Abstract
Human cells encounter dynamic mechanical cues in healthy and diseased tissues, which regulate their molecular and biophysical phenotype, including intracellular mechanics as well as force generation. Recent developments in bio/nano materials and microfluidics permit exquisitely sensitive measurements of cell mechanics, as well as tight control over external mechanical stimuli to regulate cell behavior. In this review, we consider the mechanobiology of cells interacting bi-directionally with their surrounding microenvironment, and the potential relevance for translational medicine. We first introduce key fundamental concepts underlying the mechanics of living cells as well as the extracelluar matrix. We then focus on case studies based on: 1) microfluidic measurements of non-adherent cell deformability, 2) cell migration on micro/nano topographies, 3) traction measurements of cells in 3D matrix, 4) mechanical programming of organoid morphogenesis, as well as 5) active mechanical stimuli for potential therapeutics. These examples highlight the promise of disease diagnosis using mechanical measurements, a systems-level understanding linking molecular with biophysical phenotype, as well as therapies based on mechanical perturbations. We close with a critical discussion of these emerging technologies and future directions at the interface of engineering, biology, and medicine.
Keywords: cell mechanics, mechanobiology, biomaterials, microfluidics, extracellular matrix, directed migration, traction force microscopy, deformability cytometry
Graphical Abstract
The dynamic interplay of cell mechanics with extracellular stimuli drives new opportunities for potential translational applications. This review focuses on the most recent advances on fundamental understanding of mechanobiology, and emphasizes on its application including diagnosis of disease based on mechanical measurements and therapeutic treatments that utilize mechanical stimuli and perturbations.
1. Introduction
Human cells experience forces and flows from the surrounding microenvironment, which can influence both intracellular signaling as well as force transduction back to neighboring cells and the extracellular matrix (ECM).[1] The field of cellular mechanobiology can be defined as “any aspect of cell biology in which mechanical force is generated, imparted, or sensed, leading to alterations in cellular function.” [2] Many tissues and organs can be understood as complex systems which are functionally affected by physical cues at cellular scales. Thus, a deeper understanding of mechanobiology at cellular scales is crucial to understand homeostasis in normal healthy tissues. Moreover, dysregulation of this biophysical cause and effect is implicated in a wide range of disease states, such as the aberrant stiffening of ECM during fibrosis, aging and cancer, or the hindrance of immune or stem cell recruitment to sites of injury. Recent developments in molecular and cell biology have revealed new insights into how diverse mechanical and biochemical cues regulate multicellular form and function. In parallel, technological innovations in nanofabrication, microfluidics, and biomaterials have enabled precision measurement of cell and ECM mechanics, as well as higher throughput experiments for screening many conditions rapidly. Finally, massive improvements in computational power are driving biomedical “data science,” which can be analyzed using machine learning and so-called artificial intelligence. Altogether, these fundamental and technological advances have been extremely powerful for our basic understanding of mechanobiology, but the translation to clinical medicine remains nascent.
D’Arcy Wentworth Thompson famously postulated that the (biological) “form of an object is a diagram of forces.”[3] For instance, each cell within a tissue is in physical contact with the ECM, and often mechanically adhered by clustering of integrins at the cell surface to various biochemical ligands in the ECM.[4] These integrins are linked intracellularly to the actin cytoskeleton, which acts as a mechanical scaffold for the cell, and coordinates with myosin motor proteins to exert contractile tensions.[5] Similarly, cells can also mechanically interact with neighboring cells via cell-cell junctions such as cadherins, which also are linked to the cytoskeleton.[6] This “diagram of forces” is modulated by the relative strength of cell-matrix and cell-cell adhesions, so that stiffer ECM or more contractile neighbors will directly impact cytoskeletal architecture and tension, intracellular molecular crowding, and biomolecular condensation in the cytoplasm.[7,8] More recently, mechanosensing and mechanotransduction is understood to occur through switch-like processes (e.g. ion channels) as well as dynamic processes (e.g. focal adhesions) that can respond to cyclic stimuli and adapt to changing conditions [9]. These integrated forces are transmitted by the cytoskeleton to the nucleus, driving downstream signaling for cell proliferation, differentiation, migration, or apoptosis.[10] It should be noted that the mechanics of cells and ECM vary considerably across different tissues, and are a functional biomarker for stem cells, tissue regeneration, and cancer cells.[11-14] Indeed, in various tissue-specific lineages, this variation in cell mechanics is indicative of the differentiation potential of stem cells.[15,16] Consequently, cells may extend protrusions and alter their morphology using actin polymerization, shaped by local ECM architecture, particularly topography and porosity.[17] Further, cells can exert forces back onto their surrounding matrix and neighboring cells, but also deposit and modify their extracellular matrix (i.e. dynamic reciprocity).[18] This feedback mechanism results in a stiffened ECM, elevated osmotic pressure, as well as extrinsic shear, tensile, and compressive forces in the ECM. Therefore, various mechanical cues can directly regulate cell functions and behaviors through a membrane-to-nucleus signaling transduction axis, and can regulate cell physiology including the epigenetic state, equilibrium of intracellular biochemical reactions, and cellular phase separation through changing cell mechanics.[7] Owing to the complexity of this mechanical crosstalk, it is essential to understand how each type of mechanical cue regulates cell function, and the impact of cell mechanics on tissue development and regeneration.
From a technological perspective, mechanobiology was historically conducted in vitro using tissue culture plates (i.e. 2D monolayer culture) or using small animal models. Modern nanofabrication techniques can form planar structures with highly controlled resolution at subcellular length scales, which can be replicated into elastomeric materials via soft lithography.[19] This approach has been widely utilized for controlled patterning of proteins, as well as facile preparation of microfluidic devices.[20] Compliant elastomeric and hydrogel materials have also been used as deformable planar substrates to measure cell-generated tractions, as well as to manipulate cell adhesion, migration and differentiation.[21] Ultrasensitive imaging and measurement techniques have been established for single molecule biophysics.[22] Lastly, ex vivo culture of stem cells or tissue explants (“organoids”) has been widely investigated using natural and synthetic biomaterials, often recapitulating some tissue architecture and physiological function.[23] These technologies are now being integrated in microphysiological “organ-on-a-chip” systems that permit highly defined microenvironmental conditions and are also scalable. Nevertheless, the increased complexity of these approaches also decreases ease of use, which has limited their adoption in a clinical setting.
In this review, we consider the reciprocity of cell mechanics with extracellular stimuli with potential translational applications (Scheme 1). In particular, we focus on case studies based on our own expertise at the interface of experimental cell biology with solid and statistical mechanics. In particular, we address 1) diagnosing disease based on mechanical measurements, 2) fundamental understanding of mechanobiology in the context of molecular signaling pathways (i.e. omics measurements), and 3) therapeutic treatments that utilize mechanical stimuli and perturbations. We envision that mechanobiology will thus complement existing biochemical or genetic approaches for medical diagnosis and treatment. The following sections include a primer on cell mechanics, microfluidic measurements of cell size and stiffness, a primer on extracellular mechanical stimuli, cell migration on microfabricated topographies, traction force microscopy in 3D matrix, mechanical programming of organoid morphogenesis, and active mechanical stimuli. We close with a critical discussion of these emerging technologies and future directions for the field.
Scheme 1.
Schematic illustration of reciprocity of cell mechanics with extracellular stimuli for various biomedical regenerative applications. Cell mechanics contains four characteristics including viscoelasticity, poroelasticity, physical properties, and active material behaviors. Extracellular stimuli contains four formulas of stresses, including solid stress, fluid pressure, ECM architecture, and ECM mechanics. The cell mechanics interplay with extracellular stimuli. The study of the interplays benefits fundamental mechanobiology, biophysical disease diagnosis, and therapeutic mechanical stimuli. Reproduced with permission.[11,24-31] Viscoelasticity: Copyright 2020, The American Society for Cell Biology; Poroelasticity: Copyright 2013, Nature Publishing Group; Physical Properties: Copyright 2019, Royal Society of Chemistry; Active materials: Copyright 2014, Cell Press; Extracellular stimuli: Copyright 2019, Nature Publishing Group; Solid stress: Copyright 2020, Elsevier; Fluid pressure: Copyright 1998, Elsevier. Architecture: Copyright 2020, National Academy of Sciences; Mechanics: Copyright 2020, Royal Society of Chemistry; Cell Mechanics: Copyright 2020, Cell Press.
2. Cell mechanics
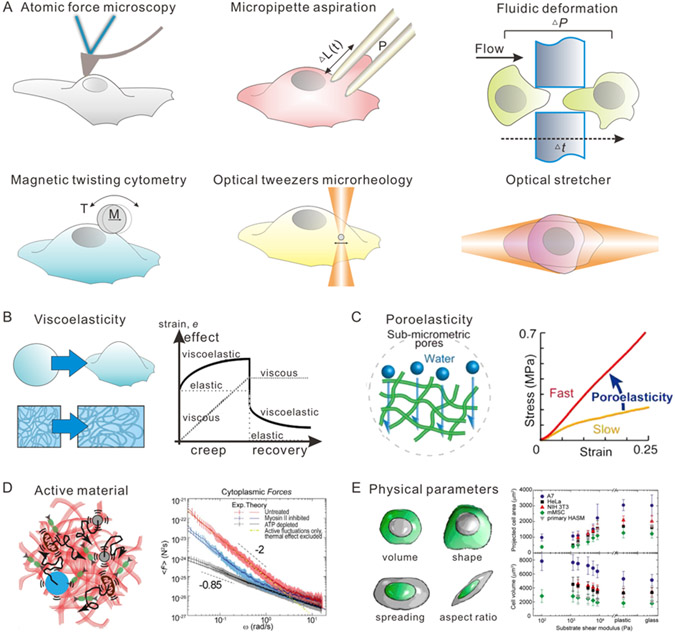
Understanding cell mechanics and its interplay with ECM mechanics is essential to design new engineering approaches to perturb local cell mechanics and guide its growth, differentiation, and regeneration. Elastic and viscoelastic properties of cells and matrix can be measured by mechanical tests including rheology, tensile measurement, AFM, optical tweezers, magnetic twist cytometry, traction force microscopy, micro-aspiration, and microfluidics at either microscale or mesoscale (Figure 1A, Table 1).[32,33] Early historical work investigated the mechanical properties of non-adherent blood cells using micropipette aspiration, revealing that their rheological behavior could be approximated as viscoelastic, with discrete contributions from the nucleus and cortical actin layer [34]. It should be noted that these techniques typically require a mechanical probe to be in direct contact with the cell, and are better suited for either non-adherent cells in solution or adherent cells on a planar substrate. In the latter scenario, adherent cells typically adopt a more flattened conformation since the cytoskeleton is under increased mechanical tension, coupling the nucleus to the underlying substrate [34]. In comparison, cells embedded within a tri-dimensional biomaterial or a living tissue are more spatially confined by their surroundings, but also less accessible to mechanical probes, making them more difficult to characterize [35].
Figure 1. Schematic illustration of different aspects of cell mechanics and tools for measurement of cell mechanics.
(A) Schematic illustration of tools for measurements of cell mechanics, including atomic force microscopy, micropipette aspiration, fluidic deformation, magnetic twisting cytometry, optical tweezer microrheology, and optical stretcher. (B) Schematic illustration of viscoelasticity of cells. (C) Schematic illustration of poroelasticity of cells. Reproduced with permission.[50] Copyright 2018, Elsevier. (D) Schematic illustration of cells as active materials. Reproduced with permission.[26] Copyright 2014, Elsevier. (E) Schematic illustration of physical parameters of cells, including but not limited to volume, shape, spreading, and aspect ratio. Reproduced with permission.[51] Copyright 2014, American Chemical Society.
Table 1.
Comparison of microfluidic methods for measuring cell mechanics.
| Method | Types of cell |
Readout | Strain rate, ε (kHz) |
Stres s, σ (kPa) |
Force sensitivity |
Throughput |
|---|---|---|---|---|---|---|
| Atomic force microscopy | Adherent cells | Elastic and viscoelastic response of a local region or whole cell | 10−5 – 10−2 | pN - μN | 1-10 cells / hour | |
| Micropipette Aspiration | Non-adherent or adherent cells | Elastic and viscoelastic response of a local region or whole cell | 10−5 – 10−3 | pN - nN | 1-10 cells / hour | |
| Particle-Tracking Rheology | Adherent cells | Elastic and viscoelastic response of a local region | pN | 10-100 cells / hour | ||
| Magnetic-Twisting Rheology | Adherent cells | Membrane /surface elasticity | 10−3 – 10−1 | pN - nN | 1000 cells / hour | |
| Optical Tweezers / Traps | Adherent or non-adherent cells | Elastic and viscoelastic response of a local region, or whole cell deformability | 10−3 – 103 | fN - pN | 1-10 cells / hour | |
| Suspended Microchannel Resonator | Suspended cells | The inverse of Passage time; Cell buoyant mass; Apparent elastic moduli | ~the order of 10−2; | The order of 1 | N.A. | 1 cells / s |
| Real-Time Deformability Cytometry | Suspended cells | 1-Circularity; Young’s moduli; Viscosity | ~the order of 0.1 | The order of 1 | N.A. | 100 cells / s |
| Deformability cytometry | Suspended cells | Aspect ratio | the order of 10; can <2 | ~6 | N.A. | 1000 cells / s |
| 2D traction force microscopy | Adherent cells | Matrix deformation | N.A. | N.A. | nN | 10-1000 cells / hour |
| 3D traction force microscopy | Embedded cells within 3D matrix | Matrix deformation | N.A. | N.A. | nN | 1-100 cells / hour |
Cell mechanics exhibit considerable complexity due to the dynamic biophysics of various intracellular components, even when non-adherent to a substrate.[8] For example, cortex mechanics, crowding of cytoplasm, and condensation of nucleus of individual cells contribute differently to cellular functions, which may or may not be causally linked. In addition, the cell morphological parameters, such as cell volume, spreading area, and aspect ratio, also impact the mechanical behavior of cells. The cytoplasm may be understood as a cytoskeletal network composed of actin, microtubules, and intermediate filaments immersed in a motor-driven active fluid [36], and a variety of physical models have revealed new quantitative insights. We briefly summarize these models below, and refer the interested readers to more comprehensive reviews elsewhere [7,37,38].
Historically, cells have been characterized as viscoelastic materials, behaving as a viscous liquid under slow deformations (low frequency) and an elastic solid under rapid deformations (high frequency) (Figure 1B). The dynamic shear modulus G*(ω) captures the frequency-dependent ratio of stress to strain, and can be expressed as: G*(ω) = G′(ω) + iG″(ω), where G′(ω) and G″(ω) are the elastic and loss moduli, respectively, and i is the unit imaginary number. In living cells, it is found that G′(ω) and G″(ω) crossover at frequencies ~0.3 Hz, where G′(ω) dominates over G″(ω) at higher frequencies suggesting a liquid-like to solid-like transition. Similarly, a crude measurement of relaxation time for cells (in response to constant stress) is approximately seconds, based on a simplistic treatment using a single exponential function.[39] Nevertheless, cellular viscoelastic behavior depends on the state of the cytoplasm, which varies among different types of cells, and during development and disease. Recent studies have revealed that different combinations of the cytoskeletal components significantly change the cellular viscoelastic behavior (e.g. the presence or absence of intermediate filaments).[40,41] This may be responsible for changes in mechanics of diseased cells. For example, nontransformed and transformed (cancer) cell lines exhibit different frequency-dependencies of the complex moduli.[42]
Empirically, the viscoelastic response of cells is poorly described by a single characteristic timescale, indicating that classical linear models based on linear combinations of limited number of springs and dashpots are inadequate [38]. Instead, the complex modulus can be fit to a power law of the form ∣G*(ω)∣~ωβ, where β = 0 represents purely elastic response and β = 1 represents purely viscous response. For living cells, β has been empirically measured to be between 0.1 to 0.5 [26,43-45], and a variety of physical models have been proposed to explain this. For instance, the sol-gel hypothesis is based on intriguing similarities of this power law scaling with reconstituted cytoskeletal networks, although it should be noted that these are passive systems in thermodynamic equilibrium [46]. Alternatively, the soft glassy rheology model also gives a power law scaling, which is attributed to intracellular components existing in a complex energy landscape with deep “wells” that are traversed by rare “hopping” events with energetic costs appreciably greater than thermally driven fluctuations [38]. More recently, a self-similar hierarchical model has been proposed which captures the universal power-law rheology behavior of cells.[47] Nevertheless, these models also do not address active cellular behaviors driven by ATP via molecular motors and other enzymatic activities.
The cytoplasm can also exhibit poroelastic behavior due to the microscale pore size of cytoskeletal networks and intracellular solvent flow (Figure 1C).[30,39,48] Cells can behave like a poroelastic gel at short timescales that are smaller than the poroelastic relaxation time (~L2/Dp), where L is the characteristic length scale of the deformation and Dp is the poroelastic diffusion coefficient. Hu et al. systematically investigated contributions of poroelasticity and viscoelasticity to the cytoplasmic mechanical response using optical tweezers to drag tracer particles of varying size and speed.[39] They presented a cytoplasmic phase diagram which shows a fluid to solid transition as the ratio between the viscoelastic relaxation timescale and the experimental time scale increases, and a transition from compressible to incompressible material as the ratio between the poroelastic relaxation timescale and the experimental time scale increases. The poroelasticity can also varies in different types of cells, as the cellular water content and cytoskeletal components can be changed in response to stimuli or during cell fate transition. For example, poroelasticity has been found to play a critical role in governing cartilage mechanics.[49]
Finally, the active, non-equilibrium contributions of ATP-driven molecular motors to cell mechanics have been increasingly recognized (Figure 1D). These intracellular forces are generated by the operation of molecular motors such as kinesin, dynein, and myosin II motors. These molecular motors are responsible for different cellular functions, including directional cargo transportations along the microtubules and active contraction of actin filaments. In addition to the motor-generated directional forces, the biochemical reactions and cell metabolism can also induce active fluctuations, which exhibit similar frequency spectra to Brownian motion but are considerably greater in magnitude. This active force fluctuation also varies in different types of cells or during disease progression; for example, malignant cancer cells exhibit greater active force fluctuations compared to their non-transformed counterparts.[26] Interestingly, recent optical tweezers measurements show that cell cytoplasm can still be approximated as a passive material in thermal equilibrium over short timescales (less than 0.1 s) or high frequencies (greater than 10 Hz). However, active contributions must be considered over longer timescales (greater than 0.1 s) or lower frequencies (less than 10 Hz). These active forces inside the cytoplasm cooperatively drive critical functions at the single cell level, such as contraction, division, and migration.
On a whole-cell level, the physical parameters, including volume, area, aspect ratio and etc., also affect cellular mechanical response to extracellular mechanical stimuli (Figure 1E).[52] For example, cells undergo volume change during many biological processes including cell cycling, stem cell differentiation, and cancer invasion.[53,54] This volume changes can be attributed to not only the changes of the amount of intracellular materials such as proteins and DNA, but also the influx/efflux of intracellular water content. The changes of cell volume, especially by water influx/efflux, lead to the varying concentrations of macromolecules in cells, thus altering the equilibrium and rate of biochemical reactions. Recently, YAP/TAZ signaling activation and Wnt/beta-catenin signaling have been found to be regulated by cell volume, which are further be related to gene expression and stem cell self-renewing.[15,55] In addition to cell volume, other parameters like area and aspect ratio are also import regulator of cell mechanics. Cell spreading area increases significantly for cells cultured on stiffer substrates. At the same time, as the cell spreading area increasing, the cell volume decreases and increased the stiffness of both the cell cortex and cytoplasm. Cell aspect ratio is also an important morphological character of the cells. An unrestricted spreading cell usually have an aspect ratio of 1, exhibiting an isotropic cell mechanical behavior. When cells undergo migration or are stimulated by extracellular directional stimuli, such as flow shear, stretching, confinement, the cells break symmetry and exhibit significant anisotropy in cytoplasmic mechanics and dynamics. These anisotropic mechanical behaviors can be attributed to the reorganization of cytoskeleton, which also accompanies with anisotropic intracellular dynamics of motors. These above-mentioned physical parameters have also been reported to be biomarker for certain cell type or process, as reliable as biochemical markers. For example, large nucleus to cytoplasm ratio has been considered to be a stemness marker of embryonic stem cells;[14] whereas large aspect ratio and small cell volume were a representative morphology of intestinal stem cells in crypt-to-villi epithelium;[15] different morphological categories could indicate different differentiation potentials of mesenchymal stem cells.[13]
In this section, we provide a brief primer on relevant mechanical principles and highlight the mechanical properties of cells that can be modified on single cells. We subsequently consider how to leverage these cellular phenotypes for engineering multicellular tissues, discovering potential diagnostic markers, and developing disease treatments.
3. High Throughput Measurements of Nonadherent Single Cell Mechanics
Cellular components of biofluid samples (e.g. blood, pleural fluid, bone marrow aspirate, cerebrospinal fluid, etc.) encode a wealth of information about patient health and disease.[56] Classical “gold standards” of diagnostic cytopathology are based on complete blood counts (CBC), visual inspection of cell shape (e.g. pathology), or characterization of surface biomarker expression via flow cytometry.[57] It is becoming increasingly appreciated that altered intracellular mechanics are indicative of cell-specific dysregulation of subcellular structure (e.g. nucleus, cytoskeletal proteins).[58] For instance, cancer cells have been widely reported to be softer / more deformable than normal (non-transformed) cells, although the underlying mechanism remains unclear.[59] In contrast, aging is associated with increased cell stiffness and diminished recoverability after large deformation.[60] Lastly, immune cell activation results in dramatic alterations of the cytoskeleton, altering cell size and shape.[61] Historically, precision measurements of single non-adherent cells have utilized atomic force microscopy, micropipette aspiration, or optical stretching, which are labor- and time-intensive techniques. Probing cells one by one is inadequate when there is appreciable phenotypic heterogeneity, which can only be resolved by rapidly sampling large numbers of cells within a given population.[62] A promising alternative is to use microfluidic devices for automated, standardized, and miniaturized analyses of single cells.[58,63-66]
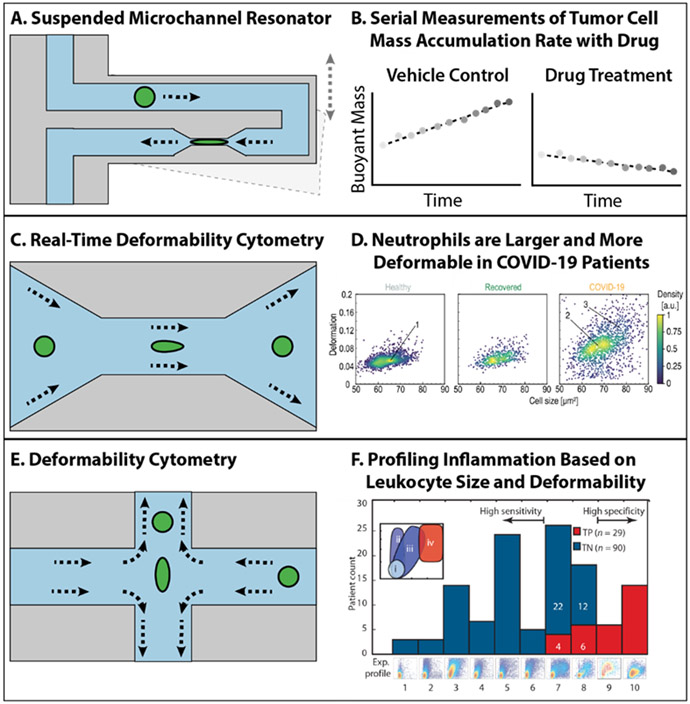
Manalis’ group pioneered the use of microfluidic channels machined within a resonating silicon microcantiever, which can “weigh” cells as they traverse these channels and alter the resonant frequency of the cantilever.[67] One variant of this suspended microchannel resonators incorporates a constricted region (6 μm wide) to deform cells over time (Figure 2A), revealing that some cancer cells with greater metastatic potential exhibit increased deformability and decreased friction (against the channel walls), relative to cells with weaker metastatic potential.[68] More recently, automated microfluidic processing was used to sort and dispense single cells from clinical specimens extracted from brain tissue and tumor resections.[69] Each single cell was repeatedly measured over time by flowing successively through an array of independent cantilevers, revealing its mass accumulation rate (“growth”).[70] Proliferating tumor cells treated with vehicle control typically exhibited an increase in buoyant mass, while cells responsive to certain targeted drugs exhibited constant or decreasing buoyant mass (e.g. recurrent glioblastoma or breast metastasis sensitive to CDK inhibitor abemaciclib) (Figure 2B). Such differences were not observed for drug resistant tumor cells (e.g. glioblastoma treated with TMZ), or non-proliferating cells isolated from healthy brain tissue. Although this approach is exquisitely sensitive for probing single cell mechanics of minute sample volumes, it should be noted that the overall throughput of hundreds of cells per hour may be inadequate to sample highly heterogeneous populations.
Figure 2. Schematic illustration of high-throughput measurement of nonadherent single cell mechanics.
(A) Schematic illustration of suspended microchannel resonator. (B) Serial measurements of tumor cell mass accumulation rate with drug using suspended microchannel resonator. (C) Schematic illustration of real-time deformability cytometry. (D) Testing deformability of neutrophils in health people and COVID-19 patients using real-time deformability cytometry. Reproduced with permission.[71] Copyright 2021, Elsevier. (E) Schematic illustration of deformability cytometry. (F) Profilling inflammation based on leukocytes and deformability using deformability cytometry. Reproduced with permission.[74] Copyright 2013, The American Association for the Advancement of Science.
Alternatively, purely hydrodynamic flows can be used to deform cells within different microfluidic channel geometries, with no physical contact between cells and the walls. Guck et al. demonstrated real-time deformability cytometry, where cells enter a channel with reduced cross-sectional area (e.g. 20 μm × 20 μm, 30 μm × 30 μm), and their deviations from a perfectly circular profile (e.g. deformability) are imaged using high-speed camera (Figure 2C).[72] Single cells were characterized based on their area and deformability, and their phenotypic distribution was visualized using a flow cytometry-like scatter plot (Figure 2D). Subsequent work profiled distinct cell types within a diluted blood sample, revealing unique signatures of erythrocytes, neutrophils, lymphocytes, monocytes, etc. based on deformation, size, and brightness.[73] This approach detected the stiffening of erythrocytes in disease states such as spherocytosis (e.g. ankyrin and spectrin mutation) as well as malaria (in vitro exposure to the parasite Plasmodium falciparum), relative to healthy donors. Further, immune cells exhibited increased size and deformability for patients with acute lung injury (due to bacterial origin) and viral repiratory tract infection. Most recently, this group analyzed blood from COVID-19 patients, showing that neutrophils were larger and more deformable, indicative of an activated state (Figure 2D).[71] Moreover, erythrocytes were stiffened, lymphocytes were more deformable, and monocytes exhibited increased size in COVID-19 patients relative to healthy donors and recovered patients. Troublingly, these alterations in deformability of neutrophils and erythrocytes appeared to persist in some recovered patients. A likely driver of these changes is reorganization of the cytoskeleton, due to viral interactions or inflammatory signaling.
Di Carlo’s group utilized a different cross-slot channel geometry, so that immune cells were stretched with an extensional flow, and their aspect ratio and initial diameter were imaged with a high speed camera (Figure 2E).[62] This technology was applied to pleural effusions, revealing an increase in aspect ratio for immune cells isolated from patients with acute inflammation relative to healthy donors (Figure 2F,i,ii).[74] In comparison, chronic inflammation resulted in greater deformability and cell size (Figure 2F,iii), while malignant diseases included a subpopulation of large and highly deformable “atypical” cells (Figure 2F,iv). Subsequent work showed that granulocytes (but not lymphocytes) exhibited enhanced deformability during sepsis, and was highly predictive of disease severity.[75] It should be noted that both deformability cytometry methods exhibit relatively high throughput (100-1000 cells per second). A recent comparison of all three of these technologies using matched cell lines revealed that they all are sensitive to differences in cytoplasmic packing, but that only the Manalis and Guck technologies are sensitive to alterations in actin cytoskeletal organization.[76] In comparison, the Di Carlo technology also probes nuclear mechanics, likely due to differences in strain rate.[62] It is advantageous that these three technologies are all non-destructive, and it will be intriguing to further integrate these with emerging single cell -omics technologies for deeper molecular phenotyping.[77] To systematically compare the available technologies for mechanically probing ECM and cells, we included a table containing different parameters for these technologies (Table 1).
There are some other technologies can be applied to quantify the mechanical/physical parameters of individual cells or even on molecular levels. For example, Young et al. developed interferometric scattering microscopy for quantification of the mass of single biomolecules in solution with 2% sequence mass accuracy with a resolution of 19-kDa and a precision of 1-kDa[78]. These technologies on molecular level could potentially be integrated with single cell analysis technologies to obtain more comprehensive physical/mechanical properties of cells in the future.
4. Extracellular Mechanical Stimuli
Cells inhabit a complex microenvironment that presents dynamic biochemical cues and mechanical stimuli.[79] The mechanical microenvironments in native tissues not only help maintaining structural integrity and homeostasis, but also govern the morphogenesis of the organs and tumors. Conceptually, this mechanism can also be understood in terms of mechanosensing (e.g. local ECM mechanics via integrins and the actomyosin cytoskeleton), driving cellular mechanotransduction (e.g. altering gene expression, survival and proliferation, as well as ECM synthesis or degradation), finally resulting in mechanoregulation of tissue form and function [80]. These mechanical cues can be conceptually understood within four categories, which includes solid stress, fluid pressure, microarchitecture of the ECM, and ECM mechanics.
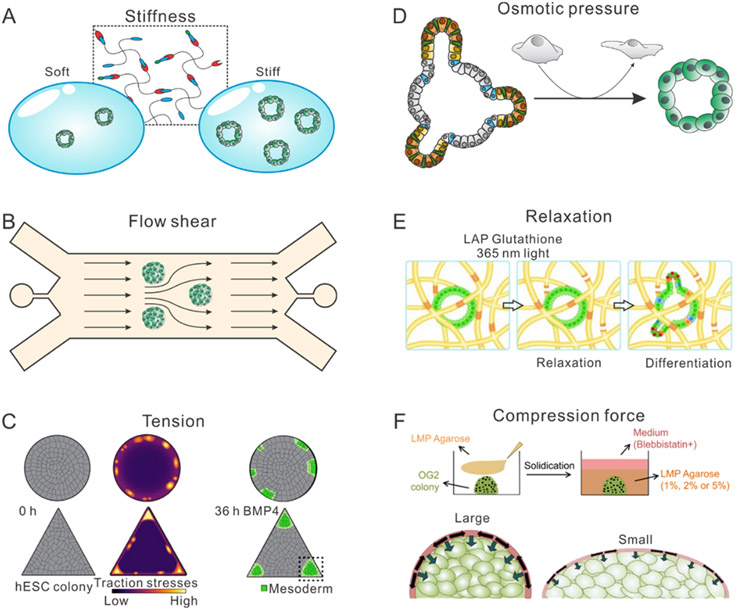
Solid stresses are usually generated in highly-proliferative regions of tissues (Figure 3A). These regions could be a normal intestinal epithelium where the cellular layers turnover every few days, or uncontrolled proliferation of a tumor to push and stretch solid components of the surrounding stroma.[81-88] Tension is another type of solid stress, which is often localized at the interface of different types of tissues, including the interface between the tumor and stroma.[89] Tension in tumors can be actively generated by invading tumor cells or cancer-associated myofibroblasts. These cells actively migrate within the ECM with their high contractility. The cell-induced stretch and strain results in a higher tension that may affect ECM alignment, tumor invasion, and angiogenesis. In normal tissues, mechanical tension, occurring via cell-cell or cell-matrix adhesions is a significant regulator during the morphogenesis of embryos and organs. The spatially non-uniform tension during development has been revealed to be essential for tissue folding, cell fate decision, and formation of multiple organs such as myocardial wall.
Figure 3. Schematic illustration of extracellular mechanical stimuli including.
(A) solid stress, Reproduced with permission.[92] Copyright 2014, Ivyspring International Publisher, (B) fluid stress, Reproduced with permission.[92] Copyright 2014, Ivyspring International Publisher, (C) microarchitecture, Reproduced with permission.[93] Copyright 2019, WILEY-VCH, and (D) mechanics, Reproduced with permission.[94] Copyright 2017, Intechopen.
Fluid pressure includes hydrostatic pressure, osmotic pressure, and shear force (Figure 3B).[90,91] The hydrostatic pressures can be caused by both hypertension diseases and tumor progressions. Blood pressure is determined by both the amount of pumped blood and the hydrostatic resistance of blood vessel to the blood flow. Hypertension causes a higher hydrostatic pressure applied to the endothelial cells and surrounding tissue, which may eventually lead to various heart diseases.[95] Another example is solid tumor progression, where the leakage of plasma from tumor-surrounded blood vessels and insufficient lymphatic drainage leads to elevated interstitial fluid pressure. This elevated interstitial fluid pressure drives both a higher hydrostatic pressure and a higher osmotic pressure.[55] Both of these mechanisms perturbs the physical and molecular phenotype of cells within the tumor microenvironment. Similarly, elevated osmotic pressure can occur in homeostatic healthy tissues. In human intestine, daily digestion and consumption result in a cyclic variation in osmolarity in the range from 300 mOsm to 450 mOsm.[15] Moreover, the kidney is known to experience hyperosmotic stress, as kidney medulla cells encountering a higher osmolarity in microenvironment than cortical cells.[55] Finally, shear force in the human body is often generated by blood flow, which constantly stimulate the endothelial cells.[90,91] Different types of shear force have distinct consequences for cell fate and disease. For example, disturbed flow in blood vessels is associated with vascular inflammation and focal distribution of atherosclerotic lesions.[96,97] In comparison, the steady laminar unidirectional shear force is anti-inflammatory and atheroprotective.
Matrix architecture and mechanics are also important extracellular mechanical cues that stimulate the embedded cells (Figure 3C).[98-100] Conceptually, these can be understood as non-living mechanical cues that act on cells, unlike solid stress and fluid pressure (although ECM can be remodeled by cells over longer timescales.[101,102] Thus, the effects of matrix architecture and mechanics are primarily dependent on the cellular capability to sense their own native microenvironment.
One widely used readout of matrix mechanics is stiffness (linear elasticity), which represents the resistance of the matrix to deformation (Figure 3D).[103,104] The stiffness is an intrinsic characteristic of the matrix and the tissue. In human body, different types of tissues and organs exhibit a wide variation of stiffness. For example, fat and brain are believed to have a relatively low stiffness (Young’s modulus) among all the tissue in the range of 500 Pa to 1 kPa. Meanwhile, bone exhibits the highest stiffness of 2-4 GPa as compared to other tissues. It is known that cells and their nuclei also adapt their stiffness to that of their localized matrix. Interestingly, by manipulating the stiffness of cells to match the stiffness of their related microenvironment, cells are prone to differentiate towards specific lineages that typically reside under those mechanical conditions. Matrix stiffness is also a hallmark for many tumors, which can be used as either a diagnostic marker or a prognostic factor. Indeed, matrix stiffening has been considered to promote tumor progression in breast, colorectal, brain, liver, and pancreatic tumors. Recent studies have shown that both the cancer cells and their surrounding stroma cells are sensitive to the stiffened matrix. A stiff matrix can enhance cell proliferation, disrupt apicobasal cell polarity, and promote invasion.[105] In cancer stroma, there are many cells that are regulated by stiff matrix, including cancer-associated fibroblasts (CAFs), immune cells, adipocytes, and endothelial cells. Immune cell infiltration and recruitment are impeded by matrix stiffness;[106] CAFs could be transferred to myofibroblasts, which change the architecture of matrix and further affect cancer invasion;[107-109] the complex interaction between the adipocytes and tumors can also be affected by their local stiffness;[110] in addition, endothelial cells are sensitive to varying local stiffness, which leads to altered angiogenesis and vessel permeability.[111]
In addition to the stiffness of the ECM (e.g. linear elasticity), other mechanical properties such as nonlinear elasticity, viscoelasticity, plasticity, and poroelasticity, have been increasingly recognized as important. Having briefly reviewed these topics in the context of cell mechanics, we highlight here selected mechanical features of ECM. First, nonlinear elasticity has been observed in many mammalian tissue and biopolymers, and attributed to the fibrillar structure of many ECM components (e.g. collagen I). Briefly, the differential modulus of a nonlinear elastic polymer network increases with the applied stress or strain.[101,112,113] By employing nonlinear elasticity, the matrix in tissues stiffen to prevent tissue damage under severe stresses or large deformations. For example, hierarchical helical structures provide the biopolymer fibrous network nonlinear elasticity in muscle and tendon, which prevent cells from damage under large deformations. In blood vessel walls, the blood flow shear stress stiffens the vessel walls to prevent vessel rupture. Second, most biological tissues and matrices are viscoelastic and exhibit stress relaxation behavior. Recent studies using synthetic viscoelastic materials have shown that stress relaxation is important in several developmental and tumorigenic processes including cell spreading, migration, cartilage formation, and neuron progenitor cell self-renewing.[103,114-118] Third, plasticity occurs when mechanical loading results in an irreversible deformation of a material. Although viscoelasticity and plasticity are rigorously defined in classical mechanics of materials, it is often challenging to decouple and tune them individually in ECM due to the complex molecular structure of biopolymers. Finally, a poroelastic material exhibits a time-dependent mechanical response due to fluid redistribution when the material is subjected to volumetric deformations. Water is the most abundant molecule in cells and tissues; it has been shown that water content plays an important role in cell mechanics and functions.[7,11,15,53-55,110,119] Indeed, water redistribution and the resultant pore pressure homogenization lead to a poroelastic stress relaxation in both cells and the extracellular matrix. This behavior can be understood through a simple scaling law Dp ~ Eξ2/μ, where E and ξ are the drained elastic modulus and the characteristic pore size of the matrix, and μ is the fluid viscosity.
Multicellular systems are self-organized by multiple types of interacting cells and matrix into a 3D architecture that maintains mechanical integrity and homeostasis. For example, the intestinal epithelium self-folds into 3D crypt and villi architecture from a 2D cell layer with a gradient of morphogens and stresses from the bottom of the crypt to the top of the villi.[15] Beyond cellular organization, the structure of the matrix, which includes the pore size of matrix, alignment and orientation of the fibers, can non-mechanically influence tissue function. For instance, the pore size of the biopolymer network is an important parameter determine biomolecular diffusion and information exchange.[120,121] Thus, the porosity can bias the molecular transport and reaction of inflammatory cytokines. The matrix architecture also plays a central role in cancer progression and therapy,[122] and the collagen organization was shown to be a prognostic biomarker.
Altogether, the matrix architecture can affect cell fate through different molecular pathways, which includes integrin, matrix metalloproteinase (MMP), YAP/TAZ, and beta-catenin. These integrated mechanical and biochemical stimuli are transduced to the cell nucleus to regulate downstream phenotypic function. As an example, ECM pore size can confine migrating cells, such as cancer cells, leukocytes, and primary mesenchymal cells. These constraints on cell nucleus can lead to severe compression and deformation, which eventually affect the integrity of the nuclear envelope, herniation of chromatin, DNA damages, chromosomal aberration, and genomic instabilities.[123-130] During the migration of cells, the cell membrane also experience shearing and stretching, which can also lead to the influx of efflux of ions by opening the mechanosensitive ion channels such as Piezo.[131-133] Overall, these myriad extracellular mechanical stimuli cannot be neglected for potential biomedical diagnostics and therapies.
5. Interrogating Cell Migration Dynamics for Diagnostics and Therapeutics
Directed cell migration occurs in response to matrix topography and other asymmetric cues during tumor invasion, leukocyte recruitment, and wound healing.[134] However, cell migration is classically measured using Transwell assays (Boyden chamber)[135] or scratch assays[136] which lack micro/nano topographies and may result in artifacts. These assays are also sensitive to initial cell seeding densities, which can further increase experimental variability. Modern fabrication techniques enable highly controlled topographic features comparable in size to the fibrillar diameter and spacing of the ECM.[137] Alternatively, cells can be fully confined within elastomeric microchannels prepared using soft lithography, where the width and height of these channels only permit single file migration.[138]
Glioblastoma multiforme (GBM) tumors diffusely invade brain tissue along blood vessels and nerve tracks[139], and are extraordinarily therapy resistant, resulting in 95% patient fatality within 5 years of diagnosis.[140] Levchenko’s group in collaboration with Quinones-Hinojosa’s group investigated the migration of patient-derived GBM cells on nanogrooved substrates that mimic brain ECM architecture, with raised features that were 350 nm wide and tall, spaced apart by 1.5 μm, coated with laminin (Figure 4A).[141] These GBM cells exhibited a multipolar morphology with slower migration on tissue culture plastic (Figure 4Ai). In comparison, these cells were more elongated with faster migration on nanogrooved substrates (Figure 4Aii), similar to their behavior within tri-dimensional reconstituted basement membrane (Matrigel) and brain slice culture. Further, some GBM patient cells on nanogrooved substrates exhibited migration that was responsive to platelet-derived growth factor (PDGF), relative to limited sensitivity on flat substrates. Indeed, this could be attributed to phenotypic heterogeneity, with some more migratory subset of the cells responsive to PDGF relative to the bulk population. These PDGF-responsive behaviors were recapitulated in an orthotopic mouse model, and also were predictive of poor prognosis in the patient cohort. This assay reveals exceptional migration phenotypes that self-sort based on speed and directional persistence, permitting increased sensitivity relative to bulk assays that average over the entire population.
Figure 4. Schematic illustration of Interrogating Cell Migration Dynamics for Diagnostics and Therapeutics.
(A) Schematic illustration showed that glioma cell migration is sensitive to PDGF on nanogrooved substrates. (B) Schematic illustration showed that T-cell directional migration is regulated by contractility and microtubule (MT) instability. (C) Schematic illustration showed that metastatic potential of breast cancer cells can be indicated by invasion of branched channels. (D) Schematic illustration showed that active neutrophils from septic patients exhibit spontaneous, sporadic migration.
T cell recruitment to solid tumors for immunotherapy is often impeded by fibrotic ECM architecture.[142] Immune cells can adapt their migration phenotype from a more adhesive, traction-driven mode (“mesenchymal”) to a more contractile and propulsive mode (“amoeboid”) based on the stiffness of the surrounding ECM. Provenzano’s group investigated T cells on patterned polyacrylamide (PA) hydrogels of varying stiffness (e.g. G’ = 2.3 – 1000 kPa) with raised grooves of 600 nm height and 800 nm width, separated at 800 nm spacing and coated with ICAM1 or fibronectin (Figure 4Bi).[143] T cells on soft hydrogels exhibited a more amoeboid phenotype with microtubule-rich pseudopods inserted into the nanogrooves, but on stiffer hydrogels exhibited a more mesenchymal-like phenotype that was spread across the nanogrooves without any pseudopods. This T cell mechanosensitivity was elucidated based on a microtubule-dependent mechanism to regulate contractile response to substrate stiffness and topography. Indeed, microtubule destabilization using Nocodazole enhanced an amoeboid mode with pseudopod insertion and directional migration relative to vehicle control (Figure 4Bii), while microtubule stabilization using Taxol diminished these behaviors and biased towards a mesenchymal-like mode. These T cell migration behaviors were further validated using primary mouse and human T cells in a 3D collagen-fibronectin matrix. These studies suggest that classical chemotherapeutics (e.g. taxanes) intended to disrupt microtubule spindle activity during cancer cell division could inadvertently inhibit T cell recruitment. Nevertheless, it should be noted that the role of microtubules in immune synapse formation remains unclear, so some care is needed when pharmacologically perturbing microtubule function.
Solid tumor invasion and metastasis are the primary driver of patient fatalities, and this systemic disease is extremely difficult to treat successively.[144] Konstantopoulos’s group investigated how breast cancer cells with varying metastatic potential invaded into confined PDMS “feeder” microchannels that were 20 μm wide, 10 μm high, and coated with collagen I.[145] Each channel then bifurcated into two channels, one that was 10 μm wide and the other that was only 3 μm wide (Figure 4C). Cells that only entered the first “feeder” microchannel were classified as “non-migratory,” while cells that traveled further into either bifurcation were classified as “migratory” (Figure 4Ci,ii). This classification was highly predictive of breast cancer cell lines with low or high metastatic potential (in mice), which could be further refined based on a biomarker for cell proliferation (e.g. Ki-67). This assay was used to select for a migratory subpopulation that successfully entered the bifurcations, which exhibited considerably increased metastatic colonization of bone, liver, and lung in a mouse model. Gene expression profiling of this migratory subpopulation revealed upregulation of PI3K-Akt and Ras-MAPK pathways, suggesting candidate treatments based on targeted inhibitors. Further, these behaviors were recapitulated in patient-derived cells that were highly metastatic in xenograft models.
Systemically dysregulated immune response after infection (i.e. sepsis) can result in organ dysfunction and death, but remains challenging to diagnose early since the physiological patterns can resemble other disease pathologies.[146] Sepsis is associated with aberrant neutrophil function, including diminished sensitivity to chemokines as well as anti-microbial activity.[147] Irimia’s group investigated how neutrophils isolated blood samples of human patients with and without sepsis migrated within confined PDMS microchannels that were 10 μm wide, 4 μm high, and coated with fibronectin (Figure 4D).[148] Since there was no exogeneous chemokine stimulation, neutrophils from non-septic patients remained largely inactivated and rarely entered the channels (Figure 4Di). Interestingly, neutrophils from septic patients often migrated “spontaneously” into the microchannels, with highly erratic behaviors (Figure 4Dii). For instance, some neutrophils would migrate up a channel, then immediately reverse its migration back down an adjacent channel. These neutrophils also exhibited oscillatory motion up and down a single channel, as well as frequent pausing. By considering all of these dynamical patterns, an unsupervised machine learning classifier was constructed that was 97% sensitive and 98% specific for septic disease in patients.
Overall, tracking the migration of various cell types isolated from patient samples permits unique insights into disease state, complementing existing measurements of molecular biomarkers. In their current form, these technologies are semi-automated and require some specialized training to use. For instance, plating cells at controlled initial density typically requires some manual dilution and pipetting, although Irimia’s device can operate directly from a droplet of blood.[148] Moreover, accurate detection and tracking of single cells from time-lapse images requires some human supervision, although this may be addressed by advances in computer vision and machine learning.[149] Indeed, some care is needed when comparing experimental conditions, since cells may migrate out of the field of view, divide, or die, resulting in changing population size over time. Ideally, readouts of cell migration should be as simple as possible, and downstream classifiers should be fully transparent to aid in data interpretation. Lastly, dynamic transitions between migration phenotypes in confined spaces, e.g. amoeboid to mesenchymal migration[150,151] or collective to individual migration are of increasing fundamental interest,[152,153] and represent a promising avenue for future investigations.
6. Traction Force Microscopy in 3D Matrix
Cells embedded within natural or synthetic biomaterials can exhibit tissue-like form and function (“organoids”), recapitulating some aspects of embryonic development and disease.[23] One potential advantage of such ex vivo culture is to culture patient-derived human cells, whose response to drugs and genetic manipulation could predict therapeutic efficacy.[154] Moreover, these platforms represent a versatile testbed to investigate the bi-directional interactions between cells and ECM. Early work by Chen’s group investigated how individual fibroblasts deformed synthetic hydrogels (e.g. polyethylene glycol diacrylate), based on the displacement of fluorescent tracer particles embedded throughout the hydrogel (e.g. traction force microscopy).[155] For these experiments, the synthetic hydrogel exhibited a linear elastic response, so that the cell-generated stresses could be extracted from the matrix strain field based on a well-defined constitutive equation. However, this approach is not necessarily applicable for naturally-derived biomaterials which exhibit nonlinear viscoelastic response, and can be irreversibly remodeled by the cells.[156]
Fabry and Wu’s groups have investigated how individual cancer cells deform highly fibrous collagen I hydrogels,[157,158] showing that cells may locally perceive very non-uniform mechanical conditions, due to the variability of the random fiber architecture.[159] Interestingly, highly contractile cells may locally align collagen fibers to stiffen the matrix, permitting longer ranged force transmission relative to synthetic biomaterials comprised of spatially homogeneous flexible polymers. Both these publications utilize constitutive equations that account for discrete fiber architectures,[157,158] but were extrapolated from rheological measurements on cell-free hydrogels, which do not include contributions from cells. Franck’s group proposed an alternative approach for neutrophils that only considers the displacement field of the 3D matrix, revealing functional information about cell contractility, volume changes, and rotation.[160] In principle, this approach can infer cell-matrix interactions based on the geometry and displacement of the cell-matrix boundary, with minimal information about cell morphology. However, the scalar metrics used for cell and matrix deformations were spatially averaged and do not account for spatially non-uniform cell behaviors.
Guo’s group utilized optical tweezers to directly probe matrix rheology near individual cells (Figure 5A,i).[101] For example, latex microparticles (4.5 μm diameter) were embedded within a collagen I hydrogel (1.5 mg/mL) (Figure 5,A,ii), and each bead was manipulated with a controlled displacement x using a tightly focused 1,064 nm laser, resulting in a resistant force F that scales with the local matrix stiffness (Figure 5A,iii). The matrix stiffness was systematically measured at varying distances from the cell (Figure 5A,iv). For highly contractile cells (e.g. mesenchymal cancer cells, MDA-MB-231), their surrounding matrix were found to be significantly stiffer as compared to remote locations, particularly parallel to the cell axis; this is because cell contraction triggers nonlinear matrix stiffening by inducing local fiber alignment and buckling . Analogous trends were observed for reconstituted basement membrane (e.g. Matrigel) as well as fibrin 3.0 mg/mL). Thus, nonlinear rheology and cell-mediated remodeling can be highly pronounced for naturally-derived biomaterials, and must be carefully considered in elucidating mechanobiology in 3D matrix.
Figure 5. Traction force microscopy in 3D matrix.
(A) i. Representative image of a MDA-MB-231 cell (blue) in a 3D collagen network (green). (Scale bar, 10 μm.) (ii–iv) Scheme of the force–displacement measurement with laser tweezers and the relation between matrix stiffening (blue potential wells) and the cell-generated stress field in the cell contraction direction. Reproduced with permission.[101] Copyright 2018. (B) i. Experimental setup for high-throughput imaging to measure cell-induced matrix deformations. ii. Multicellular clusters were grown inside a silk–collagen matrix with embedded 1-μm red fluorescent tracer particles in a 96-well-plate setup. To achieve high-throughput imaging, clusters were imaged by using a spinning-disk confocal microscopy with a low-NA air objective. iii. The 3D cell-induced matrix deformations recovered by directly tracking tracer particles. Reproduced with permission.[164] Copyright 2020. (C) Optical coherence microscopy of multicellular spheroids of mammary epithelial cells alone (MCF10at1) and in co-culture with wild type adipose stem cells (WT-ASC) or obese adipose stem cells (ob/ob ASC). Reproduced with permission.[167] Copyright 2020, Wiley-VCH.
Groups of cells can exhibit emergent behaviors such as coordinated tractions and collective migration that are not observed with individuals.[161-163] Wong’s group has profiled how multicellular clusters alter their matrix adhesions as they disorganize and disseminate.[164] This technology was implemented on a 96-well plate scale using a composite silk-collagen hydrogel,[165] permitting higher throughput measurements of cell-matrix interactions with several different drug perturbations. Using topology-based tracer particle tracking algorithms, the displacement of tracer particles could be visualized with submicron resolution,[166] revealing localized patterns of protrusive and contractile matrix displacement. As a case study, mammary epithelial cells (MCF-10A) underwent controlled induction of the Snail transcription factor, a master regulator of the epithelial-mesenchymal transition (EMT). As a negative control, epithelial clusters typically exhibited compact morphologies with several regions of protrusive and contractile matrix displacement. In comparison, clusters induced to a transitory EMT state extended protrusions and applied more extensive contractile matrix displacements to their surroundings. Lastly, mesenchymal clusters exhibited highly elongated morphologies with localized contractile displacement and minimal protrusive displacement. Thus, accounting for spatial non-uniformity in cell-matrix interactions also reveals meaningful information about functional phenotype. This approach is compatible with a wide variety of biomaterials and cell types, which may enable preclinical testing of human patient samples to inform personalized treatments.
Adie’s group has demonstrated a label-free approach based on optical coherence tomography, which can detect both cells as well as collagen fiber remodeling.[168] Using temporal speckle contrast, this approach can directly visualize collagen matrix deformation as well as degradation over time. This approach was applied to investigate multicellular spheroids of oncogene transfected mammary epithelial cells (MCF-10AT1) with or without co-culture with adipose stem cells.[167] After embedding in dense collagen I hydrogel (6 mg/mL), mammary epithelial cells alone exhibited minimal invasion into the matrix. However, spheroids that included adipose stem cells exhibited collective invasion into the matrix, which was particularly pronounced with adipose stem cells isolated from obese mice. These coordinated behaviors could be suppressed by inhibition of matrix degradation (e.g. MMP inhibitor Batimastat) or cellular contractility (e.g. Y27632). This visualization approach is highly promising for future traction measurements, particularly to elucidate the crosstalk of mechanobiology and metabolism in a controlled ex vivo setting.
Overall, profiling cell-matrix interactions in 3D biomaterials is highly promising to elucidate tissue disorganization in disease as well as drug response. Ongoing challenges include the role of nonlinear matrix response and the local fibrillar architecture, which exhibit greater spatial non-uniformity in vivo due to the presence of cell-sized spaces.[169] Moreover, ECM mechanics and architecture will continue to evolve over time as different cell types deposit and remodel their surroundings, particularly during disease states. The co-culture of epithelial cells with various stromal or immune cells represents one approach to reverse engineer these microenvironmental interactions from the “bottom-up.” Recent advances in structured illumination could further improve spatiotemporal resolution while minimizing phototoxicity [170,171]. Nevertheless, further work is needed to make these biophysical techniques more turnkey, so that they can be implemented in a clinical or biological lab setting with reduced training and instrumentation requirements.
7. Mechanical programming of organoid morphogenesis
Epithelial tissues fulfil a protective role at the surface of many organs, and must withstand harsh external environments including such as shear, tension, and scratch.[172-175] To do so, there are niches for epithelial stem cells that actively proliferate to replenish and repair wounds and replace the cells that die from the wear and tear. At the same time, the regeneration of epithelial tissues from stem cells is mechanoresponsive, and can be triggered by either the mechanical forces directly or the loss of tissue integrity.[176-179] Recent fundamental studies on epithelial development have focused on the fundamental role of stem cells and their progeny, with potential applications for for organ regeneration, and even replacement therapy by ex vivo tissue grafts.[180-184] The intestine has been widely studied as a representative model of stem cell plasticity and differentiation. Several recent papers have revealed that cells from non-stem cell pools can be mobilized after ablation of intestinal stem cells in the niche.[182,183] Intestine injury was introduced by using irradiation or cytotoxic drugs (such as 5-fluorouracil (5-FU) and doxorubicin. After injury, a rapid repair and replenishment of crypt unit have been observed, implying that plasticity of differentiated mature cells contributes to the regenerative process. One possible mechanism is that exposure to niche signals such as Wnt3a can revert differentiated cells to multipotent stem cells. The plasticity of stem cells has also been confirmed in other epithelial tissues, such as liver, hair follicles, epidermal, mammary gland, sweat gland, sebaceous glands, and lung alveolus.[180-184] In several epithelial tissue types, the transitions of mature cells to stem cells have been found to be related to mechanoresponsive pathways such as YAP/TAZ signaling and Wnt/beta-catenin signaling.[182] However, it should be noted that the injuries are often associated with the disruption of the tissue mechanical integrity. The remaining cells are exposed to various types of mechanical cues such as altered osmolarity, tension, contraction, and changes in matrix stiffness. Thus, targeting the local mechanical microenvironment of organ epithelium could be possible as a therapeutic strategy.
Manipulation of mechanical cues is a highly promising approach to engineering organoid growth, complementing other experimental systems in vivo.[188-191] An organoid is a miniaturized recapitulation of a certain type of organ that developed ex vivo or in vitro with three dimensional microanatomy of native organ. Organoids have great relevance for fundamental developmental biology, modelling diseases, facilitating drug development, and enabling tissue/organ replacement therapy. Benefitting from advances in material science and microtechnology, microenvironments with varying mechanical cues can be constructed to study the impact of extracellular stimuli on organoid growth.[186,192-198] In recent researches, engineered hydrogels with varying stiffness haven been proved to regulate the growth of intestinal organoids, kidney organoids, and liver organoids (Figure 6A). For instance, Gjorevski et al. have shown that PEG RGD hydrogels with higher stiffness promote the colony formation ratio of intestinal stem cell spheroid as compared to softer hydrogels.[199] Using a similar hydrogel, Sorrentino et al. studied the effect of matrix stiffness on liver organoid formation, demonstrating a promotion effect on liver organoid formation using a stiffer hydrogel.[200] Both these two studies using hydrogels of varying stiffness in a range from 300 Pa to 1.7 kPa, which is slightly larger than the stiffness of the Matrigel. In another study, Garreta et al. induced kidney organoids from human pluripotent stem cells.[201] By exposing the organoids to a polyacrylamide hydrogel with different stiffness (1kPa and 60kPa), they showed that a soft microenvironment promoted the growth and differentiation of implanted kidney organoids. Synthetic matrix has also used to construct pancreatic ductal adenocarcinoma organoids. In this study, Below et al. developed an eight-arm PEG-based hydrogel system, where adhesion-linker pre-functionalized vinyl sulfone-activated PEG macromers (f-PEG-VS) are cross-linked via peptides sensitive to matrix metalloproteinase to form hydrogel.[202] Their results showed that, by increasing hydrogel stiffness to mimic pancreatic cancer microenvironment, the organoids can engage mechano-sensing pathways and express tumorigenic genes. The matrix stiffness also impacts the growth of 2D multicellular organoids. Pérez-González et al. showed that, in 2D intestinal enteroid, the size of the crypt region containing stem cells varies according to the extracellular-matrix stiffness and endogenous cellular contractility.[203] They showed that a stiffer substrate leads to a smaller stem cell niche area and smaller numbers of stem cells. In addition to the stiffness, the relaxation has been reported to be an important regulator of the morphogenesis of the crypt structure (Figure 6E), which suggested that other aspects of matrix mechanics were also sensed by the organoid during its growth.
Figure 6. Schematic illustration of mechanical programming of organoid morphogenesis.
(A) Schematic illustration of synthetic hydrogel for organoid culturing with varying stiffness. (B) Schematic illustration of microfluidic device for inducing flow shearing force to cultured organoid. (C) Schematic illustration of micropatterning technology for inducing local tension and contractility for cultured organoids and embryos. Reproduced with permission.[185] Copyright 2020, Elsevier. (D) Schematic illustration of osmotic compression to mimic local fluid pressure in native tissue for organoid culturing. Reproduced with permission.[15] Copyright 2021, Elsevier. (E) Schematic illustration of photo-degradable hydrogel for mimicking tissue stress relaxation for organoid culturing. Reproduced with permission.[186] Copyright 2020, Wiley-VCH. (F) Schematic illustration of hydrogel confinement for mimicking compression force for organoid and embryo. Reproduced with permission[187]. Copyright 2019, Elsevier.
Traction force and tension are also important mechanical regulators of organoids (Figure 6C). Xue et al. reported that generation of neuroectoderm tissue from human pluripotent stem (hPS) cells self-generated patterned traction stresses, where larger traction stresses were observed on the boundary of the patterned hPS cells.[204] This organized traction stress was highly correlated with BMP-SMAD signaling and together guide the differentiation and formation of neural plate border. In a more recent study, Muncie et al reported that issue geometries can generate cell-mediated tension to direct the spatial patterning of the BMP4-dependent ‘gastrulation-like’ phenotype by enhancing phosphorylation and junctional release of b-catenin to promote Wnt signaling and mesoderm specification.[185]
In addition to synthetic materials and micropatterning technology, microfluidic devices also provide a powerful tool to control extracellular stimuli for organoid development, especially for controlling the fluid stresses (Figure 6B). Microfluidic chips enable improved regulation of organoid formation by defining local biochemical cues, matrix mechanics, initial seeding cells, and growth architectures. The microfluidic devices have been reported to generate homogeneous synthetic embryo formation and morphology-predefined intestinal organoids.[205] Thus, microfluidic chips have significant potential to control and study the effect of the extracellular stimuli on organoid growth. For instance, Homan et al. reported that cellular polarity and adult gene expression of vascularized kidney organoids can be enhanced when cultured under microfluidic flow as compared to static controls to generate more mature podocyte and tubular compartments.[206] Alternatively, our previous study controlled the local extracellular osmotic pressure (using hypertonic medium) and mechanical stress (compression by weight on microdevice) (Figure 6D).[15] We show that the volumetric compression can promote the self-renewing of intestinal stem cells via stabilizing the formation of LRP6 signalosome and elevating Wnt/beta-catenin signaling, which eventually facilitate growth of intestinal organoids. In addition to osmotic compression, an agarose-based mechanical compression device was reported to compress the embryo, while also regulate the differentiation of stem cells (Figure 6F). With the capability to engineer epithelial organoids to recapitulate multiple aspects of real organs, advances in microfluidics and materials enable organoids to model diseases for precision medicine, and transplantation for tissue replacement therapy.
The application of organoid engineering can also model immune response by mimicking either healthy lymphoid/tonsil tissue or tumor microenvironment[207]. Immune responses vary dramatically from spices to spices. However, the majority of our knowledge about adaptive immunity were gained from animal models such as mice, which have questionable physiological relevance for human patients [207]. A biobank of patient-derived organoids that mimic the native healthy lymphoid or tonsil tissue could be an ideal platform to confirm our knowledge obtained from mouse and discover the human unique adaptive immune behaviors. Wagar et al., developed a functional organotypic system from human tonsils to mimic several key germinal center features in vitro, including producing antibodies, somatic hypermutation and affinity maturation, plasmablast differentiation and class-switch recombination[207]. This study demonstrates that the ex vivo organoid technology provides new opportunity to study the immune behaviors in human genetic background. The immune response in tumor system is also species dependent, which means that we also require human resourced organotypic systems. Tumor organoids have been widely used for modeling diseases and test drug responses. The patient-derived tumor organoids are required to be co-cultured with immune cells, such as tumor-infiltrating lymphocytes (TILs), to recapitulate the immune response of human system. Neal et al. recently employed air-liquid interface (ALI) method propagated patient-derived organoids with native embedded immune cells (T cells, B cells, NK cells and macrophages) [208]. Their results demonstrated that PDO TILs accurately preserved the original tumor T cell receptor spectrum and recapitulated immune checkpoint blockade with anti-PD-1 or anti-PD-L1, which expanded and activated tumor antigen-specific TILs to elicit tumor cytotoxicity. Together, these recent studies demonstrate the promising potential of organoid technology for both fundamental and translational immunology, and the future researches can take the cell mechanics and their mechanical microenvironment into account for better ex vivo engineering.
8. Active Mechanical Stimuli
External mechanical forces apply additional stress and strain on cells and tissues within native microenvironment. Extracellular mechanical force and their consequential strain are integral parts of the cellular microenvironment to regulate cellular proliferation, regeneration, differentiation, and migration.[209] These mechanical stimuli transduced to biochemical signals that activate the genes and signaling pathways that is critical for organ development, tissue regeneration, wound healing, and inhibiting fibrosis during wound closure. Cancerogenesis and tumor metastasis are also highly correlated with abnormal mechanical properties applied to tissues.[210-213] Thus, external application of active mechanical stimuli into the native tissue could enable new mechanotherapies for many diseases. In in vitro systems, recapitulating different modulus of mechanical stimuli can develop better ex vivo multicellular model systems. The mechanical stimuli commonly actively applied is categories as compression, stretching, and shearing. Additionally, building on advances in materials science, some passive mechanical parameters in ECM can be actively manipulated.
The active mechanical stimuli are enabled by current developments in several areas including microfabrication, soft robotic, materials, and microfluidics.[214,215] The devices for active mechanical stimuli can be categorized by soft robots, mechanical hardware, stimuli responsive polymers, microfluidic devices, magnetic materials, dynamic crosslinking of hydrogels. The in vivo actuation can be achieved by embedding soft robots. In a recent work, Dolan et al. reported a milliscale dynamic soft reservoir, which can be implanted into the tissue to actively modulate the biomechanics of the biotic-abiotic interface by changing strain, fluid flow, and even the cellular activity of surrounding tissue.[221] One advantage is that this soft robot significantly inhibits the induced fibrosis as compared to similar device without actuation. In addition, by coupling this soft robot with a porous reservoir, they could enhance the transportation of therapy analogs, and promote their pharmacokinetics. The development of soft robotics could also help with regenerating heart function. Soft robotic cardiac assist devices have been developed that adhere to the external surface of the heart to augment cardiac function (Figure 7A). Alternatively, in vivo mechanical stimuli could utilize multiple types of mechanical hardware. Nia et al. developed an in vivo compression device to simulates the solid mechanical forces to mimic the compressive force generated by a hyperproliferating tumor (Figure 7B).[84] This device was fabricated by adapting standard transparent cranial windows in mice to include a turnable screw for controllably appliying compressive force to the brain tissue. This device was used to actively compress the cerebellar cortex while allowing longitudinal imaging of the brain tissue. Instead, Poling et al. employed compressed nitinol springs to generate uniaxial strain to impact transplanted human intestinal organoids for enhancement of growth and maturation of the organoids.[85]
Figure 7. Development of new technologies for inducing active mechanical stimuli.
(A) Schematic illustration of soft robot for inducing cyclic mechanical compression. Reproduced with permission.[216] Copyright 2018, Nature Publish Group. (B) Schematic illustration of compressive cranial window for mimicking solid stress generated by brain tumor. Reproduced with permission.[84] Copyright 2020, Nature Publish Group. (C) Schematic illustration of stimuli responsive polymer for inducing local force for cells on substrate. Reproduced with permission.[217] Copyright 2017, Nature Publish Group. (D) Schematic illustration of microfluidic devices for inducing flow shearing force for cells. Reproduced with permission.[218] Copyright 2018, Wiley-VCH. (E) Schematic illustration of organ-on-a-chip for inducing stretching to cultured cells. Reproduced with permission.[219] Copyright 2012, Royal Society of Chemistry. (F) Schematic illustration of magnetic nanomaterials for inducing local forces in mice. Reproduced with permission.[220] Copyright 2015, Nature Publish Group.
Magnetic nanomaterials are advantageous for remotely inducing mechanical forces. Fernandez-Sanchez et al. developed a method to mimic a mechanical pressure in intestinal crypt by using intravenous injection of stable ultra-magnetic liposomes (UML) encapsulating super-paramagnetic iron oxide nanocrystals (Figure 7F).[220] They also inserted a 3-mm cylindric magnet subcutaneously in adjacent to the colon to induce mechanical load into the colon tissue with the UML. Using this system, this study revealed the possible mechanical activation of the tumorigenic beta-catenin pathway, which might eventually contribute to tumor propagation. Another example is the usage of 3D magnetic hyaluronic hydrogel, which offers noninvasive neuromodulation via inducing magnetically-induced mechanical stimulation to primary dorsal root ganglion neurons.[222] The authors also demonstrated that mechanoresponsive ion channels, such as TRPV4 and PIEZO2, played an important role in regulating mechano-induced calcium influx in DRG neurons.
Several types of devices have also been developed for local mechanical stimuli of in vitro and ex vivo systems. In principle, these in vitro mechanical devices can generate more controlled stimuli relative to those utilized for in vivo systems. For example, these stimuli generated can not only apply forces on mesoscale tissues, but also stimulate cells in subcellular region. To do so, Sutton et al. reported a type of 2D active cell culture material, which permits directional, remotely controlled, and highly localized surface deformation in the uN force range (Figure 7C).[217] To fabricate this 2D active material, a passive array of microstructures was embedded in a stimuli-responsive hydrogel layer. These temperature-responsive PNIPAAm hydrogel layers encapsulating light-sensitive gold nanorods (AuNRs) enabled remote control of localized microstructure actuation to apply forces to cells.
Microfluidic devices are also a powerful tool to control the dynamic mechanical microenvironments for both the cultured in vitro cells and ex vivo organoids (Figure 7D). Cells are usually cultured inside the microchannels or microchambers, while subjected to a flow to control its medium exchange. By simply adjusting the flow rate of the liquid, microfluidic device can actively control the shear force and its load frequency. In a recent work, Jin et al. proposed a microfluidic-based cell culture device with a continuous dynamic flow of media.[218] The exchange of medium and the shear force that applied to the cells were controlled by a rocker system. The rocker system assisted achieving a gravity-driven flow that mimics inside the microfluidic device to mimic blood recirculation. By mimicking blood flow shearing, the 3D vascularized liver organoid in device exhibited strengthened cell-cell interaction, metabolic activity, and hepatic functionality. Another example of a microfluidic device to control mechanical stimuli is the organ-on-a-chip system (Figure 7E).[223] In lung-on-a-chip or gut-on-a-chip systems, cells are cultured on a porous membrane to mimic alveolar capillary interface of the human lung or the structure of intestinal villi. In particular, for the lung-on-a-chip system, the cyclic strain that usually applied to human lung during breathing was recapitulated by employing two side channels that connected to computer-controlled vacuum. This vacuum can achieve strain ranging from 5% to 15% to match normal levels of strain observed in alveoli within whole lung in vivo. They showed that the toxic and inflammatory responses to silica nanoparticles and the cellular uptake of nanoparticles were enhanced by the cyclic mechanical strains. Thus, soft and active devices applying programmed mechanical stimuli can control cell behaviors and fate decisions for potential biomedical applications.
9. Conclusions and Outlook
Cells dynamically sense and respond to mechanical stimuli in healthy tissues, but the dysregulation of these processes is associated with various disease states. Clinically, these functional alterations have been inferred from changes in cell morphology and tissue architecture, as well as the measurement of genetic and molecular biomarkers. In this review, we first considered how mechanical measurements represent a complementary approach to diagnose disease and elucidate fundamental systems biology.
Modern micro/nano fabrication techniques enable structures with exquisitely controlled feature sizes comparable in size to living cells. For instance, microfluidic devices have been utilized for sensitive and standardized measurements of cell size and deformability under nonadherent conditions. These approaches enable non-destructive single cell measurements with varying throughput, revealing how the statistical distribution of biophysical phenotypes varies in response to drug treatment or inflammation, including COVID-19. Moving forward, we envision that mass produced microfluidic devices in plastic can be combined with low cost digital cameras and cloud-based computer vision for turnkey technological platforms that can be pushed out to clinical settings. Larger-scale clinical studies with different patient biofluids and disease states may reveal new insights into disease progression, particularly at earlier timescales than can be achieved using conventional diagnostic pathology. Moreover, the capability to sort exceptional single cells by biophysical phenotype permits additional genetic, transcriptomic, or epigenetic characterization to link function back to molecular signaling.
Individual cell migration has also been investigated using micro/nano surface topographies and confining microchannel geometries. These structural features mimic some aspects of the ECM in vivo relative to conventional 2D tissue culture on featureless flat substrates. These devices can be fabricated and manufactured in a highly reproducible fashion, so that cell-to-cell and patient-to-patient differences in migration behavior can be attributed more to biological heterogeneity rather than experimental variability. Combined with single cell tracking, these approaches are also better able to resolve exceptional outliers and the effects of drug perturbations. Moreover, complete control over feature geometry presents the interesting possibility of interrogating cellular decision making in more complicated environments, such as bifurcating channels. Nevertheless, additional technology development is needed to streamline the process of “sample-to-answer”. For instance, isolation of patient cells can be performed directly from blood, but otherwise requires extensive processing and labor-intensive preparation before transfer to the device. Moreover, the understanding of which cell migration metrics are clinically meaningful remains relatively nascent (e.g. average speed, directionality, persistence), relative to more straightforward readouts of cell size and deformability discussed previously in deformability cytometry. Thus, additional fundamental work is needed to understand how cell migration behaviors map back to molecular signaling and population heterogeneity. Nevertheless, manufacturing of microfluidic devices and inexpensive open-source microscopy should make these platforms increasingly accessible at least to biological research labs with limited engineering expertise.
Alternatively, 3D cell culture in compliant biomaterials that mimic ECM mechanics and biochemistry represent more biomimetic conditions, and have been successfully utilized to culture primary cells from patients and mice as organoids. Nevertheless, further investigations are warranted to understand why these culture conditions are effective. In particular, naturally-derived biomaterials often exhibit a nonlinear viscoelastic response and can be remodeled by cell-generated forces and secreted factors. Thus, cells are actively reprogramming their surrounding ECM to more amenable conditions, even utilizing these fibrous architectures for longer-ranged coordination. These behaviors are further complicated by collective behaviors of interacting groups of cells within a cluster or organoid, which can be highly heterogeneous. One technological limitation is the difficulty of imaging single cell behaviors and local matrix architecture with high spatial and temporal resolution, which is necessary to fundamentally understand these dynamic cell-matrix interactions. Further, multiscale computational modeling could be trained and informed by live cell imaging to gain further physical insight into how these complex behaviors emerge.
Although organoids often exhibit remarkable tissue-like architecture and cellular hierarchy when cultured in 3D biomaterials with optimized media conditions, they often exhibit appreciable variability in size and morphology. It is intriguing that additional spatial and temporal cues that mimic the in vivo microenvironment would better regulate organoid morphogenesis. For instance, dynamically tuning mechanical properties of the biomaterial, application of mechanical pressure or shear, as well as templating organoid shape can perturb organoid growth and form. Nevertheless, this represents a sizable experimental parameter space for optimizing organoid culture conditions. A predictive and quantitative understanding of how organoids develop is likely needed to rationally design improved organoid culture systems. Ideally, these platforms should also be standardized and turnkey, to facilitate experiments with different patient-derived cell types in a clinical setting. Automated, high throughput drug screening platforms have already been demonstrated at 384 well plate scale for organoids in 3D culture, and there is no technological reason why additional mechanical stimuli and readouts could not be included. Indeed, it is conceivable that human patient organoids cultured under human-mimetic mechanical and biochemical stimuli could be more predictive of the patient disease response than a xenograft model.
Lastly, this review considers how mechanical stimuli and perturbations could be utilized for therapeutic treatments. For instance, pneumatic or magnetic actuation devices can be positioned adjacent to a tissue or implanted with animal models to apply purely mechanical stimuli to cells. Gentle application of these mechanical stimuli can reduce fibrosis and promote cell differentiation for tissue repair and regeneration. However, aberrant mechanical cues are also sufficient to enhance tumor progression. These preclinical investigations are highly promising, although additional clinical trials will be needed to rigorously validate these approaches. In principle, these approaches could augment existing drug treatments since mechanics-based therapy could have diminished side-effects, require no genetic manipulation, impose less burden to metabolic system, can be implemented using FDA-approved materials, and act upon the local ecosystem / niche rather than specific receptors or cell types.
Although this review has focused on case studies based on technology development, we briefly mention several diseases that could benefit from mechanobiology-based approaches. First, aging is a process that naturally occurs and increase the risks of many diseases. Recent data suggest that the ageing microenvironment drives biophysical alterations in the ECM, including reduced deformability and more fibrillar architecture. Fibroblasts in connective tissue lose their contractility during the aging process, and results in loss of the integrity of the skin and other soft tissues. A promising therapy is to reprogram fibroblasts in connective tissue, or replace the aged cells with reprogrammed ones. Moreover, the previously described techniques for organoid culture may be useful to diagnose and inform treatments for aged patient cells.
Second, host immune cell and stem cell homing are modulated by mechanical cues including both forces and material properties. For instance, fibrotic ECM can limit how easily immune cells can access a tissue or tumor site. An intriguing prospect is to expand and prime immune cell and stem cells with controlled mechanical cues prior to reintroduction in the body. Moreover, these cells could be genetically engineered with enhanced mechanosensitivity in order to be recruited to a disease site and release a therapeutic payload. Lastly, biomaterial-based microenvironment niche enables direct manipulation of host immune cells inside the body, thus can not only reduce the cost but also rapidly enough numbers of therapeutic cells.
Another important direction is to integrate mechanobiology with the translational medicine. It remains challenging to controllably deliver and localize mechanical cues to specific tissue sites, relative to conventional systemic delivery of drugs. Future directions could address the regulation of ECM/stroma in patients and the delivery of mechanical stimuli in patients. First, to therapeutically treat the ECM/stroma, cell-based therapy in combination with synthetic matrix could be applied. The synthetic matrix can not only define the mechanics of ECM for surrounding stroma in all aspects including stiffness, viscosity, and plasticity [224-227]. The cells can be directly used for therapy with programmed behaviors that controlled by the matrix. The cells or other encapsulated reagents such as matrix metalloproteinases (MMPs) can be applied to modify the mechanics of ECM [228-232]. Second, to deliver mechanical stimuli in situ in native tissue, employment of soft robotics or stimuli-responsive injectable materials is promising [233-237]. Before its practical usage in human, biocompatibility and biosafety requires further assessment.
Overall, cellular mechanobiology is well positioned to impact translational medicine. One overarching question is to understand how single cell heterogeneity is driven by locally varying extracellular stimuli (both mechanical and biochemical) in conjunction with the “intrinsic” noise of gene expression. Another key question is to understand how the activity of myriad signaling pathways are transduced into an integrated mechanobiological phenotype (e.g. migration, proliferation, differentiation), which could link normal and diseased states across ex vivo and in vivo conditions. We believe that new insights can be revealed by integrating mechanobiology measurements with systems biology approaches, e.g. next-generation sequencing, and mass spectrometry. We also note that mechanical cues are intrinsically linked to the spatial architecture of the tissue, and this larger scale information should be retained whenever possible (e.g. Human Cell Atlas). Since the crosstalk between different cues and molecular signaling processes is so complex, we envision that “big data” and machine learning approaches will be increasingly utilized. Finally, we note that controlled physiological function at the tissue level emerges due to multicellular (mechanical) coordination of heterogeneous single cells. We argue that improved understanding and control of local mechanical cues will improve reproducibility and augment function in disease modeling, drug development and cell-based therapies. Indeed, the capability to reverse engineer a given mechanical niche ex vivo is exciting to elucidate our understanding and interrogate multiple conditions. This could occur through biofabrication technologies such as multimaterial 3D printing, which may be amenable to bespoke engineered tissues, which could further be implanted back into animal models. Thus, new mechanobiology-enabled materials and devices, integrated with advanced imaging and artificial intelligence, are well positioned to move beyond animal models towards next generation medical diagnostics and therapeutics.
Acknowledgements
We apologize to authors whose work could not be included in this review due to space constraints. We thank E.M. Darling (Brown University) and C. Franck (University of Wisconsin-Madison) for helpful discussions. This work was funded by the National Natural Science Foundation of China grants 32171248, 12102142 (to Y.L.), and the Fundamental Research Funds for the Central Universities, HUST grant 2021GCRC056 (to Y.L.). This work was also funded by the U.S. National Institutes of Health grants R01GM140108 (to M.G. and I.Y.W.), 1R01AG064064 (to M.G.), and P20GM109035 (to I.Y.W.).
Biographies
Yiwei Li is a Professor of Biomedical Engineering at Huazhong University of Science and Technology (HUST). He received a B.S. in Biological Sciences and a P.D. in Biomedical Engineering from HUST. During his PhD period, he studied as a visiting student in Applied Physics at Harvard University for 2 years. He then completed postdoctoral research at Department of Mechanical Engineering, Massachusetts Institute of Technology. His research group at HUST studies the mechanobiology and biophysics of multicellular organoids and stem cells. He also develops biotechnologies for high-throughput analysis and engineering of multicellular systems on both organ-level and single cell level.
Ian Y. Wong is an Assistant Professor of Engineering at Brown University. He received an A.B. magna cum laude in Applied Mathematics from Harvard University and a Ph.D. in Materials Science & Engineering from Stanford University. He then completed postdoctoral research at the Center for Engineering in Medicine at Massachusetts General Hospital and Harvard Medical School. His research group at Brown engineers interfacial biomaterials and microfluidics to elucidate tumor invasion, drug resistance, and heterogeneity. He also explores unconventional fabrication techniques for self-assembly and patterning of 2D nanomaterials.
Ming Guo is currently an Associate Professor in the Department of Mechanical Engineering and Physics of Living Systems Center at MIT, where he holds the Class ‘54 Career Development Professorship. Before joining MIT in 2015, Ming obtained his PhD in 2014 in Applied Physics, and MS in 2012 in Mechanical Engineering at Harvard University, and BS in Engineering Mechanics in Tsinghua University. Dr. Guo is a Sloan Fellow in Physics. His group focuses on probing and understanding how mechanics and biology impact each other in multicellular systems, and together sculpt the evolution of multicellular physiological and pathological processes.
Contributor Information
Yiwei Li, Department of Biomedical Engineering, College of Life Science and Technology, Huazhong University of Science and Technology, 1037 Luoyu Road, Wuhan, Hubei, 430074, China..
Ian Y. Wong, School of Engineering, Center for Biomedical Engineering, Joint Program in Cancer Biology. Brown University. 184 Hope St Box D, Providence RI 02912, USA.
Ming Guo, Department of Mechanical Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA..
References Cited:
- [1].Engler AJ, Humbert PO, Wehrle-Haller B, Weaver VM, Science 2009, 324, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Jacobs CR, Huang H, Kwon RY, Introduction to cell mechanics and mechanobiology, Garland Science, 2012. [Google Scholar]
- [3].D’Arcy WT, Dover, New York, 1992. [Google Scholar]
- [4].Kechagia JZ, Ivaska J, Roca-Cusachs P, Nature Reviews Molecular Cell Biology 2019, 20, 457. [DOI] [PubMed] [Google Scholar]
- [5].Fletcher DA, Mullins RD, Nature 2010, 463, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Garcia MA, Nelson WJ, Chavez N, Cold Spring Harbor perspectives in biology 2018, 10, a029181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li Y, Tang W, Guo M, Matter 2021, 4, 1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Serrano JC, Gupta SK, Kamm RD, Guo M, Annual Review of Fluid Mechanics 2021, 53, 411. [Google Scholar]
- [9].Hoffman BD, Grashoff C, Schwartz MA, Nature 2011, 475, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cho S, Irianto J, Discher DE, Journal of Cell Biology 2017, 216, 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Han YL, Pegoraro AF, Li H, Li K, Yuan Y, Xu G, Gu Z, Sun J, Hao Y, Gupta SK, Nature physics 2020, 16, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chowdhury F, Na S, Li D, Poh Y-C, Tanaka TS, Wang F, Wang N, Nature Materials 2010, 9, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lee WC, Shi H, Poon Z, Nyan LM, Kaushik T, Shivashankar G, Chan JK, Lim CT, Han J, Van Vliet KJ, Proceedings of the National Academy of Sciences 2014, 111, E4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Stice SL, Strelchenko NS, Keefer CL, Matthews L, Biology of reproduction 1996, 54, 100. [DOI] [PubMed] [Google Scholar]
- [15].Li Y, Chen M, Hu J, Sheng R, Lin Q, He X, Guo M, Cell stem cell 2021, 28, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Urbanska M, Winzi M, Neumann K, Abuhattum S, Rosendahl P, Müller P, Taubenberger A, Anastassiadis K, Guck J, Development 2017, 144, 4313. [DOI] [PubMed] [Google Scholar]
- [17].Charras G, Sahai E, Nature reviews Molecular cell biology 2014, 15, 813. [DOI] [PubMed] [Google Scholar]
- [18].Loebel C, Mauck RL, Burdick JA, Nature materials 2019, 18, 883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Folch A, Introduction to bioMEMS, CRC Press, 2016. [Google Scholar]
- [20].Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE, Annual review of biomedical engineering 2001, 3, 335. [DOI] [PubMed] [Google Scholar]
- [21].Janmey PA, Fletcher DA, Reinhart-King CA, Physiological reviews 2020, 100, 695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Miller H, Zhou Z, Shepherd J, Wollman AJ, Leake MC, Reports on Progress in Physics 2017, 81, 024601. [DOI] [PubMed] [Google Scholar]
- [23].Simian M, Bissell MJ, Journal of Cell Biology 2017, 216, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ballermann BJ, Dardik A, Eng E, Liu A, Kidney International 1998, 54, S100. [DOI] [PubMed] [Google Scholar]
- [25].Chaubet L, Chaudhary AR, Heris HK, Ehrlicher AJ, Hendricks AG, Molecular biology of the cell 2020, 31, 1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Guo M, Ehrlicher AJ, Jensen MH, Renz M, Moore JR, Goldman RD, Lippincott-Schwartz J, Mackintosh FC, Weitz DA, Cell 2014, 158, 822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gupta SK, Li Y, Guo M, Soft Matter 2019, 15, 190. [DOI] [PubMed] [Google Scholar]
- [28].Levayer R, "Solid stress, competition for space and cancer: The opposing roles of mechanical cell competition in tumour initiation and growth", presented at Seminars in cancer biology, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mandal K, Gong Z, Rylander A, Shenoy VB, Janmey PA, Biomaterials science 2020, 8, 1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Moeendarbary E, Valon L, Fritzsche M, Harris AR, Moulding DA, Thrasher AJ, Stride E, Mahadevan L, Charras GT, Nature materials 2013, 12, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Roy B, Yuan L, Lee Y, Bharti A, Mitra A, Shivashankar G, Proceedings of the National Academy of Sciences 2020, 117, 10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wu P-H, Aroush DR-B, Asnacios A, Chen W-C, Dokukin ME, Doss BL, Durand-Smet P, Ekpenyong A, Guck J, Guz NV, Nature methods 2018, 15, 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jang H, Kim J, Shin JH, Fredberg JJ, Park CY, Park Y, Lab on a Chip 2019, 19, 1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhu C, Bao G, Wang N, Annual review of biomedical engineering 2000, 2, 189. [DOI] [PubMed] [Google Scholar]
- [35].Baker BM, Chen CS, Journal of cell science 2012, 125, 3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pegoraro AF, Janmey P, Weitz DA, Cold Spring Harbor perspectives in biology 2017, 9, a022038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hoffman BD, Crocker JC, Annual review of biomedical engineering 2009, 11, 259. [DOI] [PubMed] [Google Scholar]
- [38].Kollmannsberger P, Fabry B, Annual review of materials research 2011, 41, 75. [Google Scholar]
- [39].Hu J, Jafari S, Han Y, Grodzinsky AJ, Cai S, Guo M, Proceedings of the national Academy of Sciences 2017, 114, 9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hu J, Li Y, Hao Y, Zheng T, Gupta SK, Parada GA, Wu H, Lin S, Wang S, Zhao X, Proceedings of the National Academy of Sciences 2019, 116, 17175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Guo M, Ehrlicher AJ, Mahammad S, Fabich H, Jensen MH, Moore JR, Fredberg JJ, Goldman RD, Weitz DA, Biophysical journal 2013, 105, 1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rigato A, Miyagi A, Scheuring S, Rico F, Nature physics 2017, 13, 771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Fabry B, Maksym GN, Butler JP, Glogauer M, Navajas D, Fredberg JJ, Physical review letters 2001, 87, 148102. [DOI] [PubMed] [Google Scholar]
- [44].Gupta SK, Guo M, Journal of the Mechanics and Physics of Solids 2017, 107, 284. [Google Scholar]
- [45].Hoffman BD, Massiera G, Van Citters KM, Crocker JC, Proceedings of the National Academy of Sciences 2006, 103, 10259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kasza KE, Rowat AC, Liu J, Angelini TE, Brangwynne CP, Koenderink GH, Weitz DA, Current opinion in cell biology 2007, 19, 101. [DOI] [PubMed] [Google Scholar]
- [47].Hang J-T, Kang Y, Xu G-K, Gao H, Nature Communications 2021, 12, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Charras GT, Yarrow JC, Horton MA, Mahadevan L, Mitchison T, Nature 2005, 435, 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nia HT, Han L, Li Y, Ortiz C, Grodzinsky A, Biophysical journal 2011, 101, 2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Isobe N, Kimura S, Wada M, Deguchi S, Journal of the Taiwan Institute of Chemical Engineers 2018, 92, 118. [Google Scholar]
- [51].Mosqueira D, Pagliari S, Uto K, Ebara M, Romanazzo S, Escobedo-Lucea C, Nakanishi J, Taniguchi A, Franzese O, Di Nardo P, ACS nano 2014, 8, 2033. [DOI] [PubMed] [Google Scholar]
- [52].Fu J, Wang Y-K, Yang MT, Desai RA, Yu X, Liu Z, Chen CS, Nature methods 2010, 7, 733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Guo M, Pegoraro AF, Mao A, Zhou EH, Arany PR, Han Y, Burnette DT, Jensen MH, Kasza KE, Moore JR, Proceedings of the National Academy of Sciences 2017, 114, E8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Li Y, Guo F, Hao Y, Gupta SK, Hu J, Wang Y, Wang N, Zhao Y, Guo M, Proceedings of the National Academy of Sciences 2019, 116, 9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Cai D, Feliciano D, Dong P, Flores E, Gruebele M, Porat-Shliom N, Sukenik S, Liu Z, Lippincott-Schwartz J, Nature cell biology 2019, 21, 1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Toner M, Irimia D, Annu. Rev. Biomed. Eng 2005, 7, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Naylor B, Acta cytologica 2000, 44, 709. [DOI] [PubMed] [Google Scholar]
- [58].Darling EM, Di Carlo D, Annual review of biomedical engineering 2015, 17, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Alibert C, Goud B, Manneville JB, Biology of the Cell 2017, 109, 167. [DOI] [PubMed] [Google Scholar]
- [60].Phillip JM, Aifuwa I, Walston J, Wirtz D, Annual review of biomedical engineering 2015, 17, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Vicente-Manzanares M, Sánchez-Madrid F, Nature Reviews Immunology 2004, 4, 110. [DOI] [PubMed] [Google Scholar]
- [62].Gossett DR, Henry T, Lee SA, Ying Y, Lindgren AG, Yang OO, Rao J, Clark AT, Di Carlo D, Proceedings of the National Academy of Sciences 2012, 109, 7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hu S, Liu G, Chen W, Li X, Lu W, Lam RH, Fu J, small 2016, 12, 2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Fan Z, Xue X, Perera R, Nasr Esfahani S, Exner AA, Fu J, Deng CX, Small 2018, 14, 1803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Yu Y, Fu F, Shang L, Cheng Y, Gu Z, Zhao Y, Advanced Materials 2017, 29, 1605765. [DOI] [PubMed] [Google Scholar]
- [66].Chen Z, Yu Y, Guo J, Sun L, Zhao Y, Advanced Functional Materials 2021, 31, 2007527. [Google Scholar]
- [67].Burg TP, Godin M, Knudsen SM, Shen W, Carlson G, Foster JS, Babcock K, Manalis SR, nature 2007, 446, 1066. [DOI] [PubMed] [Google Scholar]
- [68].Sano M, Kaji N, Rowat AC, Yasaki H, Shao L, Odaka H, Yasui T, Higashiyama T, Baba Y, Analytical chemistry 2019, 91, 12890. [DOI] [PubMed] [Google Scholar]
- [69].Calistri NL, Kimmerling RJ, Malinowski SW, Touat M, Stevens MM, Olcum S, Ligon KL, Manalis SR, Nature communications 2018, 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Cermak N, Olcum S, Delgado FF, Wasserman SC, Payer KR, Murakami MA, Knudsen SM, Kimmerling RJ, Stevens MM, Kikuchi Y, Nature biotechnology 2016, 34, 1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Kubánková M, Hohberger B, Hoffmanns J, Fürst J, Herrmann M, Guck J, Kräter M, Biophysical Journal 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Otto O, Rosendahl P, Mietke A, Golfier S, Herold C, Klaue D, Girardo S, Pagliara S, Ekpenyong A, Jacobi A, Nature methods 2015, 12, 199. [DOI] [PubMed] [Google Scholar]
- [73].Toepfner N, Herold C, Otto O, Rosendahl P, Jacobi A, Kräter M, Stächele J, Menschner L, Herbig M, Ciuffreda L, elife 2018, 7, e29213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Tse HT, Gossett DR, Moon YS, Masaeli M, Sohsman M, Ying Y, Mislick K, Adams RP, Rao J, Di Carlo D, Science translational medicine 2013, 5, 212ra163. [DOI] [PubMed] [Google Scholar]
- [75].Guillou L, Sheybani R, Jensen AE, Di Carlo D, Caffery TS, Thomas CB, Shah AM, Tse HT, O’Neal HR Jr, PloS one 2021, 16, e0246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Urbanska M, Muñoz HE, Bagnall JS, Otto O, Manalis SR, Di Carlo D, Guck J, Nature methods 2020, 17, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Zhu C, Preissl S, Ren B, Nature methods 2020, 17, 11. [DOI] [PubMed] [Google Scholar]
- [78].Young G, Hundt N, Cole D, Fineberg A, Andrecka J, Tyler A, Olerinyova A, Ansari A, Marklund EG, Collier MP, Science 2018, 360, 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Dizeux A, Université Pierre et Marie Curie-Paris VI, 2015. [Google Scholar]
- [80].Humphrey JD, Dufresne ER, Schwartz MA, Nature reviews Molecular cell biology 2014, 15, 802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Nia HT, Munn LL, Jain RK, Clinical Cancer Research 2019, 25, 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Nicolas-Boluda A, Silva AK, Fournel S, Gazeau F, Biomaterials 2018, 150, 87. [DOI] [PubMed] [Google Scholar]
- [83].Stylianopoulos T, Munn LL, Jain RK, Trends in cancer 2018, 4, 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Nia HT, Datta M, Seano G, Zhang S, Ho WW, Roberge S, Huang P, Munn LL, Jain RK, Nature Protocols 2020, 15, 2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Poling HM, Wu D, Brown N, Baker M, Hausfeld TA, Huynh N, Chaffron S, Dunn JC, Hogan SP, Wells JM, Nature biomedical engineering 2018, 2, 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Xue S-L, Yin S-F, Li B, Feng X-Q, Journal of the Mechanics and Physics of Solids 2018, 121, 463. [Google Scholar]
- [87].Fan Q, Liu R, Jiao Y, Tian C, Farrell JD, Diao W, Wang X, Zhang F, Yuan W, Han H, Lab on a Chip 2017, 17, 2852. [DOI] [PubMed] [Google Scholar]
- [88].Zervantonakis IK, Hughes-Alford SK, Charest JL, Condeelis JS, Gertler FB, Kamm RD, Proceedings of the National Academy of Sciences 2012, 109, 13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Priya R, Allanki S, Gentile A, Mansingh S, Uribe V, Maischein H-M, Stainier DY, Nature 2020, 588, 130. [DOI] [PubMed] [Google Scholar]
- [90].Lee HJ, Diaz MF, Price KM, Ozuna JA, Zhang S, Sevick-Muraca EM, Hagan JP, Wenzel PL, Nature communications 2017, 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Wang L, Luo J-Y, Li B, Tian XY, Chen L-J, Huang Y, Liu J, Deng D, Lau CW, Wan S, Nature 2016, 540, 579. [DOI] [PubMed] [Google Scholar]
- [92].Kobayashi H, Watanabe R, Choyke PL, Theranostics 2014, 4, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Rauti R, Renous N, Maoz BM, Israel Journal of Chemistry 2020, 60, 1141. [Google Scholar]
- [94].Aghmiuni AI, Khiavi AA, Aromatic and Medicinal Plants–Back to Nature 2017, 2016, 1. [Google Scholar]
- [95].Stewart MP, Helenius J, Toyoda Y, Ramanathan SP, Muller DJ, Hyman AA, Nature 2011, 469, 226. [DOI] [PubMed] [Google Scholar]
- [96].Cheng C, Tempel D, Van Haperen R, Van Der Baan A, Grosveld F, Daemen MJ, Krams R, de Crom R, Circulation 2006, 113, 2744. [DOI] [PubMed] [Google Scholar]
- [97].Dabagh M, Jalali P, Butler PJ, Randles A, Tarbell JM, Journal of The Royal Society Interface 2017, 14, 20170185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Wong I, Gardel M, Reichman D, Weeks ER, Valentine M, Bausch A, Weitz DA, Physical review letters 2004, 92, 178101. [DOI] [PubMed] [Google Scholar]
- [99].Shin JH, Gardel M, Mahadevan L, Matsudaira P, Weitz D, Proceedings of the National Academy of Sciences 2004, 101, 9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Kusindarta DL, Wihadmadyatami H, Tissue regeneration 2018, 65. [Google Scholar]
- [101].Han YL, Ronceray P, Xu G, Malandrino A, Kamm RD, Lenz M, Broedersz CP, Guo M, Proceedings of the National Academy of Sciences 2018, 115, 4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Münster S, Jawerth LM, Leslie BA, Weitz JI, Fabry B, Weitz DA, Proceedings of the National Academy of Sciences 2013, 110, 12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Chaudhuri O, Cooper-White J, Janmey PA, Mooney DJ, Shenoy VB, Nature 2020, 584, 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Dellacherie MO, Seo BR, Mooney DJ, Nature Reviews Materials 2019, 4, 379. [Google Scholar]
- [105].He S, Green Y, Saeidi N, Li X, Fredberg JJ, Ji B, Pismen LM, Journal of the Mechanics and Physics of Solids 2020, 137, 103860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Zhu L, Wu Y, Yoon CW, Wang Y, Current Opinion in Biotechnology 2020, 66, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Chandler EM, Seo BR, Califano JP, Eguiluz RCA, Lee JS, Yoon CJ, Tims DT, Wang JX, Cheng L, Mohanan S, Proceedings of the National Academy of Sciences 2012, 109, 9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Seo BR, Bhardwaj P, Choi S, Gonzalez J, Eguiluz RCA, Wang K, Mohanan S, Morris PG, Du B, Zhou XK, Science translational medicine 2015, 7, 301ra130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Wang K, Eguiluz RCA, Wu F, Seo BR, Fischbach C, Gourdon D, Biomaterials 2015, 54, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Li Y, Mao AS, Seo BR, Zhao X, Gupta SK, Chen M, Han YL, Shih T-Y, Mooney DJ, Guo M, Science advances 2020, 6, eaax5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Ruehle M, Eastburn E, LaBelle S, Krishnan L, Weiss J, Boerckel J, Wood L, Guldberg R, Willett N, Science advances 2020, 6, eabb6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Ahmadzadeh H, Webster MR, Behera R, Valencia AMJ, Wirtz D, Weeraratna AT, Shenoy VB, Proceedings of the National Academy of Sciences 2017, 114, E1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Li H, Zheng Y, Han YL, Cai S, Guo M, Proceedings of the National Academy of Sciences 2021, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Chaudhuri O, Gu L, Klumpers D, Darnell M, Bencherif SA, Weaver JC, Huebsch N, Lee H.-p., Lippens E, Duda GN, Nature materials 2016, 15, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Madl CM, LeSavage BL, Dewi RE, Dinh CB, Stowers RS, Khariton M, Lampe KJ, Nguyen D, Chaudhuri O, Enejder A, Nature materials 2017, 16, 1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Lee H.-p., Gu L, Mooney DJ, Levenston ME, Chaudhuri O, Nature materials 2017, 16, 1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Lou J, Stowers R, Nam S, Xia Y, Chaudhuri O, Biomaterials 2018, 154, 213. [DOI] [PubMed] [Google Scholar]
- [118].Chaudhuri O, Gu L, Darnell M, Klumpers D, Bencherif SA, Weaver JC, Huebsch N, Mooney DJ, Nature communications 2015, 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Delarue M, Brittingham GP, Pfeffer S, Surovtsev I, Pinglay S, Kennedy K, Schaffer M, Gutierrez J, Sang D, Poterewicz G, Cell 2018, 174, 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Fuhrmann G, Nature nanotechnology 2020, 15, 168. [DOI] [PubMed] [Google Scholar]
- [121].Lenzini S, Bargi R, Chung G, Shin J-W, Nature nanotechnology 2020, 15, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Leggett SE, Khoo AS, Wong IY, Biomaterials science 2017, 5, 1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Enyedi B, Jelcic M, Niethammer P, Cell 2016, 165, 1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Mistriotis P, Wisniewski EO, Bera K, Keys J, Li Y, Tuntithavornwat S, Law RA, Perez-Gonzalez NA, Erdogmus E, Zhang Y, Journal of Cell Biology 2019, 218, 4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Irianto J, Xia Y, Pfeifer CR, Athirasala A, Ji J, Alvey C, Tewari M, Bennett RR, Harding SM, Liu AJ, Current Biology 2017, 27, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Cho S, Vashisth M, Abbas A, Majkut S, Vogel K, Xia Y, Ivanovska IL, Irianto J, Tewari M, Zhu K, Developmental cell 2019, 49, 920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Earle AJ, Kirby TJ, Fedorchak GR, Isermann P, Patel J, Iruvanti S, Moore SA, Bonne G, Wallrath LL, Lammerding J, Nature materials 2020, 19, 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Shah P, Hobson CM, Cheng S, Colville MJ, Paszek MJ, Superfine R, Lammerding J, Current Biology 2021, 31, 753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Denais CM, Gilbert RM, Isermann P, McGregor AL, Te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K, Lammerding J, Science 2016, 352, 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Wei F, Xu X, Zhang C, Liao Y, Ji B, Wang N, Nature communications 2020, 11, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Cox CD, Bavi N, Martinac B, Cell reports 2019, 29, 1. [DOI] [PubMed] [Google Scholar]
- [132].Douguet D, Patel A, Xu A, Vanhoutte PM, Honoré E, Trends in pharmacological sciences 2019, 40, 956. [DOI] [PubMed] [Google Scholar]
- [133].Pathak MM, Nourse JL, Tran T, Hwe J, Arulmoli J, Dai Trang TL, Bernardis E, Flanagan LA, Tombola F, Proceedings of the National Academy of Sciences 2014, 111, 16148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Shellard A, Mayor R, Trends in cell biology 2020. [DOI] [PubMed] [Google Scholar]
- [135].Boyden S, The Journal of experimental medicine 1962, 115, 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Ashby WJ, Zijlstra A, Integrative Biology 2012, 4, 1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Kim D-H, Provenzano PP, Smith CL, Levchenko A, Journal of Cell Biology 2012, 197, 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Paul CD, Hung W-C, Wirtz D, Konstantopoulos K, Annual review of biomedical engineering 2016, 18, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Gritsenko PG, Ilina O, Friedl P, The Journal of pathology 2012, 226, 185. [DOI] [PubMed] [Google Scholar]
- [140].Tan AC, Ashley DM, López GY, Malinzak M, Friedman HS, Khasraw M, CA: a cancer journal for clinicians 2020, 70, 299. [DOI] [PubMed] [Google Scholar]
- [141].Smith CL, Kilic O, Schiapparelli P, Guerrero-Cazares H, Kim D-H, Sedora-Roman NI, Gupta S, O’Donnell T, Chaichana KL, Rodriguez FJ, Cell reports 2016, 15, 2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Salmon H, Remark R, Gnjatic S, Merad M, Nature Reviews Cancer 2019, 19, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Tabdanov ED, Rodríguez-Merced NJ, Cartagena-Rivera AX, Puram VV, Callaway MK, Ensminger EA, Pomeroy EJ, Yamamoto K, Lahr WS, Webber BR, Nature communications 2021, 12, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Ganesh K, Massagué J, Nature Medicine 2021, 27, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Yankaskas CL, Thompson KN, Paul CD, Vitolo MI, Mistriotis P, Mahendra A, Bajpai VK, Shea DJ, Manto KM, Chai AC, Nature biomedical engineering 2019, 3, 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Cecconi M, Evans L, Levy M, Rhodes A, The Lancet 2018, 392, 75. [DOI] [PubMed] [Google Scholar]
- [147].Hotchkiss RS, Monneret G, Payen D, Nature Reviews Immunology 2013, 13, 862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Ellett F, Jorgensen J, Marand AL, Liu YM, Martinez MM, Sein V, Butler KL, Lee J, Irimia D, Nature biomedical engineering 2018, 2, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Moen E, Bannon D, Kudo T, Graf W, Covert M, Van Valen D, Nature methods 2019, 16, 1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Toyjanova J, Flores-Cortez E, Reichner JS, Franck C, Journal of Biological Chemistry 2015, 290, 3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Liu Y-J, Le Berre M, Lautenschlaeger F, Maiuri P, Callan-Jones A, Heuzé M, Takaki T, Voituriez R, Piel M, Cell 2015, 160, 659. [DOI] [PubMed] [Google Scholar]
- [152].Wong IY, Javaid S, Wong EA, Perk S, Haber DA, Toner M, Irimia D, Nature materials 2014, 13, 1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Park J, Kim D-H, Shah SR, Kim H-N, Kim P, Quiñones-Hinojosa A, Levchenko A, Nature communications 2019, 10, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Bose S, Clevers H, Shen X, Med 2021, 2, 1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Legant WR, Miller JS, Blakely BL, Cohen DM, Genin GM, Chen CS, Nature methods 2010, 7, 969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Hall MS, Long R, Feng X, Huang Y, Hui C-Y, Wu M, Experimental cell research 2013, 319, 2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Steinwachs J, Metzner C, Skodzek K, Lang N, Thievessen I, Mark C, Münster S, Aifantis KE, Fabry B, Nature methods 2016, 13, 171. [DOI] [PubMed] [Google Scholar]
- [158].Hall MS, Alisafaei F, Ban E, Feng X, Hui C-Y, Shenoy VB, Wu M, Proceedings of the National Academy of Sciences 2016, 113, 14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Beroz F, Jawerth LM, Münster S, Weitz DA, Broedersz CP, Wingreen NS, Nature communications 2017, 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Stout DA, Bar-Kochba E, Estrada JB, Toyjanova J, Kesari H, Reichner JS, Franck C, Proceedings of the National Academy of Sciences 2016, 113, 2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Lin S-Z, Ye S, Xu G-K, Li B, Feng X-Q, Biophysical journal 2018, 115, 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Trepat X, Wasserman MR, Angelini TE, Millet E, Weitz DA, Butler JP, Fredberg JJ, Nature physics 2009, 5, 426. [Google Scholar]
- [163].Yang H, Pegoraro AF, Han Y, Tang W, Abeyaratne R, Bi D, Guo M, Proceedings of the National Academy of Sciences 2021, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Leggett SE, Patel M, Valentin TM, Gamboa L, Khoo AS, Williams EK, Franck C, Wong IY, Proceedings of the National Academy of Sciences 2020, 117, 5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Khoo AS, Valentin TM, Leggett SE, Bhaskar D, Bye EM, Benmelech S, Ip BC, Wong IY, ACS biomaterials science & engineering 2019, 5, 4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Patel M, Leggett SE, Landauer AK, Wong IY, Franck C, Scientific reports 2018, 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Ling L, Mulligan JA, Ouyang Y, Shimpi AA, Williams RM, Beeghly GF, Hopkins BD, Spector JA, Adie SG, Fischbach C, Advanced Functional Materials 2020, 30, 1910650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Mulligan JA, Ling L, Leartprapun N, Fischbach C, Adie SG, Scientific reports 2021, 11, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].van Oosten AS, Chen X, Chin L, Cruz K, Patteson AE, Pogoda K, Shenoy VB, Janmey PA, Nature 2019, 573, 96. [DOI] [PubMed] [Google Scholar]
- [170].Li D, Colin-York H, Barbieri L, Javanmardi Y, Guo Y, Korobchevskaya K, Moeendarbary E, Li D, Fritzsche M, Nature communications 2021, 12, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Barbieri L, Colin-York H, Korobchevskaya K, Li D, Wolfson DL, Karedla N, Schneider F, Ahluwalia BS, Seternes T, Dalmo RA, Nature communications 2021, 12, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Lawlor KT, Vanslambrouck JM, Higgins JW, Chambon A, Bishard K, Arndt D, Er PX, Wilson SB, Howden SE, Tan KS, Nature materials 2021, 20, 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Jee JH, Lee DH, Ko J, Hahn S, Jeong SY, Kim HK, Park E, Choi SY, Jeong S, Lee JW, Stem cells international 2019, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Kratochvil MJ, Seymour AJ, Li TL, Paşca SP, Kuo CJ, Heilshorn SC, Nature Reviews Materials 2019, 4, 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Curvello R, Kerr G, Micati DJ, Chan WH, Raghuwanshi VS, Rosenbluh J, Abud HE, Garnier G, Advanced Science 2021, 8, 2002135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Giobbe GG, Crowley C, Luni C, Campinoti S, Khedr M, Kretzschmar K, De Santis MM, Zambaiti E, Michielin F, Meran L, Nature communications 2019, 10, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Rezakhani S, Gjorevski N, Lutolf M, Biomaterials 2021, 121020. [DOI] [PubMed] [Google Scholar]
- [178].Hernandez-Gordillo V, Kassis T, Lampejo A, Choi G, Gamboa ME, Gnecco JS, Brown A, Breault DT, Carrier R, Griffith LG, Biomaterials 2020, 254, 120125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Brown JW, Mills JC, Nature cell biology 2017, 19, 1307. [DOI] [PubMed] [Google Scholar]
- [180].Penkala IJ, Liberti DC, Pankin J, Sivakumar A, Kremp MM, Jayachandran S, Katzen J, Leach JP, Windmueller R, Stolz K, Cell Stem Cell 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Li W, Li L, Hui L, Trends in cell biology 2020, 30, 329. [DOI] [PubMed] [Google Scholar]
- [182].Moya IM, Halder G, Nature Reviews Molecular Cell Biology 2019, 20, 211. [DOI] [PubMed] [Google Scholar]
- [183].Blanpain C, Fuchs E, Science 2014, 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Tetteh PW, Farin HF, Clevers H, Trends in cell biology 2015, 25, 100. [DOI] [PubMed] [Google Scholar]
- [185].Muncie JM, Ayad NM, Lakins JN, Xue X, Fu J, Weaver VM, Developmental Cell 2020, 55, 679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Hushka EA, Yavitt FM, Brown TE, Dempsey PJ, Anseth KS, Advanced healthcare materials 2020, 9, 1901214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Du J, Fan Y, Guo Z, Wang Y, Zheng X, Huang C, Liang B, Gao L, Cao Y, Chen Y, Cell systems 2019, 9, 214. [DOI] [PubMed] [Google Scholar]
- [188].Kakni P, Hueber R, Knoops K, López‐Iglesias C, Truckenmüller R, Habibovic P, Giselbrecht S, Advanced Biosystems 2020, 4, 2000126. [DOI] [PubMed] [Google Scholar]
- [189].Michielin F, Giobbe GG, Luni C, Hu Q, Maroni I, Orford MR, Manfredi A, Di Filippo L, David AL, Cacchiarelli D, Cell Reports 2020, 33, 108453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Hun M, Barsanti M, Wong K, Ramshaw J, Werkmeister J, Chidgey AP, Biomaterials 2017, 118, 1. [DOI] [PubMed] [Google Scholar]
- [191].Patel S, Ishahak M, Chaimov D, Velraj A, LaShoto D, Hagan D, Buchwald P, Phelps E, Agarwal A, Stabler C, Science advances 2021, 7, eaba5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Shkumatov A, Baek K, Kong H, PloS one 2014, 9, e94764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Ng S, Tan WJ, Pek MMX, Tan M-H, Kurisawa M, Biomaterials 2019, 219, 119400. [DOI] [PubMed] [Google Scholar]
- [194].Zhang Y, Tang C, Span PN, Rowan AE, Aalders TW, Schalken JA, Adema GJ, Kouwer PH, Zegers MM, Ansems M, Advanced Science 2020, 7, 2001797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Magno V, Meinhardt A, Werner C, Advanced Functional Materials 2020, 30, 2000097. [Google Scholar]
- [196].Gjorevski N, Lutolf MP, Nature protocols 2017, 12, 2263. [DOI] [PubMed] [Google Scholar]
- [197].Cruz-Acuña R, Quirós M, Farkas AE, Dedhia PH, Huang S, Siuda D, García-Hernández V, Miller AJ, Spence JR, Nusrat A, Nature cell biology 2017, 19, 1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Valdoz JC, Franks NA, Cribbs CG, Jacobs DJ, Dodson EL, Knight CJ, Poulson PD, Garfield SR, Johnson BC, Hemeyer BM, bioRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez-Morán P, Clevers H, Lutolf MP, Nature 2016, 539, 560. [DOI] [PubMed] [Google Scholar]
- [200].Sorrentino G, Rezakhani S, Yildiz E, Nuciforo S, Heim MH, Lutolf MP, Schoonjans K, Nature communications 2020, 11, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Garreta E, Prado P, Tarantino C, Oria R, Fanlo L, Martí E, Zalvidea D, Trepat X, Roca-Cusachs P, Gavaldà-Navarro A, Nature materials 2019, 18, 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Below CR, Kelly J, Brown A, Humphries JD, Hutton C, Xu J, Lee BY, Cintas C, Zhang X, Hernandez-Gordillo V, Nature Materials 2021, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Pérez-González C, Ceada G, Greco F, Matejčić M, Gómez-González M, Castro N, Menendez A, Kale S, Krndija D, Clark AG, Nature Cell Biology 2021, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Xue X, Sun Y, Resto-Irizarry AM, Yuan Y, Yong KMA, Zheng Y, Weng S, Shao Y, Chai Y, Studer L, Nature materials 2018, 17, 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Zheng Y, Xue X, Shao Y, Wang S, Esfahani SN, Li Z, Muncie JM, Lakins JN, Weaver VM, Gumucio DL, Nature 2019, 573, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Homan KA, Gupta N, Kroll KT, Kolesky DB, Skylar-Scott M, Miyoshi T, Mau D, Valerius MT, Ferrante T, Bonventre JV, Nature methods 2019, 16, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Wagar LE, Salahudeen A, Constantz CM, Wendel BS, Lyons MM, Mallajosyula V, Jatt LP, Adamska JZ, Blum LK, Gupta N, Nature Medicine 2021, 27, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, Liu IH, Chiou S-H, Salahudeen AA, Smith AR, Cell 2018, 175, 1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Fan Q, Zheng Y, Wang X, Xie R, Ding Y, Wang B, Yu X, Lu Y, Liu L, Li Y, Angewandte Chemie International Edition 2021, 60, 11858. [DOI] [PubMed] [Google Scholar]
- [210].Vasudevan J, Lim CT, Fernandez JG, Advanced Functional Materials 2020, 30, 2005383. [Google Scholar]
- [211].Chen J, Cao X, An Q, Zhang Y, Li K, Yao W, Shi F, Pan Y, Jia Q, Zhou W, Nature communications 2018, 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Panciera T, Citron A, Di Biagio D, Battilana G, Gandin A, Giulitti S, Forcato M, Bicciato S, Panzetta V, Fusco S, Nature materials 2020, 19, 797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Yang B, Wolfenson H, Chung VY, Nakazawa N, Liu S, Hu J, Huang RY-J, Sheetz MP, Nature materials 2020, 19, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Sun L, Yu Y, Chen Z, Bian F, Ye F, Sun L, Zhao Y, Chemical Society Reviews 2020, 49, 4043. [DOI] [PubMed] [Google Scholar]
- [215].Pavesi A, Adriani G, Rasponi M, Zervantonakis IK, Fiore GB, Kamm RD, Scientific reports 2015, 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Horvath MA, Varela CE, Dolan EB, Whyte W, Monahan DS, Payne CJ, Wamala IA, Vasilyev NV, Pigula FA, Mooney DJ, Annals of biomedical engineering 2018, 46, 1534. [DOI] [PubMed] [Google Scholar]
- [217].Sutton A, Shirman T, Timonen JV, England GT, Kim P, Kolle M, Ferrante T, Zarzar LD, Strong E, Aizenberg J, Nature communications 2017, 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Jin Y, Kim J, Lee JS, Min S, Kim S, Ahn DH, Kim YG, Cho SW, Advanced Functional Materials 2018, 28, 1801954. [Google Scholar]
- [219].Kim HJ, Huh D, Hamilton G, Ingber DE, Lab on a Chip 2012, 12, 2165. [DOI] [PubMed] [Google Scholar]
- [220].Fernandez-Sanchez ME, Barbier S, Whitehead J, Béalle G, Michel A, Latorre-Ossa H, Rey C, Fouassier L, Claperon A, Brullé L, Nature 2015, 523, 92. [DOI] [PubMed] [Google Scholar]
- [221].Dolan EB, Varela C, Mendez K, Whyte W, Levey R, Robinson S, Maye E, O’dwyer J, Beatty R, Rothman A, Science Robotics 2019, 4. [DOI] [PubMed] [Google Scholar]
- [222].Tay A, Sohrabi A, Poole K, Seidlits S, Di Carlo D, Advanced Materials 2018, 30, 1800927. [DOI] [PubMed] [Google Scholar]
- [223].Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE, Science 2010, 328, 1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Rosales AM, Anseth KS, Nature Reviews Materials 2016, 1, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Mosiewicz KA, Kolb L, Van Der Vlies AJ, Martino MM, Lienemann PS, Hubbell JA, Ehrbar M, Lutolf MP, Nature materials 2013, 12, 1072. [DOI] [PubMed] [Google Scholar]
- [226].Khatua C, Min S, Jung HJ, Shin JE, Li N, Jun I, Liu H-W, Bae G, Choi H, Ko MJ, Nano letters 2020, 20, 4188. [DOI] [PubMed] [Google Scholar]
- [227].Choi H, Bae G, Khatua C, Min S, Jung HJ, Li N, Jun I, Liu HW, Cho Y, Na KH, Advanced Functional Materials 2020, 30, 2001446. [Google Scholar]
- [228].Mao AS, Shin J-W, Utech S, Wang H, Uzun O, Li W, Cooper M, Hu Y, Zhang L, Weitz DA, Nature materials 2017, 16, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Purcell BP, Lobb D, Charati MB, Dorsey SM, Wade RJ, Zellars KN, Doviak H, Pettaway S, Logdon CB, Shuman JA, Nature materials 2014, 13, 653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Darnell M, O’Neil A, Mao A, Gu L, Rubin LL, Mooney DJ, Proceedings of the National Academy of Sciences 2018, 115, E8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Wang L-L, Zhang B, Zheng M-H, Xie Y-Z, Wang C-J, Jin J-Y, Current Topics in Medicinal Chemistry 2020, 20, 2459. [DOI] [PubMed] [Google Scholar]
- [232].Piperigkou Z, Manou D, Karamanou K, Theocharis AD, Methods Mol Biol 2018, 1731, 325. [DOI] [PubMed] [Google Scholar]
- [233].Kilinc D, Dennis CL, Lee GU, Advanced materials 2016, 28, 5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Holle AW, Young JL, Van Vliet KJ, Kamm RD, Discher D, Janmey P, Spatz JP, Saif T, Nano letters 2018, 18, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Zhu M, Do TN, Hawkes E, Visell Y, Soft robotics 2020, 7, 179. [DOI] [PubMed] [Google Scholar]
- [236].Payne CJ, Wamala I, Abah C, Thalhofer T, Saeed M, Bautista-Salinas D, Horvath MA, Vasilyev NV, Roche ET, Pigula FA, Soft robotics 2017, 4, 241. [DOI] [PubMed] [Google Scholar]
- [237].Nims RJ, Pferdehirt L, Guilak F, Current opinion in biotechnology 2022, 73, 374. [DOI] [PMC free article] [PubMed] [Google Scholar]