Abstract
Purpose
Researchers have identified gut microbiota that interact with brain regions associated with emotion and mood. Literature reviews of those associations rely on rigorous systematic approaches and labor-intensive investments. Here we explore how knowledge graph, a large scale semantic network consisting of entities and concepts as well as the semantic relationships among them, is incorporated into the emotion-probiotic relationship exploration work.
Method
We propose an end-to-end emotion-probiotics relationship exploration method with an integrated medical knowledge graph, which incorporates the text mining output of knowledge graph, concept reasoning and evidence classification. Specifically, a knowledge graph for probiotics is built based on a text-mining analysis of PubMed, and further used to retrieve triples of relationships with reasoning logistics. Then specific relationships are annotated and evidence levels are retrieved to form a new evidence-based emotion-probiotic knowledge graph.
Results
Based on the probiotics knowledge graph with 40,442,404 triples, totally 1453 PubMed articles were annotated in both the title level and abstract level, and the evidence levels were incorporated to the visualization of the explored emotion-probiotic relationships. Finally, we got 4131 evidenced emotion-probiotic associations.
Conclusions
The evidence-based emotion-probiotic knowledge graph construction work demonstrates an effective reasoning based pipeline of relationship exploration. The annotated relationship associations are supposed be used to help researchers generate scientific hypotheses or create their own semantic graphs for their research interests.
Keywords: Emotion-probiotic relationship, Knowledge graph, Evidence, Semantic reasoning, Annotation
Introduction
Emotion is a crucial indicator of the mental state influenced by environmental cognition, health, and intention [1]. In recent years, studies have highlighted the influence of the gut microbiota on the gut-brain axis, and its potential role in central nervous system (CNS)-related conditions and neuropsychiatric disorders [2, 3]. Accumulating evidence in human and animal studies suggest a role for the gut microbiota in brain function, including for anxiety and mood disorders [4, 5]. On a clinical level, studies have demonstrated a change in emotion and mood in both human and rodent subjects with and without mood disorders [6, 7].
Of the various studies to explore the link between emotion and probiotics, literature reviews and meta-analyses are the most popular ways to get a systematic view of those relationships [8, 9]. However, due to the explosive growth of evidence emerging in publications, it is hard for individual scientists to keep up with all the topics through labor-intensive investments. In recent years, existing literature-based discovery methods and tools are more likely to use text-mining techniques to extract non-specified relationships between two concepts [10, 11], and researchers grow to adopt semantic web techniques for relation discovery [12].
Of the existing text mining tools and semantic web products, knowledge graph (KG), a large scale semantic network consisting of entities and concepts as well as the semantic relationships among them, has a great potential for exploring new relationships. A knowledge graph is expressed in triples which include object, relation and subject. The key idea is to embed components of a knowledge graph including entities and relations into continuous vector spaces, so as to simplify the manipulation while preserving the inherent structure of the knowledge graph [13]. Those entity and relation embeddings can further be used to benefit relation extraction [14–16]. In recent years, automatically constructing knowledge graphs from articles is less labor-consuming as the rapid progress of big data and natural language processing (NLP) technologies. A series of literature mining methods have utilized knowledge graph to discover complex associations. Malas et al. [17] leverage knowledge graph features such as the total number of intermediate concepts, the number of different semantic categories, and the predicates connecting a drug-disease pair to predict novel drug-disease associations. Bakal and Talari [18] exploit simple paths connecting biomedical entities as features of logistic regression model to discover drugs. Sang et al. [12, 19] develop a knowledge graph embedding based method for drug discovery from biomedical literatures.
Despite the success of the knowledge graph assisted relation discovery models, there still lacks an efficient and systematic procedure to build end-to-end relation discovery applications. To extract relationships from unstructured text with higher recall rates, it is recommended to initially rely on automated methods to obtain sentences at an acceptable recall level, then incorporate manual curation as a way to fix or remove irrelevant results [20].
Thereby, this study proposed an end-to-end emotion-probiotics relationship exploration method with an integrated medical knowledge graph. To get a higher recall rate, we used a semi-automated framework and evidence levels were added to make data visualizations more accessible to readers. The main contributions of this work are: (1) We proposed a knowledge graph based approach of exploring complex relationships from unstructured text with less human efforts; (2) We showed a reasoning process of concept retrieval in a knowledge graph, which incorporated knowledge to the text mining process. (3) We provided evidenced associations among emotions and probiotics to be validated by further studies.
The rest of this article is presented as follows: Sect. 2 introduces the pipeline of knowledge graph construction and application. It also introduces the definition of emotion models. Section 3 describes the problem formulation. In Sect. 4, current approaches used for detecting emotion probiotic relations are outlined. Section 5 presents the experimental results and analysis. Section 6 concludes the paper and points the future directions of the research.
General approach
Pipeline of knowledge graph construction and application development
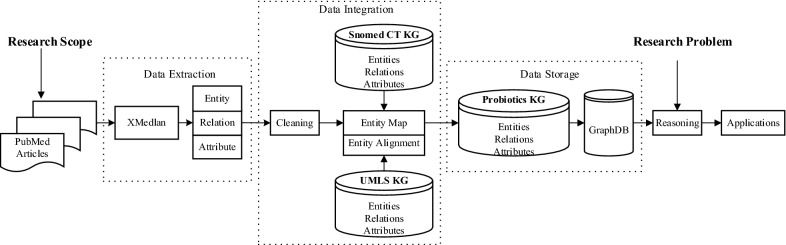
To explore relationship between emotion and probiotics with knowledge graphs, we use the seven-step-approach as described in [21], and was further developed in the work of the construction of Knowledge Graphs of Depression [22] and the construction of Knowledge Graphs of Kawasaki Disease [23]. That seven-step-approach consists of the following seven steps: identification, transformation, cleaning, integration, storage/indexing, query/reasoning and applications. An overview of our framework is illustrated in Fig. 1, which consists of the following parts: (1) Data source identification. PubMed abstracts associated with research scope (e.g. probiotics) are retrieved and downloaded; (2) Data extraction. Entities, relations, and attributes are extracted from the abstracts by XMedian, a mature NLP tool; (3) Data integration. The extracted triples are further cleaned to filter errors and inconsistencies. We use the ontology mapping tools and alignment tools to detect the semantic connection among different entities/concepts. All the entities/concepts are mapped to knowledge graphs of SNOMED CT (Systematized Nomenclature of Medicine – Clinical Terms) and UMLS (Unified Medical Language System), (4) Data storage. We use GraphDB, a semantic platform, to store those well-integrated data. The indexing system of GraphDB provides the possibility for efficient semantic search. (5) Reasoning. According to the detailed research problem, SPARQL queries are generated and further used to get targeted information from the large scale of knowledge graph data. In our work, the research problem is to explore the emotion-probiotics relationships, so the queries are emotion related concepts. (6) Application. The application interface is designed and developed for different scenarios of the applications for the knowledge graphs.
Fig. 1.
The pipeline of knowledge graph construction and application development
Distinguished from the other knowledge bases, a knowledge graph acquires and integrates information into an ontology and applies a reasoner to derive new knowledge [11]. To make the research problem more specific, we figure out the emotion concepts with emotion models. And the development of the semantic applications of knowledge graph is defined as knowledge graph based relation discovery.
Emotion models
Many emotion related tasks, such as emotion detection [24], make the definition of emotions as the research premise. Generally, emotion is defined as the manifestation or readout of motivational potential when activated by a challenging stimulus [25], and emotion models define how emotion states are represented and distinguished. Existing emotion models pay more attention on the classification of emotions [26, 27], while SNOMED provides linkages of emotions to mental disorders which meets our research problem. Therefore, “emotional state”, a child node of “mental state” in the SNOMED tree was chosen as the root concept of emotion.
Problem formulation
In this section, we describe the notations and formulate medical annotation knowledge graph based relation detection problem.
Definition 1
Medical Knowledge Graph: Let be a knowledge graph, where , and are the entity set, the relation set and the subject-relation-object triple set, respectively. The relation triples are presented as , which describes a relationship from the head node to the tail node .
Definition 2
Medical Annotation Knowledge Graph: Let be a knowledge graph, where is the probiotic related PubMed article set, is the set of links. Each article contains sentences, , and each sentence contains words, . We perform entity linking to build the token-entity alignment set , where means that word in the vocabulary can be linked with an entity in the entity set. If an article has an entity , the link will be denoted as .
To exploit linkages between entities, a path between and is denoted as . To get the specific linkages between entities, we further define the target knowledge graph.
Definition 3
Emotion-probiotic Knowledge Graph: Let be a knowledge graph, where F is the strength of the relation, the relation set is defined as {positive, positive(tbd), negative, negative(tbd)}. For example, if the entity “Lactobacillus” has a positive(tbd) relation with entity “Depression”, “Lactobacillus” is supposed to be positive to “Depression” but is not verified by the current article.
Such a knowledge path can add credibility to the article mentioning both the emotion and probiotic entities. With the above notations and definitions, we formulate the knowledge graph based relation exploration task as follows:
Problem medical knowledge graph based relation exploration.
Given a set of emotion concepts , the probiotic-related medical graph , and the evidence levels of a set articles, the goal is to retrieve the candidate emotion-probiotic relations through semantic reasoning and learn emotion-probiotic triples to construct an emotion-probiotic knowledge graph .
Methodology
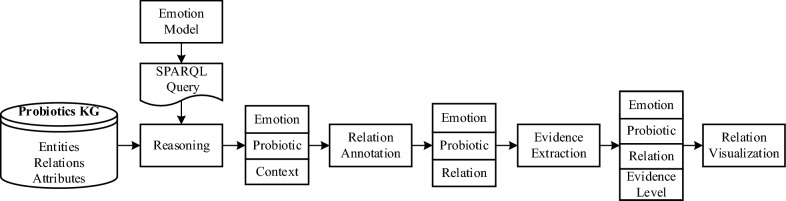
As shown in Fig. 2, following the pipeline of knowledge graph construction and application development, a knowledge graph for probiotics (KGP) is pre-constructed from probiotic and prebiotic related PubMed articles. With the emotion model defined by SNOMED concept tree, SPARQL query is generated to do the reasoning work. Next, the emotion-probiotic relationships are annotated to four classes according to context of the emotion-probiotic pairs. The evidence levels are further extracted to enrich the relationships. Finally, the detected emotion-probiotic relationships and their evidence classes are retrieved to construct a new knowledge graph and make visualizations.
Fig. 2.
The flowchart of emotion-probiotic relation detection
Knowledge graph construction
Following the seven-step-approach of knowledge graph construction framework, a probiotic knowledge graph was constructed. Firstly, we used PubMed (https://www.ncbi.nlm.nih.gov/) to retrieve and download MEDLINE [24] abstracts related to probiotics. Figure 3 shows the data schema of KGP. KGP integrates 5 different knowledge resources including UMLS, SNOMED CT, PubMed, DrugBank and PubMed annotations. KGP consists of the RDF/NTriple representations of the above knowledge resources. UMLS and SNOMED CT integrate and distribute key terminology, classification and coding standards. PubMed contains authors, title, journal name, publication date, abstract, PubMed ID (PMID), DOI and MeSH terms of the PubMed articles. DrugBank combines detailed drug data with comprehensive drug target information including sequence, structure and pathway. To integrate concepts of different resources, Xerox’s NLP tool XMedlan for semantically annotating medical text (both concept identification and relation extraction) with medical terminologies such as SNOMED CT and UMLS was used.
Fig. 3.
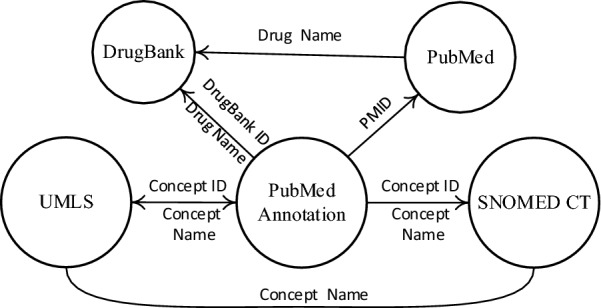
Integration of knowledge graphs for probiotics
Relationship retrieval with SPARQL queries
The top concept of emotion was clarified according to the SNOMED concept tree. The emotion concept was defined as “emotional state” with a SNOMED ID “106,126,000”, and the microbiota was defined as “bacteria” with the SNOMED ID “409,822,003”.
As shown in the appendix, SPARQL reasoning queries based on those concept IDs were designed to retrieve the PubMed Id of the entry, the source of the entry (title/abstract), the microbiota term, the context of the microbiota mention, the emotion term, the context of the emotion mention, and the microbiota/emotion candidate labels retrieved from the knowledge graph “ztone”, which was built on the main stream dictionaries including SNOMED CT and UMLS. In detail, the reasoning query means when a child node of the concept is searched, for each of the child nodes of the concept, all the children are traversed and listed.
Relationship annotation
The relationships were classified to positive, positive(tbd), negative, negative(tbd).
positive: the microbiota is positive to the emotion.
positive(tbd): the microbiota is supposed to be positive to the emotion but not verified.
negative: the microbiota is negative to the emotion.
negative(tbd): the microbiota is supposed to be negative to the emotion but not verified.
We recruited three annotators graduated from medical schools, all of whom had a medical training background and curation experience. Each article was annotated independently by two annotators (i.e., double annotation). Differences were resolved by a third and senior annotator.
To assess the consistency of the entity relationship annotation, we measured pairwise agreement of duplicate annotations using the accuracy score.
Evidence detection
In biomedical domain, levels of evidence of original articles are important factors to evaluate the creditability of the relations. To systematically overview the updated studies of the association between emotion and probiotics, related articles were identified by PubMed search with Scholarscope, an add-on of browsers like Chrome and Firefox. All the IFs and article types (Review/Randomized Controlled Trial/Meta-Analysis/Guideline/Editorial/Controlled Clinical Trial/Comparative Study/Comment/Clinical Trial/Case Reports) were automatically retrieved.
A fundamental tool for evidence-based practice has been the evidence pyramid, which depicts the hierarchy or levels of evidence from lowest to highest [28]. Since we have detailed article types, we adopted the 9 levels of evidence pyramid. Levels of evidence are arranged in increasing order of internal validity from bottom to top, with in-vitro and animal studies placed at the lowest level, followed by opinions, case reports, observational studies, RCTs, systematic reviews, and meta-analyses at the top.
Following Ting Liu’s work [29], we classified the article types to levels A-E and scores those levels to 1–9 with reference to the evidence pyramid. Detailed classifications and evidence levels are listed in Table 1.
Table 1.
Classification of conclusions based on evidence analysis
| Article type | Evidence level | The evidence level |
|---|---|---|
| Review | A | 9 |
| Meta-Analysis | A | 9 |
| Guideline | A | 9 |
| Randomized Controlled Trial | B | 8 |
| Controlled Clinical Trial | B | 7 |
| Clinical Trial | B | 7 |
| Comparative Study | C | 6 |
| Case Reports | C | 4 |
| Comment | D | 3 |
| Editorial | D | 3 |
| Others | E | 1 |
Emotion-probiotic relationship graph construction
The annotated relationships were converted to RDF triples and imported to the semantic graph database GraphDB. And the triples were further imported to Cytoscape to make network visualization.
Results
Retrieval of emotion and probiotics concepts
In the resulting knowledge graph, 75,288 concepts in articles 29,492 articles that related to prebiotics and probiotics researches were recognized by doing automatic concept annotations. Among them, 16,613 concepts from UMLS and 58,675 concepts covered by SNOMED CT. In total, 40,442,404 triples were retrieved.
After the reasoning retrieval of the emotion concept with a SNOMED ID “106126000”, we got totally 484 unique concepts and 1186 candidate labels. Likewise, the probiotic concepts were retrieved with a SNOMED ID “409822003”. Totally 11,804 unique concepts and 37,757 candidate labels were retrieved.
Retrieval of articles both have emotion and probiotics
The retrieved emotion and probiotic concepts were further used to match the articles which mention both the two kinds of concepts. In total, 143,037 entries in 1453 PubMed articles meet the requirement. As shown in Table 2, the PubMed Id of the entry, the source of a specific annotation (title/abstract), the probiotic term, the context of the probiotic mention, the emotion term, the context of the emotion mention, and the probiotic/emotion candidate labels were all collected. To focus on those entries having co-context, 13,128 entries in which the context of the probiotic mention is the same with the one of the emotion mention, had been collected. And those entries were further separated to the co-context-title set and the co-context-abstract set to make the annotation work well set out. The co-context-title set includes entries with contexts exist in titles only, while the co-context-abstract set includes entries with contexts exist in abstracts.
Table 2.
Examples of retrieved entries
| Item | Content |
|---|---|
| PubMed URL | https://www.ncbi.nlm.nih.gov/pubmed/32258258 |
| Source | Title |
| Emotion term | Depression |
| Context of emotion/probiotic term | Towards a psychobiotic therapy for depression: Bifidobacterium breve CCFM1025 reverses chronic stress-induced depressive symptoms and gut microbial abnormalities in mice |
| Candidate labels of emotion | Sadness |
| Probiotic term | Bifidobacterium breve |
| Candidate labels of probiotics | Bifidobacterium breve (organism) |
Levels of evidence were retrieved according to detailed article types. Of the 1453 articles, only 328 articles had been annotated with article types by Scholarscope. The numbers of articles leveled by evidence A-D are 177, 96, 50 and 5 respectively. Higher levels of evidence has a much larger proportion in the entire data set, which shows the high quality of the retrieved articles.
The distributions of emotional state are listed in Table 3. It is shown that negative emotional states like depression, sensitivity and anxiety are the most common ones in the whole data set. Rare positive emotional states like happiness, good mood and optimistic exist in the data set.
Table 3.
Statistics of emotion-probiotic relationship annotations
| Emotional state | Freq | Emotional state | Freq |
|---|---|---|---|
| Stress | 4008 | Indifference | 8 |
| Sensitivity | 1365 | Anger | 6 |
| Depression | 531 | Optimistic | 6 |
| Anxiety | 529 | Symptoms of stress | 6 |
| Depressive symptoms | 102 | Anxiety and fear | 4 |
| Psychological stress | 86 | Happiness | 4 |
| Excitability | 80 | Mood changes | 4 |
| Repression | 68 | Frustration | 3 |
| Agitation | 59 | Griess reaction | 3 |
| Irritability | 57 | Worry | 3 |
| Depressed | 56 | Changes in mood | 2 |
| Sad mood | 39 | Emotional stress | 2 |
| Relief | 35 | Feeding low | 2 |
| Hypersensitivity | 28 | Fussiness | 2 |
| Fear | 26 | Good mood | 2 |
| Tension | 23 | Levels of anxiety | 2 |
| Life stress | 21 | Mental stress | 2 |
| Bitterness | 15 | Mood disturbances | 2 |
| Low mood | 15 | Social anxiety | 2 |
| Decreased stress | 13 | Apprehension | 1 |
| Feeling high | 13 | Exaptations | 1 |
| Mania | 10 | Feeling of sadness | 1 |
| Alterations in food | 9 | Parental anxiety | 1 |
Relation annotation statistics
In the first step, totally 2959 entries in the co-context-title set were annotated. Over a half of the co-context-title set (65.9%) were excluded for the ambiguity of terms. For example, annotations with the term “stress” were excluded for the reason that their terms were not mentioned as emotion but “Stress on Intestinal Barrier Function”. It was also found that most annotations were stress, depression related. To cover more categories of emotions, we filtered the co-context-abstract set by excluding stress, depression related terms related annotations, and annotated the left 1172 entries in co-context-abstract set.
As shown in Table 4, for the first batch of annotation, the inter-annotation agreement (IAA) is 0.866, and the second batch 0.901. Those IAAs suggest the high quality of the annotations. It is also shown that negative emotional states like depression, sensitivity and anxiety are the most common ones in the whole data set. Rare positive emotional states like happiness, good mood and optimistic exist in the data set.
Table 4.
Statistics of microbiota-emotion relationship annotations
| 1st batch | 2nd batch | |
|---|---|---|
| IAA | 0.866 | 0.901 |
| unique PMIDs | 41 | 31 |
| IF distributions | 1.424–12.568(Median:3.945) | 0.429–38.637(Median:3.37) |
| Number of Evidence Level A | 1 | 12 |
| Number of Evidence Level B | 15 | 9 |
| Number of relationships in label level | 1008 | 809 |
| Number of relationships in term level | 66 | 42 |
| Number of positive relationships | 56 | 29 |
| Number of positive (tbd) relationships | 8 | 11 |
| Number of negative relationships | 2 | 1 |
| Number of negative (tbd) relationships | 0 | 1 |
Visualization of relationships
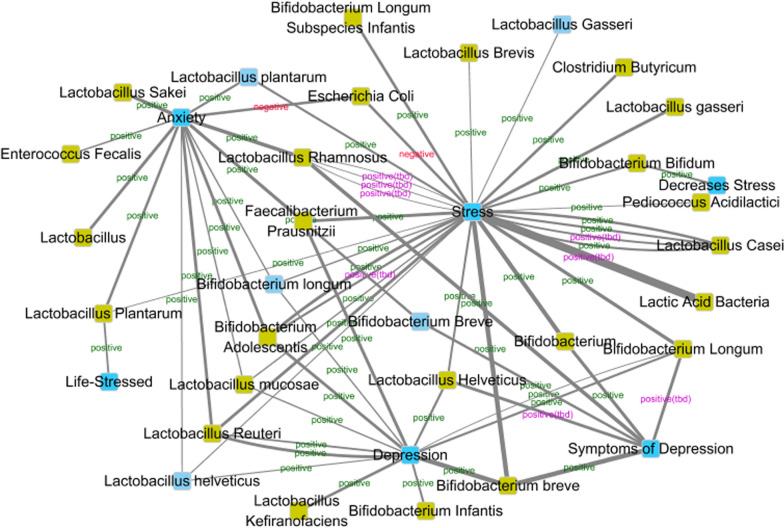
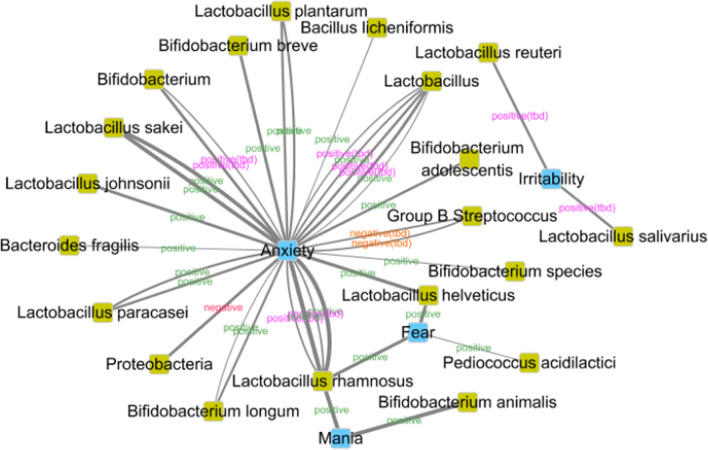
Current evidence suggest that this visualization method can clearly show different relationships of the same emotion and probiotics. As shown in Fig. 4, the emotional states in the co-context-title set are stress, depression and anxiety related. The three emotional states were associated to a network through bacteria. Escherichia Coli is a negative bacteria to both stress and anxiety. As shown in Fig. 5, anxiety are related to mania and fear, while irritability is isolated. Group B Streptococcus is a candidate negative bacteria to anxiety. Take Lactobacillus Casei as an example, three articles take it as positive while one article take it as candidate positive, which means still more work should be done to make the positive relation more solid.
Fig. 4.
Emotion-probiotic relationships visualization of co-context-title set. Knots in blue indicate emotions; knots in green indicate probiotics. Labels indicate relations between two knots. Labels in green: positive relations; labels in purple: positive(tbd) relations; labels in red: negative relations; labels in orange: negative(tbd) relations
Fig. 5.
Emotion-probiotic relationships visualization of co-context-abstract set. Knots in blue indicate emotions; knots in green indicate probiotics. Labels indicate relations between two knots. Labels in green: positive relations; labels in purple: positive(tbd) relations; labels in red: negative relations; labels in orange: negative(tbd) relations
Different bacteria have different evidence strengths. For example, “Lactic Acid Bacteria” is a candidate probiotic for the emotion “stress” with a strong evidence level while “Lactobacillus rhamnosus” with a weaker evidence level.
Moreover, these evidence-based visualizations can make a distinction between contradictory relationships. Takes the relationship between the emotion “anxiety” and the probiotic “Laxtobacillus rhamnosus” as an example, two lines indicate the relationship positive while the other two lines indicating positive(tbd). Considering that the positive lines have a larger width compared to those positive(tbd) lines, the positive relationship is more convincible.
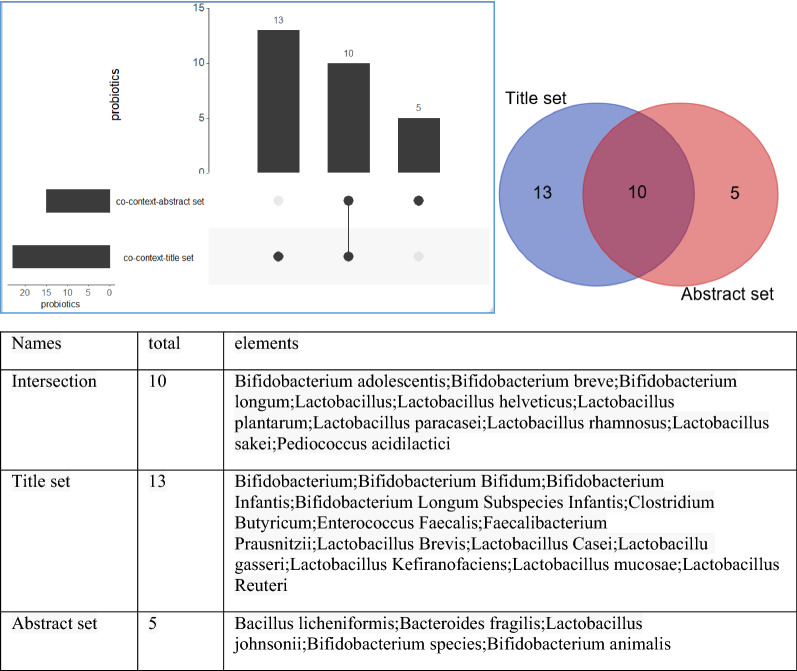
The intersections of the two sets of probiotics are also analyzed. As shown in Fig. 6, 10 bacteria both exist in the two sets, which construct the emotion linkages among anxiety, stress and depression. The unique probiotics in co-context-title set are supposed to be related with stress and depression, while the unique probiotics in co-context-abstract set are related with some other emotions like fear.
Fig. 6.
Venn presentation of the probiotics
We try to figure out the characteristics of probiotics by surveying. Bifidobacterium and Lactobacillus are the most popular probiotics for emotions, the former one is subclass of anaerobic non-sporing Gram-positive bacillus, while the later one subclass of Gram-positive bacterium. All the positive probiotics are subclass of anaerobic Gram-positive bacterium. There are also some special case required attention. For example, Enterococcus Fecalis, a probiotics for animals while a human intestinal commensal can cause various complicated infections [28], was also annotated as a positive microbiota for it can alleviate anxiety-like behavior in mice [29]. Not all Lactobacillus and Bifidobacterium were considered as probiotics, which were also annotated as positive(tbd) in some articles. In the abstract set, Lactobacillus salivarius was assumed to relate to irritability but no evidence found, which requires further researches.
Conclusions and Discussions
In our work, we proposed a concept-centered knowledge graph based complex relationship exploring method, which incorporated the text mining output of knowledge graph, concept reasoning and evidence classification. With the explicit definition of emotion, emotion related probiotics and relevant information for relationship annotation were retrieved through concept reasoning. Totally 484 unique concepts and 1186 candidate labels of emotional states were retrieved, and 143,037 entries in 1453 PubMed articles were further retrieved. Two data sets were annotated, one with 2959 entries where emotion and probiotics appears both in a title and the other one with 1172 entries where emotion and probiotics appears both in a sentence of an abstract. The detected emotion-probiotic relationships and their evidence classes were visualized by Cytoscape and the triples were stored as a new KG. These associations significantly matched current knowledge of microbiota-emotion relationships. Moreover, these associations are supposed be used to help researchers generate scientific hypotheses/designs of therapeutics or create their own semantic graphs for their research interests.
To our knowledge, it is the first time to systemically incorporate standard concept space, reasoning mechanism and evidence classification during the whole relationship exploration process. Such method makes the relationship exploration work more efficient by fully employing both the data mining results of knowledge graph and the semantic reasoning on the concept level. On the one hand, scientists can review all the topics subclass of a top topic for one time without defining inclusion and exclusion criteria as the meta-analysis does. On the other hand, the entities and their context make the relation annotation work more easy and concise. When a new batch of PubMed articles retrieved, the final relationship graph will be updated with a minimal effort of annotation. Also, the integrative method may serve as a novel approach to detect other microbiota related relationships.
The relationship exploring work provides a corpus for relation annotation, with basic entity annotation information required. The mentions, terms and context, both in a sentence or different sentences, could be further applied to the short or long distance relation detection or associated topics extraction [30]. And the annotated relationships and their context can serve as a benchmark dataset for relating text mining tasks with a natural distribution of emotions mentioned with the probiotics. When corpora annotated for relation extraction, topics of articles are often chosen in a random way for it is hard to simulate the real topic distributions [31]. Since the original PubMed annotation knowledge graph is built through the retrieval of all the probiotic and prebiotic concepts in SNOMED CT, the resulting corpus itself implies the research focus of scientists. The resulting corpus is usually imbalanced for data mining and machine learning applications, imbalanced data researches [32, 33] are thus required.
The current research scope is defined as probiotics, only a part of microbiota, which includes bacteria, archaea, protists, fungi and viruses. In the future, a study of emotion-microbiota will be preformed following the same framework. And a deep learning method for relationship detection [23] or a framework combining feature-engineering based approach and deep-learning based solution [34] will be used based on the annotated corpus of this study.
Moreover, detailed mechanisms of the impact of the probiotics on emotions should be further studied. Gut microbiota affects mental health by regulating the levels of neurotransmitters. For example, Lactobacillus plantarum was reported to increase the level of neurotransmitter Serotonin [35, 36], Dopamine [35], GABA [37] and Acetylcholine [38]. In the future work, articles related to those associations will be collected and annotated to get a scientific explanation of those emotion related probiotics.
Acknowledgements
The data has been presented previously in ISAIMS 2021 (Proceedings of the 2nd International Symposium on Artificial Intelligence for Medicine Sciences). The authors would like to thank Zidu Xu and Yaowen Gu for manually annotating the relationships, who are graduate students of Institute of Medical Information, Chinese Academy of Medical Sciences/Peking Union Medical College. The authors also would like to thank Xinjian Xie for his technical support.
Abbreviations
- KG
Knowledge graph
- CNS
Central nervous system
- NLP
Natural language processing
- SNOMED CT
Systematized nomenclature of medicine—clinical terms
- UMLS
Unified medical language system
- RDF
Resource description framework
- SPARQL
SPARQL protocol and RDF query language
- IF
Impact factor
- IAA
Inter-annotation agreement
Appendix
An example of SPARQL reasoning query to retrieve relationships.
Authors’ contributions
YS designed and conducted the experiments, analyzed the data. ZH and LH proposed the idea and supervised the whole experiment design and conduction. ZX took charge of the data quality. LH, JL and YL revised the manuscript and provided feedbacks. All the authors wrote and revised the manuscript, all the authors have read and approved the final manuscript.
Funding
The research was funded by the National Social Science Foundation for Young Scientists of China (Grant No. 18CTQ024), the Fundamental Research Funds for the Central Universities (Grant No. 3332019088), the National Key Technology Research and Development Program of China (Grant No. 2016YFC0901901), the program of China Knowledge Center for Engineering Sciences and Technology (Medical Knowledge Service System) (Grant No. CKCEST-2021-1-6), the medical knowledge service program of the Key Laboratory of Knowledge Technology for Medical Integrative Publishing.
Data availability
We have made the semantic query codes and the relationship annotations freely available at GitHub and results are also available here.
Declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
Not applicable.
Informed consent
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yueping Sun, Email: sun.yueping@imicams.ac.cn.
Jiao Li, Email: li.jiao@imicams.ac.cn.
Zidu Xu, Email: xu.zidu@imicams.ac.cn.
Yan Liu, Email: liu.yan@imicams.ac.cn.
Li Hou, Email: hou.li@imicams.ac.cn.
Zhisheng Huang, Email: z.huang@vu.nl.
References
- 1.Shao Z, et al. An analytical system for user emotion extraction, mental state modeling, and rating. Expert Syst Appl. 2019;124:82–96. doi: 10.1016/j.eswa.2019.01.004. [DOI] [Google Scholar]
- 2.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13(10):701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 3.Foster JA, McVey Neufeld KA. Gut-brain axis: How the microbiome influences anxiety and depression. Trends Neurosci. 2013;36(5):305–312. doi: 10.1016/j.tins.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Rieder R, et al. Microbes and mental health: a review. Brain Behav Immun. 2017;66:9–17. doi: 10.1016/j.bbi.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 5.Messaoudi M, et al. Assessment of psychotropic-like properties of a probiotic formulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in rats and human subjects. Br J Nutr. 2011;105(5):10. doi: 10.1017/S0007114510004319. [DOI] [PubMed] [Google Scholar]
- 6.Adikari A, Appukutty M, Kuan G. Effects of daily probiotics supplementation on anxiety induced physiological parameters among competitive football players. Nutrients. 2020 doi: 10.3390/nu12071920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor AM, et al. Associations among diet, the gastrointestinal microbiota, and negative emotional states in adults. Nutr Neurosci. 2020;23(12):983–992. doi: 10.1080/1028415X.2019.1582578. [DOI] [PubMed] [Google Scholar]
- 8.Kane L, Kinzel J. The effects of probiotics on mood and emotion. JAAPA. 2018;31(5):1–3. doi: 10.1097/01.JAA.0000532122.07789.f0. [DOI] [PubMed] [Google Scholar]
- 9.Ng QX, et al. A meta-analysis of the use of probiotics to alleviate depressive symptoms. J Affect Disord. 2018;228:13–19. doi: 10.1016/j.jad.2017.11.063. [DOI] [PubMed] [Google Scholar]
- 10.Gupta S, et al. miRiaD: a text mining tool for detecting associations of microRNAs with diseases. J Biomed Semant. 2016;7(1):9. doi: 10.1186/s13326-015-0044-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei CH, et al. Assessing the state of the art in biomedical relation extraction: overview of the BioCreative V chemical-disease relation (CDR) task. Database (Oxford), 2016. [DOI] [PMC free article] [PubMed]
- 12.Sang S, et al. SemaTyP: a knowledge graph based literature mining method for drug discovery. BMC Bioinf. 2018;19(1):193. doi: 10.1186/s12859-018-2167-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q, Mao Z, Wang B, Guo L. Knowledge graph embedding: a survey of approaches and applications. IEEE Trans Knowl Data Eng. 2017;29(12):2724–2743. doi: 10.1109/TKDE.2017.2754499. [DOI] [Google Scholar]
- 14.Weston J, Bordes A, Yakhnenko O, Usunier N. Connecting language and knowledge bases with embedding models for relation extraction. In: Proc. conf. empirical methods natural language process; 2013. pp. 1366–1371.
- 15.Riedel S, Yao L, Mccallum A, Marlin BM. Relation extraction with matrix factorization and universal schemas. In: Proc. conf. North Amer. chapter Assoc. Comput. Linguistics: Human Language Technol; 2013. pp. 74–84.
- 16.Zhang N, Deng S, Sun Z, et al. Long-tail relation extraction via knowledge graph embeddings and graph convolution networks. arXiv:1903.01306; 2019.
- 17.Malas TB, et al. Drug repurposing using a semantic knowledge graph. Tech. Rep.
- 18.Bakal G, et al. Exploiting semantic patterns over biomedical knowledge graphs for predicting treatment and causative relations. J Biomed Inform. 2018;82:189–199. doi: 10.1016/j.jbi.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sang S, et al. GrEDeL: a knowledge graph embedding based method for drug discovery from biomedical literatures. IEEE Access. 2019;7:8404–8415. doi: 10.1109/ACCESS.2018.2886311. [DOI] [Google Scholar]
- 20.Nicholson DN, Greene CS. Constructing knowledge graphs and their biomedical applications. Comput Struct Biotechnol J. 2020;18:1414–1428. doi: 10.1016/j.csbj.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fensel D, et al. Towards LarKC: a platform for Web-Scale reasoning. In: 2008 IEEE international conference on semantic computing. 2008.
- 22.Huang Z, et al. Constructing disease-centric knowledge graphs: a case study for depression (short version). In Conference on artificial intelligence in medicine in Europe. New York: Springer; 2017.
- 23.Huang ZS, et al. Knowledge graphs of Kawasaki disease. Health Inf. Sci Syst. 2021;9(1):11. doi: 10.1007/s13755-020-00130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Acheampong FA, Wenyu C, Nunoo-Mensah H. Text-based emotion detection: advances, challenges, and opportunities. Affect Comput. 2020;2(7):e12189. [Google Scholar]
- 25.Borod JC. The neuropsychology of emotion. Oxford: Oxford University Press; 2000. [Google Scholar]
- 26.Ekman P. Handbook of cognition and emotion. New York: Wiley; 1999. pp. 45–60. [Google Scholar]
- 27.Canales L, Martínez-Barco P. Emotion detection from text: a survey. In: Proceedings of the workshop on natural language processing in the 5th information systems research working days (JISIC). Quito, Ecuador: Association for Computational Linguistics. 2014.
- 28.Mulimani PS. Evidence-based practice and the evidence pyramid: a 21st century orthodontic odyssey. Am J Orthod Dentofac Orthop. 2017;152(1):1–8. doi: 10.1016/j.ajodo.2017.03.020. [DOI] [PubMed] [Google Scholar]
- 29.Liu T, et al. Influence of gut microbiota on mental health via neurotransmitters: a review. J Artif Intell Med Sci. 2020 doi: 10.2991/jaims.d.200420.001. [DOI] [Google Scholar]
- 30.Jiang Haixin, Zhou Rui, Zhang Limeng, Wang Hua, Zhang Yanchun. Sentence level topic models for associated topics extraction. World Wide Web. 2019;22(6):2545–2560. doi: 10.1007/s11280-018-0639-1. [DOI] [Google Scholar]
- 31.Sun Y, et al. RCorp: a resource for chemical disease semantic extraction in Chinese. BMC Med Inform Decis Mak. 2019;19(5):234. doi: 10.1186/s12911-019-0936-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Hu, Wang Ye, Wang Hua, Zhou Bin. Multi-window based ensemble learning for classification of imbalanced streaming data. World Wide Web. 2017;20(6):1507–1525. doi: 10.1007/s11280-017-0449-x. [DOI] [Google Scholar]
- 33.Yin Jiao, Tang MingJian, Cao Jinli, Wang Hua, You Mingshan, Lin Yongzheng. Vulnerability exploitation time prediction: an integrated framework for dynamic imbalanced learning. World Wide Web. 2022;25(1):401–423. doi: 10.1007/s11280-021-00909-z. [DOI] [Google Scholar]
- 34.He Jinyuan, Rong Jia, Sun Le, Wang Hua, Zhang Yanchun, Ma Jiangang. A framework for cardiac arrhythmia detection from IoT-based ECGs. World Wide Web. 2020;23(5):2835–2850. doi: 10.1007/s11280-019-00776-9. [DOI] [Google Scholar]
- 35.Liu W-H, et al. Alteration of behavior and monoamine levels attributable to Lactobacillus plantarum PS128 in germ-free mice. Behav Brain Res. 2016;298:202–209. doi: 10.1016/j.bbr.2015.10.046. [DOI] [PubMed] [Google Scholar]
- 36.Toy N, Özogul F, Özogul Y. The influence of the cell free solution of lactic acid bacteria on tyramine production by food borne-pathogens in tyrosine decarboxylase broth. Food Chem. 2015;173:45–53. doi: 10.1016/j.foodchem.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 37.Siragusa S, et al. Synthesis of γ-aminobutyric acid by lactic acid bacteria isolated from a variety of Italian Cheeses. Appl Environ Microbiol. 2007;73(22):7283. doi: 10.1128/AEM.01064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stanaszek PM, Snell JF, Neill JJ. Isolation, extraction, and measurement of acetylcholine from Lactobacillus plantarum. Appl Environ Microbiol. 1977;34(2):237. doi: 10.1128/aem.34.2.237-239.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We have made the semantic query codes and the relationship annotations freely available at GitHub and results are also available here.