Abstract
Stem cells are undifferentiated cells that have multi-lineage differentiation. The transition from self-renewal to differentiation requires rapid and extensive gene expression alterations. Since different stem cells exhibit diverse non-coding RNAs (ncRNAs) expression profiles, the critical roles of ncRNAs in stem cell reprogramming, pluripotency maintenance, and differentiation have been widely investigated over the past few years. Hence, in this current review, the two main categories of ncRNAs, microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), are discussed. While the primary way by which miRNAs restrict mRNA transcription is through miRNA–mRNA interaction, lncRNAs have a wide range of effects on mRNA functioning, including interactions with miRNAs. Both of these ncRNAs participate in the post-transcriptional regulation of crucial biological mechanisms, such as cell cycle regulation, apoptosis, aging, and cell fate decisions. These findings shed light on a previously unknown aspect of gene regulation in stem cell fate determination and behavior. Overall, we summarized the key roles of miRNAs (including exosomal miRNAs) and lncRNAs in the regulation of stem cell populations, such as cardiac, hematopoietic, mesenchymal, neural, and spermatogonial, as well ncRNAs’ influence on malignancy through modulating cancer stem cells, which might significantly contribute to clinical stem cell therapy and in regenerative medicine.
Keywords: MicroRNAs, Gene regulation, Stem cell populations, Cancer stem cells, Pluripotency, Therapeutics
Introduction
Stem cells are specialized undifferentiated human cells that have the ability to differentiate into any cell type of the human body. They are the organizational elements of biological systems that allow evolution through natural selection (Bacakova et al. 2018; Zhou et al. 2022). The research about stem cells has grown throughout the years due to their capacity for self-renewal, functional reconstitution of their tissue of origin, and differentiation into one or more cell lineages (Zakrzewski et al. 2019; Zhou et al. 2022). Interestingly, stem cell divisions occur in an asymmetric manner, where a stem cell can produce an exact copy of itself but also can produce a daughter cell that leaves the stem cell niche to differentiate and create multipotent progenitors. These, in turn, can generate differentiated cells and committed progenitors (López-Lázaro 2018).
Different specialty levels exist in stem cells, and each level limits their development potential. For example, totipotent stem cells can proliferate and develop into any cell in the body. They can form the embryo and extraembryonic structures, representing the highest differentiation potential (Xu et al. 2022). In contrast, pluripotent stem cells can give rise to cells of any germ layers but not to extraembryonic structures. Multipotent stem cells can differentiate into various cells belonging to specific cell lineages, and their differentiation capacities are limited to cells of their lineage (Fus-Kujawa et al. 2021). Thus, it is a narrower spectrum of differentiation, whereas unipotent stem cells, which can repeatedly divide and only form one type of cell, possess the narrowest differentiation abilities (Zakrzewski et al. 2019).
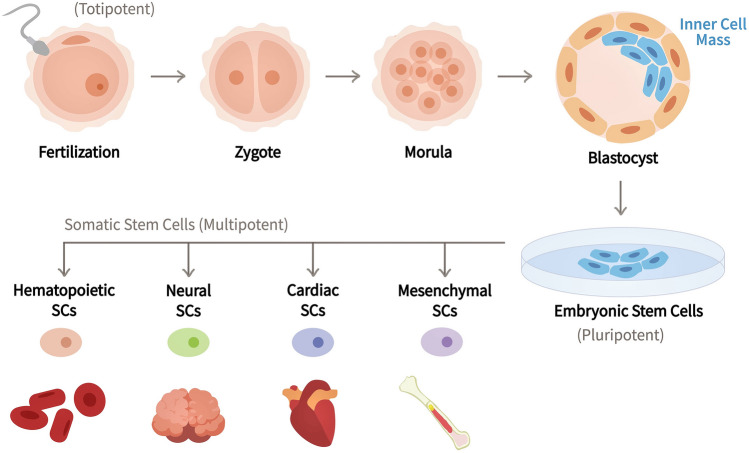
Stem cells are classified into two broad groups: embryonic stem cells (ESCs) and tissue-specific or somatic stem cells (Fig. 1) (Sada 2022). ESCs are pluripotent cells that remain undifferentiated indefinitely and present only in the early phases of development. On the contrary, adult stem cells, also known as tissue-specific stem cells (TSSCs) or somatic stem cells, are multipotent stem cells that are solely able to differentiate into cell types of their tissue or organ of origin, such as mesenchymal stem cells (MSCs), hematopoietic stem cells (HSCs), cardiac stem cells, neural stem cells (NSCs), etc. (Mens and Ghanbari 2018). Apart from ESCs and TSSCs, spermatogonial stem cells (SSCs) represent a different subtype of stem cells that possess a unique characteristic of being able to transfer genetic information to the next generations (Khanehzad et al. 2021). Intriguingly, cancer stem cells (CaSCs) comprise a completely different subtype of stem cell responsible for tumorigenesis, tumor progression, metastasis, relapse, and resistance, influencing cancer aggressiveness (Huang et al. 2020; Yoshida and Saya 2021).
Fig. 1.
Schematic representation of stem cell hierarchy. Stem cells are distinguished by their potential to differentiate into a specialized cell type. The totipotent stem cell, which emerges from the fertilization of an egg, remains at the top of the stem cell hierarchy. The totipotent stem cell produces a blastocyst after a series of divisions, containing ESCs at the inner cell mass. ESCs are pluripotent and can thus give rise to all cell types in our body, including somatic stem cells or adult stem cells, which, on the other hand, are multipotent and have a more limited ability to differentiate, being committed to a specific lineage
The exclusive characteristics of stem cells are controlled by epigenetics, extrinsic signaling, and post-transcriptional regulations (Kahney et al. 2019). Growing evidence has considered ncRNAs as critical regulators of the cell cycle of stem cells, having implications for processes such as differentiation, development, transcription, self-renewal, apoptosis, proliferation, and cell metabolism (Mens and Ghanbari 2018; Jothimani et al. 2022a; Li et al. 2022). It has been stated that ncRNAs could critically regulate the stem cells’ properties post-transcriptionally by repressing selected target transcripts, as evidenced in embryonic stem cells, germline stem cells, and various somatic tissue stem cells (Mahabadi et al. 2019; Li et al. 2022). Specifically, stem cell senescence has been reported to be controlled by miRNAs via targeting genes responsible for epigenetic modifications, metabolism, and DNA damage (Choi et al. 2017). The two main families of ncRNAs, long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), control a variety of biological processes involving gene expression (Lanzillotti et al. 2021). Novel attributions of stem cell-derived extracellular vesicles containing miRNAs have also been discovered in recent years. These attributions include the modulation of processes occurring in the normal and cancerous microenvironment, participation in signaling and cell–cell communication between neighboring cells, and alteration of the phenotypes of nearby and recipient cells (Ghafouri-Fard et al. 2020).
Therefore, this review overviews some recent exciting findings regarding the functional implications of miRNAs and lncRNAs in controlling stem cell reprogramming, pluripotency maintenance, and cell differentiation to comprehensively understand the relationship between ncRNAs expression and stem cell fate. All of this might aid in considering the functional implications of these molecules in therapeutic approaches to treating chronic.
Biogenesis of miRNAs and lncRNAs
MiRNAs are short non-coding RNA expressed endogenously and have a length of 20–24 nucleotides. They usually fine-tune crucial biological processes (Ruiz-Manriquez et al. 2022b). Since discovered in Caenorhabditis elegans (Lee et al. 1993), it has been observed that they are ubiquitously expressed in plants, animals, and viruses, demonstrating their evolutionary importance (Mens and Ghanbari 2018). Moreover, the regulatory role of miRNAs in various biological processes, including cell proliferation, differentiation, and apoptosis, has been extensively studied (Lao and Le 2020; Annese et al. 2020).
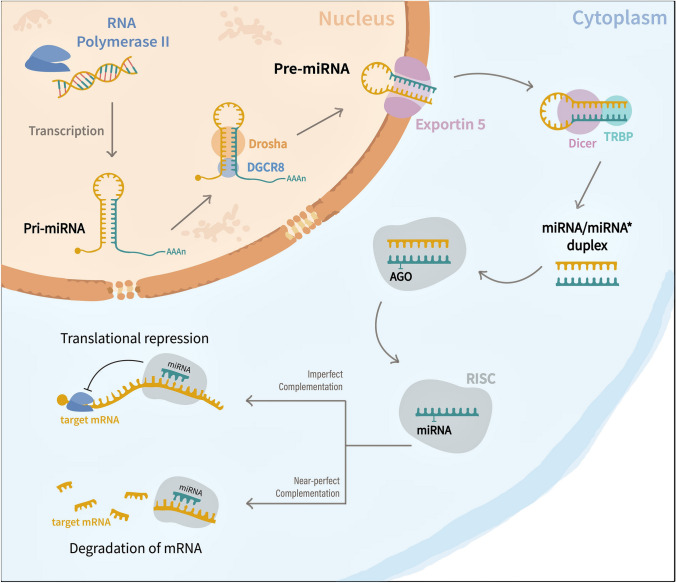
MiRNA biogenesis begins in the nucleus, where RNA polymerase II transcribes miRNA genes as long stem–loop structures known as primary miRNA transcripts (pri-miRNAs) (Fig. 2). Subsequently, the Drosha-DGCR8 microprocessor complex processes the pri-miRNAs to generate precursor miRNAs (pre-miRNAs) with shorter hairpin structures. These resulting pre-miRNAs are then exported to the cytoplasm by exportin-5 (EXP 5), where their terminal loops are cleaved by RNA III Dicer and transactivation-responsive RNA-binding protein (TRBP) to form ~ 20 to 22 nucleotide miRNA/miRNA* duplexes. The mature miRNAs from miRNA duplexes are then loaded into Argonaute proteins (Ago protein) to generate RNA-induced silencing complexes (RISCs), which are involved in regulating gene expression either by mRNA degradation or translation blocking (Zhao et al. 2019; Lao and Le 2020).
Fig. 2.
Canonical miRNA biogenesis pathway. miRNA biogenesis occurs in the nucleus when RNA Polymerase II transcribes miRNA genes into pri-miRNAs. Subsequently, a microprocessor complex made up of Drosha and DGCR8 enzymes trims pri-miRNA into pre-miRNA, which has been transported to the cytoplasm from the nucleus by Exportin 5. In the cytoplasm, pre-miRNA is then converted into mature miRNA/miRNA* duplexes by the RNase III endonuclease Dicer and the RNA-binding protein TRBP. Afterward, a helicase cleaves the duplex, and the resultant guide strand is integrated into the RNA Induced Silencing Complex (RISC) with the aid of an argonaute protein (AGO). Finally, the RISC–miRNA complex detects specific mRNAs based on the sequence complementarity, resulting in mRNA degradation or translational inhibition
LncRNAs are a different class of non-coding RNA ncRNAs that are distinguished from small RNA by having more than 200 nucleotides. They function as important regulators in a wide range of biological processes, such as chromatin remodeling, epigenetic and transcriptional modification, and nuclear trafficking (Wu et al. 2019b; Ruiz-Manriquez et al. 2022a). LncRNAs exert a significant influence on the control and fine-tuning of the translation machinery by altering the essential actions of other ncRNAs, such as miRNAs (Dahariya et al. 2019). Numerous lncRNAs have been found that regulate a wide range of cellular functions, are functionally related to both normal development and the pathophysiology of a number of disorders, and have been the subject of extensive investigation over many years (Cipolla et al. 2018).
The diversity of the biogenesis mechanisms of lncRNA owns to their specificity in being controlled and the capacity of responding to stimuli in regards to cell type and cellular stage (Ruiz-Manriquez et al. 2022a). Many classes of lncRNAs, despite their heterogeneity, converge in being transcribed by RNA polymerase II from different loci in the genome. The 5′ end of the premature lncRNAs is then capped using methyl-guanosine, the 3′ end is polyadenylated, and they are commonly expressed as alternatively spliced variants (Lanzillotti et al. 2021).
MiRNAs and stem cell regulation
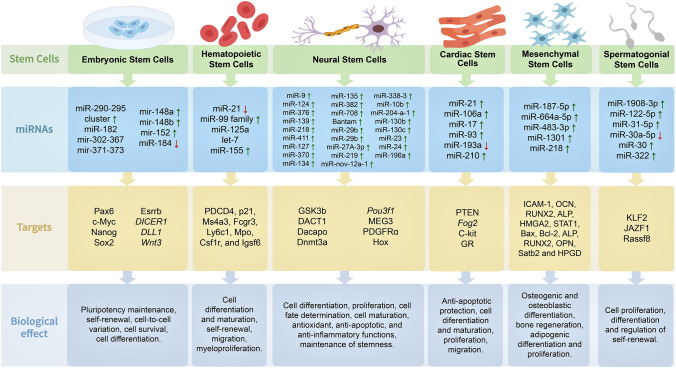
Recent studies regarding the functional implications of miRNAs in regulating different types of stem cells (Fig. 3) are discussed below and summarized in Table 1.
Fig. 3.
MiRNA-mediated regulation of stem cell populations. The biological effects of regulation induced by miRNAs and their corresponding targets are shown. Differential expression of each miRNA is shown by red (downregulated) and green (upregulated) arrows
Table 1.
MiRNAs involved in stem cells regulation, with relevant in vivo or in vitro evidence
| Stem cell type | miRNA | Regulation/expression | Target | Biologic effect/function | References |
|---|---|---|---|---|---|
| Embryonic stem cells | miR-290–295 cluster | Upregulated | Pax6 and c-Myc | Regulation of ESC pluripotency maintenance, self-renewal, and reprogramming of somatic cells to an ESC-like state | Yuan et al. (2017) and Kaspi et al. (2013) |
| miR-182 | – | Nanog, Sox2, and Esrrb | Cell-to-cell variation allows them to diversify into new states | Chakraborty et al. (2020) | |
| miR-302–367 and miR-371–373 | – | DICER1 | Promotion of cell survival and self-renewal | Teijeiro et al. (2018) | |
| miR-148a, miR-148b and miR-152 | Upregulated | DLL1 | Promotion of cardiomyocyte differentiation | Fang et al. (2019) | |
| miR-184 | Downregulated | Wnt3 | Promotion of cardiomyocyte differentiation | Liu et al. (2020) | |
| Somatic stem cells | |||||
| Hematopoietic stem cells | miR-21 | Downregulated | PDCD4, p21, Ms4a3, Fcgr3, Ly6c1, Mpo, Csf1r, and Igsf6 | Disrupts HSPC differentiation, and reduction of quiescence and long-term repopulation potential | Hu et al. (2021) |
| miR-99 family | Upregulated | Mpl, Hoxa9, Meis1, Tie1, Angpt1 and HOXA1 | Differentiation in HSPCs, LSCs and AML cell lines | Gerrits et al. (2012) and Khalaj et al. (2017) | |
| miR-125a | Upregulated | C-kit | HPSC trade, self-renewal, migration, and clone’s lifespan | Wojtowicz et al. (2019) | |
| let-7 | – | Alox5 | HPSC maturation and migration | Jiang et al. (2017) | |
| miR-155 | Upregulated | Ship1, Cebpb, and Pu.1 | Promotion of myeloproliferation | Wallace et al. (2017) | |
| Neural stem cells | miR-9 | Upregulated | GSK3b | Maturation of neurons and the formation of neuron’s branching fibers | Gizak et al. (2020) |
| miR-124 | Upregulated | DACT1 and GSK3b | Proliferation of NSCs and induction of neuronal differentiation | Gizak et al. (2020) | |
| miR-376, miR-139, miR-218, miR-411, miR-127, miR-134, miR-370, miR-135, miR-382, miR-708, and miR-124 | Upregulated | DRP2, NVL, CNOT4, PTBP1, TLE1, and SFXN5 | Synergic regulation in neuronal differentiation and rescue of neurogenesis | Pons-Espinal et al. (2017) | |
| Bantam | Upregulated | Dacapo | Maintenance and mediation of the proliferative potential of neuroblasts | Banerjee and Roy (2017) | |
| miR-29b | Upregulated | Dnmt3a and Pou3f1 | Regulation of cell fate determination. Inhibition progeny of NC cells at the initial stage of neural differentiation | Xi et al. (2017) | |
| miR-219, miR-218–2, miR-nov-12a-1, miR-338–3, miR-10b, miR-204-a-1, miR-130b, miR-130c, miR-23, miR-24, and miR-196a | Upregulated |
miR-219 target gene: PDGFRα miR-10b and miR-196a target genes: Hox |
Prevention of premature NC cell migration and possible participation in NCCs multipotency and maintenance of stemness phenotype | (Weiner 2018) | |
| Cardiac stem cells | miR-106a, miR-17 and miR-93 | Upregulated | Fog 2 gene | Differentiation of CSCs | Zhang et al. (2018) |
| miR-193a | Downregulated | C-kit | CSCs’ proliferation and migration | Sun et al. (2018) | |
| miR-210 | Upregulated | GR | Proliferation, maturation, and differentiation during cardiac fetal development | Meng et al. (2020) | |
| Mesenchymal stem cells | miR-187-5p | Upregulated | ICAM-1 | Osteogenic differentiation and bone regeneration | Sun et al. (2020) |
| miR-664a-5p | Upregulated | OCN, RUNX2, ALP, and HMGA2 | Osteogenic and osteoblastic differentiation | Zhang et al. (2020b) | |
| miR-483-3p | Upregulated | STAT1 | Osteogenic differentiation and bone formation | Xiao et al. (2019) | |
| miR-1301 | Upregulated | Bax, Bcl-2, ALP, RUNX2, OPN, and Satb2 | Adipogenic and osteogenic differentiation and proliferation | Kong et al. (2020) | |
| miR-218 | Upregulated | HPGD | Chondrogenic differentiation | Chen et al. (2019) | |
| Spermatogonial stem cells | miR-1908-3p | Upregulated | KLF2 | Promotion of proliferation and DNA synthesis of SSCs, and suppression of SSC apoptosis | Chen et al. (2020a) |
| miR-122-5p | Upregulated | CBL | Control of fate determinations of SSCs, promotion of proliferation and DNA synthesis of SSCs, and reduction of SSC early apoptosis | Zhou et al. (2020a) | |
| miR-31-5p | Upregulated | JAZF1 | Inhibition of proliferation and DNA synthesis of SSCs, and promotion of early and late SSC apoptosis | Fu et al. (2019) | |
| miR-30a-5p | Downregulated | Stra8, C-kit, GFRa1, PLZF, and ID4 | Promotion of differentiation of SSCs and suppression of self-renewal and proliferation of SSCs | Khanehzad et al. (2021) | |
| miR-30 | – | ID4 and C/kit | Regulation of self-renewal and differentiation of SSCs | Khanehzad et al. (2020) | |
| miR-322 | Upregulated | Rassf8 | Promotion of proliferation of SSCs and regulation of self-renewal and differentiation of SSCs | Wang et al. (2019) | |
| Cancer stem cells | miR-302a/d | Downregulated | E2F7 and AKT/-catenin/CCND1 signaling pathway | Associated with the cell cycle and functions of self-renewal | Ma et al. (2018) |
| miR-148a | Downregulated | SMAD2, AKT2, and KLF4 | Regulation of the TGF-β signaling pathway, inhibition of cell proliferation and EMT properties, and suppression of cytoprotective autophagy | Mohamed et al. (2019) | |
| miR‑486‑5p | Downregulated | Sirt1 | Maintenance of hepatic CSCs' self-renewal and tumorigenic capacity | Yan et al. (2019) | |
| miR-1 | Downregulated | RPPRC protein, MINOS1 and GPD2 | Regulation of mitochondrial morphology | Zhang et al. (2019a) | |
| miR-28-5p | Downregulated | IGF-1 | Regulation of programmed cell proliferation, differentiation, and apoptosis | Xia et al. (2019) | |
| miR-101 | Downregulated | ANXA2 | Cell cycle acceleration and promotion of cell migration and invasion abilities | Ma et al. (2021) | |
| miR-140-5p | Downregulated | Wnt1 | Promotion of cell cycle, migration, and survival | Wu et al. (2019a) | |
| miR-135b | Upregulated | JADE-1 | Promotion of proliferation, migration, and invasion. Inhibition of cell apoptosis | Zhou et al. (2020b) | |
| miR-188 | Downregulated | MDK | Promotion of cancer stem cell malignancy and xenograft development | Yang et al. (2020) | |
| miR-302c | Downregulated | CARF | Promotion of proliferation, migration, and invasion. Inhibition of cell apoptosis | Zhang et al. (2021) | |
| Exosomal miRNAs in stem cells | |||||
| Mesenchymal stem cells | miR-125a-3p | – | – | Reduction of effector T-cell differentiation and proliferation | Asgarpour et al. (2020) and Muralikumar et al. (2021) |
| miR-145 | – | Nestin, glial fibrillary acidic protein, microtubule-associated protein-2, and oligodendrocyte transcription factor 2 mRNA | Implications in neurogenesis | Soni et al. (2021) | |
| miR-132, miR-145, and miR-125 | – | – | Induction of angiogenesis | Soni et al. (2021) | |
| Dental pulp stem cells | miR-27a-5p | – | TGFβ1/Smad signaling cascade | Induction of odontogenic differentiation | Jin et al. (2020) |
| Urine-derived stem cells | miR-16-5p | – | – | Attenuation of the progression of type I diabetic kidney issues | Zhang et al. (2017) |
| Adipose-derived stem cells | miR-215-5p | Upregulated | – | Beneficial effect on diabetic nephropathy prevention | Zhang et al. (2017) |
Embryonic stem cells (ECSs)
ESCs are generated in epiblast tissue from the inner cell mass of a blastocyst or earlier morula stage embryos (Li et al. 2017). ESCs display two excellent properties: pluripotency, defined as the capacity to produce both somatic and germline lineages that can form the early embryo; and self-renewal, defined as the ability to form a complex range of cells with various gene expression states indefinitely, including the cells of the adult vertebrate (Divisato et al. 2020). Since the successful isolation of ESCs, extensive research has been done to demonstrate the critical transcriptional pluripotency regulatory network, including pluripotency genes such as Pou5f1 (Oct4), Sox2, and Nanog, as well as some specific miRNAs (Lee et al. 2013).
It has been noticed that embryonic stem cell-specific miRNAs play a critical role in the embryo’s early development (Müller et al. 2022). For example, the miR-290–295 cluster is the most abundant miRNA cluster found in mouse embryonic stem cells (mESCs). It is active in various biological mechanisms maintaining embryonic stem cell pluripotency, self-renewal, and converting somatic cells to resemble embryonic stem cells (Yuan et al. 2017). In both undifferentiated and developing mESCs, the loss of miR-290–295 activity de-represses paired-box transcription factor 6 (Pax6), a neuroectoderm identity regulator, and accelerates ectoderm specification at the cost of endoderm and mesoderm lineages (Kaspi et al. 2013). It has also been stated that overexpression of the miR-290–295 family increased the c-Myc level, a downstream target of the Wnt-signaling pathway necessary for ensuring stem cell pluripotency (Rasmussen et al. 2018). Besides, the miR-290–295 cluster is implicated in tumorigenesis and senescence and has a latent pro-survival role in embryonic stem cells (Yuan et al. 2017).
Chakraborty et al. (2020) showed that in mESCs, miRNAs such as miR-182 organize inherent variation into cell states associated with embryonic development. This miRNA exhibited action on genetic subcircuits of specific developmental factors like Nanog, Sox2, and Esrrb that differ between states, structuring intrinsic fluctuations in cells at a higher level. When cells were divided, this produced intrinsic heterogeneity or the capacity of cells to repopulate certain cell states. Manipulating the level of miR-182 in mESCs directly impacted cell-to-cell variation, allowing them to diversify into new states, suggesting its essential role in coordinating variations across gene networks to assist in forming various expression states (Chakraborty et al. 2020; Mackay et al. 2020).
According to Teijeiro et al. (2018), the development of DICER1 knockout on human embryonic stem cells (hESCs) lines provided factual data in evaluating the role of miRNAs in human pluripotency. DICER1 appears to be an essential requirement for hESC self-renewal, and its removal caused an increase in death receptor (DR)-mediated apoptosis and hESC self-renewal failure. Based on a targeted miRNA detection strategy, the essential roles favor the survival of miRNAs containing ESC-specific cell-cycle-regulating (ESCC) seed sequences from the mir-302–367 and mir-371–373 clusters were discovered. These clusters enhance cell survival in DICER1 knockout hESC (Teijeiro et al. 2018).
Liu et al. (2020) found that during the differentiation of human embryonic stem cells (hESCs) into cardiomyocytes, the miR-148a family, including miR-148a, miR-148b, and miR-152, was progressively enhanced. The NOTCH ligand Delta-like 1 (DLL1) has been confirmed as a target of miR-148a, and its knockdown promotes cardiomyocyte differentiation (Fang et al. 2019). In contrast, miR-184 expression is reduced when ESCs differentiated into cardiac mesoderm and subsequently increased as they differentiate into cardiomyocytes, a process negatively linked with the expression of Wnt3, a vital component of the Wnt-signaling pathway (Liu et al. 2020).
Undeniably, ESCs are an excellent model for studying the impact of miRNAs; nowadays, using novel sequencing technologies, researchers have identified some miRNAs in human and mouse embryonic stem cells involved in the proliferation and differentiation processes of ESCs. Moreover, numerous miRNAs have also been discovered in ectoderm, mesoderm, and endoderm cells. However, since the differentiation and self-renewal of ESCs mediated by miRNAs are intrinsically complicated, further research is needed to elucidate precisely.
Tissue-specific stem cells (TSSCs)/somatic stem cells
Adult somatic stem cells are self-renewing clusters of cells found in tissues and organs that can create particular lineages of precursor cells, which then develop into differentiated cell offspring (Tweedell 2017). Explicit miRNA expression profiles in distinct somatic stem cell lines state that complex interactions between miRNAs, transcription factors, and cell cycle-mediated components occur after the differentiation of multipotent stem cells into progenitor and mature cells, regulating gene expression (Mens and Ghanbari 2018). Moreover, the functional implications of miRNAs in controlling the biological processes of different somatic stem cells, including hematopoietic, neural, cardiac, and mesenchymal stem cells, have mainly been elucidated in this review.
Hematopoietic stem cells (HSCs)
HSCs are responsible for making every type of blood cell, as they have the ability to self-renew and differentiate into diverse multi-, oligo-, or unipotent progenitors, which subsequently proliferate and differentiate into a vast array of entirely functional mature cells, whether in the bone marrow (BM) or other hematopoietic or lymphoid organs (Brown et al. 2018). The hematopoietic system has an immense cell proliferation and differentiation capacity to cover the immune cell production and blood demand, all along with stress, adaptation, and homeostasis, controlled by both intrinsic and extrinsic signals from its microenvironment (Wei et al. 2018). Nevertheless, hematopoiesis is a rigorously regulated multi-step differentiation process responsible for generating mature blood cells, and several studies have demonstrated that a rigidly integrated network of miRNAs and transcription factors substantially controls this process (Weiss and Ito 2017; Kotaki et al. 2017; Krivdova et al. 2022).
In this context, Hu et al. (2021) conducted a study using a conditional knockout murine model to establish a relationship between miR-21 and HSC populations. Their study demonstrated that the induced inactivation of miR-21 in the hematopoietic compartment disrupts hematopoietic stem and progenitor cells’ (HSPC) differentiation, suggesting that this miRNA is crucial for steady-state hematopoiesis. Moreover, miR-21 deficiency in HSCs of mice BM causes a significant increase in the expression levels of myeloid differentiation genes (Ms4a3, Fcgr3, Ly6c1, Mpo, Csf1r, and Igsf6), NF-κB genes, such as p21, and PDCD4 (a direct target of miR-21). As a result, the NF-κB pathway was inhibited, which plays a crucial function in maintaining hematopoietic homeostasis and function (Sun et al. 2021). It was also found that ectopic expression of PDCD4 caused a reduction in quiescence and long-term repopulation potential of HSCs and biased differentiation (Hu et al. 2021).
Resulting in studies of the miR-99 family (miR-99a, miR-99b, and miR-100) has shown to be significantly involved in HSPCs maintenance by suppressing their differentiation. MiR-99 inhibition depletes genes recognized in HSC signatures, such as Mpl, Hoxa9, Meis1, Tie1, and Angpt1, indicating that miR-99 conserves HSCs and leukemic stem cells (LSCs) in an undifferentiated state as this miRNA functions as a key element in preventing cells from leaving the stem cell stage. The influence of miR-99 in the maintenance of human acute myeloid leukemia (AML) cells and the suppression of differentiation among human AML blasts was further elucidated, and it was confirmed that miR-99 mediates the differentiation phenotype via homeobox A1 (HOXA1) in HSPCs, LSCs, and AML cell lines (Gerrits et al. 2012; Khalaj et al. 2017).
Wojtowicz et al. (2019) observed that an increased expression of miR-125a significantly enhances clonal contribution to hematopoiesis, clone size, and longevity during long-term serial transplantations. The identification of cells with clonogenic potential in the peripheral blood (PB) of miR-125a transplanted mice further indicated that miR-125a overexpression could increase HSC trade from the BM to its surroundings and enhance the self-renewal, migration, and lifespan of HSPC clones through down-regulation of C-Kit expression. Likewise, implications of let-7 in the hematopoietic niche of the Drosha knockout mouse model were also elucidated, discovering a new stage of action that takes place after HSPC clusters emergence but before their mobilization to the fetal liver (FL). Deletion of Drosha leads to anemia and death, as well as an accumulation of HSPC in the dorsal aorta with failure of FL colonization; the depletion of the let-7 family primarily causes this defect, since it activates leukotriene B4 (LTB4) signaling and induces α4β1-integrin (Itg) cell adhesion complex in HSPC. Therefore, α4β1-Itg regulation is essential for HSPC interaction with the vascular microenvironment through cell adhesion and HSPC maturation via signaling (Jiang et al. 2017).
Using a genetic mouse model and human AML cells, Wallace et al. (2017) highlighted the relationship between miR-155 and FMS-like tyrosine kinase-3 (FLT3) internal tandem duplication (ITD) in promoting hematologic malignancy. They demonstrated that miR-155 is fundamental for robust myeloid progenitor pool maintenance. Its overexpression increases FLT3-ITD-mediated myeloproliferative disease (MDP) by enhancing the proliferation of LKS and myeloid progenitor cells among the hematopoietic compartments. Promoting FLT3-ITD + myeloid cell expansion by miR-155 was sufficient evidence of the hematopoietic cell-intrinsic role of this miRNA. Moreover, they observed that the absence of miR-155 significantly increases the expression of Ship1, Pu.1, Cebpb (direct targets of miR-155), and interferon (IFN). These findings demonstrate that miR-155 represses IFN response in FLT3-ITD+ mice HSPC and human AML cells and serves as a critical promoter of myeloproliferation in the BM, spleen, and blood of mice with FLT3-ITD mutation (Wallace et al. 2017).
There is growing evidence that miRNAs act as crucial regulators of several HSC processes, such as differentiation, proliferation, migration, and homeostasis, among others. However, the available information is mainly obtained from experimental mice models; thus, further studies in human stem cells regarding these interactions will be compelling for clinical approaches.
Neural stem cells (NSCs)
Billions of neurons that make up the neural circuits in the central nervous system (CNS) are mainly produced during the embryonic development of the mammalian brain in the germinal zones of the neural tube’s ventral and dorsal telencephalon, where NSCs give rise to adult NSCs and several other subtypes of neurons (di Marco et al. 2020). NSCs (subclassification of TSSCs) are multipotent and mitotically active and capable of differentiation, self-renewal, generation, and regeneration of brain tissue in both the embryonic and adult CNS. Several miRNAs are known to mediate fundamental biological processes in the CNS, such as brain development, neuronal growth, morphogenesis of dendrites, synaptic plasticity, stress response, and neurogenesis (Paul et al. 2020; Gizak et al. 2020; Li et al. 2022).
MiR-9 and miR-124, two of the most abundant miRNAs in the mammalian neural tissue, have been identified as regulatory molecules in the maturation of neurons and the formation of neuron’s branching fibers in a multi-step process known as dendrite arborization (Lanoue and Cooper 2019; Gizak et al. 2020). As a result of synergistic action, these two miRNAs upregulate glycogen synthase kinase-3 (GSK3b) expression, a protein active in fundamental steps of neuronal morphogenesis. Moreover, the precise role of miR-124 in the proliferation of NSCs and neuronal differentiation comprises the induction of β-catenin expression, involved in the neurogenesis of the developing and adult brain, by targeting the Dishevelled binding antagonist of beta-catenin 1 (DACT1) and GSK3b (Gizak et al. 2020).
Pons-Espinal et al. (2017) identified 11 miRNAs, including miR-376, miR-139, miR-218, miR-411, miR-127, miR-134, miR-370, miR-135, miR-382, miR-708, miR-124, and nine newly characterized miRNAs, which act synergistically in the neurogenic fate determination of adult murine hippocampal stem cells (aNSCs). Altogether these 11 miRNAs were found to rescue neurogenesis impairment after ablation of Dicer in aNSCs, unlike when tested individually. Authors further stated their synergic role in the nervous system development and neuronal differentiation by predicting 26 putative targets, including dysregulated proteins such as DRP2, NVL, CNOT4, PTBP1, TLE1, and SFXN5. Additionally, at the subgranular zone (SGZ) and the granule cell layer (GCL) of the adult murine hippocampus, the death of newborn neurons and the loss of differentiation and maturation of surviving cells after the in vivo and in vitro conditional ablation of the Dicer gene confirmed Dicer-dependent miRNAs are vital for neurogenesis in NSCs (Pons-Espinal et al. 2017).
Through Dicer-1 protein depletion experiments in the Drosophila model, Banerjee and Roy (2017) also demonstrated the significant regulatory role of specific miRNAs in NSCs. The downregulation of Dicer-1 protein in Drosophila larval neural stem cells suggested the role of Bantam miRNA in the maintenance and mediation of the proliferative potential of neuroblasts progenitor cells of the Drosophila CNS with stem cell characteristics. The results exhibited a reduced expression of Bantam miRNA occurring concurrently with the depletion of mitotically active cells and a reduction in size and number of type I and II neuroblasts in the central nervous system of third-instar larvae. More specifically, the ablation of Dicer-1 assessments reported the loss of neuroblasts proliferation as a response to the consequent down-regulation of Bantam miRNA, which further leads to the prevention of neuroblasts from transitioning to the G1- S phase due to the sequestration of Cyclin E via Dacapo, a gene whose premature expression is acknowledged to be responsible for G1 phase early arrest in Drosophila (Banerjee and Roy 2017).
The participation of miRNAs is reported to be crucial in the Neural Crest (NC) cells neurogenesis, a transitory, multipotent stem cell-like population that forms along the edge of the growing neural tube early in development. Hereby, miRNAs function as regulators of induction, specification, delamination, and differentiation (Xi et al. 2017; Ward et al. 2018; Weiner 2018). For instance, in ESCs, miR-29b was found to regulate neuronal differentiation by inhibiting progeny of NC cells; furthermore, miR-29 b’s function in cell fate determination was elucidated via targeting DNA methyltransferase 3a (Dnmt3a) and transcription factor Pou3f1 genes by Xi et al. (2017).
Experimentally, Xenopus laevis is a standard embryonic vertebrate model used to figure out the regulatory role of miRNAs in the NC (Luu and Storey 2015; Shah et al. 2016; Ward et al. 2018). For instance, miR-427 was the most prevalently expressed in the developing nervous system of Xenopus laevis, confirming its role in the primary stages of embryonic growth (Ward et al. 2018). Moreover, differential miRNA expression analysis demonstrated a significant upregulation of 11 miRNAs (miR-219, miR-218–2, miR-nov-12a-1, miR-338–3, miR-10b, miR-204-a-1, miR-130b, miR-130c, miR-23, miR-24, and miR-196a) in the NC tissue as compared to neural tissue, indicating their possible implication in NC maturation (Weiner 2018). Ward et al. (2018) explored miRNA’s multipotency function in the Xenopus laevis’s NC through quantification assays, resulting in the overexpression of miR-301 and miR-338 in the tissues of the NC and the blastula, suggesting that they might participate in NSCs regulation at embryonic development (Ward et al. 2018; Weiner 2018). Nevertheless, additional research is needed to support these miRNAs’ role in maintaining stemness.
Neurogenesis, in both the NSCs and neural progenitor cells settled at the distinct neuronal niches of the CNS, is the most studied miRNA-mediated biological process (Stappert et al. 2018). Moreover, valuable information regarding the participation of miRNAs apart from neurogenesis has also deciphered their involvement in activities such as neuroprotection, survival, and conservation of stemness traits (Gorabi et al. 2019b). However, further studies on their implication in other biological processes should be conducted. Results from dicer depletion have emerged, both embryonic and adult stages, implying that miRNAs have vital participation in NSCs at different stages of development (Ward et al. 2018; Weiner 2018; Esteves et al. 2020). Nevertheless, further research is required to elucidate the metabolic pathways, biochemical cascade reactions, and the specific targets of the newly identified miRNAs involved in NSCs.
Cardiac stem cells (CSCs)
Cardiac stem cells (CSCs) are adult stem cell subtypes that govern early stage myocardial development and possess the repair and regeneration capacity of the injured heart (Li et al. 2019a; Gorabi et al. 2019a). Cardiac progenitor cells (CPCs), which are endogenous stem cells with low proliferative capacity, manifest traits of multipotency and self-renewability and are able to differentiate into cardiac cell progeny of cardiomyocytes (CMs), endothelial cells (ECs), and smooth muscle cells (SMCs) (Amini et al. 2017; Gorabi et al. 2019a). Reports suggest that miRNAs are the key regulators in proliferation, differentiation, cell fate determination, migration, and apoptosis processes of cardiac tissue stem cells for normal heart development, and their dysregulation leads to congenital cardiac diseases (Li et al. 2019a; Paul et al. 2021; Maleki et al. 2022).
The vital role of miRNAs in cardiogenesis survival and cardiac health at both early and postanal stages of growth by the effect of Dicer deletion assessments in cardiac progenitor cells is reported (Tian 2018). Interestingly, Dicer gene knockout was found to induce embryonic death, cardiac failure at the embryonic stage, myocardial defects and embryonic lethality at the middle stage of gestation, and cardiomyopathy. Cardiac development is controlled by mechanisms, such as genetic cascades, molecular signals, patterns of gene expression, and transcriptional and post-transcriptional interactions, in which participation of miRNAs is substantial (Tian 2018; Li et al. 2019a; Tan and Lewandowski 2020).
Zhang et al. (2018) discovered the individual role of a number of miRNAs, including 106a, miR17, and miR93 (miR-17 family), in cardiac differentiation, using ESCs as a model system. Moreover, employing the CRISPR/Cas 9 technique, they also suggested that these miRNAs described above work semi-autonomously in cardiac development by downregulating the target cardiac suppressor gene Fog 2. Apart from differentiation of CSCs, Sun et al. (2018) elucidated the role of miR-193a in the proliferation and migration of C-kit positive CSCs by Insulin-like Growth Factor (IGF-1), a hormone established to induce CSCs proliferation and migration, as well as upregulation of C-kit in CSCs. A qPCR analysis indicated down-regulation of miR-193a in IGF-1 stimulated CSCs and a negative correlation between the expression of miR-193 and C-kit cardiac stem cell marker. The transfection of CSCs with miR-193a engineered lentivirus assay suggests that the proliferation and migration of CSCs result from the epigenetic silencing of miR-193a by IGF-1 upregulation of C-kit stem cell marker via the activation of the signaling pathway PI3K/AKT/DNMT (Zhang et al. 2018; Xia et al. 2019).
Hypoxia during gestation significantly negatively impacts fetal heart development (Gao et al. 2019). In this context, Meng et al. (2020) showed a novel regulatory mechanism of miR-210 in hypoxic stem cell antigen-1 (Sca-1+) CPCs during fetal cardiac development. Experimental induction of hypoxic conditions demonstrated differentiation and proliferation alterations in CPCs and a depleted cardiomyocyte maturation capacity. Moreover, the expression of miR-210 was triggered by hypoxia and impeded the differentiation of Sca-1+ CPCs. Silencing miR-210 through LNA-anti-miR-210 mediated Sca-1 + CPC transfection resulted in a drastic rescuing of Sca-1+ CPC differentiation capacity into cardiomyocytes. It has also been shown previously that the glucocorticoid receptor (GR) is a downstream target of miR-210, which plays a vital role in cardiomyocyte formation and cardiac function. Hence, miR-210-mediated GR inhibition enhances cardiomyocyte apoptosis. Although the detailed molecular mechanisms and the signaling pathways implicated remain elusive, this study suggests miR-210’s regulatory role in CPC proliferation and differentiation under hypoxia, as well as its potential as a biomarker for cardiomyopathy (Martinez et al. 2017).
Together, these findings suggest that miRNAs are strongly implicated in several CSCs and CPCs cellular processes crucial for cardiovascular health. However, further studies are required to precisely explicate miRNA/mRNA interaction during cardiogenesis.
Mesenchymal stem cells (MSCs)
MSCs are primitive heterogeneous cells initially discovered in adult bone marrow stroma (Sui et al. 2020). They are defined as adult progenitor cells and are also known as mesenchymal stromal cells (Jiang and Xu 2020). MSCs are derived from diverse tissues, such as adult bone marrow, adipose tissues, skin, and neonatal tissues like the placenta and umbilical cord (Naji et al. 2019). It has been demonstrated that these cells possess important biological characteristics like extensive proliferation ability, self-renewal, and multipotency, and can produce different cell lineages, including adipocytes, osteoblasts, and chondrocytes (Huang et al. 2017; Jothimani et al. 2022b). Moreover, they are known for their crucial role in homeostasis and regenerative repair of tissues and organs derived from endoderm, mesoderm, and ectoderm (Sui et al. 2020). It has been elucidated that diverse miRNAs modulate the MSCs’ differentiation process into different cell lineages, and dysregulation of miRNA levels could lead to aberrations in MSC functions, such as dysfunctional niche modulation, biased differentiation, and impaired proliferation (Sui et al. 2020). Recently, the specific targets of these miRNAs have been determined, and their roles are progressively being enlightened (PENG et al. 2016; Huang et al. 2017; Jiang and Xu 2020).
In this context, osteoblastic differentiation of bone marrow mesenchymal stem cells (BMSCs) studies has brought new insights into the treatment of osteoporosis, a widespread metabolic bone disease with a fundamentally irreversible mechanism (Bravo Vázquez et al. 2021). MiR-187-5p has been discovered to play a critical role in osteoblast development, and its expression is considerably elevated during BMSCs’ osteogenic differentiation in mice. Upregulation of miR-187-5p improved the effectiveness of new bone development by targeting intracellular adhesion molecule 1 (ICAM-1), linked to osteogenic differentiation and bone regeneration, and suggested a prospective therapeutic target for bone metabolic disorders (Sun et al. 2020). Likewise, Zhang et al. (2020b) found that miR-664a-5p was substantially overexpressed during BMSCs’ osteogenic differentiation, along with some osteogenesis-related genes, such as osteocalcin (OCN), runt-related transcription factor 2 (RUNX2), and alkaline phosphatase (ALP). In this work, it was also shown that miR-664a-5p directly targets high-mobility group A2 (HMGA2), a transcription factor that has been related to osteoblastic differentiation and the upregulation of miR-664a-5p markedly decreased HMGA2 level. Hence, the findings regarding the miR-664a-5p-HMGA2 pathway elucidate the mechanisms that cause BMSCs to differentiate into osteogenic cells, providing new insights for better and more effective methods to treat osteoporosis (Zhang et al. 2020b).
Xiao et al. (2019) noticed that miR-483-3p was remarkably overexpressed during osteogenic differentiation, which enhanced mineralized nodule formation, ALP activity, OCN secretion, Type I collagen RUNX2, and osterix expression levels. This evidence suggests that this miRNA promotes osteogenic differentiation of BMSCs. Additionally, miR-483-3p directly targets STAT1, which has been reported as an osteogenic differentiation inhibitor and a key element in bone formation. Thus, miR-483-3p regulates osteogenic differentiation through the miR-483-3p/STAT1/RUNX2 axis. Kong et al. (2020) conducted a study to determine the role of miR-1301 in BMSC adipogenic and osteogenic differentiation. They observed that the upregulation of miR-1301 promoted BMSCs proliferation. This overexpression also affected the expression of apoptosis and anti-apoptotic genes, downregulating Bax and upregulating Bcl-2. In addition, they found that miR-1301 upregulation enhanced ALP activity and the expression of the osteogenic genes RUNX2 and OPN. Whereas FABP4 and PPAR γ2, which are adipogenic-related genes, were downregulated. Moreover, animal trials and clinical investigations have confirmed that exosomes released by miRNAs on BMSCs have a therapeutic impact on spinal cord damage (SCI) (Yu et al. 2019).
Articular cartilage (AC) represents a unique avascular tissue characterized by restricted self-healing and regeneration capacities after injuries or degenerative diseases. Interestingly, miRNAs have been shown to play a crucial role in regulating MSCs’ chondrogenic differentiation (Chen et al. 2019). For instance, Chen et al. (2019) found that miR-218 was significantly upregulated in synovium-derived MSCs (SDSCs) and promoted early chondrocytes formation via SDSCs’ differentiation. They also proposed that miR-218 plays a key role in regulating 15-hydroxyprostaglandin dehydrogenase (HPGD), an enzyme vital for chondrogenic differentiation, indicating that miR-218 directly modulates the expression of HPGD in SDSCs through the miR-218–HPGD pathway.
Currently, miRNAs have been shown to play a crucial role in regulating several MSCs processes; however, a better understanding of the mechanisms underlying these differentiations might open a new therapeutics arena against bone ailments, such as osteoporosis and other tissue-related diseases (Bravo Vázquez et al. 2021).
Spermatogonial stem cells (SSCs)
SSCs account for only 0.02–0.03% of the germ cell population and can be located across the basement of the seminiferous tubules (Wang et al. 2019; Fu et al. 2019); they play a crucial role in spermatogenesis, a complex developmental process that occurs in the mammalian reproductive system and gives rise to highly specialized and mature sperm (Khanehzad et al. 2021). SSCs are unique stem cells that possess exclusive characteristics compared to other stem cell populations highlighting the transfer of genetic information to the next generations. Furthermore, they can self-renew throughout life and differentiate into cell lineage due to their pluripotency ability (Chen et al. 2020b; Khanehzad et al. 2021). SSCs maintain stemness by self-renewing and can differentiate into mature spermatids and spermatocytes (Zhou et al. 2020a). Several studies have demonstrated that genetic mediators and epigenetic factors, such as miRNAs, regulate SSC self-renewal, proliferation, differentiation, and apoptosis. Besides these properties, miRNAs play a crucial role in spermatogenesis (Chen et al. 2020a; Zhou et al. 2020a; Khanehzad et al. 2021; Lv et al. 2022).
Chen et al. (2020a) found that miR-1908-3p was highly expressed in human spermatogonia and the SSC line, indicating its crucial participation in regulating the fate determinations of SSCs. The artificial overexpression of miR-1908-3p achieved by miR-1908-3p mimics promoted SSC line proliferation, accelerated the DNA synthesis of the SSC line, and suppressed SSC apoptosis. Moreover, miR-1908-3p directly targets KLF2, a major mechanosensitive transcription factor in vascular homeostasis regulation, which enhances apoptosis and suppresses DNA synthesis and proliferation of the SSC cell line. These results better understand the epigenetic regulatory mechanisms underlying SSCs’ cell cycle and spermatogenesis, providing a fundamental target for male infertility prognosis and treatment. Zhou et al. (2020b) observed that miR-122-5p plays a crucial role in controlling the fate determinations of SSCs. They artificially overexpressed miR-122-5p via lentivirus transfection and found that this upregulation increased human SSC line proliferation, stimulated division and DNA synthesis of SSCs, and reduced early apoptosis of human SSCs. Intriguingly, miR-122-5p directly targets CBL, an E3 ubiquitin ligase that promotes the degradation of proteins related to migration and cell growth. By downregulating the CBL expression, miR-122-5p lentivirus enhances proliferation and inhibits early apoptosis of the SSC line. Moreover, a negative correlation between lncRNA CASC7 and miR-122-5p was also noticed.
It has been demonstrated that P21-activated kinase 1 (PAK1) is expressed in the human SSC line and controls DNA synthesis, apoptosis, and proliferation of the SSC line. Additionally, patients with nonobstructive azoospermia (NOA) present low levels of PAK1 (Fu et al. 2018). To elucidate the epigenetic regulation of PAK1 in human SSCs, Fu et al. (2019) conducted a study. They found that PAK1 knockdown triggers the miR-31-5p expression, suggesting that PAK1 negatively regulates miR-31-5p in SSCs. Moreover, employing miR-31-5p mimics, they also clarified that this miRNA inhibits proliferation and DNA synthesis of SSCs and enhances early and late apoptosis of the SSC line. Furthermore, miR-31-5p targets the transcription factor JAZF1 and negatively regulates the cell cycle-related protein cyclin A2. Regarding mouse SSCs, Khanehzad et al. (2021) observed that miR-30a-5p significantly affects SSC's fate determinations. For instance, miR-30-5p inhibition remarkably enhanced the expression of Stra8 and C-kit, which are differentiation markers, suggesting that miR-30-5p inhibition induces differentiation in mouse SSCs. Additionally, the levels of GFRa1, PLZF, and ID4, proliferation-related proteins, were considerably reduced after transfection with miR-30-5p inhibitor, indicating that miR-30a down-regulation inhibits self-renewal and proliferation in SSCs. Previously, Khanehzad et al. (2020) also found that under miR-30 mimic exposure, ID4 levels considerably increased, while the expression of C-kit protein remarkably decreased, indicating that miR-30 is a critical regulator of self-renewal and differentiation in SSCs. These results might be helpful for the development of innovative therapeutic strategies for infertility cases.
SSCs have been shown to play an essential role in spermatogenesis, and it has been elucidated that diverse miRNAs regulate mouse and human SCCs. Even though considerable progress has been made in elucidating the molecular mechanisms underlying spermatogenesis in rodents, the epigenetic regulation in male germ cells remains largely unknown. Hence, studying these interactions in human SSCs might provide novel tools to treat male infertility.
Cancer stem cells (CaSCs)
Carcinogenesis, or cancer formation, is a multi-step process that accumulates genetic, epigenetic, biochemical, and histological alterations that eventually lead to pathological manifestations (Khan et al. 2019). Although cancer begins with a single mutant cell, it can spread to include a wide range of differentiated and proliferative cells. Tumor formation, recurrence, metastasis, and treatment resistance are caused by this heterogeneity. The presence of CaSCs is crucial among the several cell types that account for cancer heterogeneity (Ayob and Ramasamy 2018). CaSCs are cancer cells with the potential for self-renewal and survival in tumors as distinct cancer cell populations that can cause relapse and metastasis by re-emerging or forming new tumors. It has been demonstrated that miRNAs regulate CaSCs in malignant tumors and are implicated in the modulation of EMT and the Notch-signaling pathway (Xu et al. 2017; Zhou et al. 2022).
Initiation and progression of hepatocellular carcinoma (HCC) are believed to be aided by liver cancer stem cells (LCSCs) (Lv et al. 2021). Several miRNAs linked to pluripotency maintenance and differentiation have been discovered to be major regulators of HCC carcinogenesis and drug resistance signaling networks. LCSCs are one of the main determinants of treatment failure and disease recurrence (Khan et al. 2019). For instance, the miR-302 family has a crucial role in carcinogenesis and progression; this family was discovered to be poorly expressed in both LCSCs and HCC tumors during tumorsphere formation to enrich LCSC cells, and patients with lower miR-302a/d expression exhibited shorter overall and progression-free survival (Ma et al. 2018). Moreover, overexpression of miR-302a/d has been shown to hinder self-renewal and cell cycle entrance of LCSCs via inhibiting its target gene E2F7 and the downstream AKT/-catenin/CCND1 signaling pathway, which are closely associated with the cell cycle and possess many regulatory functions of self-renewal. Hence, targeting LCSCs and understanding the fundamental mechanisms of miRNA in LCSCs might help improve HCC diagnostic and therapeutic strategies (Ma et al. 2018).
MiR-148a subnetwork has shown intriguing insights in the context of HCSC pathology, which is the key controller for initiating HCC. The function of miR-148a in pathogenic processes associated with the CaSC, such as chemosensitivity and EMT, suggests that it might be used as a therapeutic tool for HCC stemness (Mohamed et al. 2019). Yan et al. (2019) examined the miRNA profiles in HCC samples and neighboring non-tumor tissues, finding that miR-486 was considerably downregulated in HCC tissues and inhibited CaSC self-renewal and invasion in vitro as well as tumorigenesis in vivo. Furthermore, sirtuin 1 (Sirt1) was identified as a direct target of miR-486, essential in maintaining LCSCs’ self-renewal and tumorigenic capacity. These results indicated that the miR-486-Sirt1 axis inhibited CaSC characteristics and tumor growth in HCC.
MiR-28-5p has been demonstrated to have a key role in HCC cell sensitivity to sorafenib, the most commonly prescribed first-line targeted drug for advanced HCC patients. Furthermore, IGF-1, a significant regulator of programmed cell proliferation, differentiation, and apoptosis, has been identified as a direct target of miR-28-5p in LCSCs (Xia et al. 2019). More recently, another miRNA, miR-101, has been downregulated in LCSCs, and its induction inhibits metastasis and tumorigenic capacity in LCSCs. Interestingly, ANXA2, essential for cell cycle acceleration and LCSCs’ promotion of migration and invasion abilities, was identified as a novel target of miR-101. Subsequently, the extracellular signal-regulated kinase (ERK) expression was enhanced due to increased ANXA2, and the phosphorylation of ERK decreased the early growth response 1 (EGR1) level, which reduced miR-101 transcription. In vivo tests validated the effects of miR-101 overexpression, which has substantial inhibitory effects on LCSC xenograft tumor development and ANXA2 knockdown. These data shed light on the miR-28-5p/IGF-1 axis and miR-101/ANXA2/EGR1 regulatory loop as potential therapeutic targets for HCC (Xia et al. 2019; Ma et al. 2021).
The overexpression of miR-1 in CaSCs was found to have the potential to damage mitochondria by binding leucine-rich pentatricopeptide-repeat-containing (RPPRC) protein and targeting the genes mitochondrial inner membrane organizing system 1 (MINOS1) and glycerol-3-phosphate dehydrogenase 2 (GPD2) (both of which are required for the mitochondrial organization). This further elucidates the miRNA-mediated regulation of mitochondrial morphology in CaSCs (Zhang et al. 2019a).
Wu et al. (2019a) reported that miR-140-5p is usually downregulated in breast cancer stem cells (BCSCs), and its mimics could block these cells from proliferating by targeting the expression of Wnt1. Moreover, through the Wnt1/ABCB1 pathway, miR-140 might improve the susceptibility of BCSCs to anticancer drugs like doxorubicin (Dox) both in vitro and in vivo. Wnt1 has been reported to be associated with cancer development by promoting the cell cycle, migration, and survival, whereas ABCB1 has been linked to therapeutic resistance in various malignancies. Additionally, it has been demonstrated that Wnt-signaling pathways influence the production of ROS that may contribute to the growth of cancer by promoting cell proliferation and differentiation (Samatha Jain et al. 2022). These findings propose a novel treatment method using miR-140-5p in conjunction with anticancer medicines to overcome acquired multi-chemoresistance by targeting the Wnt1 signaling pathway.
According to Zhou et al. (2020b), miRNAs can act as essential regulators during the onset and progression of pancreatic cancer (PC). They noticed that a high miR-135b in PCSC tissues affects stemness and significantly promotes tumor growth by negatively regulating the target gene JADE-1. Moreover, the overexpression of this miRNA promoted PCSC proliferation, migration, and invasion, inhibited cell apoptosis, and increased expression of stem-related factors such as Sox-2, Oct-4, Nanog, Aldh1, and Slug.
Further studies have illustrated the relevance of miRNAs in colon and lung cancer. The effect of miR-188 on lung cancer stem cells was explored, demonstrating that the miR-188 level was low in clinical samples of lung cancer patients. When overexpressed, it suppresses lung cancer stem cells' biological characteristics by blocking the MDK-mediated Hippo pathway (Yang et al. 2020). Additionally, Zhang et al. (2021) reported that miR-302c expression was significantly decreased in colon cancer cells compared to normal cells. In vitro and in vivo analysis displayed that the artificial overexpression of miR-302c could reduce the ability of colon CaSCs to stem, proliferate, invade, and migrate while triggering apoptosis through interacting directly with CARF and thereby preventing Wnt/-catenin signaling.
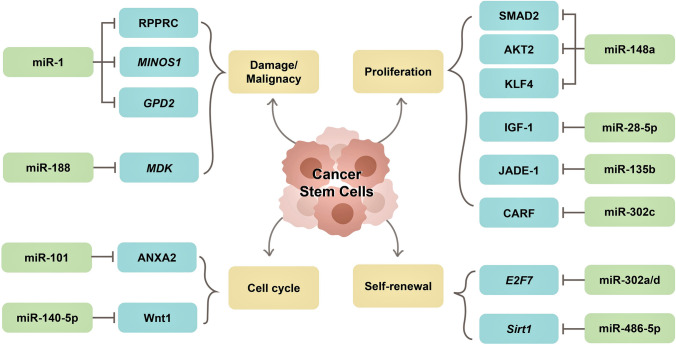
Altogether, these findings exhibit that CaSCs is the primary cause of cancer progression, metastasis, and drug resistance. Even though various mechanisms for maintaining stemness features and the functioning of CaSCs attributed to their oncogenic potential has been proposed, further studies are required. Nevertheless, miRNAs are essential molecules in promoting oncogenic potential and therapeutic hindrances caused by CaSCs (Fig. 4), making them an attractive target in cancer therapy.
Fig. 4.
MiRNAs in the regulation of cancer stem cells. MiRNAs have been associated with the different characteristics that involve cancer stem cells such as functions of self-renewal, promotion of proliferation, migration, and invasion, cell cycle acceleration, mitochondrial damage, and other malignancies
Exosomal miRNAs in stem cells
Exosomes are a subclass of extracellular vesicles that have been identified to contain miRNAs linked to proliferation, migration, and differentiation. Different types of stem cells, including MSCs, NSCs, and HSCs, produce exosomes (Muralikumar et al. 2021). Exosomes generated from MSCs contain miRNAs such as miR-125a-3p, which can reduce effector T-cell differentiation and proliferation, regulating immunological response (Asgarpour et al. 2020). According to Soni et al. (2021), many miRNAs associated with neurogenesis, including miR-145, were found in human MSC-derived exosomes, resulting in increased levels of Nestin, glial fibrillary acidic protein, microtubule-associated protein-2, and oligodendrocyte transcription factor 2 mRNA expression. Exosomes from human mesenchymal stem cells in Wharton’s jelly and bone marrow have been shown to induce angiogenesis with the help of miR-132, miR-145, and miR-125 (Soni et al. 2021). MiR-27a-5p containing exosomes play a crucial role in inducing odontogenic differentiation of dental pulp stem cells derived from humans by regulating the TGFβ1/Smad signaling cascade (Jin et al. 2020). Human urine-derived stem cells were reported to attenuate the progression of type I diabetic kidney issues in rats by the action of exosomes carrying miRNA -16-5p (Zhang et al. 2020c).
An enhanced expression of the miR-215-5p by adipose-derived stem cells exosomes was observed that benefit diabetic nephropathy prevention by promoting high-glucose-induced podocyte migration (Zhang et al. 2017). Other sources of stem cells, like hypothalamic stem cells, greatly influence age-related hypothalamic exosomal miRNAs in the cerebrospinal fluid (Zhang et al. 2017). Recent research suggests that instead of using stem cells, stem-derived compartments or the extracellular enriched with growth factors and miRNAs could be used as practical tools for managing and monitoring various diseases (Muralikumar et al. 2021).
Interaction of miRNAs with long non-coding RNAs (lncRNAs) in stem cells
LncRNAs comprise another family of non-coding RNAs with lengths greater than 200 nucleotides and are mostly found in the nucleus. Unlike miRNAs, lncRNAs are incapable of being translated into proteins (Chen et al. 2017; Zhou et al. 2022). They exhibit poor evolutionary conservation and have lower levels of expression in cells than protein-coding genes (Aich and Chakraborty 2020). Notably, both lncRNAs and miRNAs regulate gene expression by building a vast and complex regulatory network of interdependent connections. While the main mechanism by which miRNAs act is through interactions with mRNAs repressing their expression, lncRNAs have a wide range of effects on mRNA functioning, including interactions with miRNAs in a variety of biological processes involving stem cells (Lanzillotti et al. 2021) that have been summarized in Table 2.
Table 2.
The interaction of miRNA with lncRNAs in stem cell regulation
| Stem cell type | lncRNA | Regulation/expression | Related miRNA | Target | Biologic effect/function | References |
|---|---|---|---|---|---|---|
| Cancer stem cells | MBNL1-AS1 | Downregulated | miR-301b-3p | TGFBR2 | Development and progression of NSCLC, proliferation, invasion and drug resistance | Li et al. (2019b) |
| miR-412-3p | MYL9 | Cell proliferation, invasion, and migration | Zhu et al. (2020) | |||
| LINC00887 | Upregulated | miR-613, miR-206, and miR-1 | FN1, MET, and SMAD4 | Cell invasion, progression and development of stem cell traits | Tian et al. (2019) | |
| DPP10-AS1 | Downregulated | miR-127-3p | ADCY1 | Inhibition of colon CaSC proliferation, migration, and invasion | Liu et al. (2021) | |
| PART1 | Upregulated | miR-190-3p, miR-937-5p, miR-22-5p, miR-30b-3p and miR-6870-5p | BICC1, MYO5A and ZHX2 | Promotion of stemness | Cruickshank et al. (2021) | |
| HOTAIR | Upregulated | miR-206 | TBX3 | Promotion of stemness | Zhang et al. (2020a) | |
| BCAR4 | Upregulated | miR-665 | STAT3 | Promotion of self-renewal, ALDH activity, and stemness | Ouyang et al. (2019) | |
| Mesenchymal stem cells | ENST00000563492 | Upregulated | miR-205-5p | CDH11 and VEGF | Osteogenic differentiation, induction of angiogenesis, and bone regeneration | Ouyang et al. (2020) |
| TINCR | Upregulated | miR-761 | Wnt2 | Cell migration | Zheng et al. (2020) | |
| SNHG1 | Upregulated | miR-15a | SMURF1 | Suppression of liver fibrosis to prevent cirrhosis | Sun et al. (2022) | |
| NEAT1 | Downregulated | miR-29b-3p | BMP1 | Osteogenic differentiation | Zheng et al. (2020) | |
| Neural stem cells | H19 | Upregulated | miR-675-5p and miR-675-3p | TGF-β1 | Post-stroke NSCs’ neurogenesis | Fan et al. (2020) |
| MEG3 | Downregulated | miR-493–5p | MIF | NSCs’ proliferation | Zhao et al. (2021) | |
| NEAT1 | Upregulated | miR-124 | Wnt/β-catenin genes | Neuronal differentiation, survival, and migration of SP-NPCs Therapeutic recovery from spinal cord injury | Cui et al. (2019) | |
| Cardiac stem cells | CAREL | Downregulated | miR-296 | Trp53inp1 and Itm2a | CSCs replication and heart regeneration | Cai et al. (2018) |
| CRNDE | Upregulated | miR-181a | LYRM1 | Proliferation and migration | Li et al. (2020) |
LncRNAs have been demonstrated to be important regulators of cancer biology and to significantly affect cell division, metastasis, and apoptosis (Zhu et al. 2020). Different lncRNAs play a role in cancer stemness, and their dysregulation may serve as a biomarker for cancer diagnosis, prognosis, and target treatment (Chen et al. 2017; Zhou et al. 2022). LncRNA MBNL1 antisense RNA 1 (MBNL1-AS1) has been downregulated in non-small cell lung cancer (NSCLC) tissues and cells. LncRNA MBNL1-AS1 restoration may slow down the development and progression of NSCLC, demonstrating its capacity to act as a miR-301b-3p sponge to inhibit CaSC proliferation, invasion, drug resistance, and sphere formation in NSCLC by upregulating transforming growth factor beta receptor II (TGFBR2) (Li et al. 2019b). In this matter, Zhu et al. (2020) investigated the function of MBNL1-AS1 in colon cancer development, demonstrating as well as downregulated expression. When MBNL1-AS1 expression is restored, myosin regulatory light chain 9 (MYL9) may be activated by miR-412-3p suppression to protect CaSC cells against proliferation, invasion, and migration in colon cancer. The MBNL1-AS1-miR-412-3p-MYL9 network enables a novel approach to treating lung and colon cancer patients.
Tian et al. (2019) also demonstrated the functional specificity and therapeutic utility of lncRNAs in NSCLC patients. According to the experimental finding, high levels of long intergenic non-protein-coding RNA 00887 (LINC00887) substantially increased lung cancer cells' ability to invade. Numerous miRNAs can interact with LINC00887, among which miR-613, miR-206, and miR-1–2 are the most relevant. The miR-613, miR-206, and miR-1 grew when LINC00887 was silenced, demonstrating an inversely proportional link. It has been shown that LINC00887–miRNA interaction modulates downstream genes such as FN1, MET, and SMAD4, which are linked to lung cancer progression and the development of stem cell traits.
More recently, Liu et al. (2021) showed that DPP10-AS1, a little-known lncRNA, is poorly expressed in colon cancer. Therefore, the induction of DPP10-AS1 inhibited the development of colon CaSCs into tumors and their proliferation via negatively regulating miR-127. Binding sites between miR-127-3p and the adenylate cyclase (ADCY1) gene were also found, suggesting that DPP10-AS1 participates in the inhibition of colon CaSC proliferation, migration, and invasion as well as the stimulation of apoptosis through the regulation of ADCY1 through miR-127-3p. These findings point to DPP10-AS1 as a promising new molecular target for colon cancer therapy.
Regarding triple-negative breast cancer (TNBC), Cruickshank et al. (2021) found that lncRNA PART1 is overexpressed in TNBC cells, including CaSCs’ populations. This lncRNA is associated with stemness, contributes to mammosphere formation, and exerts oncogenic effects. PART1 has multiple effects on gene expression, including down-regulation of BICC1, MYO5A and ZHX2, cancer-promoting genes. Since PART1 is located in the cytoplasm of TNBC cells, it can act as a miRNA sponge. For instance, it regulates miR-190-3p, miR-937-5p, miR-22-5p, miR-30b-3p, and miR-6870-5p in TNBC. PART1 can increase and decrease miRNAs; nevertheless, only some of these interactions are explained by PART1-mediated binding, suggesting that this lncRNA can alter the miRNA landscape by indirect mechanisms. However, the results on the miRNAs and mRNAs that PART1 regulates can differ between cell lines and depend on the cellular context.
According to Zhang et al. (2020a), lncRNA HOTAIR is upregulated in ovarian cancer stem cells (OCSCs), promoting the stemness of these cells. HOTAIR targets TBX3, a transcription factor that modulates PTEN and NANOG expression, and positively regulates its expression to maintain OCSCs’ stemness. The sponge-like activity of HOTAIR promotes TBXc3 expression with miR-206, which naturally inhibits TBX3 and NANOG expression and induces PTEN expression. It was also demonstrated that the circulating HOTAIR expression level was higher among ovarian cancer patients compared to benign ovarian epithelial tumor samples. These findings place lncRNA HOTAIR as a potential diagnostic biomarker in ovarian cancer and provide the HOTAIR/miR-206/TBX3 axis as a molecular mechanism behind OCSCs’ regulation and maintenance.
In the study by Ouyang et al. (2019), lncRNA BCAR4 was remarkably upregulated in colorectal CaSCs, promoting self-renewal, ALDH activity, and colorectal CaSCs stemness. BCAR4 interacts with miR-665, a crucial regulator in cancers, which simultaneously targets STAT3, an essential transcription factor in the maintenance of SCs stemness and closely related to colon cancer. In colorectal CaSCs, BCAR4 inhibits the miR-665 expression and promotes STAT3 expression; this signaling pathway is crucial in colon cancer cell migration, self-renewal, and survival. Experimentation in vivo indicated that inhibition of BCAR4 effectively limited tumor growth and decreased STAT3 expression and phosphorylation. Hence, the BCAR4/miR-665/STAT3 ceRNA network could serve as a diagnostic marker of colorectal cancer and a novel therapeutic target for this cancer treatment.
Besides CaSCs, lncRNAs can regulate other types of stem cells (Cui et al. 2019; Ouyang et al. 2020; Fan et al. 2020; Zhao et al. 2021). For example, Ouyang et al. (2019) conducted a study to elucidate the mechanisms underlying bone nonunion, a frequent orthopedic condition that occurs in large bone defects and after fractures. They found that lncRNA ENST00000563492 is highly expressed in normal fractured tissue compared with nonunion tissue; this elevated expression was mainly observed during the early stage of osteogenic differentiation of BMSCs. Likewise, lncRNA ENST00000563492 directly regulates osteogenic differentiation of BMSCs by acting as a competing endogenous RNA (ceRNA) for miR-205-5p to modulate CDH11 and VEGF, and indirectly modulates angiogenesis of HUVECs. In vivo experiments showed that lncRNA ENST00000563492 could promote bone regeneration. Thus, low expression of lncRNA ENST00000563492 represents a significant cause of bone nonunion and appears as a novel therapeutic strategy for the treatment of nonunion.
Other pathologies in which the interactions of lncRNAs with the mechanisms of BMSCs have been involved include liver cirrhosis and fibrosis (He et al. 2020). A recent study by Sun et al. (2022) revealed that lncRNA small nucleolar RNA host gene 1 (SNHG1) affected the HLC differentiation of BMSCs, implicating it in the development of cirrhosis. Thus, it was shown that silencing SNHG1 lncRNA could enhance miR-15a expression while lowering SMURF1, reducing UVRAG ubiquitination, and promoting the ATG5/Wnt5a axis, accelerating HLC development of BMSCs, and suppressing liver fibrosis to prevent cirrhosis.
In the study of Zheng et al. (2020), it is reported that the upregulation of lncRNA-TINCR can increase migration in rat MSCs (rMSCs). TINCR can serve as a ceRNA and sponge miR-761, an miRNA that can inhibit rMSCs’ migration ability and wound healing when upregulated. Wnt2 is a migration-related gene directly targeted by miR-761; thus, TINCR modulates rMSCs’ migration through Wnt2 and its Wnt2 signaling pathway. Zhang et al. (2019b) found that with osteoporosis, lncRNA NEAT1 is overexpressed in human BMSCs (hBMSCs). NEAT1 is mainly located in hBMSCs cytoplasm, indicating that NEAT1 regulates hBMSCs at a post-transcriptional level. This lncRNA interacts with miR-29b-3p, which targets BMP1, a bone morphogenic protein that was found to promote osteogenic differentiation of hBMSCs. The expression of NEAT1 positively regulates BMP1 and negatively regulates miR-29b-3b. This ceRNA pathway (NEAT1/miR-29b-3p/BMP1) regulates osteogenic differentiation; hence, this may contribute to a better understanding of the pathogenesis of osteoporosis and the possible therapeutic targets for its treatment.
In post-stroke-induced neurogenesis, Fan et al. (2020) evaluated the role of H19 lncRNA along with the lncRNA–mRNA coexpression network in a rat ischemic model. The knockdown of H19 in ischemic NSCs by the CRISPR-CAS9 genome editing technique confirmed a correlation between H19 depletion, low proliferation, and high apoptosis rates. A positive correlation between the upregulation of H19 and high expression levels of miR-675-5p and -3p after stroke, along with inverted expression levels of miR-675 and TGF-1, further revealed the miR675/TGF-1 as the regulatory axis through which H19 modulates neurogenesis (Fan et al. 2020).
Under ischemic stroke (IS) conditions, Zhao et al. (2021) explored the lncRNA MEG3 and miR-493–5p regulatory synergic function on NSCs proliferation. Knockdown of lncRNA MEG3 via transfection of si-MEG3 resulted in the blocking of miR-493-5p and the recovery of normal proliferation levels of NSCs, accompanied by restored upregulation of MIF. Transfection of IS NSCs with miR-493–5p inhibitor resulted in NSCs proliferation repair, confirming MEG3/miR-493-5p/ MIF as a molecular regulatory axis on NSCs’ proliferation.
Cui et al. (2019) evaluated the therapeutic function of miR-124 in spinal cord injury and lncRNA NEAT1´’s role in neuronal differentiation in spinal cord neural progenitor cells (SP-NPCs). While NEAT1 in SC-NPCs alone was found to regulate neuronal differentiation, survival, and migration, the NEAT1 and miR-124 regulatory pairing role was elucidated by a correlated upregulation of miR-124 and highly expressed levels of NEAT1, along with therapeutic recovery from spinal cord injury. Furthermore, quantitative real-time PCR assay evidenced relative expression levels of lncRNA NEAT 1 and Wnt/β-catenin related genes suggesting NEAT1 mediation of Wnt/β-catenin, signaling pathway activated by miR-124. All this evidence suggests miR-124-Neat1-Wnt/β-catenin as a regulatory signaling axis of SP-NPCs.
Cai et al. (2018) elucidated the regulatory consequence of CAREL lncRNA on CSCs’ replication and heart regeneration after myocardial infarction by acting as a miR-296 competitor. Increased expression levels of CAREL were detected in cardiomyocytes lacking regenerative capacity after cardiac injury. Furthermore, over-regulation of CAREL resulted in diminished cardiac division and proliferative potential in parallel with dull cardiac regeneration. The knockdown of CAREL, on the contrary, resulted in improved cardiac function and regeneration. LncRNA CAREL was found to compete with miR-296 as an endogenous RNA to inhibit the expression of Trp53inp1 and Itm2a target genes. Overexpression of miR-296 resulted in rescued cardiac regenerative capacity and increased cardiomyocyte replication. Altogether, these results confirmed that lncRNA CAREL regulates cardiomyocyte proliferation and heart regeneration via competing with miR-296, targeting Trp53inp1 and Itm2a genes.
Under hypoxic conditions, the modulatory function of lncRNA CRNDE in cardiac progenitor cells’ proliferation and migration via the miR-181a/LYRM1 axis was evaluated by Li et al. (2020). The knockdown of CRNDE under hypoxia resulted in suppressed cardiac reproduction and migration. However, the miR-181a inhibitor’s protective effect was elucidated by restored reproduction and migration after CRNDE knockdown via miR-181a’s target gene LYRM1. LYRM1 knockdown furthermore resulted in blocked miR-181a inhibitor’s rescuing effects of regular cardiac progenitor cell functions under hypoxia (Li et al. 2020).
LncRNAs and other non-coding RNAs serve important functions in various stem cell types. Here, we explore the several regulatory mechanisms through which miRNA and lncRNAs control stem cells' biological status and functional activity. Thus, interactive networks of lncRNA, miRNA, and mRNA may offer more details on potential new targets and mechanisms for disease management.
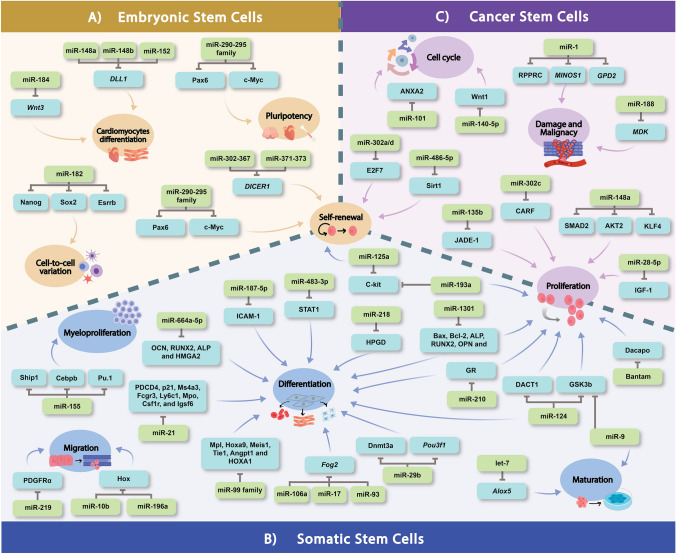
Concluding remarks and future perspectives
MiRNAs are post-transcriptional regulatory molecules that are functionally implicated in controlling several biologic processes in stem cell populations by acting either individually or synergically. Additionally, lncRNAs are another type of epigenetic non-coding RNA molecule that participate along with miRNAs as key regulators of the functions of stem cells, particularly MSCs, NSCs, CSCs, and CaSCs via specific molecular regulatory axes and targeting genes. Thorough knowledge about the roles of miRNAs and lncRNAs in stem cell fate and differentiation might open a new therapeutic arena to fight against chronic human diseases, including cancer. This review presents the recent research regarding the crucial modulatory mechanisms of miRNAs and lncRNAs in stem cell populations via distinct signaling pathways and the target genes they interact with (Fig. 5), some of them being shared by different ncRNAs, especially in the biological processes of self-renewal (Fig. 5A–C), differentiation (Fig. 5B), and proliferation (Fig. 5B, C), while other miRNAs are found interacting with specific genes for the regulation of other processes including the maintenance of pluripotency traits, regulation of survival, cell-to-cell variation, progeny differentiation, cell cycle, maturation, as well as regulatory functions in cancer cell stemness of damage and malignancy such as metastasis, apoptosis, tumor progression, and cellular drug susceptibility. Due to their capacity to target hundreds of mRNAs, miRNAs can rapidly shift cell fate and fine-tune genome expression, usually accompanied by the participation of lncRNAs. Although several miRNA-based treatments for various human ailments are currently under trial, one of the most difficult challenges remains the delivery of therapeutic miRNAs to target tissues without losing stability. Hence, miRNA therapy requires the development of authentic organ-specific delivery strategies along with the reduction of toxicity and off-target adverse effects. Nonetheless, we believe that the information presented in this review will strengthen the research arena of the biological implications of miRNAs and lncRNAs in stem cell regulation, which might help develop novel disease management strategies.
Fig. 5.
A MiRNAs and the target gene interactions are reported to regulate embryonic stem cells’ biological processes. Cardiomyocyte differentiation is regulated by miR-184 a, b and miR-152 via targeting DLL1, and miR-184 through Wnt3 gene target. Pluripotency is reported to be regulated by the family miR-290–295 through gene targets Pax6 and c-Myc. Cell-to-cell variation was reported to be mediated by the interaction between Nanog, Sox2, and Esrrb genes targeted by miR-182. Self-renewal is reported to be mediated by miR-302–367 and miR-371–373 targeting the gene DICER 1, as well as by miR-290–295 family via targeting Pax 6 and c-Myc genes. B MiRNAs and the target gene interactions are reported to regulate somatic stem cells’ biologic processes. Myeloproliferation, differentiation, migration, maturation, proliferation, and self-renewal are the biologic processes regulated by different miRNAs and target gene networks in somatic stem cells, in which miR-125a and miR-93 share c-kit as a target gene, as well as miR-9 and mir-124 share the target gene GSK3b. MiR-210, miR-124 and miR-1301 target genes are involved in the regulation of more than one biologic process of somatic stem cells. C MiRNAs and the target gene interactions are reported to regulate cancer stem cells’ biologic processes. The cell cycle is a biologic process in cancer stem cells reported to be regulated by miR-101 and miR-140-5p via targeting ANXA2 and Wnt1 genes, respectively. Damage and malignancy are regulated by miR-188 via the MDK target gene and, by miR-1 through RPPRC, MINOS1, and GPD2 target genes. Proliferation is regulated by miR-135b, miR-302c, and miR-28-5p by targeting a single gene, while miR-148a regulates proliferation via targeting 3 different genes. Self-renewal is regulated by miR-302a/d and miR-486-5p through E2F7 and SiRT1 target genes, respectively
Funding
This research receives no external funding.
Data accessibility
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Aich M, Chakraborty D. Role of lncRNAs in stem cell maintenance and differentiation. Elsiver; 2020. pp. 73–112. [DOI] [PubMed] [Google Scholar]
- Amini H, Rezaie J, Vosoughi A, et al. Cardiac progenitor cells application in cardiovascular disease. J Cardiovasc Thorac Res. 2017;9:127–132. doi: 10.15171/jcvtr.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annese T, Tamma R, de Giorgis M, Ribatti D. microRNAs biogenesis, functions and role in tumor angiogenesis. Front Oncol. 2020;10:581007. doi: 10.3389/fonc.2020.581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgarpour K, Shojaei Z, Amiri F, et al. Exosomal microRNAs derived from mesenchymal stem cells: cell-to-cell messages. Cell Commun Signal. 2020;18:149. doi: 10.1186/s12964-020-00650-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayob AZ, Ramasamy TS. Cancer stem cells as key drivers of tumour progression. J Biomed Sci. 2018;25:1–18. doi: 10.1186/S12929-018-0426-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacakova L, Zarubova J, Travnickova M, et al. Stem cells: their source, potency and use in regenerative therapies with focus on adipose-derived stem cells—a review. Biotechnol Adv. 2018;36:1111–1126. doi: 10.1016/j.biotechadv.2018.03.011. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Roy JK. Dicer-1 regulates proliferative potential of Drosophila larval neural stem cells through bantam miRNA based down-regulation of the G1/S inhibitor Dacapo. Dev Biol. 2017;423:57–65. doi: 10.1016/j.ydbio.2017.01.011. [DOI] [PubMed] [Google Scholar]
- Bravo Vázquez LA, Moreno Becerril MY, Mora Hernández EO, et al. The emerging role of microRNAs in bone diseases and their therapeutic potential. Molecules. 2021;27:211. doi: 10.3390/molecules27010211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G, Tsapogas P, Ceredig R. The changing face of hematopoiesis: a spectrum of options is available to stem cells. Immunol Cell Biol. 2018;96:898–911. doi: 10.1111/IMCB.12055. [DOI] [PubMed] [Google Scholar]
- Cai B, Ma W, Ding F, et al. The long noncoding RNA CAREL controls cardiac regeneration. J Am Coll Cardiol. 2018;72:534–550. doi: 10.1016/j.jacc.2018.04.085. [DOI] [PubMed] [Google Scholar]
- Chakraborty M, Hu S, Visness E, et al. MicroRNAs organize intrinsic variation into stem cell states. Proc Natl Acad Sci USA. 2020;117:6942–6950. doi: 10.1073/PNAS.1920695117/-/DCSUPPLEMENTAL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhu J, Wang F, et al. LncRNAs and their role in cancer stem cells. Oncotarget. 2017;8:110685–110692. doi: 10.18632/oncotarget.22161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Xu Z, Shao J, et al. MicroRNA-218 promotes early chondrogenesis of mesenchymal stem cells and inhibits later chondrocyte maturation. BMC Biotechnol. 2019;19:6. doi: 10.1186/s12896-018-0496-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Cui Y, Liu B, et al. Hsa-miR-1908-3p mediates the self-renewal and apoptosis of human spermatogonial stem cells via targeting KLF2. Mol Ther. 2020;20:788–800. doi: 10.1016/j.omtn.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, He L, Pang M, et al. Melatonin promotes neuroprotection of H2O2-induced neural stem cells via lncRNA MEG3/miRNA-27a-3p/MAP2K4 axis. Neuroscience. 2020;446:69–79. doi: 10.1016/j.neuroscience.2020.06.026. [DOI] [PubMed] [Google Scholar]
- Choi SW, Lee JY, Kang K-S. miRNAs in stem cell aging and age-related disease. Mech Ageing Dev. 2017;168:20–29. doi: 10.1016/j.mad.2017.08.013. [DOI] [PubMed] [Google Scholar]
- Cipolla GA, de Oliveira JC, Salviano-Silva A, et al. Long non-coding RNAs in multifactorial diseases: another layer of complexity. Non-Coding RNA. 2018;4:13. doi: 10.3390/NCRNA4020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshank BM, Wasson M-CD, Brown JM, et al. LncRNA part1 promotes proliferation and migration, is associated with cancer stem cells, and alters the miRNA landscape in triple-negative breast cancer. Cancers (basel) 2021;13:2644. doi: 10.3390/cancers13112644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Yin Y, Xiao Z, et al. LncRNA Neat1 mediates miR-124-induced activation of Wnt/β-catenin signaling in spinal cord neural progenitor cells. Stem Cell Res Ther. 2019;10:400. doi: 10.1186/s13287-019-1487-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahariya S, Paddibhatla I, Kumar S, et al. Long non-coding RNA: classification, biogenesis and functions in blood cells. Mol Immunol. 2019;112:82–92. doi: 10.1016/j.molimm.2019.04.011. [DOI] [PubMed] [Google Scholar]
- di Marco B, Crouch EE, Shah B, et al. Reciprocal interaction between vascular filopodia and neural stem cells shapes neurogenesis in the ventral telencephalon. Cell Rep. 2020;33:108256. doi: 10.1016/j.celrep.2020.108256. [DOI] [PubMed] [Google Scholar]
- Divisato G, Passaro F, Russo T, Parisi S. The key role of microRNAs in self-renewal and differentiation of embryonic stem cells. Int J Mol Sci. 2020;21:6285. doi: 10.3390/ijms21176285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves M, Serra-Almeida C, Saraiva C, Bernardino L. New insights into the regulatory roles of microRNAs in adult neurogenesis. Curr Opin Pharmacol. 2020;50:38–45. doi: 10.1016/J.COPH.2019.11.003. [DOI] [PubMed] [Google Scholar]
- Fan B, Pan W, Wang X, et al. Long noncoding RNA mediates stroke-induced neurogenesis. Stem Cells. 2020;38:973–985. doi: 10.1002/stem.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Miao S, Yu Y, et al. MIR148A family regulates cardiomyocyte differentiation of human embryonic stem cells by inhibiting the DLL1-mediated NOTCH signaling pathway. J Mol Cell Cardiol. 2019;134:1–12. doi: 10.1016/j.yjmcc.2019.06.014. [DOI] [PubMed] [Google Scholar]
- Fu H, Zhang W, Yuan Q, et al. PAK1 promotes the proliferation and inhibits apoptosis of human spermatogonial stem cells via PDK1/KDR/ZNF367 and ERK1/2 and AKT pathways. Mol Ther. 2018;12:769–786. doi: 10.1016/j.omtn.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Zhou F, Yuan Q, et al. miRNA-31-5p mediates the proliferation and apoptosis of human spermatogonial stem cells via targeting JAZF1 and cyclin A2. Mol Ther. 2019;14:90–100. doi: 10.1016/j.omtn.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fus-Kujawa A, Mendrek B, Trybus A, et al. Potential of induced pluripotent stem cells for use in gene therapy: history, molecular bases, and medical perspectives. Biomolecules. 2021;11:699. doi: 10.3390/BIOM11050699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Dasgupta C, Huang L, et al. Multi-omics integration reveals short and long-term effects of gestational hypoxia on the heart development. Cells. 2019;8:1608. doi: 10.3390/cells8121608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrits A, Walasek MA, Olthof S, et al. Genetic screen identifies microRNA cluster 99b/let-7e/125a as a regulator of primitive hematopoietic cells. Blood. 2012;119:377–387. doi: 10.1182/blood-2011-01-331686. [DOI] [PubMed] [Google Scholar]
- Ghafouri-Fard S, Niazi V, Taheri M. Role of miRNAs in conveying message of stem cells via extracellular vesicles. Exp Mol Pathol. 2020;117:104569. doi: 10.1016/j.yexmp.2020.104569. [DOI] [PubMed] [Google Scholar]
- Gizak A, Duda P, Pielka E, et al. GSK3 and miRNA in neural tissue: from brain development to neurodegenerative diseases. Biochim Biophys Acta Mol Cell Res. 2020;1867:118696. doi: 10.1016/j.bbamcr.2020.118696. [DOI] [PubMed] [Google Scholar]
- Gorabi AM, Bianconi V, Matteo P, Banach M, Sahebkar A. Regulation of cardiac stem cells by microRNAs: State-of-the-art. Biomed Pharmacother. 2019;120:109447. doi: 10.1016/j.biopha.2019.109447. [DOI] [PubMed] [Google Scholar]
- Gorabi AM, Kiaie N, Barreto GE, et al. The therapeutic potential of mesenchymal stem cell-derived exosomes in treatment of neurodegenerative diseases. Mol Neurobiol. 2019;56:8157–8167. doi: 10.1007/S12035-019-01663-0. [DOI] [PubMed] [Google Scholar]
- He Z, Yang D, Fan X, et al. The roles and mechanisms of lncRNAs in liver fibrosis. Int J Mol Sci. 2020;21:1482. doi: 10.3390/ijms21041482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Lu Y, Zeng H, et al. MicroRNA-21 maintains hematopoietic stem cell homeostasis through sustaining the NF-κB signaling pathway in mice. Haematologica. 2021;106:412–423. doi: 10.3324/haematol.2019.236927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Geng J, Jiang S. MicroRNAs in regulation of osteogenic differentiation of mesenchymal stem cells. Cell Tissue Res. 2017;368:229–238. doi: 10.1007/S00441-016-2462-2/FIGURES/1. [DOI] [PubMed] [Google Scholar]
- Huang T, Song X, Xu D, et al. Stem cell programs in cancer initiation, progression, and therapy resistance. Theranostics. 2020;10:8721–8743. doi: 10.7150/thno.41648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Xu J. Immune modulation by mesenchymal stem cells. Cell Prolif. 2020;53:e12712. doi: 10.1111/CPR.12712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Hawkins JS, Lee J, et al. Let-7 microRNA-dependent control of leukotriene signaling regulates the transition of hematopoietic niche in mice. Nat Commun. 2017;8:128. doi: 10.1038/s41467-017-00137-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Wang Y, Zhao L, et al. Exosomal miRNA-215-5p derived from adipose-derived stem cells attenuates epithelial-mesenchymal transition of podocytes by inhibiting ZEB2. Biomed Res Int. 2020;2020:1–14. doi: 10.1155/2020/2685305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothimani G, Bhatiya M, Pathak S, et al. Tumor suppressor microRNAs in gastrointestinal cancers: a mini-review. Recent Adv Inflamm Allergy Drug Discovery. 2022 doi: 10.2174/2772270816666220606112727. [DOI] [PubMed] [Google Scholar]
- Jothimani G, Pathak S, Dutta S, et al. A comprehensive cancer-associated microRNA expression profiling and proteomic analysis of human umbilical cord mesenchymal stem cell-derived exosomes. Tissue Eng Regen Med. 2022 doi: 10.1007/S13770-022-00450-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahney EW, Snedeker JC, Chen X. Regulation of Drosophila germline stem cells. Curr Opin Cell Biol. 2019;60:27–35. doi: 10.1016/j.ceb.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspi H, Chapnik E, Levy M, et al. Brief report: miR-290-295 regulate embryonic stem cell differentiation propensities by repressing Pax6. Stem Cells. 2013;31:2266–2272. doi: 10.1002/stem.1465. [DOI] [PubMed] [Google Scholar]
- Khalaj M, Woolthuis CM, Hu W, et al. miR-99 regulates normal and malignant hematopoietic stem cell self-renewal. J Exp Med. 2017;214:2453–2470. doi: 10.1084/jem.20161595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AQ, Ahmed EI, Elareer NR, et al. Role of miRNA-regulated cancer stem cells in the pathogenesis of human malignancies. Cells. 2019;8:840. doi: 10.3390/cells8080840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanehzad M, Abolhasani F, Hassanzadeh G, et al. Determination of the excitatory effects of microRNA-30 in the self-renewal and differentiation process of neonatal mouse spermatogonial stem cells. Galen Med J. 2020;9:e1829. doi: 10.31661/gmj.v9i0.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanehzad M, Nourashrafeddin SM, Abolhassani F, et al. MicroRNA-30a-5p promotes differentiation in neonatal mouse spermatogonial stem cells (SSCs) Reprod Biol Endocrinol. 2021;19:85. doi: 10.1186/s12958-021-00758-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J, Wan L-P, Liu Z-M, Gao S-T. MiR-1301 promotes adipogenic and osteogenic differentiation of BMSCs by targeting Satb2. Eur Rev Med Pharmacol Sci. 2020;24:3501–3508. doi: 10.26355/eurrev_202004_20809. [DOI] [PubMed] [Google Scholar]
- Kotaki R, Koyama-Nasu R, Yamakawa N, Kotani A. miRNAs in normal and malignant hematopoiesis. Int J Mol Sci. 2017;18:1495. doi: 10.3390/ijms18071495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivdova G, Voisin V, Schoof EM, et al. Identification of the global miR-130a targetome reveals a role for TBL1XR1 in hematopoietic stem cell self-renewal and t(8;21) AML. Cell Rep. 2022;38:110481. doi: 10.1016/j.celrep.2022.110481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanoue V, Cooper HM. Branching mechanisms shaping dendrite architecture. Dev Biol. 2019;451:16–24. doi: 10.1016/j.ydbio.2018.12.005. [DOI] [PubMed] [Google Scholar]
- Lanzillotti C, de Mattei M, Mazziotta C, et al. Long non-coding RNAs and MicroRNAs interplay in osteogenic differentiation of mesenchymal stem cells. Front Cell Dev Biol. 2021;9:742. doi: 10.3389/FCELL.2021.646032/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao TD, Le TAH. MicroRNAs: biogenesis, functions and potential biomarkers for early screening, prognosis and therapeutic molecular monitoring of nasopharyngeal carcinoma. Processes. 2020;8:966. doi: 10.3390/pr8080966. [DOI] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- Lee J, Chae H, June Y, et al. Graphene oxide nanoplatelets composite membrane with hydrophilic and antifouling properties for wastewater treatment. J Membr Sci. 2013;448:223–230. doi: 10.1016/j.memsci.2013.08.017. [DOI] [Google Scholar]
- Li N, Long B, Han W, et al. microRNAs: important regulators of stem cells. Stem Cell Res Ther. 2017;8:110. doi: 10.1186/s13287-017-0551-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Meng X, Zhang L. microRNAs and cardiac stem cells in heart development and disease. Drug Discov Today. 2019;24:233–240. doi: 10.1016/j.drudis.2018.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Xing W, Xu J, et al. microRNA-301b-3p downregulation underlies a novel inhibitory role of long non-coding RNA MBNL1-AS1 in non-small cell lung cancer. Stem Cell Res Ther. 2019;10:144. doi: 10.1186/s13287-019-1235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhang Y, Tang Y, et al. LncRNA CRNDE modulates cardiac progenitor cells’ proliferation and migration via the miR-181a/LYRM1 axis in hypoxia. J Thoracic Dis. 2020;12:2614–2624. doi: 10.21037/jtd.2020.03.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wang S-S, Shan B-Q, et al. miR-103-3p targets Ndel1 to regulate neural stem cell proliferation and differentiation. Neural Regen Res. 2022;17:401. doi: 10.4103/1673-5374.317987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yang Y, Wang X, et al. MiR-184 directly targets Wnt3 in cardiac mesoderm differentiation of embryonic stem cells. Stem Cells. 2020;38:1568–1577. doi: 10.1002/stem.3282. [DOI] [PubMed] [Google Scholar]
- Liu G, Zhao H, Song Q, et al. Long non-coding RNA DPP10-AS1 exerts anti-tumor effects on colon cancer via the upregulation of ADCY1 by regulating microRNA-127–3p. Aging. 2021;13:9748–9765. doi: 10.18632/aging.202729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Lázaro M. The stem cell division theory of cancer. Crit Rev Oncol Hematol. 2018;123:95–113. doi: 10.1016/j.critrevonc.2018.01.010. [DOI] [PubMed] [Google Scholar]
- Luu BE, Storey KB. Dehydration triggers differential microRNA expression in Xenopus laevis brain. Gene. 2015;573:64–69. doi: 10.1016/j.gene.2015.07.027. [DOI] [PubMed] [Google Scholar]
- Lv D, Chen L, Du L, et al. Emerging regulatory mechanisms involved in liver cancer stem cell properties in hepatocellular carcinoma. Front Cell Dev Biol. 2021;9:1993. doi: 10.3389/FCELL.2021.691410/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv W, Yu M, Su Y. miR-22-5p regulates the self-renewal of spermatogonial stem cells by targeting EZH2. Open Medicine. 2022;17:556–565. doi: 10.1515/med-2022-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y-S, Lv Z-W, Yu F, et al. MicroRNA-302a/d inhibits the self-renewal capability and cell cycle entry of liver cancer stem cells by targeting the E2F7/AKT axis. J Exp Clin Cancer Res. 2018;37:252. doi: 10.1186/s13046-018-0927-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Cheng J, Wang H, et al. A novel regulatory loop miR-101/ANXA2/EGR1 mediates malignant characteristics of liver cancer stem cells. Carcinogenesis. 2021;42:93–104. doi: 10.1093/carcin/bgaa055. [DOI] [PubMed] [Google Scholar]
- Mackay BS, Marshall K, Grant-Jacob JA, et al. Heterogeneity and ‘memory’ in stem cell populations. Phys Biol. 2020;17:065013. doi: 10.1088/1478-3975/ABBA85. [DOI] [PubMed] [Google Scholar]
- Mahabadi JA, Sabzalipoor H, Nikzad H, et al. The role of microRNAs in embryonic stem cell and induced pluripotent stem cell differentiation in male germ cells. J Cell Physiol. 2019;234:12278–12289. doi: 10.1002/jcp.27990. [DOI] [PubMed] [Google Scholar]
- Maleki B, Alani B, Tamehri Zadeh SS, et al. MicroRNAs and exosomes: cardiac stem cells in heart diseases. Pathology. 2022;229:153701. doi: 10.1016/j.prp.2021.153701. [DOI] [PubMed] [Google Scholar]
- Martinez SR, Ma Q, Dasgupta C, et al. MicroRNA-210 suppresses glucocorticoid receptor expression in response to hypoxia in fetal rat cardiomyocytes. Oncotarget. 2017;8:80249. doi: 10.18632/ONCOTARGET.17801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Zhang P, Zhang L. Fetal hypoxia impacts on proliferation and differentiation of Sca-1+ cardiac progenitor cells and maturation of cardiomyocytes: a role of microRNA-210. Genes (basel) 2020;11:328. doi: 10.3390/genes11030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mens MMJ, Ghanbari M. Cell cycle regulation of stem cells by microRNAs. Stem Cell Rev Rep. 2018;14:309–322. doi: 10.1007/s12015-018-9808-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed RH, Abu-Shahba N, Marwa M, Abdelfattah AMH, et al. Co-regulatory network of oncosuppressor miRNAs and transcription factors for pathology of human hepatic cancer stem cells (HCSC) Sci Rep. 2019;9:5564. doi: 10.1038/s41598-019-41978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Fäh T, Schaefer M, et al. AGO1 regulates pericentromeric regions in mouse embryonic stem cells. Life Sci Alliance. 2022;5:e202101277. doi: 10.26508/lsa.202101277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralikumar M, Manoj Jain S, Ganesan H, et al. Current understanding of the mesenchymal stem cell-derived exosomes in cancer and aging. Biotechnol Rep. 2021;31:e00658. doi: 10.1016/j.btre.2021.e00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naji A, Eitoku M, Favier B, et al. Biological functions of mesenchymal stem cells and clinical implications. Cell Mol Life Sci. 2019;76:3323–3348. doi: 10.1007/S00018-019-03125-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Zhou X, Chen Z, et al. LncRNA BCAR4, targeting to miR-665/STAT3 signaling, maintains cancer stem cells stemness and promotes tumorigenicity in colorectal cancer. Cancer Cell Int. 2019;19:72. doi: 10.1186/s12935-019-0784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Tan T, Zhang X, et al. LncRNA ENST00000563492 promoting the osteogenesis–angiogenesis coupling process in bone mesenchymal stem cells (BMSCs) by functions as a ceRNA for miR-205-5p. Cell Death Dis. 2020;11:486. doi: 10.1038/s41419-020-2689-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Reyes PR, Garza BS, Sharma A. MicroRNAs and child neuropsychiatric disorders: a brief review. Neurochem Res. 2020;45:232–240. doi: 10.1007/s11064-019-02917-y. [DOI] [PubMed] [Google Scholar]
- Paul S, Ruiz-Manriquez LM, Ledesma-Pacheco SJ, et al. Roles of microRNAs in chronic pediatric diseases and their use as potential biomarkers: a review. Arch Biochem Biophys. 2021;699:108763. doi: 10.1016/j.abb.2021.108763. [DOI] [PubMed] [Google Scholar]
- Peng S, Gao D, Gao C, et al. MicroRNAs regulate signaling pathways in osteogenic differentiation of mesenchymal stem cells (review) Mol Med Rep. 2016;14:623–629. doi: 10.3892/mmr.2016.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons-Espinal M, de Luca E, Marzi MJ, et al. Synergic functions of miRNAs determine neuronal fate of adult neural stem cells. Stem Cell Reports. 2017;8:1046–1061. doi: 10.1016/j.stemcr.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen ML, Ortolano NA, Romero-Morales AI, Gama V. Wnt signaling and its impact on mitochondrial and cell cycle dynamics in pluripotent stem cells. Genes. 2018;9:109. doi: 10.3390/GENES9020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Manriquez LM, Estrada-Meza C, Benavides-Aguilar JA, et al. Phytochemicals mediated modulation of microRNAs and long non-coding RNAs in cancer prevention and therapy. Phytother Res. 2022;36:705–729. doi: 10.1002/ptr.7338. [DOI] [PubMed] [Google Scholar]
- Ruiz-Manriquez LM, Ledesma Pacheco SJ, Medina-Gomez D, et al. A brief review on the regulatory roles of microRNAs in cystic diseases and their use as potential biomarkers. Genes (basel) 2022;13:191. doi: 10.3390/genes13020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sada TS. The potential of stem cells in regenerative medicine. Dis Thera Res. 2022;10:1. doi: 10.11648/J.CB.20221001.11. [DOI] [Google Scholar]
- Samatha Jain M, Makalakshmi MK, Deka D, et al. Therapeutic strategies targeting Wnt/-catenin signaling pathway in stem cells for ROS-induced cancer progression. Handbook Oxidat Stress Cancer. 2022 doi: 10.1007/978-981-16-1247-3_104-1. [DOI] [Google Scholar]
- Shah V, Soibam B, Ritter RA, et al. Data on microRNAs and microRNA-targeted mRNAs in Xenopus ectoderm. Data Brief. 2016;9:699–703. doi: 10.1016/j.dib.2016.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni N, Gupta S, Rawat S, et al. MicroRNA-enriched exosomes from different sources of mesenchymal stem cells can differentially modulate functions of immune cells and neurogenesis. Biomedicines. 2021;10:69. doi: 10.3390/biomedicines10010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappert L, Klaus F, Brüstle O. MicroRNAs engage in complex circuits regulating adult neurogenesis. Front Neurosci. 2018;12:707. doi: 10.3389/FNINS.2018.00707/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui BD, Zheng CX, Li M, et al. Epigenetic regulation of mesenchymal stem cell homeostasis. Trends Cell Biol. 2020;30:97–116. doi: 10.1016/j.tcb.2019.11.006. [DOI] [PubMed] [Google Scholar]
- Sun Y, Xu R, Huang J, et al. Insulin-like growth factor-1-mediated regulation of miR-193a expression promotes the migration and proliferation of c-kit-positive mouse cardiac stem cells. Stem Cell Res Ther. 2018;9:41. doi: 10.1186/s13287-017-0762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Wang X, Chen G, et al. miRNA-187-5p regulates osteoblastic differentiation of bone marrow mesenchymal stem cells in mice by targeting ICAM1. Biomed Res Int. 2020;2020:6139469. doi: 10.1155/2020/6139469. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sun Q, Fu Y, Zhu X, et al. Continuous NF-κB pathway inhibition promotes expansion of human phenotypical hematopoietic stem/progenitor cells through metabolism regulation. Exp Cell Res. 2021 doi: 10.1016/j.yexcr.2020.112468. [DOI] [PubMed] [Google Scholar]
- Sun J, Sun X, Hu S, et al. Long noncoding RNA SNHG1 silencing accelerates hepatocyte-like cell differentiation of bone marrow-derived mesenchymal stem cells to alleviate cirrhosis via the microRNA-15a/SMURF1/UVRAG axis. Cell Death Discovery. 2022;8:77. doi: 10.1038/s41420-022-00850-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CMJ, Lewandowski AJ. The transitional heart: from early embryonic and fetal development to neonatal life. Fetal Diagn Ther. 2020;47:373–386. doi: 10.1159/000501906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijeiro V, Yang D, Majumdar S, et al. DICER1 is essential for self-renewal of human embryonic stem cells. Stem Cell Reports. 2018;11:616–625. doi: 10.1016/j.stemcr.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y. MicroRNAs in cardiac development and function. In: Vasan RS, Sawyer DB, editors. Encyclopedia of cardiovascular research and medicine. Oxford: Elsevier; 2018. pp. 340–348. [Google Scholar]
- Tian Y, Yu M, Sun L, et al. Long non-coding RNA00887 reduces the invasion and metastasis of non-small cell lung cancer by causing the degradation of miRNAs. Oncol Rep. 2019 doi: 10.3892/or.2019.7228. [DOI] [PubMed] [Google Scholar]
- Tweedell KS. The adaptability of somatic stem cells: a review. J Stem Cells Regen Med. 2017;13:3–13. doi: 10.46582/jsrm.1301002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace JA, Kagele DA, Eiring AM, et al. miR-155 promotes FLT3-ITD-induced myeloproliferative disease through inhibition of the interferon response. Blood. 2017;129:3074–3086. doi: 10.1182/blood-2016-09-740209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li X, Gong X, et al. MicroRNA-322 regulates self-renewal of mouse spermatogonial stem cells through Rassf8. Int J Biol Sci. 2019;15:857–869. doi: 10.7150/ijbs.30611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward NJ, Green D, Higgins J, et al. microRNAs associated with early neural crest development in Xenopus laevis. BMC Genomics. 2018;19:59. doi: 10.1186/s12864-018-4436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Zhang Y, Gao X, et al. Multilayered graphene oxide membranes for water treatment: a review. Carbon NY. 2018;139:964–981. doi: 10.1016/j.carbon.2018.07.040. [DOI] [Google Scholar]
- Weiner AMJ. MicroRNAs and the neural crest: from induction to differentiation. Mech Dev. 2018;154:98–106. doi: 10.1016/j.mod.2018.05.009. [DOI] [PubMed] [Google Scholar]
- Weiss CN, Ito K. Chapter three—a macro view of microRNAs: the discovery of microRNAs and their role in hematopoiesis and hematologic disease. In: Galluzzi L, Vitale I, editors. International review of cell and molecular biology. Academic Press; 2017. pp. 99–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtowicz EE, Christina BMJ, Weersing E, Dinitzen A, et al. MiR-125a enhances self-renewal, lifespan, and migration of murine hematopoietic stem and progenitor cell clones. Sci Rep. 2019;9:4785. doi: 10.1038/s41598-019-38503-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Zhang J, Lu Y, et al. miR-140-5p inhibits the proliferation and enhances the efficacy of doxorubicin to breast cancer stem cells by targeting Wnt1. Cancer Gene Ther. 2019;26:74–82. doi: 10.1038/s41417-018-0035-0. [DOI] [PubMed] [Google Scholar]
- Wu Z, Liang S, Kuai W, et al. MicroRNAs and long noncoding RNAs: new regulators in cell fate determination of mesenchymal stem cells. RSC Adv. 2019;9:37300–37311. doi: 10.1039/C9RA06563F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi J, Wu Y, Li G, et al. Mir-29b mediates the neural tube versus neural crest fate decision during embryonic stem cell neural differentiation. Stem Cell Reports. 2017;9:571–586. doi: 10.1016/j.stemcr.2017.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Q, Han T, Yang P, et al. MicroRNA-28-5p regulates liver cancer stem cell expansion via IGF-1 pathway. Stem Cells Int. 2019;2019:8734362. doi: 10.1155/2019/8734362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Guo Q, Jiang T, et al. miR-483-3p regulates osteogenic differentiation of bone marrow mesenchymal stem cells by targeting STAT1. Mol Med Rep. 2019 doi: 10.3892/mmr.2019.10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y-F, Hannafon BN, Ding W-Q. microRNA regulation of human pancreatic cancer stem cells. Stem Cell Investig. 2017;4:5. doi: 10.21037/sci.2017.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhao J, Ren Y, et al. Derivation of totipotent-like stem cells with blastocyst-like structure forming potential. Cell Re. 2022;32:513–529. doi: 10.1038/s41422-022-00668-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Liu X, Wang Z, et al. MicroRNA-486-5p functions as a tumor suppressor of proliferation and cancer stem-like cell properties by targeting Sirt1 in liver cancer. Oncol Rep. 2019;41:1938–1948. doi: 10.3892/or.2018.6930. [DOI] [PubMed] [Google Scholar]
- Yang X, Wang B, Chen W, Man X. MicroRNA-188 inhibits biological activity of lung cancer stem cells through targeting MDK and mediating the Hippo pathway. Exp Physiol. 2020;105:1360–1372. doi: 10.1113/EP088704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida GJ, Saya H. Molecular pathology underlying the robustness of cancer stem cells. Regen Ther. 2021;17:38–50. doi: 10.1016/j.reth.2021.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Zhao C, Hou S, et al. Exosomes secreted from miRNA-29b-modified mesenchymal stem cells repaired spinal cord injury in rats. Braz J Med Biol Res. 2019;52:e8735. doi: 10.1590/1414-431x20198735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K, Ai W-B, Wan L-Y, et al. The miR-290-295 cluster as multi-faceted players in mouse embryonic stem cells. Cell Biosci. 2017;7:38. doi: 10.1186/s13578-017-0166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewski W, Dobrzyński M, Szymonowicz M, Rybak Z. Stem cells: past, present, and future. Stem Cell Res Ther. 2019;10:68. doi: 10.1186/s13287-019-1165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kim MS, Jia B, et al. Hypothalamic stem cells control aging speed partly through exosomal miRNAs. Nature. 2017;548:52–57. doi: 10.1038/nature23282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ursin R, Mahapatra S, Gallicano GI. CRISPR/CAS9 ablation of individual miRNAs from a miRNA family reveals their individual efficacies for regulating cardiac differentiation. Mech Dev. 2018;150:10–20. doi: 10.1016/j.mod.2018.02.002. [DOI] [PubMed] [Google Scholar]
- Zhang S, Liu C, Zhang X. Mitochondrial damage mediated by miR-1 overexpression in cancer stem cells. Mol Ther Nucleic Acids. 2019;18:938–953. doi: 10.1016/j.omtn.2019.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen B, Li D, et al. LncRNA NEAT1/miR-29b-3p/BMP1 axis promotes osteogenic differentiation in human bone marrow-derived mesenchymal stem cells. Pathology. 2019;215:525–531. doi: 10.1016/j.prp.2018.12.034. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Guo J, Cai E, et al. HOTAIR maintains the stemness of ovarian cancer stem cells via the miR-206/TBX3 axis. Exp Cell Res. 2020;395:112218. doi: 10.1016/j.yexcr.2020.112218. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu Y, Wu M, et al. MicroRNA-664a-5p promotes osteogenic differentiation of human bone marrow-derived mesenchymal stem cells by directly downregulating HMGA2. Biochem Biophys Res Commun. 2020;521:9–14. doi: 10.1016/j.bbrc.2019.09.122. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang J, Yang B, et al. Transfer of microRNA-216a-5p from exosomes secreted by human urine-derived stem cells reduces renal ischemia/reperfusion injury. Front Cell Dev Biol. 2020;8:1577. doi: 10.3389/FCELL.2020.610587/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Meng H, Guo K. Inhibition of microRNA-302c on stemness of colon cancer stem cells via the CARF/Wnt/β-catenin axis. Dig Dis Sci. 2021;66:1906–1915. doi: 10.1007/s10620-020-06435-8. [DOI] [PubMed] [Google Scholar]
- Zhao C, Sun X, Li L. Biogenesis and function of extracellular miRNAs. ExRNA. 2019;1:1–9. doi: 10.1186/s41544-019-0039-4. [DOI] [Google Scholar]
- Zhao F, Xing Y, Jiang P, et al. LncRNA MEG3 inhibits the proliferation of neural stem cells after ischemic stroke via the miR-493–5P/MIF axis. Biochem Biophys Res Commun. 2021;568:186–192. doi: 10.1016/j.bbrc.2021.06.033. [DOI] [PubMed] [Google Scholar]
- Zheng J, Huang Y, Li Y, et al. lncRNA-TINCR functions as a competitive endogenous RNA to regulate the migration of mesenchymal stem cells by sponging miR-761. Biomed Res Int. 2020;2020:1–10. doi: 10.1155/2020/9578730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Chen W, Cui Y, et al. miRNA-122–5p stimulates the proliferation and DNA synthesis and inhibits the early apoptosis of human spermatogonial stem cells by targeting CBL and competing with lncRNA CASC7. Aging. 2020;12:25528–25546. doi: 10.18632/aging.104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang H, Che J, et al. Silencing of microRNA-135b inhibits invasion, migration, and stemness of CD24+CD44+ pancreatic cancer stem cells through JADE-1-dependent AKT/mTOR pathway. Cancer Cell Int. 2020;20:134. doi: 10.1186/s12935-020-01210-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R-T, Ni Y-R, Zeng F-J. The roles of long noncoding RNAs in the regulation of OCT4 expression. Stem Cell Res Ther. 2022;13:383. doi: 10.1186/s13287-022-03059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K, Wang Y, Liu L, et al. Long non-coding RNA MBNL1-AS1 regulates proliferation, migration, and invasion of cancer stem cells in colon cancer by interacting with MYL9 via sponging microRNA-412-3p. Clin Res Hepatol Gastroenterol. 2020;44:101–114. doi: 10.1016/j.clinre.2019.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.