Abstract
Antimicrobial resistance is a global health challenge caused by the ability of microorganisms including bacteria, fungi, protozoans and viruses to survive the effects of drugs that hitherto were effective against them. This study sought to investigate the presence of antibiotic-resistant bacteria and their corresponding molecular determinants in fish farms of the Central and Western Regions of Ghana. Management practices and antibiotic use at the fish farms were obtained through the administration of a questionnaire. Coliform and Gram-positive bacterial loads of catfish (Clarias gariepinus), tilapia (Oreochromis niloticus) intestinal microbiota, and pond water samples recovered on MacConkey Agar and Mannitol Salt Agar were determined. Bacterial isolates were identified using various biochemical assays. Antibiotic resistance profiles and possible responsible genes of bacterial isolates were determined using the disc diffusion and Polymerase Chain Reaction (PCR) methods respectively. The study revealed that none of the fish farm managers admitted using antibiotics for prevention and treatment of diseases and no major disease outbreak had ever been recorded. Bacterial loads of pond water exceeded the acceptable level of ≤100 E. coli and <10 coliforms per mL for wastewater recommended for use in fish farming. In all, 145 bacterial isolates comprising 99 Gram negative and 46 Gram-positive bacteria were stored and identified. Most isolates were resistant to at least an antibiotic. Both Gram-negative and Gram-positive bacteria were highly resistant to beta-lactam antibiotics with a corresponding high percentage detection of the blaTEM gene compared to other classes of antibiotics. This study has revealed the presence of various molecular determinants of antibiotic resistance including blaTEM, cmIA, qnrS, tetB and blaCTX-M, in multidrug-resistant bacteria at some fish farms in Ghana. There is the need to increase awareness about risks associated with the misuse and overuse of antibiotics by humans and the potential risk of spread of multi-drug resistant-bacteria in the environment.
Keywords: Antibiotics, Fish farms, Resistance genes, Coliforms, E. coli, Fish species, Aquaculture
Antibiotics; Fish farms; Resistance genes; Coliforms; E. coli; Fish species; Aquaculture.
1. Introduction
There is no doubt that antibiotics are important drugs that play an essential role in the survival of humans against various infectious diseases and as such these drugs have over time ingrained medicine, agriculture and industries (McEwen and Collignon, 2017; WHO, 2018). This over-exploitation of antibiotics has over the years led to the fast emergence of multidrug-resistant bacteria or “superbugs”. The latter unfortunately continue to threaten humanity by rendering the treatment of bacterial diseases much more difficult (Smith et al., 2021). Multidrug-resistant bacteria are resilient to three or more antibiotics to which they were previously susceptible but due to a number of possible factors, eventually developed resistance to these drugs. There are several mechanisms that bacteria explore in order to escape the effect of antibiotics. These mechanisms have broadly been categorized into two: genetic strategies exhibited to resist the inhibitory effect of the antibiotic and the use of alternate biochemical pathways (Munita and Arias, 2016). While the genetic basis of antibiotic resistance is directed by mutational resistance and acquisition of foreign DNA through horizontal gene transfer, the metabolic strategies involve the modification of the antibiotic molecule, decreased antibiotic absorption and efflux, changes in target sites as well as global cell adaptations (Munita and Arias, 2016). In addition, the presence of antibiotics residues even in low concentrations in the environment as a result of the persistent abuse and misuse of antibiotics leads to a selective pressure among bacteria that result in the emergence of multidrug-resistant bacteria (Donkor et al., 2018; Okocha et al., 2018). It is regrettable to note that antibiotics that have been labelled as last-resort drugs used in human medicine have been reported to be extensively used in the fish culture industry (World Health Organization & WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR), 2017). Recently, some major antibiotic-resistant coliforms and Gram-positive bacterial pathogens were isolated from some fish farms which have been reported to be a threat to human health (Shrivastava et al., 2018; World Health Organization & WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR), 2017).
Fish farming, has in the past few years rapidly grown (Golub and Varma, 2014; Okocha et al., 2018) with exports from developing countries reported to have increased over the past two decades (Assefa and Abunna, 2018). In Ghana, fish farming started in the 1950s during the colonial rule to boost the livelihood of communities and to improve nutrition (Tall and Failler, 2012). Nile tilapia (Oreochromis niloticus) and African catfish (Clarias gariepinus) are two main fish types produced on a large scale in the country. With a rise from less than 11,000 tons in 2010 to over 57,000 tons in 2017, aquaculture production significantly contributes to the country's economy. The increase observed in this sector of the fisheries industry is largely due to an enhanced production from private commercial operators which essentially account for 75% of Ghana's aquaculture output (Amenyogbe et al., 2018; Doku et al., 2018).
Despite the prospects of fish farming in Ghana, unsanitary conditions at fish farms and the occurrence of “superbug” bacteria in fisheries products and waters have been reported as worrisome observations (Agoba et al., 2017; Apenteng et al., 2017). This could pose threat to human health, especially, at a time where the demand for tilapia and catfish as a source of quality animal protein in Ghana, seems to be on the ascendency (Amenyogbe et al., 2018). Infections caused by these multidrug-resistant bacteria as a result of handling or consumption of improperly cooked contaminated fishery products could be difficult to treat leading to a long stay in the hospital, increased death rates and increased medical expenses (Dadgostar, 2019). The presence of multidrug-resistant bacteria at various fish farms in the Ashanti, Central and Greater Accra regions of the country has been reported with a focus on the phenotypic characterization of antibiotic resistance (Adinortey et al., 2020; Apenteng et al., 2017; Agoba, 2016). There is, however, generally a paucity of information on the status of antibiotic use as well as the genotypic characterization of antibiotic resistance in aquaculture settings in Ghana. The current study, therefore, sought to address these issues by obtaining information on farming practices including the use of antibiotics at fish farms and investigate the level of bacterial contamination of water and fishery products. The presence of multidrug-resistant Gram-negative and Gram-positive bacteria, and the detection of antibiotic-resistance genes were also investigated with a focus on the Central and Western Regions of Ghana.
2. Materials and methods
2.1. Study areas
The Central and Western Regions of Ghana are situated in the southern part of the country. These two regions contribute to the country's economy owing to abundance of industrial minerals, agriculture, tourism and fishing activities (Finegold et al., 2010). Indigenes of the coastal areas rely heavily on fishing from the sea for a living (Finegold et al., 2010) while in-land fish farming is practised to supplement harvest from the capture fishery (Amenyogbe et al., 2018).
2.2. Ethical approval
Ethical clearance for the study with the certificate number RPN 002/CSIR-IACUC/2020, was obtained from the Institutional Animal Care and Use Committee (IACUC) of the Council for Scientific and Industrial Research (CSIR), Accra. Consent was also obtained from all farm managers who took part in the study.
2.3. Questionnaire administration
Farmers whose fish farms were in active production were specifically recruited in this study. Farms that did not consent to the research were excluded. Sample collection mainly consisted of questionnaire administration as well as the collection of fish and water samples from each farm. The questionnaire was developed to gain insights into fish farming practices, including the use of antibiotics, types of antibiotics used, history of disease outbreaks and type of feed used at the fish farms. The questionnaire form has been provided in the Supplementary Materials section.
2.4. Sample collection
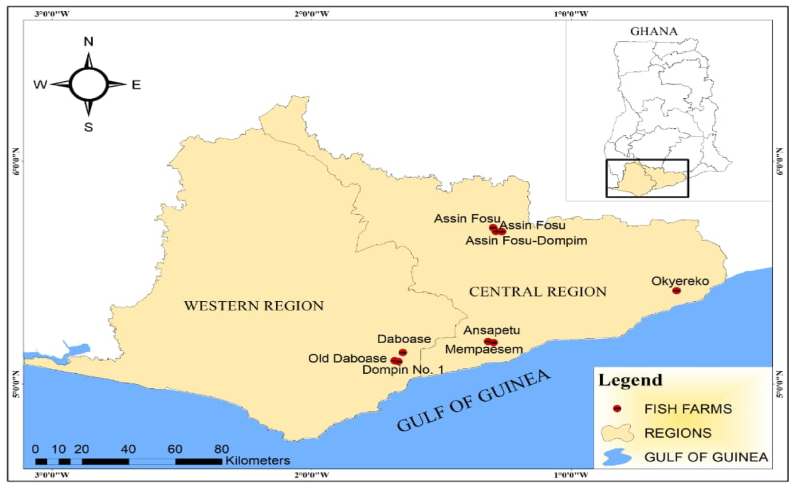
Sample collection spanned from October 2019 to February 2020. Purposive and snow-ball sampling methods were adopted to locate the fish farms. Water and fish samples were also obtained from nine farms located in townships of the two regions as shown on Figure 1.
Figure 1.
Geographic locations of fish farms where samples were obtained for the study.
On the site of each farm, fish and water samples were obtained from three collection points from the pond through fish farmers’ assistance between the hours of 9:00 to 14:00 GMT and as previously described (Adinortey et al., 2020). At each of the three collection points (the inflow, the middle and the outflow area of the pond), 100 mL of water was collected into sterile bottles, 50 cm deep below the water surface. Casting and scoop nets were used to obtain fish samples from earthen and concrete ponds respectively. Five apparently healthy fish samples were randomly selected from each catch and were immediately placed in a sterile polyethylene bag. All samples were labelled accordingly, placed on ice and transported to the laboratory for microbiological and molecular analyses. Fishes were stunned through cold shock.
Details on the nine fish farms sampled designated with alphabetic letters and the corresponding types of samples obtained have been provided (Table 1).
Table 1.
Location of fish farms and types of samples collected.
| Region | Farm | Location | Sample collected |
||
|---|---|---|---|---|---|
| Water | Tilapia | Catfish | |||
| Central Region | A | Mempaesem | ✓ | × | ✓ |
| B | Ansapetu | ✓ | × | ✓ | |
| C | Okyereko | ✓ | ✓ | × | |
| D | Assin Fosu | ✓ | ✓ | × | |
| E | Assin Fosu | ✓ | ✓ | ✓ | |
| F | Assin Fosu-Dompim | ✓ | × | ✓ | |
| Western Region | G | Dompim No. 1 | ✓ | × | ✓ |
| H | Daboase | ✓ | × | ✓ | |
| I | Old-Daboase | ✓ | ✓ | ✓ | |
✓ = sample collected; × = sample not collected because the type of fish was not cultured at those farms.
2.5. Sample processing prior to microbiological investigations
A pooled water sample was prepared into a sterile test tube by combining 3 mL aliquots of the water samples obtained from three different collection points of each pond (Huys, 2003). The resultant 9 mL-pooled water sample was thoroughly mixed and tenfold-serially diluted with a sterile 0.9 % saline solution up to the 5th dilution factor.
With regard to fish samples, the surface of the skin was disinfected with 70% ethanol and intestinal content was excised from the gut. Approximately 0.5g of the intestinal sample of the fish was transferred into a test tube containing 4.5 mL sterile 0.9 % saline solution. This test tube was vortexed and 3 mL of the intestinal samples was set aside. The same procedure was followed for fishes obtained from all three collection points. Subsequently, a 9 mL-pooled intestinal sample for every cultured fish type at each farm was obtained when all three 3mL-aliquots of the intestinal samples were put together into an additional sterile test tube. The resulting 9 mL-pooled intestinal samples were tenfold-serially diluted up the 6th dilution factor (Huys, 2003).
2.6. Bacteria isolation and determination of bacterial loads of water and fish samples
Using the pour-plate method, 1mL of the diluted suspension was inoculated on MacConkey Agar (MacA) (Oxoid Ltd., England) and Mannitol Salt Agar (MSA) (Oxoid Ltd., England) media for isolation of total coliform bacteria (based on lactose fermentation) as well as Gram-positive bacteria of the genus Staphylococcus and other related bacteria respectively. Inoculated plates were incubated at 35°C for 16–24 h.
Following the incubation period and by means of a colony counter (Stuart Scientific, UK), the bacterial load was assessed by counting all colony-forming units (CFUs) of bacteria within a range of 30–300 that grew on the culture plates. The counts were recorded based on samples from which they were recovered and mean total bacterial load of each sample was calculated as the mean value of the total bacterial load found from triplicates (Cheesbrough, 2006; Huys, 2003).
2.7. Storage and identification of bacterial isolates
About 3 to 4 morphologically distinct bacterial colonies were randomly selected from MacA and MSA plates for each sample. They were purified by subculturing and individually storing them in sterile nutrient agar slants. These were then incubated at 35 °C for 16–24 h and subsequently stored at 4 °C for further analysis.
All stored bacterial isolates were identified using Gram staining and standard biochemical tests, including Triple Sugar Iron (TSI) (Liofilchem s. r.l. Bacteriology Products, Italy) agar tests, citrate, indole and urea tests. Additional tests which include catalase, gelatinase, oxidase and coagulase tests were conducted on bacteria that were isolated using MSA (Cheesbrough, 2006).
2.8. Antibiotic susceptibility testing
The Kirby-Bauer disk diffusion method was used to determine the antibiotic susceptibility pattern as described by (Tenover, 2014). The sensitivity or resistance of all coliforms and Gram-positive bacterial isolates was determined using two sets of eight antibiotics (Abtek Biologicals Limited) that shared five antibiotics in common – namely: Cotrimoxazole (25μg), Gentamicin (10μg), Cefuroxime (30μg), Ampicillin (10μg) and Tetracycline (10μg). The remaining three antibiotics comprised Flucloxacillin (5μg), Erythromycin (5μg) and Penicillin (1.5 i. u) for testing Gram-positive bacteria while Chloramphenicol (10μg), Ceftriaxone (30μg), Cefotaxime (30μg) were used against coliform bacteria. The bacterial isolates were then classified as susceptible or resistant based on the Clinical and Laboratory Standard Institute guidelines (CLSI, 2018). Staphylococcus aureus (ATCC 662813) and Escherichia coli (ATCC 25922) were used as controls.
2.9. DNA extraction and confirmation of DNA samples as bacterial DNA
DNA extraction was done using the MaglistoTM 5M DNA Extraction kit (Bioneer Corporation, USA) according to the manufacturers' instructions. Consequently, 2 mL of fresh overnight Luria Bertani broth culture of each isolate was transferred into a sterile Eppendorf tube. Bacterial cells were recovered by centrifugation and were subsequently used for DNA extraction. All DNA samples were confirmed to be of bacterial origin using the 16S rRNA primers; 27F (5′-GAGTTTGATCCTGGCTCAG-3′) and 1492R (′5-GGTTACCTTGTTACGACTT-3’) (Bioneer Cooperation, South Korea) (Lane, 1991). Briefly, 1 μL each of 1 pmol of forward and reverse primers, 5 μL of DNA template and 13 μL of sterile molecular biology grade water were added to a PCR tube containing AccuPower PCR premix (Bioneer Corporation, South Korea) in a total reaction volume of 20 μL. The amplification programme was carried out using T100™ thermal cycler (Bio-Rad Laboratories, USA) and comprised an initial denaturation for 3 min at 95 °C; 35 cycles at 95 °C for 30 s, annealing at 55 °C, extension at 72 °C for 1 min; and a final extension at 72 °C for 5 min.
2.10. Screening for molecular determinants of antibiotic resistance
A list of 14 pairs of primer sequences used in the detection of antibiotic-resistance molecular determinants or genes is as displayed on Table 2. Based on their annealing temperature and size of the expected amplicons, the primers were grouped into five comprising four triplex and one duplex PCR assays. Triplex PCR assays were conducted by adding the following to an AccuPower Multiplex PCR premix (Bioneer Corporation, South Korea)-containing microtube designed for a single 20 μL reaction mixture: 1 μL each of 1 pmol of forward and reverse of each of 3 primers, 5 μL of DNA template and 9 μL of sterile molecular biology grade water. In the case of the duplex PCR, however, 11 μL of sterile molecular biology grade water was added to the microtube instead of 9 μL. The amplification programme comprised an initial denaturation for 3 min at 95 °C; 35 cycles at 95 °C for 30 s, annealing at 55 °C, extension at 72 °C for 1 min; and a final extension at 72 °C for 5 min.
Table 2.
Primer groupings used in PCR analysis.
| PCR Assay No. | Primer name | Primer sequence (5′–3′) | Amplicon size (bp) | References |
|---|---|---|---|---|
| 1 | cmIA |
F GGCCTCGCTCTTACGTCATC R GCGACACCAATACCCACTAGC |
662 | Ma et al., 2007 |
| Cat1 |
F AACCAGACCGTTCAGCTGGAT R CCTGCCACTCATCGCAGTAC |
549 | Zhao et al., 2001 | |
| 2 | blaTEM |
F GAGTATTCAACATTTCCGTGTCGC R TACCAATGCTTAATCAGTGAGGC |
865 | Zhang et al., 2011 |
| qnrS |
F ACGACATTCGTCAACTGCAA R TAAATTGGCACCCTGTAGGC |
417 | Robicsek, 2006 | |
| gyrA |
F CGACCTTGCGAGAGAAAT R GTTCCATCAGCCCTTCAA |
626 | Hossain, 2017 | |
| 3 | blaEBC |
F TCGGTAAAGCCGATGTTGCGG R CTTCCACTGCGGCTGCCAGTT |
302 | Perez-Perez, 2002 |
| Sul3 |
FCAGATAAGGCAATTGAGCATGCTCTGC R GATTTCCGTGACACTGCAATCATT |
569 | Arabi et al., 2015 | |
| Sul1 |
F CGGCGTGGGCTACCTGAACG R GCCGATCGCGTGAAGTTCCG |
432 | Arabi et al., 2015 | |
| 4 | blaTEM-1 |
F CCAATGCTTAATCAGTGAGG R ATGAGTATTCAACATTTCCG |
858 | Domínguez-Pérez et al., 2018 |
| qnrB |
F GATCGTGAAAGCCAGAAAGG R ACGATGCCTGGTAGTTGTCC |
469 | Wang et al., 2008 | |
| tetB |
F CAGTGCTGTTGTTGTCATTAA R GCTTGGAATACTGAGTGTTAA |
571 | Ma et al., 2007 | |
| 5 | blamecA |
F AAAATCGATGGTAAAGGTTGGC R AGTTCTGCAGTACCGGATTTG |
533 | Azimain, 2012 |
| tetA |
F TTGGCATTCTGCATTCACTC R GTATAGCTTGCCGGAAGTCG |
494 | Ma et al., 2007 | |
| blaCTX-M |
F ACGCTGTTGTTAGGAAGTG R TTGAGGCTGGGTGAAGT |
857 | Seyedjavadi, 2016 |
After the amplification process, 5 μL each of all amplicons were loaded onto a 2 % (w/v) ethidium bromide-stained agarose gel and a 100 bp DNA ladder (Bioneer Corporation. Korea) was used as a molecular weight marker. The amplicons were separated through gel agarose electrophoresis at 90 V for 60 min. Each gel was subsequently examined under an ultraviolet transilluminator (UVP products, UK) and photographic records taken using a digital camera.
2.11. Statistical analysis
Data sets recorded were entered into Microsoft Excel and then moved to GraphPad Prism software version 6. Data obtained from the questionnaire, antibiotic resistance profile and detection of antibiotic resistant genes were interpreted using descriptive statistics. Analysis of variance (ANOVA) and independent t-test were used to compare the mean of bacterial loads of water and fish samples. Confidence interval was set at 95 % and the probability value p ≤ 0.05 was considered statistically significant.
3. Results
3.1. Farming practices adopted by farm managers
The major findings obtained from the questionnaires are presented in Table 3. All nine fish farms sampled in this study made use of formulated feed purchased from different suppliers. It was also observed that three of these farms (i.e., farm C, E, and I) each operated a hatchery and supplied fingerlings to other fish farmers. Although none of the farm managers reported to have witnessed a disease outbreak and all of them with no exception emphatically stated that antibiotics were not used in any way at all stages of the culturing period, they generally reported a few occasions of deaths which they attributed to overcrowding and poor water quality.
Table 3.
Management practices adopted at various fish farms.
| Farming practices | Response | Number of Farms (Specific farms in brackets) | |
|---|---|---|---|
| Source of pond water | Borehole | 2 (F and G) | |
| Tap water | 2 (A and C) | ||
| Streams | 7 (B, C, D, E, G, H and I) | ||
| Use of antibiotics | None of the farms | 0 | |
| Incidence of disease outbreak | None of the farms | 0 | |
| Frequency of water change | Every 1–3 months and before restocking | 3 (C, G and H) | |
| Varies from farm to farm based on stocking density, quality of water or type of fish feed applied | 6 (A, B, D, E, F and I) | ||
| Farmers' reasons for changing pond water | Significant change in water quality and | 9 (All farms) | |
| observation of lethargic movement of fish | |||
| Methods used for disposing waste water from pond | Waste channeled into nearby stream | 8 (A, C, D, E, F, G, H and I) | |
| Waste channeled through drains into a temporary pond prior to use in irrigation | 1 (B) | ||
| Culture type | Monoculture | Catfish | 5 (A, B, F, G and H) |
| Tilapia | 2 (C and D) | ||
| Polyculture | Catfish + Tilapia | 2 (E and I) | |
This study revealed that sources of water used for rearing fishes comprised tap water, boreholes or nearby streams. Generally, the frequency of changing the pond water in the farms varied depending on the stocking density, quality of water and type of fish feed applied. The observation of significantly abnormal colour changes in the water required the need for a change of the water. It was observed that farmers at farms C, G and H changed the pond water every one to three months while for the other farms, the frequency of changing the pond water varied from one farm to another. For all farms except farm B, the waste generated on the site was disposed by channelling it to nearby streams. At Farm B, farm managers not only disposed of waste water from the pond into drains linked to a nearby stream but also alternatively, based on the volume, directed the waste water into a pond specifically created for receiving waste water that was subsequently used to irrigate plantations around the farm.
Tilapia (Oreochromis niloticus) and Catfish (Clarias gariepinus) were the main types of fish species commonly cultured. Seven fish farms were observed rearing only one type of fish (monoculture) while the remaining two cultured both catfish and tilapia (polyculture) (Table 3).
3.2. Characteristics of farms and bacterial loads of water and fish samples
The mean bacterial loads of water and fish samples collected from all nine farms, comprising four fish farms with concrete tanks and five farms with earthen ponds are displayed on Table 4. The fish holding-facilities used at the fish farms were either of the dug-out type, concrete type or both. It is worthy to note that farms that had both types holding facilities predominantly used their concrete tanks as hatcheries. Since only adult fishes were sampled at each farm in this study, hatchery ponds were excluded from the sample collection.
Table 4.
Total coliform and Gram-positive bacterial loads of fish and water samples from the various farms.
| Farm | Type of fish holding facility | Mean total coliform loads of underlisted samples |
Mean Gram-positive bacterial loads underlisted samples |
||||
|---|---|---|---|---|---|---|---|
| Water (x 104 cfu/mL) | Tilapia (x 104 cfu/g) | Catfish (x 104 cfu/g) | Water (x 104 cfu/mL) | Tilapia (x 104 cfu/g) | Catfish (x 104 cfu/g) | ||
| A | Concrete tank | 2.47 ± 0.01a | _ | 0.94 ± 0.02 | 0.15 ± 0.01d | _ | 0.03 ± 0.01 |
| B | Earthen pond | 154.00 ± 2.30a | _ | 2.08 ± 0.12 | 0.29 ± 0.04 | ||
| C | Concrete tank | 8.00 ± 2.03b | 0.32 ± 0.04 | _ | 0.34 ± 0.07 | 0.19 ± 0.01 | _ |
| D | Earthen pond | 25.30 ± 8.15b | 0.58 ± 0.03 | _ | 15.00 ± 5.29e | 0.15 ± 0.01 | _ |
| E | Earthen pond | 44.60 ± 8.51 | 43.70 ± 4.00g | 29.10 ± 6.10 | 11.80 ± 3.67 | 6.80 ± 0.61i | 14.30 ± 4.71j |
| F | Concrete tank | 137.00 ± 7.17a | _ | 31.30 ± 2.02 | 0.10 ± 0.01 | _ | 0.70 ± 0.25 |
| G | Concrete tank | 28.30 ± 2.03a | _ | 0.31 ± 0.01 | 0.95 ± 0.04d | _ | 0.18 ± 0.06 |
| H | Earthen pond | 87.50 ± 3.50a | _ | 0.18 ± 0.014 | 0.87 ± 0.07 | _ | 0.07 ± 0.12 |
| I | Earthen pond | 167.50 ± 13.50c | 2.20 ± 0.21 | 58.00 ± 4.36h | 2.36 ± 0.22f | 0.80 ± 0.09 | 0.64 ± 0.12 |
- type of fish not available at the farm.
Significant difference between mean total coliform load of water and catfish in farm A, B, F, G and H (p = 0.0001).
Significant difference between mean total coliform load of water and tilapia in farm C and D (p = 0.0001).
Significant difference between mean total coliform load of water, tilapia and catfish in farm I (p = 0.0001).
Significant difference between mean total Gram-positive bacterial load of water and catfish in farm A and G (p = 0.0001).
Significant difference between mean Gram-positive bacterial load of water and tilapia in farm D. (p = 0. 0485).
Significant difference between mean Gram-positive bacterial load of water, tilapia and catfish in farm I (p = 0.0002).
Significant difference between mean total coliform load of tilapia in farm E and their counterparts from other farms (p = 0.0001).
Significant difference between mean total coliform load of catfish in farm I and those from other farms (p = 0.0001).
Significant difference between mean Gram-positive bacterial load of tilapia in farm E and those from other farms (p = 0.0001).
Significant difference between the mean Gram-positive bacterial load of catfish in farm E and those from other farms (p = 0.0005).
The total coliform loads were significantly higher than Gram-positive bacterial loads for all sample type and across all nine fish farms (p = 0.0001). The total coliform and Gram-positive bacterial loads of water samples were also significantly higher than that of fish samples (p = 0.0001) except for farm E where tilapia samples recorded a total coliform load of 43.70 ± 4.00 × 104 CFU/mL which was similar to its corresponding water samples (44.60 ± 8.51) x 104 CFU/mL). Even though tilapia samples obtained from this farm recorded lower levels of Gram-positive bacteria than that of the water samples, the level of Gram-positive bacteria in catfish were significantly higher (p = 0.0005).
The total coliform and Gram-positive bacterial loads of the water samples ranged from (2.47 ± 0.01 to 167.50 ± 13.50 and 0.10 ± 0.01 to 15.00 ± 5.30) x 104 CFU/mL respectively. Farm I recorded the highest total coliform load for the water samples, followed by farm B, while Farm A recorded the least total coliform load (p = 0.0001). Among all fish samples, the highest total coliform load was again recorded in catfish obtained from farm I as (58.00 ± 4.36) x104 CFU/g while (43.70 ± 4.00) x 104 CFU/g was recorded for tilapia obtained from farm E (p = 0.0001). Farm C and H recorded the least values of (0.32 ± 0.04) x 104 CFU/g and 0.18 ± 0.01 × 104 CFU/g for tilapia and catfish respectively (Table 4).
Additionally, the levels of bacterial contamination of water samples obtained from concrete tanks were significantly lower than that of earthen ponds (p = 0.0303) except for farm F which recorded a significantly higher total bacterial load of (137.00 ± 7.17) x104 cfu/mL (p = 0.0001) even though the samples were obtained from a concrete tank. There was, however, no statistical difference between the total coliform loads of water samples from farms D and G even though the fish holding facilities at those farms were earthen and concrete ponds respectively (p = 0.2594) (Table 4).
3.3. Distribution of bacterial isolates in fish farms
A total of 145 bacterial isolates comprising 99 Gram-negative and 46 Gram-positive bacteria were recovered from all nine fish farms and identified as belonging to 22 different genera. Citrobacter freundii, was the most predominant Gram-negative bacterium with a total of 38 (26.20 %) followed by Klebsiella pneumoniae representing 22 (15.17 %) across all the farms. The least represented Gram-negative bacteria were Serratia marcescens, Escherichia coli, Edwardsiella tarda, Citrobacter diversus, Shigella sonnei.
Among Gram positive bacteria, Staphylococcus aureus was the most predominant with 25 isolates (17.24%) whiles Athrobacter sp., Staphylococcus saprophyticus, Staphylococcus capitis and Staphylococcus intermedius were the least represented with each recording only one isolate (Table 5).
Table 5.
Distribution of bacterial species according to the nine fish farms sampled.
| Type of bacteria | Bacterial species | Number of bacterial isolates recorded in fish farms |
Total (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | |||
| Gram-negative bacteria (n = 99) | Serratia marcescens | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (0.69) |
| Edwardsiella tarda | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 (0.69) | |
| Citrobacter freundii | 1 | 6 | 3 | 1 | 4 | 1 | 4 | 9 | 9 | 38 (26.20) | |
| Klebsiella oxytoca | 4 | 0 | 0 | 2 | 1 | 0 | 2 | 0 | 2 | 11 (7.58) | |
| Klebsiella pneumoniae | 3 | 2 | 0 | 1 | 2 | 4 | 2 | 4 | 4 | 22 (15.17) | |
| Salmonella sp. | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 3 | 0 | 5 (3.40) | |
| Escherichia coli | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (0.69) | |
| Proteus mirabilis | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 (1.37) | |
| Citrobacter diversus | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 (0.69) | |
| Salmonella paratyphi ‘A’ | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 2 (1.40) | |
| Shigella sonnei | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 (0.69) | |
| Enterobacter aerogenes | 0 | 0 | 1 | 3 | 1 | 2 | 2 | 2 | 0 | 11 (7.58) | |
| Salmonella enterica | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 3 (2.10) | |
| Gram-positive bacteria (n = 46) | Staphylococcus aureus | 3 | 4 | 2 | 2 | 1 | 1 | 2 | 6 | 4 | 25 (17.24) |
| Streptococcus sp. | 0 | 2 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 6 (4.10) | |
| Cellobiococcus sp. | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | 4 (2.80) | |
| Micrococcus sp. | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 1 | 5 (3.40) | |
| Athrobacter sp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 (0.69) | |
| Staphylococcus xylosus | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 2 (2.10) | |
| Staphylococcus saprophyticus | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 (0.69) | |
| Staphylococcus capitis | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 (0.69) | |
| Staphylococcus intermedius | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 (0.69) | |
| Total | 13 | 14 | 10 | 13 | 14 | 10 | 12 | 33 | 26 | 145 (100) | |
3.4. Antibiotic resistance profile of Gram-negative and Gram-positive bacterial isolates from the fish farms
All Gram-negative bacteria, with the exception of Edwardsiella tarda, showed resistance to at least one antibiotic. Apart from Salmonella enterica that showed resistance to only ampicillin, all other Gram-negative bacteria showed resistance to two or more antibiotics. All Gram-negative bacterial species except Edwardsiella tarda were highly resistant to ampicillin (100.00 %). Klebsiella pneumoniae, Citrobacter freundii, Klebsiella oxytoca and Enterobacter aerogenes recorded 100.00, 97.37, 90.91 and 90.91 % resistance respectively against cefuroxime. Most Gram-negative bacteria recorded relatively lower percentage resistance against Cotrimoxazole, chloramphenicol, ceftriaxone, cefotaxime and tetracycline. Generally, Gram-negative bacteria recorded low antibiotic resistance percentages against gentamicin and K. pneumoniae topped that category with 13.64 % resistance against gentamycin (Table 6a).
Table 6a.
Antibiotic resistance profile of Gram-negative bacteria isolated from fish farms.
| Bacterial isolate | Number of isolates (n = 99) | Number of bacterial isolates resistant to the under listed 8 antibiotics (percentages (%) in brackets) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| COT | GEN | CRX | CHL | CTR | CTX | AMP | TET | ||
| Citrobacter freundii | 38 | 7 (18.42) | 2 (5.26) | 37 (97.37) | 22 (57.89) | 17 (44.74) | 29 (76.32) | 38 (100.00) | 20 (52.63) |
| Klebsiella pneumoniae | 22 | 7 (31.82) | 3 (13.64) | 22 (100) | 12 (54.55) | 8 (36.36) | 12 (54.55) | 22 (100.00) | 12 (54.55) |
| Klebsiella oxytoca | 11 | 6 (54.55) | 1 (9.09) | 10 (90.91) | 7 (63.64) | 5 (45.45) | 5 (45.45) | 11 (100.00) | 8 (72.73) |
| Enterobacter aerogenes | 11 | 4 (36.36) | 0 (0.00) | 10 (90.91) | 4 (36.36) | 4 (36.36) | 5 (45.45) | 11 (100.00) | 9 (81.82) |
| Salmonella sp. | 5 | 3 (60.00) | 0 (0.00) | 1 (20.00) | 1 (20.00) | 1 (20.00) | 0 (0.00) | 4 (80.00) | 3 (60.00) |
| Salmonella enterica | 3 | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 3 (100.00) | 0 (0.00) |
| Proteus mirabilis | 2 | 0 (0.00) | 0 (0.00) | 2 (100.00) | 0 (0.00) | 1 (50.00) | 1 (50.00) | 2 (100) | 1 (50.00) |
| Salmonella paratyphi 'A’ | 2 | 0 (0.00) | 0 (0.00) | 2 (100.00) | 0 (0.00) | 1 (50.00) | 0 (0.00) | 2 (100.00) | 0 (0.00) |
| Edwardsiella tarda | 1 | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Shigella sonnei | 1 | 0 (0.00) | 0 (0.00) | 1 (100.00) | 1 (100) | 1 (100) | 1 (100.00) | 1 (100.00) | 1 (100.00) |
| Citrobacter diversus | 1 | 1 (100.00) | 0 (0.00) | 1 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (100.00) | 1 (100.00) |
| Serratia marcescens | 1 | 1 (100.00) | 0 (0.00) | 1 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (100.00) | 1 (100.00) |
| Escherichia coli | 1 | 0 (0.00) | 0 (0.00) | 1 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (100.00) | 1 (100.00) |
COT = Cotrimoxazole, GEN = Gentamicin, CRX = Cefuroxime, CHL = Chloramphenicol, CTR = Ceftriaxone, CTX = Cefotaxime, AMP = Ampicillin, TET = Tetracycline.
Similar to Gram-negative bacteria, all Gram-positive bacteria showed various percentage resistance to antibiotics. The percentage resistance of most Gram-positive isolates including S. aureus, Micrococcus sp. and Cellobiococcus sp to flucloxacillin, penicillin and ampicillin was 100.0 % each. Most Gram-positive bacteria recorded relatively lower percentage of resistance against Cotrimoxazole, cefuroxime, erythromycin, and tetracycline respectively recorded. Only 2 out of the 46 Gram-positive isolates (Streptococcus sp and Staphylococcus capitis) were resistant to gentamicin (Table 6b).
Table 6b.
Antibiotic resistance profile of Gram-positive bacteria isolated from fish farms.
| Bacterial isolate | Number of isolates (n = 46) | Number of bacterial isolates resistant to the under listed 8 antibiotics (percentages (%) in brackets) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| COT | GEN | CRX | PEN | FLX | ERY | AMP | TET | ||
| Staphylococcus aureus | 25 | 1 (4.00) | 0 (0.00) | 11 (44.00) | 25 (100.00) | 25 (100.00) | 15 (60.00) | 25 (100.00) | 6 (24.00) |
| Streptococcus sp. | 6 | 0 (0.00) | 1 (16.67) | 2 (33.33) | 6 (100.00) | 5 (83.33) | 2 (33.33) | 5 (83.33) | 1 (16.67) |
| Micrococcus sp. | 5 | 0 (0.00) | 0 (0.00) | 3 (60.00) | 5 (100.00) | 4 (80.00) | 3 (60.00) | 5 (100.00) | 0 (0.00) |
| Cellobiococcus sp. | 4 | 3 (75.00) | 0 (0.00) | 4 (100.00) | 4 (100.00) | 4 (100.00) | 4 (100.00) | 4 (100.00) | 2 (50.00) |
| Staphylococcus xylosus | 2 | 1 (50.00) | 0 (0.00) | 1 (50.00) | 2 (100.00) | 2 (100.00) | 2 (100.00) | 2 (100.00) | 1 (50.00) |
| Staphylococcus saprophyticus | 1 | 0 (0.00) | 0 (0.00) | 1 (100.00) | 1 (100.00) | 1 (100.00) | 1 (100.00) | 1 (100.00) | 0 (0.00) |
| Athrobacter sp. | 1 | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (100.00) | 1 (100.00) | 0 (0.00) | 1 (100.00) | 0 (0.00) |
| Staphylococcus capitis | 1 | 0 (0.00) | 1 (100.00) | 0 (0.00) | 1 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Staphylococcus intermedius | 1 | 0 (0.00) | 0 (0.00) | 1 (100.00) | 1 (100.00) | 1 (100.00) | 1 (100.00) | 1 (100.00) | 1 (100.00) |
COT = Co-trimoxazole, GEN = Gentamicin, CRX = Cefuroxime, PEN = Penicillin, FLX = Flucloxacillin, ERY = Erythromycin, AMP = Ampicillin, TET = Tetracycline.
3.5. Distribution of molecular determinants of antibiotic resistance
The distributions of antibiotic resistance genes (molecular determinants) in 99 Gram-negative and 46 Gram-positive bacteria isolated from all nine fish farms sampled are presented on Tables 7a and 7b respectively. Thirteen out of fourteen resistance genes were identified in Gram-negative and Gram-positive bacteria with various levels of detection. The tetA gene was not detected in any of the bacterial isolates while TEM was the most represented gene across all bacterial isolates with 70 out of 99 Gram-negative bacteria. Among Gram-negative bacteria, 33 out of 38, 20 out of 22 and 7 out of 11 Citrobacter freundii, K. pneumoniae and K oxytoca respectively were found to harbour the TEM gene. Aside the blaEBC gene which was detected in 11 out of 38 C. freundii isolates, all other antibiotic resistance genes were in 6 or less of each Gram-negative bacterial species (Table7a).
Table 7a.
Distribution of antibiotic resistance genes among Gram-negative bacteria.
| Bacterial isolate | Number of isolates | Number of isolates in which the under listed antibacterial genes were detected |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| blaTEM | blaTEM-1 | blaEBC | blaCTX-M | blamecA | Sul1 | Sul3 | Cat1 | cmIA | gyrA | qnrB | qnrS | tetA | tetB | ||
| Citrobacter freundii | 38 | 33 | 6 | 11 | 0 | 0 | 6 | 3 | 2 | 0 | 4 | 4 | 0 | 0 | 0 |
| Klebsiella pneumoniae | 22 | 20 | 4 | 5 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Klebsiella oxytoca | 11 | 7 | 2 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Enterobacter aerogenes | 11 | 0 | 0 | 1 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Salmonella sp. | 5 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Salmonella enterica | 3 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Proteus mirabilis | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Salmonella paratyphi ‘A’ | 2 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Escherichia coli | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Edwardsiella tarda | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Serratia marcescens | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Citrobacter diversus | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Shigella sonnei | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total number of isolates | 99 | 70 | 12 | 20 | 0 | 0 | 16 | 3 | 2 | 0 | 4 | 6 | 0 | 0 | 0 |
Also, 34 out of 46 Gram-positive bacteria harboured the TEM gene with the detection of the gene made in 22 out of 25, 4 out of 6, 2 out of 5 and 4 out of 4 S. aureus, Streptococcus sp, Micrococcus sp and Cellobiococcus sp isolates respectively. blaTEM-1 was the next commonly detected antibiotic resistance gene among Gram-positive bacteria followed by gyrA and Sul1 while blaCTX-M, Cat1, cmIA, qnrS and tetB were each detected in only one S. aureus isolate (Table7b).
Table 7b.
Distribution of antibiotic resistance genes among Gram-positive bacteria.
| Bacterial isolate | Number of isolates | Number of isolates in which the under listed antibacterial genes were detected |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| blaTEM | blaTEM-1 | blaEBC | blaCTX-M | blamecA | Sul1 | Sul3 | Cat1 | cmIA | gyrA | qnrB | qnrS | tetA | tetB | |||
| Staphylococcus aureus | 25 | 22 | 15 | 8 | 1 | 6 | 7 | 3 | 1 | 0 | 15 | 1 | 1 | 0 | 1 | |
| Streptococcus sp. | 6 | 4 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | |
| Micrococcus sp. | 5 | 2 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Cellobiococcus sp. | 4 | 4 | 2 | 1 | 0 | 1 | 2 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | |
| Staphylococcus xylosus | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Athrobacter sp. | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |
| Staphylococcus. intermedius | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| Staphylococcus saprophyticus | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Staphylococcus capitis | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Total number of isolates | 46 | 34 | 23 | 9 | 1 | 8 | 12 | 5 | 1 | 1 | 19 | 3 | 1 | 0 | 1 | |
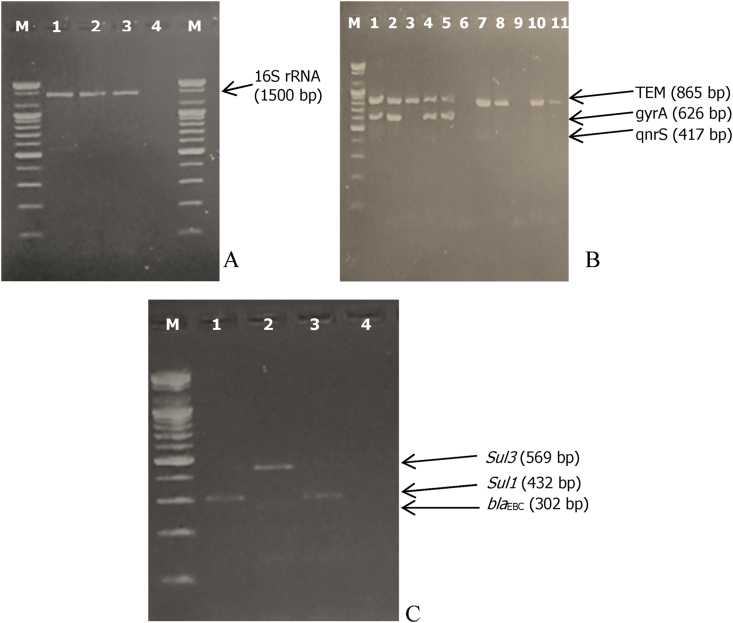
Some representative agarose gels showing the confirmation of DNA samples as of bacterial origin and the detection or otherwise of various antibiotic resistance genes have been provided in Figure 2.
Figure 2.
2.0 % ethidium bromide-stained agarose gel showing various PCR products and molecular weight markers (Lane M, 100 bp molecular ladder). (A: Confirmation of DNA samples as bacterial DNA using 16S rDNA primers. Lane 1, 2, and 3 - presence of 16S rRNA gene, Lane 4 - Molecular grade water (negative control); B: Detection of blaTEM, qnrS and gyrA genes. Lanes 1, 2, 3, 4, 5, 7, 8, 10 and 11, presence of blaTEM; Lane 6 and 9 - absence of blaTEM; lanes 1, 2, 4 and 5, presence of gyrA gene; lanes 3, 6, 7, 8, 9, 10, and 11 - absence of gyrA gene; lanes 1 to 11 - absence of qnrS; C: Detection of blaEBC, sul3 and sul1 genes. Lane 2 - presence of sul1; lane 1, 3, 4 - absence of sul1; lanes 1 and 3, presence of blaEBC and lanes 2 and 4 - absence of blaEBC; lane 1to 4 - absence of sul3.
4. Discussion
The widespread use of antibiotics in fish farming for prophylactic or therapeutic purposes, has globally been reported to contribute significantly to the emergence of antibiotic resistance (Watts et al., 2017). In this study, however, the information obtained from farm managers through the interviews conducted suggested that none of them directly used antibiotics. There are a number of welfare factors that might affect the health of the fish. It was commonly recorded across all nine farms that aside a few instances of stress-related death which they attributed to overcrowding or poor water quality, no major disease outbreak occurred since the establishment of the farms and throughout the entire study period. Consequently, there were no need for the use of antibiotics. This might have been made possible through the use of good quality healthy fingerlings used for stocking the ponds at various fish farms. The patronage of commercially formulated feed by most farm managers is also advantageous since they are of good quality and farmers can directly monitor the feeding behaviour of their fish stock. Furthermore, provision of proper fish nutrition promotes growth and also enhances the overall health status of fish stock (Craig et al., 2017). High stocking density and poor water quality are important factors that cause stress to fish and subsequently weaken their immune system (Ojonugwa and Solomon, 2017). In the present study, however, stocking density and parameters that are used to assess the quality of pond water, such as pH, temperature and dissolved oxygen were not investigated. The recommended for tilapia and catfish are 3–8 and 2–10 fingerlings/m2 respectively (Ragasa et al., 2022; Micha, 1975) and periodic checks on water quality at the farms must routinely be conducted so as to limit stress to fish under culture.
The different genera of bacterial pathogens in the gastrointestinal tract of fish represent the diversity and abundance of microorganisms of pond water (Banu et al., 2001; Cahill, 1990). The lower levels of bacterial loads in fish samples compared to water samples could be as a result of the action of beneficial microbes found on the mucosal surface of the gastrointestinal tract of the fish. These beneficial bacteria colonize the gastrointestinal tract and therefore prevent the attachment and establishment of pathogens to the mucosal surface through the secretion of extracellular enzymes that kill these pathogens (Banerjee and Ray, 2016). The bacterial loads of pond water samples recorded in this study exceeded the acceptable level of ≤100 E. coli and <10 coliforms per mL for wastewater recommended for use in fish farming (World Health Organization, 1989). All fish sampled in this study however had bacterial loads lower than the acceptable limit of 5 × 105 CFU/g at 37°C for fresh fish suggesting that they were wholesome provided, they are properly washed and thoroughly cooked (ICMSF, 1986). Microbial contamination of various fish farms could be linked to a number of contributing factors, including the source of water used for culturing the fish, on-site poor sanitary practices and the type of fish-holding facility. The handling of such contaminated fishery products by farmers and the consumption of improperly cooked contaminated fish could be dangerous as it poses a public health threat (Alikunhi et al., 2017). It is therefore very essential that pragmatic measures such as proper personal hygiene and on-site good sanitary practices are ensured by fish farmers in order to reduce contamination.
Though in this study bacterial loads of samples obtained from earthen ponds were generally higher to that of concrete ponds, there were a few exceptions in farm F, D and G. The sediment at the bottom of earthen ponds ordinarily harbours a wide range of microorganisms (Segovia et al., 2015) unlike concrete tanks that are more or less closed system and not easily accessed by bacteria from the surrounding soils and runoff water. Also, concrete ponds are periodically fully discharged and refilled with fresh water. It was therefore surprising to note the significantly high bacterial load recorded for the concrete tank holding facility sampled at farm F. The difference between the total coliform loads of water samples from farms D and G however was not significant (p = 0.2594) even though the fish holding facilities at those farms were earthen and concrete ponds respectively. The discrepancies observed in concrete ponds, on one hand could be linked to the non-adherence to good on-site sanitary practices. On the other hand, the relatively lower level of bacterial contamination observed in the earthen pond at farm D could suggest that farm managers have adopted healthy waste disposal strategies and other good sanitary practices. Examples of such interventions include the periodic treatment of pond water using ozone and ultraviolet light or the use of lime preparation prior to stocking the ponds with fingerlings (Boyd and Massaut, 1999).
The presence of Gram-negative and Gram-positive bacteria in fish and water sampled in this study, including coliforms indicates poor sanitary conditions at the farms and the potential risk to the health of fish, fish farmers and consumers. This is because these are potentially harmful pathogenic bacteria (Pepper and Gerba, 2015). Among coliforms, Citrobacter freundii was most commonly isolated bacterial species across all farms, followed by Klebsiella pneumoniae while the least represented were Escherichia coli, Citrobacter diversus, Shigella sonnei which was a single bacterial isolate. A single bacterial isolate each of two additional Gram-negative bacteria (Serratia marcescens and Edwardsiella tarda) was also recorded. Among Gram-positive bacteria, Staphylococcus aureus was the most represented across all farms, followed by Streptococcus sp. and Micrococcus sp. Staphylococcus aureus identified in this study as predominant among Gram positive bacteria, is one of the major bacterial agents causing food borne diseases in humans worldwide (Leuschner et al., 2010; Loir et al., 2003). The present study is consistent to a similar study conducted in the Greater Ashanti region of Ghana which reported S. aureus as the most isolated bacterial species in fish farms sampled (Agoba et al., 2017).
Although farm managers across all farms never admitted to using antibiotics, various levels of antibiotic resistance were recorded for all bacteria isolated from their farm. This suggests a possible antecedent exposure of these bacteria to antibiotics released into the environment through their use in human and veterinary medicine and their presence in pharmaceutical wastewaters (Manyahi et al., 2016).
Some studies have suggested that antibiotics are often used as an additive in animal feed (Van et al., 2020; Agoba et al., 2017a, Agoba et al., 2017b). However, as observed in this study, most fish farmers presumed that commercially formulated feeds did not contain antibiotics, specifically because detailed information on the exact constituents of the fish feed was usually not fully disclosed on the feed label. Even though it was not possible in this study to access the fish feed at various fish farms so as to ascertain the presence and the levels of antibiotic residues in the fish feed, a study conducted at some fish farms in the Ashanti region of Ghana revealed the presence of multidrug-resistant bacteria which was attributed to the application of antibiotics such as tetracycline and chloramphenicol to the fish feed (Agoba et al., 2017). Such findings were made despite the ban placed on the use of antibiotics in animal feed for prophylactic purposes upon recommendations by the World Health Organization (WHO) on measures to adopt to curb antimicrobial resistance (U.S. Department of Health and Human Services, 2012). According to WHO guidelines, antibiotics should only be used in treatment of infections in animals (World Health organization, 2015).
In Ghana, antibiotics are easily obtained over the counter; hence abuse and improper disposal of these antibiotics are common (Donkor et al., 2012). Some studies have reported the presence in waste water and landfill sites of varying concentrations of antibiotic residues (Azanu et al., 2018) which has been linked to the possible influx of antibiotics into aquatic environments. This might explain the multiple resistance to beta-lactam antibiotics such as penicillin and penicillin-derived antibiotics (ampicillin and flucloxacillin) on one hand and cephalosporins such as cefuroxime, cefotaxime and ceftriaxone on the other hand as exhibited by Gram-negative and Gram-positive bacteria in this study. This suggests that these antibiotic-resistant bacteria may have emerged in the environment as these drugs are commonly used in the country. The development of acquired antibiotic resistance can result from mutations in chromosomal genes or be caused by the gain of external genetic determinants of resistance from antibiotic-resistant organisms in the surrounding environment (Munita and Arias, 2016). Though not investigated in this study, the presence of various levels of antibiotic residues in water, food, landfill sites and other environmental samples in Ghana has been reported as evidenced in various studies (Addo et al., 2011; Darko et al., 2017; Azanu et al., 2018; Borquaye et al., 2019).
In this study, varying percentages of detection of antibiotic-resistant genes were recorded for all isolates. Among genes that confer resistance to beta-lactams including blaTEM, blaTEM-1, blamecA, blaCTX-M and blaEBC, blaTEM was the most commonly detected gene in Gram-negative bacteria and Gram-positive bacteria isolated from all fish farms sampled. This confirms the phenotypic expression of resistance of isolates to beta-lactam antibiotics. Consequently, most Gram-positive and Gram-negative bacteria in this study recorded 100% resistance to ampicillin, penicillin and flucloxacillin while relatively lower percentage resistance were recorded for the second-generation cephalosporin cefuroxime followed by the third-generation cephalosporins cefotaxime and ceftriaxone. blaCTX-M and blamecA genes were not detected in any of the Gram-negative bacteria while they were detected respectively in a single S. aureus isolate and 8 Gram-positive bacteria comprising 6 S. aureus and one isolate each of Micrococcus sp. and Cellobiococcus sp. The blaCTX-M result observed runs contrary to other studies where high detection of blaCTX-M gene in enterobacteria were reported (Richter et al., 2019; Hackman, 2015). The blaEBC gene, however was detected in 20 and 9 of Gram-negative and Gram-positive bacteria respectively.
An increasing number of beta-lactam variants have been discovered that differs in sequences of amino acid and their catalytic activity against β-lactam antibiotics (Bush and Jacoby, 2010). Generally, Gram-negative bacteria produce β-lactamases enzymes that render antibiotics useless. β-lactamases confer resistance to β-lactam antibiotics by splitting the four-membered ring of such antibiotics with the release of an inactive product (Toth et al., 2016). Gram-positive bacteria on the other hand also use a target modification mechanism to confer resistance to beta-lactam antibiotics. This mechanism ensures structural changes to specific structures within the Gram-positive bacteria leading to β-lactam antibiotics becoming inactive against these bacteria (Ogawara, 2015). This could account for detection of diverse variant genes of both coliforms and Gram-positive bacteria to most beta-lactams drugs. A study conducted in Egypt showed that Gram-negative bacteria isolated from some fish farms showed phenotypic resistance to beta-lactams with the corresponding detection of blaTEM-1, blaTEM-104, blaCTX-M-15, and blaSHV-89 resistant genes (Ishida et al., 2010).
In the current study, genes that confer resistance to tetracycline (tetA and tetB) recorded low levels of detection in Gram-negative and Gram-positive bacteria respectively. Even though, 45.5% phenotypic resistance to tetracycline was expressed among Gram-negative bacteria, the inability to detect these specific resistant genes could suggest resistance may be due to other tetracycline-resistant genes such as tetC, tetO or tetW. Similarly, even though, moderate percentages of phenotypic resistance to chloramphenicol were observed among Gram-negative bacteria, the chloramphenicol-resistant genes, Cat1 and cmIA were detected in two Citrobacter freundii isolates and none of the Gram-negative bacteria respectively. This may also suggest that the resistance could be attributed to other chloramphenicol-resistant genes and not necessarily Cat1 and cmIA.
Cotrimoxazole is a blend of sulfamethoxazole and trimethoprim antibiotics. In the current study, the sulfamethoxazole-resistant genes Sul1and Sul3 were also detected. Sul1 gene was respectively detected in 16 Gram-negative and 12 Gram-positive bacteria. Sul3, however, was detected in 3 Gram-negative and 5 Gram-positive bacteria respectively. In a study conducted in Tanzania, as many 98.4% of Gram-positive bacteria were found to harbour as Sul (1, 2 and 3) (Manyahi et al., 2016) which surpassed by far the percentages recorded in this study. Another study conducted in Chile reported on a high detection rate of Sul 1 gene with no detection of the Sul 3 gene (Domínguez et al., 2019). Considering the relatively low percentage of detection of Sul1and Sul3 genes as compared to the phenotypic percentage resistance to cotrimoxazole in this study, it could be postulated that resistance to co-trimoxazole could be due to other variants of the sulfamethoxazole-resistant genes or variants of trimethoprim-resistance genes.
Even though antibiotics belonging to the class of quinolones were not used in the susceptibility testing, qnrS, qnrB and gyrA genes were detected. Quinolones such as ciprofloxacin, oxonilic acid and nalidixic acid are not commonly used in fish farming, especially in Ghana (Apenteng et al., 2017).
Multiple antibiotic-resistance among isolates and detection of genes that confer resistance suggests widespread of resistance in aquatic environments indicating that fish farms may be a potential source for the dissemination of antimicrobial resistance genes.
The growing menace of antimicrobial resistance is unfortunately further exacerbated by the misuse and overuse of disinfectants such as alcohol-based hand sanitizers, especially at a time when the world is faced with the global health crisis of the dreadful COVID-19 pandemic. Serious concerns have been raised about the potential contribution of the inappropriate use of sanitizers as well as the use of sub-standard products sold on the market especially in developing countries, in accelerating antimicrobial resistance (Assefa and Melaku, 2021). It is also worth noting that in the heat of the pandemic, most patients with COVID-19 inappropriately received antibiotics despite the fact that bacterial co-infections were rare and this possibly led to health complications, including antibacterial resistance (Calderón-Parra et al., 2021).
Antibiotic resistance is a world-wide public health threat requiring that efforts are consolidated for sustainable mitigation. First hand measures to prevent the further emergence and spread of this menace are therefore crucially needed. Most developed nations, including Norway, Italy and New Zealand have adopted guidelines for rational use of antibiotics (Hillerton et al., 2017). Though some progress has been made in developing countries such as Ghana, more efforts are needed for effective policy implementation. In 2017, Ghana instituted a five-year National Action Plan (NAP) on antimicrobial resistance with its objectives being focusing on the enhanced surveillance of antibiotic-resistant infections and maximization of antimicrobial treatment in animal and human health as well as crop production (Ministry of Health, Ministry of Food and Agriculture, Ministry of Environment, Science, Technology and Innovation, 2017). As recommended by WHO, infection prevention and control measures must also be strengthened (World Health organization, 2015). Clearly, the results of this study suggest that public education through various platforms including TV programs and social media providing freely accessible information on the impact of antibiotic resistance must be intensified.
5. Conclusion and recommendations
None of the farm managers admitted using antibiotics and no record of a major disease outbreak was recorded at the farms throughout the study period. The contamination of fish and pond water with a wide variety of Gram-positive and Gram-negative bacteria including, coliforms suggest poor sanitary conditions at the farms. The majority of bacterial isolates recorded high percentage resistance to beta-lactam antibiotics and this was confirmed by the high percentage detection of blaTEM genes. An integrated approach involving all stakeholders should be put in place to properly regulate the use of antibiotics that will discourage their abuse and ultimately minimize the emergence of antibiotic resistance. The implementation of guidelines provided in the Nation Action Plan on antimicrobial resistance involving all stakeholders including the Food and Drugs Authority and the Fisheries Commission to ensure prudent use of antibiotics must be strictly adhered to. Periodic training of fish farmers and consumers on food safety must also be carried out by the stakeholders to reduce microbial contamination of fish products from fish culture industry in the country. Further research on the spread of antibiotic resistance in other areas of the country is needed to identify the scope of this menace in aquaculture settings. Also, there is the need to further investigate the presence of antibiotic residues at fish farms including the fish feed to probe into the common no-antibiotic usage assertion of farmers as recorded in this study.
Declarations
Author contribution statement
Rosemary Agbeko: Performed the experiments, Analyzed and interpreted the data; Wrote the paper.
Denis W. Aheto: Contributed reagents, materials, analysis tools or data.
Daniel K. A. Asante; Noble K. Asare: Analyzed and interpreted the data.
Alex A. Boateng: Performed the experiments.
Cynthia A. Adinortey: Conceived and designed the experiments; Wrote the paper.
Funding statement
Dr Cynthia A. Adinortey was supported by USAID/UCC Fisheries and Coastal Management Capacity Building Support Project [PIL No.: 641-A18-FY14-IL#007].
Data availability statement
Data associated with this study has been deposited at University of Cape Coast Institutional repository available at: https://ir.ucc.edu.gh/xmlui/bitstream/handle/123456789/6480/AGBEKO%2C%202020. pdf?sequence=1&isAllowed=y.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
Authors hereby express their appreciation to fish farm managers for their co-operation during this research.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Addo K.K., Mensah G.I., Aning K.G., Nartey N., Nipah G.K., Bonsu C., Akyeh M.L., Smits H.L. Microbiological quality and antibiotic residues in informally marketed raw cow milk within the coastal savannah zone of Ghana. Trop. Med. Int. Health. 2011;16(2):227–232. doi: 10.1111/j.1365-3156.2010.02666.x. [DOI] [PubMed] [Google Scholar]
- Adinortey C.A., Aheto D.W., Boateng A.A., Agbeko R. 2020. Multiple Antibiotic Resistance-Coliform Bacteria in Some Selected Fish Farms of the Central Region of Ghana; p. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agoba E.E. Kwame Nkrumah Universityof Science and Technology; 2016. THE USE OF ANTIBIOTICS AND RESISTANCE PATTERNS OF BACTERIAL ISOLATES FROM SELECTED FISH FARMS IN THE ASHANTI REGION OF GHANA A. [Google Scholar]
- Agoba E.E., Adu F., Agyare C., Boamah V.E. Antibiotic use and practices in selected fish farms in the Ashanti region of Ghana. J. Infect. Dis. Treat. 2017;3(2) [Google Scholar]
- Agoba E.E., Adu F., Agyare C., Boamah V.E., Boakye Y.D. Antibiotic resistance patterns of bacterial isolates from hatcheries and selected fish farms in the Ashanti region of Ghana. J. Microbiol. Antimicrob. 2017;9(4):35–46. [Google Scholar]
- Alikunhi N.M., Batang Z.B., Aljahdali H.A., Aziz M.A.M., Al-suwailem A.M. Culture-dependent bacteria in commercial fishes : qualitative assessment and molecular identification using 16S rRNA gene sequencing. Saudi J. Biol. Sci. 2017;24(6):1105–1116. doi: 10.1016/j.sjbs.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amenyogbe E., Chen G., Wang Z., Lin M., Lu X., Atujona D., Abarike E.D. Vol. 9. 2018. A Review of Ghana’ S Aquaculture Industry. (8) [Google Scholar]
- Apenteng J.A., Osei-Asare C., Eshun O.E., Amihere I., Hafiz M.Y. Antibiotic sensitivity patterns of microbial isolates from fish ponds: a study in the Greater Accra Region of Ghana. Afr. J. Pharm. Pharmacol. 2017;11(28):314–320. [Google Scholar]
- Assefa A., Abunna F. Veterinary Medicine International; 2018. Maintenance of Fish Health in Aquaculture : Review of Epidemiological Approaches for Prevention and Control of Infectious Disease of Fish; p. 10. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assefa D., Melaku T. Commercial hand sanitizers use amid COVID-19 pandemic : the concerns of antimicrobial resistance. Infect. Drug Resist. 2021;14:2183–2185. doi: 10.2147/IDR.S317767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azanu D., Styrishave B., Darko G., Juhl J., Clement R. Science of the Total Environment Occurrence and risk assessment of antibiotics in water and lettuce in Ghana. Sci. Total Environ. 2018;622–623:293–305. doi: 10.1016/j.scitotenv.2017.11.287. [DOI] [PubMed] [Google Scholar]
- Banerjee G., Ray A.K. 2016. Bacterial Symbiosis in the Fish Gut and its Role in Health and Metabolism. Symbiosis, Nayak; p. 2010. [Google Scholar]
- Banu A.N.H., Islam M.A., Chowdhury M.B.R. Bacterial load in pond water and different organs of a Indian major carp Cirrhinus mrigala Ham. Bangl. J. Fish. Res. 2001;5(1):53–58. [Google Scholar]
- Borquaye L.S., Ekuadzi E., Darko G., Ahor H.S., Nsiah S.T., Lartey J.A., Mutala A.H., Boamah V.E., Woode E. Occurrence of antibiotics and antibiotic-resistant bacteria in landfill sites in Kumasi, Ghana. J. Chem. 2019 2019. [Google Scholar]
- Boyd C.E., Massaut L. Vol. 20. 1999. Risks Associated with the Use of Chemicals in Pond Aquaculture; pp. 113–132. [Google Scholar]
- Bush K., Jacoby G.A. Vol. 54. 2010. MINIREVIEW Updated Functional Classification of  -Lactamases; pp. 969–976. (3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill M.M. 1990. Bacterial Flora of Fishes : A Review; pp. 21–41. [DOI] [PubMed] [Google Scholar]
- Calderón-Parra J., Muiño-Miguez A., Bendala-Estrada A.D., Ramos-Martínez A., Muñez-Rubio E., Carracedo E.F., Montes J.T., Rubio-Rivas M., Arnalich-Fernandez F., Pérez J.L.B., Bruñén J.M.G., del Corral Beamonte E., Fontan P.M.P., del Mar Carmona M., Martínez R.F.M., García A.G., Mosteiro C.S., de Almeida C.T., Moraleja J.G., et al. Inappropriate antibiotic use in the COVID-19 era: factors associated with inappropriate prescribing and secondary complications. Analysis of the registry SEMI-COVID. PLoS One. 2021;16(5 May):1–15. doi: 10.1371/journal.pone.0251340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheesbrough M. second ed. Vol. 2. Cambridge University Press; 2006. (District Laboratory Practice in Tropical Countries Part). [Google Scholar]
- CLSI . Journal of Services Marketing (28th Editi, Vol. 38, Issue 3). Wayne, PA: Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- Craig S., College V., Medicine V., Tech V. 2017. Understanding Fish Nutrition , Feeds , and Feeding. [Google Scholar]
- Dadgostar P. Antimicrobial resistance : implications and costs. Infect. Drug Resist. 2019;2019(12):3903–3910. doi: 10.2147/IDR.S234610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darko G., Borquaye L.S., Acheampong A., Oppong K. Veterinary antibiotics in dairy products from Kumasi, Ghana. Cogent Chem. 2017;3(1) [Google Scholar]
- Doku B.N.A., Chen S., Alhassan E.H., Abdullateef Y., Rahman M.M. Vol. 3. 2018. Fisheries Resources of Ghana: Present Status and Future Direction; pp. 35–41. (4) [Google Scholar]
- Domínguez M., Miranda C.D., Fuentes O., Fuente M. De, Llewellyn M.S., Miranda C.D. Vol. 10. 2019. Occurrence of Transferable Integrons and Sul and Dfr Genes Among Sulfonamide-And/or Trimethoprim-Resistant Bacteria Isolated from Chilean Salmonid Farms; pp. 1–14. (April) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkor E.S., Anim-Baidoo I., Fei E., Amponsah C., Olu-Taiwo M., Nana-Adjei D., Owusu E., Forson A.O. Occurrence of antibiotic residues and antibiotic-resistant bacteria in nile Tilapia sold in some markets in Accra, Ghana: public health implication. J. Food Res. 2018;7(6):129. [Google Scholar]
- Donkor E.S., Tetteh-quarcoo P.B., Nartey P., Agyeman I.O. 2012. Self-Medication Practices with Antibiotics Among Tertiary Level Students in Accra , Ghana : A Cross-Sectional Study; pp. 3519–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegold C., Gordon A., Mills D., Curtis L., Pulis Alan. 2010. Western Region Fisheries Sector Review, Final Report December 2010 (Issue December) [Google Scholar]
- Golub S., Varma A. United Nations Conference on Trade and Development; 2014. Fishing Exports and Economic Development of Least Developed Countries : Bangladesh , Cambodia , Comoros , Sierra Leone and Uganda.https://www.swarthmore.edu/sites/default/files/assets/documents/user_profiles/sgolub1/UNCTAD.fisheries.final.pdf February, 75. [Google Scholar]
- Hackman H.K. University of Ghana; 2015. PHENOTYPIC AND MOLECULAR CHARACTERIZATION OF EXTENDED- SPECTRUM ΒETA -LACTAMASES IN KLEBSIELLA PNEUMONIAE AND ESCHERICHIA COLI ISOLATES IN ACCRA , GHANA By. [Google Scholar]
- Hillerton J.E., Irvine C.R., Bryan M.A., Scott D., Merchant S.C. Use of antimicrobials for animals in New Zealand, and in comparison with other countries. N. Z. Vet. J. 2017;65(2):71–77. doi: 10.1080/00480169.2016.1171736. [DOI] [PubMed] [Google Scholar]
- Huys G. 2003. Standard Operating Procedure: Sampling and Sample Processing Procedures for the Isolation of Aquaculture-Associated Bacteria; pp. 1–15. [Google Scholar]
- ICMSF . The International Commission on Microbiological Specifications for Foods of the International Union of Biological Societies. 1986. Micro-organisms in foods 2: sampling for microbiological analysis; principles and specific applications; p. 131. [Google Scholar]
- Ishida Y., Ahmed A.M., Mahfouz N.B., Kimura T., El-khodery S.A. 2010. Molecular Analysis of Antimicrobial Resistance in Gram-Negative Bacteria Isolated from Fish Farms in Egypt. [DOI] [PubMed] [Google Scholar]
- Lane D. In: Nucleic Acid Techniques in Bacterial Systematic. Stackebrandt E., Goodfellow M., editors. John Wiley & Sons; 1991. 16S/23S rRNA sequencing; pp. 115–175. [Google Scholar]
- Leuschner R.G.K., Robinson T.P., Hugas M., Cocconcelli P.S., Richard-Forget F., Klein G., Licht T.R., Nguyen-The C., Querol A., Richardson M., Suarez J.E., Thrane U., Vlak J.M., von Wright A. Qualified presumption of safety (QPS): a generic risk assessment approach for biological agents notified to the European Food Safety Authority (EFSA) Trends Food Sci. Technol. 2010;21(9):425–435. [Google Scholar]
- Loir Y. Le, Baron F., Gautier M. Vol. 2. 2003. Staphylococcus aureus and Food Poisoning; pp. 63–76. (1) [PubMed] [Google Scholar]
- Manyahi J., Ndugulile F., Moyo S.J. 2016. Molecular Characterization of Cotrimoxazole Resistance Genes and Their Associated Integrons; pp. 1–7. [DOI] [PubMed] [Google Scholar]
- McEwen S.A., Collignon P.J. Antimicrobial resistance: a one health perspective. Microbiol. Spectr. 2017;6(2):1–26. doi: 10.1128/microbiolspec.arba-0009-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micha J.C. BULLETIN FRANÇAIS DE PISCICULTURE, QUARANTE-S; 1975. SYNTHESE DES ESSAIS DE REPRODUCTION, D’ALEVINAGE ET DE PRODUCTION CHEZ UN SILURE AFRICAIN : Clarias Lazera Val; pp. 77–87. (256) [Google Scholar]
- Ministry of Health, Ministry of Food and Agriculture, Ministry of Environment, Science, Technology and Innovation . Ministry of Health Ministry of Food and Agriculture Ministry of Environment, Science, Technology and Innovation Minist. 1st. 2017. National action plan (NAP) for antimicrobial use and resistance in Ghana Ghana national action plan for antimicrobial use and resistance. M. of F. and A. D. [Google Scholar]
- Munita J.M., Arias C.A. Mechanisms of antibiotic resistance. Microbiol. Spectr. 2016;4(2):1–37. doi: 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawara H. 2015. Penicillin-binding Proteins in Actinobacteria; pp. 223–245. October 2014. [DOI] [PubMed] [Google Scholar]
- Ojonugwa E.B., Solomon R.J. Effects of over stocking on the growth rate of Clarias gariepinus. J. Animal Sci. Vet. Med. 2017;2(August):84–95. [Google Scholar]
- Okocha R.C., Olatoye I.O., Adedeji O.B. 2018. Food Safety Impacts of Antimicrobial Use and Their Residues in Aquaculture; pp. 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper I.L., Gerba C.P. Environmental Biology. Elsevier; 2015. Cultural methods. [Google Scholar]
- Ragasa C., Koranteng S., Asmah R., Mensah E.T., Amewu S., Oyih M. Accelerating pond aquaculture development and resilience beyond COVID : ensuring food and jobs in Ghana. Aquaculture. 2022;547(October 2020) [Google Scholar]
- Richter L., Plessis E. M. Du, Duvenage S., Korsten L. Occurrence, identification, and antimicrobial resistance profiles of extended-spectrum and AmpC b -Lactamase-Producing enterobacteriaceae from fresh vegetables retailed in gauteng province, South Africa. FOODBORNE PATHOGENS AND DISEASE. 2019;XX(Xx):1–7. doi: 10.1089/fpd.2018.2558. [DOI] [PubMed] [Google Scholar]
- Segovia B.T., Pereira D.G., Bini L.M. 2015. The Role of Microorganisms in a Planktonic Food Web of a Floodplain Lake; pp. 225–233. [DOI] [PubMed] [Google Scholar]
- Shrivastava S.R., Shrivastava P.S., Ramasamy J. 2018. L Etter to Editor World Health Organization Releases Global Priority List of Antibiotic - Resistant Bacteria to Guide Research , Discovery , and Development of New Antibiotics. 2017–2018. [Google Scholar]
- Smith R.M., Lautenbach E., Omulo S., Araos R., Call D.R., Kumar G.C.P., Chowdhury F., McDonald C.L., Park B.J. Human colonization with multidrug-resistant organisms: getting to the bottom of antibiotic resistance. Open Forum Infect. Dis. 2021;8(11):1–3. doi: 10.1093/ofid/ofab531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tall A., Failler P. 2012. Fishery and Aquaculture Industry in Ghana. Series Report N°1 of the Review of the Fishery and Aquaculture Industry in the 22 ATLAFCO Member States; p. 52. October 2012. [Google Scholar]
- Tenover F.C. In: Antibiotic Susceptibility Testing. fourth ed. Schaechter M., editor. 2014. Encyclopedia of microbiology. [Google Scholar]
- Toth M., Antunes N.T., Stewart N.K., Frase H., Smith C., Vakulenko S., Lightsource S.R., Park M. Vol. 12. 2016. HHS Public Access; pp. 9–14. (1) [Google Scholar]
- U.S. Department of Health and Human Services . 2012. Guidance for Industry: the Judicious Use of Medically Important Antimicrobial Drugs in Food-Producing Animals. U.S. Department of Health and Human Services F. and D. A. C. for V. M. (Issue #209) [Google Scholar]
- Van T.T.H., Yidana Z., Smooker P.M., Coloe P.J. Journal of global antimicrobial resistance antibiotic use in food animals worldwide , with a focus on Africa : pluses and minuses. Integr. Med. Res. 2020;20:170–177. doi: 10.1016/j.jgar.2019.07.031. [DOI] [PubMed] [Google Scholar]
- Watts J.E.M., Schreier H.J., Lanska L., Hale M.S. Marine Drugs. Vol. 15. MDPI AG; 2017. The rising tide of antimicrobial resistance in aquaculture: sources, sinks and solutions; pp. 1–16. Issue 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2018. Critically Important Antimicrobials for Human Medicine.http://www.who.int/foodborne_disease/resistance/cia/en/#.UiMEZ7zmSDA.mendeley [Google Scholar]
- World Health organization Global action plan on antimicrobial resistance. Microbe. 2015;10(9):354–355. doi: 10.7196/samj.9644. [DOI] [PubMed] [Google Scholar]
- World Health Organization . 1989. Health Guidelines for the Use of Wastewater in Agriculture and Aquaculture. [Google Scholar]
- Critically Important Antimicrobials for Human Medicine: Ranking of Antimicrobial Agents for Risk Management of Antimicrobial Resistance Due to Non-human Use (5th rev.) World Health Organization; 2017. World health organization & WHO advisory Group on integrated surveillance of antimicrobial resistance (AGISAR)https://apps.who.int/iris/handle/10665/255027 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at University of Cape Coast Institutional repository available at: https://ir.ucc.edu.gh/xmlui/bitstream/handle/123456789/6480/AGBEKO%2C%202020. pdf?sequence=1&isAllowed=y.