Graphical abstract
Keywords: Protease, MALT1, Mucosa-associated lymphoid tissue lymphoma translocation protein 1, CBM, Proteolytic processing, Proteolysis, ZC3H12B, ZC3H12D, TAB3, TANK, CILK1, ILDR2, CASP10, NF-kB, Prediction, GO-2-Substrates, Signalling
Abstract
We developed a bioinformatics-led substrate discovery workflow to expand the known substrate repertoire of MALT1. Our approach, termed GO-2-Substrates, integrates protein function information, including GO terms from known substrates, with protein sequences to rank substrate candidates by similarity. We applied GO-2-Substrates to MALT1, a paracaspase and master regulator of NF-κB signalling in adaptive immune responses. With only 12 known substrates, the evolutionarily conserved paracaspase functions and phenotypes of Malt1–/– mice strongly implicate the existence of undiscovered substrates. We tested the ranked predictions from GO-2-Substrates of new MALT1 human substrates by co-expression of candidates transfected with the oncogenic constitutively active cIAP2-MALT1 fusion protein or CARD11/BCL10/MALT1 active signalosome. We identified seven new MALT1 substrates by the co-transfection screen: TANK, TAB3, CASP10, ZC3H12D, ZC3H12B, CILK1 and ILDR2. Using catalytically inactive cIAP2-MALT1 (Cys464Ala), a MALT1 inhibitor, MLT-748, and noncleavable P1-Arg to Ala mutant versions of each substrate in dual transfections, we validated the seven new substrates in vitro. We confirmed the cleavage of endogenous TANK and the RNase ZC3H12D in B cells by Western blotting and mining TAILS N-terminomics datasets, where we also uncovered evidence for these and 12 other candidate substrates by endogenous MALT1. Thus, protein function information improves substrate predictions. The new substrates and other high-ranked MALT1 candidate substrates should open new biological frontiers for further validation and exploration of the function of MALT1 within and beyond NF-κB regulation.
1. Introduction
Knowledge of the substrate repertoire of proteases is essential for understanding their biological roles [1]. With recent exceptions [2], [3], [4], protease cleavage site prediction algorithms are typically designed for predictions within one or a small number of targeted query proteins. Indeed, most predictor algorithms are neither designed nor scalable for proteome-wide ranking of candidate substrates. Machine-learning-based tools perform well for cleavage site prediction but are trained for only a limited range of protease clades to date and require many known cleavage sites—rather than knowledge of the protein substrates themselves—for training. Indeed, for accurate predictions of protease cleavage sites, more than ∼ 30 unique sites are required to learn protease specificity [5]. Consequently, no existing algorithm can be successfully applied for most proteases having only a few known substrates.
Sequence information derived from specificity profiling of denatured peptide libraries and known native protein cleavage sites is traditionally used as a foundation for substrate predictions [6]. Accessibility of scissile bonds [7] and substrate-binding exosites [8] also influence protein cleavage, and several approaches utilize these features to help predict cleavage sites and substrates [4], [9]. However, experimental structural information is not available for all proteins. In contrast, knowledge of protein function, localization and protein–protein interaction (PPI) data is more widespread. Even though a protease often regulates several proteins in a pathway or process, such features are not typically used for substrate prediction except to evaluate prediction quality [10] or, very recently, to refine experimentally-derived candidates [11]. Thus, we considered that gene ontology (GO) annotations of proteins, PPI and evolutionary information could be leveraged to improve substrate predictions. Here, we describe GO-2-Substrates, a proteome-wide bioinformatics substrate discovery workflow that integrates knowledge of protein function with sequence information to yield precise predictions of candidate substrates rather than just cut sites within a nominated protein or cohort.
To identify new substrates and test the precision of our workflow, we selected the paracaspase mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1). To our knowledge, no existing algorithm [10], [2], [3], [4], [12], [13], [14], [15], [16], [17] can be readily implemented to predict MALT1 substrates. As MALT1 has just 14 known cleavage sites in 12 substrates, this limits algorithm training, necessitating new approaches to predict MALT1 substrates accurately. Furthermore, a significant hurdle for experimental MALT1 substrate screening is the technical barrier associated with direct biochemical assays that require nonphysiological conditions to drive MALT1 activation artificially [18]. This is due to the unusually complex MALT1 activation mechanism. For MALT1 activation, diverse cell receptor signal pathways lead to the association of MALT1 with B cell lymphoma/leukemia 10 (BCL10) and one of the coiled-coil linked Caspase Recruitment Domain-containing proteins (CARD) 11, 14, 10 or 9 to form a family of cell-specific CBM signalosomes [19], [20]. The signalosome transduces upstream signals to activate MALT1 and relay NF-kB signalling in parallel with c-Jun N-terminal kinase (JNK) [21], [22]. Alternatively, MALT1 activation is induced upon binding to overexpressed TRAF6 [23] or when constitutively active as an oncogenic gene-fusion product with cIAP2 [24].
By co-transfection of the CARD11-CBM or cIAP2-MALT1 with 34 ranked candidate substrates, we screened, identified and biochemically validated seven new human substrates of MALT1: TRAF family member Associated NF-κB activator (TANK), TAK1 binding protein 3 (TAB3), Caspase-10 (CASP10), Zinc finger CCCH domain-containing protein 12D (ZC3H12D), Zinc finger CCCH domain-containing protein 12B (ZC3H12B), Immunoglobulin-Like Domain Containing Receptor 2 (ILDR2), and Ciliogenesis Associated Kinase 1 (CILK1). Using Western blotting and mining of proteomics datasets, we validated the cleavage of TANK and ZC3H12D in B lymphocyte cells by endogenous MALT1. Additionally, we found evidence for MALT1 cleavage of 12 other candidates in the same proteomics data. This success supports the value of using protein function to assist substrate discovery. Moreover, revealing new substrate function diversity opens new vistas for exploring MALT1 roles in NF-κB signalling and beyond.
2. Results
2.1. Limitations in sequence logos to predict MALT1 cleavage sites
For protein cleavage, the proximal amino acid residues on both the non-prime (P) side and distal prime (P’) side of the scissile bond must be accommodated in the protease catalytic S and S’ subsites, respectively (reviewed in [25]). To determine the sequence specificity of MALT1, we first consulted MEROPS [26], the definitive knowledge base for peptidases and their substrates. However, we found that the substrate information used to generate the MALT1 cleavage site logo was incomplete. Therefore, we manually curated the literature and found that the most highly conserved positions of the MALT1 cleavage sites in the 12 known human and mouse substrates are from P4 to P1′ (Fig. 1a, b, c). We derived a position-specific scoring matrix (PSSM) of the relative frequency occurrence of amino acids at these positions (Fig. 1d). Using the PSSM, we scanned the entire human proteome with the Find Individual Motif Occurrences (FIMO) search algorithm [27] to score amino acid sequences based on their similarity to the known MALT1 protein substrate cut sites (Fig. 1e). By ranking all P4–P1′ sequences in the human proteome according to their FIMO score, we detected 276 sequences (cut sites) closely matching the MALT1 consensus cleavage motif, including seven sites in six known substrates (Fig. 1f; sensitivity < 0.5). However, over 20 % of the human proteome, i.e., 5,570 sequences within 4,344 proteins, ranked equal to or better than the lowest-ranked known MALT1 substrate cleavage site, which was one of two cut sites in the LIM domain and actin-binding protein 1 (LIMA1) (Fig. 1f; sensitivity = 1). Thus, the sheer number of candidate substrates identified by an approach relying on amino acid sequence logos alone is unacceptably high for predicting MALT1 cleavage sites and substrates with high confidence.
Fig. 1.
MALT1 cleavage site analysis and FIMO proteome analyses. a Alignment of human and mouse P5 – P5′ cleavage site sequences of MALT1 substrates reported in the literature. Downward arrow indicates the site of the scissile bond. The relevant PubMed Identification (PMID) reference for each experimentally determined cleavage site is shown. Red shading indicates species-specific homology in the human and mouse cleavage site sequences. b, c Sequence logo representation of MALT1 cleavage sites in human (b) and mouse (c) substrates, generated using the ggseqlogo package in R. d Schematic of the workflow we developed to generate the Position Specific Scoring Matrix (PSSM) from the published MALT1 substrate cleavage sites. The PSSM was inputted into FIMO (Find Individual Motif Occurrences) to identify sequences in all human proteins that most closely matched the PSSM. e PSSM derived from human and mouse MALT1 substrate cleavage sites. Light to dark red colour range represents the increasing relative frequency of each amino acid from P4 – P1′. f Scatter plot showing the ranking of FIMO scores of known MALT1 P4 – P1′ cleavage site sequences found in the human protein substrates versus sensitivity. Locally weighted scatterplot smoothing was used to generate the line of best fit. Red: proteins reported to be cleaved by the CARD–BCL10–MALT1 (CBM) complex; green: proteins reported to be cleaved by cIAP2-MALT1 without evidence of CBM cleavage yet published. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.2. Improving substrate prediction rates by protein and function features
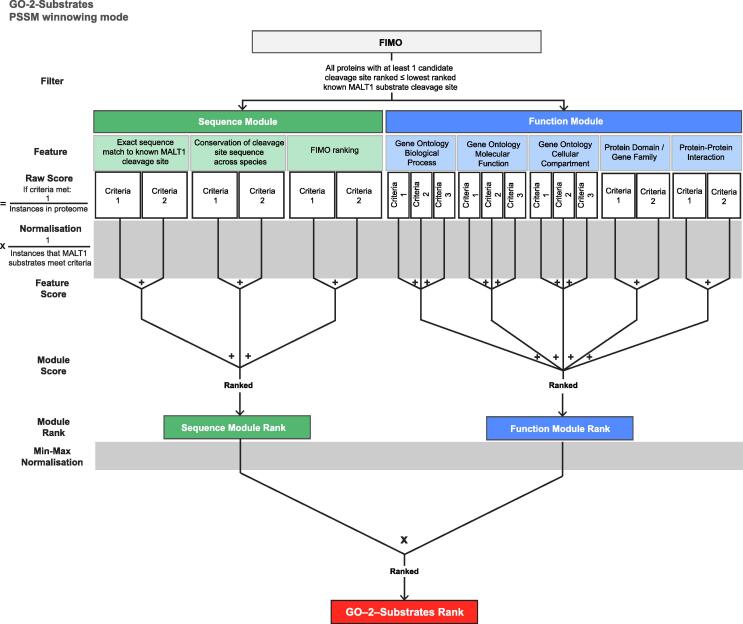
We utilized orthogonal information to winnow the 4,344 candidate protein substrates identified by FIMO. We term this process ‘PSSM winnowing mode’. As outlined in Fig. 2, the ‘Sequence Module’ prioritizes candidate human protein sequences that are identical or highly conserved in their mouse ortholog. Next, human protein sequences that matched known cleavage sites and therefore equally fitting the catalytic S and S’ subsites were considered more likely to be substrates. Finally, proteins were prioritized on their highest-ranked FIMO sequence. A normalized score was derived for each protein for each feature criteria, and the sum of these was combined as the rank for each candidate MALT1 substrate.
Fig. 2.
GO-2-Substrates integrates sequence, GO and protein features to rank candidate substrates. Schematic of the GO-2-Substrates bioinformatic workflow we developed to predict substrate cleavage sites. Features that we considered valuable in predicting cleavage were assigned to two modules, either the Sequence Module or the Function Module. Feature-specific criteria were used to derive raw scores for each protein that were normalized to the proportion of published MALT1 substrates meeting each criterion. Criteria scores were summed to yield Feature Scores, which in turn were summed to yield Module scores, which were then ranked and min–max normalized. Finally, the ranked product of normalized Sequence and Function Module ranks was derived and designated as the GO-2-Substrates rank.
We next integrated protein functional enrichment and PPI information into our workflow. GO annotations were categorized by their strength of enrichment (Supplementary Fig. 1), which prioritized candidate MALT1 substrates that possess matching annotations in their UniProtKB entries. Next, we used PPI data from BioGRID [28] to identify proteins that physically interact with MALT1 or its known substrates, which we considered superior substrate candidates to other proteins. A combined ‘Function Module’ rank was then derived for each candidate MALT1 substrate (Fig. 2).
The performance of the Function and Sequence modules in their ranking of the known MALT1 substrates at the protein level was compared with FIMO alone (Supplementary Fig. 2). Significance testing for differences in ranks was performed using the Wilcoxon signed rank test. Compared to FIMO, which ranked the known substrates within the top 3,176 proteins (Supplementary Fig. 2a, b), the Function Module significantly narrowed the ranking of the known substrates to within the top 724 proteins (Supplementary Fig. 2a), p = 4.9 × 10-3. This occurred despite the lowest-ranked substrate (LIMA1) sharing little obvious functional similarity with the other substrates (Supplementary Fig. 3). In parallel, the Sequence Module ranked MALT1 substrates within the top 28 proteins (Supplementary Fig. 2b), p = 4.9 × 10-4.
Finally, we combined the Sequence and Function Modules ranks to yield a composite “GO-2-Substrates” rank (Fig. 2, Fig. 3a, Supplementary Table 1), which identified those proteins ranked highly by both modules and so are more likely to be undescribed MALT1 substrates. Our approach also improved the identification of candidate substrates that ranked exceptionally well by either the Function or Sequence Modules but unremarkably by the other module. In this way, predictions based on a high GO-2-Substrates rank would not exclude the discovery of MALT1 substrates in novel pathways or at non-consensus cleavage sites. The significant (p = 4.9 × 10-4) compression in ranks of the known substrates to ≤ 15 by GO-2-Substrates from 4,344 by FIMO alone reveals that our workflow effectively winnows the best candidate substrates to a realistic number compatible with targeted screening.
Fig. 3.
A cell-based screen for validation of GO-2-Substrates predictions. a The top 30 GO-2-Substrates ranked proteins, their corresponding FIMO, Function and Sequence Module ranks and best ranked candidate cleavage site. Known substrates are coloured. Proteins of boxed gene names were tested for MALT1 cleavage in co-transfection assays. b, c Scatter plots visualizing the distribution of proteins selected (red) for co-transfection screen in terms of their Sequence and Function rank (b), or GO-2-Substrates rank (c). d Schematic of cDNA expression constructs used in the co-transfection screen encoding: constitutively active cIAP2-MALT1; inactive cIAP2-MALT1 (C464A), and e CARD11 (L251P), BCL10 and MALT1 that assemble active CBM. Myc, FLAG and 6 × His C-terminal tags are as indicated. f Schematic of HOIL1 (RBCK1) positive control cDNA expression construct used for co-transfection with the previously reported cut site and molecular weights of MALT1 cleavage products shown. g Western blot analysis of lysates from HEK293 cells co-transfected with RBCK1. Full-length proteins are indicated with a black arrow; red arrow indicates C-terminal cleavage product of RBCK1 (C-RBCK1). β-actin, loading control was detected by rabbit β-actin antibody. Positions of electrophoretic mobility of molecular weight markers are as shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Proteomic detection of 13 neo-N-termini of substrates predicted by GO-2-Substrates. Proteomics datasets of PMA / ionomycin stimulated B lymphocytes [22] were mined for experimental evidence to support GO-2-Substrates predictions. N-terminally TMT-modified peptide spectrum matches (PSMs) were matched to the predicted neoN-terminal peptides that would be generated upon cleavage of candidate substrates by MALT1. Matched peptides included one of the newly described in vitro MALT1 substrates, ZC3H12D (highlighted and shown in bold). PSMs corresponding to the known MALT1 cleavage site in RBCK1 were used as a positive control for our data processing workflow (shown in bold).
| GO-2-Substrates |
Protein | Gene | Candidate Cleavage |
Candidate Cleavage |
TMT-labelled Neo-N-terminal peptide | Number of PSMs | PeptideProphet Probability |
Dataset(s) Where |
|---|---|---|---|---|---|---|---|---|
| Rank | Site Position (P1′) | Site (P4↓P4′) | (Best PSM) | Peptide Observed | ||||
| 4 | Q9BYM8 | RBCK1 | 166 | LQPR↓GPLE | n[2 3 0]GPLEPGPPKPGVPQEPGR | 5 | 0.9995 | PXD006723 |
| 6 | A2A288 | ZC3H12D | 62 | LVPR↓GSCG | n[2 3 0]GSCGVPDSAQR | 1 | 1 | PXD008421 |
| 575 | P22626 | HNRNPA2B1 | 214 | GDSR↓GGGG | n[2 3 0]GGGGNFGPGPGSNFR | 6 | 0.9979 | PXD006723 |
| 984 | Q12778 | FOXO1 | 317 | FRPR↓TSSN | n[2 3 0]TSSNASTISGR | 1 | 0.9693 | PXD008421 |
| 1425 | P23588 | EIF4B | 340 | LKPR↓STPK | n[2 3 0]STPKEDDSSASTSQSTR | 2 | 0.9806 | PXD008421 |
| 1444 | Q99459 | CDC5L | 427 | LTPR↓SGTT | n[2 3 0]SGTTPKPVINSTPGR | 1 | 0.9983 | PXD008421 |
| 1458 | P08758 | ANXA5 | 277 | MVSR↓SEID | n[2 3 0]SEIDLFNIR | 3 | 0.9977 | PXD006723, PXD008421 |
| 1517 | P49368 | CCT3 | 450 | VIPR↓TLIQ | n[2 3 0]TLIQNCGASTIR | 1 | 0.9964 | PXD008421 |
| 1705 | Q14137 | BOP1 | 111 | PCPR↓TEMA | n[2 3 0]TEMASAR | 1 | 0.9108 | PXD008421 |
| 1921 | P08133 | ANXA6 | 282 | MVSR↓SELD | n[2 3 0]SELDMLDIR | 5 | 0.985 | PXD008421 |
| 1966 | P20042 | EIF2S2 | 286 | HTCR↓SPDT | n[2 3 0]SPDTILQKDTR | 1 | 0.9932 | PXD008421 |
| 4314 | P32322 | PYCR1 | 312 | LLPR↓SLAP | n[2 3 0]SLAPAGKD | 1 | 0.9054 | PXD008421 |
| 4888 | P06734 | FCER2 | 254 | PTSR↓SQGE | n[2 3 0]SQGEDCVMMR | 1 | 0.8459 | PXD006723 |
| 5505 | Q9Y388 | RBMX2 | 229 | PKSR↓TAYS | n[2 3 0]TAYSGGAEDLER | 2 | 0.9984 | PXD008421 |
2.3. Cell-based screens for validation of GO-2-Substrates predictions
Predictions generated by other algorithms are typically validated and evaluated at the cleavage site level by comparisons with knowledge held within MEROPS [26]. As the most comprehensive repository in the field, MEROPS includes substrates identified by various experimental methods and under different conditions. Consequently, for a given protease, the methods used for substrate validation are inconsistent and negative data (i.e., proteins that are not cleaved) are not included to allow the calculation of false positive / true negative predictions at the protein level. Therefore, we designed a standardized screen for the experimental assessment of our predictions.
In lymphocytes, canonical MALT1 activation requires the assembly of a protein complex of phospho-CARD11 and BCL10 with MALT1, which has not been experimentally reconstituted in vitro. To circumvent this limitation, MALT1 activation can occur in the presence of high molarity (0.8 M) kosmotropic salts to favour protein order. However, these nonphysiological conditions can introduce assay artifacts by altering the interaction of substrate and protease or lowering the Michaelis constant (KM) to favour the cleavage of non-susceptible proteins. Therefore, we established a co-transfection cell-based screen to validate the performance of GO-2-Substrates in predicting substrates for MALT1. Despite presenting its own challenges, a cell-based assay has advantages over these recombinant protein assays. All candidates can be assayed for which expression plasmids are available, which now encompass virtually the entire proteome, in the more relevant cellular milieu. Moreover, the eukaryotic-expressed candidate substrates display natural post-translational modifications that might modify cleavage susceptibility.
For screening, 34 proteins spanning a wide range of high and low probability substrates were selected to assess the sensitivity, specificity, and precision of GO-2-Substrates (Supplementary Table 2, Fig. 3b, c). We co-transfected FLAG-tagged cDNA expression constructs of the candidate substrates with plasmids encoding constitutively active cIAP2-MALT1 [29] (Fig. 3d). In the follow-up secondary screen, we assembled the CBM in Human Embryonic Kidney (HEK293) cells, which lacks detectable endogenous MALT1 activity, by transfection of a single plasmid encoding constitutively active CARD11 (Leu251Pro), BCL10, and MALT1 (Fig. 3e). A positive hit was defined by substrate cleavage by either form of MALT1.
To validate the assay, we co-transfected cIAP2-MALT1 with a C-terminal FLAG-tagged version of RBCK1 (also known as HOIL1), which we previously discovered as a novel MALT1 substrate [30] (Fig. 3f, g). We detected C-RBCK1 by α-FLAG immunoblotting, and when the CBM components were co-expressed with RBCK1, we observed cleavage as characterized previously. As negative controls, we co-transfected catalytically inactive cIAP2-mutant MALT1 (Cys464Ala) with RBCK1 or treated the CBM-transfected cells with the MALT1-specific inhibitor MLT-748 [22]. In both cases, significantly reduced cleavage of RBCK1 was apparent (Fig. 3g).
Positive validation outcomes were defined by (1) a decrease in the amount of full-length C-terminal FLAG-tagged candidate substrate, or (2) generation of a cleavage product with the predicted molecular weight and with α-FLAG immunoreactivity. To be confirmed as an in vitro substrate, these indicators of cleavage had to be reduced in N ≥ 2 independent experiments when the inactive cIAP2-mutant MALT1 (Cys464Ala) was expressed or when CBM-expressing cells were treated with MLT-748. Transfected MALT1 expression was confirmed by western blotting (Fig. 3g, Supplementary Fig. 4, Fig. 5, Fig. 6).
Fig. 4.
RNases ZC3H12D and ZC3H12B are novel MALT1 substrates. a, d Schematic of ZC3H12B and ZC3H12D cDNA expression constructs used for co-transfection with the predicted cut sites and molecular weights of MALT1 cleavage products shown. Myc and FLAG C-terminal tags are as indicated. c-f, Western blot analysis of lysates from HEK293 cells co-transfected with either ZC3H12D (b,c) or ZC3H12B (e, f), together with active or inactive forms of MALT1, or active MALT1 in the presence of a specific MALT1 inhibitor (MLT-748). Full-length proteins are indicated with a black arrow; red arrows indicate C-terminal cleavage products of ZC3H12D and ZC3H12B. c, f Cleavage of ZC3H12D and ZC3H12B are virtually eliminated where the MALT1 cleavage site is mutated to (c) ZC3H12D (R64A) and (f) ZC3H12B (R165A). * Denotes minor cleavage products of ZC3H12B generated at a different site to MALT1 by unknown protease. Mouse α-FLAG antibody was used to detect the proteins as indicated. β-actin, loading control was detected by rabbit β-actin antibody. Positions of electrophoretic mobility of molecular weight markers are as shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
Immune signalling proteins TAB3 and CASP10 are cleaved by MALT1. a, e Schematic of TAB3 (a) and CASP10 (e) cDNA expression constructs used for co-transfection showing the predicted cut sites and molecular weights of MALT1 cleavage products. Myc and FLAG C-terminal tags are as indicated. b-d, f-h Western blot analysis HEK293 cells lysates co-transfected with either TAB3 (b) or CASP10 (f); together with active or inactive forms of MALT1, or with active MALT1 and the allosteric MALT1 inhibitor (MLT-748). N = 3 independent biological experiments are shown; g, h cleavage by MALT1 in the CBM is shown in triplicate n = 3. Full-length proteins are indicated with a black arrow; C-terminal cleavage product of TAB3 (C-TAB3) is indicated by red arrow. c C-TAB3 was only detected upon proteasome inhibition with MG-132. d, h Cleavage of TAB3 and CASP10 are eliminated where the predicted MALT1 cleavage site was mutated to TAB3 (R605A) (d) and CASP10 (R254A) (h). Mouse α-FLAG antibody was used to detect the proteins as indicated.β-actin, loading control was detected by rabbit β-actin antibody. The positions of electrophoretic mobility of molecular weight markers are as shown. Imaged immunoblots are displayed at K = 0, except as indicated when K = 1. K value refers to the curve applied to the pixel intensity histogram in Image Studio Lite. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
Validation of GO-2-Substrates prediction of CILK1 and ILDR2 as novel MALT1 substrates. Schematic of CILK1 (a) and ILDR2 (e) used for co-transfection with the predicted cut sites and molecular weights of MALT1 cleavage products shown. Myc and FLAG C-terminal tags are as indicated. b-g Western blot analysis of lysates from HEK293 cells co-transfected with either CILK1 (b) or ILDR2 (f), together with active (cIAP2-MALT1, CBM) or inactive (cIAP2-MALT1 C464A) forms of MALT1, or with active MALT1 in the presence of a specific MALT1 inhibitor (MLT-748). Full-length proteins are indicated with a black arrow; C-terminal cleavage products of CILK1 (C-CILK1) and ILDR2 (C-ILDR2) are indicated by red arrows. c Inhibition of CILK1 cleavage is greater in cells treated with the irreversible active site inhibitor of MALT1 (z-VRPR-fmk) compared with a reversible allosteric inhibitor (MLT-748). d Cleavage of CILK1 is eliminated where the MALT1 cleavage site is mutated to CILK1 (R407A). g ILDR2 is cleaved by CBM proportional to increased MALT1 cDNA transfected from 1.25 µg to 3.25 μg as indicated by a black-filled triangle. Mouse α-FLAG antibody was used to detect the proteins as indicated. β-actin, loading control was detected by rabbit β-actin antibody. Positions of electrophoretic mobility of molecular weight markers are as shown. Imaged immunoblots are displayed at K = 0, except as indicated when K = 1. K value refers to the curve applied to the pixel intensity histogram in Image Studio Lite. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.4. RNases ZC3H12D and ZC3H12B are cleaved in vitro by MALT1
Four RNases, ZC3H12A, RC3H1, RC3H2, and N4BP1 are known MALT1 substrates [31], [32], [33]. Among the proteins with the highest GO-2-Substrates ranks were RNases ZC3H12D and ZC3H12B, ranked 10 and 21, respectively, suggesting these may also be cleaved by MALT1. Human (h) ZC3H12D displays a predicted MALT1 cleavage site at Leu-Val-Pro-Arg61↓Gly (Supplementary Fig. 15c), which is identical to the MALT1 cleavage site in human and mouse (m) ZC3H12A and mRELB. The mouse orthologue of ZC3H12D has a homologous site, Leu-Ile-Pro-Arg64↓Gly. Co-transfection of ZC3H12D with cIAP2-MALT1 led to a reduction in the amount of full-length protein, together with the appearance of a C-terminal FLAG-tagged cleavage fragment (C-ZC3H12D) of the predicted size (Fig. 4a, b, Supplementary Fig. 4, Fig. 5, Fig. 6) (N = 7). Co-transfection of catalytic inactive cIAP2-MALT1 (Cys464Ala) with ZC3H12D did not generate the FLAG-tagged cleavage fragment and did not change the amount of full-length ZC3H12D compared to the control transfectants without MALT1 (Fig. 4b) (N = 7).
To confirm these results, we co-transfected ZC3H12D with the CBM. This too, resulted in a reduction in ZC3H12D protein coincident with the detection of C-ZC3H12D (Fig. 4b, Supplementary Fig. 4, Fig. 5, Fig. 6) (N = 7). Transfectants cultured in the presence of the MLT-748 inhibitor showed reduced cleavage, demonstrating that the processing of ZC3H12D was MALT1-dependent. Finally, we performed site-directed mutagenesis to ablate the predicted cleavage site in ZC3H12D from Leu-Ile-Pro-Arg64↓Gly to Leu-Ile-Pro-Ala64↓Gly. Neither cIAP2-MALT1 nor the CBM cleaved the mutant ZC3H12D (Arg64Ala) (Fig. 4c), confirming that ZC3H12D was processed and at the predicted MALT1 cleavage site (N = 4).
The related RNase ZC3H12B was similarly assayed (Fig. 4d, e), revealing MALT1 cleavage by cIAP2-MALT1 and the CBM, with near-total loss of the full-length protein (N = 6). Mutating the putative MALT1 cleavage site in mZC3H12B (Arg165Ala) (N = 3) (Fig. 4f) confirmed the cleavage site to be Leu-Leu-Pro-Arg165↓Gly, which is homologous to the human ZC3H12B sequence Leu-Val-Pro-Arg166↓Gly. The processing of ZC3H12B at two additional sites (indicated by * in Fig. 4e, f), even in the catalytic mutant transfected cells or with MLT-748, suggests minor cleavage by an unrelated cell protease. Thus, GO-2-Substrates identified two RNases as new substrates of MALT1, rendering the six RNase MALT1 substrates the largest substrate class to date.
2.5. TAB3 and CASP10 are in vitro MALT1 substrates relevant to NF-κB signalling
With a predicted cleavage site at Leu-Gln-Ser-Arg601↓Gly (identical to mouse Leu-Gln-Ser-Arg605↓Gly), TAB3 was highly ranked by GO-2-Substrates (Fig. 5a, Supplementary Fig. 15a). TGF-β activated kinase 1 (TAK1/MAP3K7) complexes with TAB3 and TRAF2 or TRAF6 [34] to activate NF-κB and JNK, making TAB3 a biologically-compelling candidate substrate. Upon co-transfection with cIAP2-MALT1 or the CBM, we observed the loss of full-length TAB3 protein, which did not occur with the inactive cIAP2-MALT1 (Cys464Ala) or with MLT-748 treatment of CBM-expressing cells (Fig. 5b) (N = 4). However, cleavage fragments were not detected. As the proteasome turns over TAB3 [35], we hypothesized that rapid proteasomal degradation of the FLAG-tagged C-TAB3 fragment occurs following MALT1 cleavage. We inhibited proteasome activity with the inhibitor MG-132, which stabilized full-length TAB3 and, after cleavage by cIAP2-MALT1, C-TAB3 was now detected (Fig. 5c) (N = 3). The location of the MALT1 cleavage site was confirmed by site-directed mutagenesis of TAB3 (Arg605Ala), which abolished TAB3 processing by cIAP2-MALT1 and the CBM (Fig. 5d) (N = 2).
Our understanding of the precise role of caspase 10 in immune cell fate is incomplete. Caspase 10 mediates T cell apoptosis in death-receptor signalling [36], [37]. However, other evidence suggests that caspase 10 switches the cellular response favouring NF-κB activation and cell survival [38]. We selected caspase 10 for validation (Fig. 5e, Supplementary Fig. 15f) and found that full-length caspase 10 levels were reduced, and the C-terminal cleavage product (C-CASP10) was detected in cells co-transfected with CBM in N = 5 independent biological replicate experiments, but not in cells transfected with cIAP2-MALT1 (N = 3) (Fig. 5f, Supplementary Fig. 7). We also observed cleavage of caspase 10 by MALT1 in a CBM-concentration dependent manner (Supplementary Fig. 7) (N = 2). Both mutating the predicted MALT1 cleavage site in caspase 10 (Leu-Val-Ser-Arg254↓Gly) with an Arg254Ala replacement (N = 3, n = 5) and inhibition of MALT1 with MLT-748, (N = 5, n = 9), reduced cleavage (Fig. 5f – h). Because caspase 10 is activated by auto-processing at Ile-Glu-Ala-Asp415↓Ala [39], we repeated the co-transfection assays with 20 μM SCP0094, a caspase 10 inhibitor, which did not alter MALT1 cleavage of caspase 10 and confirmed that cleavage was not due to autoprocessing (Supplementary Fig. 7).
2.6. New substrates beyond the NF-κB pathway
Despite lacking any overt link to known MALT1 biology, CILK1 was ranked highly by GO-2-Substrates (#28), with identical cleavage sites Leu-Ile-Ser-Arg↓Ser predicted in human and mouse at Arg410 and Arg407, respectively (Fig. 6a, Supplementary Fig. 15g). As a widely-expressed kinase, CILK1 is essential in cilia formation [40]. This intriguing hint of potential new biological roles for MALT1 outside NF-κB activation with a near consensus MALT1 cleavage site triggered further investigation. Co-transfection of CILK1 with cIAP2-MALT1 or the CBM led to efficient cleavage of CILK1, reducing the amount of full-length CILK1 protein coincident with the detection of C-CILK1 (Fig. 6a-d) (N = 8). MALT1-dependent cleavage did not occur in cells co-transfected with the catalytic inactive MALT1 or when the non-cleavable CILK1 (Arg407Ala) mutant was expressed (Fig. 6d) (n = 2). Inhibition of CILK1 cleavage was more significant in cells treated with an active site inhibitor of MALT1 (z-VRPR-fmk) (n = 2) compared with an allosteric inhibitor (MLT-748) (Fig. 6c).
ILDR2 has a lower GO-2-Substrates rank (#196) than the substrates described above but has three candidate cleavage sites (Fig. 6e, Supplementary Fig. 15e). Using the same approaches, we showed that ILDR2 was cleaved in a MALT1-dependent manner by cIAP2-MALT and the CBM (Fig. 6f) (N = 4). Co-transfection of ILDR2 with an increasing quantity of cIAP2-MALT1 expression plasmid led to a concentration-dependent decrease in full-length ILDR2 and increased detection of C-ILDR2 (517–642) (Fig. 6g) (n = 3). The catalytic mutant cIAP2-MALT1 (Cys464Ala) and MLT-748 reduced cleavage (Fig. 6f, g), confirming that ILDR2 is also an in vitro MALT1 substrate.
In addition to the six new MALT1 substrates that we conclusively identified, four other candidates—CRADD, USP10, TNFRSF25, and ZFPL1 yielded experimental evidence for cleavage by one or more of our criteria yet were inconclusive or not reliably reproducible N ≥ 2 (Supplementary Fig. 8). In the interest of high stringency, these four proteins were considered 'null' outcomes of the screen, but future studies may confirm their cleavage.
2.7. Performance assessment of GO-2-Substrates predictions
A predictive model's performance assessment can include a measure of feature importance to determine the most predictive features. However, our cellular screens limit the scale of such analyses due to the number of candidate substrates that are feasible to test by co-transfection with the positive and negative controls for validation. Nonetheless, we expanded our screen with substrate candidates spanning a wide range of GO-2-Substrates ranks, which we expected would include true negatives for improved algorithm performance assessment (Fig. 3b, c, Supplementary Table 1). Of these, 24 proteins were not cleaved, including CARD14, CFLAR, IL17RA, NLRP3, TRIM56 and a chemokine receptor, CXCR3, all of which have immune functions and so were reasonable to screen (Supplementary Fig. 9 – 11). Interestingly, the sequence logo of the candidate cleavage sites from the proteins that screened negative is similar to the consensus cleavage site logos of the known substrates (Fig. 1b, c, Supplementary Fig. 11 h). The inability of MALT1 to cleave these proteins may be due to cleavage site inaccessibility within the folded protein or the inability of these proteins to interact with substrate-binding exosites [8] on the CBM complex.
With 34 proteins tested in the co-transfection assays, Receiver Operating Characteristic (ROC) plots could now assess the sensitivity and specificity (Fig. 7a) of predicted screening outcomes for each module separately and then combined as used in GO-2-Substrates. As a measure of prediction performance, the Area Under the Curve (AUC) of the candidate substrate ranks from the Function Module, 0.698 (Fig. 7c), Sequence Module, 0.816 (Fig. 7d), and GO-2-Substrates, 0.84 (Fig. 7e) all surpassed sequence analysis alone by FIMO, 0.632 (Fig. 7b). Therefore, the ROC analyses validated our new criteria that increase the power of substrate predictions.
Fig. 7.
GO-2-Substrates rank is predictive of cleavage by MALT1. a The equations used to generate receiver operating characteristic (ROC) curves. True positives (TP), false positives (FP), true negatives (TN) and false negatives (FN). b-e ROC curves and associated area under the curve (AUC) were calculated using precrec package in R to measure the performance of FIMO (b), the Function Module (c), the Sequence Module (d), or GO-2-Substrates (e) in the classification of outcomes from the co-transfection screen. Outcomes were defined as 'positive’ (n = 6) or ‘negative’ (n = 24) where evidence of cleavage was reproducibly detected or absent, respectively. In terms of AUC, the Function and Sequence Modules both surpassed the performance of FIMO for classification. GO-2-Substrates was the best classifier overall. f Definition of precision used to generate Precision-Recall (PR) curves. g-j PR curves and associated PR-AUC were calculated using the precrec package in R to measure the performance of FIMO (g), the Function Module (h), the Sequence Module (i), or GO-2-Substrates (j) in the classification of outcomes from the co-transfection screen. When applied alone or combined with the Substrate Module as part of GO-2-Substrates, the Function Module increased precision at most sensitivity thresholds versus FIMO or the Sequence Module, respectively. k Scatterplot showing the experimentally derived classification precision of GO-2-Substrates, at the thresholds of all proteins included in the co-transfection screen, up to GO-2-Substrates rank 196 (sensitivity = 1). l Precision at the GO-2-Substrates rank thresholds of all positive outcomes from the co-transfection screen. A precision of 0.5 was determined for classifications at the GO-2-Substrates rank threshold of 40.
As our screen yielded an imbalanced set of outcomes (6 positives, 24 negatives, and 4 nulls), Precision-Recall (PR) (Fig. 7f) plots can better predict future classification performance [41]. The PR-AUC values for FIMO, 0.245 (Fig. 7g), the Function Module, 0.290 (Fig. 7h), the Sequence Module, 0.566 (Fig. 7i), and GO-2-Substrates (0.616) (Fig. 7j) showed the increased prediction precision by incorporating the Function Module at most sensitivity thresholds versus FIMO or the Sequence Module alone. Thus, protein function improves substrate predictions.
To more intuitively interpret the likely outcomes of future MALT1 substrate screens using GO-2-Substrates, we compared prediction precision versus the GO-2-Substrates rank at thresholds corresponding to all experimentally tested proteins up to the last positive outcome (i.e., sensitivity = 1, rank 196) (Fig. 7k, l). Up to a GO-2-Substrates rank of 40, five candidate substrates were cleaved and five were not, rendering a Precision = 0.5 (Fig. 7l). With 18 untested candidates in the top 40 proteins ranked by GO-2-Substrates (Supplementary Table 1), we predict that approximately 50 % will likely be new MALT1 substrates, a notable success rate. Indeed, during the preparation of this manuscript, CARD10 (rank 14) was validated as a novel MALT1 substrate [42]. To our knowledge, our work represents the first method to rank human proteins as potential MALT1 substrates by proteome-wide analysis. Thus, a direct comparison of GO-2-Substrates performance with other existing algorithms is precluded.
2.8. Function Module surpasses PSSM-based prediction of substrates for other proteases
Despite the clear sequence specificity of MALT1, our predictions were improved by incorporating the Function Module. We hypothesized that the Function Module would be even more valuable for substrate prediction of proteases with looser sequence specificity, such as matrix metalloproteinases (MMPs) [43]. We obtained substrate cleavage data from MEROPS for MMP9 and neutrophil elastase (also known as human leukocyte elastase, HLE), which are representative of the metallo- and serine protease classes, respectively. Unlike for MALT1, the high number of known physiologically relevant substrates of these proteases (MMP9, N = 51 proteins; HLE, N = 54 proteins) allowed us to split protein substrates into training (75 %) and test (25 %) datasets and assess the performance of the Function Module versus FIMO. Training data for each protease was used as input for FIMO or the Function Module to rank all human proteins. These ranks were then compared using the Mann-Whitney-Wilcoxon test. In both cases, the Function Module outperformed FIMO by ranking known substrates in the test dataset significantly higher (Supplementary Fig. 12) (HLE, p = 5.8 × 10-5; MMP9, p = 6.6 × 10-3). Thus, even when agnostic of cleavage site sequence information, the Function Module is predictive of substrates for non-generalist proteases of different clades.
2.9. Expansion of sequence space searches identifies TANK as a MALT1 substrate
The GO-2-Substrates analyses reported above for MALT1 in PSSM winnowing mode were restricted to the subset of the human proteome containing a P4–P1′ sequence having a FIMO rank higher than the lowest-ranked known MALT1 substrate cleavage site (Fig. 8a). Our experimental outcomes showed that by leveraging functional knowledge with sequence information, the precision of candidate screening is dramatically improved over predictions based on FIMO rank alone. Buoyed by this, we expanded the scope of GO-2-Substrates from these 4,321 proteins to now search the entire proteome to capture novel substrates with cleavage sites diverging even more from the known MALT1 substrates. We identified TANK as a promising candidate by this expanded GO-2-Substrates analysis termed ‘whole proteome mode’ (Fig. 8b, Supplementary Table 2, Supplementary Fig. 15d). By sequestering TRAF1, 2 and 3 and regulating IκB-kinase, TANK (Supplementary Fig. 16d) is a negative regulator of NF-κB and notably forms complexes with the substrate ZC3H12A. TANK ranked exceptionally high in the Function Module (#21), which led to a high GO-2-Substrates rank (#98) despite a poor Sequence rank (#11,251) (Fig. 8c) for a potential His-Ile-Pro-Arg394↓Val cleavage site (Fig. 8d, Supplementary Table 3).
Fig. 8.
Expansion of analysis identifies TANK as a novel MALT1 substrate. GO-2-Substrates can be analyzed in two modes, which rank a different range of proteins according to their potential for cleavage by MALT1. a 'PSSM winnowing mode' considers only proteins containing a candidate cleavage site with FIMO ranking better than the last known MALT1 substrate cleavage site, or b 'Whole proteome mode' that includes every human protein. c TANK was identified as one of the top 15 functionally ranked candidate substrates of GO-2-Substrates analyzed in Whole proteome mode. d, Schematic of TANK cDNA expression construct used in the MALT1 co-transfection screen, together with the predicted cut-site and MALT1 cleavage products if cut. e Western blot showing TANK cleavage by cIAP2-MALT1, which was not observed where TANK was co-transfected with inactive cIAP2-MALT1 (C464A) or using noncleavable MALT1 (R215K). f TANK was cleaved by co-transfected active CBM. g Apparent molecular weights of TANK cleavage products predicted from cleavage at the indicated cut-site in TANK. h TANK in vitro cleavage by MALT in 0.8 M Na-citrate, 0.1 mM EGTA, 0.05 % CHAPS, 1 mM DTT, 200 mM Tris-HCl, pH 7.4, for 2 h at 37 °C; cleavage was proportional to MALT1 concentration. i Mutagenesis of TANK (R125K) eliminated MALT1 cleavage by the CBM in the co-transfection assay. j TBK1 was not cleaved by co-transfected CBM. k Endogenous TANK cleavage in SSK41 and RAJI cells stimulated with PMA/ionomycin for the times shown and in the presence of MG-132. Positive controls, cleavage fragments (Δ) of RELB and CYLD. β-actin and β-tubulin loading controls were detected by rabbit β-actin and mouse β-tubulin antibodies. Positions of electrophoretic mobility of molecular weight markers are shown.
In co-transfection experiments, TANK was cleaved by cIAP2-MALT1 (Fig. 8e) and the CBM (Fig. 8f). However, the cleavage site was uncertain as the apparent molecular weight of the N-terminal FLAG-product (∼30 kDa) differed markedly from the predicted 45.3-kDa N-terminal product (Fig. 8d). This was unlikely to be due to another protease as transfection of the inactive mutant of cIAP2-MALT1 (Cys464Ala) (Fig. 8e) or CBM containing MALT1 (Cys464Ala) showed no cleavage (Fig. 8f). Sequence inspection revealed a second potential cleavage site at Val-Thr-Pro-Arg215↓Gly. To help clarify this without any other possible proteases present, we incubated recombinant MALT1 with C-terminally tagged recombinant TANK alone in kosmotropic salt buffer (Fig. 8g, h). The similar sizes of the cleavage product in blots probed with two different antibodies supported cleavage at Arg215↓Gly. Indeed, Arg215Lys site-directed mutagenesis ablated TANK cleavage by cIAP2-MALT1 and the CBM (Fig. 8e, i), confirming this was the MALT1 cleavage site. The higher-ranked Arg394↓Val site may be inaccessible due to structural constraints, or FIMO may cease to be predictive at lower thresholds. TBK1 (TANK binding kinase 1) forms part of the same signalling complex as TANK, but TBK1 has a much lower Function (#808) and GO-2-Substrates (#1,839) rank than TANK. We checked TBK1 for cleavage by the CBM, but it was not cut (Fig. 8j).
Screening of B and T cell lines revealed relatively high levels of endogenous TANK in the SSK41 and RAJI human B-lymphoma cell lines but not A20 murine B-lymphoma or Jurkat human T-lymphoma cells (Supplementary Fig. 13). To confirm cleavage of endogenous TANK, SSK41 and RAJI cells were treated with 200 ng ml−1 phorbol myristate acetate (PMA) and 1 µg ml−1 ionomycin to induce endogenous MALT1 activation (Fig. 8k). After 30-min stimulation, we detected TANK cleavage to a ∼ 26-kDa fragment at Arg215↓Gly. Product accumulation increased at 90 min. MALT1-dependent cleavage of the NF-κB inhibitor RelB and the ubiquitin carboxyl-terminal hydrolase CYLD were positive controls.
2.10. Proteomic detection of cleaved substrates predicted by GO-2-Substrates
The variable expression of TANK that we observed in different B-lymphoma cell lines exemplified one of the many challenges in identifying the precise cell type, context, and stimulus where cleavage of the new in vitro MALT1 substrates may naturally occur. Therefore, to detect substrates predicted by GO-2-Substrates using a proteomic approach, we mined two of our published proteomics datasets (PRIDE accessions PXD008421 [30] and PXD006723 [22]) of PMA/ionomycin stimulated human B lymphocytes. We originally generated these datasets to discover novel MALT1 substrates by our N-terminomics substrate discovery method, TAILS [44], where we were the first to describe RBCK1 (also known as HOIL1) as a new substrate [22], [30]. Using a more advanced data analysis program (MSFragger [45]), the raw mass spectrometry data were searched again. Tandem mass tag (TMT)-labelled neo-N-termini generated by protease activity after PMA/ionomycin cell stimulation for two hours were identified by peptide spectrum matches (PSMs). Five PSMs of MALT1-cleaved RBCK1 and 26 PSMs from the neo-N-termini of 13 MALT1 substrate candidates predicted by GO-2-Substrates (Table 1, Fig. 9a and Supplementary Fig. 14) were detected, including ZC3H12D, which we validated in Fig. 4 as a new substrate. To orthogonally validate the mass spectrometry data, we analyzed lysates of PMA/ionomycin stimulated human B lymphocytes by Western blotting. Using an antibody raised against the N-terminus of ZC3H12D, we observed a decrease in ZC3H12D at two hours post-stimulation in three independent B cell lines, two being presented in Fig. 9b-d (N = 3, n = 8). The quantity of ZC3H12D detected in stimulated cells cultured with MLT-748 inhibitor was equal to unstimulated controls, demonstrating that the processing of ZC3H12D was MALT1-dependent. As ZC3H12D and RBCK1 are cleaved by endogenous MALT1 in activated human B lymphocytes, this analysis supports GO-2-Substrates predictions of the other proteins listed in Table 1 being high confidence substrate candidates worthy of further validation.
Fig. 9.
Endogenous ZC3H12D is processed by MALT1 in two independent normal human B lymphocyte cell lines. a Raw mass spectrometry data from proteomic and TMT-TAILS analysis of PMA/ionomycin-stimulated human B lymphocytes [22] were searched against the human proteome using MSFragger. TMT-labelled neo-N-termini generated by protease activity were identified by PSMs, which were compared in sequence with the predicted neo-N-termini of GO-2-Substrates candidate substrates. Annotated fragment spectrum of a ZC3H12D PSM at the predicted MALT1 cleavage site is shown. b Schematic of human ZC3H12D, with the validated and candidate MALT1 cut sites (solid / dashed arrows, respectively) shown. The location of the TMT-labelled neoN-terminal peptide identified by TAILS N-terminomics is shown in red. The peptide spanning the MALT1 cleavage site that was used to raise the 24991–1-AP antibody to ZC3H12D is depicted by a blue line. c, d Cleavage of endogenous ZC3H12D in normal human B lymphocyte cell lines derived from two different donors after stimulation with PMA/ionomycin, 2 h, performed in triplicate (n = 3) for each donor. Cleavage of ZC3H12D was not detected by loss of intact protein when the cells were treated with the MLT-748 inhibitor. The antibody was raised to a peptide spanning one of the two cleavage sites of MALT1, and so it was not unexpected that cleavage fragments of ZC3H12D were not detected. β-tubulin loading control was detected by mouse β-tubulin antibody. Positions of electrophoretic mobility of molecular weight markers are shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3. Discussion
To our knowledge, no current algorithm can be readily implemented to predict MALT1 substrates with measurable precision due to the low number (12) of known substrates that limit training. The GO-2-Substrates approach differs from most cleavage site predictors by integrating features encoding knowledge of protein function and sequence information to rank candidate substrates. Thereby, we discovered and validated seven new in vitro MALT1 substrates — ZC3H12D, ZC3H12B, CILK1, TAB3, CASP10, ILDR2 and TANK in the cellular context and detected cleavage of endogenous TANK and ZC3H12D in B lymphocytes upon stimulation.
Informed by the known substrates of a protease, GO-2-Substrates dynamically scales feature weighting based on their prevalence in the proteome. In so doing, a feature's relative contribution to predictions evolves with each newly identified substrate and with newly-deposited database annotations. On retraining the GO-2-Substrates predictions by including the seven new MALT1 substrates, we generated a revised ranking of candidate MALT1 substrates with an estimated precision of ∼ 50 % for the top 40 predictions. We present Supplementary Table 4 of these high-confidence substrate candidates as a community resource to inspire targeted MALT1 substrate research.
During the preparation of this manuscript, Israël and coworkers [42] described the MALT1 cleavage of CARD10. Despite an unusual Ala at P2 in its MALT1 cleavage site (Leu-Leu-Ala-Arg587↓Gly), GO-2-Substrates correctly ranked CARD10 as an exceptionally strong candidate substrate—mainly due to its functional similarity to known substrates. Indeed, sequence-agnostic analysis of substrates of MMP9 and neutrophil elastase revealed that functional features of known substrates are predictive of cleavage for other ‘non-generalist’ proteases. These results further highlight the utility of knowledge beyond sequence and structural information for protein-level substrate prediction. Future integration of protein structure and knowledge from protein isoforms, post-translational modifications, animal model phenotypes, and genetic variants linked to human disease will be valuable for further improving GO-2-Substrates. Conversely, the integration of protein function features should improve the precision of existing machine-learning algorithms designed to predict the substrates of other proteases.
By sequential cleavage of negative and then positive regulators of NF-κB signalling, MALT1 is proposed to function as a cellular clock for the temporal synchronization of NF-κB signalling initiation and termination [30]. Cleavage of TANK, TAB3 and CASP10 might be cogs in this mechanism. Interestingly, CASP10, TAB3, TANK, and also ZC3H12D are highly connected to protein complexes containing other MALT1 substrates (Supplementary Fig. 17a), which may facilitate this coordination. In response to interleukin-1 or TNF receptor stimulation, TANK dampens NF-κB by association with TRAF6 and TAK1 protein complexes (Supplementary Fig. 15, 16) [46]. Thus, TANK cleavage should increase NF-κB activity by removing TANK’s inhibition of the E3 ligase activity of TRAF6. Indeed, in Tank –/– mice, canonical NF-κB signalling increases in B lymphocytes and macrophages [47]. Conversely, cleavage of TAB3 likely results in a reduction of the NF-κB and JNK signalling flux, evidenced by numerous studies showing the critical role of TAB–TAK complexes in integrating signals to activate these pathways [34], [48]. Notably, the cleavage product N-CASP10 (1–254) lacks the canonical C-terminal domain and mimics the 10-G isoform of CASP10 (1–273) (Supplementary Fig. 15) that induces NF-κB in lymphocytes [49]. Likewise, N-CASP10 may exert a similar role in NF-κB induction.
Given the high homology of ZC3H12B and ZC3H12D with the known RNase substrate ZC3H12A (regnase-1), it was not surprising that ZC3H12B and ZC3H12D were ranked in the top 0.02 % of candidates. We confirmed both RNases as MALT1 substrates, and despite being ‘obvious’ by retrospective consideration, neither had been investigated as substrates before. This highlights the effectiveness of an unbiased workflow, which does not require the user to hold prior knowledge to identify and evaluate candidates. MALT1 cleavage of ZC3H12B and ZC3H12D should increase the stability of their proinflammatory target mRNAs and increase their products' coordinated expression. Indeed, ZC3H12D is a negative regulator of cytokine expression in memory T cells [50] that is relieved upon clonal expansion and activation of these cells. Yang et al. also recently described a role for ZC3H12D in the nuclear transport of nex-IL1β-mRNA, which regulates anti-apoptotic gene expression, migration and interferon-γ production in natural killer cells [51]. MALT1 is activated in natural killer cells via the CARD11-CBM [52]. Thus, cleavage of ZC3H12D by MALT1 might also alter its roles in mRNA transport.
Investigation of ILDR2 and CILK1 cleavage is an exciting new frontier for MALT1 research. CILK1 and another MALT1 substrate (LIMA1) both interact with the regulatory associated protein of mTOR (RPTOR) (Supplementary Fig. 16, 17). Notably, MALT1 has been implicated in the regulation of mTOR signalling [53], which controls cell growth, survival and metabolic flux. Thus, we speculate that MALT1 might sculpt protein complexes regulating mTOR pathway flux. However, validation of substrate cleavage by endogenously expressed MALT1 will be an essential step toward uncovering any functional consequences. The precise cell types and stimuli under which cleavage of ZC3H12B, CASP10, TAB3, ILDR2 and CILK1 may naturally occur, remain to be discovered. Our success in mining N-terminomics proteomic data to identify endogenous cleavage of ZC3H12D and the twelve other candidates highlights the potential to utilize targeted proteomics to complement bioinformatics predictions. By analyzing different primary human cell types, it should be possible to perform high throughput screening of candidate substrates under physiologically relevant stimuli, at endogenous protein levels, and within endogenous complexes.
3.1. Caveats
GO-2-Substrates is a bioinformatics approach and, like all predictive algorithms, has inherent limitations and bias. The precision of our approach is based on the number of substrates known, which was 12 when our study began. This low input number could neither be used for machine learning nor to predict proteins with completely unknown functions or similarities to any known substrate. Thus, our method was designed for predictions for proteases with low numbers of known substrates but is less suited for substrates that differ markedly from known substrates in every feature utilized in predictions. The different activation platforms of MALT1 (cIAP2-MALT1, TRAF6-MALT1 and CARD9, 10, 11 and 14 CBM) may result in the cleavage of some substrates more efficiently than others in specific contexts. This may affect the performance of our predictions and the ability to validate substrates in physiological conditions when even the cell type to investigate may be unknown. By co-transfecting constitutively active MALT1 with candidate substrates, our cell-based screen circumvented the variable factor of MALT1 activation and avoided the limitations associated with the high molarity salt in vitro assays. However, our screen does not identify the stimuli and physiological conditions under which substrate cleavage occurs. Furthermore, some of the candidates that yielded a negative outcome may be false negatives. For example, they may not be cleaved in the screening cells as isolated proteins and instead might require context or cell-specific PPI or CARD 9, 10 or 14 CBM complexes. Transfection could also result in overexpression or mislocalization of proteins, which may introduce false positive or negative outcomes to the screen. Nonetheless, a biological false positive in this screen still confirms the validity of the method, just not the biological context such a substrate might be cleaved. Despite these limitations, we consider that our approach was the most appropriate to achieve a high confidence ranking and validation of new MALT1 substrates.
4. Methods
4.1. Data collection and FIMO
Experimentally validated substrates of human MALT1 were manually curated from the literature (accessed on 15-Nov-2020). Mouse orthologs of human MALT1 substrates were included if any of the following conditions were met: 1) MALT1 cleavage of the mouse ortholog has been experimentally demonstrated; 2) the mouse ortholog contains an exact sequence match to an experimentally determined MALT1 cleavage site from P4–P1′ of any protein (human or mouse). The consensus cleavage sites were visualized using ggseqlogo within R [54]. Amino acid sequences spanning P5–P5′ of human and mouse cleavage sites were converted to a combined PSSM using Script G. The PSSM was used as the input for FIMO (MEME v5.3.3) [27], which we applied to search human proteome sequences (Reviewed entries of UP000005640; 07-Mar-2021 release) for instances of motif occurrence. The FIMO output scores were then ranked. FIMO parameters were set to defaults except for the following: count = 0.01, output threshold = 0.01.
4.2. Features and criteria utilized by GO-2-Substrates
To rank proteins for their MALT1 cleavage potential, we integrated information from various sources using Script A. Proteins were assigned a score when the following qualifying criteria were met.
i) Sequence Module. Exact sequence match to a known MALT1 cleavage site. Criteria 1: The protein contains an exact sequence match to P4 – P2′ of a human or mouse MALT1 cleavage site sequence. Criteria 2: As for Criteria 1, except sequence matches P4 – P1′.
ii) FIMO ranking. Criteria 1: The sensitivity of identification of known MALT1 substrates by FIMO was modelled relative to FIMO rank by local regression (LOESS). Criteria 1: The protein contains a candidate cleavage site ranked ≤ 50 % of the known human MALT1 cleavage sites. Criteria 2: As Criteria 1, except a candidate cleavage site is ranked ≤ 80 %.
iii) Conservation of cleavage sites across species. Orthology between human and mouse proteins was exported from the OMA orthology database using the online genome pair orthology tool [55]. From this output, their Ensemble IDs were converted to UniProt entry names using Script C. Criteria 1: Human and mouse protein orthologs both contain a sequence exactly matching a human or mouse MALT1 cleavage site from P4 – P1′ sequence. Criteria 2: Mouse ortholog of a human protein contains a sequence matching a candidate MALT1 cleavage site from P4 – P1′ with a FIMO rank ≤ 80 % of known human MALT1 cleavage sites.
iv) Function Module. Gene Ontology. UniProt entry names of published MALT1 substrates were input to the STRING (v11.0) [56] web interface for functional enrichment analysis (parameters used: query proteins only; background whole genome). Enriched Biological Process, Molecular Function, and Cellular Compartment terms were exported. For each category, enriched terms were ordered by their 'strength' of enrichment (i.e., log10 of the number of MALT1 substrates annotated with a GOTERM/total number of proteins annotated with that term). Terms with lesser strength contain lower information content. Therefore, to differentiate between more and less informative enriched GOTERMS, we derived the optimal linear model for the lowest ranking 5 % of enriched GOTERM strengths vs the number of proteins possessing that annotation (Supplementary Fig. 1). The y-intercept of this model was used to define the criteria for GOTERM scoring as follows. Criteria 1: The protein is annotated with a GOTERM enriched among MALT1 substrates and possesses a strength of enrichment ≥ median strength of all enriched GOTERMS above the linear model y-intercept. Criteria 2: As Criteria 1, except enrichment strength is ≥ the linear model y-intercept. Criteria 3: As Criteria 1, except enrichment strength is ≤ the linear model y-intercept.
v) Protein domain/gene family: Criteria 1: Protein is annotated with ≥ 1 InterPro feature enriched among known MALT1 substrates. Criteria 2: Protein is annotated with ≥ 1 InterPro feature in common with annotations of any known MALT1 substrate.
vi) Protein-Protein interactions: All known protein–protein interactions present in BioGRID (Homo Sapiens v4.2.191) were exported (15-Nov-2020), and physical interactors of both MALT1 and experimentally validated MALT1 substrates were filtered. Criteria 1: Protein interacts with MALT1. Criteria 2: Protein interacts with ≥ 1 MALT1 substrate.
4.3. Feature scoring
We used Script A to identify proteins meeting the criteria set for each feature comprising the Sequence and Biological Modules. Proteins meeting each criterion were assigned a raw score equal to the reciprocal number of instances in the proteome where that criterion was met. This raw score was then normalized as the product of the proportion of published MALT1 substrates meeting the same criteria (Fig. 2). This yielded a 'Criteria Score' for each protein; the Criteria Scores were summed for each feature to yield a 'Feature Score'.
4.4. Module scoring, ranking and calculation of GO–2–Substrates rank
The sum of Feature Scores for each module were ranked to generate 'Sequence Module' and 'Function Module' ranks for each candidate MALT1 substrate. As a consequence of how tied ranks were handled (value of tied ranks assigned to first instance) and the nature of the data, the maximum rank differed for each module. Hence, module ranks were normalized using min–max normalization to improve the direct comparison of ranks between modules and prevent skewing of GO-2-Substrates ranks towards any given module. Finally, a GO-2-Substrates rank was calculated for each protein as the ranked product of normalized Biological and Sequence module ranks.
4.5. Construct design
Expression plasmids encoding cIAP2-MALT1 and cIAP2-MALT1 Cys464Ala were obtained from the BCCM/LMBP collection (#5537 and #5538). Human CBM components were encoded in a single expression plasmid-encoding FLAG-tagged MALT1 (NM_006785.3), BCL10 (NM_003921.4), and an active oncogenic mutant of CARD11 (Leu251Pro) (NM_032415.5). Expression constructs encoding candidate substrates with a C-termini myc and FLAG-tag (Supplementary Table 2) were from (Origene, GeneCopoeia, and GenScript). Noncleavable human TANK (Arg215Lys) was made using Phusion Site-Directed mutagenesis (Thermo Fisher Scientific); all other substrate P1-Arg to Ala cleavage-resistant constructs were generated using Quick-Change XL (Agilent Technologies) site-directed mutagenesis. Mutagenesis primers are listed in Supplementary Table 2. All DNA sequences were verified by Sanger sequencing.
4.6. Co-transfection screen for MALT1 substrates
HEK293 epithelial cells (verified mycoplasma free) (ATCC, CRL-1573, RRID: CVCL_0045) were maintained in Dulbecco's Modified Eagle Medium (DMEM) high glucose (Sigma), supplemented with 10 % (v/v) fetal bovine serum (FBS) (Sigma-Aldrich), 2 mM l-glutamine, 100 U ml−1 penicillin and 100 µg−1 streptomycin (Gibco). Cells were transfected with 1.25 µg plasmid DNA encoding the candidate MALT1 substrates, wild type or catalytic inactive human MALT1 (Cys464Ala) using lipofectamine 3000 (Thermo Fisher Scientific) according to manufacturer instructions in 6-well plates. For MALT1 inhibitor experiments, cells were treated with either 2 µM MLT-748 [22] or 75 µM z-VRPR-phenyl methyl ketone (fmk) for 30 min before transfection and again after 24 h. MG-132 treatment (10 µM) was initiated 8 h before cell lysis. Cell culture media was removed 48-h post-transfection, and the cells were washed with ice-cold phosphate-buffered saline before lysis and sonication in 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 % NP-40, 1 × Protease Inhibitor Cocktail (Bimake), 50 µM PR-619 (LifeSensors Inc.). Lysates were clarified by centrifugation at 14,000 × g for 15 min, and the protein concentration was measured by BCA assay. Lysates (20 µg) were analyzed by Western blotting with the following primary antibodies: mouse anti-FLAG-M2 (1:4000, Sigma, F1804), mouse or rabbit anti-MALT1 (1:500, Abcam; clone 50, Serotec; polyclonal H-300 from Santa Cruz, or polyclonal PA5-79622 from ThermoFisher, respectively), mouse anti-myc-tag (Clone 9E10) (Millipore), rabbit anti-β-actin antibodies (1:200, Abcam, ab115771), and mouse anti-β-tubulin (Sigma, 1:2000, TUB 2.1), 2 h, 20 °C; secondary antibodies used were: donkey anti-mouse Alexa-Fluor 680 (Invitrogen) and donkey-anti rabbit IRDye 800CW (LI-COR Biosciences) for 1 h, 20 °C. Western blots were imaged using an Odyssey scanner (LI-COR Biosciences) or by enhanced chemiluminescence (Amersham) using Kodak SB5 radiographic film. Scanned images of immunoblots were cropped in the final figures for clarity and conciseness. Immunoblots imaged using Odyssey scanner were analyzed using Image Studio Lite (v5.2.5) and are displayed at K = 0, except as indicated by a box when K = 1. K value refers to the curve applied to the pixel intensity histogram in Image Studio Lite.
4.7. Statistical performance measurement
Significance testing for differences between FIMO, Function Module and Sequence Module ranks was performed using the Wilcoxon signed rank test in R. The outcomes of the co-transfection screen were assessed by Receiver Operator Characteristic curves, Area Under the Curve measurements, Precision-Recall curves and Precision-Recall Area Under the Curve measurement, which were generated and calculated using precrec in R [57].
4.8. MALT1 in vitro cleavage assays
Full-length human MALT1 protein was expressed and purified as described previously [58]. Full-length human TANK protein with a C-terminal Myc/DDK tag was obtained from Origene Technologies (TP309759). TANK (0.05 µg μL−1) was incubated with different concentrations of MALT1 in kosmotropic salt assay buffer (200 mM Tris-HCl, 0.8 M Na-citrate, 0.1 mM EGTA, 0.05 % CHAPS, 1 mM DTT, pH 7.4) for 2 h at 37 °C. Assay products were separated on 4–12 % Bis-Tris SDS-PAGE gradient gels (Life Technologies), and TANK cleavage was confirmed by immunoblotting using antibodies to TANK (rabbit anti-TANK; #2141, Cell Signalling Technology) and the DDK tag (mouse anti-FLAG-M2, Sigma). MALT1 protein was detected using a mouse anti-MALT1 antibody (clone 50, Bio-Rad AbD Serotec ltd).
4.9. Cleavage of endogenous TANK in SSK41 and RAJI cells
The RAJI cell line was obtained from Deutsches Zentrum für Mikroorganismen und Zellkulturen (DSMZ); Dr. Martin S. J. Dyer (University of Leicester, U.K.) generously provided the SSK41 cell line. Single-nucleotide polymorphism profiling authenticated the cell lines, and all cells were mycoplasma negative. The suspension cells were grown in RPMI containing 10 % fetal bovine serum, 2 mM l-glutamine, 100 U ml−1 penicillin, and 100 µg ml−1 streptomycin (Amimed). B cell lymphoma cells were stimulated for 30 or 90 min using 200 ng ml−1 PMA and 1 µM ionomycin (Sigma-Aldrich). All cells were treated for 120 min with 5 µM MG-132 (Sigma) to stabilize cleavage products from proteasomal degradation. For cell lysis, cells were harvested by centrifugation, washed in phosphate-buffered saline and the cell pellets lysed in NP-40 lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1.0 % NP-40) supplemented with complete protease inhibitor cocktail and phosSTOP phosphatase inhibitor cocktail (Roche Life Science) and clarified by centrifugation. After boiling in 1 × Laemmli buffer, proteins were separated on 4–12 % Bis-Tris SDS-PAGE gradient gels (Life Technologies). Proteins were detected by immunoblotting using the following antibodies: mouse anti-CYLD (E-10) and mouse anti-TANK (D-2) from Santa Cruz Biotechnology; mouse anti-MALT1 (MCA2801, clone 50) from Bio-Rad AbD Serotec; mouse anti-β-tubulin (TUB 2.1) from Sigma-Aldrich; rabbit anti-RelB (C1E4) from Cell Signalling Technology. Horseradish peroxidase-coupled donkey anti-rabbit and sheep anti-mouse secondary antibodies were from G.E. Healthcare.
4.10. Cleavage of endogenous ZC3H12D in human B cells
The University of British Columbia/Children’s and Women’s Health Centre of British Columbia Research Ethics Board approved the research protocols for studies on human samples. Written informed consent and assent from minors for participation in this study were obtained. Normal human B cells (EBV-immortalized) [30] (5 × 105 cells) were cultured in RPMI media as described for SSK41 and RAJI cells. Cells were stimulated with 50 nM PMA and 1 µM ionomycin for 2 h, with or without 1 h pre-treatment with 2 µM MLT-748. Cells were washed with ice-cold phosphate-buffered saline before lysis and sonication in 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 % NP-40, 1 × Protease Inhibitor Cocktail (Bimake), 50 µM PR-619 (LifeSensors Inc.). Lysates were clarified by centrifugation at 14,000 × g for 15 min, and the protein concentration was measured by BCA assay. Lysates (20 µg) were analyzed by Western blotting with the following primary antibodies: rabbit anti-ZC3H12D (1:500, ProteinTech, 24991–1-AP) or mouse anti-β-tubulin (1:2000, AbLab, BT7R), 16 h, 4 °C; secondary antibodies used were: donkey anti-rabbit Alexa-Fluor 680 (Invitrogen) and donkey-anti mouse IRDye 800CW (LI-COR Biosciences) for 1 h, 20 °C. Western blots were imaged using an Odyssey scanner (LI-COR Biosciences).
4.11. Mass spectrometry data analysis
Raw mass spectrometry data (PRIDE accessions PXD008421 and PXD006723) of TMT-tagged peptides labelled before trypsin digestion at the whole protein level and then enriched by TAILS N-terminomic analyses of normal human B cells (EBV-immortalized) stimulated with 50 nM PMA and 1 µM ionomycin for 2 h, as previously described in full [30], were searched using MSFragger [45] (v3.4) within FragPipe (v.17.1). Search criteria included 20 ppm tolerance for MS1 and 0.6 Da for MS2 with optimised mass calibration, fixed TMT modification on lysine (+229.1629 Da), fixed iodoacetylation on cysteine (+57.0215 Da Da), variable Met oxidation (+15.9949 Da), variable cyclization of N-terminal (i) glutamine (Gln → pyro-Glu; −17.0266 Da) and (ii) glutamate (Glu → pyro-Glu; −18.0106 Da), and the following variable N-terminal modifications: TMT (+229.1629 Da), acetylation (+42.0106 Da), methylation (+14.0157 Da), dimethylation (+28.0313 Da) and trimethylation (+42.0797 Da). Up to one missed cleavage was allowed and a single enzymatic terminus was required (N-terminal nonspecific). A false discovery rate (FDR) cut off < 0.01 was applied for all PSMs by PeptideProphet within FragPipe. N-terminally modified PSMs were compared with the GO-2-Substrates predicted neoN-terminal peptides that would be generated upon cleavage by MALT1 in R (v4.0.3).
4.12. Protein-protein interaction and Pathfinder analysis
All known PPIs present in BioGRID (v4.3.196) were downloaded (Accessed on 15-Nov-2020). PPIs were filtered to include only ‘physical’ interactions between human proteins using Script H. We calculated the number of instances that each PPI has been observed and the number of experimental methods validating each PPI using Script H. PathLinker [59] (v1.4.3) within Cytoscape [60] calculated the top 100 paths connecting all MALT1 substrates based on the confidence of PPIs, which was based on the number of observations of each PPI. Unconnected MALT1 substrates in these Top 100 paths were analyzed in isolation by PathLinker. The top 10 paths connecting all MALT1 substrates to ILDR2, CILK1, ZC3H12B and LIMA1 alone were calculated and visualized.
4.13. Computer code
Code and accompanying documentation are available for download as Supplementary data, or from https://github.com/OverallLab/go-2-substrates.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We thank M. Gold for insightful comments and discussion, T. Klein for assistance with proteomic datasets and Marvin Festag for expert technical support. This work was supported by the Canada Research Chairs program to C.M.O. (950-20-3877) and S.E.T., the Canadian Institutes of Health Research (CIHR) Foundation grant program (FDN-148408 to C.M.O.), the Michael Smith Foundation for Health Research (MSRF) (IN-NPG-00105 to C.M.O.), Genome British Columbia (SIP007 to S.E.T.), a CIHR grant (PJT 178054 to S.E.T.), a German Academic Exchange Service (DAAD) PROMO scholarship (to S.S.), a Frederick Banting and Charles Best Canada Graduate Scholarships Doctoral Awards fellowship (to H.Y.L.), a Killam Doctoral Scholarship (to H.Y.L.), a Friedman Award for Scholars in Health (to H.Y.L.), and a University of British Columbia Four Year Doctoral Fellowship (to H.Y.L.).
Author contributions
P.A.B. wrote all the computer code, designed and performed all experiments except as listed, analyzed all data, prepared all figures and tables, wrote and edited the paper, and conceived the study. P.A.B. and S.S. designed and performed co-transfection screens and analyzed data. P.A.B. and C.P. performed CASP10 and CILK1 co-transfection experiments and analyzed data. F.R. designed and performed TANK co-transfection experiments, recombinant and endogenous TANK cleavage experiments, and analyzed data. P.A.B., S.S. and H.L. performed endogenous ZC3H12D cleavage experiments and analyzed data. S.E.T. analyzed data and edited the manuscript. F.B provided input into experimental design, analyzed data, provided expression constructs and edited the manuscript. C.H.R. designed the TANK experiments, analyzed data, and edited the manuscript. C.M.O. designed experiments, analyzed the data, wrote and edited the paper, and conceived and supervised the research study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.08.021.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Overall C.M., Blobel C.P. In search of partners: linking extracellular proteases to substrates. Nat Rev Mol Cell Biol. 2007;8:245–257. doi: 10.1038/nrm2120. [DOI] [PubMed] [Google Scholar]
- 2.Li F., et al. DeepCleave: a deep learning predictor for caspase and matrix metalloprotease substrates and cleavage sites. Bioinformatics. 2020;36:1057–1065. doi: 10.1093/bioinformatics/btz721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott B.M., Lacasse V., Blom D.G., Tonner P.D., Blom N.S. Predicted coronavirus Nsp5 protease cleavage sites in the human proteome. BMC Genom Data. 2022;23:25. doi: 10.1186/s12863-022-01044-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li F., et al. Procleave: Predicting Protease-specific Substrate Cleavage Sites by Combining Sequence and Structural Information. Genomics Proteomics Bioinformatics. 2020;18:52–64. doi: 10.1016/j.gpb.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schauperl M., et al. Characterizing Protease Specificity: How Many Substrates Do We Need? PLoS ONE. 2015;10:e0142658. doi: 10.1371/journal.pone.0142658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li F., et al. Twenty years of bioinformatics research for protease-specific substrate and cleavage site prediction: a comprehensive revisit and benchmarking of existing methods. Brief Bioinform. 2019;20:2150–2166. doi: 10.1093/bib/bby077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turk B., Turk V., Turk D. Structural and functional aspects of papain-like cysteine proteinases and their protein inhibitors. Biol Chem. 1997;378:141–150. [PubMed] [Google Scholar]
- 8.Overall C.M. Molecular determinants of metalloproteinase substrate specificity: matrix metalloproteinase substrate binding domains, modules, and exosites. Mol Biotechnol. 2002;22:51–86. doi: 10.1385/MB:22:1:051. [DOI] [PubMed] [Google Scholar]
- 9.Zhou J., et al. Deep profiling of protease substrate specificity enabled by dual random and scanned human proteome substrate phage libraries. Proc Natl Acad Sci U S A. 2020;117:25464–25475. doi: 10.1073/pnas.2009279117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song J., et al. iProt-Sub: a comprehensive package for accurately mapping and predicting protease-specific substrates and cleavage sites. Brief Bioinform. 2019;20:638–658. doi: 10.1093/bib/bby028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uliana F., et al. Mapping specificity, cleavage entropy, allosteric changes and substrates of blood proteases in a high-throughput screen. Nat Commun. 2021;12:1693. doi: 10.1038/s41467-021-21754-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilkins M.R., et al. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 13.Wang M., et al. Cascleave 2.0, a new approach for predicting caspase and granzyme cleavage targets. Bioinformatics. 2014;30:71–80. doi: 10.1093/bioinformatics/btt603. [DOI] [PubMed] [Google Scholar]
- 14.Piippo M., Lietzén N., Nevalainen O.S., Salmi J., Nyman T.A. Pripper: prediction of caspase cleavage sites from whole proteomes. BMC Bioinf. 2010;11:320. doi: 10.1186/1471-2105-11-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wee L.J., Tan T.W., Ranganathan S. CASVM: web server for SVM-based prediction of caspase substrates cleavage sites. Bioinformatics. 2007;23:3241–3243. doi: 10.1093/bioinformatics/btm334. [DOI] [PubMed] [Google Scholar]
- 16.Cieplak P., Strongin A.Y. Matrix metalloproteinases - From the cleavage data to the prediction tools and beyond. Biochim Biophys Acta Mol Cell Res. 2017;1864:1952–1963. doi: 10.1016/j.bbamcr.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozols M., et al. Predicting Proteolysis in Complex Proteomes Using Deep Learning. Int J Mol Sci. 2021;22 doi: 10.3390/ijms22063071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hachmann J., et al. Mechanism and specificity of the human paracaspase MALT1. Biochem J. 2012;443:287–295. doi: 10.1042/BJ20120035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruland J., Hartjes L. CARD-BCL-10-MALT1 signalling in protective and pathological immunity. Nat Rev Immunol. 2019;19:118–134. doi: 10.1038/s41577-018-0087-2. [DOI] [PubMed] [Google Scholar]
- 20.Zotti T., Polvere I., Voccola S., Vito P., Stilo R. CARD14/CARMA2 Signaling and its Role in Inflammatory Skin Disorders. Front Immunol. 2018;9:2167. doi: 10.3389/fimmu.2018.02167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staal J., et al. Ancient Origin of the CARD-Coiled Coil/Bcl10/MALT1-Like Paracaspase Signaling Complex Indicates Unknown Critical Functions. Front Immunol. 2018;9:1136. doi: 10.3389/fimmu.2018.01136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quancard J., et al. An allosteric MALT1 inhibitor is a molecular corrector rescuing function in an immunodeficient patient. Nat Chem Biol. 2019;15:304–313. doi: 10.1038/s41589-018-0222-1. [DOI] [PubMed] [Google Scholar]
- 23.Bardet M., et al. MALT1 activation by TRAF6 needs neither BCL10 nor CARD11. Biochem Biophys Res Commun. 2018;506:48–52. doi: 10.1016/j.bbrc.2018.10.029. [DOI] [PubMed] [Google Scholar]
- 24.Kingeter L.M., Schaefer B.C. Malt1 and cIAP2-Malt1 as effectors of NF-kappaB activation: kissing cousins or distant relatives? Cell Signal. 2010;22:9–22. doi: 10.1016/j.cellsig.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein T., Eckhard U., Dufour A., Solis N., Overall C.M. Proteolytic Cleavage-Mechanisms, Function, and “Omic” Approaches for a Near-Ubiquitous Posttranslational Modification. Chem Rev. 2018;118:1137–1168. doi: 10.1021/acs.chemrev.7b00120. [DOI] [PubMed] [Google Scholar]
- 26.Rawlings N.D., et al. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018;46:D624–D632. doi: 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant C.E., Bailey T.L., Noble W.S. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oughtred R., et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019;47:D529–D541. doi: 10.1093/nar/gky1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noels H., et al. A Novel TRAF6 binding site in MALT1 defines distinct mechanisms of NF-kappaB activation by API2middle dotMALT1 fusions. J Biol Chem. 2007;282:10180–10189. doi: 10.1074/jbc.M611038200. [DOI] [PubMed] [Google Scholar]
- 30.Klein T., et al. The paracaspase MALT1 cleaves HOIL1 reducing linear ubiquitination by LUBAC to dampen lymphocyte NF-κB signalling. Nat Commun. 2015;6:8777. doi: 10.1038/ncomms9777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uehata T., et al. Malt1-induced cleavage of regnase-1 in CD4(+) helper T cells regulates immune activation. Cell. 2013;153:1036–1049. doi: 10.1016/j.cell.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 32.Jeltsch K.M., et al. Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote T(H)17 differentiation. Nat Immunol. 2014;15:1079–1089. doi: 10.1038/ni.3008. [DOI] [PubMed] [Google Scholar]
- 33.Yamasoba D., et al. N4BP1 restricts HIV-1 and its inactivation by MALT1 promotes viral reactivation. Nat Microbiol. 2019;4:1532–1544. doi: 10.1038/s41564-019-0460-3. [DOI] [PubMed] [Google Scholar]
- 34.Xu Y.R., Lei C.Q. TAK1-TABs Complex: A Central Signalosome in Inflammatory Responses. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.608976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian Y., et al. RBCK1 negatively regulates tumor necrosis factor- and interleukin-1-triggered NF-kappaB activation by targeting TAB2/3 for degradation. J Biol Chem. 2007;282:16776–16782. doi: 10.1074/jbc.M701913200. [DOI] [PubMed] [Google Scholar]
- 36.Wang J., Chun H.J., Wong W., Spencer D.M., Lenardo M.J. Caspase-10 is an initiator caspase in death receptor signaling. Proc Natl Acad Sci U S A. 2001;98:13884–13888. doi: 10.1073/pnas.241358198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kischkel F.C., et al. Death receptor recruitment of endogenous caspase-10 and apoptosis initiation in the absence of caspase-8. J Biol Chem. 2001;276:46639–46646. doi: 10.1074/jbc.M105102200. [DOI] [PubMed] [Google Scholar]
- 38.Horn S., et al. Caspase-10 Negatively Regulates Caspase-8-Mediated Cell Death, Switching the Response to CD95L in Favor of NF-κB Activation and Cell Survival. Cell Rep. 2017;19:785–797. doi: 10.1016/j.celrep.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wachmann K., et al. Activation and specificity of human caspase-10. Biochemistry. 2010;49:8307–8315. doi: 10.1021/bi100968m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaya T., Omori Y., Kuwahara R., Furukawa T. ICK is essential for cell type-specific ciliogenesis and the regulation of ciliary transport. EMBO J. 2014;33:1227–1242. doi: 10.1002/embj.201488175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saito T., Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS ONE. 2015;10:e0118432. doi: 10.1371/journal.pone.0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Israël L., et al. CARD10 cleavage by MALT1 restricts lung carcinoma growth in vivo. Oncogenesis. 2021;10:32. doi: 10.1038/s41389-021-00321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eckhard U., et al. Active site specificity profiling of the matrix metalloproteinase family: Proteomic identification of 4300 cleavage sites by nine MMPs explored with structural and synthetic peptide cleavage analyses. Matrix Biol. 2016;49:37–60. doi: 10.1016/j.matbio.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 44.Kleifeld O., et al. Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat Biotechnol. 2010;28:281–288. doi: 10.1038/nbt.1611. [DOI] [PubMed] [Google Scholar]
- 45.Kong A.T., Leprevost F.V., Avtonomov D.M., Mellacheruvu D., Nesvizhskii A.I. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat Methods. 2017;14:513–520. doi: 10.1038/nmeth.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang W., et al. TRAF Family Member-associated NF-κB Activator (TANK) Inhibits Genotoxic Nuclear Factor κB Activation by Facilitating Deubiquitinase USP10-dependent Deubiquitination of TRAF6 Ligase. J Biol Chem. 2015;290:13372–13385. doi: 10.1074/jbc.M115.643767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kawagoe T., et al. TANK is a negative regulator of Toll-like receptor signaling and is critical for the prevention of autoimmune nephritis. Nat Immunol. 2009;10:965–972. doi: 10.1038/ni.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato S., et al. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat Immunol. 2005;6:1087–1095. doi: 10.1038/ni1255. [DOI] [PubMed] [Google Scholar]
- 49.Wang H., et al. Cloning and characterization of a novel caspase-10 isoform that activates NF-kappa B activity. Biochim Biophys Acta. 2007;1770:1528–1537. doi: 10.1016/j.bbagen.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 50.Emming S., et al. A molecular network regulating the proinflammatory phenotype of human memory T lymphocytes. Nat Immunol. 2020;21:388–399. doi: 10.1038/s41590-020-0622-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomita T., et al. Extracellular mRNA transported to the nucleus exerts translation-independent function. Nat Commun. 2021;12:3655. doi: 10.1038/s41467-021-23969-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gross O., et al. Multiple ITAM-coupled NK-cell receptors engage the Bcl10/Malt1 complex via Carma1 for NF-kappaB and MAPK activation to selectively control cytokine production. Blood. 2008;112:2421–2428. doi: 10.1182/blood-2007-11-123513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hamilton K.S., et al. T cell receptor-dependent activation of mTOR signaling in T cells is mediated by Carma1 and MALT1, but not Bcl10. Sci Signal. 2014;7:ra55. doi: 10.1126/scisignal.2005169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wagih O. ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics. 2017;33:3645–3647. doi: 10.1093/bioinformatics/btx469. [DOI] [PubMed] [Google Scholar]
- 55.Altenhoff A.M., et al. The OMA orthology database in 2018: retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucleic Acids Res. 2018;46:D477–D485. doi: 10.1093/nar/gkx1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Szklarczyk D., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saito T., Rehmsmeier M. Precrec: fast and accurate precision-recall and ROC curve calculations in R. Bioinformatics. 2017;33:145–147. doi: 10.1093/bioinformatics/btw570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiesmann C., et al. Structural determinants of MALT1 protease activity. J Mol Biol. 2012;419:4–21. doi: 10.1016/j.jmb.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 59.Gil, D. P., Law, J. N. & Murali, T. M. The PathLinker app: Connect the dots in protein interaction networks. F1000Res 6, 58 (2017). 10.12688/f1000research.9909.1. [DOI] [PMC free article] [PubMed]
- 60.Shannon P., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.