Abstract
A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone) at an extremely low concentration triggers streptomycin production and aerial mycelium formation in Streptomyces griseus. A-factor induces the expression of an A-factor-dependent transcriptional activator, AdpA, essential for both morphological and physiological differentiation by binding to the A-factor receptor protein ArpA, which has bound and repressed the adpA promoter, and dissociating it from the promoter. Nine DNA fragments that were specifically recognized and bound by histidine-tagged AdpA were isolated by cycles of a gel mobility shift-PCR method. One of them was located in front of a gene encoding an extracytoplasmic function ς factor belonging to a subgroup of the primary ς70 family. The cloned gene was named AdpA-dependent sigma factor gene (adsA), and the gene product was named ςAdsA. Transcription of adsA depended on A-factor and AdpA, since adsA was transcribed at a very low and constant level in an A-factor-deficient mutant strain or in an adpA-disrupted strain. Consistent with this, transcription of adsA was greatly enhanced at or near the timing of aerial hyphae formation, as determined by low-resolution S1 nuclease mapping. High-resolution S1 mapping determined the transcriptional start point 82 nucleotides upstream of the translational start codon. DNase I footprinting showed that AdpA bound both strands symmetrically between the transcriptional start point and the translational start codon; AdpA protected the antisense strand from positions +7 to +41 with respect to the transcriptional start point and the sense strand from positions +12 to +46. A weak palindrome was found in the AdpA-binding site. The unusual position bound by AdpA as a transcriptional activator, in relation to the promoter, suggested the presence of a mechanism by which AdpA activates transcription of adsA in some unknown way. Disruption of the chromosomal adsA gene resulted in loss of aerial hyphae formation but not streptomycin or yellow pigment production, indicating that ςAdsA is involved only in morphological development and not in secondary metabolic function. The presence of a single copy in each of the Streptomyces species examined by Southern hybridization suggests a common role in morphogenesis in this genus.
The gram-positive bacterial genus Streptomyces shows characteristic morphological differentiation resembling that of filamentous fungi (8, 9). Early in the life cycle of a streptomycete on solid medium, it grows as a branching, multinucleoid substrate mycelium mainly by cell wall extension at the hyphal tips. As older parts of the substrate mycelium produce aerial mycelium, most cells in the substrate mycelium die (61). After septa have been produced at regular intervals along the hyphae to form uninucleoid compartments, long chains of spores are formed. This genus is also characterized by its ability to produce a wide variety of secondary metabolites. Some regulatory steps for morphological differentiation and secondary metabolism share common genes and common metabolites. For example, Streptomyces coelicolor A3(2) bld mutants are defective in both aerial mycelium formation and antibiotic production, depending on the carbon source of medium (9). A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone) is another example that acts as a switch for aerial mycelium formation and secondary metabolite formation in Streptomyces griseus (18–20). Downstream from the common regulatory pathway, there must be a regulatory pathway specific to morphological differentiation and secondary metabolism.
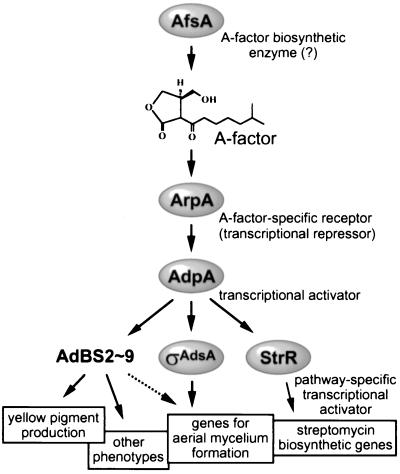
We have long studied A-factor that triggers, at an extremely low concentration, aerial mycelium formation and streptomycin production in S. griseus and recently revealed a major regulatory cascade leading to streptomycin production (40; see also Fig. 9). A-factor is gradually accumulated in a growth-dependent manner by the action of AfsA probably condensing a glycerol derivative and a β-keto acid. When the concentration of A-factor reaches a certain critical level, it binds an A-factor-specific receptor ArpA, which has bound and repressed the promoter of adpA, and dissociates ArpA from the promoter, thus leading to transcription and translation of a transcription factor AdpA. AdpA then activates transcription of strR by binding an upstream activation sequence. A pathway-specific transcriptional activator StrR induced in this way activates transcription of most of the streptomycin biosynthetic genes by binding multiple sites in the gene cluster. Of the members in this cascade, AfsA, ArpA, and AdpA are common to both morphological differentiation and secondary metabolite formation, because mutation and disruption of any of these genes simultaneously influence both aerial mycelium and streptomycin formation (21, 40, 41). We then started to detect genes that are controlled by AdpA, on the assumption that one or some of such genes are involved in aerial mycelium formation as a member just downstream of AdpA in the hierarchy of the A-factor regulatory cascade. AdpA belonging to the AraC/XylS family of regulators is 405 amino acid residues long and contains a helix-turn-helix DNA-binding motif in the middle of the protein (40). We have succeeded in isolating such a gene by first cloning a DNA fragment that is bound by AdpA by a gel mobility shift-PCR method and then analyzing its transcription in response to AdpA.
FIG. 9.
The A-factor regulatory cascade leading to ςAdsA. The A-factor signal is transmitted ςAdsA via ArpA (A-factor-specific receptor serving as a transcriptional repressor for adpA) and AdpA (transcriptional activator for multiple genes). Of genes with the binding sites (AdBS2 to AdBS9) that are controlled by AdpA, there may be a gene controlling yellow pigment production. In addition to adsA, one or more genes controlled by AdpA should be involved in aerial mycelium formation since introduction of adsA into ΔadpA strains does not restore aerial mycelium formation. AdpA also activates strR encoding a pathway-specific transcriptional activator for streptomycin biosynthetic genes. See the text for details.
This study deals with a gene encoding an extracytoplasmic function (ECF) ς factor that has been identified as a target of AdpA. The ECF ς subgroup of the primary ς70 family is a class of environmentally responsive transcriptional regulators (30, 62). Transcriptional analysis of the cloned ς gene and gene disruption experiments show that this ECF ς factor, as a member in the A-factor regulatory cascade, concerns only with aerial mycelium formation and not with secondary metabolism. Distribution of DNA sequences homologous with the ς gene in a wide variety of Streptomyces species suggests its important role in morphological development in general.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The wild-type strain S. griseus IFO13350 and an A-factor-deficient mutant strain HH1 were described previously (21). S. griseus ΔadpA was also described (40). The other Streptomyces strains were obtained from the Institute of Fermentation, Osaka, Japan, and American Type Culture Collection (ATCC). Bacillus subtilis ATCC 6633 was used for the bioassay of streptomycin. Escherichia coli JM109 and pUC19 (63) for DNA manipulation were purchased from Takara Shuzo. E. coli JM110 containing dam and dcm mutations was used for preparing nonmethylated Streptomyces DNA used for gene disruption. Media and growth conditions for E. coli were described by Maniatis et al. (31). Plasmid pET26b(+) and E. coli BL21(DE3) (Novagen) were used for producing histidine-tagged AdpA. The low-copy-number E. coli-Streptomyces shuttle vector, pKU209, carrying the thiostrepton and ampicillin resistance genes and the SCP2*-replication origin (24), was obtained from H. Ikeda, Kitasato University, Tokyo, Japan. pIJ486 with a copy number of 40 to 100 per genome, carrying the neomycin and thiostrepton resistance genes, was obtained from D. A. Hopwood, John Innes Centre (60). YMPD medium (40), Bennett medium without maltose (21), and minimal medium (38) for S. griseus were as described earlier. Ampicillin and kanamycin at final concentrations of 50 μg/ml were used for E. coli, when necessary. For S. griseus, thiostrepton and neomycin were added at final concentrations of 50 and 15 μg/ml, respectively, when necessary.
General recombinant DNA studies.
Restriction enzymes, T4 DNA ligase, and other DNA-modifying enzymes were purchased from Takara Shuzo. [α-32P]dCTP (110 TBq/mmol) for DNA labeling with the Takara BcaBest DNA labeling system and [γ-32P]ATP (220 TBq/mmol) for end labeling at 5′ ends with T4 polynucleotide kinase were purchased from Amersham Pharmacia Biotech. DNA was manipulated in Streptomyces (17) and in E. coli (2, 31), as described earlier. Nucleotide sequences were determined by the dideoxy chain termination method (50) with the Thermo Sequenase fluorescence-labeled primer cycle sequencing kit (Amersham) or the DNA sequencing kit (ABI Prizm) on an automated DNA sequencer.
Production and purification of histidine-tagged AdpA.
The adpA sequence was amplified by PCR with S. griseus chromosomal DNA as a template and the following two primers; N-Eco-Nde: 5′-TATgaattcCATATGAGCCAGGACTCCGCCGCC-3′ (the italic letters indicate the start codon of adpA; the lowercase letters and underlining indicate the EcoRI and NdeI sites, respectively) and C-Xho-Bam: 5′-TATggatccTCGAGCGGGGCACTCCGCTGTCCCGG-3′ (the italic letters indicate the terminal Pro-405 codon of adpA; the lowercase letters and underlining indicate the BamHI and XhoI sites, respectively). The amplified fragment was digested with EcoRI plus BamHI and cloned in pUC19. After no errors in PCR had been checked by nucleotide sequencing, the adpA sequence was excised with NdeI and XhoI and cloned in pET26b(+), generating pET-adpA. This plasmid contained the entire adpA sequence-CTC-GAG-(CAC)6-TGA under the control of the T7 promoter and the lac operator in pET-26b(+).
We examined culture conditions of E. coli BL21(DE3) harboring pET-adpA to produce the histidine-tagged AdpA (AdpA-H) as a soluble form as much as possible, because AdpA-H was found in both soluble and insoluble fractions. After cultivation temperature, medium, inoculation size, induction of the T7 promoter via the lac operator with isopropyl-β-d-thiogalactopyranoside (IPTG) had been examined, we established the following cultivation conditions. E. coli BL21(DE3) harboring pET-adpA was cultured at 37°C for 7 h in 2× YT medium containing 50 μg of kanamycin per ml, and 50 μl of the seed culture was transferred to 10 ml of Luria-Bertani broth containing the same concentration of kanamycin and cultured overnight at 30°C without induction with IPTG. The cells were harvested from 20 ml of culture by centrifugation and disrupted by sonication. A soluble fraction obtained by centrifugation at 20,000 × g of the sonicate was applied to the Ni-NTA Spin Column (Qiagen), and AdpA-H was eluted according to the manual of the supplier. From 20 ml of culture, about 0.2 mg of AdpA-H was obtained.
Gel mobility shift assay.
The gel mobility shift assay was performed essentially according to the method of Vujaklija et al. (58). For the binding assay, 3,000 cpm of 32P-labeled probe DNA was incubated with AdpA-H (0.2 to 1 μg) at 30°C for 30 min in a buffer containing 50 mM sodium phosphate buffer (pH 8.0), 10% glycerol, 1 μg of poly(dI-dC)–poly(dI-dC), and 0.01% bovine serum albumin (BSA) in a total volume of 40 to 50 μl. BSA was used as a stabilizer of AdpA-H. After incubation, complexes and free DNA were resolved on nondenaturing 4 or 6% polyacrylamide gels (mono/bis, 79:1) with a running buffer containing 40 mM Tris-HCl (pH 7.8), 20 mM sodium acetate, and 1 mM EDTA. Gels were dried and subjected to autoradiography with a Du Pont Cronex intensifying screen.
For determination of the ability of AdpA-H to bind the upstream activation sequence of strR (58), the binding site, from positions −288 to −189 with respect to the transcriptional start point of strR, was prepared by PCR with 5′-TCGAAGAGAATCAGCC-3′ and 5′-CCATCAAAATAAACCGCA-3′ as primers and their 5′ ends were labeled with [γ-32P]ATP and T4 polynucleotide kinase. For determination of the binding between AdBS1 (see below) and AdpA-H, the AdBS1 sequence was excised with EcoRI from the recombinant pUC19 plasmid and the 5′ ends were similarly 32P labeled.
Gel mobility shift-PCR for isolation of protein-bound DNA fragments.
A library of small DNA fragments with a linker at both ends was constructed according to the method of Kinzler and Vogelstein (29). Chromosomal DNA of S. griseus IFO13350 was partially digested with HaeIII, and fragments of ca. 300 to 500 bp were prepared by agarose gel electrophoresis with low-melting-temperature agarose and the QIAquick gel extraction kit (Qiagen). The linker to be attached to the ends was prepared by annealing catch A (5′-gagTAGAATTCTAATATctc-3′) and catch B (5′-gagATATTAGAATTCTActc-3′). The catch linker contained an EcoRI site (underline) and self-ligated linkers were cleaved with XhoI (lowercase letters). About 3 μg of DNA (12 pmol) was ligated with the catch linkers (1,500 pmol), and the ligated sample was treated with XhoI. The linker-attached DNA fragments were isolated by agarose gel electrophoresis as described above. The 300- to 500-bp fragments with the catch linker were incubated with AdpA-H under the conditions described above and subjected to 6% polyacrylamide gel electrophoresis. A gel piece was cut out on the basis of the positions of 300- to 500-bp fragments bound to AdpA-H, determined as follows, and DNA was extracted by the method of Beutel and Gold (4). As a control experiment, the positions of the 300- and 500-bp DNA fragments bound to AdpA-H on polyacrylamide gel electrophoresis under the same conditions were determined with the binding site upstream of strR; 5′-TCGAAGAGAATCAGCCGCCGTG-3′ (primer A) and 5′-GAGCAATGCTTTCGCACTTCGC-3′ (primer B) were used as the primers to generate a 32P-labeled 306-bp fragment (positions −288 to +18 with respect to the transcriptional start point of strR). For generation of a control 500-bp fragment, primer A and 5′-CGACATCCTCGCCGGCACTG-3′ was used to generate a 506-bp fragment (positions −288 to +218) containing the AdpA-binding site close to one end. A 500-bp fragment (positions −482 to +18) containing the AdpA-binding site in the middle was also generated with 5′-GCGGCACGTATGGCCTCCAG-3′ and primer B were used. The DNA extracted from the gel slice was amplified by PCR with the catch A and B linkers as primers. This procedure was one cycle of the gel mobility shift-PCR method. After the fourth cycle, DNA fragments able to bind AdpA-H were isolated by cloning them in pUC19 by use of the EcoRI site in the linker. Nine DNA fragments, AdBS1 to AdBS9, were obtained by this procedure.
Cloning of a DNA fragment containing AdBS1.
AdBS1 was excised with EcoRI from the recombinant pUC19 plasmid, purified by agarose gel electrophoresis with the GeneClean III kit (Bio 101), and 32P labeled with [α-32P]dCTP and the BcaBest labeling kit (Takara). By standard DNA manipulation, a 4.7-kb PstI fragment giving a signal by Southern hybridization with the 32P probe was cloned in pUC19, generating pBS1-Pst5k. After restriction mapping, the nucleotide sequence of the 4.7-kb fragment was determined and analyzed by Frame Plot analysis (6, 23).
Construction of plasmids containing adsA.
Two primers were used to amplify the whole adsA and its promoter sequence by PCR with pBS1-Pst5k as the template: sigprm-F, 5′-tttgaattcaagcttGCTGACCCGCACCCCTTCCG-3′ (the underlining indicates the termination codon of serB, see Fig. 3A; the lowercase letters indicate a linker sequence containing an EcoRI and HindIII sites), and BHdsig-R, 5′-tttggatccaagcttGATGATCGGACCAGTGCGTGACG-3′ (the lowercase letters indicate a linker sequence containing a BamHI and HindIII sites; the capital letters indicate the sequence 45 to 23 bp downstream of the termination codon of adsA). After PCR under the standard conditions, the amplified fragment was digested with EcoRI and BamHI and cloned in pUC19. No errors in PCR were confirmed by nucleotide sequencing. The cloned fragment was excised with HindIII and then cloned in pKU209, generating pKU209-adsA. For construction of pIJ486-adsA, the cloned fragment was excised with EcoRI plus BamHI and ligated with pIJ486 digested with the same enzymes.
FIG. 3.
Restriction map of the cloned 4.7-kb PstI fragment and the positions and directions of ORFs in the fragment (A), amino acid alignment of ςAdsA with other ECF ς factors (B), and nucleotide sequence of part of adsA (C). (A) The extents and directions of ORFs predicted by Frame Plot analysis of the nucleotide sequence are indicated by arrows. The position of AdBS1 is also shown. The space between the termination codon of serB and the start codon of adsA is 395 bp. S. coelicolor A3(2) contains these genes in the same organization (http://www.sanger.ac.uk/Projects/S_coelicolor/). The percentages of identical amino acid residues of corresponding gene products are shown. (B) The amino acid sequence of ςAdsA (SgAdsA) is aligned with the ςAdsA homologue (ScECFς) of S. coelicolor A3(2), CarQ (MxCarQ) responsible for the light-inducible biosynthesis of carotenoid in Myxococcus xanthus (36), and SigX (BsSigX) responsible for survival at high temperature in B. subtilis (22). Highly conserved amino acid residues among these ς factors are boxed. Dashes indicate gaps introduced for alignment. (C) The transcriptional start point at position +1, indicated by an open triangle, of adsA (see Fig. 4B) and the AdpA-binding site (see Fig. 6) are shown. A probable −10 sequence is underlined. Primer sequences sigS1L-F and sigS1L-R used for preparing the probe for low-resolution S1 mapping of adsA (Fig. 4A), sigS1H-F and sigS1H-R used for preparing sigS1H probe for high-resolution S1 mapping of adsA (Fig. 4B) and for DNase I footprinting (Fig. 6), and sigFP-F and sigFP-R used for preparing sigFP probe for DNase I footprinting (Fig. 6) are also shown.
S1 nuclease mapping.
Methods for RNA preparation from cells grown on cellophane on the surface of agar medium and S1 nuclease mapping were as described by Kelemen et al. (26). Hybridization probes were prepared by PCR with a pair of 32P-labeled and nonlabeled primers. For low-resolution S1 mapping for hrdB, 5′-TCGGCCCATTTCGTCACGTATGAG-3′ (from positions −121 to −98 with respect to the transcriptional start point of hrdB [52]) and 5′-TCGATGAGCGCCATCACAGACTCG-3′ (positions +193 to +170) labeled at the 5′ end were used. For low-resolution S1 mapping of adsA, sigS1L-F (5′-CCCGGCCACAACACGTCGCC-3′; positions −167 to −148 with respect to the transcriptional start point of adsA; see Fig. 3C) and sigS1L-R (5′-CTGCGTTCGGCCAGGGCGTAG-3′; positions +237 to +217) labeled at the 5′ end were used. For high-resolution S1 mapping of adsA, sigS1H-F (5′-GATCAATAAACGGTCACCATGTGC-3′; positions −121 to −98) and sigS1H-R (5′-GACTCCCAGAGGCAGAGCTTCC-3′; positions +80 to +59) labeled at the 5′ end were used. Protected fragments were analyzed on 6% DNA sequencing gels by the method of Maxam and Gilbert (35).
DNase I footprinting.
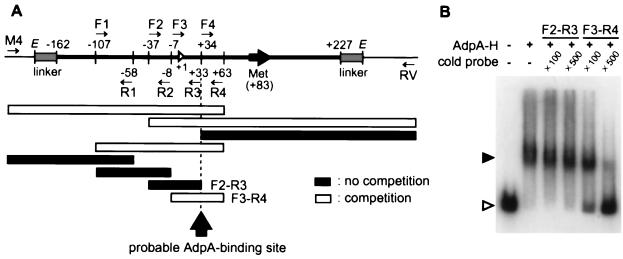
Before DNase I footprinting, an approximate location of the AdpA-binding site in AdBS1 was determined by means of competition in the gel mobility shift assay. DNA fragments of various lengths were prepared by PCR and used as a competitor in the binding between AdpA-H and AdBS1 (see Fig. 5A). For this purpose, the following primers were used: 5′-GTTTTCCCAGTCACGACGTT-3′ (M4); 5′-CAGGAAACGGCTATGACCAT-3′ (RV); 5′-CACCATGTGCTACTTGATAC-3′ (F1; positions −107 to −88); 5′-TGTAGCGCTGCCTGCACG-3′ (F2; positions −37 to −20); 5′-CCTCAGACGCATCACGGAAC-3′ (F3; positions −7 to +13); 5′-TGACGCTTTTCGAACTGCTC-3′ (F4; positions +34 to +53); 5′-GTTCCGTCGCTTCTGTTATC-3′ (R1; positions −58 to −77); 5′-AGAATAACGCTTCGTGCAGGCAG-3′ (R2; positions −8 to −30); 5′-CTCGTGGTAGCGAGTGGCGG-3′ (R3; positions +33 to +14); and 5′-CTTCCATCACGAGCAGTTCG-3′ (R4; positions +63 to +44). An excess amount (×100 and ×500) of these fragments was added to the binding mixture, and its ability to compete the binding between AdpA-H and AdBS1 was examined by the gel mobility shift assay.
FIG. 5.
Determination of the approximate AdpA-H-binding site in AdBS1. (A) The ability of each of the indicated fragments to compete the binding between AdpA-H and AdBS1 was examined by gel mobility shift assay. The nucleotide numbers are shown, taking the transcriptional start point of adsA as +1. Short arrows indicate the locations of primer sequences used for preparing 32P-labeled probes. The fragments that competed the binding are shown by open bars, and those that did not are shown by solid bars. (B) Excess amounts (×100 and ×500) of nonlabeled probe F3-R4 compete the binding between AdpA-H (0.2 μg) and AdBS1, whereas those of nonlabeled probe F2-R3 did not.
For preparation of the antisense strand for DNase I footprinting, sigS1H-F and sigS1H-R (see Fig. 3C) used for the above-described high-resolution S1 mapping of adsA were used. The sense strand was prepared with sigFP-F (5′-TAGCGCTGCCTGCACGAAGCG-3′; positions −35 to −15 with respect to the transcriptional start point of adsA; see Fig. 3C) labeled at the 5′ end and sigFP-R (5′-GACGAAGCCGCGCAAGCGGTC-3′; positions +163 to +143) were used. The reaction mixture (50 μl) contained 10 kcpm 32P-labeled DNA probe, 0.2 to 0.4 μg of AdpA-H, 25 mM HEPES-KOH (pH 7.9), 0.5 mM EDTA-NaOH (pH 8.0), 50 mM KCl, and 10% glycerol. After incubation of the mixture for 30 min at 25°C, DNase I was added at a final concentration of 20 μg/ml, and the mixture was further incubated for 1 min. The digestion was stopped by adding 100 μl of stop solution (100 mM Tris-HCl, pH 8.0; 100 mM NaCl; 1% sodium N-lauroyl sarcosinate; 10 mM EDTA-NaOH, pH 8.0; 25 μg of salmon sperm DNA per ml) and 300 μl of phenol-CHCl3 (1:1). After ethanol precipitation, the pellet was washed with 80% ethanol, dissolved in 6 μl of the formamide-dye mixture (35), and run on 6% polyacrylamide gel.
Gene disruption.
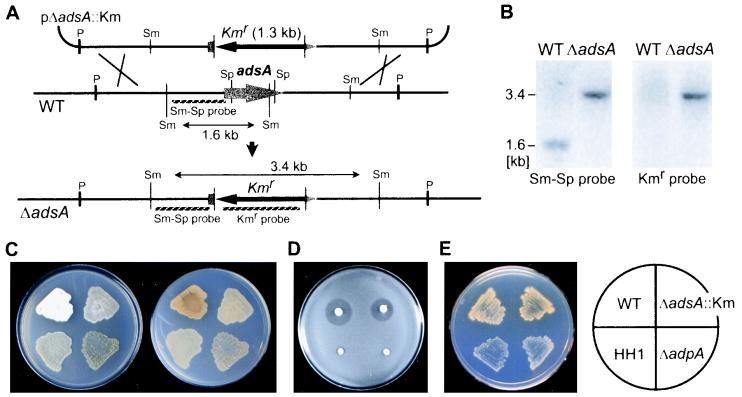
pBS1-Pst5k was digested with SplI, and the ends were flush ended with Klenow fragment. The 3.4-kb kanamycin resistance (Kmr) determinant obtained by digestion of Tn5 (3) with HindIII was cloned in the HindIII site of pUC19, and then a 1.3-kb Kmr gene was excised with SmaI. The SmaI fragment was then ligated with the SplI-digested plasmid. Plasmid pΔadsA::Km in which the kanamycin resistance gene was inserted in the opposite direction to that of adsA was selected. pΔadsA::Km was once propagated in E. coli JM110 to prepare nonmethylated plasmid DNA. The plasmid was cut with DraI, alkali denatured (39), and introduced by protoplast transformation into S. griseus IFO13350. Among kanamycin-resistant S. griseus transformants, true adsA disruptants were selected by Southern hybridization with two probes; one was the kanamycin resistance gene (the Kmr probe shown in Fig. 7A), and the other was a 960-bp SmaI-SplI fragment (Sm-Sp probe).
FIG. 7.
Phenotypes of S. griseus ΔadsA. (A) Schematic representation of the strategy used for disruption of adsA. (B) Southern hybridization analysis against the SmaI-digested chromosomal DNA from an adsA disruptant to confirm correct gene replacement by homologous recombination. When the indicated 960-bp SmaI-SplI fragment (Sm-Sp probe) was used as 32P probe, a signal of 1.6-kb in the wild-type strain (WT) and of 3.4 kb in an adsA disruptant were observed. When the kanamycin resistance determinant from Tn5 (Kmr probe) was used as a probe, no signal in the wild-type strain was observed, but a 3.4-kb signal in the disruptant was observed. (C) Loss of aerial hyphae formation by adsA disruption. S. griseus IFO13350 (WT), an A-factor-deficient mutant (HH1), an adpA disruptant (ΔadpA), and an adsA disruptant (ΔadsA::Km) were grown at 28°C for 5 days on YMPD medium. The upper (left) and lower (right) surfaces were photographed. (D) No effect of adsA mutations on streptomycin production. The indicated S. griseus strains were grown at 28°C for 4 days, and B. subtilis spores suspended in nutrient agar were overlaid and incubated at 28°C overnight. The wild-type strain and the adsA disruptant produced streptomycin, as judged from growth inhibition of the indicator around the colonies, whereas strain HH1 or the adpA disruptant produced no streptomycin. (E) No effect of adsA mutations on yellow pigment production was seen. The S. griseus strains were grown at 28°C for 5 days on phosphate-depleted minimal medium. The yellow pigment was produced by the wild-type and ΔadsA::Km strains but not by HH1 or ΔadpA strains.
Streptomycin production by the wild-type and adsA-disrupted strains was determined by overlaying spores of B. subtilis suspended in nutrient agar (0.35%) on colonies grown at 28°C for 4 days on Bennett medium without maltose. Aerial mycelium formation was examined on YMPD medium. Yellow pigment production was examined on phosphate-depleted minimal medium.
Southern hybridization for examining distribution of adsA.
BamHI-digested chromosomal DNAs (5 μg each) of Streptomyces species were separated on agarose gel electrophoresis and transferred to a positively charged nylon membrane. 32P-labeled probe was prepared by PCR with ENdsig-F (5′-tttgaattccatatgTACCCACACGTCGGGGTTG-3′; lowercase letters indicate a linker sequence, and capitals indicate a sequence from +86 to +104 with respect to the transcriptional start point of adsA; underlining indicates the Tyr-2 codon of adsA; see Fig. 3C) and BHdsig-R (used for construction of adsA-containing plasmids).
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been deposited to the DDBJ, EMBL, and GenBank DNA databases under accession no. AB039273.
RESULTS
Production and purification of histidine-tagged AdpA.
The AdpA protein purified from S. griseus was very unstable, forming inactive aggregates. This is a feature common to the AraC/XylS family (15). For production of AdpA in a large amount in E. coli and in a convenient form for rapid purification, we placed adpA in pET26b(+) to fuse a histidine tag to the COOH-terminal end of the whole AdpA sequence. The recombinant plasmid pET-adpA would direct the synthesis of AdpA-Leu-Glu-His6. We examined culture conditions, such as culture temperature, culture period, and induction with IPTG, to obtain AdpA-H as a soluble form as much as possible. Under the conditions we established, a considerable amount of AdpA-H was produced in a soluble fraction (Fig. 1A, lane 2). We purified the protein from the soluble fraction by using His-bind resin (lane 3).
FIG. 1.
Purification of AdpA-H (A) and binding of AdpA-H to the upstream activation sequence of strR (B). (A) Appropriate amounts of the insoluble (lane 1) and soluble (lane 2) fractions prepared from E. coli cells harboring pET-adpA and about 0.2 μg of protein of the sample (lane 3) purified with His-bind resin were run. (B) A 32P-labeled 100-bp fragment, including the AdpA-binding site upstream of strR, was prepared by PCR and used in the gel mobility shift assay. The positions of AdpA-H-bound (solid triangle) and free (open triangle) probes are shown. The position of the gel well is also indicated by an arrow. The retarded signals become stronger with an increase in the amount of AdpA-H. The amounts of AdpA-H were 0.04 μg (lane 2), 0.2 μg (lane 3), and 1 μg (lane 4). Lane 1 is a control lane in which no AdpA-H was contained.
The ability of AdpA-H obtained in this way to bind an upstream activation sequence of strR was examined by gel mobility shift assay (Fig. 1B). The 100-bp fragment (58), from positions −288 to −189 with respect to the transcriptional start point of strR, was shifted by AdpA-H. This shift was competed by an excess of nonlabeled probe (data not shown). When the amount of AdpA-H increased, most AdpA-H-bound fragments remained in the gel well, probably because AdpA-H readily formed aggregates. We thus judged that AdpA-H, showing specific binding to the upstream activation sequence of strR, could be used for screening of DNA fragments specifically bound by this protein by means of the following gel mobility shift-PCR method.
Isolation of DNA fragments that are bound by AdpA.
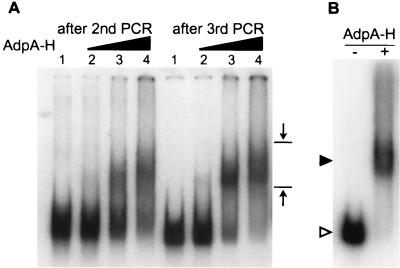
The principle of the gel mobility shift-PCR method for isolation of DNA fragments bound by AdpA-H was to separate AdpA-H-bound DNA fragments from free fragments by polyacrylamide gel electrophoresis, to amplify the separated fragments by PCR by use of the primer sites at the ends of the fragments, and to clone each of the amplified fragments into an E. coli vector. Because of the high G+C composition of the Streptomyces DNA, we partially digested chromosomal DNA of S. griseus by HaeIII with a recognition sequence of GGCC and fragments of ca. 300 to 500 bp were recovered by agarose gel electrophoresis. After attachment of primer sites at the ends of the DNA fragments and trimming of the ends, these were incubated with AdpA-H and subjected to polyacrylamide gel electrophoresis to separate AdpA-H-bound fragments from free fragments. As with the preliminary gel mobility shift experiments beforehand, we prepared 32P-labeled 300- and 500-bp DNA fragments containing the AdpA-H-binding site for strR and determined their shifted positions by polyacrylamide gel electrophoresis. On the basis of the positions determined by the earlier experiments, a gel slice probably containing AdpA-H-bound DNA fragments was excised, as shown in Fig. 2A, and they were extracted from the gel piece, amplified by PCR with the linkers at both ends as primers, and purified by agarose gel electrophoresis. This is one cycle of the gel mobility shift-PCR method. For enrichment of the DNA fragments actually bound by AdpA-H, the cycle was repeated four times, and the AdpA-H-bound DNA fragments were cloned in pUC19. We obtained 141 E. coli transformants after the fourth cycle. The nucleotide sequence of each of the DNA fragments in the recombinant pUC19 plasmids was determined and classified. Of the 141 transformants, 114 (82%) contained the same region in the cloned fragments. We chose one of these fragments and named it AdBS1 (Fig. 2B). In addition to AdBS1, eight different DNA fragments (named AdBS2 to AdBS9) that were found to be recognized and bound by AdpA-H by gel mobility shift assay were obtained.
FIG. 2.
Gel mobility shift-PCR for isolation of DNA fragments recognized and bound by AdpA-H (A) and gel mobility shift of AdBS1 caused by AdpA-H (B). (A) The S. griseus chromosomal DNA of 300 to 500 bp obtained after HaeIII digestion was sandwiched by the catch linkers, 32P-labeled, mixed and incubated with AdpA-H, and run on a polyacrylamide gel. The amounts of AdpA-H used were 0.02 μg (lane 2), 0.2 μg (lane 3), and 1 μg (lane 4). Lane 1 is a control lane in which there was no AdpA-H. The DNA fragments retarded were recovered and subjected to a second cycle and further cycles. The mobility shift patterns after the second and third cycles are presented, showing the presence of retarded signals. The opposing arrows show the area from which DNA was extracted; the upper position was determined with a 500-bp DNA fragment, including the AdpA-binding site for strR, and the lower position was determined with a 300-bp fragment including the same AdpA-binding site, as described in Materials and Methods. (B) AdBS1 was excised by EcoRI digestion of the recombinant pUC19 plasmid, 32P-labeled, and subjected to gel mobility shift assay. In the presence of AdpA-H (0.2 μg), AdBS1 is shifted. The positions of AdpA-H-bound (solid triangle) and free (open triangle) probes are shown.
Cloning and nucleotide sequence of a gene as a target of AdpA.
A 4.7-kb PstI fragment covering AdBS1 was cloned by standard DNA manipulation, including colony hybridization, and its nucleotide sequence was determined. Analysis of the nucleotide sequence by Frame Plot analysis predicted the presence of four complete open reading frames (ORFs) and one truncated ORF (Fig. 3A). Just downstream of AdBS1, an ORF of 258 amino acids was found. This ORF showed high end-to-end similarity to ECF ς factors (Fig. 3B). As described below, this gene was controlled by AdpA, and we named it adsA (AdpA-dependent sigma factor) and the gene product ςAdsA. A computer-aided search in the databases showed that S. coelicolor A3(2) contains similar genes in the same organization in cosmid E68 [the S. coelicolor A3(2) genome project, Sanger Centre]. The identity in amino acid sequence between ςAdsA and the corresponding protein of S. coelicolor A3(2) was 91%. The other gene products were also very similar (Fig. 3A).
ECF ς factors belonging to a subfamily of the sigma 70 class are involved in the regulation of gene expression in response to various extracellular changes (30, 37, 62). Examples include ςE responsible for normal cell wall integrity (44, 46) and ςR responsible for the response to various oxidants (45) in S. coelicolor A3(2), ςX responsible for survival at high temperature in B. subtilis (22), AlgU responsible for the biosynthesis of alginate in Pseudomonas aeruginosa (33), PbrA responsible for iron uptake in Pseudomonas fluorescens (51), CarQ responsible for the biosynthesis of carotenoid in Myxococcus xanthus (34, 36), and ςE responsible for heat shock response and protein folding in the periplasm in E. coli (11). ECF ς factors share similarity with ς70 in two (regions 2 and 4) of four conserved regions but have a shorter region 3 and lack most of region 1 that prevents free ς factors from binding directly to the promoter (13, 30). ςAdsA also lacks most of region 1 but contains an additional sequence of 92 amino acids at its NH2-terminal end (Fig. 3B). The ECF ς factors so far known do not contain such an additional sequence at their NH2-terminal ends. In some of the ECF ς factors, including ςR in S. coelicolor A3(2) (25) and CarQ in M. xanthus (16), the activity is modulated by a cognate protein, serving as an anti-sigma factor, which is encoded close to the respective ς gene. No such ORFs are present near the ςAdsA gene.
A-factor- and AdpA-dependent transcription of adsA.
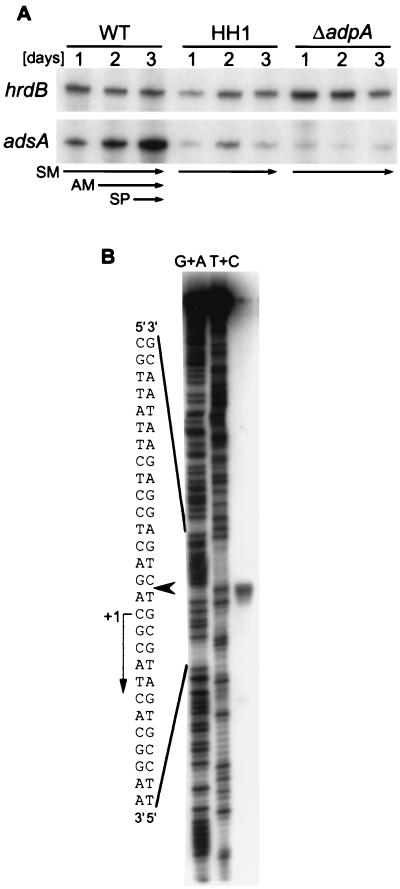
AdpA was expected to control transcription of adsA, since it acts as a transcriptional activator for strR (40, 58, 59). We therefore examined the time course of adsA transcription in the wild-type S. griseus strain and an adpA-disrupted strain S. griseus ΔadpA by low-resolution S1 nuclease mapping with RNAs prepared from cells that were grown on agar medium (Fig. 4A). hrdB that encodes ςHrdB and is transcribed throughout growth (52) was used to monitor the quantity and quality of the mRNA used. In the wild-type strain, adsA was transcribed from a single start site and the transcript increased with growth. After 2 days, the wild-type cells grew as a mixture of aerial hyphae and substrate mycelium and, after 3 days, they grew as a mixture of spores and aerial hyphae. From the intensities of the signals, we judged that transcription of adsA to be markedly increased at or just before the onset of aerial mycelium formation. On the other hand, adsA was transcribed at a very low level throughout growth in the ΔadpA strain. In an A-factor-deficient mutant strain HH1 in which transcription of adpA is repressed constitutively by ArpA, adsA was also repressed, as expected.
FIG. 4.
Low-resolution S1 nuclease mapping of adsA in S. griseus strains (A) and determination of the transcriptional start point of adsA by high-resolution S1 mapping (B). (A) RNA was prepared from cells grown at 28°C for the indicated number of days on solid medium from the wild-type S. griseus IFO13350 (WT), an A-factor-deficient mutant strain (HH1), and an adpA disruptant (ΔadpA). The wild-type strain grew as substrate mycelium (SM) on day 1, as a mixture of aerial and substrate mycelium (AM) on day 2, and as a mixture of aerial hyphae and spores (SP) on day 3. Strains HH1 and ΔadpA grew only as substrate mycelium. (B) RNA prepared from the wild-type cells grown for 3 days on solid medium was used. The arrowhead indicates the position of the S1-protected fragment. The 5′ terminus of the mRNA was assigned to the indicated position because the fragments generated by the chemical sequencing reactions migrate 1.5 nucleotides further than the corresponding fragments generated by S1 nuclease digestion of the DNA-RNA hybrids (half a residue from the presence of the 3′-terminal phosphate group and one residue from the elimination of the 3′-terminal nucleotide) (53).
The transcriptional start point of adsA was determined by high-resolution S1 mapping to be the C that was 82-bp upstream of the translational start codon (Fig. 4B). In front of the transcriptional start point, a TATTCT sequence, very similar to a typical −10 sequence TATAAT found in Streptomyces spp. as well as other bacteria, is present (Fig. 3C). However, no sequence similar to a −35 consensus sequence, TTGACA for many bacteria and TTGACR (R, A or G) for Streptomyces spp. (55), is present at an appropriate position.
Determination of the AdpA-binding site in adsA.
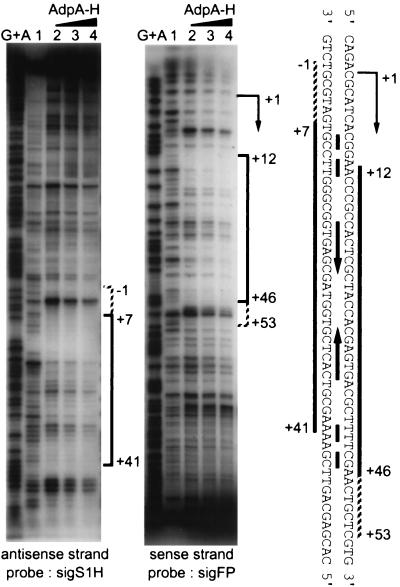
The cloned fragment in AdBS1 was 389 bp in length (nucleotide no. −162 to +227 with respect to the transcriptional start point of adsA; Fig. 5A), which was sandwiched between the primer sequences at both ends. For more precise mapping of the AdpA-binding site in the 389-bp region, we added various parts in this region to the AdpA-AdBS1 mixture to determine which part served as a competitor in the gel mobility shift assay. The results are summarized in Fig. 5. For example, probe F3-R4 competed the AdpA-H and AdBS1 binding, whereas probe F2-R3 did not. The probable AdpA-binding site was therefore expected to be at or near 50 bp upstream of the translational start codon of adsA and 33 bp downstream of the transcriptional start site. This competition experiment also showed that no other binding site was present within the fragment examined. We then used probes sigS1H and sigFP, both of which covered the AdpA-binding site (Fig. 3C), for the following DNase I footprinting assays.
AdpA-H protected a sequence (from positions +7 to +41 with respect to the transcriptional start point of adsA) from DNase I digestion, when probe sigS1H with 32P at the 5′ end was used (Fig. 6). When the amount of AdpA-H was increased, seven additional nucleotides from positions +6 to −1 appeared to be protected. Similar DNase I protection assay with probe sigFP with 32P at the 5′ end showed that a sequence from positions +12 to +46 was protected from DNase I digestion. An increased amount of AdpA-H also protected seven additional nucleotides from positions +47 to +53. Thus, we concluded that AdpA-H bound a region just downstream of the transcriptional start point of adsA. A palindrome-like sequence is present in this region. The pattern of protection observed with the DNase I footprinting shows that AdpA-H binds both strands symmetrically, supporting the idea that the inverted repeat is important for AdpA-H to recognize the binding site. The symmetrical protection pattern also suggests that AdpA-H is a dimer or tetramer, as is observed with many other transcriptional factors. Our attempt to determine the subunit structure of AdpA purified from S. griseus or AdpA-H purified from E. coli by gel filtration column chromatography failed, because they readily formed aggregates and passed through the column. Most of the transcriptional factors in the AraC/XylS family are highly insoluble (15).
FIG. 6.
Analysis of AdpA-H binding to the region downstream of the adsA promoter by DNase I footprinting assays. Probe sigS1H for the antisense strand was prepared by PCR with primers sigS1H-F and sigS1H-R (Fig. 3C). Probe sigFP for the sense strand was similarly prepared with primers sigFP-F and sigFP-R. The antisense strand from positions +7 to +41 with respect to the transcriptional start point of adsA is protected, and an additional sequence from positions −1 to +6 is also weakly protected. The sense strand from positions +12 to +46 is protected, and a sequence from positions +47 to +53 is also weakly protected. An inverted repeat within the AdpA-H-binding site is indicated between the both strands, together with the transcriptional start point of adsA.
Phenotypes of adsA mutants.
adsA was expected to regulate morphological and/or physiological development, since ςAdsA induced by AdpA was a member in the hierarchy of the A-factor regulatory cascade. To compare phenotypes with those of the wild-type strain, we generated adsA disruptants by replacing the adsA coding sequence (Ala-32 to Ala-245) by a kanamycin resistance gene by means of double crossover (Fig. 7A). Correct disruption was checked by Southern hybridization (Fig. 7B). Six adsA disruptants obtained in this way grew normally, but they formed few aerial hyphae (Fig. 7C). Even after prolonged incubation, the ΔadsA strains still formed few aerial hyphae, whereas the ΔadpA strains or the A-factor-deficient mutant strain HH1 produced no detectable aerial hyphae. Introduction of adsA on a low-copy-number plasmid pKU209 (plasmid pKU209-adsA) into the ΔadsA mutants restored the defect. On the other hand, the adsA mutation caused almost no effect on streptomycin production; the ΔadsA mutants produced almost the same amount of streptomycin when assayed by bioautography with B. subtilis as an indicator (Fig. 7D). Furthermore, the adsA mutation neither affected yellow pigment production, whereas strains HH1 and ΔadpA produced no yellow pigment (Fig. 7E). In the assay of yellow pigment production, strains were grown on phosphate-depleted minimal medium because production of this pigment was repressed by phosphate in the medium (our unpublished observation). These findings clearly show that adsA concerns only aerial mycelium formation and not secondary metabolic function.
We introduced adsA on a high-copy-number plasmid pIJ486 (plasmid pIJ486-adsA) into the ΔadpA strain and the A-factor-deficient mutant strain HH1. These transformants grew as substrate mycelium and did not form aerial hyphae. As discussed below, we assume that some additional gene products, which are induced by AdpA, are also required for normal development of aerial hyphae.
Distribution of adsA in Streptomyces spp.
We examined distribution of adsA in Streptomyces species by Southern hybridization by using an adsA sequence as 32P-labeled probe. In all the 14 species examined, signals were detected, indicating wide distribution of adsA among Streptomyces spp. (Fig. 8). Although multiple ECF ς factors are supposedly present in a given Streptomyces strain, a single signal is detected for each species. The signals detected under these conditions therefore represent the ς factors structurally and functionally similar to ςAdsA.
FIG. 8.
Wide distribution of nucleotide sequence homologues with adsA among actinomycetes. Lane 1, S. albus IFO3710; lane 2, S. antibioticus IFO3126; lane 3, S. blastmyceticus IFO12747; lane 4, S. coelicolor A3(2) M130; lane 5, S. collinus IFO12759; lane 6, S. cyaneofuscatus IFO13190; lane 7, S. flaveolus IFO3408; lane 8, S. fradiae ATCC21096; lane 9, S. griseus IFO13350; lane 10, S. lividans HH21; lane 11, S. sindenensis IFO12915; lane 12, S. zaomyceticus IFO13348; lane 13, Actinomyces citreofluorescens IFO12853; and lane 14, A. fluorescens IFO12861.
DISCUSSION
adsA encoding ςAdsA belonging to an ECF subfamily of the ς70 class was found to be a direct target of the A-factor-dependent transcriptional factor AdpA. ECF ς factors are responsible for the response of a variety of extracellular signals. S. griseus uses an ECF ς factor to form aerial hyphae in response to both the internal and external A-factor signal. A single substrate hypha develops into an aerial hypha and a chain of spores when the intrahyphal concentration of A-factor reaches a critical level and when it accepts A-factor produced by a different hypha in the neighborhood. Although it is still unclear whether or how afsA responsible for A-factor biosynthesis is induced by external stimuli, it is reasonable to assume that A-factor serves as both intracellular and extracellular signals in the ecosystem. This may be one of the strategies for S. griseus to survive in the environment; simultaneous sporulation of a group of hyphae that accepts A-factor from a neighboring hypha is advantageous to survival in the ecosystem, rather than piecemeal sporulation.
Of the regulatory proteins in the A-factor regulatory cascade (18, 40), afsA probably encoding an A-factor biosynthetic enzyme, arpA encoding a repressor for adpA in the absence of A-factor, and adpA induced by A-factor are common to secondary metabolism and morphological development (Fig. 9). In this cascade, there should be a branching point of the signal relay, from which the regulatory cascade common to morphological development and secondary metabolism is divided into two parts: one for the developmental process and the other for secondary metabolism. Since ςAdsA is concerned only with aerial mycelium formation and not with secondary metabolism, it serves as one of the proteins after the branch point. In the cascade leading to streptomycin production after the branch point, StrR, which serves as a pathway-specific transcriptional activator for the streptomycin biosynthetic gene cluster (49), is one of the direct targets of AdpA. It is quite conceivable that ςAdsA together with RNA polymerase core enzyme transcribes multiple genes required for and specific to aerial mycelium formation. ςAdsA [and its counterpart, BldN, in S. coelicolor A3(2); see below] is the only sigma factor that has so far been found to control commitment from substrate mycelium to aerial mycelium, although ςWhiG belonging to a subgroup of flagellar sigma factors controls commitment from aerial mycelium to spores (27, 57) and SigF similar to the way B. subtilis SigB belonging to a subgroup of heat shock sigma factors controls a late stage in sporulation (47). ςWhiG activates the early-sporulation whi genes, of which the translated proteins together with ςWhiG itself are necessary for transcription of sigF. It is tempting to speculate that, as is found in the developmental decisions during sporulation in the aerial mycelium in Streptomyces (10) and also in endospore formation in B. subtilis (14, 54, 62), a sigma cascade in which one or more ς factors function downstream of ςAdsA in the hierarchy for the developmental decisions in substrate mycelium is present.
We have isolated nine different DNA fragments, including AdBS1 controlling adsA, that are all recognized and bound by AdpA-H. The upstream activation sequence for strR was not included in this library. This means that further gel mobility shift-PCR experiments will yield more DNA fragments bound by AdpA-H. One of such DNA fragment is expected to control the gene expression required for yellow pigment production, since the ΔadsA mutation caused no effect on secondary metabolite formation. The presence of many genes, all of which are simultaneously activated by AdpA at a specific point in the growth phase, means that the signal from A-factor is greatly amplified at this regulatory step via AdpA as an amplifier. ςAdsA is also an amplifier of the A-factor signal since it supposedly transcribes multiple genes required for ordered development. Simultaneous and ordered expression of a group of genes makes it possible for the cells to adequately adapt to rapid physiological and environmental changes. The failure of adsA on a high-copy-number plasmid to recover the defect in aerial hyphae formation in the ΔadpA and HH1 strains can be ascribed to this amplification system; multiple gene products induced by AdpA, in addition to ςAdsA, are required for aerial mycelium formation. Thus, we catch a glimpse of the mechanism by which the A-factor signal is amplified to commit aerial mycelium formation and secondary metabolite formation.
AdpA-H binds the DNA sequence from positions +7 to +46 just downstream of the transcriptional start point of adsA. When the amount of AdpA-H in the DNase I footprinting assay was increased, the binding site extended from positions −1 to +53. In most gene expression, this site is usually a place where a repressor sits and inhibits the initiation of transcription by RNA polymerase holoenzyme. However, we can also say that this site is the place for AdpA to interact directly with RNA polymerase that sits on the promoter sequence. The 3′ side of the binding site of bacterial RNA polymerase is known to extend to +1 with respect to the transcriptional start point (32, 48). A speculative, very simple model to explain the function of AdpA is that it recruits RNA polymerase to the promoter sequence (48). This recruiting function of AdpA may compensate for the absence of a −35-like sequence in the adsA promoter. On the other hand, the upstream activation sequence for strR, to which AdpA binds, is about 270 nucleotides from its transcriptional start point (40, 58). In addition, in front of the transcriptional start point of strR, TTGGCC for −35 and TACTAT for −10 are present (12, 59). Nevertheless, it is possible that AdpA activates transcription of strR by recruiting RNA polymerase to its promoter. We also have to take into consideration the observation that the promoters controlled by regulators in the AraC/XylS family usually contain more than one binding site for the regulation (15), although a second AdpA-binding site was not found in the 389-bp fragment covering the adsA promoter (Fig. 5). For elucidation of the mechanism by which AdpA activates transcription of these promoters, in vitro transcription assays are absolutely necessary. Characterization of AdBS2 to AdBS9 will help to deduce a consensus sequence which AdpA recognizes.
S. coelicolor A3(2) also contains adpA- and adsA-like genes (adpA-c and adsA-c), and the gene organizations around these genes are the same as those in S. griseus (40; unpublished data). adsA-c has been identified as a gene that complements a whi mutation and is named bldN because of a Bld phenotype of disruptants (7). Thus, two groups have reached the same gene, adsA in S. griseus and bldN in S. coelicolor A3(2), through different approaches. Involvement of γ-butyrolactone-type regulators in S. coelicolor A3(2) is not adequately understood, although three ArpA-like proteins controlling both morphological and physiological differentiation (43; E. Takano and M. J. Bibb, personal communication) and a series of A-factor-like compounds (1, 5) have been reported. However, a sequence similar to the consensus sequence bound by the γ-butyrolactone receptor proteins, such as ArpA (42), CprA (56), CprB (56), and BarA (28), is neither present in front of adpA-c nor is present a sequence similar to the AdpA-binding site in front of bldN. Our preliminary gel retardation experiments showed that AdpA-H did not bind the corresponding region for bldN; only a large amount of AdpA-H gave a very faint retarded signal (data not shown). A computer-aided search in the databases reveals the presence of more than eight AdpA homologues in S. coelicolor A(3). One of them may activate transcription of bldN. Although no information about a signal relay to adpA-c or about regulation of bldN by AdpA-c is available, we speculate that AdpA-c induced by a still-unknown mechanism activates bldN for the substrate mycelium to develop into aerial hyphae. γ-Butyrolactones may participate in the regulation of these genes, as a tuner but not as a switch. In general in Streptomyces species, the presence of a single copy of adsA-like sequence in a given Streptomyces species suggests its common role in aerial hyphae formation in this genus.
ACKNOWLEDGMENTS
This work was supported, in part, by the Waksman Foundation of Japan, by the “Research for the Future” Program of the Japan Society for the Promotion of Science, and by the Bio Design Program of the Ministry of Agriculture, Forestry, and Fisheries of Japan (BDP-00-VI-2-1).
REFERENCES
- 1.Anisova L N, Blinova I N, Efremenkova O V, Koz'min Y P, Onoprienko V V, Smirnova G M, Khokhlov A S. Regulators of the development of Streptomyces coelicolor A3(2) Izv Akad Nauk SSSR Ser Biol. 1984;1:98–108. [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingstone R E, Moore D O, Seidman J S, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 3.Beck E, Ludwig G, Auerswald A, Reiss B, Schaller H. Nucleotide sequence and exact localisation of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982;19:327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- 4.Beutel B A, Gold L. In vitro evolution of intrinsically bent DNA. J Mol Biol. 1992;228:803–812. doi: 10.1016/0022-2836(92)90865-h. [DOI] [PubMed] [Google Scholar]
- 5.Bibb M J. The regulation of antibiotic production in Streptomyces coelicolor A3(2) Microbiology. 1996;142:1335–1344. doi: 10.1099/13500872-142-6-1335. [DOI] [PubMed] [Google Scholar]
- 6.Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984;30:157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- 7.Bibb M J, Molle V, Buttner M J. ςBldN, an extracytoplasmic function RNA polymerase sigma factor required for aerial mycelium formation in Streptomyces coelicolor A3(2) J Bacteriol. 2000;182:4606–4616. doi: 10.1128/jb.182.16.4606-4616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chater K F. Sporulation in Streptomyces. In: Smith I, Slepecky R A, Setlow P, editors. Regulation of procaryotic development: structural and functional analysis of bacterial sporulation and germination. Washington, D.C.: American Society for Microbiology; 1989. pp. 277–299. [Google Scholar]
- 9.Chater K F. Genetics of differentiation in Streptomyces. Annu Rev Microbiol. 1993;47:685–713. doi: 10.1146/annurev.mi.47.100193.003345. [DOI] [PubMed] [Google Scholar]
- 10.Chater K F. Developmental decisions during sporulation in the aerial mycelium in Streptomyces. In: Brun Y V, Shimkets L J, editors. Prokaryotic developments. Washington, D.C.: American Society for Microbiology; 2000. pp. 33–48. [Google Scholar]
- 11.Connolly L, De Las Penas A, Alba B M, Gross C A. The response to extracytoplasmic stress in Escherichia coli is controlled by partially overlapping pathways. Genes Dev. 1997;11:2012–2021. doi: 10.1101/gad.11.15.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Distler J, Ebert A, Mansouri K, Pissowotzki K, Stockmann M, Piepersberg W. Gene cluster for streptomycin biosynthesis in Streptomyces griseus: nucleotide sequence of three genes and analysis of transcriptional activity. Nucleic Acids Res. 1987;15:8041–8056. doi: 10.1093/nar/15.19.8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dombroski A J, Walter W A, Gross C A. Amino-terminal amino acids modulate sigma-factor DNA-binding activity. Genes Dev. 1993;7:2446–2455. doi: 10.1101/gad.7.12a.2446. [DOI] [PubMed] [Google Scholar]
- 14.Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallegos M-T, Schleif R, Bairoch A, Hofmann K, Ramos J L. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham H C, McGowan S J, Robson P R, Hodgson D A. Light-induced carotenogenesis in Myxococcus xanthus: light-dependent membrane sequestration of ECF sigma factor CarQ by anti-sigma factor CarR. Mol Microbiol. 1996;19:171–186. doi: 10.1046/j.1365-2958.1996.360888.x. [DOI] [PubMed] [Google Scholar]
- 17.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 18.Horinouchi S. Streptomyces genes involved in aerial mycelium formation. FEMS Microbiol Lett. 1996;141:1–9. doi: 10.1111/j.1574-6968.2010.02177.x. [DOI] [PubMed] [Google Scholar]
- 19.Horinouchi S, Beppu T. Autoregulatory factors and communication in actinomycetes. Annu Rev Microbiol. 1992;46:377–398. doi: 10.1146/annurev.mi.46.100192.002113. [DOI] [PubMed] [Google Scholar]
- 20.Horinouchi S, Beppu T. A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol Microbiol. 1994;12:859–864. doi: 10.1111/j.1365-2958.1994.tb01073.x. [DOI] [PubMed] [Google Scholar]
- 21.Horinouchi S, Kumada Y, Beppu T. Unstable genetic determinant of A-factor biosynthesis in streptomycin-producing organisms: cloning and characterization. J Bacteriol. 1984;158:481–487. doi: 10.1128/jb.158.2.481-487.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang X, Decatur A, Sorokin A, Helmann J D. The Bacillus subtilis ςX protein is an extracytoplasmic function ς factor contributing to survival at high temperature. J Bacteriol. 1997;179:2915–2921. doi: 10.1128/jb.179.9.2915-2921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishikawa J, Hotta K. Frame Plot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol Lett. 1999;174:251–253. doi: 10.1111/j.1574-6968.1999.tb13576.x. [DOI] [PubMed] [Google Scholar]
- 24.Kakinuma S, Takada Y, Ikeda H, Tanaka H, Omura S. Cloning of large DNA fragments, which hybridize with actinorhodin biosynthesis genes, from kalafungin and nanaomycin A methyl ester producers and identification of genes for kalafungin biosynthesis of the kalafungin producer. J Antibiot. 1991;44:995–1005. doi: 10.7164/antibiotics.44.995. [DOI] [PubMed] [Google Scholar]
- 25.Kang J-G, Paget M S B, Seok Y-J, Hahn M Y, Bae J-B, Hahn J-S, Kleanthous C, Buttner M J, Roe J-H. RsrA, an anti-sigma factor regulated by redox change. EMBO J. 1999;18:4292–4298. doi: 10.1093/emboj/18.15.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelemen G H, Brian P, Flärdh K, Chamberlin L, Chater K F, Buttner M J. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2) J Bacteriol. 1998;180:2515–2521. doi: 10.1128/jb.180.9.2515-2521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelemen G H, Brown G L, Kormanec J, Potuckova L, Chater K F, Buttner M J. The position of the sigma-factor genes, whiG and sigF, in the hierarchy controlling the development of spore chains in the aerial hyphae of Streptomyces coelicolor A3(2) Mol Microbiol. 1996;21:593–603. doi: 10.1111/j.1365-2958.1996.tb02567.x. [DOI] [PubMed] [Google Scholar]
- 28.Kinoshita H, Tsuji T, Ipposhi H, Nihira T, Yamada Y. Characterization of binding sequences for butyrolactone autoregulator receptors in streptomycetes. J Bacteriol. 1999;181:5075–5080. doi: 10.1128/jb.181.16.5075-5080.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinzler K W, Vogelstein B. Whole genome PCR: application to the identification of sequences bound by gene regulatory proteins. Nucleic Acids Res. 1989;17:3645–3653. doi: 10.1093/nar/17.10.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lonetto M A, Brown K L, Rudd K E, Buttner M J. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase ς factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci USA. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 32.Marqués S, Gallegos M T, Ramos J L. Role of ςS in transcription from the positively controlled Pm promoter of the TOL plasmid of Pseudomonas putida. Mol Microbiol. 1995;18:851–857. doi: 10.1111/j.1365-2958.1995.18050851.x. [DOI] [PubMed] [Google Scholar]
- 33.Martin D W, Schurr M J, Yu H, Deretic V. Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: relationship to ςE and stress response. J Bacteriol. 1994;176:6688–6696. doi: 10.1128/jb.176.21.6688-6696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez-Argudo I, Ruiz-Vazquez R M, Murillo F J. The structure of an ECF-sigma-dependent, light-inducible promoter from the bacterium Myxococcus xanthus. Mol Microbiol. 1998;30:883–893. doi: 10.1046/j.1365-2958.1998.01129.x. [DOI] [PubMed] [Google Scholar]
- 35.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 36.McGowan S J, Gorham H C, Hodgson D A. Light-induced cartenogenesis in Myxococcus xanthus: DNA sequence analysis of the carR region. Mol Microbiol. 1993;10:713–735. doi: 10.1111/j.1365-2958.1993.tb00943.x. [DOI] [PubMed] [Google Scholar]
- 37.Missiakas D, Raina S. The extracytoplasmic function sigma factors: role and regulation. Mol Microbiol. 1998;28:1059–1066. doi: 10.1046/j.1365-2958.1998.00865.x. [DOI] [PubMed] [Google Scholar]
- 38.Neumann T, Piepersberg W, Distler J. Decision phase regulation of streptomycin production in Streptomyces griseus. Microbiology. 1996;142:1953–1963. [Google Scholar]
- 39.Oh S H, Chater K F. Denaturation of circular or linear DNA facilitates targeted integrative transformation of Streptomyces coelicolor A3(2): possible relevance to other organisms. J Bacteriol. 1997;179:122–127. doi: 10.1128/jb.179.1.122-127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohnishi Y, Kameyama S, Onaka H, Horinouchi S. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol Microbiol. 1999;34:102–111. doi: 10.1046/j.1365-2958.1999.01579.x. [DOI] [PubMed] [Google Scholar]
- 41.Onaka H, Ando N, Nihira T, Yamada Y, Beppu T, Horinouchi S. Cloning and characterization of the A-factor receptor gene from Streptomyces griseus. J Bacteriol. 1995;177:6083–6092. doi: 10.1128/jb.177.21.6083-6092.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Onaka H, Horinouchi S. DNA-binding activity of the A-factor receptor protein and its recognition DNA sequences. Mol Microbiol. 1997;24:991–1000. doi: 10.1046/j.1365-2958.1997.4081772.x. [DOI] [PubMed] [Google Scholar]
- 43.Onaka H, Nakagawa T, Horinouchi S. Involvement of two A-factor receptor homologues in Streptomyces coelicolor A3(2) in the regulation of secondary metabolism and morphogenesis. Mol Microbiol. 1998;28:743–753. doi: 10.1046/j.1365-2958.1998.00832.x. [DOI] [PubMed] [Google Scholar]
- 44.Paget M S B, Chamberlin L, Atrih A, Foster S J, Buttner M J. Evidence that the extracytoplasmic function sigma factor ςE is required for normal cell wall structure in Streptomyces coelicolor A3(2) J Bacteriol. 1999;181:204–211. doi: 10.1128/jb.181.1.204-211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paget M S B, Kang J-G, Roe J-H, Buttner M J. ςR, an RNA polymerase sigma factor that modulates expression of the thioredoxin system in response to oxidative stress in Streptomyces coelicolor A3(2) EMBO J. 1998;17:5776–5782. doi: 10.1093/emboj/17.19.5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paget M S B, Leibovitz E, Buttner M J. A putative two-component signal transduction system regulates ςE, a sigma factor required for normal cell wall integrity in Streptomyces coelicolor A3(2) Mol Microbiol. 1999;33:97–107. doi: 10.1046/j.1365-2958.1999.01452.x. [DOI] [PubMed] [Google Scholar]
- 47.Potuckova L, Kelemen G H, Findlay K C, Lonetto M A, Buttner M J, Kormanec J. A new RNA polymerase sigma factor, ςF, is required for the late stage of morphological differentiation in Streptomyces spp. Mol Microbiol. 1995;17:37–48. doi: 10.1111/j.1365-2958.1995.mmi_17010037.x. [DOI] [PubMed] [Google Scholar]
- 48.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 49.Retzlaff L, Distler J. The regulator of streptomycin gene expression, StrR, of Streptomyces griseus is a DNA binding protein with multiple recognition sites. Mol Microbiol. 1995;18:151–162. doi: 10.1111/j.1365-2958.1995.mmi_18010151.x. [DOI] [PubMed] [Google Scholar]
- 50.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sexton R, Gill P R, Jr, Dowling D N, O'Gara F. Transcriptional regulation of the iron-responsive sigma factor gene pbrA. Mol Gen Genet. 1996;250:50–58. doi: 10.1007/BF02191824. [DOI] [PubMed] [Google Scholar]
- 52.Shinkawa H, Hatada Y, Okada M, Kinashi H, Nimi O. Nucleotide sequence of a principal sigma factor gene (hrdB) of Streptomyces griseus. J Biochem. 1995;118:494–499. doi: 10.1093/oxfordjournals.jbchem.a124935. [DOI] [PubMed] [Google Scholar]
- 53.Sollner-Webb B, Reeder R H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979;18:485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- 54.Sonenshin A L. Endospore-forming bacteria: an overview. In: Brun Y V, Shimkets L J, editors. Prokaryotic development. Washington, D.C.: American Society for Microbiology; 2000. pp. 133–150. [Google Scholar]
- 55.Strohl W R. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992;20:961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sugiyama M, Onaka H, Nakagawa T, Horinouchi S. Site-directed mutagenesis of the A-factor receptor protein: Val-41 important for DNA-binding and Trp-119 important for ligand-binding. Gene. 1998;222:133–144. doi: 10.1016/s0378-1119(98)00487-9. [DOI] [PubMed] [Google Scholar]
- 57.Tan H, Yang H, Tian Y, Wu W, Whatling C A, Chamberlin L C, Buttner M J, Nodwell J, Chater K F. The Streptomyces coelicolor sporulation-specific sigma WhiG form of RNA polymerase transcribes a gene encoding a ProX-like protein that is dispensable for sporulation. Gene. 1998;21:137–146. doi: 10.1016/s0378-1119(98)00152-8. [DOI] [PubMed] [Google Scholar]
- 58.Vujaklija D, Horinouchi S, Beppu T. Detection of an A-factor-responsive protein that binds to the upstream activation sequence of strR, a regulatory gene for streptomycin biosynthesis in Streptomyces griseus. J Bacteriol. 1993;175:2652–2661. doi: 10.1128/jb.175.9.2652-2661.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vujaklija D, Ueda K, Hong S-K, Beppu T, Horinouchi S. Identification of an A-factor-dependent promoter in the streptomycin biosynthetic gene cluster of Streptomyces griseus. Mol Gen Genet. 1991;229:119–128. doi: 10.1007/BF00264220. [DOI] [PubMed] [Google Scholar]
- 60.Ward J M, Janssen G R, Kieser T, Bibb M J, Buttner M J, Bibb M J. Construction and characterization of a series of multicopy plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol Gen Genet. 1986;203:468–478. doi: 10.1007/BF00422072. [DOI] [PubMed] [Google Scholar]
- 61.Wildermuth H. Development and organization of the aerial mycelium in Streptomyces coelicolor. J Gen Microbiol. 1970;60:43–50. doi: 10.1099/00221287-60-1-43. [DOI] [PubMed] [Google Scholar]
- 62.Wösten M M S M. Eubacterial sigma-factors. FEMS Microbiol Rev. 1998;22:127–150. doi: 10.1111/j.1574-6976.1998.tb00364.x. [DOI] [PubMed] [Google Scholar]
- 63.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]