Abstract
Background:
The impact of breastfeeding on certain childhood respiratory illnesses remains controversial.
Objective:
To examine the effect of exclusive breastfeeding on the early-life upper respiratory tract (URT) and gut microbiome, the URT immune response in infancy, and the risk of common pediatric respiratory diseases.
Methods:
We analyzed data from a birth cohort of healthy infants with prospective ascertainment of breastfeeding patterns and common pediatric pulmonary and atopic outcomes. In a subset of infants, we also characterized the URT and gut microbiome using 16S ribosomal RNA sequencing and measured 9 URT cytokines using magnetic bead-based assays.
Results:
Of the 1,949 infants enrolled, 1,495 (76.71%) had 4-year data. In adjusted analyses, exclusive breastfeeding 1) had an inverse dose-response on the α-diversity of the early-life URT and gut microbiome, 2) was positively associated with the URT levels of IFN-α, IFN-γ, and IL-17A in infancy, and 3) had a protective dose-response on the development of a lower respiratory tract infection (LRTI) in infancy, 4-year current asthma, and 4-year ever allergic rhinitis (AR) (OR [95% CI] for each 4 weeks of exclusive breastfeeding = 0.95 [0.91–0.99], 0.95 [0.90–0.99], and 0.95 [0.92–0.99], respectively). In exploratory analyses, we also found that the protective association of exclusive breastfeeding on 4-year current asthma was mediated through its impact in the gut microbiome (p=0.03).
Conclusions:
Our results support a protective causal role of exclusive breastfeeding on the risk of developing an LRTI in infancy and asthma and AR in childhood. They also shed light on potential mechanisms of these associations, including the effect of exclusive breastfeeding on the gut microbiome.
Keywords: 16S rRNA sequencing, airway, allergic rhinitis, asthma, breastfeeding, bronchiolitis, cytokines, food sensitization, gut, immune response, infancy, lower respiratory tract infection, microbiome, nasopharynx, respiratory
Capsule Summary:
Our results support a protective causal role of exclusive breastfeeding on the risk of developing a lower respiratory tract infection in infancy and asthma and allergic rhinitis in childhood. They also shed light on potential mechanisms of these associations, including the effect of exclusive breastfeeding on the gut microbiome.
Introduction:
Breastfeeding has numerous advantages for the infant, the mother, and society; including improving nutritional, neurocognitive, and socioeconomic outcomes.1, 2 Whereas most studies have also shown a protective effect of breastfeeding on lower respiratory tract infections (LRTIs) in early life,3 those examining the association of breastfeeding on the onset of other common childhood pulmonary and atopic outcomes (such as food sensitization, asthma, or allergic rhinitis [AR]) have yielded conflicting results, including reports of detrimental effects.4–8 In addition, the interpretation of available data has been difficult, as many of the prior studies in this field have not taken into account the duration or exclusivity of breastfeeding, have not examined the presence of dose-response associations, have had a cross-sectional or retrospective design, have not addressed the possibility of reverse causation, or have not controlled for key confounders of these associations.7–10 Furthermore, our understanding on whether breastfeeding is associated with other important determinants of pediatric respiratory health (such as the developing microbiome or immune response) is still limited.
To address the above limitations, we examined the effect of exclusive breastfeeding on 1) the upper respiratory tract (URT) microbiome, gut microbiome, and URT immune response in infancy, and 2) the development of common childhood respiratory and atopic outcomes (including an LRTI in infancy, one-year food sensitization, and childhood asthma and AR) in a large (n=1,949), population-based, birth cohort of healthy infants with careful characterization of breastfeeding patterns, close longitudinal follow-up, and detailed assessment of multiple confounders of these associations. Using a causal mediation framework for multidimensional data, we further explored whether the effect of exclusive breastfeeding on clinical outcomes is mediated by the early-life URT microbiome, gut microbiome, or URT immune response (eFigure 1).
Methods:
Full details are available in the eMethods section of the Online Supplement.
Overview of the Study Population and Design
This was a sub-study of the Infant Susceptibility to Pulmonary Infections and Asthma Following Respiratory Syncytial Virus Exposure study (INSPIRE) (n=1,949).11 Eligible infants were healthy, term, and enrolled near birth (eTable 1). For those infants enrolled during the 2nd year of the study (n=1,088), the enrollment visit included the collection of nasal filters and stool samples.11–13 To capture all acute respiratory infections during each infant’s first winter viral season, we conducted intensive passive and active surveillance during November to March of their first year, which included in-person respiratory illness visits and viral testing for those meeting pre-specified criteria (see the eMethods section of the Online Supplement). Yearly follow-up for the development of common pediatric respiratory diseases is ongoing. The Institutional Review Board of Vanderbilt University approved this study and parents provided written informed consent. The detailed methods for INSPIRE have been previously reported.11
Definition of Exposures and Outcomes
Our main exposures were parental report of type of feeding at enrollment (exclusive breast milk, combined, or exclusive formula) and the duration of exclusive breastfeeding (in weeks).14 To make our results comparable to those of prior studies in this field, we also created different categorical variables taking into account the infant’s age and breastfeeding pattern. These included: 1) ever any breastfeeding (yes vs. no), 2) current any breastfeeding at enrollment (yes vs. no), 3) current any breastfeeding at age one year (yes vs. no), and 4) type of milk at age one year (only breast milk, some breast milk, or no breast milk).
Our initial outcomes included several metrics of the URT and gut microbiome, as well as the levels of 9 a priori selected URT cytokines, which were all assessed in the nasal filters or stool samples collected at enrollment from a subset of infants. The URT cytokines were selected based on their known anti-viral, pro-inflammatory, or pro-allergic functions. These included: IFN-α, IFN-γ, IL-4, IL-5, IL-13, IL-17A, IL-33, TNF-α, and TSLP. Our clinical outcomes included the development of an LRTI in infancy, one-year food sensitization, 4-year current asthma, and 4-year ever AR, which were ascertained using common epidemiological definitions (see the eMethods section of the Online Supplement).
Characterization of the Early-life Microbiome and Upper Respiratory Tract Immune Responses
The methods used to characterize the URT and gut microbiome and to measure URT cytokines in infants enrolled in INSPIRE have been previously described.12, 13, 15–18 In brief, we amplified the V4 region of the 16S ribosomal RNA (rRNA) gene, sequenced the libraries on an Illumina MiSeq platform, and grouped sequences using an amplicon sequence variant-based pipeline.19–21 To measure the levels of the 9 a priori selected cytokines in the nasal filters, we used multiplex magnetic bead-based assays and ultimately processed the data using the method described by Won et al,22 which uses median fluorescence intensities (MFIs) of individual beads instead of the usual standard curve-based data-processing method to increase the sensitivity and accuracy of high-throughput immunoassays.17, 22–24
Statistical Analyses
The framework for our statistical analyses is shown in eFigure 1. These were all performed using SAS and R.25
For the statistical analyses of the URT and gut microbiome, we first rarefied the processed datasets to the lowest library size of all samples (n=1,019 for nasal filters and n=1,179 for stool samples) and calculated common α-diversity (observed taxa, exponentiated Shannon, and inverse Simpson indices) and β-diversity (Bray-Curtis dissimilarity) metrics at the amplicon sequence variant level. To compare α-diversity metrics between groups, we used Kruskal-Wallis tests and linear regression. To evaluate the differences in β-diversity between groups, we used non-metric-multidimensional scaling and permutational multivariate analysis of variance.26 Next, using a non-rarefied processed dataset, we used the DESeq2 method to compare the abundance of genera between groups.27, 28 The p-values were corrected for multiple comparisons using the Benjamini-Hochberg method (q-values).29
The URT cytokines and clinical outcomes were first compared between groups using Mann–Whitney U, Pearson chi-squared, or Kruskal-Wallis tests, as appropriate. We then used linear or logistic regression, as applicable, to obtain estimates of the associations. For the statistical analyses of the type of feeding at enrollment with the clinical outcomes, we also conducted sensitivity analyses using propensity score (PS) matching with a 1:1 nearest neighbor algorithm and no replacement within a pre-specified caliper distance (0.2) to address the possibility of residual confounding by balancing covariates between the exclusive breast milk and exclusive formula groups. To assess whether the matching had been successful, we calculated standardized mean differences between groups. Last, we assessed the association of exclusive breastfeeding with each of the clinical outcomes in the PS-matched dataset using logistic regression.30, 31
Based on our results and to investigate the pathways through which exclusive breastfeeding exerts an effect on pediatric respiratory health, we also conducted exploratory analyses by building causal mediation models that included exclusive breastfeeding as the exposure, either the early-life URT or gut microbiome β-diversity as the mediator, and each of the clinical outcomes as the outcome. We then tested the global hypothesis that the overall structure of these microbial communities mediate the association between the exposure and the outcome using multivariate omnibus distance mediation analyses.32 Likewise, we built similar models including the levels of URT cytokines in infancy as separate and joint mediators (eFigure 1).
For all statistical analyses, we created unadjusted and adjusted models including covariates selected based on available literature.4, 8–10 For the association of the duration of exclusive breastfeeding with the clinical outcomes, we also tested the presence of interactions with selected covariates in the models and built supplementary models by including additional covariates. The URT cytokines were all generalized-log-transformed prior to statistical analyses.17 Statistical significance was defined as a p- or q-value ≤0.05.
Results:
Participants Characteristics and Follow-up
The median (interquartile range [IQR]) age at enrollment was 55 (16–78) days and the majority of infants were males, White non-Hispanic, and born by vaginal delivery. There were multiple differences in baseline characteristics of infants by the type of feeding at enrollment (Table 1).
Table 1.
| Baseline characteristic | All (n=1,949) | Type of feeding at enrollment | ||
|---|---|---|---|---|
| Exclusive breast milk (n=667) | Combined (n=353) | Exclusive formula (n=929) | ||
| Age at enrollment (days) | 55 (16–78) | 27 (14–65) | 28 (14–64) | 64 (26–115)‡ |
| Female sex | 929 (47.67%) | 327 (49.03%) | 172 (48. 73%) | 430 (46.29%) |
| Race or ethnicity | ||||
| Black non-Hispanic | 343 (17.60%) | 65 (9.75%) | 70 (19.83%) | 208 (22.39%)‡ |
| White non-Hispanic | 1,269 (65.11%) | 513 (76.91%) | 201 (56.94%) | 555 (59.74%) |
| Hispanic | 171 (8.77%) | 46 (6.90%) | 51 (14.45%) | 74 (7.97%) |
| Other | 166 (8.52%) | 43 (6.45%) | 31 (8.78%) | 92 (9.90%) |
| Gestational age (weeks) | 39 (39–40) | 39 (39–40) | 39 (39–40) | 39 (38–40)‡ |
| Birth weight (grams) | 3,405 (3,120–3,740) | 3,490 (3,206–3,802) | 3,433 (3,093–3,717) | 3,348 (3,064–3,660)‡ |
| Birth by cesarean section | 612 (31.40%) | 180 (26.99%) | 116 (32.86%) | 316 (34.02%)‡ |
| Daycare attendance at enrollment | 590 (33.56%) | 253 (41.21%) | 101 (31.46%) | 236 (26.68%)‡ |
| Presence of another child age ≤6 years living in the same home at enrollment | 985 (50.54%) | 357 (53.52%) | 146 (41.36%) | 482 (51.88%)‡ |
| Exposure to tobacco smoke in utero or prior to enrollment | 426 (21.89%) | 50 (7.51%) | 56 (15.85%) | 320 (34.52%)‡ |
| Exposure to antibiotics in utero or prior to enrollment | 964 (49.46%) | 288 (43.18%) | 180 (50.99%) | 496 (53.39%)‡ |
| Maternal asthma | 379 (19.46%) | 108 (16.19%) | 61 (17.28%) | 210 (22.63%)‡ |
| Type of insurance at enrollment | ||||
| Federal or state | 1,061 (54.44%) | 213 (31.93%) | 190 (53.82%) | 658 (70.83%)‡ |
| Private | 873 (44.79%) | 450 (67.47%) | 161 (45.61%) | 262 (28.20%) |
| Other | 15 (0.77%) | 4 (0.60%) | 2 (0.57%) | 9 (0.97%) |
Data presented as median (interquartile range) for continuous variables or number (%) for categorical variables. The total sample size of each group (n) is also shown.
Data calculated for infants with complete data.
p-value ≤0.05 for the comparison between groups using Kruskal-Wallis or Pearson chi-squared tests, as appropriate.
Of the 1,949 infants enrolled, 1,495 (76.71%) had 4-year data available. Children without 4-year data had a higher birth weight and were more likely to have been exposed to tobacco smoke and to have private insurance at enrollment than those with 4-year data (eTable 2).
Exclusive Breastfeeding and the Early-life Upper Respiratory Tract and Gut Microbiome
Four hundred fifty-nine infants had nasal filters available for the characterization of the URT microbiome and 445 (96.95%) of these were included in the statistical analyses after quality control. Six hundred and forty-seven infants had stool samples available for the characterization of the gut microbiome and 637 (98.45%) of these were included in the statistical analyses after quality control. The median (IQR) high-quality sequence count per nasal filter and stool sample was 13,506 (8,026–20,065) and 8,685 (6,565–11,611), respectively. There were minimal differences between infants included and not included in the statistical analyses of the URT and gut microbiome (eTables 3–4).
There was a dose-response effect of the type of feeding at enrollment on the α-diversity (richness ± evenness) of both the URT and gut microbiome, with infants who were exclusively breastfed at enrollment having the lowest values for all α-diversity metrics examined in both unadjusted and adjusted analyses (Figure 1 and eTables 6–7). The β-diversity (overall structure) of the URT and gut microbiome also differed by type of feeding at enrollment in both unadjusted and adjusted analyses (p<0.01 for all comparisons) (eFigure 2). Likewise, there were multiple differences in the abundance of URT and gut genera by the type of feeding at enrollment in both unadjusted and adjusted analyses, with 5 URT genera (including Granulicatella, Gemella, Acinetobacter, Streptococcus, Corynebacterium 1) and 3 gut genera (including Escherichia-Shigella, Bifidobacterium, and Clostridium innoccuum group) being differentially abundant in adjusted analyses (eTables 8–9), several of which appeared to have a dose-response association (Figure 2). Including other covariates in these adjusted models (e.g., tobacco smoke exposure in utero or prior to enrollment, daycare attendance, or type of insurance) did not change our results (data not shown).
Figure 1:
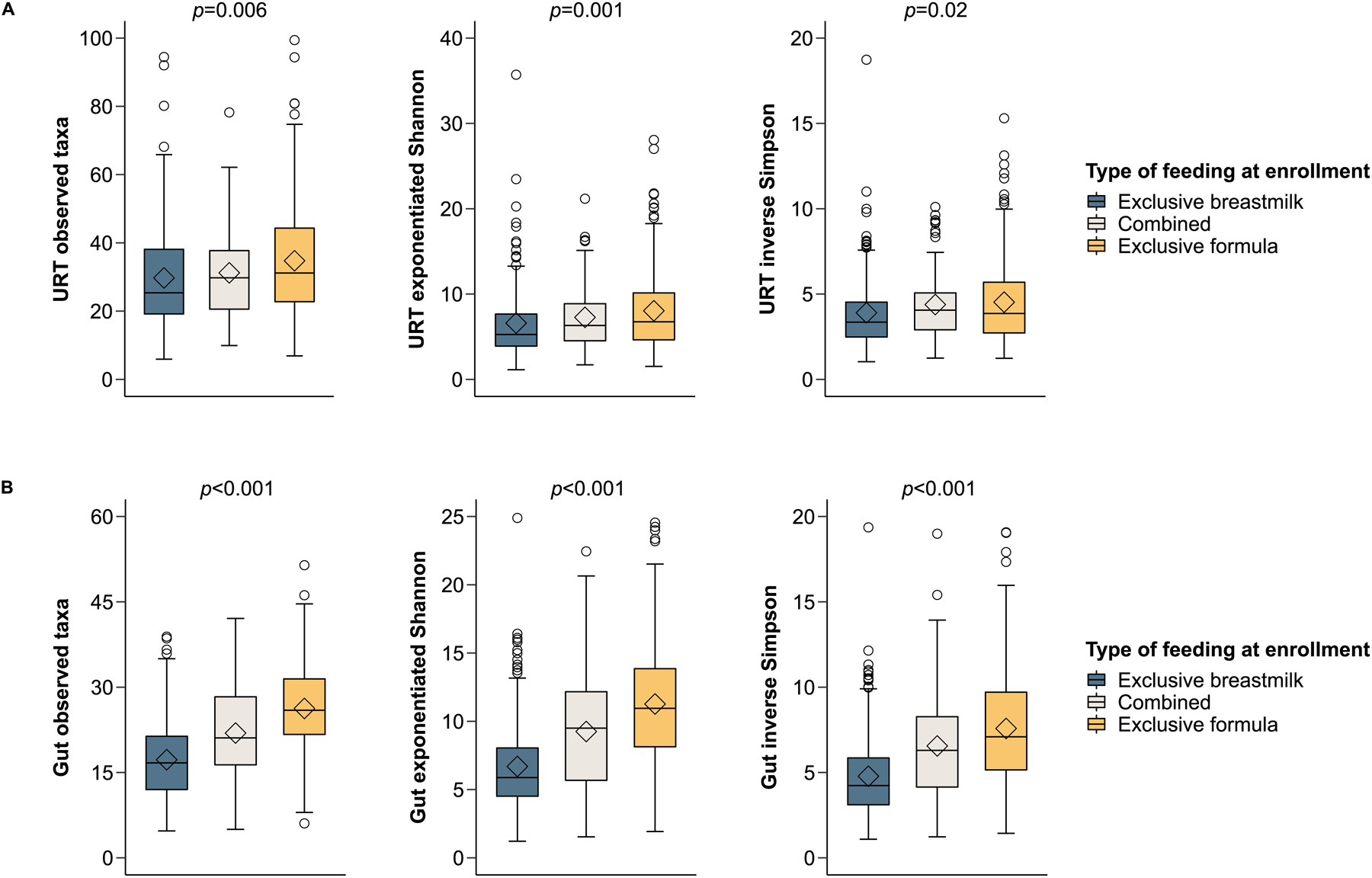
The association of type of feeding with the α-diversity of the upper respiratory tract (URT) (1A) and gut (1B) microbiome in infancy. The box-and-whisker plots show the mean (diamond), median (middle bar), 1st quartile (lower bar), 3rd quartile (upper bar), minimum observation above the lowest fence (lower whisker), maximum observation below the upper fence (upper whisker), and outliers (circles) of common α-diversity indices by type of feeding (all assessed at enrollment near birth [median age (interquartile range) = 55 (16–78) days in all infants enrolled]). The p-values for the comparison between groups using Kruskal-Wallis tests are also shown.
Figure 2:
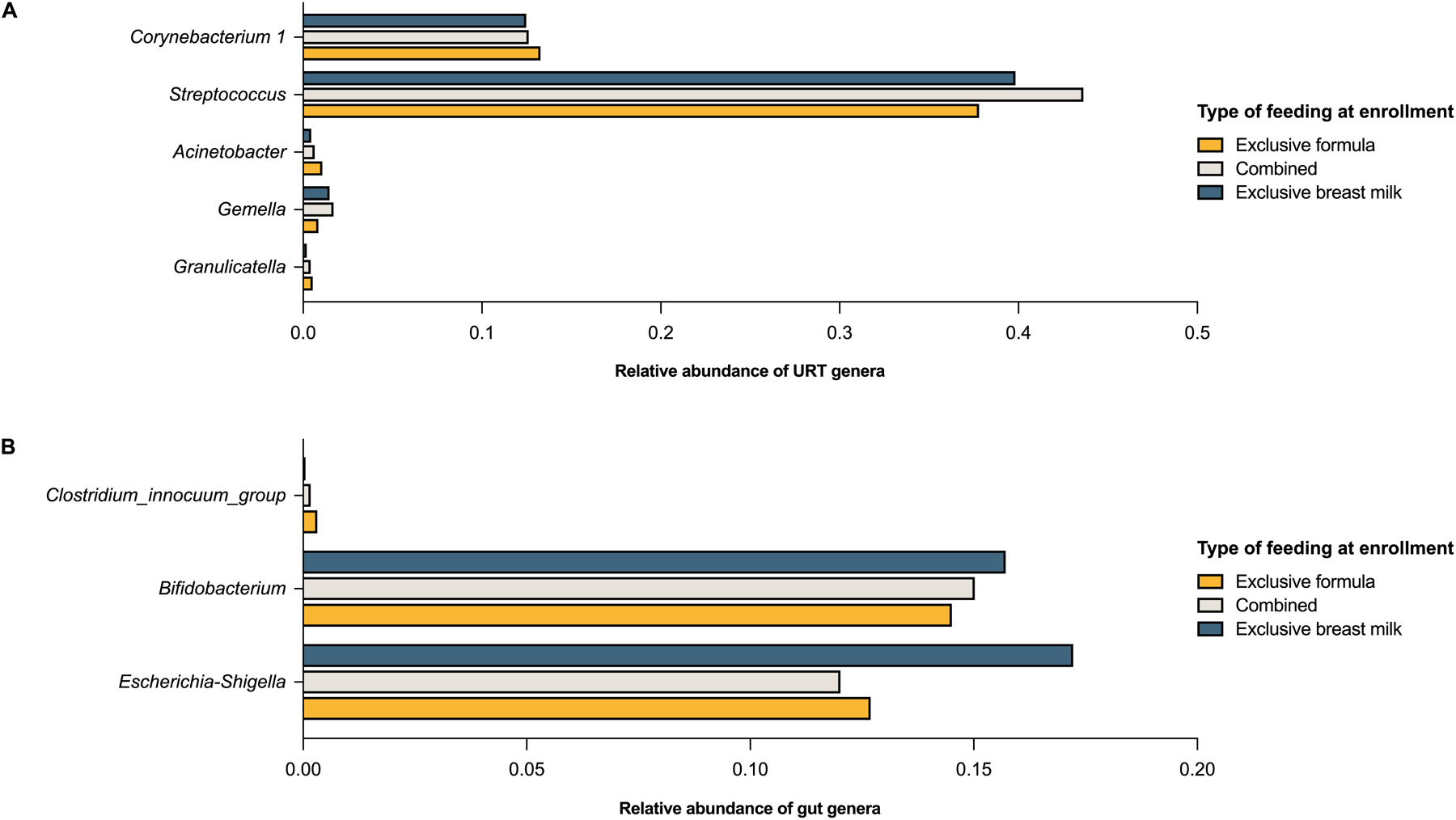
The association of type of feeding with the abundance of genera of the upper respiratory tract (URT) (2A) and gut (2B) microbiome in infancy. The bar graphs show the mean relative abundance of each genus by type of feeding (all assessed at enrollment near birth [median age (interquartile range) = 55 (16–78) days in all infants enrolled]). Only genera with a q-value ≤0.05 in DESeq2 models including the infant’s age at enrollment, mode of delivery, and exposure to antibiotics in utero or prior to enrollment as covariates are shown.
Exclusive Breastfeeding and the Upper Respiratory Tract Immune Response in Infancy
Four hundred and forty-two infants had nasal filters available for the measurement of URT cytokines and all of these were included in the statistical analyses of at least one cytokine after quality control. There were minimal differences between infants included and not included in the statistical analyses of the URT immune response (eTable 5).
In unadjusted analyses, the type of breastfeeding was associated with the levels of antiviral and pro-inflammatory cytokines (IFN-α, IFN-γ, TNF-α, and IL-17A), but not with levels of pro-allergic cytokines (IL-4, IL-5, IL-13, IL-33, or TSLP) (eTable 10). With the exception of TNF-α, these associations remained significant in adjusted analyses, with exclusive breastfeeding at enrollment being associated with higher URT levels of IFN-α, IFN-γ, and IL-17A (Figure 3 and eTable 11).
Figure 3:
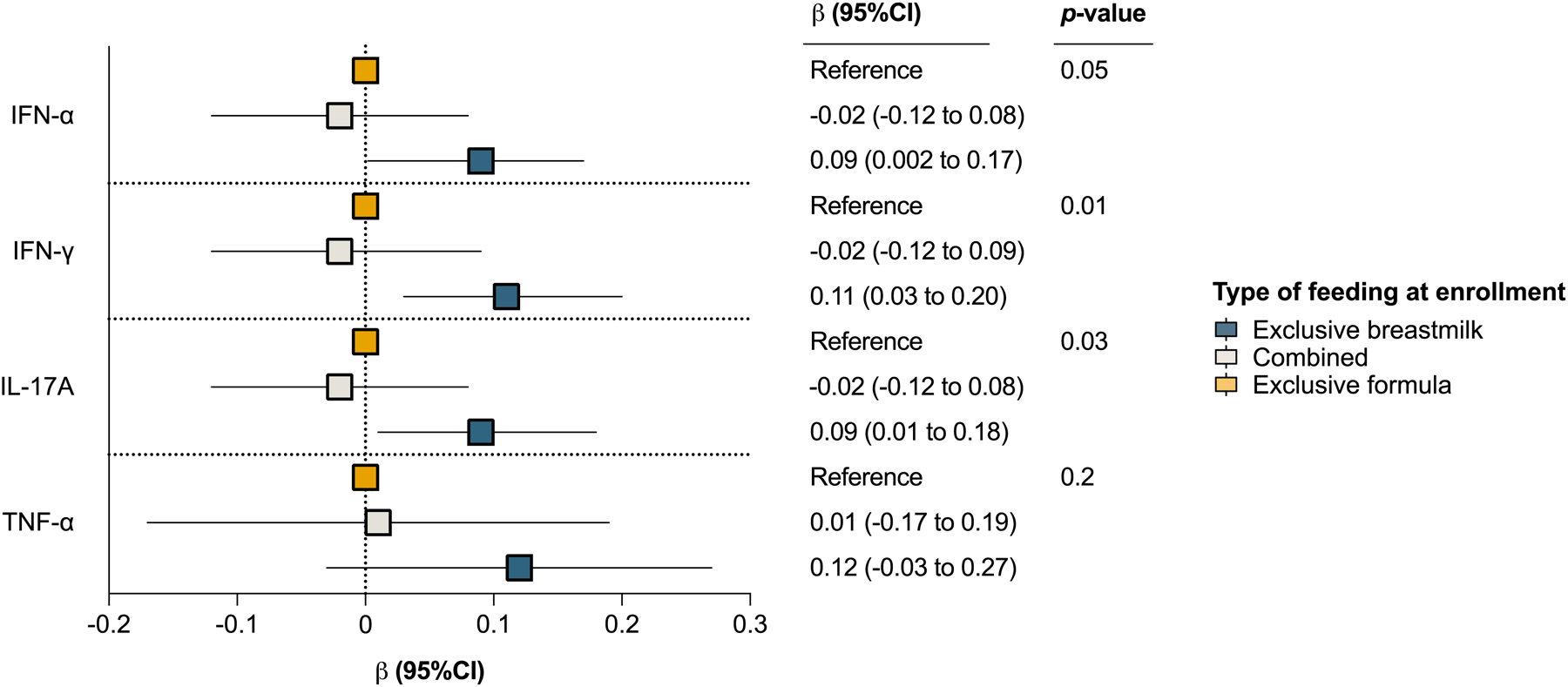
The association of type of feeding with the median fluorescence intensity (MFI) of upper respiratory tract cytokines (URT) in infancy. The plot estimates were obtained from linear regression models including the infant’s age at enrollment, sex, and maternal asthma as covariates. The β coefficient (squares), corresponding 95% CI (adjacent lines), and p-value for each type of feeding are shown for each model. Exclusive formula was used as the reference category in all models and the URT cytokine MFIs were generalized log transformed prior to analyses (all assessed at enrollment near birth [median age (interquartile range) = 55 (16–78) days in all infants enrolled]). Only URT cytokines with a q-value ≤0.05 in unadjusted analyses are shown.
Exclusive Breastfeeding and Common Childhood Respiratory Illnesses
The median (IQR) duration of exclusive breastfeeding in the 1,949 infants enrolled was 6 (0–20) weeks. The number (%) of children with an LRTI in infancy, one-year food sensitization, 4-year current asthma, and 4-year ever AR were 440 (22.58%), 209 (10.72%), 286 (14.67%), and 516 (26.48%), respectively.
There was a dose-response effect of the type of feeding at enrollment on an LRTI in infancy and 4-year current asthma in both unadjusted and adjusted analyses, with infants who were exclusively breastfed at enrollment having the lowest odds of developing these clinical outcomes (eFigures 3–4 and Figure 4). There was no association of the type of feeding at enrollment with one-year food sensitization or 4-year ever AR in unadjusted or adjusted analyses (eFigures 5–7 and Figure 4).
Figure 4:
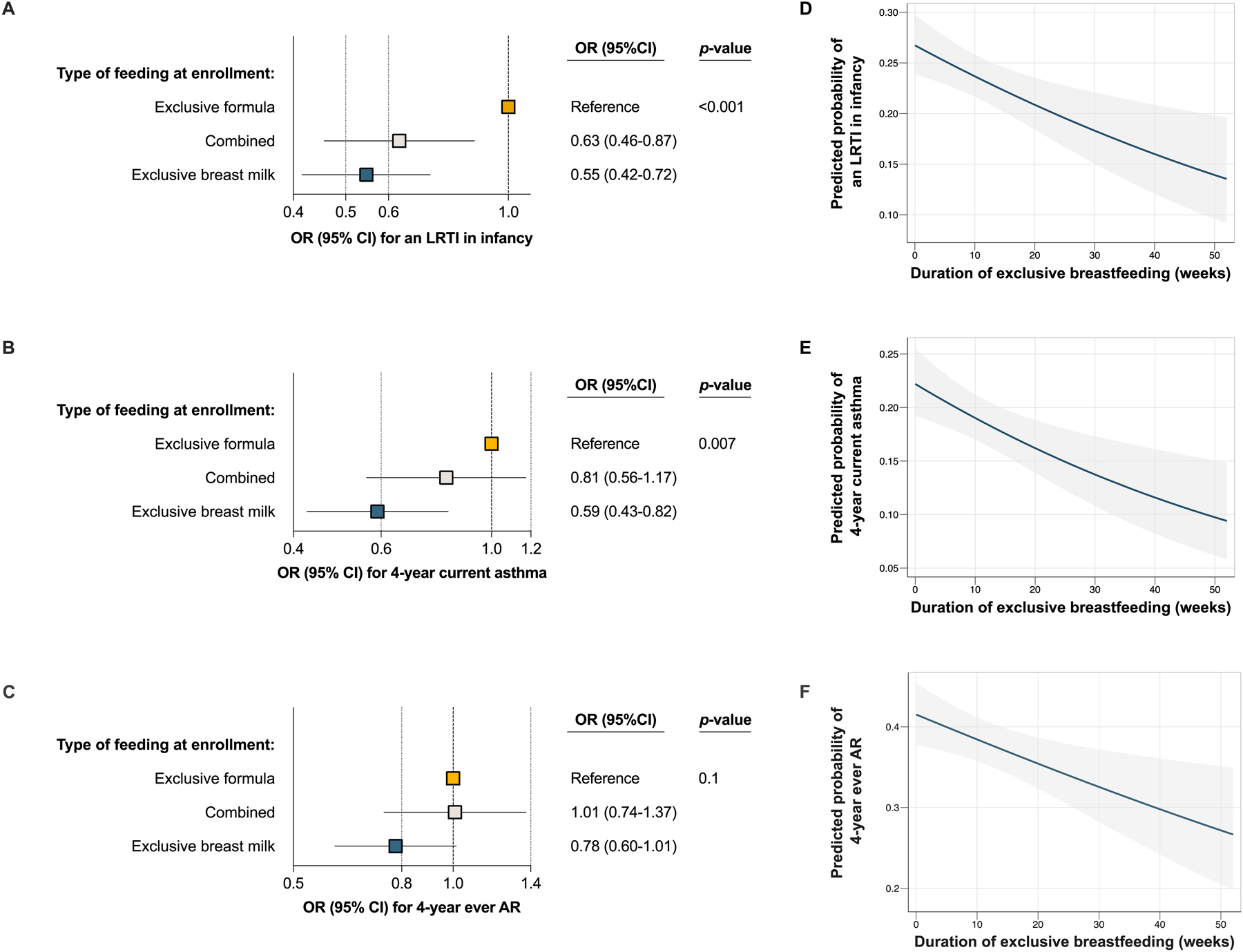
The association of type of feeding (4A-C) and of the duration of exclusive breastfeeding (4D-F) with a lower respiratory tract infection (LRTI) in infancy, 4-year current asthma, and 4-year ever allergic rhinitis (AR). 4A-C: The plot estimates were obtained from logistic regression models including the infant’s sex, race and ethnicity, mode of delivery, daycare attendance at enrollment, maternal asthma, exposure to tobacco smoke in utero or prior to enrollment, and (for the outcome of 4-year current asthma) exposure to antibiotics in utero or prior to enrollment as covariates. Type of feeding was assessed at enrollment near birth [median age (interquartile range) = 55 (16–78) days in all infants enrolled). The OR (squares), corresponding 95% CI (adjacent lines), and p-value are shown for each model. 4D-F: The solid blue lines represent the predicted probability values of developing the clinical outcome by the duration of exclusive breastfeeding. The shaded gray bands indicate the corresponding lower and upper 95% CIs. The plot estimates were obtained from unadjusted logistic regression models.
The duration of exclusive breastfeeding had a protective dose-response effect of on an LRTI in infancy, 4-year current asthma, and 4-year ever AR in both unadjusted and adjusted analyses (eFigure 8, Figure 4, and eTables 12–14). In adjusted models, each 4 weeks of exclusive breastfeeding decreased the odds of an LRTI in infancy, 4-year current asthma, and 4-year ever AR by ~5% (OR=0.95, 95% CI=0.91–0.99, p=0.03; OR=0.95, 95% CI=0.90–0.99, p=0.03; and OR=0.95, 95% CI=0.92–0.99, p=0.02; respectively). We obtained similar estimates in adjusted models that included additional covariates (eTables 12–14). There were no significant interactions between the duration of exclusive breastfeeding and infant’s sex, race or ethnicity, or maternal asthma in any of these models (p≥0.2 for all interaction terms) (eTable 15). The duration of exclusive breastfeeding was associated with one-year food sensitization in unadjusted analyses (eFigure 8), but this association did not remain significant in adjusted models (eTable 16).
Overall, we found similar results when examining the effects of different categorical variables that took into account the infant’s age and breastfeeding pattern on the clinical outcomes in both unadjusted and adjusted analyses (eFigures 3–7).
Through PS matching, we were able to balance all the baseline characteristics between infants who were exclusively breastfed and those who were exclusively formula fed at enrollment (eTables 17–19). Using the PS-matched datasets, exclusively breastfed infants also had similar lower odds of an LRTI in infancy, 4-year current asthma, and 4-year ever AR in both unadjusted and adjusted analyses, although the association of exclusive breastfeeding and 4-year ever AR was only borderline significant (eTable 20).
The type of feeding at enrollment remained associated with the development of an LRTI in infancy after including the respiratory virus as a covariate in adjusted models performed in the subset of infants with at least one in-person respiratory illness visit (n=1,047) (eTable 21). The interaction between the type of feeding at enrollment and the viral etiology was not significant in this model (p= 0.8 for the interaction term).
In exploratory analyses, there was a significant mediating effect of the β-diversity of the gut microbiome, but not of that of the URT microbiome, on the association of exclusive breastfeeding on 4-year current asthma (eTable 22). There were no mediating effects of the β-diversity of the early-life URT or gut microbiome β-diversity on the associations of exclusive breastfeeding with other clinical outcomes (p>0.05 for all mediation effects). There were also no mediating effects of the URT cytokines in infancy on the association of exclusive breastfeeding with any of the clinical outcomes (p>0.05 for all mediation effects).
Discussion:
Using a combination of prospective data from a population-based birth cohort, multiple statistical approaches to account for the possibility of confounding or bias, and next-generation sequencing, we found that exclusive breastfeeding 1) has an inverse dose-response on the α-diversity of on 2 different microbial communities in infancy (the URT and gut microbiome), 2) is associated with antiviral and pro-inflammatory URT immune responses in infancy, and 3) has a protective dose-response on the risk of developing an LRTI in infancy and asthma and AR in childhood. Furthermore, by using novel bioinformatics and causal mediation modeling of multidimensional data, our results also suggest that the gut microbiome is an important mediator of the effect of exclusive breastfeeding on childhood asthma. Our findings of a protective dose-response effect of exclusive breastfeeding on the risk of our multiple outcomes suggest a causal effect and support adding important benefits of breastfeeding on childhood respiratory health to the list of its multiple non-respiratory benefits when educating families.1, 2 Our study was not designed to identify specific thresholds of different types of breastfeeding patterns that could exert a protective effect on common childhood respiratory illnesses. However, our results suggest that whereas longer durations of exclusive breastfeeding likely offer the highest protection, even non-exclusive breastfeeding during the first few months could offer some benefit.
Few other studies have found a similar dose-response effect of exclusive breastfeeding on the gut microbiome (also demonstrating a lower gut microbiome α-diversity in exclusively breastfed infants), but —to our knowledge— this has not been shown for the URT microbiome.33
Biesbroek et al found exclusively breastfed infants to have a lower nasal α-diversity and a different β-diversity than exclusively formula-fed infants, which is comparable to what we found.34 Yet, their results for the comparisons of α-diversity and abundance of URT taxa between these 2 groups differed from ours. For example, the authors found exclusive breastfeeding to be associated with lower nasal abundance of Gemella and a higher nasal abundance of Corynebacterium, while we found the opposite. The study by Biesbroek et al was smaller, had a retrospective design, did not include infants receiving both breast milk and formula, utilized a different 16S rRNA region and bioinformatic pipeline, and used a less stringent false discovery rate, which may explain the discrepant results. These factors could also explain the dissimilarities between our results and those from other studies examining the effect of exclusive breastfeeding on the gut microbiome, several of which have found the abundances of Bacteroides, Eubacterium, Veillonella, and Megasphaera to be lower in exclusively breastfed infants,33 whereas we did not find the abundance of these genus to differ by exclusive breastfeeding. However, our results are consistent with multiple other studies that have found Bifidobacterium to be a predominant taxon of both human breast milk and the early-life gut microbiome, an important determinant of homeostasis and immunity in the infant gut, and a protective factor for the development of atopic diseases (including childhood asthma).35–37
The literature demonstrating that exclusive breastfeeding protects against the incidence or prevalence of early-life LRTIs is extensive, although most prior studies have been conducted in low-to-medium-income countries and it has been suggested that many of these studies (particularly those in high-income countries) may have overestimated their results.2, 3 This is in part due to the fact that many of the prior studies in this field have not addressed the possibility of reverse causation, as an LRTI in infancy may lead to the modification or interruption of a mother’s breastfeeding patterns. However, we still found strong protective effects of exclusive breastfeeding on an LRTI in infancy when only including type of feeding data captured at enrollment, when a minority of infants had developed an LRTI (n=80 [4.10%]). Excluding those infants from statistical analyses did not change our results, which also makes reverse causation unlikely (data not shown).
To date, studies examining whether particular breastfeeding patterns are associated with the onset of food sensitization, asthma, or AR in childhood have yielded conflicting results, with some of these even showing a detrimental effect of breastfeeding on these outcomes.38–43 The results from published meta-analyses have also been difficult to interpret,7, 8 as many of the prior studies have lacked a detailed ascertainment of the duration or exclusivity of breastfeeding, have been at risk of recall bias due to their non-prospective study design, have only used categorical breastfeeding variables, have inadequately adjusted for confounders, or have used outcome definitions that did not include objective data.4–10 Our study addresses all of these limitations. Limiting the effect of confounders has been especially problematic in this field, as breastfeeding is more common in mothers of certain backgrounds.2, 10 To address this, we 1) created primary and secondary models adjusting for multiple confounders of the association identified based on available literature,4, 8–10 and 2) conducted sensitivity analyses using PS matching to balance covariates between groups. Our results remained similar across the different statistical analyses, which suggests that confounding by measured covariates is unlikely.
The pathways through which exclusive breastfeeding exerts a beneficial effect on pediatric respiratory health are likely multifaceted. Our results provide novel insights into these pathways by showing that the protective association of exclusive breastfeeding on childhood asthma is mediated, at least in part, by its impact in the gut microbiome. In spite of the strong effects of exclusive breastfeeding on the URT microbiome and URT cytokines, as well as on an LRTI in infancy and childhood AR, we did not find any significant effects in other causal mediation models. This could be explained by a lack of power, the fact that we only assessed these at a single point in time, or because the association of exclusive breastfeeding with these other clinical outcomes is mediated by pathways not examined in our study (such as epigenetic modification).1, 2 The development of statistical techniques for mediation analyses using microbiome data is an ongoing area of research and, due to the inherent limitations of the method we used, we were unable to identify the precise gut taxa that could be acting as mediators, so our mediation analyses should be considered exploratory.
Our study has numerous strengths, but we should acknowledge additional limitations. First, we had limited data on the specific types of formula being used and lacked data on the type of solid food initially introduced; therefore, we could not consider these in statistical analyses.44 Second, the proportion of children with missing data for some of the study outcomes (e.g., food sensitization) was relatively large, so we may have been underpowered to test some associations. In addition, there were certain differences between those included and not included in statistical analysis, which could suggest some degree of selection bias, and we did not have a replication cohort. Third, we only had data on preschool asthma (more commonly driven by respiratory viruses) and lacked data on school-aged asthma (more commonly atopic). Our results support the idea of breastfeeding being protective against both virus- and allergen-drive childhood asthma phenotypes, although likely more for the former, as we found it to have a protective effect against preschool AR (a risk factor for atopic asthma), but to only be associated with anti-viral and anti-inflammatory cytokines and not with pro-allergic cytokines. Because the burden of preschool asthma is substantial, our results are still highly relevant even if exclusive breastfeeding only protects against childhood asthma in the first few years of life. Fourth, while the results of some studies suggest a modification of the effect of breastfeeding on certain common pediatric respiratory diseases based on maternal asthma, we did not find this to be present, although we may have been underpowered to test interactions in our statistical analyses. Fifth, because standard curve-based data processing methods often lead to undetectable levels of analytes and a large proportion of missing data when measuring URT cytokines in infancy, we opted to use MFIs instead of concentrations. This is an approach that has been used and validated in several other studies.22–24 However, MFIs may only provide a relative quantification of the analyte and could be impacted by plate-to-plate variability.22 Last, despite controlling for multiple comparisons, type-1 error can impact differential abundance analyses of microbiome data. We also didn’t have serial samples for the analyses of the early-life microbiome or the immune response, so we were unable to evaluate whether the effect of exclusive breastfeeding on these other outcomes persists beyond infancy. Other studies have shown that the effect of breastfeeding on the gut microbiome persists after 6 months of age,33 but if this is true for the URT microbiome or immune response is not known.34 The first few months of life are a critical period of immune development and lung development, so it is plausible that even subtle alterations during this short period could have an important effect on the later origins of common pulmonary diseases.
In summary, our results support a protective causal role of exclusive breastfeeding on the risk of developing an LRTI in infancy and childhood asthma and AR, with dose-response effects not only on the risk of these common pediatric respiratory diseases, but also on both the early-life URT and gut microbiome. In addition, we found exclusive breastfeeding to be associated with antiviral and pro-inflammatory URT immune responses in infancy. Furthermore, our results suggest that the gut microbiome is an important mediator of the effect of exclusive breastfeeding on childhood asthma. For its respiratory and multiple non-respiratory benefits for the infant, the mother, and society, exclusive breastfeeding should continue to be strongly recommended to all families.1, 2
Supplementary Material
Key Messages:
Our results support a protective causal role of exclusive breastfeeding on the risk of developing a lower respiratory tract infection in infancy and asthma and allergic rhinitis in childhood.
In addition, they shed light on potential mechanisms of these associations, including the effect of exclusive breastfeeding on the gut microbiome.
For its respiratory and multiple non-respiratory benefits for both the infant and the mother, exclusive breastfeeding should continue to be strongly recommended to all families.
Declaration of All Sources of Funding:
This work was supported by funds from the National Institute of Allergy and Infectious Diseases (under award numbers U19AI095227, K24AI77930, HHSN272200900007C, R21AI142321, U19AI110819, R21AI154016, and R21AI149262); the National Heart, Lung, and Blood Institute (under award numbers K23HL148638, K01HL149989, T32HL087738, and R01HL146401); the Office of the Director of the National Institutes of Health (under award number UG3OD023282); the National Institute of General Medical Sciences (under award number R01GM140464); the Parker B. Francis Fellowship Program; the Vanderbilt Institute for Clinical and Translational Research (grant support from the National Center for Advancing Translational Sciences under award number UL1TR000445); the Department of Pediatrics at Vanderbilt University Medical Center (grant support from the Eunice Kennedy Shriver National Institute of Child Health and Human Development under award number K12HD087023); the Vanderbilt Building Interdisciplinary Research Careers in Women’s Health K12 program (grant support from the Eunice Kennedy Shriver National Institute of Child Health and Human Development under award number K12HD04348318); and the Vanderbilt Technologies for Advanced Genomics Core (grant support from the National Institutes of Health under award numbers UL1RR024975, P30CA68485, P30EY08126, and G20RR030956).
Declaration of Conflict of Interests:
LJA has done paid consultancies on respiratory syncytial virus (RSV) vaccines for Bavarian Nordic, Novavax, Daiichi-Sankyo, ClearPath Development Company, and Pfizer. He receives funding from Pfizer through Emory University for laboratory surveillance studies of RSV infection in adults. He is a co-inventor on several Centers for Disease Control and Prevention patents on the RSV G protein and its CX3C chemokine motif relative to immune therapy and vaccine development. He is also co-inventor on a patent filing for the use of RSV platform virus-like particles with the F and G proteins for vaccines. TVH has served on RSV-related scientific advisory boards for Pfizer and Sanofi. The other authors do not have a commercial or other associations that might pose a conflict of interest.
Abbreviations:
- AR
Allergic rhinitis
- INSPIRE
Infant Susceptibility to Pulmonary Infections and Asthma Following Respiratory Syncytial Virus Exposure study
- IQR
Interquartile range
- MFI
Median fluorescence intensity
- LRTI
Lower respiratory tract infection
- PS
Propensity score
- rRNA
Ribosomal RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Notice of Prior Presentation:
Some of the results of these studies have been previously reported in the form of an abstract at the American Thoracic Society International Conference in May 2019 (Dallas, TX).
References:
- 1.Rollins NC, Bhandari N, Hajeebhoy N, Horton S, Lutter CK, Martines JC, et al. Why invest, and what it will take to improve breastfeeding practices? Lancet 2016;387:491–504. [DOI] [PubMed] [Google Scholar]
- 2.Victora CG, Bahl R, Barros AJ, Franca GV, Horton S, Krasevec J, et al. Breastfeeding in the 21st century: epidemiology, mechanisms, and lifelong effect. Lancet 2016;387:475–90. [DOI] [PubMed] [Google Scholar]
- 3.Horta BL, Victora CG, Organization WH. Short-term effects of breastfeeding: a systematic review on the benefits of breastfeeding on diarrhoea and pneumonia mortality. 2013.
- 4.Gungor D, Nadaud P, LaPergola CC, Dreibelbis C, Wong YP, Terry N, et al. Infant milk-feeding practices and food allergies, allergic rhinitis, atopic dermatitis, and asthma throughout the life span: a systematic review. Am J Clin Nutr 2019;109:772S–99S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gdalevich M, Mimouni D, Mimouni M. Breast-feeding and the risk of bronchial asthma in childhood: a systematic review with meta-analysis of prospective studies. J Pediatr 2001;139:261–6. [DOI] [PubMed] [Google Scholar]
- 6.Brew BK, Allen CW, Toelle BG, Marks GB. Systematic review and meta-analysis investigating breast feeding and childhood wheezing illness. Paediatr Perinat Epidemiol 2011;25:507–18. [DOI] [PubMed] [Google Scholar]
- 7.Lodge CJ, Tan DJ, Lau MX, Dai X, Tham R, Lowe AJ, et al. Breastfeeding and asthma and allergies: a systematic review and meta-analysis. Acta Paediatr 2015;104:38–53. [DOI] [PubMed] [Google Scholar]
- 8.Dogaru CM, Nyffenegger D, Pescatore AM, Spycher BD, Kuehni CE. Breastfeeding and childhood asthma: systematic review and meta-analysis. Am J Epidemiol 2014;179:1153–67. [DOI] [PubMed] [Google Scholar]
- 9.Kramer MS. Invited commentary: Does breastfeeding protect against “asthma”? Am J Epidemiol 2014;179:1168–70. [DOI] [PubMed] [Google Scholar]
- 10.Kramer MS. Does breast feeding help protect against atopic disease? Biology, methodology, and a golden jubilee of controversy. J Pediatr 1988;112:181–90. [DOI] [PubMed] [Google Scholar]
- 11.Larkin EK, Gebretsadik T, Moore ML, Anderson LJ, Dupont WD, Chappell JD, et al. Objectives, design and enrollment results from the Infant Susceptibility to Pulmonary Infections and Asthma Following RSV Exposure Study (INSPIRE). BMC Pulm Med 2015;15:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosas-Salazar C, Shilts MH, Tovchigrechko A, Chappell JD, Larkin EK, Nelson KE, et al. Nasopharyngeal Microbiome in Respiratory Syncytial Virus Resembles Profile Associated with Increased Childhood Asthma Risk. Am J Respir Crit Care Med 2016;193:1180–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shilts MH, Rosas-Salazar C, Tovchigrechko A, Larkin EK, Torralba M, Akopov A, et al. Minimally Invasive Sampling Method Identifies Differences in Taxonomic Richness of Nasal Microbiomes in Young Infants Associated with Mode of Delivery. Microb Ecol 2016;71:233–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization. Indicators for assessing infant and young child feeding practices: conclusions of a consensus meeting held 6–8 November 2007 in Washington D.C., USA Geneva, Switzerland: World Health Organization; 2008. [Google Scholar]
- 15.Rosas-Salazar C, Shilts MH, Tovchigrechko A, Schobel S, Chappell JD, Larkin EK, et al. Nasopharyngeal Lactobacillus is Associated with Childhood Wheezing Illnesses Following Respiratory Syncytial Virus Infection in Infancy. J Allergy Clin Immunol 2018;142:1447–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shilts MH, Rosas-Salazar C, Turi KN, Rajan D, Rajagopala SV, Patterson MF, et al. Nasopharyngeal Haemophilus and local immune response during infant respiratory syncytial virus infection. J Allergy Clin Immunol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turi KN, Shankar J, Anderson LJ, Rajan D, Gaston K, Gebretsadik T, et al. Infant Viral Respiratory Infection Nasal Immune-Response Patterns and Their Association with Subsequent Childhood Recurrent Wheeze. Am J Respir Crit Care Med 2018;198:1064–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosas-Salazar C, Tang ZZ, Shilts MH, Turi KN, Hong Q, Wiggins DA, et al. Upper Respiratory Tract Bacterial-Immune Interactions during Respiratory Syncytial Virus Infection in Infancy. J Allergy Clin Immunol 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 2007;35:7188–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018;6:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Won JH, Goldberger O, Shen-Orr SS, Davis MM, Olshen RA. Significance analysis of xMap cytokine bead arrays. Proc Natl Acad Sci U S A 2012;109:2848–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breen EJ, Tan W, Khan A. The Statistical Value of Raw Fluorescence Signal in Luminex xMAP Based Multiplex Immunoassays. Sci Rep 2016;6:26996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breen EJ, Polaskova V, Khan A. Bead-based multiplex immuno-assays for cytokines, chemokines, growth factors and other analytes: median fluorescence intensities versus their derived absolute concentration values for statistical analysis. Cytokine 2015;71:188–98. [DOI] [PubMed] [Google Scholar]
- 25.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2006. [Google Scholar]
- 26.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology 2001;26:32–46. [Google Scholar]
- 27.Tovchigrechko A MGSAT - Statistical analysis of microbiome and proteome abundance matrices with automated report generation, 2015. Available from: https://github.com/andreyto/mgsat.
- 28.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological 1995;57:289–300. [Google Scholar]
- 30.Austin PC. Optimal caliper widths for propensity-score matching when estimating differences in means and differences in proportions in observational studies. Pharm Stat 2011;10:150–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Austin PC. A comparison of 12 algorithms for matching on the propensity score. Stat Med 2014;33:1057–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamidi B, Wallace K, Alekseyenko AV. MODIMA, a Method for Multivariate Omnibus Distance Mediation Analysis, Allows for Integration of Multivariate Exposure-Mediator-Response Relationships. Genes (Basel) 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho NT, Li F, Lee-Sarwar KA, Tun HM, Brown BP, Pannaraj PS, et al. Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations. Nat Commun 2018;9:4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biesbroek G, Bosch AA, Wang X, Keijser BJ, Veenhoven RH, Sanders EA, et al. The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am J Respir Crit Care Med 2014;190:298–308. [DOI] [PubMed] [Google Scholar]
- 35.Laursen MF, Sakanaka M, von Burg N, Morbe U, Andersen D, Moll JM, et al. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat Microbiol 2021;6:1367–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biagi E, Quercia S, Aceti A, Beghetti I, Rampelli S, Turroni S, et al. The Bacterial Ecosystem of Mother’s Milk and Infant’s Mouth and Gut. Front Microbiol 2017;8:1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stokholm J, Blaser MJ, Thorsen J, Rasmussen MA, Waage J, Vinding RK, et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun 2018;9:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sears MR, Greene JM, Willan AR, Taylor DR, Flannery EM, Cowan JO, et al. Long-term relation between breastfeeding and development of atopy and asthma in children and young adults: a longitudinal study. Lancet 2002;360:901–7. [DOI] [PubMed] [Google Scholar]
- 39.Matheson MC, Erbas B, Balasuriya A, Jenkins MA, Wharton CL, Tang ML, et al. Breastfeeding and atopic disease: a cohort study from childhood to middle age. J Allergy Clin Immunol 2007;120:1051–7. [DOI] [PubMed] [Google Scholar]
- 40.Fredriksson P, Jaakkola N, Jaakkola JJ. Breastfeeding and childhood asthma: a six-year population-based cohort study. BMC Pediatr 2007;7:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Midodzi WK, Rowe BH, Majaesic CM, Saunders LD, Senthilselvan A. Predictors for wheezing phenotypes in the first decade of life. Respirology 2008;13:537–45. [DOI] [PubMed] [Google Scholar]
- 42.da Costa Lima R, Victora CG, Menezes AM, Barros FC. Do risk factors for childhood infections and malnutrition protect against asthma? A study of Brazilian male adolescents. Am J Public Health 2003;93:1858–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han YY, Lee YL, Guo YL. Indoor environmental risk factors and seasonal variation of childhood asthma. Pediatr Allergy Immunol 2009;20:748–56. [DOI] [PubMed] [Google Scholar]
- 44.Simons E, Balshaw R, Lefebvre DL, Dai D, Turvey SE, Moraes TJ, et al. Timing of Introduction, Sensitization, and Allergy to Highly Allergenic Foods at Age 3 Years in a General-Population Canadian Cohort. J Allergy Clin Immunol Pract 2020;8:166–75 e10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


