Abstract
Transcription requires unwinding complementary DNA strands, generating torsional stress, and sensitizing the exposed single strands to chemical reactions and endogenous damaging agents. In addition, transcription can occur concomitantly with the other major DNA metabolic processes (replication, repair, and recombination), creating opportunities for either cooperation or conflict. Genetic modifications associated with transcription are a global issue in the small genomes of microorganisms in which noncoding sequences are rare. Transcription likewise becomes significant when one considers that most of the human genome is transcriptionally active. In this review, we focus specifically on the mutagenic consequences of transcription. Mechanisms of transcription-associated mutagenesis in microorganisms are discussed, as is the role of transcription in somatic instability of the vertebrate immune system.
Keywords: DNA damage, topoisomerase, somatic hypermutation, class-switch recombination, cytosine deamination
INTRODUCTION
The maintenance of genome integrity is usually considered in relation to the three Rs: replication, repair, and recombination. This review focuses on how the other major DNA metabolic process---transcription---affects stability of the underlying DNA template. This is of particular significance when one considers that functional genes make up only 1% of the human genome, and yet recent estimates indicate that up to 80% of the human genome may be transcriptionally active (24a). An effect of transcription on mutagenesis was first recognized in microorganisms almost a half-century ago, but the diverse causes and potential evolutionary implications of transcription-associated mutagenesis (TAM) have only recently been appreciated.
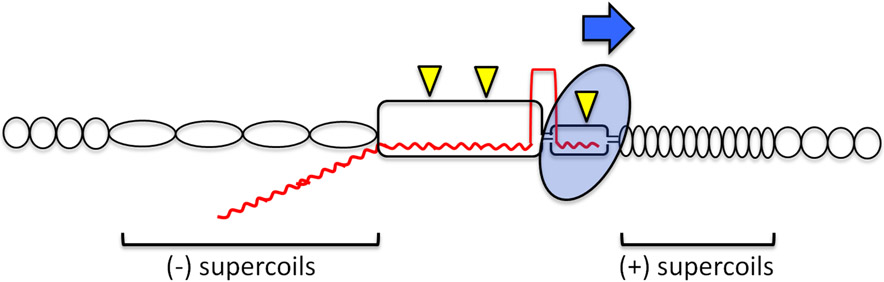
Transcription typically copies only one DNA strand, leaving the other (nontranscribed) strand in a transiently single-stranded state (32) that renders it chemically reactive and vulnerable to endogenous DNA damage. Within the transcription bubble transiently created by RNA polymerase (RNAP), only a short tract of single-stranded DNA (ssDNA) is exposed (Figure 1). Following RNA polymerization, the transcript and its complementary DNA strand exit RNAP through separate channels (99), thereby disrupting short RNA:DNA hybrids and promoting the reannealing of DNA strands. However, very long tracts of ssDNA can form if the transcript threads back and stably base pairs with its template. The resulting three-strand structure is referred to as an R-loop (Figure 1). Within R-loops the nontranscribed strand (NTS) is not only vulnerable to damage, it can also assume secondary structures that perturb or trigger other DNA metabolic processes. In prokaryotes, transcription and translation are coupled, with immediate transcript engagement by ribosomes preventing stable R-loop formation (34). In eukaryotes, where transcription and translation occur in separate cellular compartments, cotranscriptional processing of transcripts (e.g., splicing and nuclear transport) similarly discourages R-loop formation (58). Finally, transcription produces twin domains of positive and negative supercoiling (63). Positive supercoils are generated ahead of the transcription machinery and reflect overwinding of the helix as DNA strands are separated (Figure 1). Behind the machinery, the corresponding underwound state of DNA leads to the accumulation of negative supercoils. Underwinding exposes both DNA strands to endogenous damage and promotes R-loop formation, whereas overwinding can impede further strand separation. Supercoils are relaxed by topoisomerases, which nick and reseal one or both strands of DNA (97).
Figure 1.
Effects of transcription on the DNA template. The transcription bubble and a trailing R-loop are indicated as small and large rectangles, respectively. Circles indicate normal intertwining of DNA strands; compressed or extended ovals correspond to over- or underwound strands, respectively, and the regions of associated positive (+) or negative (−) supercoils are indicated. RNAP is depicted as a blue oval, and the blue arrow indicates its direction of movement on the DNA template. DNA and RNA strands are black and red, respectively; yellow triangles indicate damage to ssDNA.
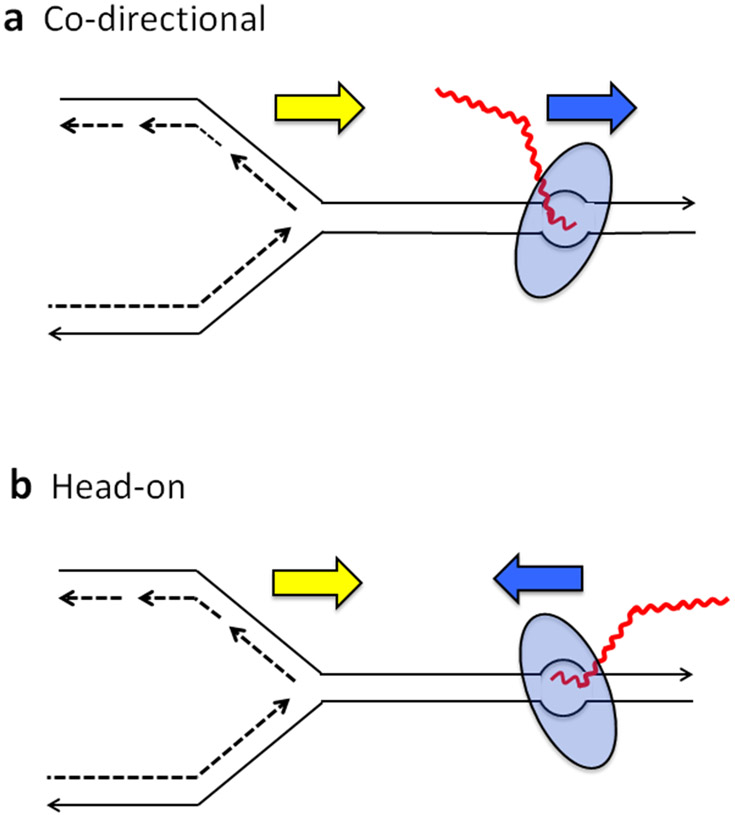
The potential effects of transcription on DNA stability become more complex if one considers that transcription occurs concurrently with and is influenced by other major DNA metabolic processes. Superimposed on top of transcription-associated DNA damage, for example, is the occurrence of transcription-coupled repair (TCR). TCR is triggered by damage in the transcribed strand (TS) that blocks RNAP and leads to recruitment of the nucleotide-excision repair (NER) machinery. This results in the preferential repair of lesions in the transcribed relative of the NTS of DNA (36). Potential conflicts between transcription and DNA replication have attracted particular attention; these conflicts are exacerbated by R-loop formation and are the major source of transcription-associated recombination (reviewed in 1). Transcription-replication conflicts are defined as codirectional if the replication fork moves in the same direction as the transcription machinery and as head-on if the two converge (Figure 2). In the codirectional orientation, the TS is the leading strand of replication; in the head-on orientation, the TS is the lagging strand of replication. Head-on conflicts are generally considered more detrimental than codirectional conflicts and have been invoked to explain the co-orientation of most bacterial genes with replication-fork movement, especially the highly transcribed ribosomal RNA operons, (65). However, because the rate of bacterial replication is approximately ten times faster than that of transcription, codirectional conflicts may also occur when the replication apparatus overtakes RNAP. In eukaryotes, the transcription and replication machineries move at similar rates, and transcription and replication are usually temporally separated within the cell cycle. Even so, transcription-replication conflicts within very long genes are inevitable and have been linked to common fragile sites in mammalian genomes (41). Here, only contributions of replication-transcription conflicts to localized mutagenesis are considered, and we refer the reader to several excellent reviews that deal with transcription-associated recombination and gross chromosome alterations (1, 40).
Figure 2.
Conflicts between the replication and transcription machineries. Movement of the replisome and RNAP in the same or opposite direction can cause (a) codirectional or (b) head-on conflicts, respectively. Red and black lines represent RNA and DNA, respectively; dashed lines depict newly synthesized DNA; blue ovals represent RNAP. Yellow and blue arrows indicate the direction of the replication fork and RNAP movement, respectively.
Although TAM clearly has pathological effects on genome integrity, it has been harnessed during evolution to drive localized and very rapid genetic change. This is particularly evident in the vertebrate immune system, where transcription is required for somatic hypermutation (SHM) and class-switch recombination (CSR) within immunoglobulin (Ig) genes. TAM also provides a potential source of replication-independent genetic change in nongrowing cells, and this has been implicated in stress-induced mutation in bacteria (103) and in trinucleotide-repeat instability in eukaryotes (59). Below, we summarize the characterized sources of TAM in bacteria, discuss specific mechanisms of TAM that have been uncovered in budding yeast, and consider the specific example of transcription-associated instability in the vertebrate immune system.
TRANSCRIPTION-ASSOCIATED MUTAGENESIS IN BACTERIA
The first suggestions of TAM date to the early 1970s, when it was reported that induction of the lac operon increased reversion caused by the frameshift mutagen ICR-191 in Escherichia coli (43). It similarly was reported that derepression of his genes increased UV-induced reversion in Salmonella typhimurium (86). Another 20 years passed, however, before a link between mutagenesis and transcription was definitively established.
The detection of TAM in bacteria (as well as in yeast; see below) has relied primarily on selective systems in which transcription can be varied at will. Most studies have used reversion assays, which are inherently limited because they detect only a subset of all possible mutations. A potential complication in reversion assays is that a transcription-driven increase in the corresponding gene product may shorten the time needed to express the selected phenotype and thereby artificially inflate the measured rate. In contrast to the functional restoration required by reversion assays, forward mutation assays select against the encoded protein. Although the spectrum of mutation types detected is much broader, elevated transcription can exacerbate an associated phenotypic lag (i.e., the wild-type gene product must be diluted out before the mutant phenotype is expressed), and this has the potential to underestimate or completely mask TAM. Finally, in addition to inherent biases associated with a given assay, the magnitude and/or mechanism of TAM may be affected by location of the reporter on a plasmid versus the chromosome, orientation of the reporter relative to replication fork movement, and the specific growth conditions used. These issues should be borne in mind in the TAM descriptions that follow.
DNA Damage and Strand-Related Asymmetries in Mutation Accumulation
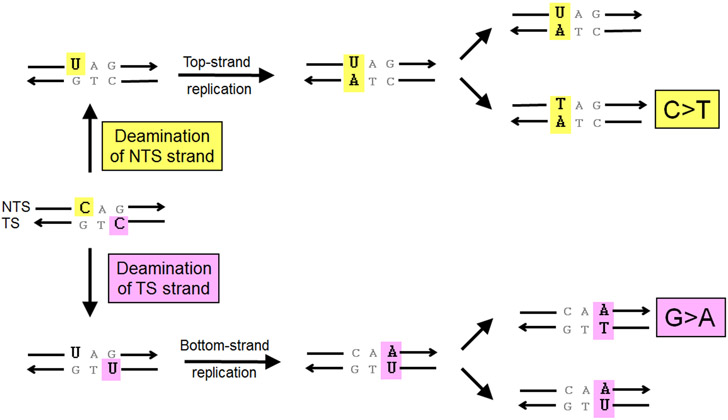
An early observation of a strand-related bias in mutation accumulation was made in E. coli, where the sequence change diagnostic of hydrolytic cytosine deamination to uracil was strongly biased to the NTS of lacI (26). In this and other experiments that have focused on a particular type of damage, strain backgrounds that are defective in its repair are often used. By convention, the sequence of the NTS, which is identical to that of the mRNA, is the sequence reported. Thus, as illustrated in Figure 3, deamination of cytosine on the NTS results in C > T mutations, whereas deamination of cytosine on the TS generates G > A sequence changes. Thus, by comparing the accumulation of C > T versus G > A changes, one can infer relative deamination of the NTS versus TS strand, respectively. Comparative analysis of genes in E. coli and S. enterica indicates that the cytosine deamination bias primarily reflects an asymmetry associated with transcription rather than replication (28). Although it was not possible to infer whether the strand-associated asymmetry reflected preferential damage of the NTS and/or biased repair of the TS via TCR, a subsequent comparison of mutation patterns in expressed versus nonexpressed DNA was more consistent with the former (29). The enhanced deamination of the NTS inferred in vivo is consistent with much faster cytosine deamination in ssDNA than in double-stranded DNA in vitro (30).
Figure 3.
Inferring strand specificity from mutation patterns associated with cytosine deamination. Yellow and pink boxes indicate consequences of cytosine deamination on the nontranscribed strand (NTS) and transcribed strand (TS), respectively.
The first direct demonstration of a correlation between transcription level and preferential deamination of cytosine on the NTS came through analyzing reversion of a missense allele under control of the highly inducible tac promoter (6). It was found that the bias for cytosine deamination on the NTS was evident only if transcription was highly activated. Importantly, the NTS bias was maintained when the direction of transcription through the reporter was reversed and was thus independent of the sequence of the NTS. That the NTS has single-strand characteristics relative to the TS is additionally supported by its enhanced sensitivity to enzymatic deamination following expression of a mammalian cytosine deaminase (see below).
In addition to the specific case of cytosine deamination, an NTS bias is also evident for spontaneous oxidative lesions (53) and for damage generated by the alkylating agent methylmethane sulfonate (MMS) (25). Oxidative damage to guanine generates 7,8-dihydro-8-oxo-guanine (8-oxoG), which mispairs frequently with adenine and gives rise to GC > TA mutations. Reversion of a TGA stop codon via 8-oxoG-associated G > T transversions was examined at a reporter inserted in both orientations relative to the strong tac promoter. Significantly, transcription from Ptac elevated G > T tranversions only if the stop codon was on the NTS, consistent with enhanced transcription-associated damage to this strand (53). Interestingly, high levels of transcription reduced reversion when the TGA was on the TS, suggesting that preferential repair of lesions on the TS via TCR may also contribute to some of the strand specificity. A strand bias of MMS-induced mutations was similarly assayed by scoring reversion of a CCA missense allele inserted in either orientation relative to Ptac (25). Inducing transcription caused mutations only at cytosines in the NTS, consistent with methylation targeted to ssDNA. Whether TAM in these systems primarily reflects ssDNA within the transcription bubble or more extensive ssDNA exposed within R-loops has not been specifically addressed.
The observation that many different types of base substitutions accumulate in a transcription-dependent fashion underscores the generality of TAM (46, 52). Mutagenesis is initiated more frequently on the NTS than on the TS strand of active genes, but whether all nucleotides on the NTS are equally mutable is unclear. It has been suggested, for example, that the folding of ssDNA into stem-loop structures exposes bases in single-strand loops to endogenous damage and renders them hypermutable. The mfd program developed by Wright and colleagues uses the free energy of all possible stem-loop structures to derive a mutability index for each base within a short stretch of ssDNA (105). Correlations have been observed between the calculated mutability index and reversion rates at specific sites in E. coli reporters (11, 87), and a similar correlation has been noted for highly mutable sites in the p53 tumor suppressor gene (107) and in Ig genes (106, 108).
Effects of Starvation/Stress on Mutagenesis
TAM is readily observed when transcription is induced to high levels in a reporter fused to a heterologous promoter, but elevated transcription is also a natural response of bacteria to amino acid starvation. Starvation generally induces/derepresses only those genes relevant to biosynthesis of the corresponding amino acid and is additionally modulated as part of the ppGpp-mediate stringent response. An effect of the stringent response was found when examining reversion of leuB and argH alleles in E. coli, and it was speculated that this could reflect an associated increase in transcription (102). A correlation between reversion and starvation-induced transcription was subsequently established (104), and the relationship between the two is linear (81). One important consequence of starvation-stimulated transcription is that mutagenesis is higher in those genes in which changes can potentially be beneficial (reviewed in 103), and it is possible that a similar phenomenon may underlie some examples of adaptive mutation (18).
A relationship between stress-associated transcription and reversion has also been reported in Bacillus subtilis. In this case, the correlation was made under prolonged starvation conditions in which mutations accumulated in a replication-independent, but time-dependent, manner (80). It has been argued that such stress-induced mutations represent an adaptive response that fosters rapid evolutionary change (reviewed in 31). Interestingly, recent work in E. coli has demonstrated the importance of R-loops in mutagenesis that occurs in stressed cells (100). In this case, a novel mechanism of transcription-initiated genetic instability was proposed in which an exposed 3′ end of the RNA within an R-loop is used to initiate origin-independent replication. A subsequent encounter of DNA polymerase with a nick on the template strand is hypothesized to generate a double-strand end that then initiates recombination-associated mutagenesis.
Replication-Transcription Conflicts
Head-on encounters between highly transcribed ribosomal RNA genes and replication forks slow DNA synthesis and affect overall fitness (92). A strong codirectional orientation bias also has been reported for a set of core genes common to diverged B. subtilis strains (75). Comparative analyses of these strains suggested that nonsynonymous changes accumulate faster in core genes with the head-on orientation and that there is a positive correlation between these changes and transcript abundance. In the few cases in which the direction of replication-fork movement on mutagenesis within a defined reporter has been directly examined, mutation rates were higher in the head-on orientation than in the codirectional orientation. In B. subtilis, for example, reversion of a hisC nonsense allele was affected by the direction of replication but only under conditions of high transcription (75). In E. coli, forward mutations in rpoB were similarly higher in the head-on orientation than in the codirectional orientation, but the specific contribution of transcription was not examined (92). Although the reason why head-on conflicts are more mutagenic than codirectional conflicts is not known, studies in yeast suggest there may be a recombination connection. In particular, recombination-associated DNA synthesis is more error prone than replicative DNA synthesis (44, 95), and head-on transcription-replication conflicts stimulate recombination more than do codirectional conflicts (79).
TRANSCRIPTION-ASSOCIATED MUTAGENESIS IN YEAST
TAM has been well documented in Saccharomyces cerevisiae, but similar reports have not emerged from the other major yeast model, Schizosaccharomyces pombe. Although all the data described below were obtained using budding yeast, there is no a priori reason to suspect that results will not be widely applicable to other eukaryotes. An early indication that TAM occurs in yeast came 20 years after initial reports in bacteria. As in bacteria, it was found that limiting a specific amino acid was associated with elevated reversion of a gene in the corresponding biosynthetic pathway, and it similarly was speculated that this might be related to starvation-associated induction of transcription (55). Definitive evidence of TAM was obtained following fusion of a forward- or reverse-mutation reporter to the highly inducible, galactose-regulated pGAL promoter (17). Subsequent studies of TAM have used pGAL or the heterologous tetracycline/doxycycline-regulated pTET promoter. As in bacterial cells, there is a direct proportionality between the transcript level and mutagenesis (48). Although head-on encounters between transcription and replication forks in yeast also slow DNA replication more than do codirectional encounters (20, 79), reversing the direction of replication through a pTET-driven frameshift reporter had no significant effect on the reversion rate. There were, however, orientation-specific effects evident in the corresponding spectra (48, 49). On a genome-wide scale, the accumulation of DNA polymerase correlates with high transcription, indicating that both head-on and codirectional conflicts slow replication in yeast (4).
A central question has been whether results obtained with a small number of reporter genes are relevant on an evolutionary timescale. This has recently been addressed through comparative analysis of S. cerevisiae and Saccharomyces paradoxus genomes, as well as by sequencing spontaneous mutations that accumulate over hundreds of generations in budding yeast. Both types of analysis revealed a positive correlation between transcription and mutagenesis (73). Importantly, analyses were confined to intronic sequences, thereby removing confounding selective constraints on the analyzed sequences. Below, we focus on the diverse mechanisms that contribute to TAM in budding yeast.
DNA Damage as a Source of Transcription-Associated Mutagenesis
The most extensive TAM studies have been done using LYS2-based frameshift reversion assays that detect either net +1 or −1 events. Genetic studies with these systems (17, 50, 66), as well as recent experiments with nonsense reversion assays (2, 51), have implicated DNA damage as a major source of TAM. Key observations have been that TAM increases when an error-free mechanism of lesion bypass (i.e., template switch or homologous recombination) is impaired, increases when either NER or base-excision repair (BER) is inactivated, and decreases in the absence of the error-prone translesion synthesis (TLS) DNA polymerase Pol ζ (for a review of repair/bypass pathways in yeast, see 9). In nonsense reversion assays, all detectable base substitutions were elevated, but a strong proportional increase in transversions at GC base pairs was noted (2, 51). Although no preferential accumulation of spontaneous damage on the NTS was evident, it should be noted that nonsense reversion assays are incapable of detecting the CG > TA mutations characteristic of cytosine deamination. In relation to possible strand specificity, enzymatic deamination of cytosine by human activation-induced deaminase (AID) was reported to target both DNA strands, suggesting that negative supercoiling behind the transcription machinery may be relevant. However, deamination occurred preferentially on the NTS when conditions favoring R-loop formation were used (33).
Topoisomerase 1 as a Mutagen in Transcriptionally Active DNA
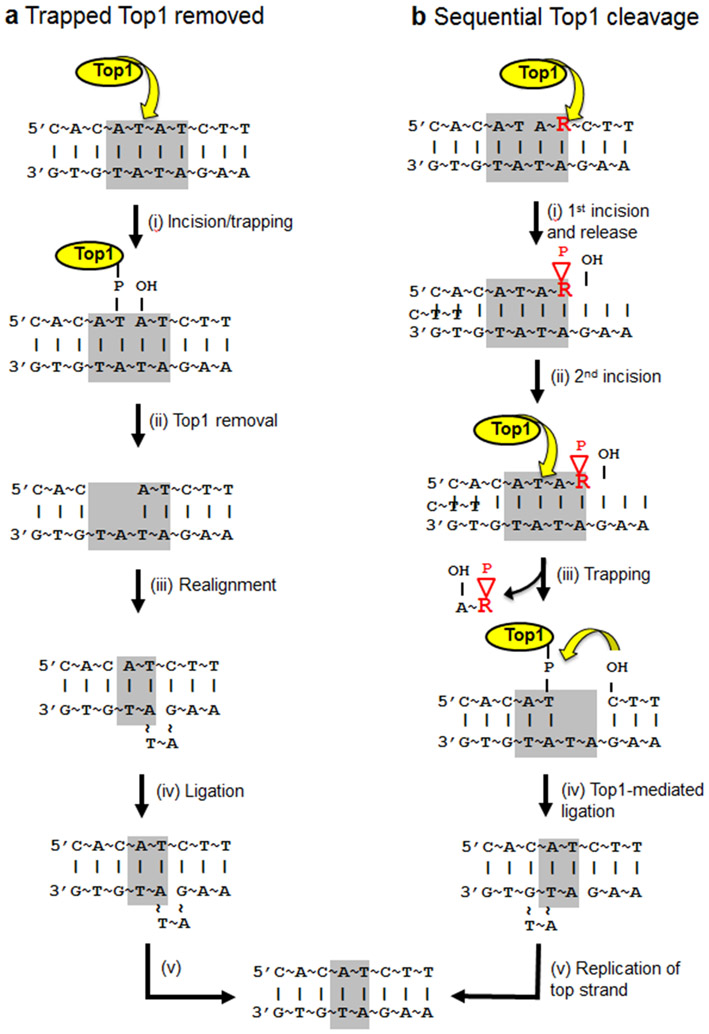
Sequence analysis in a pGAL-LYS2 forward-mutation assay revealed that most, if not all, mutation types were elevated by transcription, but established small deletions of 2--5 bp as a specific signature of TAM. These events made up ~25% of mutations if a reporter was highly transcribed but were absent if transcription occurred at very low levels (61). Subsequent analyses of mutagenesis in pGAL-CAN1 and pTET-CAN1 reporters confirmed the short-deletion TAM signature and demonstrated that events accumulate at discrete tandem-repeat hot spots (62, 96). The size of the deletion corresponded to the size of the repeat unit, and the repeat was present in only —two to four copies prior to the deletion event. Furthermore, the primary sequence of the repeat unit was highly variable, indicating that any repeat can potentially harbor a transcription-associated deletion. Significantly, short deletions were completely eliminated upon loss of topoisomerase 1 (Top1), an enzyme that resolves transcription-associated supercoils by nicking and resealing one strand of DNA. Subsequent work demonstrated that the Top1-dependent hot spots are of two distinct types: those that reflect processing of a covalently trapped Top1 cleavage complex and those that reflect incision at a ribonucleoside monophosphate (rNMP) embedded in duplex DNA (Figure 4) (15). It should be noted that this particular TAM signature is expected to be associated only with a eukaryotic-specific type 1B enzyme, which forms a 3′-phosphotyrosyl link to the nicked DNA. It is possible, however, that other types of topoisomerase-mediated damage may have mutagenic consequences that have yet to be defined.
Figure 4.
Mechanisms of Top1 mutagenesis in transcriptionally active DNA. Two distinct mechanisms of Top1-dependent mutagenesis are shown, with a hypothetical dinucleotide repeat highlighted in gray. When Top1 incision occurs, the active-site tyrosine forms a covalent linkage to the 3′-PO4 on one side of the DNA nick, leaving a 5′-OH on the other side. (a) Top1 becomes trapped as a stabilized cleavage complex (step i), and its removal by unknown proteins generates a 2-nt gap within the 2-bp tandem repeat (step ii). Realignment of the DNA strands converts the gap to a nick (step iii), which facilitates ligation and produces the mutation intermediate (step iv). (top) Replication of the newly ligated strand results in a permanent, 2-bp deletion (step v); replication of the other strand results is of no genetic consequence. (b) Top1 incises at the position of an rNMP (red R). The 2′-OH of ribose attacks the phosphotyrosyl bond, releasing Top1 and generating a 2′,3′-cyclic phosphate (red triangle; step i). A second incision by Top1 upstream of the nick (step ii) releases the intervening oligonucleotide and transiently traps the covalent enzyme-DNA intermediate (step iii). Realignment of the two DNA strands by the repeat sequence correctly orients the Top1-DNA complex and the 5′-OH, enabling efficient Top1-mediated rejoining of the ends (step iv). Replication of the top strand fixes the 2-bp deletion (step v).
RNA: DNA Hybrids Initiate Complex Mutations in Highly Transcribed DNA
The mutagenic consequences of Top1 incision at an rNMP are most evident in the absence of RNase H2, an enzyme that initiates error-free removal of 1--3 rNMPs from DNA as well as the degradation of the RNA component of R-loops (13, 91). Studies in RNase H2-deficient strains have revealed the occurrence of complex mutations, which are characterized by multiple, simultaneous sequence changes that extend identity between the arms of an imperfect inverted repeat. Although these mutations are evident only under conditions of highly activated transcription, they are mechanistically distinct from rNMP-initiated small deletions, in that they do not require Top1 activity and are strongly affected by the direction of replication-fork movement (49). The effect of replication direction coupled with multiple sequence changes suggests a template-switch mechanism, with persistent rNMPs in the DNA template being the likely trigger. An additional requirement for RNase H1, which likely only processes R-loops, for the generation of complex mutations suggests that either the RNA primers of Okazaki fragments or cotranscriptional R-loops may be the source of the relevant rNMPs (49).
Replacement of Thymine with Uracil in Transcriptionally Active DNA
Hydrolytic or enzymatic release of a base from the phosphodiester backbone generates an apurinic/apyrimidic (AP) site that is a potent block to both DNA and RNAPs. Genetic studies with a frameshift reversion assay revealed that TAM increased when AP-site repair was disrupted, indicating that AP sites are one type of damage that initiates TAM (66). Yeast has five DNA N-glyosylases that remove abnormal or damaged bases from DNA (reviewed in 9), and each was eliminated to determine its contribution to AP-site formation. The only glycosylase relevant to TAM was uracil N-glycosylase (UNG), which specifically excises uracil from DNA (50). Uracil in DNA can result from cytosine deamination, as noted previously, or can arise through use of dUTP in place of dTTP during DNA synthesis. The relevance of the latter was demonstrated by showing a reduction in TAM upon overproduction of Dut1, an enzyme that hydrolyzes dUTP to prevent its use during DNA synthesis (50). It should be noted that the assays used did not exclude introduction of uracil into transcriptionally active DNA via spontaneous cytosine deamination as well. The reason why elevated levels of uracil are incorporated into DNA under high-transcription conditions remains a subject of investigation.
Does Transcription Contribute to Mutagenesis in Nongrowing or Stressed Cells?
Budding yeast does not have a stress response analogous to the SOS system of bacterial cells, which promotes global mutagenesis through the activation of TLS polymerases (31). Nevertheless, in yeast, as in bacterial cells, starvation for a specific nutrient or provision of a specific carbon source can induce the expression of genes encoding the corresponding biosynthetic or catabolic activities (reviewed in 45). It seems likely that TAM will be relevant to at least some examples of so-called adaptive mutation in yeast (39), but this issue has not been specifically addressed. Indeed, genome-wide positions of DNA turnover in stationary-phase cells have been correlated with transcription in microarray-based analyses (19). Such replication-independent DNA synthesis likely reflects repair reactions, which in turn may reflect transcription-associated damage to the DNA template. A relevant observation may be the recent report that abnormal transcription in nondividing yeast cells contributes to trinucleotide repeat instability (114). The specific relationship between transcription and the instability of sequences that can adopt non-B secondary structures (e.g., stem-loops, triplexes, or G-quadruplexes) has been the subject of a recent review (7) and is not further considered here.
TRANSCRIPTION-ASSOCIATED MUTAGENESIS IN HIGHER EUKARYOTES
Given the universality of DNA structure and the high conservation in basic DNA metabolic processes, it seems likely that many of the TAM mechanisms documented in microorganisms will extend to higher eukaryotes. Attempts to link elevated transcription to increased forward mutation in a specific target gene have been unsuccessful, however (60, 71). This could reflect either a correspondingly lengthened phenotypic lag or, given the proportionality between transcription and mutagenesis observed in microbial systems, an insufficient level of transcription to detect an effect. More recently, a very specific case of TAM emerged when examining the strand-specificity of UV-induced mutations in the hprt gene (42). Furthermore, comparative genome analyses suggest a global link between transcription and mutagenesis (35, 78). Although a focus on TAM in individual genes has generally been unsuccessful, evolution has co-opted this process to drive maturation of the vertebrate immune system. The remainder of this review focuses on this specialized case of TAM, in which some of the lessons learned may be more generally applicable.
Features of Somatic Hypermutation and Class-Switch Recombination
Vertebrate antibody genes undergo three genetic alterations that result in antibody maturation (Figure 5): SHM , gene conversion (GC), and CSR. We summarize here recent work regarding the transcription dependence of SHM and CSR, focusing mainly on mammalian systems. More extensive discussion of relevant literature and detailed models for the role of transcription in AID-generated hypermutations may be found in other reviews (85, 93, 94).
Figure 5.
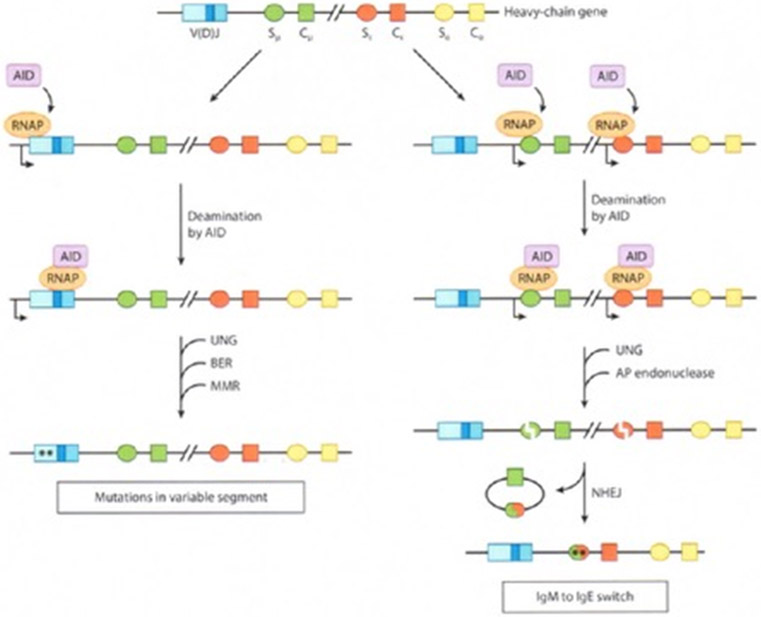
A model for somatic hypermutation (SHM) and class-switch recombination (CSR) during antibody maturation. Different sequence elements are shown as rectangles or ovals of different colors. The direction of transcription is indicated by a rightward arrow. AID is recruited at a proximal pause site for RNAP II. Following release from the pause, AID travels with RNAP II as it transcribes DNA and converts cytosine to uracil on each DNA strand. Uracil is excised by uracil N-glycosylase (UNG) and processed by error-prone base-excision repair (BER) or by mismatch repair (MMR) to introduce point mutations, which are indicated by asterisks. To initiate CSR, double-strand breaks are generated by AP endonuclease (64a) incision at UNG-generated apurinic/apyrimidinic AP sites. Broken ends are ligated by the nonhomologous end-joining (NHEJ) pathway, and the intervening DNA is released as a switch circle. Abbreviations: C, constant segment (only the μ, ε, α regions are shown); S, switch region preceding each C segment; V(D)J, variable segment.
The variable segment of an Ig molecule interacts with an antigen. SHM generates point mutations that enable antibody-antigen interactions to be fine-tuned and optimized by affinity selection. SHM occurs within variable segment of Ig genes, where it is confined to an ~1,500-bp region that begins ~150 bp downstream of Ig promoters; and within the S regions, which are targets of CSR. CSR is a region-specific recombination event that replaces the default constant segment of the Ig heavy chain (μ) with one of the other constant segments, thereby changing the functional consequence of an antibody-antigen interaction.
AID is a B-cell specific deaminase that converts cytosines to uracils in ssDNA but not in double-stranded DNA (10, 14, 21, 90) and is required to initiate SHM and CSR (3, 37, 67). During SHM, AID-generated uracils are either not repaired, leading to CG > TA transitions, or are repaired by error-prone pathways to create other types of mutations. During CSR, processing of AID-generated uracils within noncoding switch (S) regions creates double-strand breaks that initiate genetic rearrangement; collateral base substitutions are also acquired within the S regions. Both SHM and S-region mutations occur at frequencies that are several orders of magnitude higher than the normal somatic mutation frequency and both require transcription of the target sequences.
The Role of Transcription in Somatic Hypermutation and Class-Switch Recombination
AID immunoprecipitates with RNAP II (69), and ChIP-seq analysis demonstrates that AID associates with nearly 6,000 genes in stimulated murine B cells (112). Genes associated with AID have a corresponding mRNA abundance 40 times greater than that of genes that did not recruit the protein. In transcriptionally active genes, AID and RNAP II peaked at the transcription start site, and AID occupancy mirrored RNAP II density along individual genes. Although most of the non-Ig genes that recruited AID were not hypermutated in stimulated wild-type B cells, mutations did accumulate in an UNG−/− background (112). These and earlier results (8, 64, 74, 88) demonstrate that AID targets many RNAP II-transcribed genes but that the level of deamination-associated, off-target mutagenesis is much lower in other genes than in Ig genes.
A key question regarding the transcription dependence of SHM is how AID is recruited to transcribed DNA. At least three possibilities have been considered, and these are not mutually exclusive: (a) transcription promotes formation of non-B DNA structures to which AID preferentially binds, (b) transcription-associated chromatin modifications recruit AID, and (c) specific transcription-associated protein factors recruit AID. As mentioned previously, transcription can generate a variety of non-B DNA structures. It has been suggested, for example, that formation of stem-loop structures in the variable segment of Ig genes sensitizes bases in single-strand loops to deamination by AID (106, 108). Within S regions, the asymmetric distribution of guanines on the NTS promotes R-loop formation (113), with the displaced strand furthermore having the ability to form G-quadruplex DNA (22, 23). AID may interact specifically with the ssDNA within these structures, or these structures may cause the elongating RNAP II to pause or stall. AID may also interact with DNA in stalled transcription bubbles (24). One difficulty in explaining SHM based on transcription-associated, non-B DNA structures alone is that these structures are inherently asymmetric and primarily affect only one DNA strand. By contrast, AID-associated cytosine deamination lacks a strand bias (88, 109), and closely spaced nicks on both DNA strands of S regions are likely required to create the double-strand breaks required for CSR. One possibility is that negative supercoiling behind the transcription machinery renders both DNA strands accessible to AID, and this might explain the inverse correlation between Top1 level and SHM frequency (54).
There are complex changes in chrormatin regions of Ig genes during antibody maturation, and these are more fully discussed in other reviews (for example, 57). It is useful to note, however, that in heavy-chain genes, different S regions are transcribed in response to different cytokine stimuli, and only the transcribed regions undergo switching and hypermutation (109). Although activating histone marks and germ-line transcripts are found in the Sμ region even in unstimulated B cells, active hypermutation of other S regions requires cytokine activation of B cells (68). Interestingly, treatment of hypermutating cells with the histone deacetylase inhibitor, trichostatin A, increased histone H4 acetylation and resulted in hypermutation of a normally unmutated constant-segment exon (101).
Numerous proteins or protein complexes have reported interactions with AID (56). Many of the AID-associated nuclear proteins are involved in transcription and RNA processing, and it is possible that AID is part of transcripton ‘factories’ that contain many transcription-related protein factors (72). The DRB sensitivity-inducing factor (DSIF) complex, for example, causes RNAP II to pause shortly after promoter clearance and, following release from this pause, DSIF travels with the elongating polymerase (38, 77, 111). AID interacts with the Spt5 component of DSIF in vitro and in vivo and the co-occupancy of genes with AID and RNAP II depends on the presence of Spt5 (76). Although Spt5 occupancy of genes correlates with hypermutation frequencies associated with CSR (76), recent work demonstrated that Spt5 knockdown slightly increased SHM (98). With regard to SHM, it was suggested that knockdown of Spt5 reduced RNAP II processivity, promoting transcription termination and RNA degradation by the exosome.
Components of the RNA exosome complex interact with AID (5), and their knockdown reduces both CSR (5) and SHM (98). Furthermore, addition of RNA exosome-enriched extracts to an in vitro transcription system enhances the ability of AID to target both DNA strands (5). An attractive model is that degradation of pre-mRNA by the RNA exosome upon stalling of RNAP II makes both DNA strands accessible to AID. ChIP-Seq analysis has shown that replication protein A (RPA), an AID cofactor in CSR, is localized to Ig switch regions but not to most non-Ig AID-targeted genes (112). In addition, depletion of a specific isoform of the pre-mRNA splicing factor SRSF1 (serine/arginine rich splicing factor-1) has been reported to suppress SHM in chicken DT40 cells without affecting off-target mutagenesis. It was suggested that an isoform-associated reduction of splicing specifically at variable segments creates cotranscriptional R-loops, thereby generating the requisite ssDNA substrates for AID (47).
It is likely that local changes in DNA structure and chromatin remodeling, along with the help of protein chaperones and enzymes, together conspire to hypermutate a transcribed gene in an AID-dependent manner. Although the focus here has been on the specific relationship between AID and transcription during targeted mutagenesis in the vertebrate immune system, it is important to note that AID is only one member of the larger APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like) family of cytosine deaminases (89). APOBECs have been implicated in the generation of clustered mutations (“kataegis”) in tumor cells (82), with persistent ssDNA generated during double-strand break repair providing the substrate for a similar phenomenon in budding yeast (12, 70, 83, 84). It seems likely that the ssDNA component of cotranscriptional DNA structures will also be a target of APOBECs as well as endogenous DNA damage.
SUMMARY POINTS.
Transcription creates transient regions of ssDNA, which is more chemically reactive and damage accessible than duplex DNA. ssDNA exists within the transcription bubble created by RNAP, is enhanced by negative supercoiling, and is associated with R-loop structures.
In bacterial cells, TAM preferentially targets the NTS of active genes and is influenced by the direction of replication-fork movement. Importantly, transcription may be relevant to stress responses and adaptation to adverse/novel environments.
In budding yeast, transcription elevates all mutation types. Documented causes of TAM include an increase in associated DNA damage, an elevation in direct dUMP incorporation into the underlying DNA template, and recruitment of Top1 to relieve associated supercoiling.
Genetic alterations associated with SHM and CSR in the vertebrate immune system provide an example of the importance of transcription in regulated genetic instability.
Comparative genome analyses suggest that transcription modifies the mutation landscape in both prokaryotes and eukaryotes on an evolutionary timescale.
ACKNOWLEDGMENTS
A.S.B. would like to thank Ursula Storb (University of Chicago) and David Schatz (Yale University) for sharing unpublished data and for useful discussions, and Sophia Shalhout (Wayne State University) for comments on the manuscript. S.J.R. gratefully acknowledges the intellectual and experimental contributions of past and present lab members. A.S.B. has been supported by National Institutes of Health grant GM057200 and by a Grants-Plus award from Wayne State University. Transcription-related work in the S.J.R. lab has been funded by National Institutes of Health grants GM038464, GM093197. and GM101690.
ACRONYMS/DEFINITIONS
- Activation-induced deaminase (AID)
enzyme that deaminates cytosine in DNA to uracil and is required for postinfection genetic alterations in Ig genes
- Apurinic/apyrimidinic (AP) site
site in DNA that is missing a base; also referred to as an abasic site
- Base-excision repair (BER)
incises the DNA backbone adjacent to an abasic site and initiates replacement of an abasic site with a nucleotide specified by the complementary strand
- Immunoglobulin (Ig)
antigen-binding protein that comprises two heavy and two light chains; also referred to as an antibody
- Nontranscribed strand (NTS)
DNA strand that has the same sequence as the RNA transcript; also referred to as the coding strand
- Nucleotide-excision repair (NER)
removes a lesion-containing oligonucleotide, leaving a 20--25 nt gap that is filled in using the undamaged strand as template
- Replication protein A (RPA)
heterotrimeric complex that binds ssDNA and prevents pairing between complementary strands
- R-loop
three-strand structure in which RNA is base-paired with one strand of duplex DNA, leaving the other DNA strand unpaired
- RNA polymerase (RNAP)
the enzyme/complex that makes an RNA copy of a DNA template; RNAP II specifically synthesizes mRNA
- Switch (S) regions
GC-rich, repetitive regions upstream of the constant segment of Ig heavy-chain exons where class-switch recombination occurs; are several kb in length
- Topoisomerase 1 (Top1)
eukaryotic Type 1B enzyme that relaxes supercoils and forms a 3′-phosphotyrosyl linkage when it nicks DNA
- Transcribed strand (TS)
strand of DNA copied by RNAP
- Transcription-associated mutagenesis (TAM)
localized changes in DNA that are associated with transcription of the target sequence
- Transcription-coupled repair (TCR)
NER subpathway that specifically removes damage from the transcribed strand of active genes
- Translesion synthesis (TLS)
polymerization of DNA opposite lesions by specialized, low-fidelity DNA polymerases
- Uracil N-glycosylase (UNG)
enzyme that removes uracil from DNA, creating an AP site
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Aguilera A, Garcia-Muse T. 2012. R loops: from transcription byproducts to threats to genome stability. Mol. Cell 46:115–24 [DOI] [PubMed] [Google Scholar]
- 2.Alexander MP, Begins KJ, Crall WC, Holmes MP, Lippert MJ. 2013. High levels of transcription stimulate transversions at GC base pairs in yeast. Environ. Mol. Mutagen 54:44–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arakawa H, Hauschild J, Buerstedde JM. 2002. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science 295:1301–6 [DOI] [PubMed] [Google Scholar]
- 4.Azvolinsky A, Giresi PG, Lieb JD, Zakian VA. 2009. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol. Cell 34:722–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basu U, Meng FL, Keim C, Grinstein V, Pefanis E, et al. 2011. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell 144:353–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beletskii A, Bhagwat AS. 1996. Transcription-induced mutations: increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:13919–24 Demonstrated that cytosine deamination is more frequent on the NTS in E. coli.
- 7.Belotserkovskii BP, Mirkin SM, Hanawalt PC. 2013. DNA sequences that interfere with transcription: implications for genome function and stability. Chem. Rev 113:8620–37 [DOI] [PubMed] [Google Scholar]
- 8.Bemark M, Neuberger MS. 2000. The c-MYC allele that is translocated into the IgH locus undergoes constitutive hypermutation in a Burkitt’s lymphoma line. Oncogene 19:3404–10 [DOI] [PubMed] [Google Scholar]
- 9.Boiteux S, Jinks-Robertson S. 2013. DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae. Genetics 193:1025–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bransteitter R, Pham P, Scharff MD, Goodman MF. 2003. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. USA 100:4102–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burkala E, Reimers JM, Schmidt KH, Davis N, Wei P, Wright BE. 2007. Secondary structures as predictors of mutation potential in the lacZ gene of Escherichia coli. Microbiology 153:2180–89 [DOI] [PubMed] [Google Scholar]
- 12.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, et al. 2013. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 494:366–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerritelli SM, Crouch RJ. 2009. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 276:1494–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 422:726–30 [DOI] [PubMed] [Google Scholar]
- 15.Cho JE, Kim N, Li YC, Jinks-Robertson S. 2013. Two distinct mechanisms of topoisomerase 1--dependent mutagenesis in yeast. DNA Repair 12:205–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Datta A, Jinks-Robertson S. 1995. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science 268:1616–19 [DOI] [PubMed] [Google Scholar]
- 18.Davis BD. 1989. Transcriptional bias: a non-Lamarckian mechanism for substrate-induced mutations. Proc. Natl. Acad. Sci. USA 86:5005–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Morgan A, Brodsky L, Ronin Y, Nevo E, Korol A, Kashi Y. 2010. Genome-wide analysis of DNA turnover and gene expression in stationary-phase Saccharomyces cerevisiae. Microbiology 156:1758–71 [DOI] [PubMed] [Google Scholar]
- 20.Deshpande AM, Newlon CS. 1996. DNA replication fork pause sites dependent on transcription. Science 272:1030–33 [DOI] [PubMed] [Google Scholar]
- 21.Dickerson SK, Market E, Besmer E, Papavasiliou FN. 2003. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med 197:1291–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. 2004. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 18:1618–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duquette ML, Pham P, Goodman MF, Maizels N. 2005. AID binds to transcription-induced structures in c-MYC that map to regions associated with translocation and hypermutation. Oncogene 24:5791–98 [DOI] [PubMed] [Google Scholar]
- 24.Eddy J, Vallur AC, Varma S, Liu H, Reinhold WC, et al. 2011. G4 motifs correlate with promoter-proximal transcriptional pausing in human genes. Nucleic Acids Res. 39:4975–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.ENCODE Proj. Consort., Bernstein BE, Birney E, Dunham I, Green ED, et al. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fix D, Canugovi C, Bhagwat AS. 2008. Transcription increases methylmethane sulfonate-induced mutations in alkB strains of Escherichia coli. DNA Repair 7:1289–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fix DF, Glickman BW. 1987. Asymmetric cytosine deamination revealed by spontaneous mutational specificity in an Ung- strain of Escherichia coli. Mol. Gen. Genet 209:78–82 [DOI] [PubMed] [Google Scholar]
- 27.Fraenkel S, Mostoslavsky R, Novobrantseva TI, Pelanda R, Chaudhuri J, et al. 2007. Allelic “choice” governs somatic hypermutation in vivo at the immunoglobulin kappa-chain locus. Nat. Immunol 8:715–22 [DOI] [PubMed] [Google Scholar]
- 28.Francino MP, Chao L, Riley MA, Ochman H. 1996. Asymmetries generated by transcription-coupled repair in enterobacterial genes. Science 272:107–9 [DOI] [PubMed] [Google Scholar]
- 29.Francino MP, Ochman H. 2001. Deamination as the basis of strand-asymmetric evolution in transcribed Escherichia coli sequences. Mol. Biol. Evol 18:1147–50 [DOI] [PubMed] [Google Scholar]
- 30.Frederico LA, Kunkel TA, Shaw BR. 1990. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry 29:2532–37 [DOI] [PubMed] [Google Scholar]
- 31.Galhardo RS, Hastings PJ, Rosenberg SM. 2007. Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol 42:399–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. 2001. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science 292:1876–82 [DOI] [PubMed] [Google Scholar]
- 33. Gomez-Gonzalez B, Aguilera A. 2007. Activation-induced cytidine deaminase action is strongly stimulated by mutations of the THO complex. Proc. Natl. Acad. Sci. USA 104:8409–14 Examined the interplay between transcription, R-loop formation, and AID expression in yeast.
- 34.Gowrishankar J, Harinarayanan R. 2004. Why is transcription coupled to translation in bacteria? Mol. Microbiol 54:598–603 [DOI] [PubMed] [Google Scholar]
- 35.Green P, Ewing B, Miller W, Thomas PJ, NISC Comp. Seq., Green ED. 2003. Transcription-associated mutational asymmetry in mammalian evolution. Nat. Genet 33:514–17 [DOI] [PubMed] [Google Scholar]
- 36.Hanawalt PC, Spivak G. 2008. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol 9:958–70 [DOI] [PubMed] [Google Scholar]
- 37.Harris RS, Sale JE, Petersen-Mahrt SK, Neuberger MS. 2002. AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr. Biol 12:435–38 [DOI] [PubMed] [Google Scholar]
- 38.Hartzog GA, Fu J. 2013. The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation. Biochim. Biophys. Acta 1829:105–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heidenreich E 2007. Adaptive mutation in Saccharomyces cerevisiae. Crit. Rev. Biochem. Mol. Biol 42:285–311 [DOI] [PubMed] [Google Scholar]
- 40.Helmrich A, Ballarino M, Nudler E, Tora L. 2013. Transcription-replication encounters, consequences and genomic instability. Nat. Struct. Mol. Biol 20:412–18 [DOI] [PubMed] [Google Scholar]
- 41.Helmrich A, Ballarino M, Tora L. 2011. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell 44:966–77 [DOI] [PubMed] [Google Scholar]
- 42.Hendriks G, Calleja F, Besaratinia A, Vrieling H, Pfeifer GP, et al. 2010. Transcription-dependent cytosine deamination is a novel mechanism in ultraviolet light-induced mutagenesis. Curr. Biol 20:170–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herman RK, Dworkin NB. 1971. Effect of gene induction on the rate of mutagenesis by ICR-191 in Escherichia coli. J. Bacteriol 106:543–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hicks WM, Kim M, Haber JE. 2010. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science 329:82–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hinnebusch AG. 1992. General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthesis enzymes in Saccharomyces cerevisiae. In The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression, ed. Jones EW, Pringle JR, Broach JR, pp. 319–414. Cold Spring Harbor,NY: Cold Spring Harbor Lab. Press [Google Scholar]
- 46.Hudson RE, Bergthorsson U, Ochman H. 2003. Transcription increases multiple spontaneous point mutations in Salmonella enterica. Nucleic Acids Res. 31:4517–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanehiro Y, Todo K, Negishi M, Fukuoka J, Gan W, et al. 2012. Activation-induced cytidine deaminase (AID)-dependent somatic hypermutation requires a splice isoform of the serine/arginine-rich (SR) protein SRSF1. Proc. Natl. Acad. Sci. USA 109:1216–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim N, Abdulovic AL, Gealy R, Lippert MJ, Jinks-Robertson S. 2007. Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA Repair 6:1285–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim N, Cho JE, Li YC, Jinks-Robertson S. 2013. RNA:DNA hybrids initiate quasi-palindrome-associated mutations in highly transcribed yeast DNA. PLoS Genet. 9:e1003924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kim N, Jinks-Robertson S. 2009. dUTP incorporation into genomic DNA is linked to transcription in yeast. Nature 459:1150–53 Demonstrated increased incorporation of uracil into transcriptionally active yeast DNA.
- 51.Kim N, Jinks-Robertson S. 2010. Abasic sites in the transcribed strand of yeast DNA are removed by transcription-coupled nucleotide excision repair. Mol. Cell. Biol 30:3206–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klapacz J, Bhagwat AS. 2002. Transcription-dependent increase in multiple classes of base substitution mutations in Escherichia coli. J. Bacteriol 184:6866–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klapacz J, Bhagwat AS. 2005. Transcription promotes guanine to thymine mutations in the non-transcribed strand of an Escherichia coli gene. DNA Repair 4:806–13 [DOI] [PubMed] [Google Scholar]
- 54.Kobayashi M, Sabouri Z, Sabouri S, Kitawaki Y, Pommier Y, et al. 2011. Decrease in topoisomerase I is responsible for activation-induced cytidine deaminase (AID)-dependent somatic hypermutation. Proc. Natl. Acad. Sci. USA 108:19305–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Korogodin VI, Korogodin VL, Fajszi C, Chepurnoy AI, Mikhova-Tsenova N, Simonyan NV. 1991. On the dependence of spontaneous mutation rates on the functional state of genes. Yeast 7:105–17 [DOI] [PubMed] [Google Scholar]
- 56.Larijani M, Martin A. 2012. The biochemistry of activation-induced deaminase and its physiological functions. Semin. Immunol 24:255–63 [DOI] [PubMed] [Google Scholar]
- 57.Li G, Zan H, Xu Z, Casali P. 2013. Epigenetics of the antibody response. Trends Immunol. 34:460–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, Manley JL. 2006. Cotranscriptional processes and their influence on genome stability. Genes Dev. 20:1838–47 [DOI] [PubMed] [Google Scholar]
- 59.Lin Y, Hubert L Jr, Wilson JH. 2009. Transcription destabilizes triplet repeats. Mol. Carcinog 48:350–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lippert MJ, Chen Q, Liber HL. 1998. Increased transcription decreases the spontaneous mutation rate at the thymidine kinase locus in human cells. Mutat. Res 401:1–10 [DOI] [PubMed] [Google Scholar]
- 61.Lippert MJ, Freedman JA, Barber MA, Jinks-Robertson S. 2004. Identification of a distinctive mutation spectrum associated with high levels of transcription in yeast. Mol. Cell. Biol 24:4801–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lippert MJ, Kim N, Cho JE, Larson RP, Schoenly NE, et al. 2011. Role for topoisomerase 1 in transcription-associated mutagenesis in yeast. Proc. Natl. Acad. Sci. USA 108:698–703 Along with Reference 96, reported that Top1 is the major source of TAM in yeast.
- 63.Liu LF, Wang JC. 1987. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 84:7024–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, et al. 2008. Two levels of protection for the B cell genome during somatic hypermutation. Nature 451:841–45 [DOI] [PubMed] [Google Scholar]
- 64a.Masani S, Han L, Yu K. 2013. Apurinic/apyrimidinic endonuclease I is the essential nuclease during immunoglobulin calss switch recombination. Mol. Cell Biol 33:1468–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Merrikh H, Zhang Y, Grossman AD, Wang JD. 2012. Replication-transcription conflicts in bacteria. Nat. Rev. Microbiol 10:449–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morey NJ, Greene CN, Jinks-Robertson S. 2000. Genetic analysis of transcription-associated mutation in Saccharomyces cerevisiae. Genetics 154:109–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102:553–63 [DOI] [PubMed] [Google Scholar]
- 68.Nagaoka H, Muramatsu M, Yamamura N, Kinoshita K, Honjo T. 2002. Activation-induced deaminase (AID)-directed hypermutation in the immunoglobulin Sμ region: implication of AID involvement in a common step of class switch recombination and somatic hypermutation. J. Exp. Med 195:529–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nambu Y, Sugai M, Gonda H, Lee CG, Katakai T, et al. 2003. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science 302:2137–40 Demonstrated that AID coimmunoprecipitates with RNAP II.
- 70.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, et al. 2012. Mutational processes molding the genomes of 21 breast cancers. Cell 149:979–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Palombo F, Kohfeldt E, Calcagnile A, Nehls P, Dogliotti E. 1992. N-methyl-N-nitrosourea-induced mutation in human cells: effects of the transcriptional activity of the target gene. J. Mol. Biol 223:587–94 [DOI] [PubMed] [Google Scholar]
- 72.Papantonis A, Cook PR. 2013. Transcription factories: genome organization and gene regulation. Chem. Rev 113:8683–705 [DOI] [PubMed] [Google Scholar]
- 73.Park C, Qian W, Zhang J. 2012. Genomic evidence for elevated mutation rates in highly expressed genes. EMBO Rep. 13:1123–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pasqualucci L, Migliazza A, Fracchiolla N, William C, Neri A, et al. 1998. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc. Natl. Acad. Sci. USA 95:11816–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Paul S, Million-Weaver S, Chattopadhyay S, Sokurenko E, Merrikh H. 2013. Accelerated gene evolution through replication-transcription conflicts. Nature 495:512–15 Documented the evolutionary consequences of transcription-replication conflicts in bacteria.
- 76.Pavri R, Gazumyan A, Jankovic M, Di Virgilio M, Klein I, et al. 2010. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell 143:122–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peterlin BM, Price DH. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23:297–305 [DOI] [PubMed] [Google Scholar]
- 78.Polak P, Querfurth R, Arndt PF. 2010. The evolution of transcription-associated biases of mutations across vertebrates. BMC Evol. Biol 10:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Prado F, Aguilera A. 2005. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 24:1267–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pybus C, Pedraza-Reyes M, Ross CA, Martin H, Ona K, et al. 2010. Transcription-associated mutation in Bacillus subtilis cells under stress. J. Bacteriol 192:3321–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reimers JM, Schmidt KH, Longacre A, Reschke DK, Wright BE. 2004. Increased transcription rates correlate with increased reversion rates in leuB and argH Escherichia coli auxotrophs. Microbiology 150:1457–66 [DOI] [PubMed] [Google Scholar]
- 82.Roberts SA, Gordenin DA. 2014. Clustered and genome-wide transient mutagenesis in human cancers: hypermutation without permanent mutators or loss of fitness. BioEssays 36:382–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roberts SA, Sterling J, Thompson C, Harris S, Mav D, et al. 2012. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol. Cell 46:424–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sakofsky CJ, Roberts SA, Malc E, Mieczkowski PA, Resnick MA, et al. 2014. Break-induced replication is a source of mutation clusters underlying kataegis. Cell Rep. 7:1640–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Samaranayake M, Bujnicki JM, Carpenter M, Bhagwat AS. 2006. Evaluation of molecular models for the affinity maturation of antibodies: roles of cytosine deamination by AID and DNA repair. Chem. Rev 106:700–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Savic DJ, Kanazir DT. 1972. The effect of a histidine operator-constitutive mutation on UV-induced mutability within the histidine operon of Salmonella typhimurium. Mol. Gen. Genet 118:45–50 [DOI] [PubMed] [Google Scholar]
- 87.Schmidt KH, Reimers JM, Wright BE. 2006. The effect of promoter strength, supercoiling and secondary structure on mutation rates in Escherichia coli. Mol. Microbiol 60:1251–61 [DOI] [PubMed] [Google Scholar]
- 88.Shen HM, Tanaka A, Bozek G, Nicolae D, Storb U. 2006. Somatic hypermutation and class switch recombination in Msh6−/−Ung−/− double-knockout mice. J. Immunol 177:5386–92 [DOI] [PubMed] [Google Scholar]
- 89.Smith HC, Bennett RP, Kizilyer A, McDougall WM, Prohaska KM. 2012. Functions and regulation of the APOBEC family of proteins. Semin. Cell Dev. Biol 23:258–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sohail A, Klapacz J, Samaranayake M, Ullah A, Bhagwat AS. 2003. Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res. 31:2990–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sparks JL, Chon H, Cerritelli SM, Kunkel TA, Johansson E, et al. 2012. RNase H2-initiated ribonucleotide excision repair. Mol. Cell 47:980–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Srivatsan A, Tehranchi A, MacAlpine DM, Wang JD. 2010. Co-orientation of replication and transcription preserves genome integrity. PLoS Genet. 6:e1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Storb U 2014. Why does somatic hypermutation by AID require transcription of its target genes? Adv. Immunol 122:253–77 [DOI] [PubMed] [Google Scholar]
- 94.Storb U, Shen HM, Michael N, Kim N. 2001. Somatic hypermutation of immunoglobulin and non-immunoglobulin genes. Philos. Trans. R. Soc. Lond. B 356:13–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Strathern JN, Shafer B, McGill CB. 1995. DNA synthesis errors associated with double-strand-break repair. Genetics 140:965–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Takahashi T, Burguiere-Slezak G, Van der Kemp PA, Boiteux S. 2011. Topoisomerase 1 provokes the formation of short deletions in repeated sequences upon high transcription in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 108:692–97 Along with Reference 62, reported that Top1 is the major source of TAM in yeast.
- 97.Wang JC. 2002. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol 3:430–40 [DOI] [PubMed] [Google Scholar]
- 98.Wang X, Fan M, Kalis S, Wei L, Scharff MD. 2014. A source of the single-stranded DNA substrate for activation-induced deaminase during somatic hypermutation. Nat. Commun 5:4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Westover KD, Bushnell DA, Kornberg RD. 2004. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science 303:1014–16 [DOI] [PubMed] [Google Scholar]
- 100.Wimberly H, Shee C, Thornton PC, Sivaramakrishnan P, Rosenberg SM, Hastings PJ. 2013. R-loops and nicks initiate DNA breakage and genome instability in non-growing Escherichia coli. Nat. Commun 4:2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Woo CJ, Martin A, Scharff MD. 2003. Induction of somatic hypermutation is associated with modifications in immunoglobulin variable region chromatin. Immunity 19:479–89 [DOI] [PubMed] [Google Scholar]
- 102.Wright BE. 1996. The effect of the stringent response on mutation rates in Escherichia coli K-12. Mol. Microbiol 19:213–19 [DOI] [PubMed] [Google Scholar]
- 103.Wright BE. 2004. Stress-directed adaptive mutations and evolution. Mol. Microbiol 52:643–50 [DOI] [PubMed] [Google Scholar]
- 104.Wright BE, Longacre A, Reimers JM. 1999. Hypermutation in derepressed operons of Escherichia coli K12. Proc. Natl. Acad. Sci. USA 96:5089–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wright BE, Reschke DK, Schmidt KH, Reimers JM, Knight W. 2003. Predicting mutation frequencies in stem-loop structures of derepressed genes: implications for evolution. Mol. Microbiol 48:429–41 [DOI] [PubMed] [Google Scholar]
- 106.Wright BE, Schmidt KH, Davis N, Hunt AT, Minnick MF. 2008. II. Correlations between secondary structure stability and mutation frequency during somatic hypermutation. Mol. Immunol 45:3600–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wright BE, Schmidt KH, Hunt AT, Lodmell JS, Minnick MF, Reschke DK. 2011. The roles of transcription and genotoxins underlying p53 mutagenesis in vivo. Carcinogenesis 32:1559–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wright BE, Schmidt KH, Minnick MF, Davis N. 2008. I. VH gene transcription creates stabilized secondary structures for coordinated mutagenesis during somatic hypermutation. Mol. Immunol 45:3589–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xue K, Rada C, Neuberger MS. 2006. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice. J. Exp. Med 203:2085–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yamaguchi Y, Shibata H, Handa H. 2013. Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond. Biochim. Biophys. Acta 1829:98–104 [DOI] [PubMed] [Google Scholar]
- 112.Yamane A, Resch W, Kuo N, Kuchen S, Li Z, et al. 2011. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat. Immunol 12:62–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yu K, Roy D, Bayramyan M, Haworth IS, Lieber MR. 2005. Fine-structure analysis of activation-induced deaminase accessibility to class switch region R-loops. Mol. Cell. Biol 25:1730–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang Y, Shishkin AA, Nishida Y, Marcinkowski-Desmond D, Saini N, et al. 2012. Genome-wide screen identifies pathways that govern GAA/TTC repeat fragility and expansions in dividing and nondividing yeast cells. Mol. Cell 48:254–65 [DOI] [PMC free article] [PubMed] [Google Scholar]