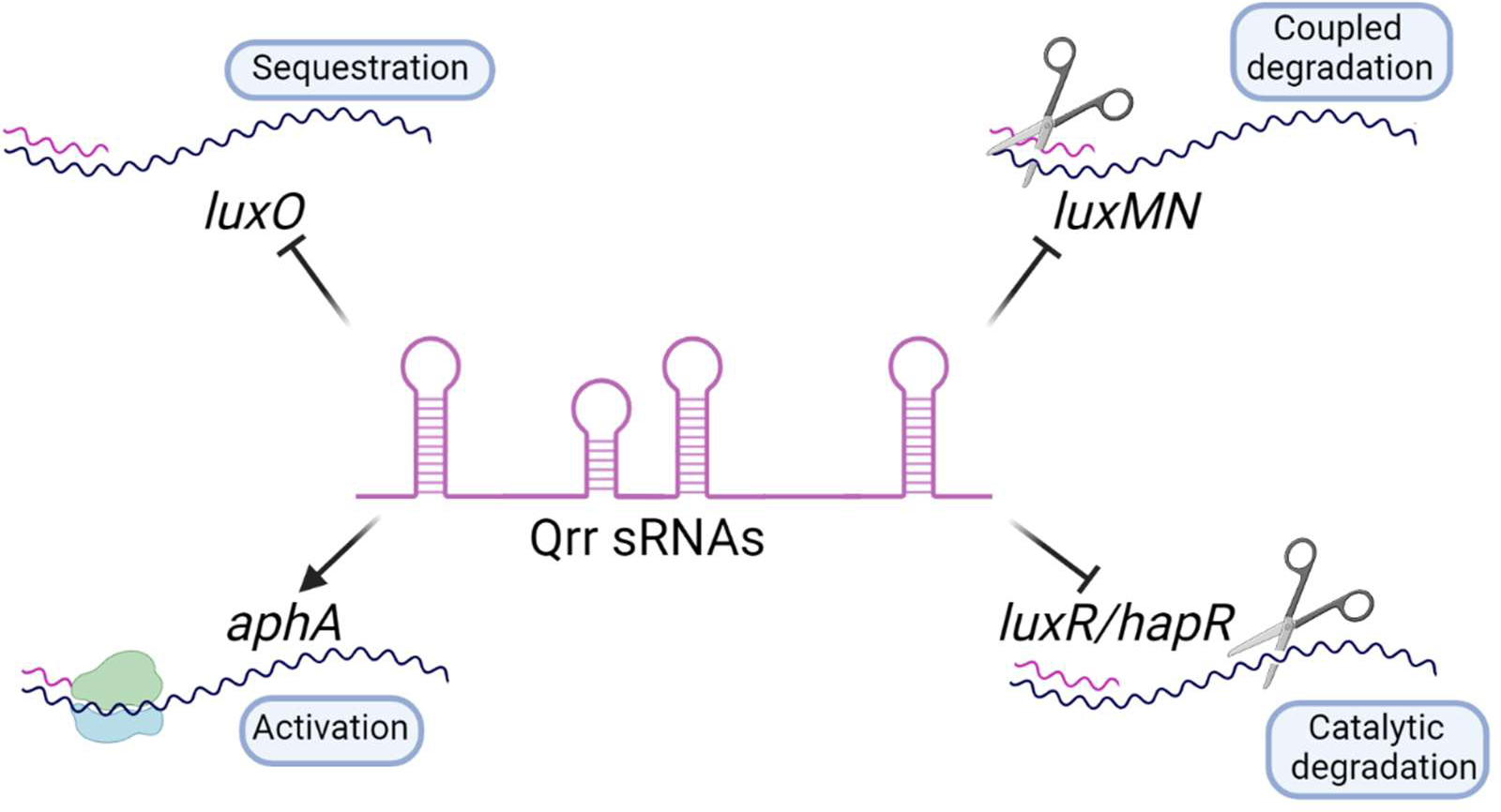
The Qrr sRNAs function by four post-transcriptional regulatory mechanisms. They activate translation by facilitating ribosome binding. They repress translation by sequestration, coupled degradation, and catalytic degradation of target mRNAs. One representative target mRNA that is controlled by each mechanism is shown. Figure created with BioRender.com.
c-di-GMP signaling fidelity
The vibrio quorum-sensing coincidence-detection mechanism enables bacterial populations to gain additional information from each ligand that is present, despite the fact that autoinducer information is relayed through a common regulatory pathway. Other converging pathways that control group behaviors are also tasked with decoding the information contained in distinct ligands. It is now apparent that evolution has produced alternative mechanisms to coincidence detection to overcome this challenge. A prominent example concerns c-di-GMP signaling. c-di-GMP levels are regulated directly by many inputs, including surface substrate properties, polyamines, oxygen, light, and arginine, among others (41). Additionally, some c-di-GMP metabolizing enzymes do not contain ligand-binding domains, and thus, may alter c-di-GMP levels through changes in their abundances, which are controlled by yet other signals, such as quorum-sensing autoinducers (30). Many bacteria, including P. aeruginosa and V. cholerae, contain dozens of c-di-GMP metabolizing enzymes. How bacteria tune their responses to each input stimulus, despite the output being the same diffusible second-messenger molecule, is a prominent area of signal-transduction research. Two main models currently contend with signaling specificity through c-di-GMP (30): 1) A global model in which individual diguanylate cyclases or phosphodiesterases contribute/remove distinct amounts of c-di-GMP to/from a global cytoplasmic pool of c-di-GMP (30). In this model, at low cytoplasmic c-di-GMP levels, only c-di-GMP-responsive effectors with the highest binding affinities for c-di-GMP are activated. As cytoplasmic c-di-GMP levels increase, effectors with lower binding affinities are engaged and detect c-di-GMP. 2) A local model for c-di-GMP signaling posits instead that diguanylate cyclases reside near specific c-di-GMP-responsive effectors, and the local pool of c-di-GMP produced by a particular diguanylate cyclase activates the proximate effector(s) before c-di-GMP diffuses away and increases the concentration of the global c-di-GMP pool (30). The colocalization of diguanylate cyclases with specific effectors commonly occurs through direct binding, although modeling studies have suggested that physical interactions are not required for local c-di-GMP signal transmission (27). Phosphodiesterases are thought to contribute to the specificity of local c-di-GMP signaling by hydrolyzing c-di-GMP from the global pool that diffuses into the vicinity of each diguanylate-cyclase-effector pair.
Evidence for both global and local c-di-GMP signaling exists. For example, c-di-GMP controls the timing of inhibition of motility and cellulose synthesis through changes to the global c-di-GMP pool in Salmonella enterica. Indeed, an over 40-fold difference in binding affinities for c-di-GMP exists between YcgR and BcsA, the effectors responsible for motility inhibition and cellulose synthesis, respectively (65). This feature is responsible for motility inhibition preceding cellulose synthesis when cytoplasmic c-di-GMP levels change. The proposed model is that some diguanylate cyclases produce an initial pool of c-di-GMP that is sufficient to suppress the motility apparatus, followed by a second set of diguanylate cyclases producing more c-di-GMP to reach a high enough concentration to activate the cellulose synthesis machinery (65). An instance of local c-di-GMP signaling was uncovered in the Lap biofilm regulatory system in Pseudomonas fluorescens (14, 34, 59). Here, the large adhesin protein LapA spans the bacterial outer membrane (55). When its external domains adhere to a surface, biofilm formation is initiated. In the absence of local c-di-GMP, the periplasmic protease LapG cleaves the N-terminus of LapA, removing LapA from the cell surface and facilitating biofilm detachment (7). When local c-di-GMP is present, the LapG protease is sequestered and inactivated by c-di-GMP-bound LapD (60). Sequestration prevents LapG-directed cleavage of LapA. Thus, P. fluorescens forms biofilms. Two diguanylate cyclases, GcbB and GcbC, transmit c-di-GMP specifically to LapD, and physical interactions between GcbC and LapD are necessary to promote LapD-mediated sequestration of LapG (14). Moreover, a chimeric protein possessing diguanylate cyclase activity and the GcbC and LapD interaction domains funneled c-di-GMP to LapD through direct physical interaction, which promoted sequestration of LapG and biofilm formation by P. fluorescens (14).
While both local and global mechanisms of c-di-GMP signaling have been demonstrated, it is possible that some diguanylate cyclases and phosphodiesterases are responsible for setting the global c-di-GMP concentration, whereas others fine tune c-di-GMP signaling by acting locally (30). In this context it is important to note that evidence for the local signaling mechanism preventing crosstalk across c-di-GMP-responsive effectors is scarce. Local c-di-GMP signal transmission may offer advantages outside of specificity, for example, increased sensitivity to ligands. Indeed, it was recently shown that local signaling is not required for signaling specificity by the V. cholerae polyamine-responsive bifunctional diguanylate cyclase/phosphodiesterase receptor called MbaA (11). Rather, the local c-di-GMP signaling mechanism enables V. cholerae to respond to nanomolar polyamine levels, as opposed to the micromolar polyamine levels that are necessary for MbaA to alter the global c-di-GMP concentration. The influence of local signaling mechanisms on features such as response sensitivity, the amplitude of the input-output response, and response specificity hinges on the abundances of the relevant receptors and effectors, which in turn, may be dynamically altered by the activities of other signaling relays. Future studies could probe how the information flow through c-di-GMP metabolizing receptors and other regulatory circuits that affect c-di-GMP dynamics influence the phenotypic outputs of the individual signaling systems.
Receptor bifunctionality
Many of the bacterial group behaviors discussed above are controlled by receptors harboring opposing catalytic activities, i.e., kinase and phosphatase or diguanylate cyclase and phosphodiesterase. Ligand binding regulates the balance between the enzymatic activities. Modeling and experimental studies of SHKs suggest that bifunctionality buffers signaling circuits against fluctuations in the concentrations of the receptors (2). Specifically, since both activities are contained in a single SHK, changes in receptor concentrations do not affect the balance of catalytic activities. Therefore, net phospho-flow is unchanged when bifunctional receptor levels fluctuate. Thus, even in the face of noise, SHKs transmit information with high fidelity. In addition, it is thought that bifunctional SHKs harboring basal phosphatase activities insulate signal transmission from crosstalk; a receptor’s basal phosphatase activity negates nonspecific phosphorylation events (70). Although not yet studied in detail, bifunctionality in c-di-GMP metabolizing proteins could endow their circuits with analogous benefits. Additionally, we posit that bifunctionality in c-di-GMP enzymes could enhance local signaling specificity. Possessing both c-di-GMP biosynthetic and catabolic capability could allow a given receptor to tightly constrain the activity of a proximal c-di-GMP-responsive effector. Acting as a diguanylate cyclase could facilitate c-di-GMP transmission directly to a particular effector. By contrast, acting as phosphodiesterase could allow the same receptor to protect its pathway from spillover input from the global c-di-GMP pool when the global pool is at high concentration.
FEEDBACK REGULATION
Multiple feedback loops are embedded in quorum-sensing circuits
From embryonic development to neurotransmission to bacterial cell-cell communication, biological signaling networks possess feedback loops that modulate features including input-output relations, response timing, response amplitude, noise reduction, and commitment to new states (8). Here, we discuss the roles of negative and positive feedback loops in the control of quorum-sensing transitions in V. harveyi/V. cholerae. We note that feedback also exists in c-di-GMP signaling networks, for example, diguanylate cyclases are inhibited by c-di-GMP product at “I-sites”, which establishes an upper bound on enzyme output (13).
Negative auto-feedback
Negative autoregulation, in which a signaling component inhibits its own activity can suppress noise, set the sensitivity to a stimulus, limit the maximum output of a pathway, and drive transient responses to an input (3, 8, 17). In the V. harveyi/V. cholerae quorum-sensing circuit, there exist, to our knowledge, six negative autoregulatory feedback loops (Fig. 5, magenta). Some negative autoregulation occurs directly; for example, LuxO, AphA, and LuxR/HapR each bind their own promoters and repress transcription (Fig. 5). Other negative autoregulatory feedback loops function by indirect mechanisms via repression of upstream activators (e.g., Qrr sRNAs inhibit luxO and AphA inhibits qrr sRNA transcription) or by activation of upstream repressors (e.g., LuxR/HapR activates qrr sRNA transcription) (Fig. 5). In all of the two-step indirect negative autoregulation cases, one component must be an activator and the other must be a repressor.
Fig. 5. Autoregulatory feedback loops in the V. harveyi/V. cholerae quorum-sensing circuit.
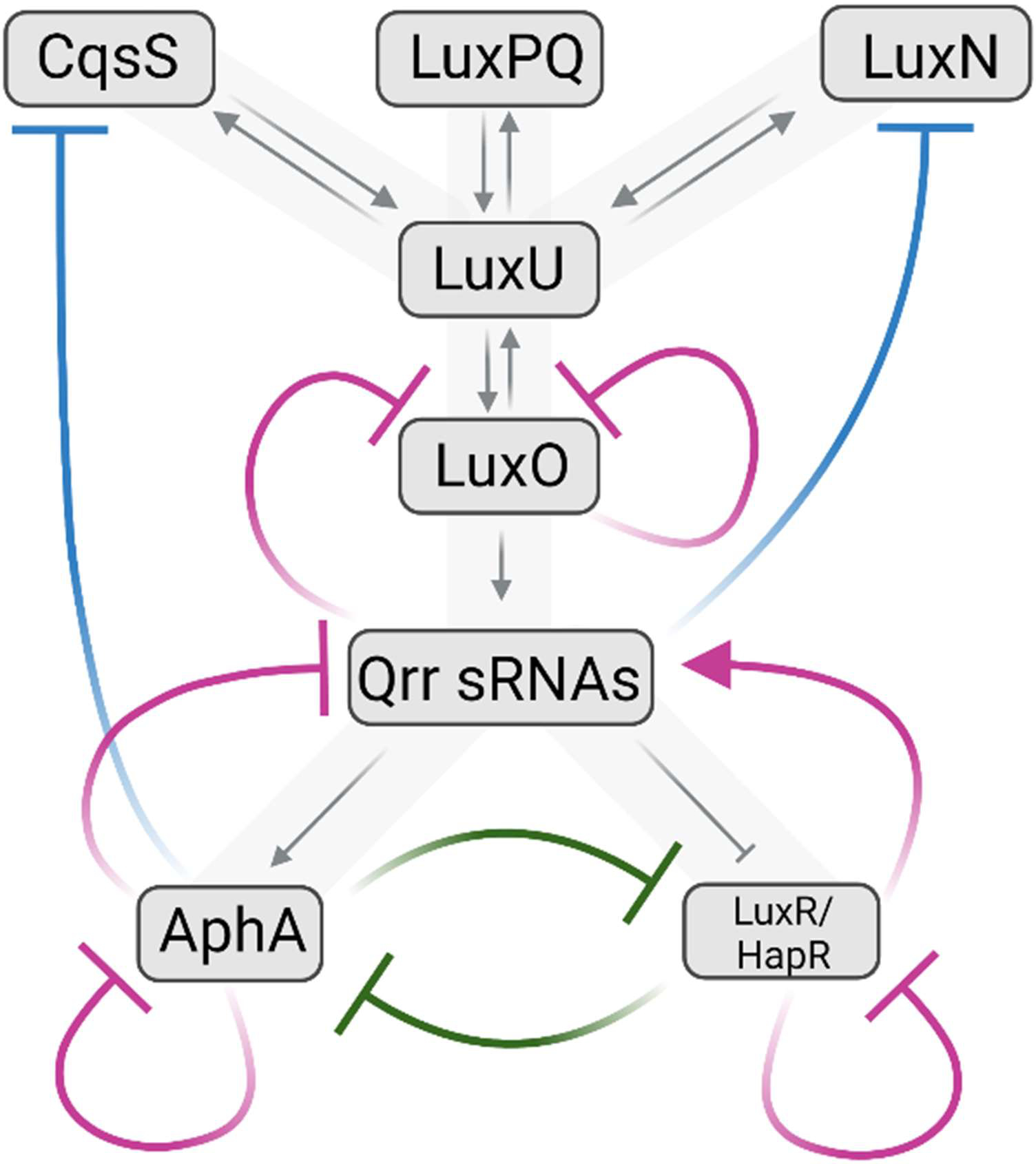
Negative autoregulatory feedback loops are colored magenta and positive autoregulatory feedback loops are colored green. Two additional regulatory loops exist that control receptor levels, shown in blue. Figure created with BioRender.com.
Why do so many negative auto-regulatory feedback loops exist in the V. harveyi/V. cholerae quorum-sensing circuit, and what consequence does operation of each loop have for signal transduction? Studies of quorum-sensing signaling in mutants defective in particular negative autoregulatory loops have delivered some insight. A summary of the findings is as follows: The two LuxO autorepression loops (direct and indirect via the Qrr sRNAs, Fig. 5) put an upper limit on LuxO production which increases sensitivity to autoinducers and dampens noise (i.e., cell-to-cell variability in quorum-sensing state) across autoinduction conditions (85, 87). The two negative autoregulatory feedback loops involving LuxR/HapR (direct and indirect via the Qrr sRNAs, Fig. 5) have distinct roles. LuxR/HapR direct autorepression limits the maximum output of the quorum-sensing cascade (49, 87). LuxR/HapR negative autoregulation through the Qrr sRNAs does not influence steady-state quorum-sensing output, but rather, controls the speed by which quorum-sensing state transitions occur. Specifically, LuxR/HapR activation of qrr sRNA gene transcription facilitates the rapid transition from the HCD to the LCD gene expression program (83). Because LuxO~P is required for Qrr sRNA production, this feedback loop is only productive when both LuxO~P and LuxR/HapR are simultaneously present, which is thought to exclusively occur immediately following dilution from the HCD state. Note that at HCD, absolute levels of LuxO are also high (as the negative feedback via Qrr sRNAs is inactive), which, following dilution, facilitates the rapid accumulation of LuxO~P. The effect is that the LuxO~P and LuxR/HapR proteins, which both remain high for several cell divisions following the transition, together boost Qrr production. The Qrr sRNAs, in turn, launch the expression of the LCD quorum-sensing regulon. Because the Qrr sRNAs repress luxO and luxR/hapR, new LuxO and LuxR/HapR proteins are not made, residual LuxO~P and LuxR/HapR diminish over time, and the feedback loop is terminated (83). The presumed ecological benefit of this arrangement is that it promotes fast phenotypic alterations when cell numbers rapidly change from high to low (i.e., perhaps following expulsion from the host or displacement from a biofilm). Finally, although not as extensively studied, the two negative autoregulatory feedback loops involving AphA (direct and indirect via the Qrr sRNAs, Fig. 5) play similar roles: (1) repression of qrr transcription by AphA implies that the rapid accumulation of Qrr sRNAs upon dilution is capped as soon as AphA levels become high enough to repress enhanced Qrr production, and (2) AphA autorepression limits noise in LCD gene expression (50, 71). Taken together, the ensemble of negative autoregulatory feedback loops in the V. harveyi/V. cholerae quorum-sensing circuit serves to establish the precise quorum-sensing input-output dynamics. By setting the signal transduction output ceiling, limiting noise across autoinduction conditions, and dictating the pace by which cell density transitions occur, negative autoregulatory feedback loops allow vibrios to make robust collective decisions in the face of fluctuating environmental conditions.
Positive Feedback
Whereas negative autoregulatory feedback loops typically fine-tune and stabilize signal transmission, positive autoregulation is associated with signal amplification and fosters dramatic changes in signaling states (8). In positive autoregulatory loops, activation or accumulation of a component leads to feedback activation of that same component. Thus, signaling pathways possessing positive autofeedback display increased sensitivity to inputs at the cost of increased noise, and positive autofeedback is often associated with all-or-none gene expression that drives bistable behaviors (8).
Much like negative autoregulation, positive feedback can occur by direct or indirect mechanisms. In the case of the V. harveyi/V. cholerae quorum-sensing pathway, only indirect positive autofeedback has been discovered. There are two known loops (Fig. 5, green) that drive mutual repression between AphA and LuxR/HapR: AphA represses luxR/hapR and LuxR/HapR represses aphA (50, 71). Thus, both LuxR/HapR and AphA are indirectly positively autoregulated (Fig. 5). Analyses of mutants defective in these two autoregulatory loops revealed that this arrangement ensures maximal AphA and minimal LuxR/HapR production at LCD, and conversely, maximal LuxR/HapR and minimal AphA production at HCD (71). Thus, the quorum-sensing positive autoregulatory feedback loops drive commitment to the LCD or HCD gene expression program in response to autoinducer levels. Nonetheless, compared to negative autofeedback, positive autofeedback appears to play a more minor role in controlling the overall vibrio quorum-sensing input-output response. Since positive feedback is associated with amplified response to small changes in signals but high noise, the near absence of positive feedback in the vibrio quorum-sensing circuits suggests that noise reduction is prioritized over sensitivity. This notion is consistent with the view that quorum sensing drives collective behaviors in which all cells participate in unison, in contrast to behaviors such as foraging (chemotaxis) or bet hedging against stresses, in which the objective is to ensure success of a subset of the population (63).
Feedback control of receptor ratios
Two additional feedback loops exist in the V. harveyi pathway that control the expression of genes encoding the two-component histidine kinase receptors CqsS and LuxN (Fig. 5, blue). Both loops operate at LCD; AphA represses cqsS and the Qrr sRNAs repress luxN (29, 38, 85). As autoinducers accumulate with increasing cell density, negative regulation is relieved because AphA and the Qrr sRNAs are not produced at HCD. Thus, increased production of the CqsS and LuxN receptors occurs at HCD. What is key is that LuxPQ levels remain constant with changing cell density, so the ratios of CqsS and LuxN to LuxPQ increase (38). The consequence is that, at HCD, input from CqsS and LuxN overpowers input from LuxPQ (38, 85). The model is that at LCD, quorum-sensing gene expression is responsive to all three autoinducers (see coincidence detection section above) while at HCD, the AI-2 signal becomes irrelevant. As a reminder, the LuxN AI-1 ligand specifies “kin”, the CqsS CAI-1 ligand specifies “closely-related” species, and the LuxPQ AI-2 ligand is produced widely among diverse bacteria. Thus, at HCD, V. harveyi pays attention to numbers of kin and ignores non-kin sources of AI-2. The rationale is that when a quorum-sensing bacterium’s kin is the majority species, a minority population of unrelated non-kin bacteria are inconsequential to the decision to commit to HCD collective processes. We note that V. cholerae does not possess LuxN and it does not make AI-1, so only the AphA-cqsS feedback loop exists. Presumably, the principles underlying its operation remain relevant because in V. cholerae, CAI-1 is the autoinducer that specifies kin. An interesting unanswered question is whether, when vibrios are a local minority, their gene expression response to AI-2 allows them to “cheat” by benefiting from public goods produced by other species.
CONCLUDING REMARKS
Bacteria display remarkable agility in adapting to fluctuating environments. In this review, we have highlighted the mechanistic principles that promote flexibility and ensure robustness in quorum-sensing and c-di-GMP signaling. Going forward, it will be critical to understand how the signaling mechanisms described here play out in authentic scenarios such as in three-dimensional biofilms, in mixed-species consortia, and in the presence of flow. Moreover, we know signaling cascades do not function in isolation. Thus, another pressing issue is how the different collective-behavior-regulating pathways are integrated to drive global changes in behavior. Germane to this review, both quorum-sensing and c-di-GMP pathways regulate biofilm formation in many pathogens. Highly quantitative mechanistic studies could provide the insight required to understand how bacteria accomplish these fascinating feats.
ACKNOWLEDGEMENTS
We thank the members of the Bassler and Wingreen groups for insightful discussions. This work was supported by the Howard Hughes Medical Institute (B.L.B.); the National Science Foundation through the Center for the Physics of Biological Function (PHY-1734030), as well as NSF grants MCB-1713731 (B.L.B.) and MCB-1853602 (B.L.B. and N.S.W.); NIH grants 1R21AI144223 (B.L.B. and N.S.W.), 2R37GM065859 (B.L.B.), GM082938 (N.S.W.), and 1K99AI158939 (A.A.B).
LITERATURE CITED
- 1.Azimi S, Klementiev AD, Whiteley M, Diggle SP. 2020. Bacterial Quorum Sensing During Infection. Annual Review of Microbiology. 74(1):201–19 [DOI] [PubMed] [Google Scholar]
- 2.Batchelor E, Goulian M. 2003. Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. Proc Natl Acad Sci U S A. 100(2):691–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becskei A, Serrano L. 2000. Engineering stability in gene networks by autoregulation. Nature. 405(6786):590–93 [DOI] [PubMed] [Google Scholar]
- 4.Bomchil N, Watnick P, Kolter R. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J Bacteriol. 185(4):1384–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bomchil N, Watnick P, Kolter R. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J Bacteriol. 185(4):1384–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bottomley MJ, Muraglia E, Bazzo R, Carfì A. 2007. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J Biol Chem. 282(18):13592–600 [DOI] [PubMed] [Google Scholar]
- 7.Boyd CD, Chatterjee D, Sondermann H, O’Toole GA. 2012. LapG, Required for Modulating Biofilm Formation by Pseudomonas fluorescens Pf0–1, Is a Calcium-Dependent Protease. Journal of Bacteriology. 194(16):4406–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandman O, Meyer T. 2008. Feedback Loops Shape Cellular Signals in Space and Time. Science. 322(5900):390–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bridges AA, Bassler BL. 2019. The intragenus and interspecies quorum-sensing autoinducers exert distinct control over Vibrio cholerae biofilm formation and dispersal. PLoS Biol. 17(11):e3000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bridges AA, Bassler BL. 2021. Inverse regulation of Vibrio cholerae biofilm dispersal by polyamine signals. eLife. 10:e65487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bridges AA, Prentice JA, Fei C, Wingreen NS, Bassler BL. 2021. Quantitative input-output dynamics of a c-di-GMP signal-transduction cascade in Vibrio cholerae [DOI] [PMC free article] [PubMed]
- 12.Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, et al. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 415(6871):545–49 [DOI] [PubMed] [Google Scholar]
- 13.Christen B, Christen M, Paul R, Schmid F, Folcher M, et al. 2006. Allosteric control of cyclic di-GMP signaling. J Biol Chem. 281(42):32015–24 [DOI] [PubMed] [Google Scholar]
- 14.Dahlstrom KM, Giglio KM, Collins AJ, Sondermann H, O’Toole GA. 2015. Contribution of Physical Interactions to Signaling Specificity between a Diguanylate Cyclase and Its Effector. mBio. 6(6):e01978–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duddy OP, Bassler BL. 2021. Quorum sensing across bacterial and viral domains. PLoS Pathog. 17(1):e1009074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, Oppenheimer NJ. 1981. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 20(9):2444–49 [DOI] [PubMed] [Google Scholar]
- 17.Elowitz MB, Levine AJ, Siggia ED, Swain PS. 2002. Stochastic Gene Expression in a Single Cell. Science. 297(5584):1183–86 [DOI] [PubMed] [Google Scholar]
- 18.Engebrecht J, Silverman M. 1984. Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. U.S.A 81(13):4154–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng L, Rutherford ST, Papenfort K, Bagert JD, van Kessel JC, et al. 2015. A qrr noncoding RNA deploys four different regulatory mechanisms to optimize quorum-sensing dynamics. Cell. 160(1–2):228–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher SL, Kim S-K, Wanner BL, Walsh CT. 1996. Kinetic Comparison of the Specificity of the Vancomycin Resistance Kinase VanS for Two Response Regulators, VanR and PhoB. Biochemistry. 35(15):4732–40 [DOI] [PubMed] [Google Scholar]
- 21.Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. 2016. Biofilms: an emergent form of bacterial life. Nature Reviews Microbiology. 14(9):563–75 [DOI] [PubMed] [Google Scholar]
- 22.Freeman JA, Bassler BL. 1999. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J Bacteriol. 181(3):899–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freeman JA, Bassler BL. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol Microbiol. 31(2):665–77 [DOI] [PubMed] [Google Scholar]
- 24.Freeman JA, Lilley BN, Bassler BL. 2000. A genetic analysis of the functions of LuxN: a two-component hybrid sensor kinase that regulates quorum sensing in Vibrio harveyi. Mol Microbiol. 35(1):139–49 [DOI] [PubMed] [Google Scholar]
- 25.Gerdt JP, Wittenwyler DM, Combs JB, Boursier ME, Brummond JW, et al. 2017. Chemical Interrogation of LuxR-type Quorum Sensing Receptors Reveals New Insights into Receptor Selectivity and the Potential for Interspecies Bacterial Signaling. ACS Chem. Biol. 12(9):2457–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grimshaw CE, Huang S, Hanstein CG, Strauch MA, Burbulys D, et al. 1998. Synergistic Kinetic Interactions between Components of the Phosphorelay Controlling Sporulation in Bacillus subtilis. Biochemistry. 37(5):1365–75 [DOI] [PubMed] [Google Scholar]
- 27.Ha D-G, O’Toole GA. 2015. c-di-GMP and its Effects on Biofilm Formation and Dispersion: a Pseudomonas Aeruginosa Review. Microbiol Spectr. 3(2):MB-0003–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hawver LA, Jung SA, Ng W-L. 2016. Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiology Reviews. 40(5):738–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haycocks JRJ, Warren GZL, Walker LM, Chlebek JL, Dalia TN, et al. 2019. The quorum sensing transcription factor AphA directly regulates natural competence in Vibrio cholerae. PLoS Genet. 15(10):e1008362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hengge R 2021. High-specificity local and global c-di-GMP signaling. Trends Microbiol. 29(11):993–1003 [DOI] [PubMed] [Google Scholar]
- 31.Hengge R, Häussler S, Pruteanu M, Stülke J, Tschowri N, Turgay K. 2019. Recent Advances and Current Trends in Nucleotide Second Messenger Signaling in Bacteria. J Mol Biol. 431(5):908–27 [DOI] [PubMed] [Google Scholar]
- 32.Henke JM, Bassler BL. 2004. Three Parallel Quorum-Sensing Systems Regulate Gene Expression in Vibrio harveyi. Journal of Bacteriology. 186(20):6902–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higgins DA, Pomianek ME, Kraml CM, Taylor RK, Semmelhack MF, Bassler BL. 2007. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 450(7171):883–86 [DOI] [PubMed] [Google Scholar]
- 34.Hinsa SM, Espinosa-Urgel M, Ramos JL, O’Toole GA. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol. 49(4):905–18 [DOI] [PubMed] [Google Scholar]
- 35.Hossain S, Heckler I, Boon EM. 2018. Discovery of a Nitric Oxide Responsive Quorum Sensing Circuit in Vibrio cholerae. ACS Chem Biol. 13(8):1964–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Høyland-Kroghsbo NM, Paczkowski J, Mukherjee S, Broniewski J, Westra E, et al. 2017. Quorum sensing controls the Pseudomonas aeruginosa CRISPR-Cas adaptive immune system. Proc Natl Acad Sci U S A. 114(1):131–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hudaiberdiev S, Choudhary KS, Vera Alvarez R, Gelencsér Z, Ligeti B, et al. 2015. Census of solo LuxR genes in prokaryotic genomes. Front Cell Infect Microbiol. 5:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hurley A, Bassler BL. 2017. Asymmetric regulation of quorum-sensing receptors drives autoinducer-specific gene expression programs in Vibrio cholerae. PLOS Genetics. 13(5):e1006826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ismail AS, Valastyan JS, Bassler BL. 2016. A Host-Produced Autoinducer-2 Mimic Activates Bacterial Quorum Sensing. Cell Host Microbe. 19(4):470–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacob-Dubuisson F, Mechaly A, Betton J-M, Antoine R. 2018. Structural insights into the signalling mechanisms of two-component systems. Nat Rev Microbiol. 16(10):585–93 [DOI] [PubMed] [Google Scholar]
- 41.Jenal U, Reinders A, Lori C. 2017. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol. 15(5):271–84 [DOI] [PubMed] [Google Scholar]
- 42.Joris PX, Smith PH, Yin TC. 1998. Coincidence detection in the auditory system: 50 years after Jeffress. Neuron. 21(6):1235–38 [DOI] [PubMed] [Google Scholar]
- 43.Jung SA, Chapman CA, Ng W-L. 2015. Quadruple quorum-sensing inputs control Vibrio cholerae virulence and maintain system robustness. PLoS Pathog. 11(4):e1004837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karatan E, Michael AJ. 2013. A wider role for polyamines in biofilm formation. Biotechnol Lett. 35(11):1715–17 [DOI] [PubMed] [Google Scholar]
- 45.Ke X, Miller LC, Bassler BL. 2015. Determinants governing ligand specificity of the Vibrio harveyi LuxN quorum-sensing receptor. Mol Microbiol. 95(1):127–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koskella B, Brockhurst MA. 2014. Bacteria–phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev. 38(5):916–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laub MT, Goulian M. 2007. Specificity in two-component signal transduction pathways. Annu Rev Genet. 41:121–45 [DOI] [PubMed] [Google Scholar]
- 48.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 118(1):69–82 [DOI] [PubMed] [Google Scholar]
- 49.Lin W, Kovacikova G, Skorupski K. 2005. Requirements for Vibrio cholerae HapR Binding and Transcriptional Repression at the hapR Promoter Are Distinct from Those at the aphA Promoter. J Bacteriol. 187(9):3013–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin W, Kovacikova G, Skorupski K. 2007. The quorum sensing regulator HapR downregulates the expression of the virulence gene transcription factor AphA in Vibrio cholerae by antagonizing Lrp- and VpsR-mediated activation. Molecular Microbiology. 64(4):953–67 [DOI] [PubMed] [Google Scholar]
- 51.McCready AR, Paczkowski JE, Henke BR, Bassler BL. 2019. Structural determinants driving homoserine lactone ligand selection in the Pseudomonas aeruginosa LasR quorum-sensing receptor. PNAS. 116(1):245–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michael B, Smith JN, Swift S, Heffron F, Ahmer BMM. 2001. SdiA of Salmonella enterica Is a LuxR Homolog That Detects Mixed Microbial Communities. Journal of Bacteriology. 183(19):5733–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller SD, Haddock SHD, Elvidge CD, Lee TF. 2005. Detection of a bioluminescent milky sea from space. PNAS. 102(40):14181–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mok KC, Wingreen NS, Bassler BL. 2003. Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. EMBO J. 22(4):870–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monds RD, Newell PD, Gross RH, O’Toole GA. 2007. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0–1 biofilm formation by controlling secretion of the adhesin LapA. Mol Microbiol. 63(3):656–79 [DOI] [PubMed] [Google Scholar]
- 56.Mukherjee S, Bassler BL. 2019. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol 17(6):371–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neiditch MB, Federle MJ, Miller ST, Bassler BL, Hughson FM. 2005. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol Cell. 18(5):507–18 [DOI] [PubMed] [Google Scholar]
- 58.Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, et al. 2006. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 126(6):1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Newell PD, Boyd CD, Sondermann H, O’Toole GA. 2011. A c-di-GMP Effector System Controls Cell Adhesion by Inside-Out Signaling and Surface Protein Cleavage. PLOS Biology. 9(2):e1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Newell PD, Monds RD, O’Toole GA. 2009. LapD is a bis-(3’,5’)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0–1. Proc Natl Acad Sci U S A. 106(9):3461–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Papenfort K, Bassler BL. 2016. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol. 14(9):576–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Papenfort K, Silpe JE, Schramma KR, Cong J-P, Seyedsayamdost MR, Bassler BL. 2017. A Vibrio cholerae autoinducer-receptor pair that controls biofilm formation. Nat. Chem. Biol 13(5):551–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park J, Dies M, Lin Y, Hormoz S, Smith-Unna SE, et al. 2018. Molecular Time Sharing through Dynamic Pulsing in Single Cells. Cell Syst. 6(2):216–229.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patel HK, Suárez-Moreno ZR, Degrassi G, Subramoni S, González JF, Venturi V. 2013. Bacterial LuxR solos have evolved to respond to different molecules including signals from plants. Front Plant Sci. 4:447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pultz IS, Christen M, Don Kulasekara H, Kennard A, Kulasekara B, Miller SI. 2012. The response threshold of Salmonella PilZ domain proteins is determined by their binding affinities for c-di-GMP. Mol Microbiol. 86(6):1424–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rahbari KM, Chang JC, Federle MJ. 2021. A Streptococcus Quorum Sensing System Enables Suppression of Innate Immunity. mBio. 12(3):e03400–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roilides E, Simitsopoulou M, Katragkou A, Walsh TJ. 2015. How Biofilms Evade Host Defenses. Microbiol Spectr. 3(3): [DOI] [PubMed] [Google Scholar]
- 68.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the First 25 Years of a Universal Bacterial Second Messenger. Microbiol Mol Biol Rev. 77(1):1–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roome CJ, Kuhn B. 2020. Dendritic coincidence detection in Purkinje neurons of awake mice. eLife. 9:e59619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rowland MA, Deeds EJ. 2014. Crosstalk and the evolution of specificity in two-component signaling. Proc Natl Acad Sci U S A. 111(15):5550–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rutherford ST, van Kessel JC, Shao Y, Bassler BL. 2011. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev. 25(4):397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schauder S, Shokat K, Surette MG, Bassler BL. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Molecular Microbiology. 41(2):463–76 [DOI] [PubMed] [Google Scholar]
- 73.Shah M, Taylor VL, Bona D, Tsao Y, Stanley SY, et al. 2021. A phage-encoded anti-activator inhibits quorum sensing in Pseudomonas aeruginosa. Mol Cell. 81(3):571–583.e6 [DOI] [PubMed] [Google Scholar]
- 74.Shikuma NJ, Fong JCN, Odell LS, Perchuk BS, Laub MT, Yildiz FH. 2009. Overexpression of VpsS, a hybrid sensor kinase, enhances biofilm formation in Vibrio cholerae. J Bacteriol. 191(16):5147–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Silpe JE, Bassler BL. 2019. A Host-Produced Quorum-Sensing Autoinducer Controls a Phage Lysis-Lysogeny Decision. Cell. 176(1–2):268–280.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Silpe JE, Bassler BL. 2019. Phage-Encoded LuxR-Type Receptors Responsive to Host-Produced Bacterial Quorum-Sensing Autoinducers. mBio. 10(2): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Silpe JE, Bridges AA, Huang X, Coronado DR, Duddy OP, Bassler BL. 2020. Separating Functions of the Phage-Encoded Quorum-Sensing-Activated Antirepressor Qtip. Cell Host & Microbe. 27(4):629–641.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sobe RC, Bond WG, Wotanis CK, Zayner JP, Burriss MA, et al. 2017. Spermine inhibits Vibrio cholerae biofilm formation through the NspS–MbaA polyamine signaling system. Journal of Biological Chemistry. 292(41):17025–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Srivastava D, Harris RC, Waters CM. 2011. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J Bacteriol. 193(22):6331–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Srivastava SK, Rajasree K, Fasim A, Arakere G, Gopal B. 2014. Influence of the AgrC-AgrA Complex on the Response Time of Staphylococcus aureus Quorum Sensing. J Bacteriol. 196(15):2876–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stevens AM, Fujita N, Ishihama A, Greenberg EP. 1999. Involvement of the RNA polymerase alpha-subunit C-terminal domain in LuxR-dependent activation of the Vibrio fischeri luminescence genes. J Bacteriol. 181(15):4704–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Subramoni S, Florez Salcedo DV, Suarez-Moreno ZR. 2015. A bioinformatic survey of distribution, conservation, and probable functions of LuxR solo regulators in bacteria. Frontiers in Cellular and Infection Microbiology. 5:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Svenningsen SL, Waters CM, Bassler BL. 2008. A negative feedback loop involving small RNAs accelerates Vibrio cholerae’s transition out of quorum-sensing mode. Genes Dev. 22(2):226–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Swem LR, Swem DL, Wingreen NS, Bassler BL. 2008. Deducing receptor signaling parameters from in vivo analysis: LuxN/AI-1 quorum sensing in Vibrio harveyi. Cell. 134(3):461–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Teng S-W, Schaffer JN, Tu KC, Mehta P, Lu W, et al. 2011. Active regulation of receptor ratios controls integration of quorum-sensing signals in Vibrio harveyi. Mol Syst Biol. 7:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tsai C-S, Winans SC. 2010. LuxR-type quorum-sensing regulators that are detached from common scents. Mol Microbiol. 77(5):1072–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tu KC, Long T, Svenningsen SL, Wingreen NS, Bassler BL. 2010. Negative Feedback Loops Involving Small Regulatory RNAs Precisely Control the Vibrio harveyi Quorum-Sensing Response. Molecular Cell. 37(4):567–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Valastyan JS, Kraml CM, Pelczer I, Ferrante T, Bassler BL. 2021. Saccharomyces cerevisiae Requires CFF1 To Produce 4-Hydroxy-5-Methylfuran-3(2H)-One, a Mimic of the Bacterial Quorum-Sensing Autoinducer AI-2. mBio. 12(2):e03303–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vannini A, Volpari C, Gargioli C, Muraglia E, Cortese R, et al. 2002. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 21(17):4393–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Venturi V, Ahmer BMM. 2015. Editorial: LuxR Solos are Becoming Major Players in Cell–Cell Communication in Bacteria. Frontiers in Cellular and Infection Microbiology. 5:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang B, Zhao A, Novick RP, Muir TW. 2014. Activation and inhibition of the receptor histidine kinase AgrC occurs through opposite helical transduction motions. Mol Cell. 53(6):929–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Watve S, Barrasso K, Jung SA, Davis KJ, Hawver LA, et al. 2020. Parallel quorum-sensing system in Vibrio cholerae prevents signal interference inside the host. PLoS Pathog. 16(2):e1008313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wellington S, Greenberg EP. Quorum Sensing Signal Selectivity and the Potential for Interspecies Cross Talk. mBio. 10(2):e00146–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu L, Luo Y. 2021. Bacterial Quorum-Sensing Systems and Their Role in Intestinal Bacteria-Host Crosstalk. Frontiers in Microbiology. 12:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Young EC, Baumgartner JT, Karatan E, Kuhn ML. 2021. A mutagenic screen reveals NspS residues important for regulation of Vibrio cholerae biofilm formation. Microbiology (Reading) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang L, Li S, Liu X, Wang Z, Jiang M, et al. 2020. Sensing of autoinducer-2 by functionally distinct receptors in prokaryotes. Nat Commun. 11(1):5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang R, Pappas KM, Pappas T, Brace JL, Miller PC, et al. 2002. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature. 417(6892):971–74 [DOI] [PubMed] [Google Scholar]
- 98.Zschiedrich CP, Keidel V, Szurmant H. 2016. Molecular mechanisms of two-component signal transduction. J Mol Biol. 428(19):3752–75 [DOI] [PMC free article] [PubMed] [Google Scholar]


