Abstract
TNBC is a heterogeneous subtype of breast cancer, and only a subset of TNBC can be established as PDXs. Here, we show that there is an engraftment bias toward TNBC with low levels of immune cell infiltration. Additionally, TNBC that failed to engraft show gene expression consistent with a cancer-promoting immunological state, leading us to hypothesize that the immunological state of the tumor and possibly the state of the immune system of the host may be essential for engraftment.
Subject terms: Breast cancer, Cancer microenvironment
Introduction
Experimentally tractable in vivo models are critical for understanding tumor biology, developing therapeutic interventions, and uncovering mechanisms of therapy resistance. Better understanding of model limitations is key to improving rigor and reproducibility of basic research. The success of clinical trials also critically depends on understanding biases of preclinical models. Historically, immortalized cell lines and genetically engineered mouse models have been used primarily to study breast cancer biology. However, in vitro models lack the tumor microenvironment1, and cell lines do not faithfully recapitulate the biology of the tumor of origin2–4. Previous studies have shown that as many as 20% of cell lines are cross-contaminated, and that the repeated passage of cell lines is associated with the accumulation of mutations not seen in the primary tumor5. Genetically Engineered Mouse Models (GEMMs) better represent the tumor microenvironment of primary tumors. However, because an individual GEMM yields a relatively homogeneous set of tumors, GEMMs do not model the full diversity of human breast cancer6. A possible exception is the mouse TP53-null mammary epithelial transplantation model. This model can generate tumors representing multiple tumor types, but the relationship between mouse TP53-null tumors and human tumors of various molecular subtypes remains unclear7,8.
To address some of the shortcomings of traditional cancer models, over the last two decades there has been significant progress in the development of Patient-Derived Xenografts (PDXs). Large PDX cohorts are now available for breast cancer, as well as other tumor types (https://pdxportal.research.bcm.edu/, https://pdmr.cancer.gov/, http://www.pdxnetwork.org, https://www.pdxfinder.org/). Breast cancer PDXs have been well characterized and shown to be biologically consistent with patient tumors across multiple “omics” types including mutations, copy number alterations, transcriptomics, and proteomics6,9–15. Despite advances in PDX modeling, not all breast cancers can be engrafted successfully as PDX models. Triple Negative Breast Cancer (TNBC) has a higher engraftment rate as PDXs (~60%) than hormone positive or HER2 + tumors (~10–15%)11, and are therefore the best represented breast cancer subtype in PDX collections. TNBCs lack the expression of the ESR1 (ER) and PGR (PR) steroid hormone receptors, and do not show amplification or overexpression of oncogenic ERBB2 (HER2). TNBCs are highly heterogenous, and multiple subtypes of TNBCs have previously been identified based on histopathology16, genomic alterations17, transcriptomic profiling18, a combination of mutational profiling and transcriptomic profiling19, and by the tumor microenvironment20.
Several TNBC subtyping methods have identified an immunomodulatory subtype, and TNBC has the highest proportion of immune cell-enriched “hot” tumors of all breast cancer subtypes21. A necessary disadvantage of PDX models is that an immunocompromised host (typically mouse) is needed to ensure that the patient tumor is not rejected by the immune system. The immune system is a double-edged sword that can promote tumor growth as well as target tumor cells for elimination22. As immunodeficient PDX models lack a functional immune system, we investigated whether the immune status of human breast cancers was related to growth as a PDX.
Because immunologically “hot” and “cold” tumors were shown previously to have distinct epithelial cancer cell profiles23,24, we asked if the cancer cell fraction in PDX models mostly matches the cancer cell profiles of the immunologically “hot” or “cold” tumors in patient tumors. Ideally, the state of cancer cells within PDXs should match the cancer cell state in human tumors. The cancer cell state in PDX models is readily ascertainable against the background of the mouse cells because the cancer cell expression profile can be separated from the bulk expression by determining which mRNA-sequencing reads have human origins25. To access the cancer cell state in human primary tumors from readily available bulk mRNA gene expression profiles of TNBC tumors from The Cancer Genome Atlas (TCGA)26,27, we apply the EDec26 computational cell type deconvolution method. Immune proportions of each tumor in the TCGA collection were determined, and the “hot” tumors were defined as those with immune proportions in the top quartile, while “cold” tumors were defined as those with immune infiltration in the bottom quartile.
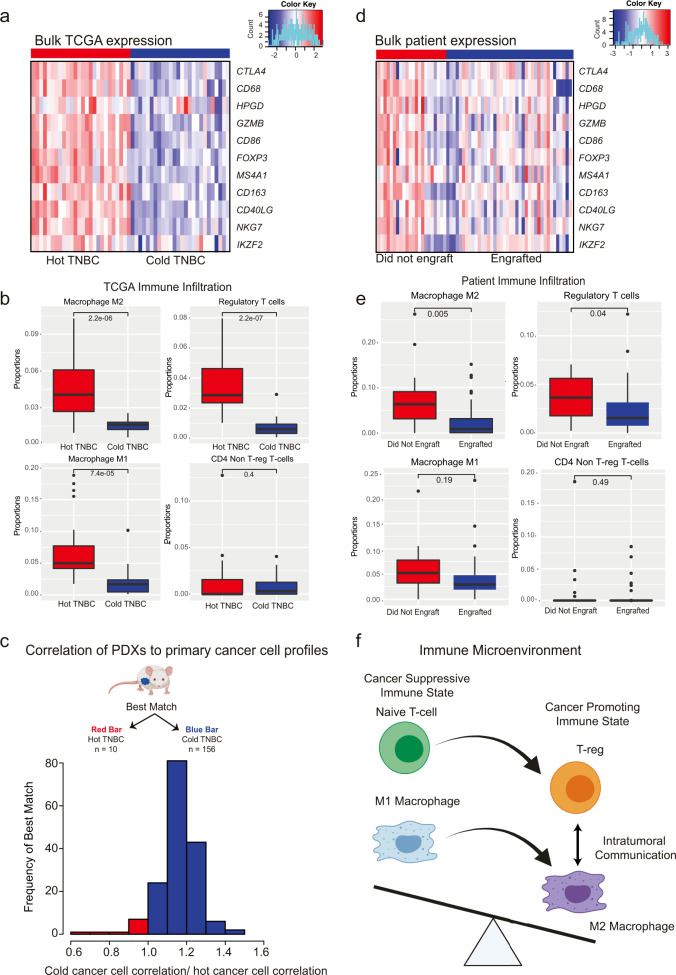
As expected, Treg markers (FOXP3, CTLA4, HPGD, IKZF2), CD4 T-cell markers (GZMB, NKG7, CD40LG), M2 macrophage markers (CD163), and M1 macrophage markers (CD68, CD86), and B-cell marker (MS4A1) were all shown to be more highly expressed in the bulk expression profiles of hot TNBC group vs the cold TNBC group (Fig. 1a).
Fig. 1. “Cold” TNBC show higher rates of engraftment in PDX models in contrast to immune enriched “Hot” TNBCs.
a Bulk expression of immune genes in TCGA TNBC. Heatmap of the bulk expression of immune gene markers in hot and cold TNBC TCGA tumors (red bars and blue bars respectively). b Immune proportions in TCGA TNBC. Proportions of M2 macrophages (top left), T-regs (top right), M1 macrophages (bottom left), and non-T-reg CD4 T-cells (bottom right) in hot and cold TCGA TNBC tumors (red bars and blue bars respectively). T-tests were used to investigate the differences between hot and cold TNBC samples, and enter lines in the boxplots indicate the median. The bounds of the box indicate the first to third quartile values, while the bounds of the lower and upper whiskers indicate the smallest observation greater than or equal to the first quartile −1.5 times the inter-quartile range and the largest observation smaller or equal to the third quartile +1.5 times the inter-quartile range respectively. c Matching the PDX cancer cell profiles to the deconvoluted TCGA cancer cell profiles. 94% of the PDX models match the cold cancer cell profile. The histogram shows the frequency of the ratios of each model’s correlation the cold cancer cell profile vs the hot cancer cell profile. The bars in red (left) indicate that the hot cancer cell profile was the best match, while the bars in blue (right) indicate that the cold cancer cell profile was the best match. d Bulk expression of immune genes in patient tumors. Heatmap of bulk expression of immune cell markers in the HCI tumors that did not engraft in PDXs vs those that did (blue and red bars respectively). e Immune proportions in patient tumors. Proportions of M2 macrophages (top left), T-regs (top right), M1 macrophages (bottom left), and non-T-reg CD4 T-cells (bottom right) in hot and cold TCGA TNBC tumors (red bars and blue bars respectively). T-tests were used to investigate the differences between TNBCs that engrafted in murine models and those that did not. Center lines in the boxplots indicate the median, and the bounds of the box indicate the first to third quartile values. The bounds of the lower and upper whiskers indicate the smallest observation greater than or equal to the first quartile −1.5 times the inter-quartile range and the largest observation smaller or equal to the third quartile +1.5 times the inter-quartile range respectively. f Immune State. M2 macrophages release exosomes that include miR-29a-3p and miR-21-5p that reprogram T-cells to T-regs. Conversely, Tregs then release interleukins including IL-10, IL-4 and IL-13 that then reprogram M1 to M2 macrophages. The presence of T-regs and M2 macrophages leads to a cancer promoting tumor state.
The cancer cell-specific gene expression profiles of hot TNBC and cold TNBC were then deconvoluted separately. The cold and hot groups also showed distinct epithelial cancer cell profiles, with 438 differentially expressed genes identified (at 5-fold difference between the cold and hot cancer cell profiles p < 0.05 thresholds, t-test). KEGG28 analysis showed that genes that were significantly upregulated in the hot vs cold cancer cell profiles were enriched for the cytokine-cytokine receptor interaction pathway (p = 0.02, hypergeometric test) (Table 1). Moreover, the GO29 analysis also showed an enrichment for pathways associated with immune activation (16/20 top pathways are associated with immune response) (Supplementary Table 1).
Table 1.
KEGG analysis (hypergeometric test).
| Pathway | Genes Total | DE genes | P value | |
|---|---|---|---|---|
| path: hsa04060 | Cytokine-cytokine receptor interaction | 295 | 3 | 0.02215201 |
| path: hsa04390 | Hippo signaling pathway | 157 | 2 | 0.04182547 |
| path: hsa04610 | Complement and coagulation cascades | 85 | 2 | 0.01331597 |
| path: hsa05150 | Staphylococcus aureus infection | 96 | 2 | 0.01678076 |
| path: hsa05417 | Lipid and atherosclerosis | 215 | 2 | 0.07321532 |
After characterizing the hot and cold primary TNBC cancer cell profiles, we then asked if the TNBC PDXs best matched the hot or cold cancer cell expression profiles. To address this question, we obtained RNA-seq data for 166 TNBC PDX models from the BCM and HCI collections, as well as other publicly available datasets (https://pdxportal.research.bcm.edu/pdxportal, https://pdmr.cancer.gov/, https://www.pdxfinder.org/, Table 2, Supplementary Table 2). The human cancer cell profiles of these PDXs were isolated by Xenome25, and the PDX cancer cell profiles were correlated to the deconvoluted cold and hot primary TCGA TNBC profiles. 94% (Fig. 1cn = 156 p < 0.001, binomial test) best matched cold cancer profile.
Table 2.
PDX Summary statistics.
| TNBC PDX models | Unique patients | |
|---|---|---|
| BCM | 50 | 44 |
| WHIM | 9 | 6 |
| HCI | 26 | 25 |
| UOM-BC | 5 | 5 |
| NKI | 6 | 6 |
| NCI PDMR | 70 | 12 |
| All Collections | 166 | 98 |
To gain insights into the immunological mechanisms that may explain the engraftment bias, we then used quanTIseq30, an immune specific in silico deconvolution method, to perform an immune cell deconvolution of the hot and cold TNBC TCGA tumors. QuanTIseq does not estimate cell type-specific genes expression profiles but does allow for a more granular deconvolution of the tumor microenvironment. In hot TNBC TCGA tumors, there was a significantly higher proportion of M2 macrophage- (p < 0.001, t-test), T-reg- (p < 0.001 t-test), and M1 macrophage-related gene expression (p < 0.001, t-test) (Fig. 1b).
To explore the potential immunological influence on engraftment further, we then obtained RNA-seq data for 62 primary patient tumors that were implanted previously in PDX engraftment attempts (HCI collection, BCM collection, https://www.pdxfinder.org/, Table 3). Of these primary tumors 40 engrafted successfully, while 22 did not. As expected, the immune-related marker genes were found to be more highly expressed in the bulk expression profiles of the tumors that did not engraft (Fig. 1d). Quantiseq was used to deconvolute the immune fraction of these tumors. Tumors that failed to engraft in PDX models had a significantly higher proportion of total immune infiltration (p = 0.024, t-test). Both the proportions of T-regs (p = 0.04, t-test) and M2 macrophages (p = 0.005, t-test) were significantly higher in the samples that failed to engraft. T-regs and alternatively activated M2 macrophages both contribute to a cancer-promoting immune state, and have been previously implicated in promotion of tumor growth and proliferation. Additionally, intratumorally signaling between T-regs and M2 macrophages is associated with both tumor progression and a cancer-promoting immune state (Fig. 1f)31–40. Because samples that were enriched in T-reg and M2 macrophage signatures failed to engraft, we hypothesize that these samples had a cancer-promoting immune state that is not recapitulated in the immunodeficient PDX models.
Table 3.
Matched patient summary statistics.
| Engrafted | Did not Engraft | |
|---|---|---|
| BCM | 30 | 15 |
| NKI | 5 | 0 |
| HCI | 5 | 7 |
| All Collections | 40 | 22 |
Taken together, our findings demonstrate that there is a bias towards the more successful engraftment of cold TNBC tumors as PDX models. Moreover, the immunologically hot tumors that do not engraft successfully as PDX models have an activated, growth-promoting immune cell state. Our result further suggests the intriguing possibility that an intact immune system of the PDX model may be essential for the growth of a majority of immunologically hot tumors. If confirmed by future research, this hypothesis would open the opportunity for exploring novel treatment options for hot TNBC by modulating the state of the host immune system. One major obstacle on that path of inquiry is that the “humanization” of mouse hosts by reconstitution of the human immune system has proven highly variable, as well as labor-intensive and expensive41. Thus, until immune system reconstitution in mice can be improved, our hypothesis of essentiality will be difficult to test.
Several other hypotheses could also explain the engraft bias towards cold TNBC tumors. Hot tumors may be targeted by the murine innate immune system, and thus be rejected despite the immunocompromised status of the host with respect to the adaptive immune system. Arguing against this hypothesis, our previous studies showed that the initial take rate (first passage in a host animal) for all breast cancer subtypes (~40%) is considerably higher than the proportion of tumors that can be established as stable PDX (~30% overall)9,11, suggesting that initial immune rejection does not completely explain lack of PDX engraftment. However, several studies have shown that superior engraftment of PDX occurs when mice lacking NK cells (e.g., NSG or SCID/Bg) are used, suggesting that at least this component of the innate immune system can limit engraftment. This was recently discussed in the context of breast cancer PDX models11. The specific roles of other innate immune cells (e.g., macrophages) in limiting PDX engraftment has not been determined.
Other systematic differences between the PDX models and “hot” TNBC tumors could also explain the engraftment bias. After engraftment, human stromal cells are replaced with murine stroma42, which was shown to be distinct from human stroma43. Cancer-Associated Fibroblasts (CAFs) promote angiogenesis and remodel the ECM. Additionally, CAFs in primary human tumors recruit T-regs and M2 macrophages and reprogram the immune system towards a cancer-promoting immune state44. Thus, differences in the murine and human stroma could also play a role in the failure of “hot” TNBC tumor engraftment.
Finally, it is also possible that engraftment may be a function of time to allow the hot donor tissue to adapt to the “cold” (lacking T-, B- and NK-cell) host environment, such that some tumors adapt quickly and grow, while others do not adapt in the timeframe of the useful lifespan of the immunocompromised host. In such a case, the lack of an immune system may or may not be the thing to which the tumor must adapt, as there are several other non-immune cell types present in the mammary gland (e.g., fibroblasts, adipocytes, vasculature) known to influence the growth of tumors. If true, serial transplantation of a donor tumor from the initial host to new hosts before the original host dies may allow growth of refractory tumor types such as immunologically hot TNBC, and perhaps also ER + and HER2 + breast cancers. Ultimately, better understanding of this modeling bias should help improve methods for engraftment, with subsequent increase in rigor, reproducibility, and translational potential of basic and pre-clinical cancer research involving mouse PDX models.
Methods
EDec deconvolution
TCGA TNBC RNA-Seq data was obtained from the UCSC Xena server (https://tcga.xenahubs.net) (Illumina HiSeq log2(x + 1) transformed RSEM normalized counts) and deconvoluted with EDec.
In Stage 1 of deconvolution, the immune proportion was determined for all TNBC tumors. This allowed for the identification of patients with low immune infiltration (bottom 25%) and high immune infiltration (top 25%). Stage 2 of EDec determines the cell type intrinsic expression profiles, and a separate Stage 2 deconvolution was performed for both the low immune component and the high immune component group. Differential gene expression was used to identify KEGG pathways that were differentially activated in the hot and cold TNBC cancer cell profiles.
QuanTISeq deconvolution
quanTISeq was used to determine immune cell proportions for both TCGA tumors as well as the BCM and HCI tumors.
Murine cancer cell profile matching
RNA-Seq data was obtained for the BCM and HCI PDX collection https://pdxportal.research.bcm.edu/ and the murine and human reads were separated with Xenome. The cancer reads were then correlated to the two cancer cell profiles obtained from the deconvolution of TGCA. RNA-seq for other PDX models was obtained from https://pdmr.cancer.gov/ and https://www.pdxfinder.org/.
Statistical analysis
A binomial test was used to determine if there was an engraftment bias for hot vs cold patient tumors. The null hypothesis was that the PDXs represent the primary tumor population equally and 64% of the PDX models should match a cold basal profile45.
T-tests were used to determine p-values of the differences in the proportions of immune cells.
Informed consent was obtained for all human participants and all relevant ethical regulations were followed. The BCM institutional review board approved the study protocol.
Supplementary information
Acknowledgements
This work was supported by NIH/NCI grants U54 CA224076 (to A.L.W., B.E.W., and M.T.L.), U24 CA226110 (to M.T.L.), a P50 supplement CA186784 (to M.T.L.), and a grant from the National Institute On Drug Abuse of the National Institutes of Health under Award Number U54DA049098 (to A.M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (to A.M.), CPRIT Core Facilities Support Grant RP170691 (to M.T.L.), and a gift from the Korell family for the study of triple negative breast cancer.
Author contributions
Conceptualization V.P., E.L.P., M.T.L., A.M.; Methodology, V.P., R.R., L.E.D., M.T.L., and A.M.; Formal Analysis, V.P., and A.M.; Resources, A.L.W., B.E.W., L.E.D., M.H.B, M.T.L, A.M.; Data Curation, V.P.; Writing V.P., M.T.L. and A.M.; Visualization, V.P., and A.M.; Supervision, A.M.; Project Administration, A.M.; Funding Acquisition, M.T.L., and A.M.
Data availability
The data that supports the finding of this study is available from the following public databases: https://tcga.xenahubs.net, http://pdxportal.research.bcm.edu, https://pdmr.cancer.gov/, http://www.pdxnetwork.org, https://www.pdxfinder.org/, https://www.envigo.com/whim-pdx-models or upon reasonable request from M.T.L (GEO:GSE183187). Supplementary Table 3 provides further details on how to access each of the datasets utilized in this study.
Competing interests
V.P., E.L., R.R., and A.M. have no outside interests to disclose. LD is employed by StemMed Ltd. M.T.L. is a founder of, and equity stake holder in, Tvardi Therapeutics, a founder of, and uncompensated Limited Partner in, StemMed LP, and an uncompensated Manager in StemMed Holdings, its General Partner. He also receives a portion of royalties if Baylor College of Medicine licenses certain PDX models to for-profit entities. The HCI PDX models are licensed by the University of Utah and may result in royalties to A.L.W. an B.E.W.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Michael T. Lewis, Aleksandar Milosavljevic.
Supplementary information
The online version contains supplementary material available at 10.1038/s41523-022-00476-0.
References
- 1.Imamura Y, et al. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015;33:1837–1843. doi: 10.3892/or.2015.3767. [DOI] [PubMed] [Google Scholar]
- 2.Mosoyan G, et al. Multiple Breast Cancer Cell-Lines Derived from a Single Tumor Differ in Their Molecular Characteristics and Tumorigenic Potential. Plos One. 2013;8:e55145. doi: 10.1371/journal.pone.0055145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang G, et al. Comprehensive comparison of molecular portraits between cell lines and tumors in breast cancer. Bmc Genomics. 2016;17:525. doi: 10.1186/s12864-016-2911-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai X, Cheng H, Bai Z, Li J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer. 2017;8:3131–3141. doi: 10.7150/jca.18457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burdall SE, Hanby AM, Lansdown MR, Speirs V. Breast cancer cell lines: friend or foe? Breast Cancer Res. 2003;5:89. doi: 10.1186/bcr577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holen I, Speirs V, Morrissey B, Blyth K. In vivo models in breast cancer research: progress, challenges and future directions. Dis. Model Mech. 2017;10:359–371. doi: 10.1242/dmm.028274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang M, et al. Intratumoral Heterogeneity in a Trp53-Null Mouse Model of Human Breast Cancer. Cancer Disco. 2015;5:520–533. doi: 10.1158/2159-8290.CD-14-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vargo-Gogola T, Rosen JM. Modelling breast cancer: one size does not fit all. Nat. Rev. Cancer. 2007;7:659–672. doi: 10.1038/nrc2193. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, et al. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res. 2013;73:4885–4897. doi: 10.1158/0008-5472.CAN-12-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savage P, et al. Chemogenomic profiling of breast cancer patient-derived xenografts reveals targetable vulnerabilities for difficult-to-treat tumors. Commun. Biol. 2020;3:310. doi: 10.1038/s42003-020-1042-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobrolecki LE, et al. Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metast Rev. 2016;35:547–573. doi: 10.1007/s10555-016-9653-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whittle JR, Lewis MT, Lindeman GJ, Visvader JE. Patient-derived xenograft models of breast cancer and their predictive power. Breast Cancer Res Bcr. 2015;17:523. doi: 10.1186/s13058-015-0523-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo, X. Y. et al. Conservation of copy number profiles during engraftment and passaging of patient-derived cancer xenografts. Biorxiv 861393 10.1101/861393 (2019).
- 14.Evrard, Y. A. et al. Systematic Establishment of Robustness and Standards in Patient-Derived Xenograft Experiments and Analysis. Biorxiv 790246 10.1101/790246 (2019). [DOI] [PMC free article] [PubMed]
- 15.Guillen KP, et al. A human breast cancer-derived xenograft and organoid platform for drug discovery and precision oncology. Nat. Cancer. 2022;3:232–250. doi: 10.1038/s43018-022-00337-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao S, Ma D, Xiao Y, Jiang Y-Z, Shao Z-M. Clinicopathologic features and prognoses of different histologic types of triple-negative breast cancer: A large population-based analysis. Eur. J. Surg. Oncol. 2018;44:420–428. doi: 10.1016/j.ejso.2017.11.027. [DOI] [PubMed] [Google Scholar]
- 17.Jiang Y-Z, et al. Genomic and Transcriptomic Landscape of Triple-Negative Breast Cancers: Subtypes and Treatment Strategies. Cancer Cell. 2019;35:428–440.e5. doi: 10.1016/j.ccell.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 18.Lehmann BD, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao Y, et al. Multi-Omics Profiling Reveals Distinct Microenvironment Characterization and Suggests Immune Escape Mechanisms of Triple-Negative Breast Cancer. Clin. Cancer Res. 2019;25:5002–5014. doi: 10.1158/1078-0432.CCR-18-3524. [DOI] [PubMed] [Google Scholar]
- 21.Disis, M. L. & Stanton, S. E. Triple-Negative Breast Cancer: Immune Modulation as the New Treatment Paradigm. Am Soc Clin Oncol Educ Book e25–e30 10.14694/edbook_am.2015.35.e25 (2015). [DOI] [PubMed]
- 22.Qiu S-Q, et al. Tumor-associated macrophages in breast cancer: Innocent bystander or important player? Cancer Treat. Rev. 2018;70:178–189. doi: 10.1016/j.ctrv.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 23.Bonaventura P, et al. Cold Tumors: A Therapeutic Challenge for Immunotherapy. Front Immunol. 2019;10:168. doi: 10.3389/fimmu.2019.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas A, et al. Tumor mutational burden is a determinant of immune-mediated survival in breast cancer. Oncoimmunology. 2018;7:1–12. doi: 10.1080/2162402X.2018.1490854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conway T, et al. Xenome—a tool for classifying reads from xenograft samples. Bioinformatics. 2012;28:i172–i178. doi: 10.1093/bioinformatics/bts236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Onuchic V, et al. Epigenomic Deconvolution of Breast Tumors Reveals Metabolic Coupling between Constituent Cell Types. Cell Rep. 2016;17:2075–2086. doi: 10.1016/j.celrep.2016.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Decamps C, et al. Guidelines for cell-type heterogeneity quantification based on a comparative analysis of reference-free DNA methylation deconvolution software. Bmc Bioinforma. 2020;21:16. doi: 10.1186/s12859-019-3307-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carbon S, et al. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2020;49:D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finotello F, et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019;11:34. doi: 10.1186/s13073-019-0638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark NM, et al. Regulatory T Cells Support Breast Cancer Progression by Opposing IFN-γ-Dependent Functional Reprogramming of Myeloid Cells. Cell Rep. 2020;33:108482. doi: 10.1016/j.celrep.2020.108482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou, J. et al. Exosomes released from tumor-associated macrophages transfer miRNAs that induce a Treg/Th17 cell imbalance in epithelial ovarian cancer. Cancer Immunol Res 6, canimm.0479.2017 (2018). [DOI] [PubMed]
- 33.Gwak JM, Jang MH, Kim DI, Seo AN, Park SY. Prognostic Value of Tumor-Associated Macrophages According to Histologic Locations and Hormone Receptor Status in Breast Cancer. Plos One. 2015;10:e0125728. doi: 10.1371/journal.pone.0125728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi J, Gyamfi J, Jang H, Koo JS. The role of tumor-associated macrophage in breast cancer biology. Histol. Histopathol. 2017;33:133–145. doi: 10.14670/HH-11-916. [DOI] [PubMed] [Google Scholar]
- 35.Yang J, et al. Tumor‐Associated Macrophages Regulate Murine Breast Cancer Stem Cells Through a Novel Paracrine EGFR/Stat3/Sox‐2 Signaling Pathway. Stem Cells. 2013;31:248–258. doi: 10.1002/stem.1281. [DOI] [PubMed] [Google Scholar]
- 36.Pan Y, Yu Y, Wang X, Zhang T. Tumor-Associated Macrophages in Tumor Immunity. Front Immunol. 2020;11:583084. doi: 10.3389/fimmu.2020.583084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohue Y, Nishikawa H. Regulatory T (Treg) cells in cancer: Can Treg cells be a new therapeutic target? Cancer Sci. 2019;110:2080–2089. doi: 10.1111/cas.14069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun W, et al. A positive-feedback loop between tumour infiltrating activated Treg cells and type 2-skewed macrophages is essential for progression of laryngeal squamous cell carcinoma. Brit J. Cancer. 2017;117:1631–1643. doi: 10.1038/bjc.2017.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tiemessen MM, et al. CD4+CD25+Foxp3+ regulatory T cells induce alternative activation of human monocytes/macrophages. Proc. Natl Acad. Sci. 2007;104:19446–19451. doi: 10.1073/pnas.0706832104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szajnik M, Czystowska M, Szczepanski MJ, Mandapathil M, Whiteside TL. Tumor-Derived Microvesicles Induce, Expand and Up-Regulate Biological Activities of Human Regulatory T Cells (Treg) Plos One. 2010;5:e11469. doi: 10.1371/journal.pone.0011469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lai Y, et al. Current status and perspectives of patient-derived xenograft models in cancer research. J. Hematol. Oncol. 2017;10:106. doi: 10.1186/s13045-017-0470-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeRose YS, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat. Med. 2011;17:1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cassidy JW, Caldas C, Bruna A. Maintaining Tumor Heterogeneity in Patient-Derived Tumor Xenografts. Cancer Res. 2015;75:2963–2968. doi: 10.1158/0008-5472.CAN-15-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu T, et al. Cancer-associated fibroblasts: an emerging target of anti-cancer immunotherapy. J. Hematol. Oncol. 2019;12:86. doi: 10.1186/s13045-019-0770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Craven KE, Gökmen-Polar Y, Badve SS. CIBERSORT analysis of TCGA and METABRIC identifies subgroups with better outcomes in triple negative breast cancer. Sci. Rep.-uk. 2021;11:4691. doi: 10.1038/s41598-021-83913-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that supports the finding of this study is available from the following public databases: https://tcga.xenahubs.net, http://pdxportal.research.bcm.edu, https://pdmr.cancer.gov/, http://www.pdxnetwork.org, https://www.pdxfinder.org/, https://www.envigo.com/whim-pdx-models or upon reasonable request from M.T.L (GEO:GSE183187). Supplementary Table 3 provides further details on how to access each of the datasets utilized in this study.