Abstract
As the bacterial multidrug resistance crisis continues, membrane-active antimicrobial peptides are being explored as an alternate treatment to conventional antibiotics. In contrast to antimicrobial peptides, which function by a nonspecific membrane disruption mechanism, here we describe a series of transmembrane (TM) peptides that are designed to act as drug efflux inhibitors by aligning with and out-competing a conserved TM4-TM4 homodimerization motif within bacterial small multidrug resistance proteins. The peptides contain two terminal tags: a C-terminal lysine tag to direct the peptides toward the negatively charged bacterial membrane, and an uncharged N-terminal sarcosine (N-methyl-glycine) tag to promote membrane insertion. While effective at inhibiting efflux activity, ostensibly through their designed mechanism of action, the impact of the peptides on the bacterial inner membrane remains undetermined. To evaluate the extant peptide-membrane interactions, we performed a series of biophysical measurements. Circular dichroism spectroscopy and Trp fluorescence showed that the peptides insert into the membrane generally in helical form. Interestingly, differential scanning calorimetry of the peptides added to bacterial-like membranes (POPE:POPG 3:1) revealed the peptides’ ability to demix the POPE and POPG lipids, creating two pools, one of which is likely a peptide-POPG conglomerate, and the other a POPE-rich component where the native POPG content has been depleted. However, dye leakage assays confirmed that these events occur without causing significant membrane disruption both in vitro and in vivo, indicating that the peptides can target the small multidrug resistance TM4-TM4 motif without nonspecific membrane disruption. In related studies, DiOC2(3) fluorescence indicated moderate peptide-mediated reduction of the proton motive force for all peptides, including control peptides that did not display inhibitory activity. The overall findings suggest that peptides designed with suitable tags, sequence hydrophobicity, and charge distribution can be directed more generally to impact proteins whose function involves membrane-embedded protein-protein interactions.
Significance
Transmembrane (TM) peptides are an expanding subclass of membrane-active peptides that target membrane-buried protein-protein interactions (PPIs). Our unique tagging method mimics the positive charge of antimicrobial peptides (AMPs) that directs them toward the bacterial membrane, where they then stably insert via an uncharged N-terminal tag without incurring disruptive effects. This membrane stability apparently remains despite peptide-mediated demixing of the negatively charged lipids from the zwitterionic lipids. The overall results indicate that the designed TM peptides can be successfully targeted toward and inserted into the bacterial membrane to outcompete oligomerization of dimeric bacterial small multidrug resistance (SMR) proteins and thereby inhibit their drug efflux activity. The results validate this approach as a platform to discover new drug targets by making membrane-buried PPIs accessible.
Introduction
Research on membrane-active peptides has seen exponential growth in the last 20 years, as the crisis for development of new antibiotics against multidrug-resistant bacteria continues (1, 2, 3). This field largely consists of antimicrobial peptides (AMPs), which cause bacterial death through disturbing membrane integrity and cell-penetrating peptides that pass through the membrane to a cytoplasmic target; however, less research has been conducted on transmembrane (TM) peptides that stably insert into the membrane (1). TM peptides are designed to mimic a TM domain directly involved in membrane protein oligomerization, thereby inserting into the membrane and disrupting the buried protein-protein interactions (PPIs) (4). This method has been variously applied, for example, as potential therapeutics against cancer (5, 6, 7) and neurodegenerative disorders (8, 9, 10, 11), with peptide designs including diverse tags, chimeras, and delivery techniques to target the peptides toward the intended PPIs. Using suitable N-terminal and C-terminal tags, our lab has applied this approach to develop several TM peptides (12, 13, 14, 15) to target the bacterial drug efflux pumps that contribute to antibiotic drug resistance (16,17).
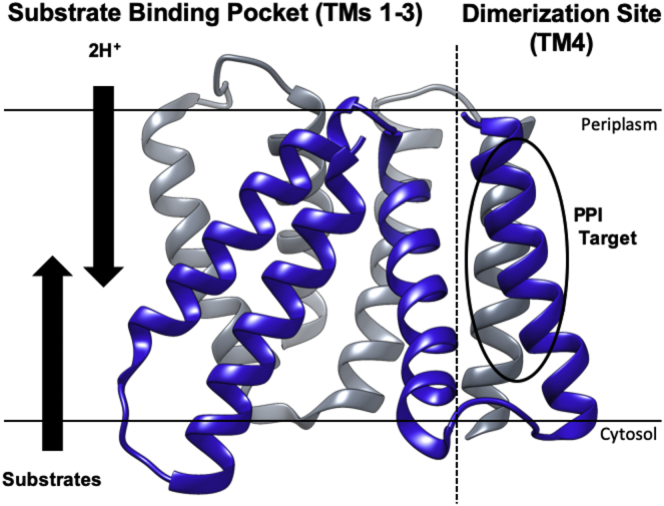
The small multidrug resistance (SMR) proteins reside on the bacterial inner membrane and use the proton motive force (PMF) to efflux toxic substrates from the cell (18,19). Each SMR monomer consists of four TM helices and dimerizes through a TM4-TM4 PPI to function (20). It was determined that a glycine heptad repeat in the SMR TM4 was necessary for dimerization (21) and, as such, this sequence became the target for development of peptide efflux inhibitors through disruption of this TM4-TM4 locus (Fig. 1). In our design, two terminal tags are added to the peptides to facilitate this process: first, a positively charged C-terminal tri-lysine tag directs the peptides toward the negativity-charged bacterial membranes. Then, upon binding to the bacterial membrane surface, the peptides pass a hydrophobicity threshold (22) and “corkscrew” into the membrane, mediated by a noncharged, N-terminal peptoid sarcosine (N-methyl-glycine) tag (23,24). After peptide insertion, the inhibitor peptides should adopt the prototypical TM α-helix, display the TM4 dimerization motif, and then competitively disrupt the native SMR TM4-TM4 dimerization site, ultimately reducing efflux pump function.
Figure 1.
Structure of the small bacterial multidrug-resistant protein from E. coli. The SMR dimerizes through a seven-residue Gly-Gly heptad (GG7) motif in TM4 (circled) that consists, for example, of 90GLALIVAG97V98 in H. salinarum (21). This membrane-embedded PPI is the target of the designed TM peptides described herein. Substrates are driven out of the bacterial cytosol by an oppositely directed PMF (indicated as 2H+). SMR monomers are depicted in blue and gray. The structure is adapted from PDB: 7JK8 (25).
Although these designed peptides have been shown to be effective in reducing SMR-driven efflux (13,15), it remains to be determined if they act through their designed mechanism of action. Whether they directly disrupt the intended SMR dimerization site, or instead operate similarly to AMPs by acting through nonspecific physical disruptive effects on the bacterial membrane per se and/or other functional aspects of the efflux process, such as disruption of the PMF (18). Here, we explore the extant peptide-membrane interactions that may occur before—and/or in concert with—direct SMR TM4-TM4 targeting. More broadly, as helix-helix interactions are key to the functioning of a wide range of both bacterial and mammalian membrane proteins, the present work assists in evaluating the general applicability of our peptide design and tagging approach to targeting membrane-embedded PPIs.
Materials and methods
Peptide synthesis and characterization
Peptides were either synthesized using methods described previously (15), or purchased from Vivitide (Gardner, MA, USA) with >95% purity. Peptides were received as a lyophilized powder, treated with 1,1,1,3,3,3-hexafluoroisopropanol, dried into a film, and dissolved in 2,2,2-trifluoroethanol to be quantified. Since the wild-type TM4 peptides do not contain a native Trp residue, one PAsmrTM4 peptide was substituted at Ile-95 with a Trp residue in a nonconserved region, and this peptide was used as a standard, as it is the same length and maintains a similar hydrophobicity to other TM4 peptides (PAsmrW; Table 1). The concentration of the peptide was determined using the tryptophan absorption at 280 nm using a quartz cuvette with a pathlength of 1 cm and an Ultrospec 3000 UV/Vis spectrophotometer (Pharmacia Biotech, Uppsala, Sweden). Various concentrations were used to generate a standard curve at 215 nm, and the data were fitted by interpolating a standard curve in GraphPad Prism 7. According to Beer’s law (A = εLc), the slope of the line was taken as the extinction coefficient, and was calculated to be 51,870 M−1 cm−1. Peptides were stored as lyophilized powders at −20°C and dissolved in dimethyl sulfoxide (DMSO) for further study. Melittin was purchased from Sigma Aldrich (St. Louis, MO, USA), purified to ≥85% by HPLC from honeybee venom.
Table 1.
Sequences of the peptides studied in this work
| Peptide | Sequencea,b | Species |
|---|---|---|
| HsmrTM4 | Ac-A-(Sar)3-VVGLALIVAGVVV-KKK-NH2 | Halobacterium salinarum |
| HsmrScr | Ac-A-(Sar)3-VVLVGIAGVALVV-KKK-NH2 | |
| EmrETM4 | Ac-A-(Sar)3-IIGMMLISAGVLI-KKK-NH2 | Escherichia coli |
| EmrEScr | Ac-A-(Sar)3-IIVLMGIASMGLI-KKK-NH2 | |
| MmrTM4 | Ac-A-(Sar)3-VVGIGLIVVGTVT-KKK-NH2 | Mycobacterium tuberculosis |
| MmrScr | Ac-A-(Sar)3-VVVLGIVGVIGVT-KKK-NH2 | |
| PAsmrTM4 | Ac-A-(Sar)3-LLGIGLIIAGVLV-KKK-NH2 | Pseudomonas aeruginosa |
| PAsmrScr | Ac-A-(Sar)3-LLVLGAIGIIGLV-KKK-NH2 | |
| PAsmrW | Ac-A-(Sar)3-LLGIGLIWAGVLV-KKK-NH2 | |
| Melittin | GIGAVLKVLTTGLPALISWIKRKRQQ-NH2 | honeybeec |
The sequences are comprised of wild-type residues 88–100 in transmembrane helix 4 of the SMR proteins in each of the indicated bacteria. The “GG7” dimerization motif reported previously (21) is bolded in the native sequences; the substituted Trp in PAsmrW is underlined. A sequence-scrambled peptide (denoted Scr) was synthesized as a control for each species.
Sar (sarcosine), N-methyl-glycine; Ac, acetylated N-terminus, NH2, amidated C-terminus.
Melittin was used as a control for an AMP, purified from the insect Apis mellifera.
Gibbs free energy of peptide partitioning to the water/bilayer interface (ΔGIF) and partitioning from interface to bilayer core/octanol (ΔGOCT-IF) were measured by the Totalizer module of Membrane Protein Explorer (MPEx) (26) (https://blanco.biomol.uci.edu/mpex/) utilizing the Wimley-White Scale (27). N-terminal acetylation and C-terminal amidation were included in the analysis, and sarcosine residues were substituted for alanine to allow for the complete peptide sequence to be assessed.
Liposome preparation
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG), and 1-palmitoyl-2-(11,12-dibromo)stearoyl-sn-glycero-3-phosphocholine lipids were purchased from Avanti Polar Lipids (Alabaster, AL, USA) and received in chloroform. Lipids were prepared with a 3:1 ratio of POPE:POPG and increasing amounts of dibrominated lipids (10, 20, and 30%) were added for tryptophan-quenching experiments. Lipids were dried into a film, water washed, and then lyophilized into a powder. The powder was brought up in a Tris-Cl buffer (10 mM Tris, 10 mM NaCl [pH 7.2]) and went through five freeze-thaw cycles (dry ice/50°C water bath). Lipids were then passed through a 0.2 μM filter 14× and kept at room temperature. Lyophilized peptides were brought up in a Tris-Cl buffer with a 1:1000 ratio of sodium dodecyl sulfate, which was added to liposomes in a 1:1000 peptide to lipid ratio. The samples were dialyzed for 3 days and then stored benchtop.
Circular dichroism spectroscopy and determination of helical content
Circular dichroism (CD) spectra of peptides were collected on a Jasco J-1500 CD spectropolarimeter using a 0.1 cm pathlength quartz cuvette. Generally, peptides were read at room temperature, using a 50 nm/s scanning speed; spectra were recorded between 190 and 250 nm with three accumulations. Graphs were plotted using GraphPad Prism 7. Peptide spectra were recorded in an aqueous environment (20 μM peptide, 10 mM Tris, 10 mM NaCl [pH 7.2]), and in 20 mM 3:1 POPE:POPG liposomes as described above. Spectra represent the average of three independent trials, where the raw millidegree (mdeg) signal from the CD has been background subtracted and converted to mean residue ellipticity (MRE) using standard formula below, where c is concentration in μM, l represents the pathlength in cm, and n is the number of amino acids.
The converted MRE value at 222 nm was then used to estimate the helical percentage for each peptide, using the standard formula below (28,29), where MRE222 is the MRE value at 222 nm and n is the number of peptide bonds.
Tryptophan fluorescence
The spectrum for PAsmrW was recorded on a Photon Technology International (Birmingham, NJ, USA) fluorimeter at room temperature using a 500 μL quartz cuvette. The spectrum was recorded in an aqueous buffer (5 μM peptide, 10 mM Tris, 10 mM NaCl [pH 7.2]), and for peptides reconstituted into a lipid bilayer, in the additional presence of 5 mM POPE:POPG. Trp was excited at 280 nm (slit width 2 nm) and the emission (slit width 4 nm) was recorded for three separate samples at 300–400 nm, with a step size of 2 nm and integration of 1 s. Signals were all background subtracted.
Tryptophan quenching by dibrominated lipids
Dibrominated lipids 1-palmitoyl-2-(11,12-dibromo)stearoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids) were mixed with POPE:POPG lipids in various molar amounts (0%, 10%, 20%, and 30%). Peptides were reconstituted into liposomes as described previously and samples were excited at 280 nm, with the emission recorded at 300–400 nm and background subtracted. Data were plotted in GraphPad Prism 7, the area under the curve was determined and used to generate a Stern-Volmer plot by plotting initial fluorescence values/fluorescence in presence of quencher (F0/F) vs. mol % brominated lipid. The slope values were determined from the line of best fit for each corresponding curve to provide information on the degree of Trp quenching. Bovine serum albumin (Sigma-Aldrich, St. Louis, MO, USA) was used as a nonmembrane interacting soluble control, and the peptide A0L5 (sequence: KKK-AAAAAALLLWLLAAAAAAA-KKK) was used as a membrane-inserting control as has been determined previously (30).
Differential scanning calorimetry
Peptides were reconstituted into liposomes as described previously in a Tris-Cl buffer (5 μM peptide, 5 mM POPE:POPG, 10 mM Tris, 10 mM NaCl [pH 7.2]). Calorimetry was performed on a Nano DSC (T A Instruments, Grimsby, Ontario, Canada). Tris-Cl buffer was manually loaded into a blank cell and reconstituted peptides were added to the sample cell. Heating experiments were performed with a scan rate of 1°C/min from 0 to 30°C, with a 10-min equilibration time. Readings were buffer subtracted and fitted to a first-order polynomial baseline using NanoAnalyze software. Peptide alone in buffer exhibited no thermal events over a temperature range of 0–30°C.
Terbium fluorescence assay
During liposome preparation, lipids were brought up in terbium buffer (10 mM Tris, 10 mM NaCl, 50 mM Tb3Cl, 85 mM sodium citrate [pH 7.4]). After extrusion, liposomes were buffer exchanged with size-exclusion buffer (10 mM Hepes, 100 mM KCl [pH 7.4]) on a size-exclusion column (Superdex 200 10/300, GE Healthcare Life Sciences, Chicago, IL, USA). Fluorescence was measured using a Photon Technology International (Birmingham, NJ, USA) fluorimeter and a 1 cm pathlength quartz cuvette. Liposomes were diluted twice in a buffer containing dipicolinic acid (100 mM DPA, 10 mM Hepes, 100 mM KCl [pH 7.4]), Tb3+ was excited at 314 nm (2 nm slit width), and the emission was recorded at 544 nm (5 nm slit width). Peptides were added in a 1:1000 peptide to lipid ratio to a stirred solution of Tb3+-loaded liposomes and fluorescence was recorded for 600 s. Readings were blank subtracted and normalized to the fluorescence of the sample treated with 0.1% Triton, representing 100% disruption.
Minimum inhibitory concentration assay
Peptide minimum inhibitory concentration (MICs) were determined at 100% reduction in growth using methods described previously (15). In brief, E. coli BL21(DE3) cells, with endogenous SMR (EmrE) and expressed Hsmr, were grown overnight in Mueller-Hinton broth. Twofold serial dilutions of peptide were made from 0–32 μM peptide and plated with 50,000 CFU/well. Plates were incubated at 37°C for 20 h and the optical density (OD) was measured at 600 nm to determine cell growth.
Propidium iodide assay
The extent of membrane disruption was determined using methods described previously (15). In brief, overnight cultures of BL21 E. coli cells expressing PAsmr were spun down and resuspended in minimal medium. Cells were inoculated into either minimal medium or 70% isopropanol to achieve “viable” and “disrupted” cells, respectively, as isopropanol permeabilizes membranes (31). Cells were incubated at room temperature for 1 h and diluted to 0.1 OD600 for consistency with prior efflux assays (15). Twofold serial dilutions of peptide were made from 0 to 32 μM peptide and plated with cells in minimal medium for 1 h. Propidium iodide (PI) was added to each well to a final concentration of 5 μM, and plates were covered and shaken for 15 min. Fluorescence was measured using a SpectraMax Gemini EM Microplate Reader and excited at 488 nm and emission recorded at 630 nm. Values were blank subtracted and compared with a standard curve generated from viable and disrupted cells using GraphPad Prism 7.
PMF disruption assay
Overnight cultures of BL21 E. coli cells expressing EmrE were spun down and resuspended in 5 mL minimal medium and incubated at 37°C, with shaking at 250 rpm for 45 min. Cells were spun down and resuspended to an OD600 of 0.4 in minimal medium. Cells were then treated with EDTA for 5 min, spun down, and resuspended to remove EDTA. Cells were then treated with DiOC2(3) to a final concentration of 30 μM and incubated in the dark for 5 min. Twofold serial dilutions of peptide were plated from 0 to 8 μM peptide and treated with cells. Cells were also treated with 20 μM CCCP and 8 μM melittin as a positive control. Plates were incubated for an hour, and fluorescence was measured using a SpectraMax Gemini EM Microplate Reader at an excitation wavelength of 450 nm and emission at 510 nm. Values were buffer subtracted and normalized to the fluorescence of the dye alone in minimal medium.
Results
Design of efflux pump inhibitor TM peptides
Here, we undertook to determine the mechanism of action of our previous inhibitor peptides designed for the H. salinarum and P. aeruginosa SMRs (13,15); also, since SMRs are almost ubiquitous across bacterial species (32), we wanted to explore the generality of the approach, and, in the longer range, to identify peptides that are active against a broad range of SMRs. Thus, additional peptides were designed to target SMRs from E coli and Mycobacterium tuberculosis, the latter which is designated as a top-threat resistant species (3). Two peptides for each species were synthesized, viz., a parent peptide containing the native TM4 sequence of residues 88–100—which includes the GG7 dimerization motif—and a “scrambled” sequence counterpart where the GG7 motif has been disrupted (Table 1). All peptides contain the aforementioned tags and include an amidated C-terminus to neutralize the negative C-terminus. The scrambled peptides serve as a control as they contain the same amino acid composition and therefore overall length and average hydrophobicity, but have no sequence complementarity for the corresponding SMR. In addition, the peptide for P. aeruginosa has an Ile residue not directly engaged in the TM4-TM4 binding motif near the middle of the sequence, which we substituted with a Trp residue (PAsmrW; W underlined in Table 1) for quantification purposes and biophysical analysis. The pore-forming AMP melittin was also used as a control in several of the biophysical assays as a comparison for antimicrobial peptide activity.
Folding of inhibitor peptides in bacterial membrane mimetics
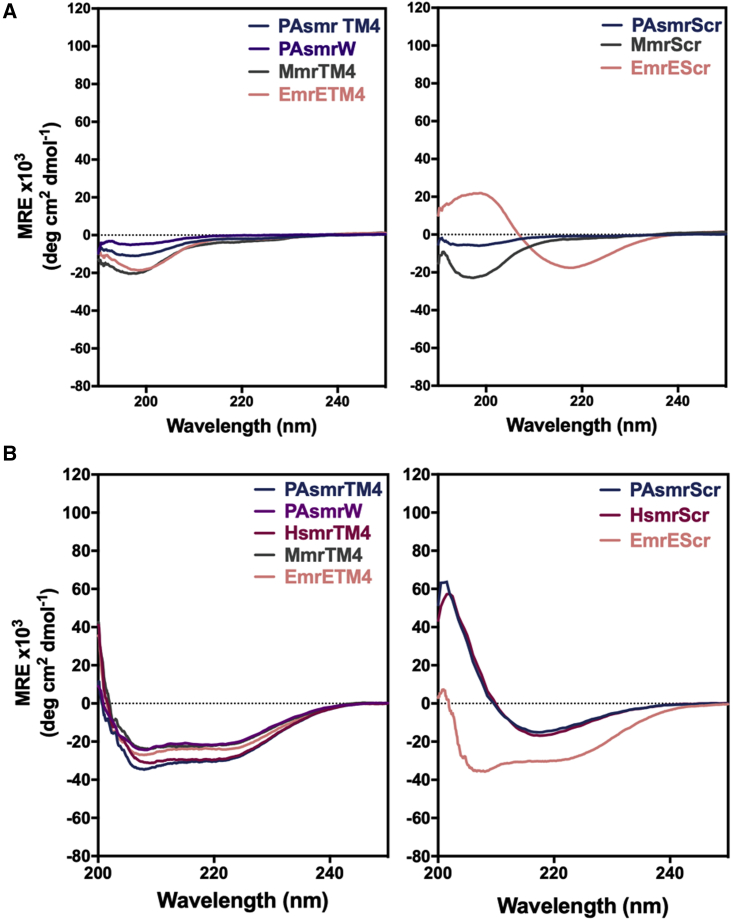
For the peptide inhibitors to reach their intended SMR target, they must not aggregate in an aqueous environment before addition to the cells. They must then insert into the membrane, forming a stable α-helix to display the correct TM4-TM4 GG7 dimerization motif. Secondary structure was therefore assessed through CD spectroscopy under various conditions (Fig. 2). In an aqueous environment, all the peptides were unstructured, with the exception of the EmrEScr, which displayed a β-sheet (Fig. 2 A). The peptides were diluted from DMSO stocks in cell-based assays to avoid potential solubility issues with EmrEScr and to maintain consistency across peptides. The peptides were then dialyzed from SDS micelles to liposomes, with the exception of MmrScr, which aggregated in the presence of SDS (Fig. 2 B). Interestingly, all the parent peptides formed an α-helix in liposomes that contain the same two primary phospholipids and in the corresponding ratio as found in typical bacterial membranes (3:1 POPE: POPG), albeit among the scrambled peptides, only EmrEScr displayed a helical conformation.
Figure 2.
Inhibitor peptides display a range of secondary structures in various local environments. Circular dichroism was performed on 20 M peptide (A) in an aqueous environment (10 mM Tris-Cl); and (B) in a model bacterial membrane (20 mM 3:1 POPE:POPG, 10 mM Tris-Cl, 1/1000 peptide to lipid [P/L] ratio). Spectra for the H. salinarum peptides were recorded previously (13). Each spectrum represents the average of at least three independent trials.
To determine if the peptides would favorably partition into the bilayer, percent helicity was first quantified by the ellipticity of each peptide at 222 nm and subsequently used by the Totalizer module of MPEx to evaluate the free energy of partitioning (Table 2). We observed that each peptide demonstrates favorable interaction with the water/bilayer interface, with spontaneous ΔGIF values ranging between −4.54 and −8.31 kcal/mol. Similarly, each peptide displays a likelihood to penetrate the bilayer core, as previous studies have indicated that ΔGOCT-IF ≤ 20 kcal/mol is favorable for peptide penetration (33).
Table 2.
Thermodynamic parameters associated with peptide-membrane interactions
| Peptide | % Helixa | ΔGIF (kcal/mol)b | ΔGOCT-IF (kcal/mol)b |
|---|---|---|---|
| HsmrTM4 | 85 | −6.35 | 8.87 |
| EmrETM4 | 71 | −7.01 | 8.21 |
| EmrEScr | 86 | −8.21 | 8.21 |
| MmrTM4 | 64 | −4.54 | 9.98 |
| PAsmrTM4 | 81 | −8.21 | 8.80 |
| PAsmrW | 63 | −8.31 | 9.37 |
% Helix calculated as the percentage of α-helix by peptide MRE at 222 nm in a model bacterial membrane (3:1 POPE:POPG).
Free energy of partitioning was measured using the Totalizer module of Membrane Protein Explorer using the Wimley-White Scale. ΔGIF represents the peptide partitioning to the water/bilayer interface and ΔGOCT-IF represents the peptide partitioning from interface to the bilayer core/octanol.
Peptide insertion into membrane environments
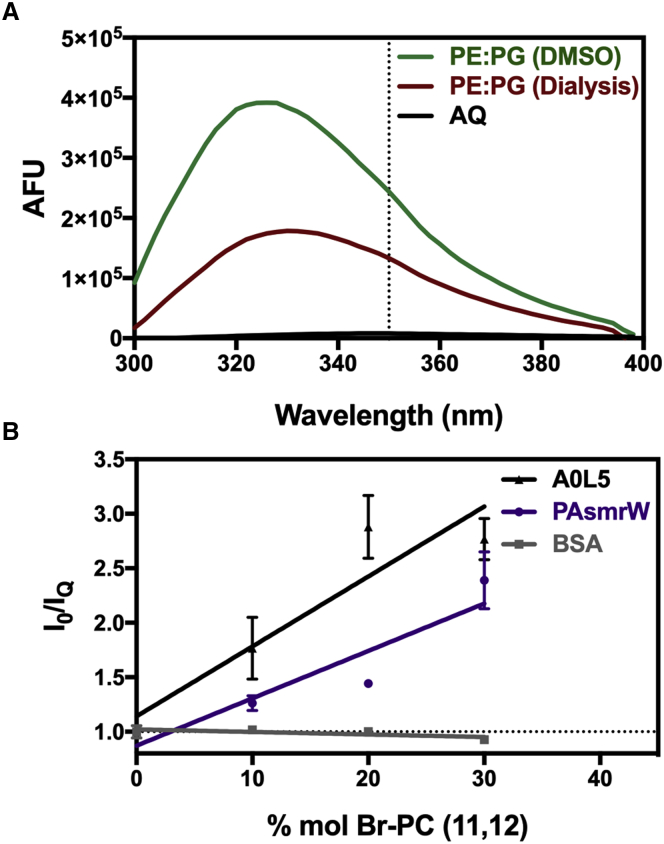
To determine orientation with respect to whether the peptide resides on the membrane surface or has achieved a measure of insertion, we used PAsmrW as a model peptide and evaluated the Trp fluorescence upon addition to a hydrophobic environment (Fig. 3 A). After dialysis into liposomes as described previously or diluted from DMSO as in cell-based assays, this peptide displayed a strong 20 nm blue shift, indicating the Trp residue resides in a hydrophobic environment. We then monitored the depth of peptide insertion through Trp quenching with dibrominated lipids (11,12-BrPC), where bromine atoms are substituted toward the end of the lipid tails. Lipid-reconstituted PAsmrW peptide displayed an increase of Trp quenching with dibrominated lipids, suggesting that the peptide is deeply inserting into the membrane (Fig. 3 B). The PAsmrW slope was comparable with the control peptide A0L5, shown previously to insert into a membrane environment (30), while the Trp fluorescence of the soluble BSA protein was not quenched in the presence of the lipid environment.
Figure 3.
Tryptophan fluorescence of PAsmrW. (A) Trp fluorescence of 5 M PAsmrW peptide in an aqueous environment denoted AQ (10 mM Tris-Cl); SDS detergent micelle (5 mM SDS, 10 mM Tris-Cl); and reconstituted into liposomes representing a bacterial membrane (5 mM POPE:POPG, 10 mM Tris-Cl) (n = 3). (B) Stern-Volmer plots were generated for PAsmrW reconstituted into POPE:POPG liposomes supplemented with increasing molar concentrations of dibrominated lipid 1-palmitoyl-2-(11,12-dibrogmo)stearoyl-sn-glycero-3-phosphocholine (n = 3). A0L5 demonstrates the curve for a known membrane-inserting peptide; BSA represents a negative control for a soluble protein (n = 2). Error bars represent standard error of the mean.
Positively charged peptide tags recruit negative lipid headgroups
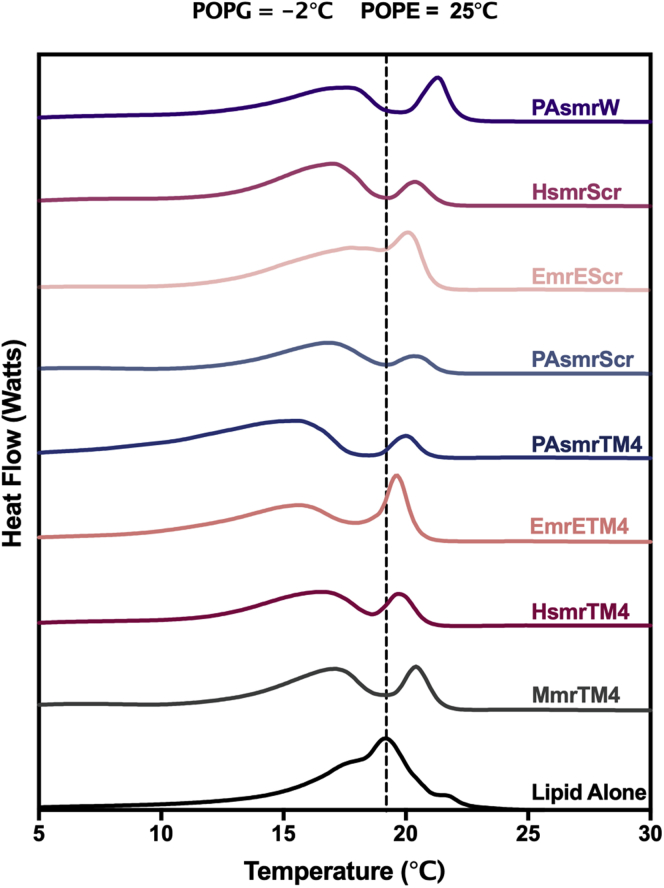
Peptide insertion may be affecting membrane integrity by modulating fluidity and lipid organization. We used differential scanning calorimetry to determine lipid packing as a function of lipid melting point (Tm) (Fig. 4). The POPE:POPG liposome Tm was 19.5°C; however, this peak was relatively broad with a slight shoulder, which could indicate incomplete mixing, as the Tm values of the individual lipids differ by >20°C. When peptides are added to the liposomes, the peptides are apparently inducing lipid demixing, as displayed by the appearance of two peaks moving in opposite directions from the pure liposome Tm (Table 3). The lower Tm likely represents peptides preferentially recruiting the negatively charged POPG lipid headgroups as mediated by their Lys tag. This will generate pools enriched in POPE, as seen by the peak increasing toward the native POPE Tm (25°C). EmrEScr, however, did not display strong separation but rather peak broadening, which may be an indication of an alternate mechanism for this peptide.
Figure 4.
Differential scanning calorimetry of inhibitor peptides in model bacterial membranes. Spectra represent a 1/1000 P/L ratio using POPE:POPG (3:1 mol/mol) liposomes in a Tris-Cl buffer. Melting points are given in Table 2. Each line represents the average of at least three individual experiments.
Table 3.
Melting points Tm (°C) of liposomes representing bacterial-like membranesa upon addition of SMR TM4 peptides
| Peptide | Tm 1b | Tm 2b |
|---|---|---|
| PAsmrW | 16.7 0.2 | 19.9 0.2 |
| HsmrScr | 17.1 0.03 | 20. 0.06 |
| EmrEScr | 17.8 0.02 | 20.2 0.1 |
| PAsmrScr | 17.0 0.2 | 20.5 0.1 |
| PAsmrTM4 | 15.6 0.03 | 20.1 0.08 |
| EmrETM4 | 16.0 0.5 | 20.2 0.5 |
| HsmrTM4 | 16.5 0.2 | 19.8 0.2 |
| MmrTM4 | 17.0 0.2 | 20.3 0.2 |
| Lipid Alone | 19.5 0.3 | – |
POPE:POPG 3:1 mol/mol lipid composition.
Mean values are reported with standard deviation.
Peptide insertion causes minimal disruption of membrane integrity in vitro
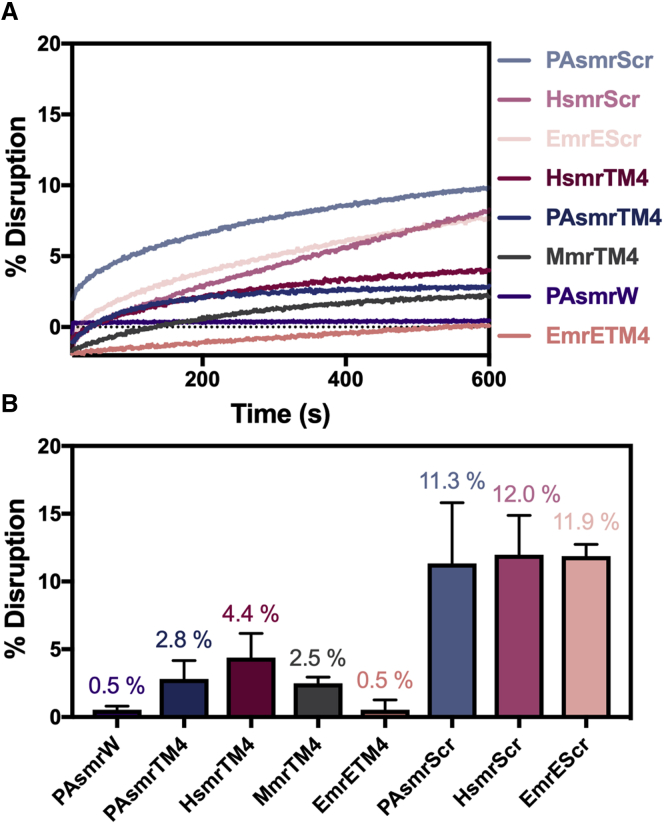
As the peptides are seen to be causing lipid reorganization upon insertion, we next assessed whether this insertion resulted in dye release due to disruption of the bacterial membrane (34). Using an in vitro liposome terbium dye release assay, we measured the effective membrane disruption of the liposomes containing peptides diluted from DMSO (Fig. 5). All the peptides displayed low disruption when compared with the positive control Triton, with parent peptides causing less than 5% disruption, and scrambled peptides displaying ∼12% disruption. Despite the PAsmrScr and HsmrScr peptides not displaying helical character in liposomes (Fig. 2), they appear to be inducing more nonspecific disruption than their parent peptide counterparts. In addition, the control PAsmrW peptide displayed very low disruption, similar to that of the EmrETM4 peptide and its PAsmrTM4 parent counterpart.
Figure 5.
Peptide-mediated dye leakage in model bacterial membranes. Terbium fluorescence was measured in POPE:POPG (3:1 mol/mol) liposomes with a 1/1000 P/L ratio. (A) Fluorescence was measured over 10 min at 544 nm and normalized to 100% disruption by Triton after treatment with each peptide and (B) quantitated at the end of the experiment (n = 3). Error bars represent standard error of the mean.
Peptide insertion causes minimal disruption of membrane integrity in vivo
To determine if the in vitro observations were comparable in vivo, we examined how disruptive the peptides were in the E. coli cells used for inhibition assays (15). First, the MIC was determined for all peptides (Table 4). The PAsmrTM4 and corresponding PAsmrScr peptides displayed the lowest MIC at 16 μM, while all other peptides did not reach their MIC below 32 μM. Melittin and DMSO were used as controls, with the finding that melittin had an MIC of 2 μM and that the DMSO vehicle conditions did not affect growth. Although only the P. aeruginosa peptide set displayed an MIC <32 μM, all other peptides did reduce the bacterial growth rate at higher peptide concentrations, suggesting that there is some nonspecific toxicity of the peptides.
Table 4.
Inhibitor TM peptide MIC values
| Peptide | MICa (μM) |
|---|---|
| PAsmrTM4 | 16b |
| PAsmrScr | >32b |
| PAsmrW | >32b |
| HsmrTM4 | >32 |
| HsmrScr | >32 |
| EmrETM4 | >32 |
| EmrEScr | >32 |
| MmrTM4 | >32 |
MIC values were determined in E. coli cells.
MIC value recorded previously (15).
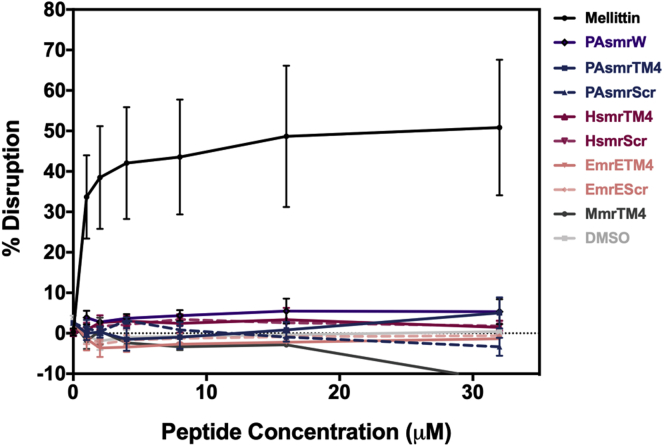
To assess the effect of the peptides on the bacterial membrane specifically, PI fluorescence was measured as peptides were added to bacterial cells and compared with those disrupted by 70% isopropanol (Fig. 6). PI is an ethidium bromide analog that is impermeable to bacterial membranes (31), and accordingly the dye will enter the cell and fluoresce only if the peptides are causing disruption. We found that there was minimal disruption by all peptides upon addition (<10%) when compared with the control melittin peptide which elicited ∼50% disruption.
Figure 6.
Measurement of peptide-mediated propidium iodide dye entry into E. coli BL21 cells. Serial dilutions of peptides were incubated with E. coli BL21 cells treated with 5 M PI for 60 min. Samples were excited at 488 nm and emission recorded at 630 nm. Values were interpolated from a standard curve calibrated by cells 100% disrupted by 70% isopropanol (n = 3). Error bars represent standard error of the mean.
Peptide addition indicates moderate disruption of the PMF
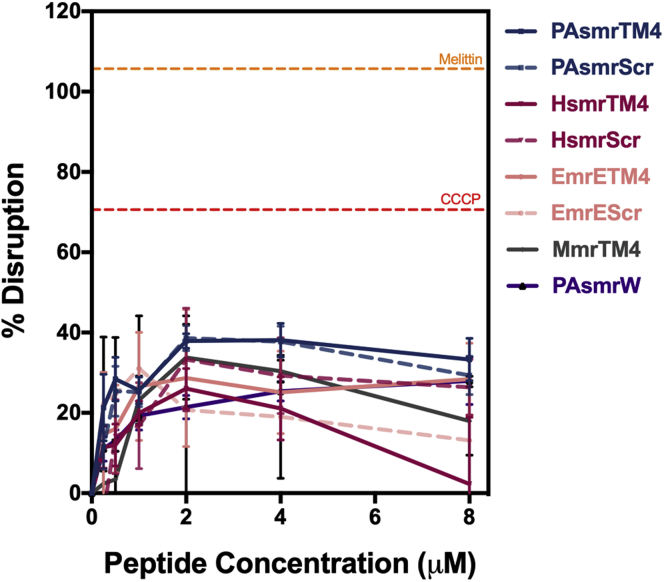
As the peptides displayed minimal membrane disruption in vivo or in vitro, we next evaluated the peptides’ effects on the PMF. The fluorescence of the dye DiOC2(3) is quenched in the presence of a PMF (35) and therefore will increase if the peptides are disrupting it. The peptides showed varying levels of disruption (Fig. 7), with the P. aeruginosa set having the highest disruption of ∼40%, which is consistent with the MIC data. These results suggest there is an effect on the PMF by the peptides, albeit less than when compared with CCCP at ∼70% and with melittin, which displayed a higher fluorescence than the baseline of the dye in minimal medium, at just over 100%.
Figure 7.
Disruption of the proton motive force by inhibitor peptides in E. coli BL21 cells. Fluorescence of the DiOC2(3)dye was recorded for cells treated with peptide for 60 min; solid lines represent parent peptides, and dashed lines represent scrambled sequences. Orange and red dashed lines represent cells treated with 8 μM melittin and 20 μM CCCP, respectively. Samples were normalized to dye fluorescence in minimal medium (n = 3). Error bars represent standard error of the mean.
Discussion
Naturally occurring cationic AMPs have a range of structures (34,36), but generally exert their disruptive function by first being targeted to bacterial membranes through electrostatic interactions between the positively charged polar region(s) of the peptide and the negatively charged bacterial lipid headgroups (37). Upon binding, the extent of peptide-mediated disruption may be a product of membrane thinning, hydrophobic mismatch, pore formation, lipid deformation, and/or general detergent-like properties (38, 39, 40). Although the relationships among these properties and whether the peptide will reside on the surface or penetrate into the membrane core are not well understood, it appears that a threshold of hydrophobicity balanced with positions of hydrophilic residues may be a determining factor (22,41,42).
These latter features, as incorporated into our designed peptides, likely promote membrane insertion without significant disruption. Thus, in the present work, the CD data confirmed that the parent inhibitor peptides and EmrEScr each acquire a helical conformation in an anionic membrane environment (Fig. 2B). The Trp fluorescence of the PAsmrW peptide, representative of the other inhibitor peptides, displayed a strong blue shift in a lipid environment, and was quenched in the presence of brominated lipids, confirming its deep insertion into the membrane core (Fig. 3). Yet the collection of TM peptides displayed minimal antimicrobial activity (Table 4), and caused less than 15% membrane disruption in vitro (Fig. 5), and less than 10% in vivo (Fig. 6). Nevertheless, the TM peptides may be disrupting the membrane sufficiently to dissipate the PMF, as seen by a 20–40% increase in the fluorescence of DiOC2(3), which may result in some cell death with higher peptide concentrations over a prolonged period (Fig. 7).
Perhaps most intriguing in our determination of the peptides’ direct effects on bacterial model membranes is their ability to demix the POPE and POPG lipids, creating two pools, one of which is likely a peptide-POPG conglomerate, and the other a POPE-rich component where the native POPG content has been depleted (Fig. 4). PE and PG lipids have previously been found to demonstrate nonideal mixing as a result of the lipid headgroup interactions (43), and this effect is exemplified by the addition of our peptides and is consistent with the peak splitting observed in Fig 4. This phenomenon was earlier reported when cations and basic peptides were found to bind to the negatively charged PG or PS lipid headgroups (44,45), and has since been expanded with examples of cationic antimicrobial peptides causing separation in mixed zwitterionic and negatively charged lipid headgroups (46, 47, 48, 49, 50). Some AMPs have been found to cause cell death through this membrane-demixing event, as the creation of such lipid domains caused significant disturbance that led to autolysis (51).
In the context of the SMRs, their structure and function have been found to be impacted based on lipid environment, with oligomeric states being reported in liposomes of varying compositions (52), and, although substrate binding was not affected, the SMR activity was improved by the presence of both PE and PG lipids (52, 53, 54). Not only may SMR function be impacted by peptide-mediated demixing, but interaction of the TM peptides with the SMR could be altered by changes in the lipid environment, as seen by other homodimerizing TMDs (55,56). Although the lipid composition is thus demonstrably important for SMR function and cell survival, it is noted that the PAsmrScr peptide also displays lipid demixing, yet had no activity in cell-based studies (15). As such, it is unlikely that the peptides are acting through the demixing mechanism, but rather that the demixing may be a fairly general consequence of cationic peptide interaction with mixed charge lipid membranes.
While our TM peptides share with AMPs the biophysical characteristic of targeting bacterial cells, the evidence presented herein suggests that these peptides lack their membrane-disruptive properties. The overall results thus minimize the presence of significant nonspecific antimicrobial activity, and support the notion that the peptides are likely acting at the intended TM4-TM4 interaction site. This conclusion is reinforced by our previous studies that have shown that peptides targeting PAsmr and Hsmr can significantly reduce efflux activity and potentiate the antimicrobial activity of several biocides (12,13,15). As with hydrophobic AMPs, common limitations for therapeutic potential for these efflux inhibitors include toxicity to mammalian cells, and susceptibility to proteolytic degradation (57). We have previously addressed the latter limitation by modifying the HsmrTM4 peptide with a hydrocarbon staple, thereby improving the peptide’s half-life in blood plasma and liver homogenates (12). Therefore, we believe that the peptide-tagging technique coupled with targeting membrane-based oligomeric sites constitutes, in principle, a viable approach to inhibiting drug efflux by bacterial efflux pumps and, more generally, to impacting the activity of many proteins that function through membrane-embedded PPIs.
Author contributions
C.JM. designed and carried out experiments and performed data analysis. T.S.J. assisted in experimental procedures, data analysis, interpretation, experimental design, and manuscript review and editing. C.M.D. and C.JM. conceptualized the project and wrote the manuscript.
Acknowledgments
This work was supported, in part, by grants to C.M.D. from the Canadian Institutes of Health Research (CIHR project grant no. 376666) and from Cystic Fibrosis Canada. C.JM. held a Restracomp Scholarship from the Hospital for Sick Children. The authors also wish to thank the Structural and Biophysical Core Facility, notably Greg Wasney and James Magnus Jorgensen, at the Hospital for Sick Children for their assistance with equipment use.
Declaration of interests
The authors declare no competing interests.
Editor: Antje Pokorny Almeida.
References
- 1.Avci F.G., Akbulut B.S., Ozkirimli E. Membrane active peptides and their biophysical characterization. Biomolecules. 2018;8:77. doi: 10.3390/biom8030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Heath Organization . 2017. Antibacterial Agents in Clinical Development. An Analysis of the Antibacterial Clinical Development Pipeline, Including Tuberculosis. [Google Scholar]
- 3.Centers for Disease Control and Prevention . 2019. Biggest Threats and Data: 2019 AR Threats Report. 2019. [Google Scholar]
- 4.Albrecht C., Appert-Collin A., et al. Bennasroune A. Transmembrane peptides as inhibitors of protein-protein interactions: an efficient strategy to target cancer cells? Front. Oncol. 2020;10:519. doi: 10.3389/fonc.2020.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennasroune A., Fickova M., et al. Hubert P. Transmembrane peptides as inhibitors of ErbB receptor signaling. Mol. Biol. Cell. 2004;15:3464–3474. doi: 10.1091/mbc.E03-10-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arpel A., Sawma P., et al. Bagnard D. Transmembrane domain targeting peptide antagonizing ErbB2/Neu inhibits breast tumor growth and metastasis. Cell Rep. 2014;8:1714–1721. doi: 10.1016/j.celrep.2014.07.044. [DOI] [PubMed] [Google Scholar]
- 7.Gamper C., Spenlé C., et al. Heinlein M. Functionalized tobacco mosaic virus coat protein monomers and oligomers as nanocarriers for anti-cancer peptides. Cancers. 2019;11:1609. doi: 10.3390/cancers11101609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harikumar K.G., Pinon D.I., Miller L.J. Transmembrane segment IV contributes a functionally important interface for oligomerization of the class II G protein-coupled secretin receptor. J. Biol. Chem. 2007;282:30363–30372. doi: 10.1074/jbc.M702325200. [DOI] [PubMed] [Google Scholar]
- 9.Borroto-Escuela D.O., Rodriguez D., et al. Carlsson J. Mapping the interface of a GPCR Dimer: a structural model of the A2A Adenosine and D2 dopamine receptor heteromer. Front. Pharmacol. 2018;9:1–16. doi: 10.3389/fphar.2018.00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallo M., Navarro G., et al. Andreu D. A2A receptor homodimer-disrupting sequence efficiently delivered by a protease-resistant, cyclic cpp vector. Int. J. Mol. Sci. 2019;20:4937. doi: 10.3390/ijms20194937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borroto-Escuela D.O., Wydra K., et al. Fuxe K. Disruption of A2AR-D2R heteroreceptor complexes after A2AR transmembrane 5 peptide administration enhances cocaine self-administration in rats. Mol. Neurobiol. 2018;55:7038–7048. doi: 10.1007/s12035-018-0887-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellmann-Sickert K., Stone T.A., et al. Deber C.M. Efflux by small multidrug resistance proteins is inhibited by membrane-interactive helix-stapled peptides. J. Biol. Chem. 2015;290:1752–1759. doi: 10.1074/jbc.M114.616185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poulsen B.E., Deber C.M. Drug efflux by a small multidrug resistance protein is inhibited by a transmembrane peptide. Antimicrob. Agents Chemother. 2012;56:3911–3916. doi: 10.1128/AAC.00158-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jesin J.A., Stone T.A., et al. Deber C.M. Peptide-based approach to inhibition of the multidrug resistance efflux pump AcrB. Biochemistry. 2020;59:3973–3981. doi: 10.1021/acs.biochem.0c00417. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell C.J, Stone T.A., Deber C.M. Peptide-based efflux pump inhibitors of the small multidrug resistance protein from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/AAC.00730-19. e00730-19-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bay D.C., Rommens K.L., Turner R.J. Small multidrug resistance proteins: a multidrug transporter family that continues to grow. Biochim. Biophys. Acta. 2008;1778:1814–1838. doi: 10.1016/j.bbamem.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 17.Bay D.C., Turner R.J. Diversity and evolution of the small multidrug resistance protein family. BMC Evol. Biol. 2009;9:140. doi: 10.1186/1471-2148-9-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson A.E., Thomas N.E., et al. Henzler-Wildman K.A. New free-exchange model of EmrE transport. Proc. Natl. Acad. Sci. USA. 2017;114:E10083–E10091. doi: 10.1073/pnas.1708671114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hussey G.A., Thomas N.E., Henzler-Wildman K.A. Highly coupled transport can be achieved in free-exchange transport models. J. Gen. Physiol. 2020;152:e201912437. doi: 10.1085/jgp.201912437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y.-J., Pornillos O., et al. Chang G. X-ray structure of EmrE supports dual topology model. Proc. Natl. Acad. Sci. USA. 2007;104:18999–19004. doi: 10.1073/pnas.0709387104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poulsen B.E., Rath A., Deber C.M. The assembly motif of a bacterial small multi drug resistance protein. J. Biol. Chem. 2009;284:9870–9875. doi: 10.1074/jbc.M900182200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glukhov E., Burrows L.L., Deber C.M. Membrane interactions of designed cationic antimicrobial peptides: the two thresholds. Biopolymers. 2008;89:360–371. doi: 10.1002/bip.20917. [DOI] [PubMed] [Google Scholar]
- 23.Melnyk R.A., Partridge A.W., et al. Deber C.M. Polar residue tagging of transmembrane peptides. Biopolymers. 2003;71:675–685. doi: 10.1002/bip.10595. [DOI] [PubMed] [Google Scholar]
- 24.Tang Y.C., Deber C.M. Hydrophobicity and helicity of membrane-interactive peptides containing peptoid residues. Biopolymers. 2002;65:254–262. doi: 10.1002/bip.10236. [DOI] [PubMed] [Google Scholar]
- 25.Shcherbakov A.A., Hisao G., et al. Hong M. Structure and dynamics of the drug-bound bacterial transporter EmrE in lipid bilayers. Nat. Commun. 2021;12:172. doi: 10.1038/s41467-020-20468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snider C., Jayasinghe S., et al. White S.H. MPEx: a tool for exploring membrane proteins. Protein Sci. 2009;18:2624–2628. doi: 10.1002/pro.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White S.H., Wimley W.C. Membrane protein folding and stability: physical Principles. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y.H., yang J.T., Chau K.H. Determination of the Helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 29.Lacroix E., Viguera A.R., Serrano L. Elucidating the folding problem of α-helices: local motifs, long-range electrostatics, lonic-strength dependence and prediction of NMR parameters. J. Mol. Biol. 1998;284:173–191. doi: 10.1006/jmbi.1998.2145. [DOI] [PubMed] [Google Scholar]
- 30.Stone T.A., Schiller N., et al. Deber C.M. Hydrophobic clusters raise the threshold hydrophilicity for insertion of transmembrane sequences in vivo. Biochemistry. 2016;55:5772–5779. doi: 10.1021/acs.biochem.6b00650. [DOI] [PubMed] [Google Scholar]
- 31.Stiefel P., Schmidt-Emrich S., et al. Ren Q. Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiol. 2015;15:36. doi: 10.1186/s12866-015-0376-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bay D.C., Turner R.J. Diversity and evolution of the small multidrug resistance protein family. BMC Evol. Biol. 2009;9:140. doi: 10.1186/1471-2148-9-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Almeida P.F., Pokorny A. Mechanisms of antimicrobial, cytolytic, and cell-penetrating peptides: from kinetics to thermodynamics. Biochemistry. 2009;48:8083–8093. doi: 10.1021/bi900914g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koehbach J., Craik D.J. The vast structural diversity of antimicrobial peptides. Trends Pharmacol. Sci. 2019;40:517–528. doi: 10.1016/j.tips.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 35.McAuley S., Huynh A., Czarny T.L., Brown E.D., Nodwell J.R. Membrane activity profiling of small molecule: B. subtilis growth inhibitors utilizing novel duel-dye fluorescence assay. Medchemcomm. 2018;9:554–561. doi: 10.1039/c8md00009c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huan Y., Kong Q., et al. Yi H. Antimicrobial peptides: classification, design, application and research progress in multiple fields. Front. Microbiol. 2020;11:582779. doi: 10.3389/fmicb.2020.582779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Epand R.M., Walker C., et al. Magarvey N.A. Molecular mechanisms of membrane targeting antibiotics. Biochim. Biophys. Acta. 2016;1858:980–987. doi: 10.1016/j.bbamem.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 38.Brogden K.A. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 39.Kabelka I., Vácha R. Advances in molecular understanding of α-helical membrane-active peptides. Acc. Chem. Res. 2021;54:2196–2204. doi: 10.1021/acs.accounts.1c00047. [DOI] [PubMed] [Google Scholar]
- 40.Sani M.A., Separovic F. How membrane-active peptides get into lipid membranes. Acc. Chem. Res. 2016;49:1130–1138. doi: 10.1021/acs.accounts.6b00074. [DOI] [PubMed] [Google Scholar]
- 41.Kabelka I., Vácha R. Optimal hydrophobicity and reorientation of amphiphilic peptides translocating through membrane. Biophys. J. 2018;115:1045–1054. doi: 10.1016/j.bpj.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin L.M., Edwards M.A., et al. Deber C.M. Roles of hydrophobicity and charge distribution of cationic antimicrobial peptides in peptide-membrane interactions. J. Biol. Chem. 2012;287:7738–7745. doi: 10.1074/jbc.M111.303602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garidel P., Blume A. Miscibility of phosphatidylethanolamine-phosphatidylglycerol mixtures as a function of pH and acyl chain length. Eur. Biophys. J. 2000;28:629–638. doi: 10.1007/s002490050003. [DOI] [PubMed] [Google Scholar]
- 44.Mosior M., McLaughlin S. Binding of basic peptides to acidic lipids in membranes: effects of inserting alanine(s) between the basic residues. Biochemistry. 1992;31:1767–1773. doi: 10.1021/bi00121a026. [DOI] [PubMed] [Google Scholar]
- 45.Garidel P., Blume A. Interaction of alkaline earth cations with the negatively charged phospholipid 1, 2-Dimyristoyl-sn-glycero-3-phosphoglycerol: a differential scanning and isothermal titration calorimetric study. Langmuir. 1999;15:5526–5534. [Google Scholar]
- 46.Hoernke M., Schwieger C., et al. Blume A. Binding of cationic pentapeptides with modified side chain lengths to negatively charged lipid membranes: complex interplay of electrostatic and hydrophobic interactions. Biochim. Biophys. Acta. 2012;1818:1663–1672. doi: 10.1016/j.bbamem.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 47.Arouri A., Dathe M., Blume A. Peptide induced demixing in PG/PE lipid mixtures: a mechanism for the specificity of antimicrobial peptides towards bacterial membranes? Biochim. Biophys. Acta. 2009;1788:650–659. doi: 10.1016/j.bbamem.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 48.Mason A.J., Martinez A., et al. Bechinger B. The antibiotic and DNA-transfecting peptide LAH4 selectively associates with, and disorders, anionic lipids in mixed membranes. Faseb. J. 2006;20:320–322. doi: 10.1096/fj.05-4293fje. [DOI] [PubMed] [Google Scholar]
- 49.Munhoz V.H.O., Ferreira C.S., et al. Verly R.M. Epimers L- and D-Phenylseptin: how the relative stereochemistry affects the peptide-membrane interactions. Biochim. Biophys. Acta Biomembr. 2021;1863:183708. doi: 10.1016/j.bbamem.2021.183708. [DOI] [PubMed] [Google Scholar]
- 50.Joanne P., Galanth C., et al. Alves I.D. Lipid reorganization induced by membrane-active peptides probed using differential scanning calorimetry. Biochim. Biophys. Acta. 2009;1788:1772–1781. doi: 10.1016/j.bbamem.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 51.Scheinpflug K., Wenzel M., et al. Strahl H. Antimicrobial peptide cWFW kills by combining lipid phase separation with autolysis. Sci. Rep. 2017;7:44332. doi: 10.1038/srep44332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mörs K., Hellmich U.A., et al. Glaubitz C. A lipid-dependent link between activity and oligomerization state of the M. tuberculosis SMR protein TBsmr. Biochim. Biophys. Acta Biomembr. 2013;1828:561–567. doi: 10.1016/j.bbamem.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 53.Curnow P., Lorch M., et al. Booth P.J. The reconstitution and activity of the small multidrug transporter EmrE is modulated by non-bilayer lipid composition. J. Mol. Biol. 2004;343:213–222. doi: 10.1016/j.jmb.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 54.Charalambous K., Miller D., et al. Booth P.J. Lipid bilayer composition influences small multidrug transporters. BMC Biochem. 2008;9:31. doi: 10.1186/1471-2091-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aisenbrey C., Salnikov E.S., Bechinger B. Solid-State NMR investigations of the MHC II transmembrane domains: topological equilibria and lipid interactions. J. Membr. Biol. 2019;252:371–384. doi: 10.1007/s00232-019-00071-8. [DOI] [PubMed] [Google Scholar]
- 56.Salnikov E.S., Aisenbrey C., et al. Bechinger B. Structure, topology, and dynamics of membrane-inserted polypeptides and lipids by solid-state NMR spectroscopy: investigations of the transmembrane domains of the DQ beta-1 subunit of the MHC II receptor and of the COP I protein p24. Front. Mol. Biosci. 2019;6:1–14. doi: 10.3389/fmolb.2019.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J., Koh J.J., Liu S., et al. Beuerman R.W. Membrane active antimicrobial peptides: translating mechanistic insights to design. Front. Neurosci. 2017;11:73. doi: 10.3389/fnins.2017.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]