Abstract
In Drosophila melanogaster, the pacemaker located in the brain plays the main role in maintaining circadian rhythms; however, peripheral oscillators including glial cells, are also crucial components of the circadian network. In the present study, we investigated an impact of oscillators located in astrocyte-like glia, the chiasm giant glia of the optic lobe, epithelial and subperineurial glia on sleep of Drosophila males. We described that oscillators located in astrocyte-like glia and chiasm giant glia are necessary to maintain daily changes in clock neurons arborizations, while those located in epithelial glia regulate amplitude of these changes. Finally, we showed that communication between glia and neurons through tripartite synapses formed by epithelial glia and, in effect, neurotransmission regulation plays important role in wake-promoting during the day.
SIGNIFICANCE STATEMENT Circadian clock or pacemaker regulates many aspects of animals' physiology and behavior. The pacemaker is located in the brain and is composed of neurons. However, there are also additional oscillators, called peripheral clocks, which synchronize the main clock. Despite the critical role of glia in the clock machinery, little is known which type of glia houses peripheral oscillators and how they affect neuronal clocks. This study using Drosophila shows that oscillators in specific glia types maintain awakeness during the day by regulating the daily plasticity of clock neurons.
Keywords: circadian clock, insect behavior, neuronal plasticity, sleep
Introduction
Glial cells are crucial for proper functions of the nervous system. They regulate development and synaptogenesis, metabolism of neurotransmitters and transport of ions and other compounds to neurons. Based on the topology and cell shape, glial cells of the central nervous system in Drosophila are divided into three main classes: surface glia (perineurial and subperineurial), cortex glia surrounding somata of neurons, ensheathing glia wrapping neural processes and astrocyte-like glia known also as neuropil glia. In the fly's visual system glial cells are highly specialized and additional classes of glia have been distinguished: fenestrated, pseudocartridge, epithelial and marginal glia (Hartenstein, 2011). Some glial cells in the brain express per and other clock genes and function as peripheral oscillators (glial clocks; Ewer et al., 1992; Damulewicz et al., 2015; Long and Giebultowicz, 2018); however, their activity can be regulated by the main pacemaker as some of them express pigment-dispersing factor (PDF) receptors (Im and Taghert, 2010; Damulewicz et al., 2015; Górska-Andrzejak et al., 2018). Glial clocks affect the circadian rhythm of the locomotor activity (Suh and Jackson, 2007; Ng et al., 2011) and probably modulate function of clock neurons by releasing gliotransmitters (for review, see Jackson et al., 2015). They are also important for maintaining rhythms in the visual system, for example in changes of size and shape of axons and dendrite of interneurons (Pyza and Górska-Andrzejak, 2004; Weber et al., 2009). Glial cells contact other glia or neurons by septate, gap, and adherent junctions, and are involved in the formation of tripartite and tetrad synapses (aminergic and glutamatergic; Meinertzhagen and O'Neil, 1991; Jackson and Haydon, 2008; Edwards and Meinertzhagen, 2010). Blocking gap junctions with octanol abolishes daily changes in cell size of L1 and L2 interneurons in the first neuropil of the optic lobe (Pyza and Górska-Andrzejak, 2004). Astrocyte-like glia modulates function of synapses by regulating glutamate re-uptake from the synaptic cleft by transporters EAAT1 and Genderblind (Grosjean et al., 2008). Moreover, glial cells can communicate with neurons using exosomes. In mammals, mRNAs, miRNAs and proteins exported from glial cells into exosomes (Smalheiser, 2007) play a role in neurotransmission (Antonucci et al., 2012), neuroprotection (Taylor et al., 2007), immune responses (Fitzner et al., 2011), and other processes (Frühbeis et al., 2012).
In Drosophila, the daily pattern of activity is very well described, with morning and evening bouts of activity, prolonged siesta time in the middle of the day and sleep during the night. This pattern is regulated by different clusters of cells, including clock neurons (Grima et al., 2004; Stoleru et al., 2004; Picot et al., 2007; Cusumano et al., 2009; Zhang et al., 2009; Yao and Shafer, 2014; Chatterjee et al., 2018; Díaz et al., 2019; Schlichting et al., 2019). The most important clock neurons are small ventral lateral neurons (sLNvs), which are necessary to maintain the normal rhythm of sleep and activity. They project to the dorsal brain where PDF, a circadian neurotransmitter, is released. The sLNv terminals show daily plasticity with more complex structure at the beginning of the day than during the night. It has been shown previously that this structural remodeling is also regulated by glial clock (Herrero et al., 2017). Remodeling of neuronal connections is fundamental to drive complex behavior. Daily changes in synapse number and size within PDF processes are involved in the sleep-wake regulation (Fernández et al., 2008; Bushey et al., 2011). LNvs cyclically express miR-210 (Chen and Rosbash, 2017), important for proper development of neuronal processes. Flies with miR-210 overexpression showed abnormal networks of the lLNvs processes, no the daily rhythm in the sLNvs terminal arborization and, in effect, increased total sleep time during the day (Cusumano et al., 2018). Fasciculation of the sLNv processes is also activity dependent (Fernandez-Chiappe et al., 2021), with a dominant role of the transcription factor Mef2 (Sivachenko et al., 2013), miR-92a (Chen and Rosbash, 2017), and dTau, which mutation affects activity pattern and decreases daytime sleep level (Abruzzi et al., 2017; Tracy and Gan, 2018; Arnes et al., 2019).
The role of glia in the sleep regulation is still not fully recognized. The most important role seems to play astrocyte-like glia which is important in sleep homeostasis through Notch (Seugnet et al., 2011) and TNF signaling (Vanderheyden et al., 2018) and modulates sleep level by the fatty acid-binding protein fabp7 (Gerstner et al., 2011), uptake and metabolism of GABA and glutamate (Chen et al., 2015; Farca Luna et al., 2017) as well as by the regulation of taurine transport (Stahl et al., 2018). Moreover, glial cells which form the hemolymph–brain barrier affect sleep through regulating endocytosis (Artiushin et al., 2018).
In the present study we examined several types of glial clocks looking for those which are important for the regulation of sleep. Our results showed that peripheral oscillators located in astrocyte-like glia, epithelial, subperineurial and chiasm giant glia are all involved in maintaining awake and sleep pattern; however, they have specific effects on sleep parameters. In addition, we showed that this effect on behavior is strictly connected with the regulation of sLNv daily neuronal plasticity.
Materials and Methods
Fly strains
The following strains of Drosophila melanogaster were used: repo-Gal4 (Gal4 expressed in all glial cell types), netB-GAL4 (expressing Gal4 in the epithelial glia and medulla chandelier glia, BDSC no. 49478; Edwards et al., 2012), alrm-Gal4 (expressing Gal4 in astrocyte-like glia, BDSC no. 67032; Doherty et al., 2009), moody-Gal4 (subperineurial and pseudocartridge glia, Gal4 on 3. Chromosome; Edwards et al., 2012), Wnt4-Gal4 (chiasm giant glia, BDSC no. 49102; Edwards et al., 2012), UAS-GFP.Valium10 (control for dsRNAi, BDSC no. 35 786), UAS-cycΔ24 (overexpression of dominant negative mutant of CYCLE causing disruption of the circadian clock; Tanoue et al., 2004), UAS-perRNAi (VDRC no. 5711), UAS-dlg1RNAi (BDSC no. 25780), UAS-Eaat1RNAi (VDRC no. 109401), UAS-ebonyRNAi (VDRC no. 104174; strains with expression of dsRNA for specific gene under control of UAS sequence), UAS-TeTx (a strain with expression of tetanus toxin under control of UAS, BDSC no. 28837) and UAS-mCD8-GFP (GFP fused with cell membrane protein, BDSC no. 5130). Driver lines were crossed with Tub-Gal80ts; TM2/TM6B (BDSC no. 7019) to obtain strains with TARGET system. In this system, Tub-Gal80ts transgene encodes a ubiquitously expressed, temperature-sensitive Gal4 repressor that is active at lower temperature (20°C), but not at restrictive temperature (28°C; McGuire et al., 2003). We checked alrm-Gal4 strain carefully as our previous locomotor activity data suggested that it may have perslih allele (Konopka and Benzer, 1971; Hamblen et al., 1998). However, per gene locus is located on X chromosome and to avoid perslih phenotype, we used only males of the alrm-Gal4 strain to cross with Tub-Gal80ts. We confirmed that the progeny obtained from these crosses has period of locomotor activity and morning anticipation similar to wild-type flies, while flies having perslih allele show longer period and no morning anticipation (Hamblen et al., 1998). Therefore, it seems that we were able to avoid this allele in the experimental flies.
Flies were maintained on a standard cornmeal medium under LD12:12 (12 h of light and 12 h of darkness) regime and at constant temperature 20°C. Two-day-old males of the crosses with Tub-Gal80ts were transferred to 29°C for 3 d to induce adult-specific expression of CYC dominant negative mutant in specific glia types. Experimental flies were compared with parental strains backcrossed to w1118 strain.
Behavioral assays
Locomotor activity was recorded at 29°C using Drosophila Activity Monitoring System (DAMS; Trikinetics, Waltham) for 3 d in LD12:12 and next for 5 d in constant darkness (DD). Activity was examined every 1 min and analyzed in Excel by using “Befly!” software (Department of Genetics, Leicester University). Lomb–Scargle normalized periodogram was used to determine rhythmic flies; individuals with period value <10 (confidence level 0.05) were regarded as arrhythmic. Flies which did not survive until the end of experiments were removed from analyses. Every experiment was repeated three times, at least 60 male flies in total were used.
Sleep analysis was performed on the third day of LD12:12, and sleep was recorded as at least 5 min of a fly immobility. Experimental and control flies were analyzed at 29°C.
Immunohistochemistry
Flies were collected at zeitgeber time ZT2 and ZT14 (where ZT0 means the time when the lights are on, ZT12, when the lights are off), their heads were fixed in 4% paraformaldehyde and brains were isolated. After washing in 0.2% PBS with Triton X-100 (PBST) and 30 min of blocking in normal goat serum (NGS) they were incubated overnight with primary antibodies: mouse anti-GFP (1:1000, Novus Biologicals), rabbit anti-PER (1:5000, kindly donated by R. Stanewsky, University of Munster), mouse anti-PDF (1:500, Developmental Studies Hybridoma Bank), rabbit anti-RFP (1:500, TakaraBio). Next, samples were washed in PBST and incubated with secondary antibodies (1:1000, anti-mouse Alexa488 and 1:500, anti-rabbit Cy3, or 1:500 anti-mouse Cy3, respectively). Whole brains were mounted in Vectashield medium (Vector) and examined with a Zeiss Meta 510 Laser Scanning Microscope.
Scholl analysis
To visualize axon projections of sLNvs whole-brain confocal images were used. Pictures were taken using 40× objective with an optical zoom of two. Galleries between 7 and 19 images were projected in the x-y-axes to obtain a reconstruction of the full trajectory of those axons. Sholl's method was used to quantify the axonal arbor in the dorsal protocerebrum. Concentric rings centered at the point where the first dorsal ramification opens up were drawn on each brain hemisphere. The number of intersections of each projection with a particular ring was counted. The total number of intersections were compared (according to Fernández et al., 2008).
Experimental design and statistical analysis
GraphPad Prism software was used for statistics and making graphs. Outliers were removed using Grubbs' test (GraphPad online software). Shapiro–Wilk's test was used to check normality in distribution. Sleep time data showed normal distribution and we used one-way ANOVA with post hoc Tukey's test for analysis. Number of sleep episodes and bout length data which lacked normal distribution were analyzed using nonparametric Kruskal–Wallis test. Total number of flies used for every experiment is provided in Table 1. Statistically significant differences are marked above bars at specific figures and detailed statistics are provided in Tables 2, 3. For analysis of the number of intersections which also had normal distribution we performed t test to compare statistically significant differences between two groups. Detailed statistics are provided in figure legends.
Table 1.
Behavioral analysis of experimental and control flies
| Glia type | Genotype | Period | % rhythmic | N |
|---|---|---|---|---|
| Pan-glial | TubGal80ts; repo> cycΔ | 23.5 | 95 | 74 |
| TubGal80ts; repo>TeTx | 23.4 | 100 | 79 | |
| repo>dlgRNAi | 23.7 | 97 | 99 | |
| TubGal80ts; repo-Gal4/+ | 23.6 | 95 | 69 | |
| repo>Valium | 24.1 | 92 | 70 | |
| Subperineurial, pseudocartridge | TubGal80ts; moody> cycΔ | 24.2 | 89 | 72 |
| TubGal80ts; moody> perRNAi | 24.2 | 93 | 81 | |
| moody>dlgRNAi | 24.2 | 99 | 72 | |
| TubGal80ts; moody-Gal4/+ | 24.2 | 99 | 67 | |
| moody>Valium | 24.0 | 88 | 69 | |
| Epithelial | TubGal80ts; netB> cycΔ | 23.7 | 75 | 61 |
| TubGal80ts; netB>perRNAi | 23.7 | 91 | ||
| TubGal80ts; netB>ebonyRNAi | 23.8 | 100 | 66 | |
| netB>dlgRNAi | 23.7 | 85 | 73 | |
| TubGal80ts; netB-Gal4/+ | 23.8 | 87 | 69 | |
| netB>Valium | 23.6 | 80 | 75 | |
| Outer giant chiasm | TubGal80ts; Wnt4> cycΔ | 23.5 | 74 | 77 |
| TubGal80ts; Wnt4>perRNAi | 23.9 | 88 | 80 | |
| Wnt4>dlgRNAi | 24.3 | 80.3 | 76 | |
| TubGal80ts; Wnt4-Gal4/+ | 23.8 | 88 | 76 | |
| Wnt4>Valium | 24.0 | 85.9 | 72 | |
| Astrocyte-like | TubGal80ts; alrm>cycΔ | 23.3 | 97 | 75 |
| TubGal80ts; alrm>eaatRNAi | 23.4 | 90 | 69 | |
| TubGal80ts; alrm>perRNAi | 23.3 | 82 | 74 | |
| alrm>dlgRNAi | 24.0 | 85 | 60 | |
| TubGal80ts; alrm-Gal4/+ | 23.4 | 81 | 78 | |
| alrm>Valium | 24.1 | 95 | 76 | |
| Control | UAS-cycΔ/+ | 23.9 | 85 | 68 |
| UAS-perRNAi/+ | 23.7 | 100 | 83 | |
| UAS-eaatRNAi/+ | 23.2 | 100 | 66 | |
| UAS-ebonyRNAi/+ | 23.1 | 100 | 66 | |
| UAS-TeTx/+ | 23.3 | 99 | 90 | |
| UAS-dlgRNAi/+ | 23.9 | 98 | 65 |
Table shows period of locomotor activity rhythm, percentage of rhythmic individuals, and total number of flies. Bold text are marked experimental groups (to distinguish from control lines).
Table 2.
Detailed statistics for sleep analysis
| Daytime sleep |
Nighttime sleep |
|||||
|---|---|---|---|---|---|---|
| GAL4 p | UAS p | F (DFn,DFd) | GAL4 p | UAS p | F (DFn,DFd) | |
| TubGal80ts;repo>cyc Δ24 | 0.0011 | <0.0001 | 15.69 (2,237) | 0.0038 | <0.0001 | 17.53 (2,237) |
| TubGal80ts;netB>cycΔ24 | <0.0001 | <0.0001 | 19.61 (2,282) | 0.0269 | <0.0001 | 64.53 (2,282) |
| TubGal80ts;moody>cycΔ24 | <0.0001 | <0.0001 | 30.14 (2,274) | 0.0549 | <0.0001 | 35.59 (2,274) |
| TubGal80ts;alrm>cycΔ24 | <0.0001 | <0.0001 | 35.52 (2,269) | <0.0001 | <0.0001 | 37.99 (2,269) |
| TubGal80ts;Wnt4>cycΔ24 | <0.0001 | <0.0001 | 50.45 (2,216) | 0.0079 | <0.0001 | 24.52 (2,216) |
| TubGal80ts;alrm>perRNAi | <0.0001 | <0.0001 | 18.25 (2,247) | 0.5582 | 0.0002 | 9.355 (2,247) |
| TubGal80ts;Wnt4>perRNAi | <0.0001 | 0.0021 | 56.02 (2,204) | 0.9322 | 0.1396 | 15.33 (2,204) |
| TubGal80ts;netB>perRNAi | <0.0001 | 0.0083 | 9.634 (2,190) | 0.5859 | 0.0817 | 5.308 (2,190) |
| TubGal80ts;moody>perRNAi | 0.3002 | 0.0019 | 5.962 (2,274) | 0.2464 | <0.0001 | 9.563 (2,274) |
| TubGal80ts;alrm>eaatRNAi | 0.5030 | <0.0001 | 17.22 (2,267) | 0.0135 | 0.9998 | 5.592 (2,267) |
| TubGal80ts;netB>ebonyRNAi | 0.0012 | 0.0006 | 8.789 (2,190) | <0.0001 | 0.9305 | 14.29 (2,190) |
| TubGal80ts;repo>TeTx | <0.0001 | 0.0034 | 30.62 (2,269) | <0.0001 | <0.0001 | 40.34 (2,269) |
| repo>dlgRNAi | <0.0001 | <0.0001 | 46.29 (2,212) | 0.0438 | <0.0001 | 84.72 (2,212) |
| netB>dlgRNAi | 0.0446 | <0.0001 | 23.44 (2,233) | <0.0001 | <0.0001 | 48.38 (2,233) |
| alrm>dlgRNAi | >0.9999 | 0.0004 | 9.456 (2,205) | <0.0001 | 0.0024 | 63.33 (2,193) |
| Wnt4>dlgRNAi | 0.9896 | 0.6915 | 0.2939 (2,193) | <0.0001 | <0.0001 | 114.8 (2,193) |
| moody>dlgRNAi | 0.3057 | <0.0001 | 22.66 (2,204) | 0.0487 | <0.0001 | 18.64 (2,207) |
Every experimental strain was compared with control strains (Gal4 and UAS) using one-way ANOVA and Tukey's test. Degrees of freedom [F (DFn,DFd)] are listed for every group.
Table 3.
Number of sleep episodes and bout length during the day and night
| Daytime |
Nighttime |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sleep episodes | Gal4 p | UAS p | Bout length | Gal4 p | UAS p | Sleep episodes | Gal4 p | UAS p | Bouth length | Gal4 p | UAS p | |
| TubGal80ts; repo>cyc24 | 4.7 ± 0.3 | 0.0037 | <0.0001 | 133.4 ± 9.8 | 0.003 | <0.0001 | 6.3 ± 0.4 | <0.0001 | 0.047 | 101.4 ± 12.9 | 0.8871 | <0.0001 |
| TubGal80ts; repo-Gal4/+ | 6.1 ± 0.3 | 87.7 ± 12.8 | 9.2 ± 0.5 | 95.7 ± 15.0 | ||||||||
| TubGal80ts; alrm>cyc24 | 7.0 ± 0.4 | 0.4177 | 0.059 | 95.1 ± 8.2 | 0.0232 | 0.0011 | 8.9 ± 0.4 | 0.3664 | <0.0001 | 79.1 ± 15.1 | <0.0001 | 0.0001 |
| TubGal80ts; alrm>perRNAi | 5.3 ± 0.3 | <0.0001 | 0.0424 | 132.5 ± 11.1 | <0.0001 | <0.0001 | 9.5 ± 0.4 | <0.0001 | 0.181 | 48.2 ± 3.8 | 0.1456 | 0.988 |
| TubGal80ts; alrm-Gal4/+ | 8.7 ± 0.5 | 66.6 ± 3.6 | 14.9 ± 0.7 | 35.3 ± 2.7 | ||||||||
| TubGal80ts; wnt4>cyc24 | 5.7 ± 0.4 | 0.0024 | 0.0171 | 132.8 ± 10.7 | <0.0001 | <0.0001 | 9.7 ± 0.4 | 0.0003 | 0.0614 | 55.9 ± 6.8 | 0.1329 | 0.1696 |
| TubGal80ts; wnt4>peRNAi | 6.6 ± 0.5 | 0.068 | 0.5856 | 101.4 ± 8.8 | <0.0001 | 0.0044 | 12.6 ± 0.8 | 0.5904 | 0.3343 | 63.1 ± 3.31 | 0.4325 | 0.6548 |
| TubGal80ts; wnt4-Gal4/+ | 8.9 ± 0.8 | 65.6 ± 3.9 | 13.5 ± 1.0 | 79.9 ± 15.7 | ||||||||
| TubGal80ts; netB>cyc24 | 7.0 ± 0.5 | 0.4202 | 0.284 | 103.9 ± 7.8 | 0.0003 | 0.0584 | 10.6 ± 0.5 | 0.0013 | 0.0535 | 82.3 ± 12.6 | 0.0002 | 0.1388 |
| TubGal80ts; netB>perRNAi | 12.2 ± 0.9 | 0.0013 | 0.001 | 59.2 ± 6.1 | 0.3271 | 0.9257 | 14.5 ± 1.1 | 0.1395 | 0.0162 | 34.1 ± 3.8 | 0.0636 | 0.4821 |
| TubGal80ts; netB-Gal4/+ | 8.31 ± 0.7 | 73.5 ± 6.5 | 12.5 ± 0.8 | 57.2 ± 15.0 | ||||||||
| TubGal80ts; moody>cyc24 | 5.4 ± 0.4 | 0.0085 | 0.0008 | 149.0 ± 14.6 | <0.0001 | <0.0001 | 10.5 ± 0.5 | 0.0064 | 0.1224 | 48.4 ± 6.8 | 0.4934 | 0.0488 |
| TubGal80ts;moody>perRNAi | 7.5 ± 0.6 | 0.5953 | 0.9877 | 49.7 ± 8.6 | 0.1932 | 0.8016 | 11.7 ± 0.9 | 0.9067 | 0.8982 | 24.3 ± 2.5 | 0.0039 | 0.5143 |
| TubGal80ts; moody-Gal4/+ | 8.4 ± 0.6 | 78.7 ± 7.1 | 12.1 ± 0.7 | 82.5 ± 16.9 | ||||||||
| UAS-cyc24/+ | 7.9 ± 0.5 | 57.4 ± 5.1 | 7.9 ± 0.7 | 32.0 ± 3.5 | ||||||||
| UAS-perRNAi/+ | 7.7 ± 0.6 | 63.9 ± 6.3 | 11.2 ± 0.8 | 46.9 ± 6.6 | ||||||||
Experimental flies with clock disruptions were compared with parental strains. Because data did not pass normality test, Kruskal–Wallis analysis was performed. Bold values define statistically significant data.
Results
Disruption of glial clocks affects sleep
To examine whether peripheral oscillators located in glial cells are involved in the regulation of rhythmic behavior we analyzed period of the locomotor activity rhythm and sleep level in flies with pan-glial (repo-Gal4) overexpression of CYCLE (CYC) (UAS-cycΔ24). This strain expresses dominant- negative form of CYC, in which a portion of its basic regions were removed to impair DNA binding while retaining the ability to form heterodimers with CLK. This genetic manipulation disrupts the molecular mechanism of the circadian clock and oscillators lose their function. Because chronic CYC overexpression may cause developmental abnormalities, we used temperature sensitive Gal80 expressed under Tubulin promoter to induce adult-specific expression. We found that Tub-Gal80ts; repo>cycΔ24 flies were still rhythmic in DD (95% rhythmic flies while in controls: 85% in UAS-cycΔ24/+, 95% in Tub-Gal80ts; repo-Gal4/+) and their periods of locomotor activity rhythm (23.5 h) were similar to control flies (Table 1). We observed, however, changes in the sleep pattern with more sleep bins during the day and night (Fig. 1A). Siesta and nighttime sleep level, measured as minutes per 12 h, were also increased comparing with control flies (daytime: p ≤ 0.0013 with Gal4, p ≤ 0.0001 with UAS, nighttime: p ≤ 0.0038 with Gal4, p ≤ 0.0001 with UAS; Fig. 1B). Interestingly, these flies had lower number of sleep episodes during the day and night [daytime: 4.7 compared with 6.1 for Gal4 (p ≤ 0.0037) and 7.9 for UAS (p ≤ 0.0001), nighttime: 6.3 compared with 9.2 for Gal4 (p ≤ 0.0001) and 7.9 for UAS (p ≤ 0.047)], but mean bout length was longer only during the day [daytime: 133.4 compared with 87.7 (p ≤ 0.003) and 57.4 (p ≤ 0.0001), nighttime: 101.4 compared with 95.7 for Gal4 (p ≤ 0.8871) and 32.0 for UAS (p ≤ 0.0001); Table 3]. We tried to perform reversed experiment, with CYCΔ expression only during the larval stage; however, we observed low level of eclosion and individual adult flies did not survive until the end of experiment, suggesting their developmental abnormalities.
Figure 1.
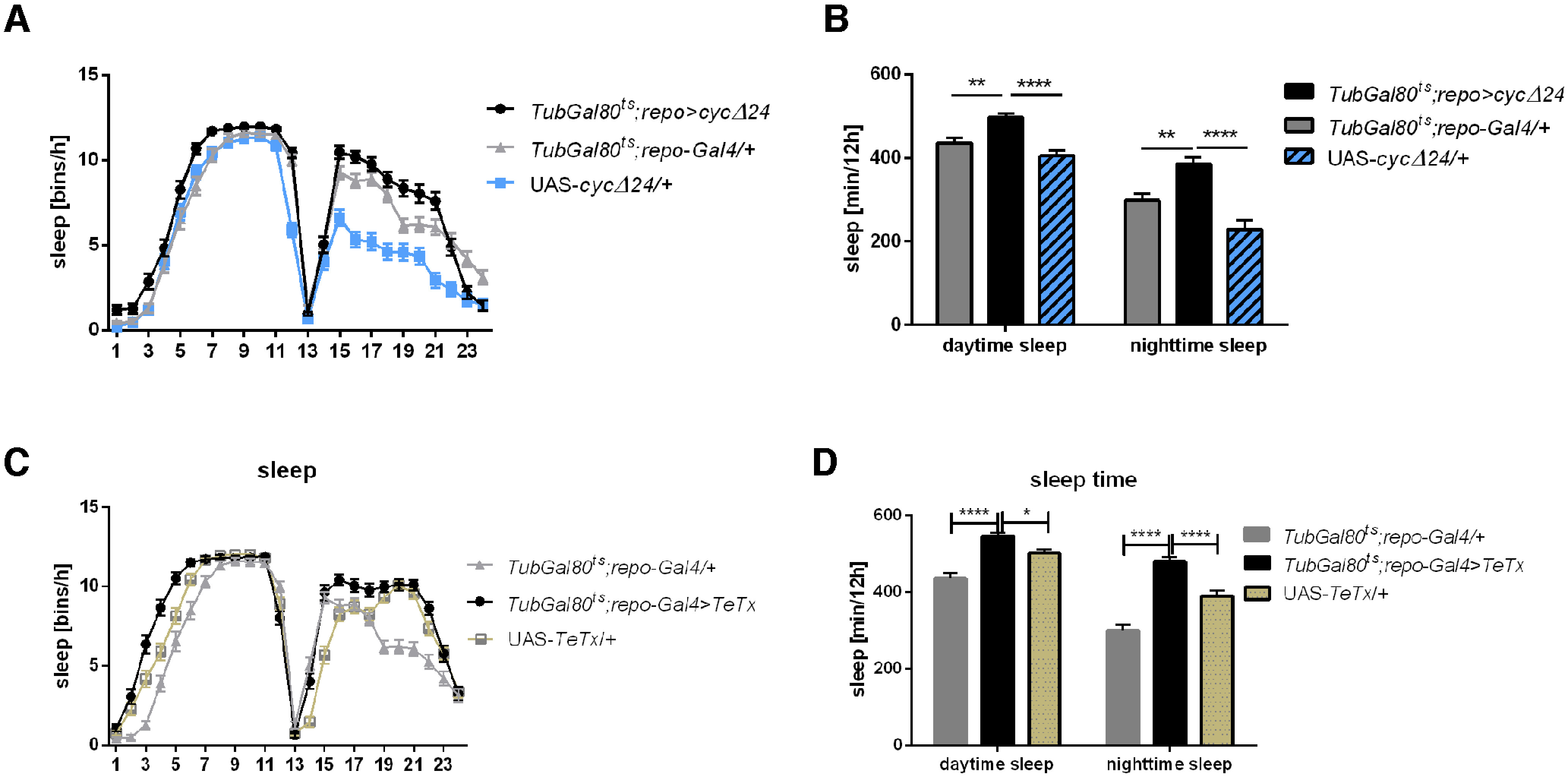
Glial clock regulates sleep amount. A, Sleep pattern within 24 h in LD12:12 measured as bins per hour. Experimental flies with adult-specific clock disruption in glial cells (TubGal80ts;repo>cycΔ24) are compared with parental lines (TubGal80ts;repo-Gal4/+, UAS-cycΔ24/+). B, Sleep amount of TubGal80ts;repo>cycΔ24 and control flies. C, D, Sleep pattern and level of flies with adult-specific exocytosis blocking in glia (TubGal80ts;repo>TeTx) compared with parental strains (TubGal80ts;repo-Gal4/+, UAS-TeTx/+). Nighttime sleep and daytime sleep were measured in minutes per 12 h of light and dark phase, respectively. Means ± SE. Statistically significant differences are marked with asterisks: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. Detailed statistics are provided in Table 2.
To check whether the regulation of sleep by glial clock is connected with gliotransmission, we forced expression of tetanus toxin in all glial cells (Tub-Gal80ts; repo>TeTx), and we observed similar changes in sleep level - amount of sleep during the day and night was increased (daytime: p ≤ 0.0001 with Gal4 and p ≤ 0.0034 with UAS, nighttime: p ≤ 0.0001 with both control strains; Fig. 1C,D; Table 2).
Because obtained data indicated that peripheral oscillators located in glial cells are involved in the regulation of sleep, we tried to identify specific glia types responsible for this phenotype. We employed different drivers to manipulate gene expression in specific types of glia.
Astrocyte-like glia
First, we used GFP expression to label astrocyte-like glia and anti-PER immunostaining to specify whether studied cells express circadian genes and we found that PER was co-localized with GFP in alrm>GFP cells (Fig. 2A).
Figure 2.
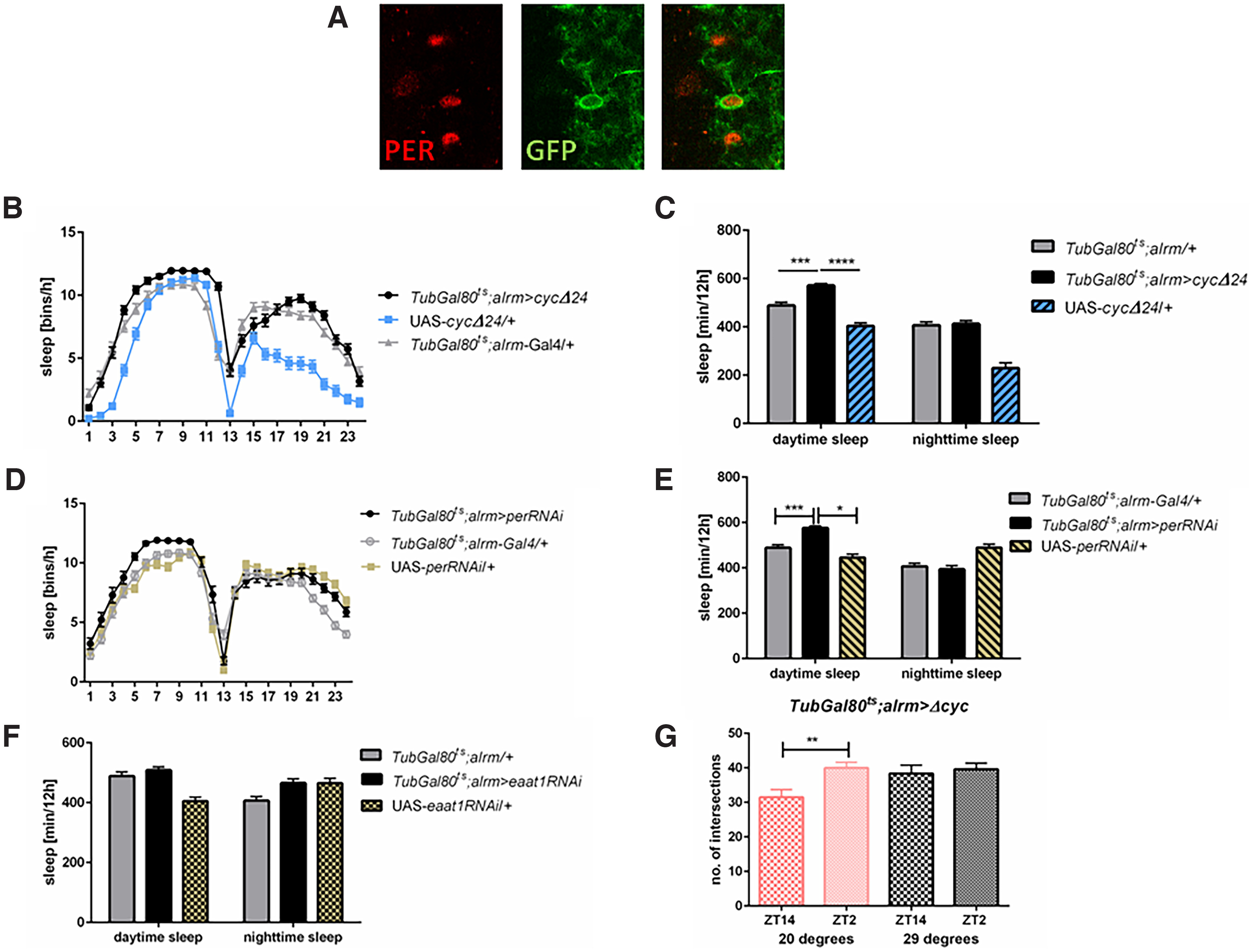
Astrocyte-like glia are involved in the sleep regulation. A, Whole mount brain immunolabeling of the alrm>GFP strain with anti-PER (red) and anti-GFP (green) antibodies confirms that clock oscillators are located in astrocyte-like glia. B, Sleep pattern of TubGal80ts;alrm>cycΔ24 is changed comparing with parental lines (TubGal80ts;alrm-Gal4/+, UAS-cycΔ24/+). C, Daytime sleep is significantly increased in flies with clock disruption in astrocyte-like glia. D, Sleep pattern is changed in the middle of the day in TubGal80ts;alrm>perRNAi flies. E, Daytime sleep amount is increased after per silencing in astrocytes. F, Glutamate re-uptake decreasing in astrocyte-like glia (TubGal80ts;alrm>Eaat1RNAi) does not affect sleep amount. Statistically significant differences are marked with asterisks: **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. Detailed statistics for C, E, F are provided in Table 2. G, Temperature-dependent clock disruption in astrocyte-like glia stops daily changes in the complexity of sLNv neurons terminals. Statistically significant differences with p = 0.0035 between time points in control group, no differences in experimental group (p = 0.6884). Means ± SE.
Clock disruption in astrocyte-like glia (Tub-Gal80ts; alrm>cycΔ24) did not affect period of the locomotor activity rhythm (Table 1). However, in these flies, the sleep pattern (Fig. 2B) and amount of sleep (Fig. 2C) were significantly affected with longer sleep time during the day (daytime: p ≤ 0.0001 with both controls, nighttime: p ≤ 0.9314 with Gal4, p ≤ 0.0001 with UAS; Table 2). Number of sleep episodes was not significantly changed [daytime: 7 compared with 8.7 in Gal4 (p ≤ 0.42) and 7.9 in UAS (p ≤ 0.059), nighttime: 8.9 compared with 14.9 (p ≤ 0.0001) in Gal4 and 7.9 (p ≤ 0.37)], but bout length was longer both during the day and night [daytime: 95.1 compared with 66.6 in Gal4 (p ≤ 0.023) and 57.4 in UAS (p ≤ 0.0011), nighttime: 79.1 compared with 35.6 in Gal4 (p ≤ 0.0001) and 32.2 in UAS (p ≤ 0.0001); Table 3]. To confirm these results we used adult-specific per silencing in astrocyte-like glia (Tub-Gal80ts; alrm>perRNAi flies) and observed a similar phenotype in files which showed a prolonged time of sleep during the day (daytime: p ≤ 0.0001 with both controls, nighttime: p ≤ 0.5582 with Gal4 and p ≤ 0.0001with UAS; Fig. 2D,E; Table 2). However, these flies showed less sleep episodes during the day [5.3 compared with 8.7 in Gal4 (p ≤ 0.0001) and 7.9 in UAS (p ≤ 0.042)] with longer mean time of bouts [132.5 compared with 66.6 in Gal4 (p ≤ 0.0001) and 63.9 in UAS (p ≤ 0.0001); Table 3].
Based on the fact, that astrocytes are involved in glutamate recycling, we checked whether the prolonged sleep behavior will be present in flies with silenced Excitatory amino acid transporter (Eaat1), in which astrocyte specific glutamate re-uptake is disrupted. However, we did not observe any changes in sleep amount or sleep pattern between Tub-Gal80ts; alrm>Eaat1RNAi and the control strains (daytime: p ≤ 0.5 with Gal4 and p ≤ 0.0001 with UAS, nighttime: p ≤ 0.01 with Gal4 and p ≤ 0.99 with UAS; Fig. 2F; Table 2).
It was previously shown that pan-glial clock disruption affects daily changes in plasticity of the sLNv terminals (Herrero et al., 2017). We investigated this phenomenon in details and found that one of the glia types involved in this process is astrocyte-like glia. A comparison between Tub-Gal80ts; alrm>cycΔ24 flies maintained at 20°C and 29°C showed that 3 d of clock disruption in astrocytes is enough to set projection complexity both during day and night on the level observed during the day in control flies (at 20°C: p ≤ 0.035, t = 3.013, df = 79, at 29°C: p ≤ 0.69, t = 0.4025, df = 82; Fig. 2G).
Chiasm glia
The second type of glia selected for our research was the chiasm giant glia. Brains immunostained for PER as well the per>stingerRFP strain showed very specific fluorescent pattern in the optic lobe. Large, immune-positive cells were arranged linearly in the inner chiasm area, between the medulla and lobula (Fig. 3A,B). Using Wnt4>GFP strain we confirmed that these PER-immunopositive cells were the giant chiasm glia (Fig. 3C). Measurements of fluorescent intensity at different times of the day showed that PER expression in the chiasm giant glial cells cycles with maximum and minimum at ZT20 and ZT12, respectively (Fig. 3D).
Figure 3.
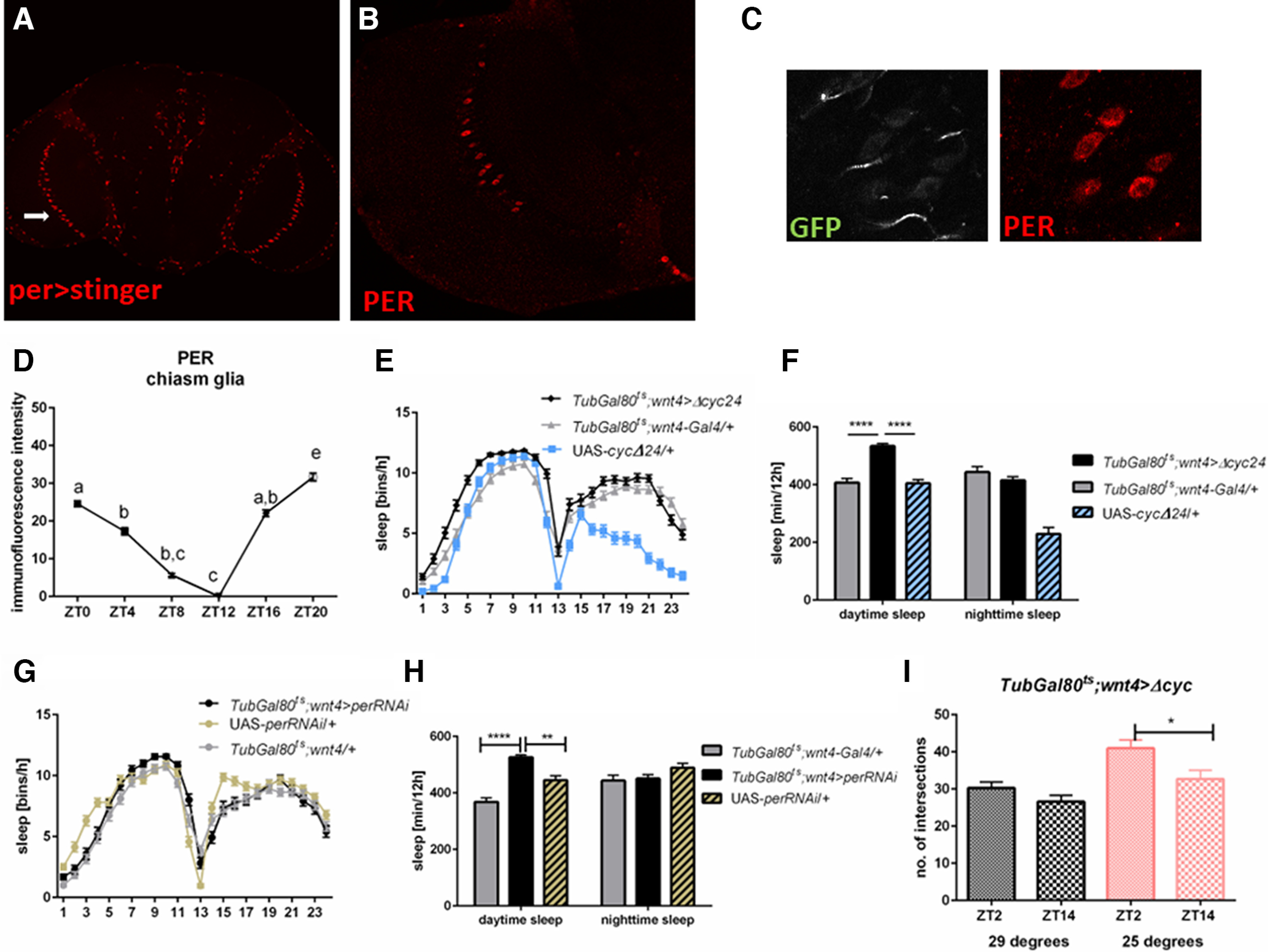
Effects of clock disruption in the chiasm giant glia on sleep in LD12:12. A, Whole mount brain immunolabeling of per>Red-Stinger, visualizes nuclei of per-expressing cells. Optical section shows signal in chiasm glia (marked with arrow). B, Whole mount brain immunolabeling of PER shows similar pattern characteristic for chiasm glia. C, Wnt4>GFP strain with marked PER protein shows co-localization. D, Measurement of fluorescence intensity of PER immunostaining in chiasm glia at different time points, sampled every 4 h in LD12:12 light regime. Statistically significant differences are marked with different letters. E, F, Sleep pattern and level of flies with clock disruption in chiasm glia (TubGal80ts; Wnt4>cycΔ24) are changed during the day comparing with parental strains (TubGal80ts; Wnt4-Gal4/+; UAS-cycΔ24/+). G, H, per silencing in chiasm glia affects sleep level during the day. Statistically significant differences are marked with asterisks: *p ≤ 0.05, ****p ≤ 0.0001. Detailed statistics are provided in Table 2. I, Temperature-dependent clock disruption in chiasm glia stops daily changes in the complexity of sLNv neurons terminals (p = 0.0135 for control and p = 0.1194 for experimental group). Means ± SE.
Our results showed that ΔCYC expression in the chiasm giant glia (Tub-Gal80ts; Wnt4>cycΔ24) did not affect the number of rhythmic flies (Table 1), while it changed the sleep pattern and increased its level during the day (Fig. 3E,F; p ≤ 0.0001 with both controls). In addition, the number of sleep episodes was lower [5.7 compared with 8.9 in Gal4 (p ≤ 0.0024) and 7.9 in UAS (p ≤ 0.0171)], and mean bout length was increased during the day [132.8 compared with 65.6 in Gal4 (p ≤ 0.0001) and 57.4 in UAS (p ≤ 0.0001)], similarly to pan-glial clock disruption (Table 3). Similar effect on sleep time was observed after silencing per in the chiasm giant glia (daytime sleep: p ≤ 0.0001 with both controls, nighttime sleep: p ≤ 0.56 with Gal4 and p ≤ 0.0002 with UAS; Tub-Gal80ts; Wnt4>perRNAi; Fig. 3G,H; Table 2).
The Scholl analysis of the sLNv terminals provided information that the chiasm glial clock is necessary to maintain daily changes in their complexity. Tub-Gal80ts; Wnt4>cycΔ24 at higher temperature showed no changes between the day and night, and the number of intersections was similar in both cases to that observed during the night at the lower temperature (at 20°C: p ≤ 0.0135, t = 2.517, df = 93, at 29°C: p ≤ 0.1194, t = 1.575, df = 76; Fig. 3I).
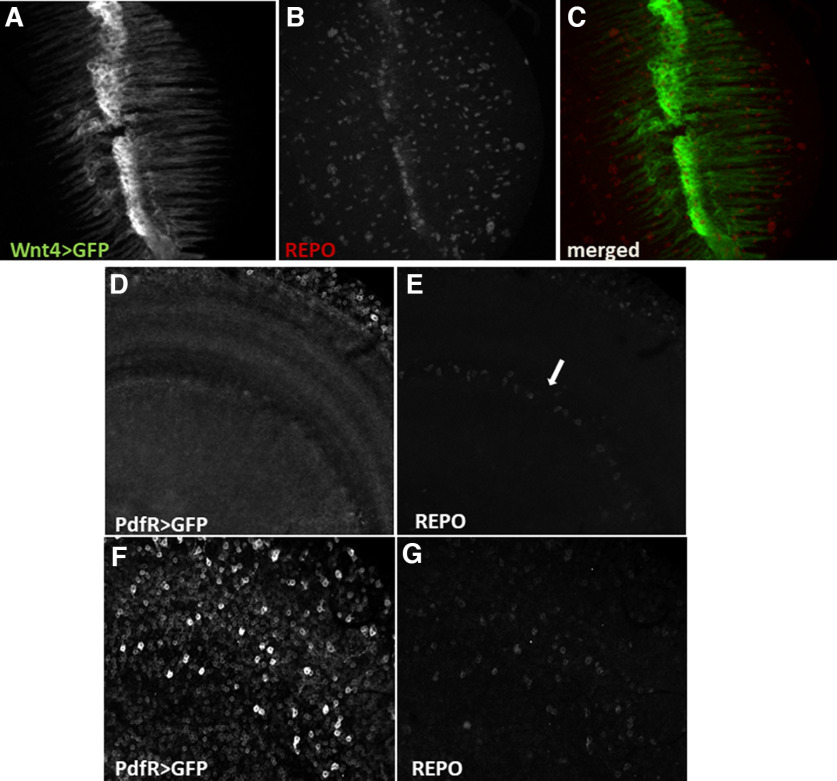
In addition, we checked that oscillators located in chiasm glia are PDF-independent as we showed that they do not have PDFR expression (Fig. 4).
Figure 4.
Chiasm glia oscillators are not regulated by PDF released from LNvs. A–C, Whole-brain immunostaining of Wnt4>GFP flies using anti-Repo antibodies visualized chiasm glia cells (A, C, green) and pan-glial nuclei (B, C, red). D–G, Whole-brain anti-Repo immunostaining of PDFR>GFP flies visualized cells expressing receptor for PDF neuropeptide (D, F) and glial cells nuclei (E, G). D, E, Optical section shows no signal in the place where chiasm glia is located (marked with arrow). F, G, Glial cells other than chiasm glia show expression of PDFR.
Epithelial glia
Clock disruption in the epithelial glia (Tub-Gal80ts; netB>cycΔ24) did not affect neither the rhythmicity nor its period (Table 1). In these flies the sleep pattern was changed with higher number of sleep bins observed during the day (Fig. 5A; Table 2) and the total sleep time was increased during the day and night (daytime sleep: p ≤ 0.0001 with both controls, nighttime sleep: p ≤ 0.027 with Gal4 and p ≤ 0.0001 with UAS; Fig. 5B; Table 2). Silencing of per in this glia type affected the sleep time only during the day (daytime sleep: p ≤ 0.0001 with Gal4 and p ≤ 0.0083 with UAS, nighttime sleep p ≤ 0.58 with Gal4 and p ≤ 0.08 with UAS; Fig. 5C,D; Table 2); however, because of cell-specific effect, we were not able to check silencing efficiency which is always below 100%.
Figure 5.
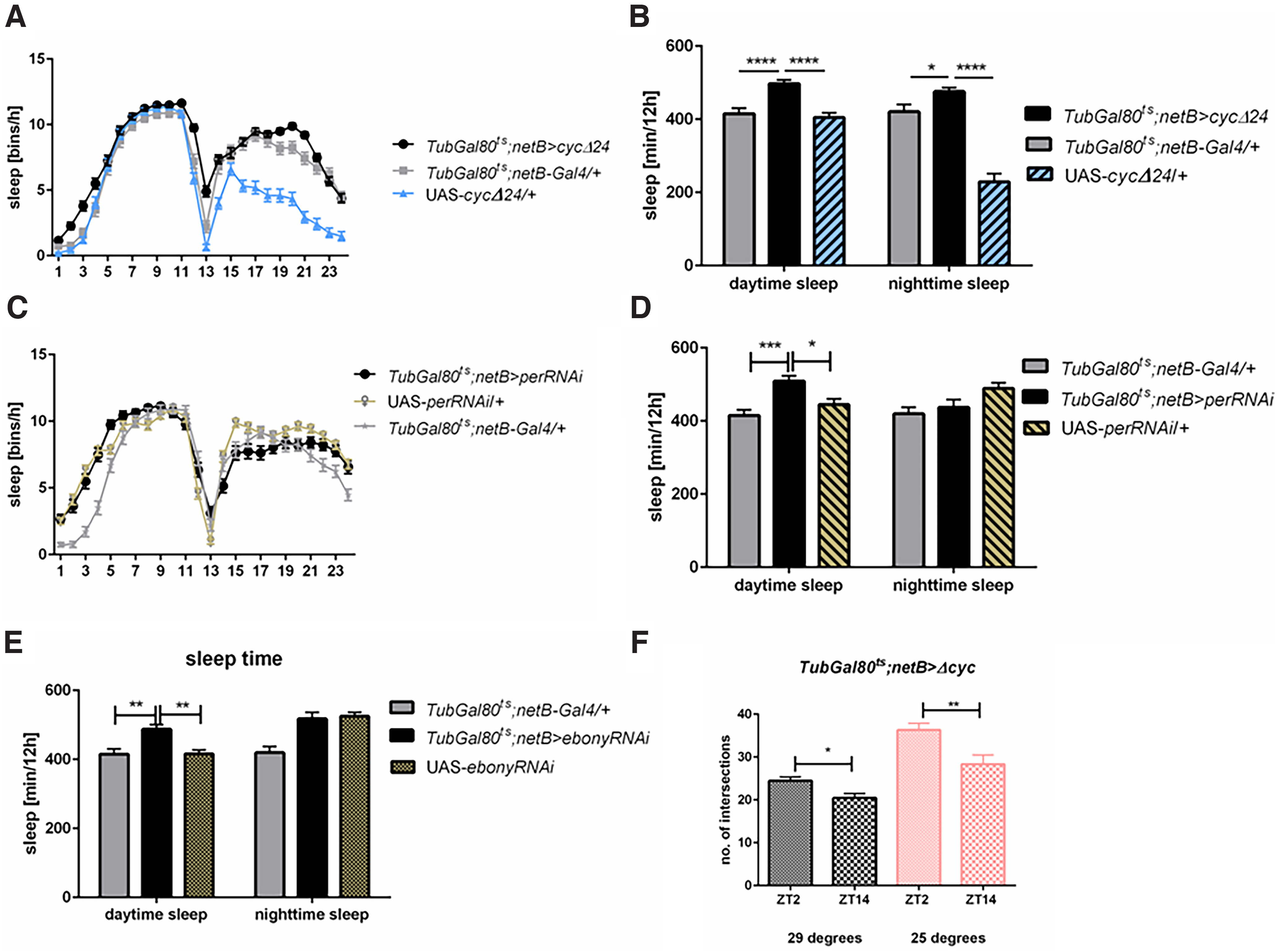
The role of peripheral oscillators located in epithelial glia in sleep regulation in LD12:12. A, B, Clock disruption in the epithelial glia (TubGal80ts;netB>cycΔ24) affects sleep during the day and night comparing with both parental strains (TubGal80ts;netB-Gal4/+, UAS- cycΔ24/+). C, D, Per silencing in the epithelial glia affects sleep time during the day. E, Adult-specific neurotransmission disruption in epithelial glia (TubGal80ts;netB>ebonyRNAi) affects sleep amount during the day. Statistically significant differences are marked with asterisks: *p ≤ 0.05, **p ≤ 0.01, ****p ≤ 0.0001. Detailed statistics are provided in Table 2. F, Temperature-dependent clock disruption in epithelial glia does not affect daily changes in the complexity of sLNv neurons terminals (differences between time points are statistically significant with p = 0.0061 for control, p = 0.0031 for experimental group), but it decreases amplitude of this oscillations. Means ± SE.
Knowing that the epithelial glia rhythmically express Ebony, which is involved in neurotransmission regulation, we suppressed its expression in this type of glia. In effect, we observed that sleep was increased during the day (p ≤ 0.0012 with Gal4 and p ≤ 0.0006 with UAS), but not affected during the night (p ≤ 0.0001 with Gal4 and p ≤ 0.93 with UAS; Fig. 5E; Table 2).
Surprisingly, daily changes in the number of intersections of the sLNv terminals in the dorsal brain were maintained after disrupting the epithelial glia clock. However, the amplitude of these changes was lower, but the pattern was the same as in control (at 20°C: p ≤ 0.061, t = 2.827, df = 72, at 29°C: p ≤ 0.0031, t = 3.066, df = 72; Fig. 5F).
Subperineurial and pseudocartridge glia
CYC overexpression in moody-positive cells (subperineurial and pseudocartridge glia) did not affect the period of the locomotor activity rhythm (Table 1). The sleep pattern and its level in TubGal80ts;moody>cycΔ24 were changed during the day with longer sleep time (p ≤ 0.0001 with both controls), lower number of sleep episodes [5.4 compared with 7.5 in Gal4 (p ≤ 0.008) and 7.9 in UAS (p ≤ 0.0008)] and longer bout length [149.0 compared with 78.7 in Gal4 (p ≤ 0.0001) and 57.4 in UAS (p ≤ 0.0001)] when compared with controls (Fig. 6A,B; Tables 2, 3). However, per silencing in this type of glia did not change sleep amount (Fig. 6C,D; Table 2), which suggests that the observed effect is not connected with the clock disruption. In addition, we did not observe changes in the sLNv projection complexity in TubGal80ts;moody>cycΔ24 flies (at 20°C: p ≤ 0.0001, t = 5.281, df = 99, at 29°C: p ≤ 0.0035, t = 3.019, df = 73; Fig. 6E).
Figure 6.
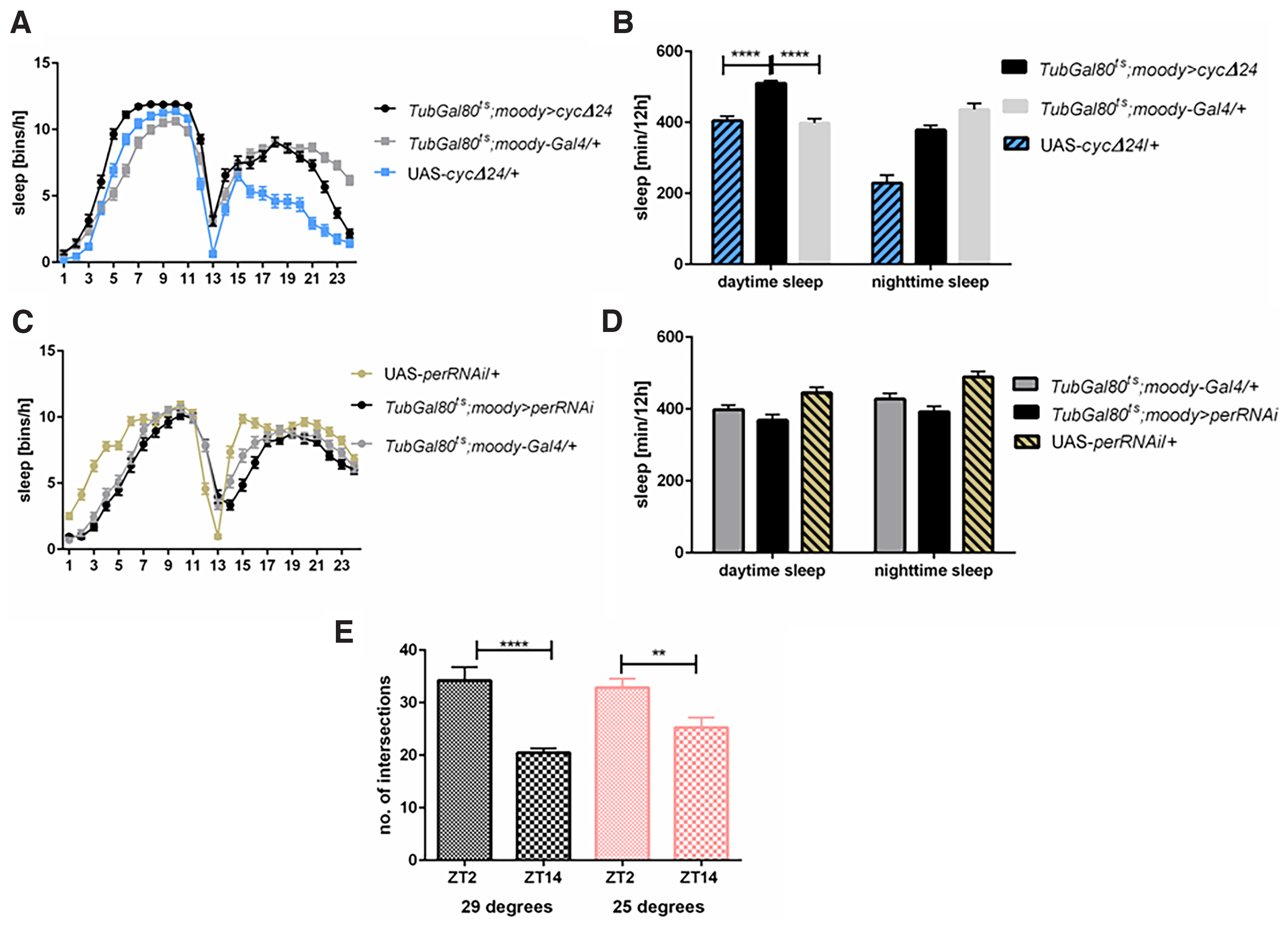
Subperineurial and pseudocartridge glia may be involved in the sleep regulation. A, B, Clock disruption in the subperineurial and pseudocardridge glia (TubGal80ts;moody>cycΔ24) increases sleep during the day comparing with both parental strains (TubGal80ts;moody-Gal4/+, UAS-cycΔ24/+). C, D, Per silencing in moody-expressing cells did not change sleep time. Statistically significant differences are marked with asterisks: **p ≤ 0.01, ****p ≤ 0.0001. Detailed statistics are provided in Table 2. E, Temperature-dependent clock disruption in moody-expressing cells does not affect daily changes in the complexity of sLNv neurons terminals (p = 0.035 for control and p ≤ 0.0001 for experimental group). Means ± SE.
Sleep regulation requires neuron-glia synaptic communication
Glial cells may participate in the formation of synaptic contacts as postsynaptic partners using DLG1 protein. We analyzed flies with pan-glial dlg1 silencing and observed that although they were rhythmic with a normal period of the locomotor activity rhythm (Table 1), their sleep pattern during the day was changed. They started siesta earlier in the morning and it was ended later (Fig. 7A). In effect, also total sleep time was increased during the day (p ≤ 0.0001 when compared with both controls; Fig. 7B; Table 2).
Figure 7.
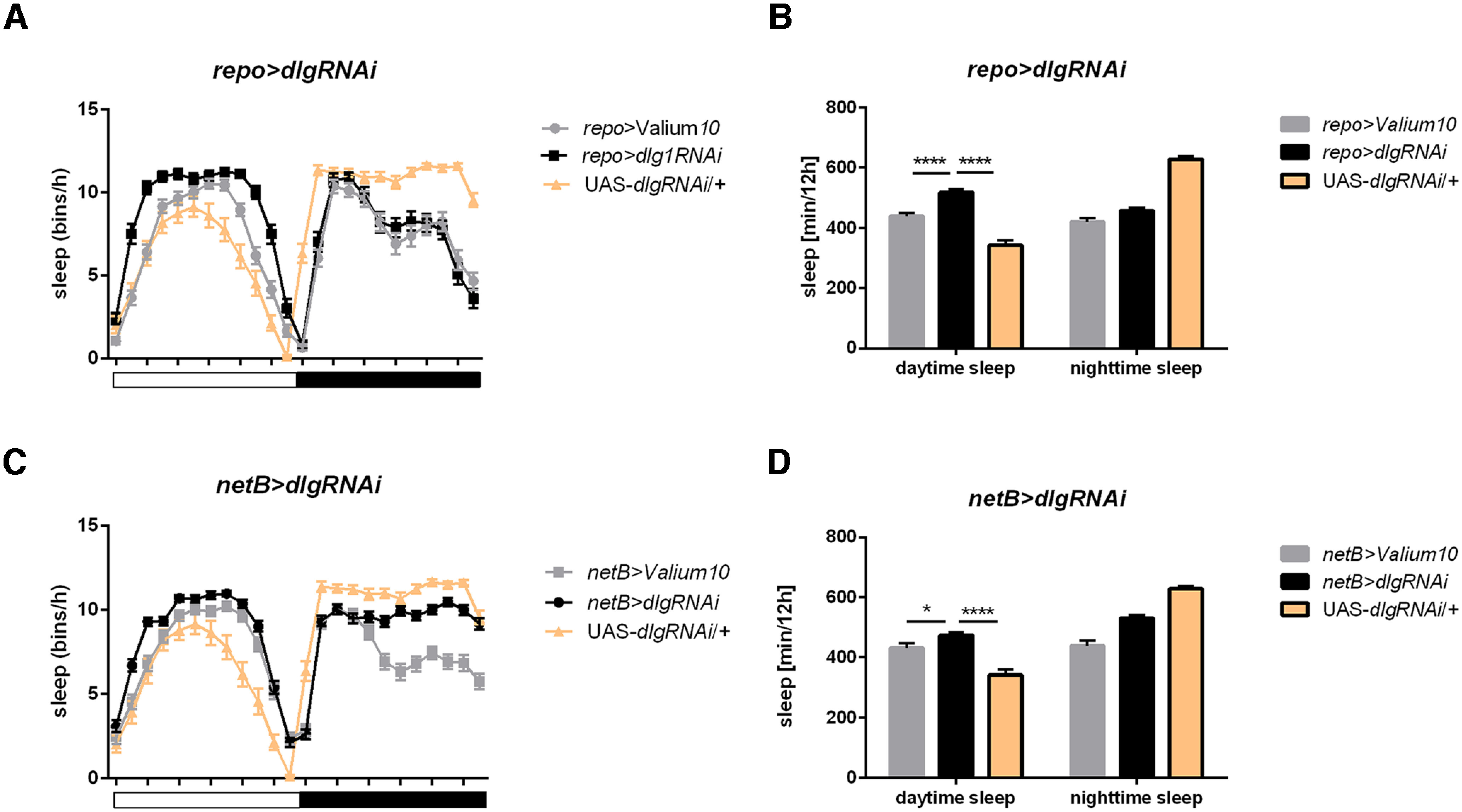
Effect of dlg1 silencing in glia on sleep pattern and level. A, B, Pan-glial dlg1 silencing causes increased sleep time during the day. C, D, Flies with dlg1RNAi expressed in epithelial glia show longer day time sleep. Means ± SE. Statistically significant differences are marked with asterisks: *p ≤ 0.05, ****p ≤ 0.0001. Detailed statistics are provided in Table 2.
Then, we tried to investigate which glia type is responsible for this phenotype, and we detected significant changes for netB>dlg1RNAi (Fig. 7C,D; p < 0.05 exp. vs netB>Val and p ≤ 0.0001 exp. vs UAS-dlg1RNAi/+).
However, the silencing of dlg1 in other types of glia: the chiasm glia, astrocyte-like glia, and subperineurial and pseudocartridge glia, did not affect neither the period of the locomotor activity rhythm nor sleep (Fig. 8A–F; Tables 1, 2).
Figure 8.
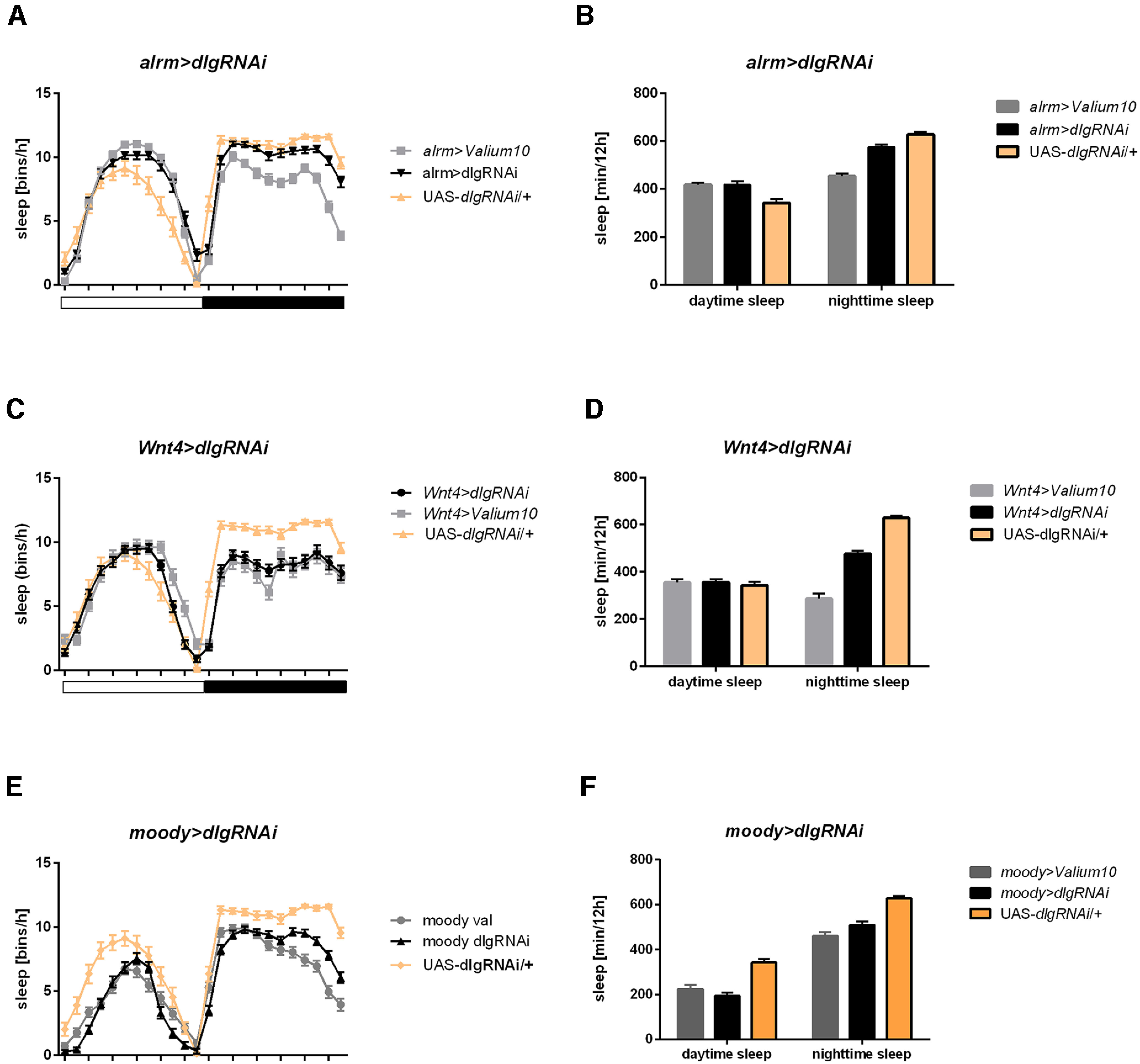
Silencing of postsynaptic dlg1 in A–B: astrocyte-like glia (alrm>dlg1RNAi), C–D: chiasm giant glia (Wnt4>dlg1RNAi), E–F: subperineurial and pseudocartridge glia (moody>dlg1RNAi) do not affect sleep pattern nor level. Detailed statistics are provided in Table 2.
Discussion
Glia are composed of heterogenous cell types with different functions involved in brain development, in neurotransmission, and in other processes, including olfaction (Liu et al., 2014), vision (Borycz et al., 2012), and memory formation (Yamazaki et al., 2014; Matsuno et al., 2015). It was previously shown that a subpopulation of glial cells has rhythmic expression of clock gene per and timeless (tim), suggesting that they play a role of peripheral oscillators (Ewer et al., 1992; Damulewicz et al., 2015; Long and Giebultowicz, 2018). However, little is known about functions of different types of glia in rhythmic processes in the brain and mechanisms of glial-neuronal communication involved in sleep regulation.
Glia play a role in the regulation of rhythmic behavior, which is modulated by releasing gliotransmitters (Ng et al., 2011) or affecting clock neuron activity by changes in PDF transport or release (Ng et al., 2011). Moreover, glial oscillators mediate the daily structural plasticity of clock neuron terminals in the dorsal brain (Herrero et al., 2017). It has been shown that glia activity is calcium dependent (Ng et al., 2011), but the mechanism of this regulation is unknown.
Our results showed that the clock disruption in glia has no effect on period of the locomotor activity rhythm and it seems not to be important for maintaining rhythmicity. This conclusion is supported by the previous data, showing that per silencing under the repo driver affects neither rhythmicity nor period of the rhythm in these flies (Ng et al., 2011). On the other hand, we observed a strong effect of glia on sleep. Pan-glial disruption of the molecular clock mechanism as well as blocking exocytosis increased sleep length during the day and night. Moreover, CYC overexpression in specific glia types affected sleep mostly during daytime. This suggests that mechanisms of nap and sleep regulation are different and may involve different glia types.
We found that peripheral oscillators located in a few types of glia are involved in the regulation of sleep/activity pattern. They include astrocyte-like, giant chiasm glia, epithelial, subperineurial and pseudocartridge glia which have been reported to express per and are regarded as circadian oscillators (Siwicki et al., 1988; Zerr et al., 1990; Suh and Jackson, 2007; Ng et al., 2016; Long and Giebultowicz, 2018).
Astrocytes regulate sleep using mechanism other than vesicular trafficking
It has been suggested that the most important type of glia involved in the regulation of behavior are astrocytes (You et al., 2018). In flies, astrocytes play a similar role as in mammals in regulating metabolism, neurotransmitter turnover, and transport. They are activated in a calcium-dependent manner (Ng et al., 2011), and their role in maintaining circadian rhythms is correlated with a proper vesicle trafficking (Ng et al., 2011, 2016; Jackson et al., 2015; Ng and Jackson, 2015). It was previously shown that glutamate transporter Eaat1 plays an important role in the modulation of fast synaptic transmission (Macnamee et al., 2016); however, our results suggest that astrocyte oscillators regulate sleep using other mechanism than glutamate signaling. It is also possible that the oscillation of glutamate release is crucial in this process. To test this possibility, we used flies with glutamate transporter silencing, which weakened but did not disrupt this signaling completely, and in effect, we did not observe any significant changes in the level and pattern of sleep. On the other hand, the previous data showed that daytime sleep is affected by Eaat1 silencing in both cortex and astrocyte-like glia; however, the amount of sleep was decreased (Farca Luna et al., 2017). Interestingly, Eaat1 reduction in these glia types did not affect glutamine synthetase level (Farca Luna et al., 2017). Thus, astrocyte-like glia seem to affect sleep by another mechanism than vesicular trafficking (Artiushin et al., 2018).
Epithelial glia regulate sleep by modulation of neurotransmission through Ebony
The epithelial glia seem to be an important element of the clock system, as well. In Drosophila these glial cells are in a close vicinity to clock neurons: sLNvs, dorsal lateral neurons (LNd), dorsal neurons DN1 and DN3 (Suh and Jackson, 2007), and to PDF-immunoreactive processes. Moreover, they change size and shape during the day in the visual system, in the reversed pattern to neurons, and participate in transmitting circadian information from the clock in the brain to neurons in the visual system (Pyza and Górska-Andrzejak, 2004). This process may depend on changes in neurotransmitter or ion concentrations in the extracellular space. Moreover, the epithelial glial cells in the first optic neuropil (lamina) exhibit rhythmic expression of the α subunit of the sodium-potassium pump (Górska-Andrzejak et al., 2009), which is under control of peripheral clocks located in the retina photoreceptors, glia and the pacemaker via PDF signaling (Damulewicz et al., 2013). In addition, the epithelial glia have rhythmic expression of ebony, which is necessary for circadian rhythmicity (Suh and Jackson, 2007), because its protein takes part in metabolism of neurotransmitters. Our results suggest that daily changes observed in the epithelial glia regulate also sleep time, especially during the day and Ebony is involved in this wake-promoting effect by daily modulation of dopamine level.
Chiasm giant glia affect sleep level
The chiasm giant glia are located between the lamina and medulla neuropils (outer chiasm, X0) and between the medulla and lobula (inner chiasm, Xi), and form an envelope around axon bundles which cross both chiasmata (Tix et al., 1997). Our study, supported by already published data showed that the chiasm giant glial cells express per cyclically, so they belong to the system of peripheral oscillators (Long and Giebultowicz, 2018). Flies with the clock disruption in the chiasm glial cells are rhythmic but their sleep is changed, which suggests that these peripheral oscillators are necessary to maintain behavioral rhythms. They may be involved in the daily synchronization of the visual system with processes in the brain and with behavior.
Subperineurial glia may be a part of glial clock network
Subperineurial glia are an important component of the hemolymph-brain barrier, which protects neurons from unregulated exchange with humoral fluids. Our data indicate that oscillators located in this type of glia might be involved in sleep regulation; however, per silencing did not show significant results. On the one hand, it is possible that ΔCYC expression caused metabolic changes in the glia which are not connected with clock functions, but on the other hand, per silencing could be not effective enough to disrupt the clock in these cells. Although an involvement of clock in subperineurial glia needs more detailed examination, the role of these cells in the regulation of sleep has already been confirmed. It was previously shown that disruption of endocytosis in moody-expressing cells enhances total and daytime sleep (Artiushin et al., 2018). Endocytosis at the hemolymph-brain barrier shows daily changes and it is higher during the early night, but there are strong evidences that it is regulated rather by sleep need than the circadian clock (Artiushin et al., 2018). However, disruption of oscillators in subperineurial glia might change rhythms in endocytosis and in effect affect sleep amount.
Involvement of epithelial glia in tripartite synapses is important for sleep regulation
In the present study, we also tried to find a signaling pathway involved in the regulation of circadian rhythms by glia. It has been shown that the fly's glia form aminergic and glutamatergic tripartite synapses (Jackson and Haydon, 2008; Edwards and Meinertzhagen, 2010). In the first optic neuropil (lamina), R1-R6 photoreceptor terminals form tetrad synapses with four postsynaptic partners including the epithelial glial cells, most probably using DLG1 as a postsynaptic protein (Hamanaka and Meinertzhagen, 2010). The other tripartite synapses were described in neuromuscular junctions formed by perisynaptic glia (Strauss et al., 2015). Moreover, astrocyte-like glia seem not to be involved in synapse formation in Drosophila, opposite to mammals (Macnamee et al., 2016).
To check whether this type of signaling is necessary to maintain the daily activity rhythm and sleep pattern, we silenced discs large (dlg1) expression using the repo driver. DLG1 is a presynaptic and postsynaptic protein (Lahey et al., 1994) involved in the formation of scaffolding postsynaptic structures (Budnik et al., 1996). Because DLG1 is involved in neurotransmission, its silencing in a specific glia type should block communication between neurons and the examined glia. We found sleep lengthening during the day after pan-glial dlg1 silencing; however, the similar effect was observed only when DLG1 level was diminished in the epithelial glia. The only known tripartite synapses involving epithelial glia are formed with the retinal R1-R6; however, disruption of neurotransmission from these photoreceptors affects sleep level during the night, but not during the day (Damulewicz et al., 2020). It seems that other presynaptic partners for the epithelial glia need to be studied in the future.
Circadian changes in the sLNv terminal complexity are maintained by astrocytes and chiasm glia
Sleep amount is regulated by many different neurons located in sleep centers and by clock neurons. Among clock neurons the most important are PDF-expressing sLNv, which send axonal terminals to the dorsal brain. These axonal ends change their complexity in the daily pattern (Cao and Nitabach, 2008; Fernández et al., 2008). It was previously shown that this daily plasticity causes changes of postsynaptic partners, in effect sLNvs contact with mushroom bodies in specific time of the day (Gorostiza et al., 2014). Structural plasticity of axonal terminals depends on circadian oscillators, in both, clock neurons and glial cells (Fernández et al., 2008; Herrero et al., 2017). It has been shown that pan-glial clock disruption abolishes structural plasticity of the sLNv terminals (Herrero et al., 2017), and now in the present study, we described specific glia types involved in this process, which are astrocytes and giant optic chiasm glia. Additionally, oscillators located in the epithelial glia regulate the amplitude of this rhythm. The lack of effects on the rhythm of PDF+ cell axonal terminals after clock disruption in subperineurial glia supports a hypothesis that this type of glia works locally and does not interacts directly with clock neurons.
To summarize, we showed that glial cells are involved in the regulation of rhythmic behavior. Peripheral oscillators in glia have a wake-promoting effect. In addition, synaptic contacts formed between the photoreceptor terminals and the epithelial glial cells activate the wake-promoting pathway. Our study showed that the most important glial oscillators which regulate sleep are located in astrocyte-like glia, the chiasm giant glia, epithelial glia; however, other glia types seem also contribute to this process.
Footnotes
This work was supported by the BioS Priority Research Area under the “Excellence Initiative–Research University” program at the Jagiellonian University in Krakow Grant U1U/P03/DO/14.81 (to M.D.). Z.B. was supported by the National Science Center in Poland through the ETIUDA Scholarship 2019/32/T/NZ3/00444. The delayed open-access publication of this article was funded by the Priority Research Area BioS under the program “Excellence Initiative – Research University” at the Jagiellonian University in Krakow.
The authors declare no competing financial interests.
References
- Abruzzi KC, Zadina A, Luo W, Wiyanto E, Rahman R, Guo F, Shafer O, Rosbash M (2017) RNA-seq analysis of Drosophila clock and non-clock neurons reveals neuron-specific cycling and novel candidate neuropeptides. PLoS Genet 13:e1006613. 10.1371/journal.pgen.1006613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonucci F, Turola E, Riganti L, Caleo M, Gabrielli M, Perrotta C, Novellino L, Clementi E, Giussani P, Viani P, Matteoli M, Verderio C (2012) Microvesicles released from microglia stimulate synaptic activity via enhanced sphingolipid metabolism. EMBO J 31:1231–1240. 10.1038/emboj.2011.489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnes M, Alaniz ME, Karam CS, Cho JD, Lopez G, Javitch JA, Santa-Maria I (2019) Role of tau protein in remodeling of circadian neuronal circuits and sleep. Front Aging Neurosci 11:320. 10.3389/fnagi.2019.00320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artiushin G, Zhang SL, Tricoire H, Sehgal A (2018) Endocytosis at the Drosophila blood–brain barrier as a function for sleep. Elife 7:e43326. 10.7554/eLife.43326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borycz J, Borycz JA, Edwards TN, Boulianne GL, Meinertzhagen IA (2012) The metabolism of histamine in the Drosophila optic lobe involves an ommatidial pathway: β-alanine recycles through the retina. J Exp Biol 215:1399–1411. 10.1242/jeb.060699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik V, Koh YH, Guan B, Hartmann B, Hough C, Woods D, Gorczyca M (1996) Regulation of synapse structure and function by the Drosophila tumor suppressor gene dlg. Neuron 17:627–640. 10.1016/s0896-6273(00)80196-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey D, Tononi G, Cirelli C (2011) Sleep and synaptic homeostasis: structural evidence in Drosophila. Science 332:1576–1581. 10.1126/science.1202839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, Nitabach MN (2008) Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J Neurosci 28:6493–6501. 10.1523/JNEUROSCI.1503-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Lamaze A, De J, Mena W, Chélot E, Martin B, Hardin P, Kadener S, Emery P, Rouyer F (2018) Reconfiguration of a multi-oscillator network by light in the Drosophila circadian clock. Curr Biol 28:2007–2017. 10.1016/j.cub.2018.04.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WF, Maguire S, Sowcik M, Luo W, Koh K, Sehgal A (2015) A neuron–glia interaction involving GABA transaminase contributes to sleep loss in sleepless mutants. Mol Psychiatry 20:240–251. 10.1038/mp.2014.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Rosbash M (2017) MicroRNA-92a is a circadian modulator of neuronal excitability in Drosophila. Nat Commun 8:14707. 10.1038/ncomms14707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusumano P, Klarsfeld A, Chélot E, Picot M, Richier B, Rouyer F (2009) PDF-modulated visual inputs and cryptochrome define diurnal behavior in Drosophila. Nat Neurosci 12:1431–1437. 10.1038/nn.2429 [DOI] [PubMed] [Google Scholar]
- Cusumano P, Biscontin A, Sandrelli F, Mazzotta GM, Tregnago C, De Pittà C, Costa R (2018) Modulation of miR-210 alters phasing of circadian locomotor activity and impairs projections of PDF clock neurons in Drosophila melanogaster. PLoS Genet 14:e1007500. 10.1371/journal.pgen.1007500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damulewicz M, Rosato E, Pyza E (2013) Circadian regulation of the Na+/K+-Atpase alpha subunit in the visual system is mediated by the pacemaker and by retina photoreceptors in Drosophila melanogaster. PLoS One 8:e73690. 10.1371/journal.pone.0073690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damulewicz M, Loboda A, Bukowska-Strakova K, Jozkowicz A, Dulak J, Pyza E (2015) Clock and clock-controlled genes are differently expressed in the retina, lamina and in selected cells of the visual system of Drosophila melanogaster. Front Cell Neurosci 9:353. 10.3389/fncel.2015.00353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damulewicz M, Ispizua JI, Ceriani MF, Pyza EM (2020) Communication among photoreceptors and the central clock affects sleep profile. Front Physiol 11:993–917. 10.3389/fphys.2020.00993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz MM, Schlichting M, Abruzzi KC, Long X, Rosbash M (2019) Allatostatin-C/AstC-R2 is a novel pathway to modulate the circadian activity pattern in Drosophila. Curr Biol 29:13–22. 10.1016/j.cub.2018.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty J, Logan MA, Taşdemir ÖE, Freeman MR (2009) Ensheathing glia function as phagocytes in the adult Drosophila brain. J Neurosci 29:4768–4781. 10.1523/JNEUROSCI.5951-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TN, Meinertzhagen IA (2010) The functional organisation of glia in the adult brain of Drosophila and other insects. Prog Neurobiol 90:471–497. 10.1016/j.pneurobio.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TN, Nuschke AC, Nern A, Meinertzhagen IA (2012) Organization and metamorphosis of glia in the Drosophila visual system. J Comp Neurol 520:2067–2085. 10.1002/cne.23071 [DOI] [PubMed] [Google Scholar]
- Ewer J, Frisch B, Hamblen-Coyle MJ, Rosbash M, Hall JC (1992) Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells' influence on circadian behavioral rhythms. J Neurosci 12:3321–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farca Luna AJ, Perier M, Seugnet L (2017) Amyloid precursor protein in Drosophila glia regulates sleep and genes involved in glutamate recycling. J Neurosci 37:4289–4300. 10.1523/JNEUROSCI.2826-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández MP, Berni J, Ceriani MF (2008) Circadian remodeling of neuronal circuits involved in rhythmic behavior. PLoS Biol 6:e69. 10.1371/journal.pbio.0060069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Chiappe F, Frenkel L, Colque CC, Ricciuti A, Hahm B, Cerredo K, Muraro NI, Ceriani MF (2021) High-frequency neuronal bursting is essential for circadian and sleep behaviors in Drosophila. J Neurosci 41:689–710. 10.1523/JNEUROSCI.2322-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzner D, Schnaars M, van Rossum D, Krishnamoorthy G, Dibaj P, Bakhti M, Regen T, Hanisch U-K, Simons M (2011) Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J Cell Sci 124:447–458. 10.1242/jcs.074088 [DOI] [PubMed] [Google Scholar]
- Frühbeis C, Fröhlich D, Krämer-Albers EM (2012) Emerging roles of exosomes in neuron-glia communication. Front Physiol 3:119. 10.3389/fphys.2012.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstner JR, Vanderheyden WM, Shaw PJ, Landry CF, Yin JCP (2011) Fatty-acid binding proteins modulate sleep and enhance long-term memory consolidation in Drosophila. PLoS One 6:e15890. 10.1371/journal.pone.0015890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorostiza EA, Depetris-Chauvin A, Frenkel L, Pírez N, Ceriani MF (2014) Circadian pacemaker neurons change synaptic contacts across the day. Curr Biol 24:2161–2167. 10.1016/j.cub.2014.07.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górska-Andrzejak J, Salvaterra PM, Meinertzhagen IA, Krzeptowski W, Görlich A, Pyza E (2009) Cyclical expression of Na+/K+-ATPase in the visual system of Drosophila melanogaster. J Insect Physiol 55:459–468. 10.1016/j.jinsphys.2009.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górska-Andrzejak J, Chwastek EM, Walkowicz L, Witek K (2018) On variations in the level of PER in glial clocks of Drosophila optic lobe and its negative regulation by PDF signaling. Front Physiol 9:230. 10.3389/fphys.2018.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B, Chélot E, Xia R, Rouyer F (2004) Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431:869–873. 10.1038/nature02935 [DOI] [PubMed] [Google Scholar]
- Grosjean Y, Grillet M, Augustin H, Ferveur J-F, Featherstone DE (2008) A glial amino-acid transporter controls synapse strength and courtship in Drosophila. Nat Neurosci 11:54–61. 10.1038/nn2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka Y, Meinertzhagen IA (2010) Immunocytochemical localization of synaptic proteins to photoreceptor synapses of Drosophila melanogaster. J Comp Neurol 518:1133–1155. 10.1002/cne.22268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblen MJ, White NE, Emery PTJ, Kaiser K, Hall JC (1998) Molecular and behavioral analysis of four period mutants in Drosophila melanogaster encompassing extreme short, novel long, and unorthodox arrhythmic types. Genetics 149:165–178. 10.1093/genetics/149.1.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V (2011) Morphological diversity and development of glia in Drosophila. Glia 59:1237–1252. 10.1002/glia.21162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero A, Duhart JM, Ceriani MF (2017) Neuronal and glial clocks underlying structural remodeling of pacemaker neurons in Drosophila. Front Physiol 8:918. 10.3389/fphys.2017.00918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SH, Taghert PH (2010) PDF receptor expression reveals direct interactions between circadian oscillators in Drosophila. J Comp Neurol 518:1925–1945. 10.1002/cne.22311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson FR, Haydon PG (2008) Glial cell regulation of neurotransmission and behavior in Drosophila. Neuron glia Biol 4:11–17. 10.1017/S1740925X09000027 [DOI] [PubMed] [Google Scholar]
- Jackson FR, Ng FS, Sengupta S, You S, Huang Y (2015) Glial cell regulation of rhythmic behavior. Methods Enzymol 552:45–73. 10.1016/bs.mie.2014.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S (1971) Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A 68:2112–2116. 10.1073/pnas.68.9.2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahey T, Gorczyca M, Jia XX, Budnik V (1994) The Drosophila tumor suppressor gene dlg is required for normal synaptic bouton structure. Neuron 13:823–835. 10.1016/0896-6273(94)90249-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhou B, Yan W, Lei Z, Zhao X, Zhang K, Guo A (2014) Astrocyte-like glial cells physiologically regulate olfactory processing through the modification of ORN-PN synaptic strength in Drosophila. Eur J Neurosci 40:2744–2754. 10.1111/ejn.12646 [DOI] [PubMed] [Google Scholar]
- Long DM, Giebultowicz JM (2018) Age-related changes in the expression of the circadian clock protein PERIOD in Drosophila glial cells. Front Physiol 8:1131. 10.3389/fphys.2017.01131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnamee SE, Liu KE, Gerhard S, Tran CT, Fetter RD, Cardona A, Tolbert LP, Oland LA (2016) Astrocytic glutamate transport regulates a Drosophila CNS synapse that lacks astrocyte ensheathment. J Comp Neurol 524:1979–1998. 10.1002/cne.24016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno M, Horiuchi J, Yuasa Y, Ofusa K, Miyashita T, Masuda T, Saitoe M (2015) Long-term memory formation in Drosophila requires training-dependent glial transcription. J Neurosci 35:5557–5565. 10.1523/JNEUROSCI.3865-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL (2003) Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302:1765–1768. 10.1126/science.1089035 [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, O'Neil SD (1991) Synaptic organization of columnar elements in the lamina of the wild type in Drosophila melanogaster. J Comp Neurol 305:232–263. 10.1002/cne.903050206 [DOI] [PubMed] [Google Scholar]
- Ng FS, Jackson FR (2015) The ROP vesicle release factor is required in adult Drosophila glia for normal circadian behavior. Front Cell Neurosci 9:256. 10.3389/fncel.2015.00256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng FS, Tangredi MM, Jackson FR (2011) Glial cells physiologically modulate clock neurons and circadian behavior in a calcium-dependent manner. Curr Biol 21:625–634. 10.1016/j.cub.2011.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng FS, Sengupta S, Huang Y, Yu AM, You S, Roberts MA, Iyer LK, Yang Y, Jackson FR (2016) TRAP-seq profiling and RNAi-based genetic screens identify conserved glial genes required for adult Drosophila behavior. Front Mol Neurosci 9:1–15. 10.3389/fnmol.2016.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picot M, Cusumano P, Klarsfeld A, Ueda R, Rouyer F (2007) Light activates output from evening neurons and inhibits output from morning neurons in the Drosophila circadian clock. PLoS Biol 5:e315. 10.1371/journal.pbio.0050315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyza E, Górska-Andrzejak J (2004) Involvement of glial cells in rhythmic size changes in neurons of the housefly's visual system. J Neurobiol 59:205–215. 10.1002/neu.10307 [DOI] [PubMed] [Google Scholar]
- Schlichting M, Weidner P, Diaz M, Menegazzi P, Dalla Benetta E, Helfrich-Förster C, Rosbash M (2019) Light-mediated circuit switching in the Drosophila neuronal clock network. Curr Biol 29:3266–3276. 10.1016/j.cub.2019.08.033 [DOI] [PubMed] [Google Scholar]
- Seugnet L, Suzuki Y, Merlin G, Gottschalk L, Duntley SP, Shaw PJ (2011) Notch signaling modulates sleep homeostasis and learning after sleep deprivation in Drosophila. Curr Biol 21:835–840. 10.1016/j.cub.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivachenko A, Li Y, Abruzzi KC, Rosbash M (2013) The transcription factor Mef2 links the Drosophila core clock to Fas2, neuronal morphology, and circadian behavior. Neuron 79:281–292. 10.1016/j.neuron.2013.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwicki KK, Eastman C, Petersen G, Rosbash M, Hall JC (1988) Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron 1:141–150. 10.1016/0896-6273(88)90198-5 [DOI] [PubMed] [Google Scholar]
- Smalheiser NR (2007) Exosomal transfer of proteins and RNAs at synapses in the nervous system. Biol Direct 2:35. 10.1186/1745-6150-2-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl BA, Peco E, Davla S, Murakami K, Caicedo Moreno NA, van Meyel DJ, Keene AC (2018) The taurine transporter Eaat2 functions in ensheathing glia to modulate sleep and metabolic rate. Curr Biol 28:3700–3708.e4. 10.1016/j.cub.2018.10.039 [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M (2004) Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431:862–868. 10.1038/nature02926 [DOI] [PubMed] [Google Scholar]
- Strauss AL, Kawasaki F, Ordway RW (2015) A distinct perisynaptic glial cell type forms tripartite neuromuscular synapses in the Drosophila adult. PLoS One 10:e0129957. 10.1371/journal.pone.0129957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh J, Jackson FR (2007) Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron 55:435–447. 10.1016/j.neuron.2007.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue S, Krishnan P, Krishnan B, Dryer S, Hardin P (2004) Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila. Curr Biol 14:638–649. 10.1016/j.cub.2004.04.009 [DOI] [PubMed] [Google Scholar]
- Taylor AR, Robinson MB, Gifondorwa DJ, Tytell M, Milligan CE (2007) Regulation of heat shock protein 70 release in astrocytes: role of signaling kinases. Dev Neurobiol 67:1815–1829. 10.1002/dneu.20559 [DOI] [PubMed] [Google Scholar]
- Tix S, Eule E, Fischbach KF, Benzer S (1997) Glia in the chiasms and medulla of the Drosophila melanogaster optic lobes. Cell Tissue Res 289:397–409. 10.1007/s004410050886 [DOI] [PubMed] [Google Scholar]
- Tracy TE, Gan L (2018) Tau-mediated synaptic and neuronal dysfunction in neurodegenerative disease. Curr Opin Neurobiol 51:134–138. 10.1016/j.conb.2018.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderheyden WM, Goodman AG, Taylor RH, Frank MG, Van Dongen HPA, Gerstner JR (2018) Astrocyte expression of the Drosophila TNF-alpha homologue, Eiger, regulates sleep in flies. PLoS Genet 14:e1007724. 10.1371/journal.pgen.1007724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P, Kula-Eversole E, Pyza E (2009) Circadian control of dendrite morphology in the visual system of Drosophila melanogaster. PLoS One 4:e4290. 10.1371/journal.pone.0004290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki D, Horiuchi J, Ueno K, Ueno T, Saeki S, Matsuno M, Naganos S, Miyashita T, Hirano Y, Nishikawa H, Taoka M, Yamauchi Y, Isobe T, Honda Y, Kodama T, Masuda T, Saitoe M (2014) Glial dysfunction causes age-related memory impairment in Drosophila. Neuron 84:753–763. 10.1016/j.neuron.2014.09.039 [DOI] [PubMed] [Google Scholar]
- Yao Z, Shafer OT (2014) The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science 343:1516–1520. 10.1126/science.1251285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You S, Fulga TA, Van Vactor D, Jackson FR (2018) Regulation of circadian behavior by astroglial microRNAs in Drosophila. Genetics 208:1195–1207. 10.1534/genetics.117.300342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerr DM, Hall JC, Rosbash M, Siwicki KK (1990) Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J Neurosci 10:2749–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lear BC, Seluzicki A, Allada R (2009) The CRYPTOCHROME photoreceptor gates PDF neuropeptide signaling to set circadian network hierarchy in Drosophila. Curr Biol 19:2050–2055. 10.1016/j.cub.2009.10.058 [DOI] [PMC free article] [PubMed] [Google Scholar]