Abstract
In Streptomyces griseus, A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone) serves as a microbial hormone that switches on many genes required for streptomycin production and morphological development. An open reading frame (Orf1) showing high sequence similarity to oligoribonucleases of various origins is present just downstream of adpA, one of the A-factor-dependent genes. Orf1 was named OrnA (oligoribonuclease A) because it showed 3′-to-5′ exo-oligoribonuclease activity, releasing [32P]CMP from ApCpC[32P]pC used as a substrate. Reverse transcription-PCR and S1 nuclease mapping analyses revealed that ornA was transcribed from two promoters; one was a developmentally regulated, A-factor-dependent promoter in front of adpA, and the other was a constitutive promoter in front of the ornA coding sequence. Transcription of ornA was thus additively enhanced at the initiation stage for secondary metabolism and aerial mycelium formation. ornA-disrupted strains grew slowly and scarcely formed aerial mycelium. ornA homologues were distributed in a wide variety of Streptomyces species, including S. coelicolor A3(2), as determined by Southern hybridization analysis. Disruption of the ornA homologue in S. coelicolor A3(2) also caused phenotypes similar to those of the S. griseus ΔornA strains. The OrnA oligoribonucleases in Streptomyces species are therefore not essential but play an important role in vegetative growth and in the initiation of differentiation.
The filamentous, soil-inhabiting, gram-positive bacterial genus Streptomyces is characterized by the ability to produce a wide variety of secondary metabolites and by complex morphological differentiation culminating in sporulation (5). In Streptomyces griseus, A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone) is required for streptomycin (Sm) production and cell differentiation (13–15). A-factor at an extremely low concentration triggers Sm production and aerial mycelium formation by binding a repressor-type receptor protein (ArpA) and dissociating it from the DNA (23, 24). Recently we identified adpA, which encodes a transcriptional activator for strR, a pathway-specific regulatory gene responsible for transcription of other Sm biosynthetic genes, as one of the target genes of ArpA (22). ArpA binds the adpA promoter and represses its transcription in the absence of A-factor during early growth phase. adpA is thus developmentally regulated by A-factor. During these studies, we found an open reading frame (Orf1) showing end-to-end similarity to the oligoribonuclease of Escherichia coli (31) only 10 bp downstream from the termination codon of adpA. Because orf1 is located just downstream of adpA and was expected to be developmentally regulated by A-factor and because little is known about RNA degradation in members of Streptomyces with a complex life cycle, we analyzed the enzyme activity of Orf1 and disrupted orf1 to examine the function of the gene product.
Exo-oligoribonuclease activity of Orf1.
Orf1 shows high sequence similarity (44% identity) to the oligoribonuclease of E. coli (31). Figure 1A shows amino acid alignment of Orf1 and homologues in prokaryotes and eukaryotes. We examined oligoribonuclease activity of Orf1 by using ApCpC[32P]pC (as a substrate) and histidine-tagged Orf1 produced in E. coli, essentially by the method of Ghosh and Deutscher (9). The expression plasmid pET-ORNA was constructed and histidine-tagged Orf1 was purified as follows. With two oligonucleotides, 5′ - GGCGAAT TCATATGAACGACCGCATGGTGTGG - 3′ (the italic and bold letters indicate an NdeI cleavage sequence for cloning into pET16b and the initiation codon of orf1, respectively; the underline indicates an EcoRI cleavage sequence) and 5′-CGCGGATCCTACGGTGCGGCCGGAGCCGAC-3′ (the bold letters indicate the termination codon of orf1, and the underline indicates a BamHI cleavage sequence), the orf1 sequence was amplified by PCR. The amplified fragment was digested with EcoRI and BamHI and cloned between the EcoRI and BamHI sites on pUC19. After the sequence was checked by sequencing, an NdeI-BamHI fragment was excised from the recombinant plasmid and ligated with NdeI-plus- BamHI-digested pET16b, resulting in pET-ORNA. The histidine-tagged Orf1 encoded by pET-ORNA had a structure of Met-Gly-His10-Ser2-Gly-His-Ile-Glu-Gly-Arg-His-Orf1. For purification of His-tagged Orf1, E. coli BL21(DE3) harboring pET-ORNA was cultured overnight at 37°C without isopropyl-β-d-thiogalactopyranoside. The cells were harvested and disrupted by sonication. Cell debris was removed by centrifugation and filtration using a Millipore filter (pore size, 0.45 μm). The cleared lysate was applied to a column with His-bind resin (Novagen), and His-tagged Orf1 was eluted with a linear gradient of 60 to 1,000 mM imidazole (Fig. 2A). The substrate was prepared as follows. [5′-32P]pCp was attached to ApCpC with T4 RNA ligase. The ApCpC[5′-32P]pCp product was treated with bacterial alkaline phosphatase, and the ApCpC[5′-32P]pC product was separated by thin-layer chromatography (TLC) on PSC-Fertigplatten cellulose (Merck) by using 1 M ammonium acetate–95% ethanol (60:40, vol/vol) as the solvent. The radioactive tetraribonucleotide was eluted with H2O from cellulose powder collected from the thin-layer chromatography plate and used directly for the oligoribonuclease assay.
FIG. 1.
Amino acid sequences of OrnA and OrnA-c (A) and gene organizations of the ornA-c and ornA loci and schematic representation of plasmids used in this study (B). (A) Amino acid alignment with the oligoribonuclease of E. coli (31) and homologues found in Schizosaccharomyces pombe (protein database accession no. CAB37438) and Homo sapiens (protein database accession no. AAD34109) is also shown. Solid boxes indicate that among five proteins, more than three amino acid residues in the alignment are identical. Between OrnA and OrnA-c, identical amino acid residues are indicated by grey boxes. Dashes indicate gaps introduced for alignment. (B) The percentages of identical amino acid residues of corresponding gene products are shown. The following abbreviations for restriction enzymes are used: Bs, BstPI; Fb, FbaI; Kp, KpnI; Pm, PmaCI; Ps, PstI; Sc, SacI; Sl, SalI; Sp, SphI; and Pv, PvuII.
FIG. 2.
Exo-oligoribonuclease activity of His-tagged OrnA. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of protein samples. Lane 1, the crude lysate from E. coli BL21(DE3) harboring pET-ORNA; lane 2, after His-bind resin column chromatography; and lane M, molecular size markers. (B) Release of [32P]CMP from ApCpC[32P]pC by His-tagged OrnA. After the product had been separated by paper chromatography, the paper was analyzed by using an image analyzer. The origin, [32P]CMP, and ApCpC[32P]pC are indicated by arrows. The concentrations (in micrograms per milliliter) of His-tagged OrnA in the reaction mixtures were 11.2 (lane 1), 5.6 (lane 2), 2.8 (lane 3), 1.4 (lane 4), and 0.7 (lane 5).
The standard assay was carried out in 100 μl of reaction mixtures containing 100 mM Tris-HCl (pH 8.0), 5 mM MnCl2, and about 3 fmol of ApCpC[5′-32P]pC. Various amounts of purified recombinant Orf1 were added, and the samples were incubated at 37°C for 30 min. The oligoribonuclease activity was analyzed by monitoring the release of [32P]CMP from ApCpC[32P]pC. The product was separated by paper chromatography on Whatman 3MM paper by using 1 M ammonium acetate–95% ethanol (60:40, vol/vol) as the solvent. The paper was analyzed with a Fujix BAS2000 image analyzer. As expected, the recombinant Orf1 showed distinct 3′-to-5′ exo-type oligoribonuclease activity, releasing [32P]CMP at the 3′ end from the substrate oligoribonucleotide (Fig. 2B). We named Orf1 oligoribonuclease A (OrnA). For determining the cation requirement of OrnA, 5 mM MgCl2, CoCl2, ZnCl2, FeCl2, CuCl2, CaCl2, NaCl, KCl, or NH4Cl instead of MnCl2 was used. Like the oligoribonuclease of E. coli (21), OrnA required Mn2+ for its activity. Mg2+, Co2+, and Zn2+ also activated the enzyme but to a smaller extent. Fe2+, Cu2+, Ca2+, or monovalent cations such as K+, Na+, and NH4+ did not activate the enzyme under the assay conditions. For determining the optimum pH, the assay was carried out at pH 7.0, 7.5, 8.0, 8.5, 9.0, 9.5, and 10.0, using Tris-HCl buffer. The optimum pH range of recombinant OrnA was 8.5 to 9.5, whereas pH 8.0 to 9.0 was reported to be optimum for the E. coli oligoribonuclease (21). In these experiments 5 mM MgCl2 was used instead of MnCl2, because MnCl2 caused precipitation of Mn(OH)2 around pH 9. For determining the optimum temperature, the assay was carried out at 30, 40, 50, and 60°C. In these experiments, the pH of each reaction mixture was adjusted at the temperature of the reaction, and 5 mM MgCl2 was used instead of MnCl2. The optimum temperature for OrnA was determined to be 40°C, whereas that for the E. coli enzyme is 50°C (21).
Transcription of ornA by two promoters.
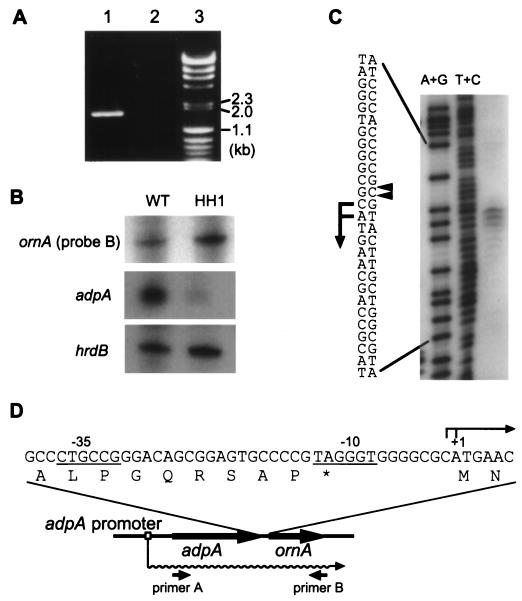
ornA is located just downstream of adpA, one of the targets of ArpA (Fig. 1B) (22). The nucleotide sequence of this region has been registered in the DDBJ, EMBL, and GenBank databases under accession no. AB023785. Because of only a 10-bp space between the termination codon, TAG, of adpA and the initiation codon, ATG, of ornA (Fig. 3D) and because of the absence of typical transcriptional terminator sequences in the region downstream from adpA, we tried to detect a possible polycistronic mRNA by reverse transcription (RT)-PCR. Two primers, 5′-AGCGTCATGAGCCAGGACTCCGCC-3′ (primer A; the underline indicates the initiation codon of adpA) and 5′-CTACGGTGCGGCCGGAGCCGACAAGCG-3′ (primer B; the underline indicates the termination codon of ornA), were used. First strand cDNA was synthesized by using primer B (10 pmol) and 3 μg of total RNA prepared from the mycelium grown at 30°C for 24 h in YMPD liquid medium (22). The RNA complementary to the cDNA was removed with RNase H. The cDNA was amplified by PCR with primers A and B (10 pmol each) at 94°C for 30 s, 58°C for 30 s, and 72°C for 3 min in a total of 30 cycles. Agarose gel electrophoresis of the RT-PCR product revealed a 1.8-kb fragment (Fig. 3A), indicating that adpA and ornA were cotranscribed from the A-factor-dependent promoter in front of adpA.
FIG. 3.
Transcriptional analyses of ornA. (A) Cotranscription of adpA and ornA analyzed by RT-PCR. A 1.8-kb fragment was amplified by RT-PCR with primers A and B when RNA prepared from the S. griseus wild-type cells was used as a template (lane 1). A control experiment with no reverse transcriptase (lane 2) confirmed that the RNA sample contained no chromosomal DNA. λ DNA digested with HindIII and φX174 DNA digested with HincII were used as size markers (lane 3). (B) Low-resolution S1 nuclease analysis of ornA. RNA was isolated from the mycelium of S. griseus IFO13350 (WT) and that of the A-factor-deficient mutant (HH1) after they were grown at 30°C for 24 h. For analysis of ornA transcription, the ornA probe B (nt −189 to +210 with respect to the transcriptional start point of ornA) was used. (C) High-resolution S1 nuclease mapping for determination of the transcriptional start point of ornA with probe A (nt −348 to +52 with respect to the transcriptional start point of ornA). Maxam-Gilbert sequencing ladders (A+G and C+T reactions) were generated with the same 32P-labeled fragment. The positions of the S1-protected fragments are shown by arrowheads, and the transcriptional start sites are assigned to the C and A residues as shown. (D) Nucleotide sequence of the ornA promoter region. The A-factor-dependent transcript from the adpA promoter (22) is also illustrated by a wavy line. Below it, the positions of the primers used for the RT-PCR experiment are shown. The termination codon is indicated by an asterisk.
We also examined the possible transcription of ornA from its own promoter by low-resolution S1 nuclease mapping with two 32P-labeled probes (nucleotide positions [nt] from −348 to +52 [probe A] and from −189 to +210 [probe B], with respect to the transcriptional start point of ornA that was later determined by high-resolution S1 mapping). A single start point was detected near the initiation codon with the two probes. This transcript was detected in both the wild-type (WT) and an A-factor-deficient mutant strain, HH1 (Fig. 3B), indicating that this promoter was independent of A-factor. Transcription of hrdB encoding ςHrdB was used to monitor the quantity and quality of the mRNA used. The transcripts of adpA and hrdB in strains WT and HH1 were analyzed by low-resolution S1 mapping, as previously described (22). The amount of the ornA mRNA relative to that of the hrdB mRNA was smaller than that of the adpA mRNA, but it was still distinct. High-resolution S1 mapping with probe A determined the transcriptional start points to be the C that was one nucleotide upstream from the initiation codon and the A of the initiation codon ATG (Fig. 3C). The assignment is based on the fact that the fragments generated by the chemical sequencing reactions migrate 1.5 nt further than the corresponding fragments generated by S1 nuclease digestion of the DNA-RNA hybrids (half a residue from the presence of the 3′-terminal phosphate group and one residue from the elimination of the 3′-terminal nucleotide) (27). In front of the start point, CTGCCG and TAGGGT with an 18-bp space, which are similar to one type (TTGACR for −35 and TAGRRT for −10; R: A or G [28]) of Streptomyces promoters, are present (Fig. 3D). Promoters of this type are believed to be active during vegetative growth (12). Although the translation of ornA mRNA presents a contrast to the conventional interaction between ribosomes and Shine-Dalgarno sequences in translational initiation in other bacteria, this transcription-translation feature is not uncommon in members of Streptomyces. For example, the 23S rRNA methylase mediating erythromycin resistance in Streptomyces erythraeus (4), the aminoglycoside phosphotransferase mediating neomycin resistance in Streptomyces fradiae (3), and the streptothricin acetyltransferase mediating streptothricin resistance in Streptomyces lavendulae (16) are translated from leaderless transcripts.
Disruption of ornA causes slow growth in S. griseus.
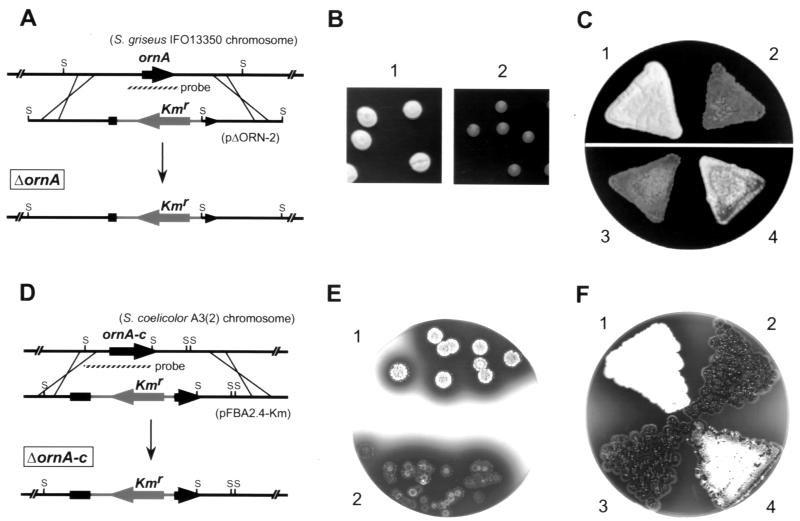
We disrupted the chromosomal ornA gene by insertion of a kanamycin resistance gene to determine the function of ornA in S. griseus (Fig. 4A). For this purpose, plasmid pΔORN-2 (Fig. 1B) was constructed as follows. DNA was manipulated in Streptomyces species (11) and in E. coli (19). The 4.5-kb SphI fragment containing the whole ornA gene was cloned on pUC19, resulting in pSPH5-1. pSPH5-1 was digested with BstPI, and the ends were flush ended with Klenow fragment. The linear plasmid was digested by BamHI, and the resultant 4.8-kb fragment (containing the whole vector and upstream and 5′-end regions of ornA) was ligated with a 1.3-kb SmaI-BamHI fragment containing the kanamycin resistance gene from Tn5 (2), resulting in pΔORN-1. pΔORN-1 had a unique KpnI site in the multicloning site of pUC19. A 2.0-kb KpnI fragment containing a 3′ end and downstream regions of ornA was excised from pSPH5-1 and inserted into the KpnI site of pΔORN-1 in the correct orientation to construct pΔORN-2, used for gene disruption. Plasmid pΔORN-2 was linearized by digestion of the vector sequence with HindIII and DraI, denatured with NaOH, and introduced by protoplast transformation into the wild-type strain S. griseus IFO13350. Correct gene replacement by means of double crossover was confirmed by Southern hybridization (data not shown). The kanamycin resistance gene was inserted between Thr-30 and Tyr-42. The ΔornA strains grew more slowly than the wild-type strain and scarcely formed aerial hyphae (Fig. 4B). Even after 3 to 4 weeks of cultivation, the ΔornA strains formed very sparse aerial hyphae and spores. We assume that the sparse aerial hyphae formation was due to disturbance of growth. Sm production by the ΔornA mutants was also repressed until 5 to 6 days, but after 2 weeks of growth the mutants produced Sm at a very low yield, assayed by bioassay with Bacillus subtilis as an indicator (17). We therefore assume that the ornA mutation does not directly affect, if at all, Sm production. The delay in Sm production appears to be due to slow growth. As described below, ΔornA mutants of Streptomyces coelicolor A3(2) produced actinorhodin almost normally. ornA on a low-copy-number (1 to 2 per genome [18]) plasmid pKU209 (plasmid pADP12LΔSP2) (Fig. 1B) recovered its growth and spore formation in the ΔornA strains (Fig. 4C). In pADP12LΔSP2, a 2.1-kb fragment, which contained a 0.9-kb upstream region of adpA, an in-frame-deleted adpA, and the intact ornA, was inserted in the BamHI site of pKU209.
FIG. 4.
Phenotypes of ΔornA mutants of S. griseus and S. coelicolor A3(2). (A) Schematic representation of the strategy used for disruption of ornA. S represents SphI. The position of the probe used for Southern hybridization is also shown. (B) Slow growth and sparse aerial mycelium formation of ΔornA. Seven-day-old colonies of wild-type (panel 1) and ΔornA (panel 2) strains grown from spores at 28°C were photographed. (C) Complementation of ΔornA phenotypes by ornA introduced on a low-copy-number plasmid. Panel 1, S. griseus IFO13350; panel 2, S. griseus ΔornA; panel 3, S. griseus ΔornA(pKU209); panel 4, S. griseus ΔornA(pADP12LΔSP2). pADP12LΔSP2 has a deletion of adpA and carries an intact ornA with the adpA promoter region (see Fig. 1B). The photograph was taken after 7 days of cultivation at 28°C. (D) Schematic representation of the strategy used for disruption of ornA-c. S represents SmaI. The position of the probe used for Southern hybridization is also shown. (E) Slow growth and sparse aerial mycelium formation of ΔornA-c. Seven-day-old colonies of parent strains (panel 1) and ΔornA-c (panel 2) grown from spores were photographed. (F) Complementation of ΔornA-c phenotypes by ornA-c introduced on a low-copy-number plasmid. Panel 1, S. coelicolor A3(2) M130; panel 2, S. coelicolor A3(2) ΔornA-c; panel 3, S. coelicolor A3(2) ΔornA-c(pKU209); panel 4, S. coelicolor A3(2) ΔornA-c(pORNAcL). pORNAcL carries ornA-c with its promoter region (Fig. 1B). The photograph was taken after 7 days of cultivation at 30°C.
Wide distribution of ornA in Streptomyces.
To search for oligoribonuclease homologues in Streptomyces, we analyzed the genomes of several Streptomyces species by Southern hybridization using a 32P-labeled 0.8-kb SalI-PvuII fragment containing ornA of S. griseus (Fig. 1B) as a probe and BamHI-digested chromosomal DNAs (data not shown). The actinomycetes examined were Streptomyces albus IFO3710, Streptomyces antibioticus IFO12652, S. coelicolor A3(2) M130, Streptomyces flaveolus IFO3408, Streptomyces fradiae ATCC 21096, Streptomyces globisorus IFO12208, Streptomyces lividans HH21, and Streptomyces viridochromogenes IFO3710. In all the eight species, signals were detected (data not shown), indicating wide distribution of ornA among members of Streptomyces. Because a single signal was detected for every strain, there must be only one copy of the ornA gene in each species.
An oligoribonuclease gene in S. coelicolor A3(2).
We cloned an ornA homologue from S. coelicolor A3(2), which is the most genetically characterized strain among Streptomyces. A 2.4-kb FbaI fragment showing a positive signal on the Southern blot was cloned in the BamHI site of pUC19 by the standard DNA probing method, including colony hybridization (plasmid pFBA2.4) (Fig. 1B). The nucleotide sequence of the cloned fragment predicted the presence of an open reading frame which showed high sequence similarity to OrnA of S. griseus and two truncated open reading frames (Fig. 1B). The alignment of the amino acid sequences of OrnA and the S. coelicolor A3(2) OrnA homologue is shown in Fig. 1A. Because 88% of amino acid residues are identical in the two sequences, the S. coelicolor A3(2) OrnA homologue is assumed to have oligoribonuclease activity. We hence designate the ornA homologue of S. coelicolor A3(2) ornA-c. A homology search using the database of the S. coelicolor A3(2) genome project in Sanger Centre revealed that part (from nt 1 to 707, starting at one of the FbaI sites; see Fig. 1B) of the determined sequence had been deposited and was the same as the 3′ end region (nt 26257 to 26963) of cosmid StC105 (Fig. 1B). The gene organization upstream of ornA in S. griseus and S. coelicolor A3(2) is identical, although downstream of ornA, an additional gene, orf4, encoding a protein very similar to a chitin binding protein, is present in S. griseus. The percent identities in amino acid sequences of corresponding gene products are shown in Fig. 1B. On the S. coelicolor A3(2) genome, an adpA homologue (adpA-c) is located upstream from the ornA homologue. Both genes are spaced by a 341-nucleotide sequence, while only 10 nucleotides intervene between adpA and ornA in S. griseus. At present, whether ornA-c and adpA-c are cotranscribed and how ornA-c is controlled are not clear. adpA encoding a transcriptional activator is controlled by a repressor-type regulator, ArpA, in S. griseus (22). Upstream of the initiation codon of adpA-c, there are no sequences resembling a consensus sequence for ArpA, CprA, or CprB binding. These proteins are specific receptors for γ-butyrolactones (24, 29). Transcriptional studies of adpA-c and ornA-c are necessary to elucidate the regulation of these genes.
Phenotypes of ΔornA-c mutants of S. coelicolor A3(2).
The S. coelicolor A3(2) chromosomal ornA-c was disrupted by inserting between His-92 and Val-93 the kanamycin resistance gene on plasmid pFBA2.4-Km, resulting in ΔornA-c strains (Fig. 4D). pFBA2.4-Km was constructed by inserting a 1.3-kb SmaI fragment containing the kanamycin resistance gene from Tn5 into the unique PmaCI site (nt 973) of pFBA2.4. Southern hybridization confirmed the correct insertion (data not shown). Like the S. griseus ΔornA mutants, the ΔornA-c strain grew slowly in comparison with the parent strain and formed sparse aerial mycelium (Fig. 4E). Despite the slow growth, no great effect of the ornA-c disruption on production of the pigmented antibiotics, actinorhodin, and undecylprodigiosin was observed. This is a contrast with the ornA mutations of S. griseus, which caused a delay in Sm production that probably resulted from slow growth. The difference may have resulted from a difference in the regulation of the secondary metabolism in the two strains. For complementation of ΔornA-c mutations, plasmid pORNAcL (Fig. 1B) was constructed by inserting a 1.6-kb HindIII-PstI fragment (nt 1 to 1551) between the PstI and HindIII sites of pKU209. When pORNAcL was introduced, the ΔornA-c strains grew normally and formed aerial mycelium (Fig. 4F), which showed that the slow growth and sparse aerial mycelium formation of the ΔornA-c strains were due solely to the ornA-c mutation.
A possible role of OrnA.
Disruption of ornA in both S. griseus and S. coelicolor A3(2) caused slow growth and sparse aerial mycelium formation. This is a contrast to the lethal effect of mutations in the E. coli oligoribonuclease (9). The E. coli oligoribonuclease is a 3′-to-5′ hydrolytic exoribonuclease specific for small oligoribonucleotides (6, 21, 30) and an essential component in the mRNA decay pathway (9). Why is the oligoribonuclease of Streptomyces not essential for growth? One possibility is that Streptomyces species have other oligoribonucleases which partially compensate for the loss of OrnA. In relation to this point, B. subtilis, which is relatively close to Streptomyces in evolution, contains no OrnA homologues. In B. subtilis, mRNA degradative activity is primarily phosphorolytic, whereas in E. coli it is primarily hydrolytic (7, 8). Ghosh and Deutscher (9) pointed out that this might account for the lack of an oligoribonuclease requirement in B. subtilis. Transcription of ornA must be greatly enhanced during the second exponential growth period because of its A-factor dependence; it is conceivable that some additional oligoribonucleases, of either the hydrolytic or phosphorolytic type, are produced during the first period of exponential growth. A poor supply of monoribonucleotides in the ΔornA mutants supposedly causes slow growth, which results in the loss of aerial mycelium formation and the delay in Sm production.
ornA appears to be transcribed by its own promoter throughout vegetative growth, and its transcription from the A-factor-dependent adpA promoter is greatly enhanced by A-factor at the initiation stage of cell differentiation and secondary metabolite production (13, 15). During this stage, OrnA supposedly supplies monoribonucleotides for mRNA synthesis by degrading unnecessary mRNA, and at the same time the rates of mRNA decay are affected by OrnA as well as by RNase E-type ribonucleases (20, 26), leading to a rapid shift of gene expression. This idea is consistent with a drastic change in physiological conditions for proliferation and development induced by A-factor. Little is known about RNA decay in members of Streptomyces with a complex life cycle, and only a few ribonucleases have been identified. RNase ES that resembles E. coli RNase E and is produced at a late stage in S. coelicolor A3(2) and S. lividans (10) and RNase III that is involved in actinorhodin production in S. coelicolor A3(2) (1, 25) are examples. Because of the complex life cycle of Streptomyces species, a variety of developmentally regulated ribonucleases are presumably present to degrade specific transcripts differentially during various growth stages.
Nucleotide sequence accession no.
The nucleotide sequence of the ornA homologue cloned from S. coelicolor A3(2) was submitted to the DDBJ, EMBL, and GenBank databases under accession no. AB036424.
Acknowledgments
This work was supported, in part, by the Waksman Foundation of Japan, by the “Research for the Future” Program of JSPS, and by the Bio Design Program of the Ministry of Agriculture, Forestry, and Fisheries of Japan (BDP-00-VI-2-2).
REFERENCES
- 1.Aceti D J, Champness W C. Transcriptional regulation of Streptomyces coelicolor pathway-specific antibiotic regulators by the absA and absB loci. J Bacteriol. 1998;180:3100–3106. doi: 10.1128/jb.180.12.3100-3106.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck E, Ludwig G, Auerswald A, Reiss B, Schaller H. Nucleotide sequence and exact localisation of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982;19:327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- 3.Bibb M J, Janssen G R. Unusual features of transcription and translation of antibiotic resistance genes in antibiotic-producing Streptomyces. In: Alacevic M, Hranueli D, Toman Z, editors. Genetics of industrial microorganisms. Karlovac, Yugoslavia: Ognjen Prica Printing Works; 1987. pp. 309–318. [Google Scholar]
- 4.Bibb M J, Janssen G R, Ward J M. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene. 1986;41:E357–E368. doi: 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- 5.Chater K F. Genetics of differentiation in Streptomyces. Annu Rev Microbiol. 1993;47:685–713. doi: 10.1146/annurev.mi.47.100193.003345. [DOI] [PubMed] [Google Scholar]
- 6.Datta A K, Niyogi S K. A novel oligoribonuclease of Escherichia coli. II. Mechanism of action. J Biol Chem. 1975;250:7313–7319. [PubMed] [Google Scholar]
- 7.Deutscher M P, Reuven N B. Enzymatic basis for hydrolytic versus phosphorolytic mRNA degradation in Escherichia coli and Bacillus subtilis. Proc Natl Acad Sci USA. 1991;88:3277–3280. doi: 10.1073/pnas.88.8.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donovan W P, Kushner S R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci USA. 1986;83:120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh S, Deutscher M P. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc Natl Acad Sci USA. 1999;96:4372–4377. doi: 10.1073/pnas.96.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagége J M, Cohen S N. A developmentally regulated Streptomyces endoribonuclease resembles ribonuclease E of Escherichia coli. Mol Microbiol. 1997;25:1077–1090. doi: 10.1046/j.1365-2958.1997.5311904.x. [DOI] [PubMed] [Google Scholar]
- 11.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 12.Hopwood D A, Bibb M J, Chater K F, Janssen G R, Malpartida F, Smith C P. Regulation of gene expression in antibiotic-producing Streptomyces. In: Booth I R, Higgins C F, editors. Regulation of gene expression—25 years on. Cambridge, England: Cambridge University Press; 1986. pp. 251–276. [Google Scholar]
- 13.Horinouchi S. γ-Butyrolactones that control secondary metabolism and cell differentiation in Streptomyces. In: Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C.: American Society for Microbiology; 1999. pp. 193–207. [Google Scholar]
- 14.Horinouchi S, Beppu T. Autoregulatory factors and communication in actinomycetes. Annu Rev Microbiol. 1992;46:377–398. doi: 10.1146/annurev.mi.46.100192.002113. [DOI] [PubMed] [Google Scholar]
- 15.Horinouchi S, Beppu T. A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol Microbiol. 1994;12:859–864. doi: 10.1111/j.1365-2958.1994.tb01073.x. [DOI] [PubMed] [Google Scholar]
- 16.Horinouchi S, Furuya K, Nishiyama M, Suzuki H, Beppu T. Nucleotide sequence of the streptothricin acetyltransferase gene from Streptomyces lavendulae and its expression in heterologous hosts. J Bacteriol. 1987;169:1929–1937. doi: 10.1128/jb.169.5.1929-1937.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horinouchi S, Kumada Y, Beppu T. Unstable genetic determinant of A-factor biosynthesis in streptomycin-producing organisms: cloning and characterization. J Bacteriol. 1984;158:481–487. doi: 10.1128/jb.158.2.481-487.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kakinuma S, Takada Y, Ikeda H, Tanaka H, Omura S. Cloning of large DNA fragments, which hybridize with actinorhodin biosynthesis genes, from kalafungin and nanaomycin A methyl ester producers and identification of genes for kalafungin biosynthesis of the kalafungin producer. J Antibiot. 1991;44:995–1005. doi: 10.7164/antibiotics.44.995. [DOI] [PubMed] [Google Scholar]
- 19.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 20.Nicholson A W. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol Rev. 1999;23:371–390. doi: 10.1111/j.1574-6976.1999.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 21.Niyogi S K, Datta A K. A novel oligoribonuclease of Escherichia coli. I. Isolation and properties. J Biol Chem. 1975;250:7307–7312. [PubMed] [Google Scholar]
- 22.Ohnishi Y, Kameyama S, Onaka H, Horinouchi S. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol Microbiol. 1999;34:102–111. doi: 10.1046/j.1365-2958.1999.01579.x. [DOI] [PubMed] [Google Scholar]
- 23.Onaka H, Ando N, Nihira T, Yamada Y, Beppu T, Horinouchi S. Cloning and characterization of the A-factor receptor gene from Streptomyces griseus. J Bacteriol. 1995;177:6083–6092. doi: 10.1128/jb.177.21.6083-6092.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onaka H, Horinouchi S. DNA-binding activity of the A-factor receptor protein and its recognition DNA sequences. Mol Microbiol. 1997;24:991–1000. doi: 10.1046/j.1365-2958.1997.4081772.x. [DOI] [PubMed] [Google Scholar]
- 25.Price B, Adamidis T, Kong R, Champness W. A Streptomyces coelicolor antibiotic regulatory gene, absB, encodes an RNase III homolog. J Bacteriol. 1999;181:6142–6151. doi: 10.1128/jb.181.19.6142-6151.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rauhut R, Klug G. mRNA degradation in bacteria. FEMS Microbiol Rev. 1999;23:353–370. doi: 10.1111/j.1574-6976.1999.tb00404.x. [DOI] [PubMed] [Google Scholar]
- 27.Sollner-Webb B, Reeder R H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979;18:485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- 28.Strohl W R. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992;20:961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugiyama M, Onaka H, Nakagawa T, Horinouchi S. Site-directed mutagenesis of the A-factor receptor protein: Val-41 important for DNA-binding and Trp-119 important for ligand-binding. Gene. 1998;222:133–144. doi: 10.1016/s0378-1119(98)00487-9. [DOI] [PubMed] [Google Scholar]
- 30.Yu D, Deutscher M P. Oligoribonuclease is distinct from the other known exoribonucleases of Escherichia coli. J Bacteriol. 1995;177:4137–4139. doi: 10.1128/jb.177.14.4137-4139.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Zhu L, Deutscher M P. Oligoribonuclease is encoded by a highly conserved gene in the 3′-5′ exonuclease superfamily. J Bacteriol. 1998;180:2779–2781. doi: 10.1128/jb.180.10.2779-2781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]