Abstract
The first COVID-19 confirmed case was reported in Wuhan, China, and spread across the globe with an unprecedented impact on humanity. Since this pandemic requires pervasive diagnosis, developing smart, fast, and efficient detection techniques is significant. To this end, we have developed an Artificial Intelligence engine to classify the lung inflammation level (mild, progressive, severe stage) of the COVID-19 confirmed patient. In particular, the developed model consists of two phases; in the first phase, we calculate the volume and density of lesions and opacities of the CT scan images of the confirmed COVID-19 patient using Morphological approaches. The second phase classifies the pneumonia level of the confirmed COVID-19 patient. We use a modified Convolution Neural Network (CNN) and k-Nearest Neighbor; we also compared the results of both models to the other classification algorithms to precisely classify lung inflammation. The experiments show that the CNN model can provide testing accuracy up to 95.65% compared with exiting classification techniques. The proposed system in this work can be applied efficiently to CT scan and X-ray image datasets. Also, in this work, the Transfer Learning technique has been used to train the pre-trained modified CNN model on a smaller dataset than the original dataset; the modified CNN achieved 92.80% of testing accuracy for detecting pneumonia on chest X-ray images for the relatively extensive dataset.
Keywords: COVID-19, Deep learning, Coronavirus detection, CT scan image, CNN, kNN, Transfer learning
Introduction
The novel COVID-19 started in Wuhan city, Hubei Province of China, in November 2019. In December 2019, the world health organization (WHO) announced that the virus could cause a severe respiratory disease characterized clinically by fever, cough, and lung inflammation. Even though the virus initially appeared in China, it has been recognized in numerous countries around the world [1, 2]. WHO announced this virus as a public health emergency on January 30, 2020. This is because of its fast spread and because most infected people have no immunity against this virus.
COVID-19 spreads among humans and animals such as cats, camels, bats, and cattle. Primary cases of COVID-19 had links with live animal shops and seafood at the epicenter in Wuhan, suggesting an animal-to-person route of transmission. Reports show a daily rise of confirmed cases since the outbreak began. Consequently, When the number of confirmed cases reached 118,000, surpassing 4000 deaths, The WHO, on March 11, 2020, announced COVID-19 as a pandemic [3]. Not long ago, Italy passes China in the number of mortality.
COVID-19 and human coronaviruses are classified below the Coronaviridae family. When these viruses infect people, it causes Severe Acute Respiratory Syndrome (SARS), or moderate cold Middle East Respiratory Syndrome (MERS) [4]. SARS is a viral respiratory disease caused by SARS-associated coronavirus (SARS-CoV); it was first reported in 2003 in Southern China and spread to several countries worldwide. Furthermore, MERS virus cases were first reported in Saudi Arabia, causing 858 deaths. This virus is believed to have appeared in bats; this belief depends on examining virus genomes [5].
The severity of coronavirus disease 2019 symptoms have been classified from very mild to severe; symptoms that may appear after the first two days of exposure are fever, headache, dry cough, shortness of breath addition to the loss of smell or taste, and symptoms might be changing after day 14 to persistent pain or pressure in the chest, confusion and breathing trouble. Many techniques apply to diagnose COVID-19, such as the Computed Tomography (CT) scan, Nucleic Acid Test (NAT), and X-ray image. The NAT is used to recognize precise nucleic acid sequence and species of organism, mainly bacteria or virus, which causes disease in tissue, blood, or urine. Even though NAT and detection kits play important roles in identifying COVID-19, CT scans and X-ray images are still the most efficient and practical methods for detecting the severity of the lung inflammation expected to be correlated with COVID-19 [6]. The embodiment of radiographic appearance of pneumonia for clinical diagnostic standards in Hubei province was approved by the National Health Commission of China [7], which confirms the importance of CT scan images in diagnosing COVID-19 pneumonia severity.
Some experts believe that congregating COVID-19 cases with others may lead to more infections, and hospitals become a hotspot of infection while waiting to have CT scan Image testing. In addition, the radiologists’ number is fewer than the number of patients, which may result in late diagnosis and quarantine of infected people and less effective treatment of patients [7]. Consequently, for instance, in Italy, hospitals have prioritized patients with more severe manifestations such as high fever and breathing difficulty over others with less severe symptoms [8].
The fast transmission of COVID-19 and the increased demand for diagnosis have encouraged researchers to evolve more intelligent, highly sensitive, and effective diagnostic methods that might help stem the spread of COVID-19 disease. The radiologists manage the diagnosis method as the manual measure of the lung infection quantity. Furthermore, Al-based automated pneumonia diagnosis distinguishes the lesions’ density, size, and opacities in COVID-19 confirmed cases. These algorithms can analyze CT scan images’ outcomes in a little while compared to other available ways [9].
The chest CT scan images utilized by the radiologist to monitor the confirmed cases vary from early to serious stages [9]. The rapid progression of lung infection demands several CT scan images, understanding and analyzing these Images takes too much time and is challenging, particularly when the lesion measurements should be done manually on the CT scan and X-ray images. Therefore, the evolvement of the intelligent algorithm is urgently needed to precisely and automatically detect COVID-19 cases. Moreover, it is essential to establish a complete and ready-to-experiment dataset for the research community.
In this paper, we propose an Artificial Intelligence (AI) engine to classify the lung inflammation level of the COVID-19 confirmed patient. In particular, the developed model consists of two phases; in the first phase, we calculate the volume and density of lesions and opacities of the CT scan images of confirmed COVID-19 patients using the Morphological approach. In the second phase, the second method classifies the pneumonia level of the confirmed COVID-19 patient. To achieve the precise classification of lung inflammation, we used a modified Convolution Neural Network (CNN) and k-Nearest Neighbor (kNN); we also compared the results for both models to the other classification algorithms. The experiments show that the utilized models can provide accuracy up to 95.65% and accuracy using the modified CNN. We also used the Transfer Learning technique to train the pre-trained modified CNN model on a smaller dataset than the original dataset; the modified CNN achieved 92.80% of testing accuracy for detecting pneumonia on chest X-ray images for the relatively extensive dataset. The proposed system in this work can be applied efficiently to CT scan and X-ray image datasets.
The rest of this paper is organized as follows. Section 2 provides the literature review of recent advances in developed AI systems for COVID-19 detection. Section 3 presents an overview of the proposed approach and details of the designed algorithm. Section 4 presents details of the dataset, materials, and experimental results. Section 5 presents an overview of the Transfer Learning technique, the experiments of training the modified CNN model with the Transfer Learning, and the results. Finally, Sect. 5 concludes the paper.
Related work
In this section, the review of state-of-the-art methods used for the diagnosis of novel COVID-19. Many methods are utilized to distinguish viral pneumonia in dubious cases.
Because of the new development of COVID-19, there are a small number of up-to-date reviews in this field, even though there is scarce literature on the diagnosis of this virus. The researchers developed an Al engine based on deep learning to identify COVID-19 using super-resolution CT images [1]. However, their suggested model depends exclusively on CT images. In reliance on the newest research [10], detecting COVID-19 outcomes is more credible when multiple methods are used together. In another try, an intelligent reading system for CT images, developed by Ping An Insurance Company of China Ltd [10], can be read and examined briefly.
As mentioned earlier, the typical way of diagnosing COVID-19 pneumonia is by using a CT scan or X-ray images. In [1], the authors have researched to build a diagnosis system depending on deep learning. The system works in diagnosing COVID-19 with super-resolution CT scan images, which helps radiologists with their work and assists in controlling the pandemic. It was fascinating that they could collect 46,096 unknown images from 106 admitted patients, including 51 cases with confirmed COVID-19 pneumonia. Also, 55 admitted control patients of other diseases have collected in the Renmin Hospital of Wuhan University in Wuhan city, Hubei Province of China. These CT scan images were collected to develop and train their suggested model. Moreover, to assess and compare the accuracy of radiologists versus COVID-19 pneumonia with the performance of their model.
It is crucial to point out that their suggested deep learning model has displayed an equal performance compared with the radiologists. Furthermore, the proficiency of radiologists could also develop while performing their clinical practice, which is important with such outbreaks status when cases rapidly increase. Therefore, we can point out an argument: owning such a diagnosing system provides multiple potentials to reduce the heavy responsibilities of radiologists and provide an in-time diagnosis of COVID-19. Moreover, early detection of such an extremely infectious pandemic will further help in the plans of isolation and remedy, and finally helps countries in restraining and ending this pandemic [1].
There was an extremely important argument in [1] related to the importance of CT Scan Images in the detection of COVID-19 cases, which can be deemed much faster compared to the traditional method utilizing nucleic acid detection. Besides its effectiveness in diagnosing the disease, it can assess the severity level of pneumonia [11]. With all 140 laboratory-confirmed COVID-19 cases, CT results were reported positive. Moreover, a CT scan could identify these positive cases even within their early stage, showing its efficiency [12, 13]. On the other hand, the 5th version of the COVID-19 diagnostic manual has been reported by the National Health and Health Commission of China; the pneumonia radiographic features are integrated with the clinical diagnostic standard in Hubei province.
After that, to highlight the effectiveness and proficiency of CT scan images in COVID-19 diagnosis, in a single day, as was reported, 14,840 direct cases of the infection were mentioned by the 13th of February 2020 in Wuhan; this number included 13332 events of clinical diagnosis. All these proofs emphasized the importance of CT scan images in COVID-19 pneumonia diagnosis.
Considering the discussion mentioned above, the researchers in [1] were primarily concentrating on accomplishing a model that could nearly diagnose COVID-19 in the same way that radiologists do, but with a shorter time. They could attain a similar performance to that of the expert radiologist with 65% less time to diagnose cases than in-clinic radiologist time. This will allow the patients and suspected cases to have a self-check system, as a result, so they save time and prevent direct contact that may consequently transmit the virus to the doctors and nurses.
On the other hand, the authors in [9] have mentioned that till the time of writing their report, there is no such automatic set of tools to clinically identify the stage of COVID-19 infection. For this goal, they have presented Deep Learning (DL)-based system to automatically segment and identify the spots of infection in COVID-19 patients and get a complete view of the lung to be isolated from the chest CT scan. To perform image segmentation, a DL-based network named VB-Net was suggested. Also, authors in [9] have proposed a 3-D convolutional neural network which corporations V-Net [14] together with the bottleneck structure presented in [15]. The adopted VB-Net is exhibited in Fig. 1, which requires two paths, the dashed boxes highlighting the utilized bottleneck elements within their V-shaped network.
Fig. 1.
Phase transition diagram to classify the severity of COVID-19 patient
In contrast, the authors in [9] proposed DL-based segmentation based on the “VB-Net” neural network to extract all COVID-19 infection areas out of CT scan images. They have trained their gadget with 249 COVID- 19 cases and validated it with 300 new COVID-19 cases. We can see that they have additionally brought a Human-in-the-loop (HITL) method to be used at some point in the coaching segment of their model to speed up the procedure of the manual definition of CT scan images. This strategy has helped radiologists by tagging some footnotes on each case’s scanned image. The proposed model was implemented and tested across the conventional way of diagnosing the COVID-19 to locate that the proposed system could minimize four minutes after three iterations of model updating, in contrast with the full guide delineation, which normally may also take time diverse between 1 and 5 h.
In another attempt, Adrian Rosebrock in [16] proposed a DL model for diagnosing COVID19 using Keras library and TensorFlow training platform. Using 50 images, which have been equally divided into 25 positive COVID-19 and 25 negative X-rays images, they have to build, educate and validate their model. Out of the performed experiments, their proposed model should diagnose COVID-19 with an average accuracy of 90–92%, which was once applied on their testing set with 100% sensitivity and 80% specificity due to the restricted data. It is also indispensable to highlight that having an exceedingly robust COVID-19 diagnoses system should be executed with a multi-modal, which processes multiple factors such as patient vitals, population density, geographical location, and others. Therefore, having a diagnosis system relying only on X-ray images will not be that reliable in ending such an excessive threat pandemic of COVID-19.
The proposed approach
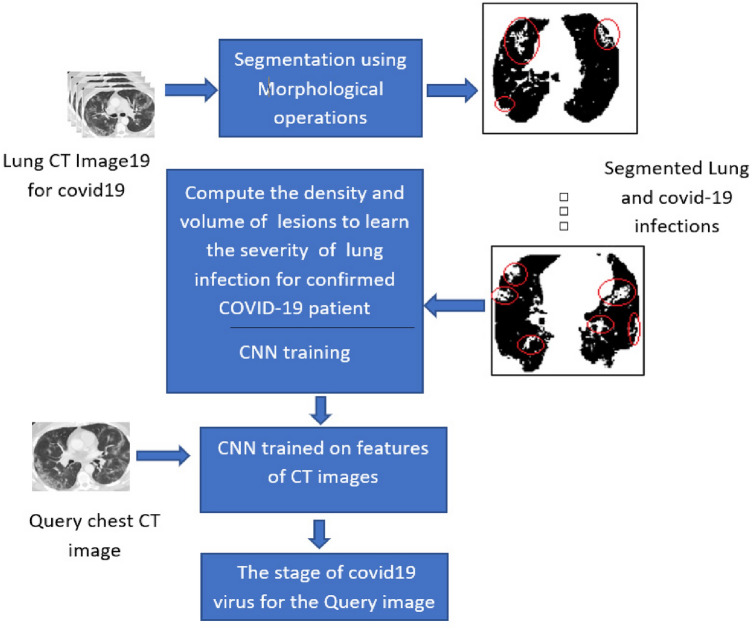
The proposed work contains two main phases: calculating the weight for the virus stages scene using a Morphological approach for the CT scan images, and the Recognition Stages of the COVID-19 virus. CNN and kNN have been built with feeding CT scan images for classification purposes. A detailed explanation of the feature extraction algorithm is proposed in the next section by considering the processing and clustering of the features.
Weight calculation phase
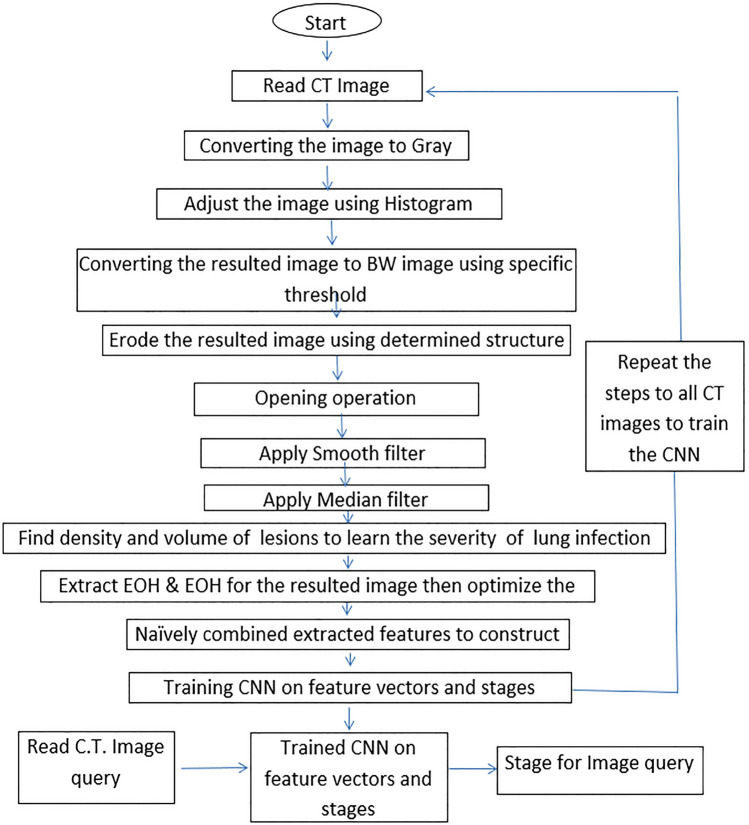
In this phase, the pre-processing and clustering of the features are calculated and fed to the CNN. This process segments the infected lung lesion and enhances the image by spotting the infected lung regions. Subsequently, extracting important features of the lungs, including lesion regions. These features Naïvely were merged to enhance the feature description of the infected lung. The steps of the proposed algorithm for extracting feature vectors for the images are illustrated in the diagram presented in Fig. 2. The process will apply to all the images needed to train the system on the cases inside the CT images to be classified the other cases based on these trained images.
Fig. 2.
Feature vectors extraction using morphological approach
The pre-processing mechanism is applied based on the morphological approach by memorizing a set of features for CT scan images. These features are useful for diagnosing the severity of lung inflammation (mild, progressive, and severe). In addition, normal CT scan cases based on weight represented by the white pixels disappeared in the two sides of the lung.
Most morphological operations have made the lung lesion regions clearer; this will affect the extraction of the feature vectors. The morphological operations on the original images and the result will round the lesion regions caused by COVID-19 to be white pixels in the black lung regions; this makes the calculations of the infected regions by the COVID-19 very trivial. Figure 3 shows the image after applying the operations of the steps, which are listed in Fig. 2. Two algorithms, Histogram of Gradient (HOG) and Edge of Histogram (EOH), are extracted the significant features to be fed to the CNN. The volume and density of the lesions in the left and right lungs depend on the duration where COVID-19 remains in the lungs. For example, the infected lung image containing small volume and density lesions indicates mild stages of the disease, while the large lesion regions of infected lungs indicate the severe stage for the confirmed COVID-19 patient.
Fig. 3.
CT scan images after Morphological Operation a the original image, b the processed image
Histogram of gradient (HOG)
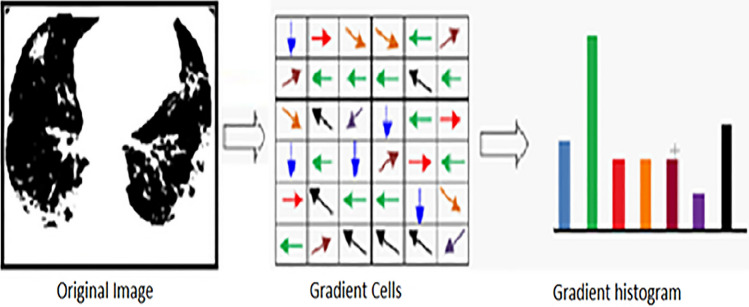
The CT scan images are subdivided into windows of images (cells); these windows represent small spatial windows or regions (cells), the histograms for each region will generate a local histogram of these partition gradient directions, which will be accumulated together. Combining histograms of the regions separately will form the whole image representation. After that, the normalization for these local histograms could be established, or some measurements on these local histograms to differentiate the local responses for the CT images considered. This leads to the greater spatial region that can be exploited for significantly normalized descriptors such as Histogram of Oriented Gradient [17]. These descriptors’ promise results and realization make the technique efficient in analyzing these CT scan images and combining them naively with EOH features after applying morphological operations to extract more distinctive descriptors for lesion positions in the lung. Figure 4 shows a representation of feature construction for the image. The CT image will be subdivided into cells into small areas connected. Then, each cell containing pixels casting channel orientation is based on the resulted value of gradient computation. From 0 to 180 degrees, the histogram channels are spreading equally. Afterward, collecting these histograms of gradient directions gives the features vector for the image.
Fig. 4.
CT scan images after Morphological Operation a the original image, b the processed image
Edge of histogram (EOH) method
The directions of edges inside the CT scan images give significant features that indicate some extra lung lesions, coming from the local orientation of edges inside the CT image, which can be used as descriptors. Edge Orientation Histograms (EOH) is exploited to extract the most important features for describing the two side of the lung. The histograms are built by starting with calculating the orientation of the edges inside the image. This will be performed by filtering the edge inside the image. This filtering process uses two kernels: And to obtain filtered CT images directed by dx and dy, respectively. In this process, two arguments are considered, the direction and the magnitude (M) for the pixel’s edges inside the image. These are computed by Eqs. 1 and 2 respectively.
| 1 |
| 2 |
The bins related to edge orientation in the histogram are equally distributed through the direction of the edge. Further, the number of histogram bins depends on the edge magnitude, then the calculation of the histogram will be related to the weighted vote [18]. Figure 5 illustrate the impact of the edge detection over COVID-19 confirmed cases and normal CT images. Combining HOG and EOH features Naively Learning visual elision recognition is constructing more variant features for the lung as expressed in Eq. 3:
| 3 |
x is the CT scan image processed morphologically, Hg is the histogram of oriented gradient, and Eh is the edge histogram.
Fig. 5.
Edge description a Processed image, b Edge detection image
Convolutional neural network (CNN)
This section presents the design of the proposed CNN algorithm used to identify the severity level of COVID-19 confirmed cases and details of proposed CNN model implementations.
The result of extracting feature vectors is converted to the square matrix to be used as input to the CNN for training. For validation and testing of CNN, CT images of the dataset are processed according to the same steps illustrated in Fig. 2, and the attained features from the morphological phase are prepared to feed CNN. In this paper, 15 layers have been used to train the CNN to identify the severity of COVID-19 cases. These layers comprise one layer for input, 3 of them as convolution layers with 3 3 filter size, two layers for max pooling, three layers for batch normalization, three layers for the rectified linear unit, one layer for fully connected, one layer for softmax, and one layer for classification.
1-Input layer This layer is responsible for reading features crop and resizing CT images to make them ready for the next layer [19]. The CT images are collected from different repositories. Thus, the preprocessing mechanism on C.T. images makes the results more accurate.
2-Convolution layers These layers are considered major building blocks of CNN that they represent the applications of filters to the input layer. Each layer has convolutions and kernels with a stable stride run over the CT complete image. Here the interior features of images will be extracted and passed to the pooling layer Eq. (4) represents the convolution operation [20]:
| 4 |
where is the convolution kernel between output y map and input x map and the sign indicates to convolution operation.
3-Max Pooling Layer This layer is an operation typically added to CNN following individual convolution layers. This layer shrinks the dimensionality of big-size CT images by reducing the number of pixels in the output of previous convolution layers. The maximum value will be retained; eventually, every feature in the window will keep the finest fits. The max-pooling process is illustrated in Eq. 5.
| 5 |
4-Rectified Linear Units Layers (ReLU) The negative numbers resulting from the pooling layer will be considered zeros in this layer. This layer makes the CNN reach a steady state. It is represented by Eq. 6:
| 6 |
5-Batch normalization layer This layer is considered an important part of CNN to reduce the number of training epochs. This subsequently stabilizes the learning process of CNN. It supplies an introduction for input feed-forwarding and calculating gradients about the parameters [21].
6-Softmax layer This function characterizes a CNN to provide a solution for a classification problem. It identifies the discrete probability P allocation for K classes. This can be indicated by . Suppose x is the activation, and is weight parameters at the softmax layer, then o is the input to the softmax layer,
| 7 |
subsequently
| 8 |
Therefore, the prediction of the class would be:
| 9 |
for
k-nearest neighbor (kNN)
kNN is a supervised machine learning algorithm; it is a simple and common method to categorize natural scene objects. The process of the model is built on one notion the similarity between observations and groups which relate to each other [22]. The model’s algorithm has two phases, which are training and testing. The training phase builds the training dataset using a set of cases holding training patterns with its related class.
The query begins with a given unlabeled point in the testing phase, and the algorithm generates a set of k closest or nearest (results) scores corresponding to the trained input patterns. The similarity results of two feature vectors are estimated using distance measures such as Euclidian or Manhattan. Finally, the classification or decision is established by tagging the class of a tested pattern based on the majority voting method.
Experimental setup
This section presents the results of the proposed CNN and kNN models. The feature vectors are constructed naively using 89 features (81 extracted from HOG, 8 retrieved from EOH).
CT scan dataset
In this work, we used a CT scan images dataset collectively available in a GitHub repository. It was constructed by a group in [23] on several confirmed COVID-19 patients. The COVID-19 dataset comprises 186 CT scan images of pneumonia Lungs. In particular, the images clearly show the variation of lung inflammation in COVID-19 patients between 1- and 21 Days. The scanned image size is 346 442 3 unit 8. We currently do not have enough COVID-19 images publicly available to the research community to conduct an intense investigation. There is an immediate need to collect more radiology images accessible by the research community.
COVID-19 severity classification using CNN
The experiments are conducted using CNN on the CT images dataset; the dataset is divided into two groups: testing and training datasets. Moreover, the Weka Multilayer perceptron’s deep learning has been used to determine the accuracy of the proposed approach. Figure 7 shows the accuracy performance measure to find the lung infection’s severity level. Further, Fig. 8 classified the query CT image as a severe level of lung inflammation. This is because the volume and density of lesions are very high.
Fig. 7.
Diagnosing the COVID-19 confirmed patient pneumonia stage
Fig. 8.
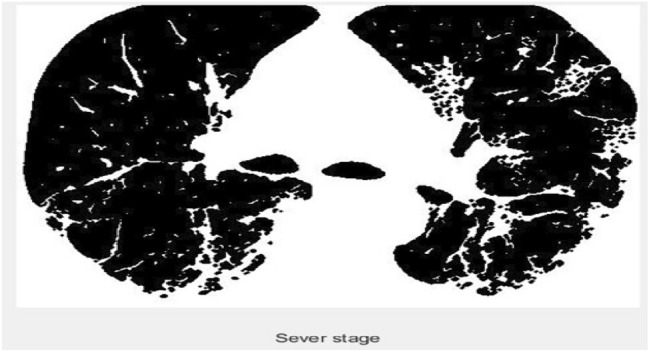
Diagnosing the COVID-19 confirmed patient pneumonia stage
COVID-19 severity classification using kNN
The experiments were also conducted using kNN on the CT images dataset. Euclidean’s distance is applied to the training and testing feature vectors. These distances are sorted using a tested CT image to determine the smallest distance. Figure 6 shows the result of checking the query test image and the results of the kNN algorithm on the data set.
Fig. 6.

The 4 distances between the Query image and the other images
The precision p of the N retrieved CT images for the query Q CT image is expressed by following equation:
| 10 |
where g(Q) represents the group stages of the virus for the query image and Ir is the retrieved CT image. The results for all created distance feature values are sorted; then, the minimum values are the best matching virus stage.
Results and discussion
In this work, the accuracy of pneumonia level detection during a course of confirmed COVID-19 patients has been calculated. To achieve this goal, morphological operations, the CNN and kNN models are applied to the extracted feature vectors using HOG and EOH. GitHub dataset (COVID-19 dataset includes a set of CT scan images for confirmed COVID-19 patients) repository is utilized to extract feature vectors. The image contains two sides of the patient’s lungs; the dataset consists of several stages (early stage, progressive, and severe). The dataset is composed of 186 CT images of size 372 556 3 unit 8. Morphological operations are applied to convert the dataset to logical images. In this operation, noises within the images are filtered out to make the lesions and opacities on the infected lungs appear more clearly—the feature vector constructed from HOG and EOH to get higher invariant features for classification. When CNN is used to find the severity stage of the lung infection, we obtained a testing accuracy of 92.307% for HOG and 95.6% for combined featured HOG and EOG, while we obtained 87.5% for kNN of HOG and combined featured; using kNN for classification indicates that combining features does not affect kNN, this is because of the nature of the data. We also applied experiments on different classification models and compared the performance to CNN and kNN. Table 1 presents the testing accuracy of classification methods applied on different feature vectors to see the effect of these features.
Table 1.
Accuracy performance of common classification models
| Model | HOG (%) | HOG + EOH (%) |
|---|---|---|
| Convolutional Neural Network (CNN) | 92.307 | 95.652 |
| k-Nearest Neighbor (kNN) | 87.50 | 87.50 |
| Decision Tree (DT) | 80.769 | 86.956 |
| Trees random forest (TRF) | 88.461 | 92.95 |
| Bagging | 76.923 | 91.304 |
| Bayes Net | 88.461 | 93.655 |
| Naïve Bayes | 80.769 | 91.304 |
Table 1 shows that The Decision Tree (DT) provides an accuracy of 80.769% for HOG and 86.956% for combined features of HOG and EOG; While in Trees Random Forest (TRF) was 88.461% for HOG and 95.63% for combined features, the situation is better than DT, the reason is that TRF creates a number of trees and take the voting results; The bagging procedure provides 76.923% for HOG and 91.304% for combined features, while the Bayes Net provides 88.461% for HOG and 93.655% for combined features, and lastly, the Bayes Net provides 80.769% for HOG and 91.304% for combined features. We can observe that the Bayes Net and Naïve Bayes provided promising results, but the two models will not provide the same results when they use only a single feature. Therefore they can not be compared to the results obtained by CNN here. However, ensemble CNN gave higher accuracy most of the time than the kNN and other classification models, with a testing accuracy of 95.652% for combined features.
Transfer learning
One of the machine learning techniques is Transfer Learning, whereby a model is developed and trained to perform one specific task and then reused on a second related task. Transfer Learning refers to learning in a specific setting that can be exploited for optimization improvement in another setting [24]. Transfer Learning can be applied when there is a dataset smaller than the original dataset used to train the pre-trained models [25].
In Sect. 4, we confirmed that the modified CNN had performed better than other classification models used in this work. Therefore, we decided to train the existing architecture of the modified CNN model proposed in Sect. 3 with Transfer Learning; the two algorithms, Histogram of Gradient (HOG) and Edge of Histogram (EOH), will extract the significant features to be fed to the modified CNN, the feature vectors are constructed naively using 89 features (81 extracted from HOG, 8 retrieved from EOH); the modified CNN model was first trained on a relatively small dataset (CT scan dataset) and reused to learn features (or transfer features) and train them on a new dataset (Chest X-Ray Images (Pneumonia)) [26], the training dataset contains around 624 chest X-ray images, 234 image was normal, which means for patients with no pneumonia, and 390 images for patients with pneumonia. All the chest X-ray images were collected as a part of the patient’s routine clinical care. To analyze the chest X-ray images, all the chest radiographs were screened for quality control by removing all the unreadable or low-quality scans.
With regards to the initial training, Transfer Learning allows us to start with features that are learned by the model on the (CT scan) dataset and adjust these features and sometimes the model’s structure to target the (Chest X-Ray Images) dataset or task instead of starting the learning process on the (Chest X-Ray Image) dataset from scratch with random weight initialization. To achieve this, we used TensorFlow to facilitate Transfer Learning of the modified CNN pre-trained model. We also study the modified CNN architecture topology to find a suitable model while changing and testing the network topology (i.e., parameters) and some characteristics related to the dataset to help us to determine which variable can affect the classification accuracy with limited computational power and time. After 100 epochs (the iterations through the entire training dataset) of the modified CNN model, the model’s training was stopped due to the absence of further improvement in accuracy and loss.
For testing, we tested our CNN model trained with Transfer Learning on 5216 chest X-ray images; 1341 image was normal, and 3875 images were for patients with pneumonia. When we trained the model, it retained high performance in accuracy, sensitivity, specificity, and the area under the Receiver Operating Characteristic curve (ROC curve) for obtaining the correct diagnosis; this shows the power of the Transfer Learning system by making an effective classification even with the relatively small or limited training dataset.
Comparing chest X-rays, which present pneumonia versus normal, we achieved a testing accuracy of 92.8%, a sensitivity of 93.2%, and specificity of 90.1%, when the area under the ROC curve for pneumonia detection from normal was 96.8% for combined features of HOG and EOG. Moreover, the Transfer Learning technique efficacy described here can potentially be employed in different medical image disciplines. Here we provide a direct illustration of its broad applicability by demonstrating its efficacy in analyzing chest X-ray or CT scan images. Figure 9 shows the overall performance results of the modified CNN model trained with Transfer Learning to detect pneumonia versus normal.
Fig. 9.

Modified CNN trained with Transfer Learning performance results
Conclusion
In this paper, CNN, kNN, and other popular classication algorithms have been used to detect the lung’s pneumonia level using CT scan images. This work is adopted to improve diagnosing lung inammation severity in conrmed COVID-19 patients. We built a complete pre-processed dataset of CT scan images from the GitHub repository. After extensive experiments, the results show that the proposed CNN with combined HOG and EOH or HOG as a single feature signicantly outperforms the others. Furthermore, when a morphological operation based on EOH is used, the result is not that promising but makes the features more invariant. This indicates that Multi-scale features for medical image recognition are better than single-scale features. The morphological operation is more beneficial to lter out noises and unnecessary features in the classication, resulting in high accuracy of lung inammation detection. Also, in this work, the Transfer Learning technique has been used to train the pre-trained model on a smaller dataset than the original dataset; the modified CNN model achieved 92.80% of testing accuracy for detecting pneumonia on chest X-ray images for the relatively extensive dataset. The proposed system in this work can be applied efficiently to CT scan and X-ray image datasets.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest. Moreover, this research was not funded by any funding agency.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Abbas M. Ali, Email: abbas.mohamad@su.edu.krd
Kayhan Ghafoor, Email: kayhan.ghafoor@knu.edu.iq.
Aos Mulahuwaish, Email: amulahuw@svsu.edu.
Halgurd Maghdid, Email: Halgurd.Maghdid@koyauniversity.org.
References
- 1.Chen J, Wu L, Zhang J, Zhang L, Gong D, Zhao Y, Hu S, Wang Y, Hu X, Zheng B, et al (2020) Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study. medRxiv, Cold Spring Harbor Laboratory Press
- 2.Holshue ML, DeBolt C, Lindquist S, et al. Novel coronavirus in the United States. N Engl J Med. 2020;31:2019. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CNN Health https://edition.cnn.com/2020/03/11/health/coronavirus-pandemic-world-health-organization/index.html. Accessed March 2020
- 4.Novel Coronavirus (2019-nCoV). World Health Organization https://www.who.int/emergencies/diseases/novel-Coronavirus-2019. Accessed March 2020
- 5.Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003 (2004) World Health Organization. https://www.who.int/csr/sars/country/table20040421/en/
- 6.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus infected pneumonia in Wuhan, China. Jama Netw J. 2020;323:1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.National Health Commission of China (2020) http://www.chinadaily.com.cn/m/chinahealth/index.html
- 8.Canada Broadcast Cooperation (2020) https://www.cbc.ca/player/play/1709650499517
- 9.Shan F, Gao Y, Wang J, Shi W, Shi N, Han M, Xue Z, Shen D, Shi Y (2020) Lung infection quantification of COVID-19 in CT images with deep learning. arXiv preprint arXiv:2003.04655
- 10.BioWorld (2020) https://www.bioworld.com/articles/433530-china-uses-ai-in-medical-imaging-to-speed-up-covid-19-diagnosis
- 11.Zu ZY, Jiang MD, Xu PP, Chen W, Ni QQ, Lu GM, Zhang LJ. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. 2020;296:E15. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milletari F, Navab N, Ahmadi S-A (2016) V-net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 fourth international conference on 3D vision (3DV), pp 565–571
- 15.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
- 16.Roquette BP, Nagano H, Marujo EC, Maiorano AC. Prediction of admission in pediatric emergency department with deep neural networks and triage textual data. Neural Netw. 2020;126:170. doi: 10.1016/j.neunet.2020.03.012. [DOI] [PubMed] [Google Scholar]
- 17.Jiang Y-G, Ngo C-W, Yang J (2007) Towards optimal bag-of-features for object categorization and semantic video retrieval. In: Proceedings of the 6th ACM international conference on Image and video retrieval, pp 494–501
- 18.Timotius IK, Setyawan I (2014) Evaluation of edge orientation histograms in smile detection. In: 2014 6th international conference on information technology and electrical engineering (ICITEE), pp 1–5
- 19.Sharma N, Jain V, Mishra A. An analysis of convolutional neural networks for image classification. Procedia Comput Sci. 2018;132:377–384. doi: 10.1016/j.procs.2018.05.198. [DOI] [Google Scholar]
- 20.Chen Z, Jacobson A, Sünderhauf N, Upcroft B, Liu L, Shen C, Reid I, Milford M (2017) Deep learning features at scale for visual place recognition. In: 2017 IEEE international conference on robotics and automation (ICRA), pp 3223–3230
- 21.Schilling F (2016) The effect of batch normalization on deep convolutional neural networks
- 22.Bin Abdullah A. Supervised learning algorithms for visual object categorization. Universiteit Utrecht; 2010. [Google Scholar]
- 23.Joseph PC, Paul M, Lan D (2020) COVID-19 image data collection. vol. 6. arXiv:2003.11597
- 24.Gao Y, Mosalam KM. Deep transfer learning for image-based structural damage recognition. Comput Aided Civ Infrastruct Eng. 2018;33(9):748–768. doi: 10.1111/mice.12363. [DOI] [Google Scholar]
- 25.Larsen-Freeman D. Transfer of learning transformed. Lang Learn. 2013;63:107–129. doi: 10.1111/j.1467-9922.2012.00740.x. [DOI] [Google Scholar]
- 26.Kermany DS, Michael G, Wenjia C, Carolina CSV, Huiying L, Sally LB, Alex M, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]