Abstract
A urease-deficient derivative of Streptococcus salivarius 57.I was constructed by allelic exchange at the ureC locus. The wild-type strain was protected against acid killing through hydrolysis of physiologically relevant concentrations of urea, whereas the mutant was not. Also, S. salivarius could use urea as a source of nitrogen for growth exclusively through a urease-dependent pathway.
Bacterial ureases are multisubunit enzymes that require Ni2+ for catalytic activity (9). Several bacterial urease gene clusters have been isolated, and high degrees of homology between species have been observed (5, 9). Most bacterial ureases consist of three subunits, α, β, and γ, encoded by ureC, -B, and -A, respectively. Other genes are present in urease clusters, i.e., ureDEFG, which encode proteins that are required for incorporation of Ni2+ into the metallocenter of the catalytic site (9). Additional gene products involved in urease biogenesis and urea metabolism include nickel and urea transporters (10, 14).
Urea is present in saliva and crevicular fluids at 3 to 10 mM in healthy individuals (7, 8), and it is hydrolyzed by ureases to generate two molecules of ammonia and one molecule of CO2. Ammonia can neutralize acids generated from bacterial glycolysis, inhibiting the initiation and progression of tooth decay. Ureolysis also creates a less acidic environment, enhancing the survival of acid-sensitive species and promoting the stability of a healthy oral flora (1). Despite the abundance of urea and ureolytic activity in the oral cavity, and the impact of ureolysis on oral health and ecology, the benefits for oral microorganisms of possessing ureases are not established.
In general, urease expression in enteric organisms is positively regulated and transcription is activated either in the absence of an assimilable nitrogen source or in the presence of urea (5, 9). Unlike in enteric bacteria, Streptococcus salivarius 57.I urease expression is derepressed at low pH and is further enhanced in the presence of excess carbohydrate (2). Based on this mode of regulation, ureolysis by S. salivarius may function primarily to protect the organisms against acid damage or the bacteria may use ureolysis to acquire nitrogen when carbohydrates are present in excess. To test these hypotheses, an otherwise isogenic, urease-deficient derivative of S. salivarius 57.I was constructed, and the behavior of this mutant and that of the wild-type strain under different growth conditions were compared.
Construction of a urease-deficient S. salivarius strain.
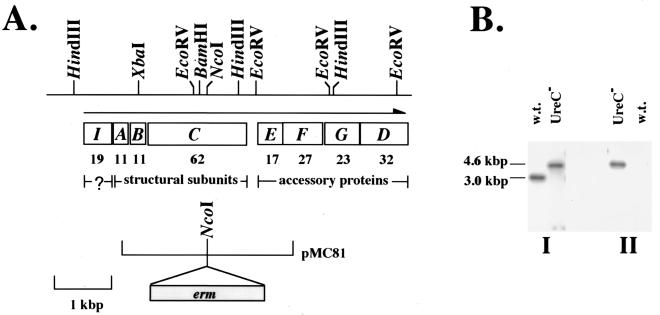
To construct a urease-deficient S. salivarius strain, a gene specifying erythromycin resistance (erm) was cloned within ureC, the gene encoding the α subunit of urease, to generate plasmid pMC81 (Fig. 1A). Plasmid pMC81 contains an Escherichia coli replicon and thus can be used as a suicide vector in streptococcal hosts. Plasmid pMC81 was introduced into wild-type 57.I by electroporation as previously described (4), and erythromycin-resistant (Emr) transformants were selected and subjected to Southern blot analysis (12). Wild-type 57.I and the Emr transformants were grown in brain heart infusion (Difco, Detroit, Mich.) medium supplemented with 20 mM dl-threonine and 10 mM glucose, without or with erythromycin (3 μg/ml), respectively, to mid-exponential phase. Chromosomal DNAs were isolated as previously described (3), the DNA was digested with HindIII, and the fragments were separated on a 0.8% agarose gel. Southern hybridization was performed at high stringency with probes internal to ureC or to erm (Fig. 1B). Finally, an antibody raised to purified, histidine-tagged UreC of S. salivarius 57.I revealed that no detectable UreC protein was produced (data not shown). A single ureC-deficient mutant generated by allelic exchange was identified and used for further analysis.
FIG. 1.
Construction and characterization of UreC-deficient S. salivarius. (A) A restriction map of the chromosomal region containing the ure cluster is shown at the top. The organization of the operon is shown, and the direction of transcription is indicated by a horizontal arrow. The molecular mass (in kilodaltons) of each open reading frame is shown below the gene. The limits of pMC81 in relation to the ure cluster and the location of erm within pMC81 are also shown. (B) Southern blot analysis of the wild-type strain and otherwise isogenic ureC-deficient derivative. Total cellular DNA was digested with HindIII and transferred onto two membranes simultaneously by a sandwich blot. The membranes were probed with a ureC-specific probe (I) or with an erm-specific probe (II).
Ureolysis can protect S. salivarius 57.I against lethal acidification.
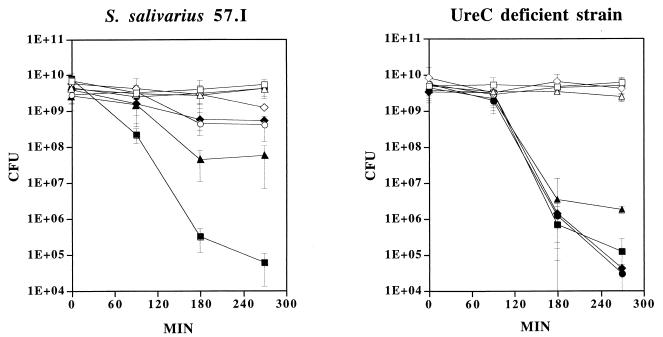
Studies by Sissons and Hancock (11) have suggested that ammonia production through ureolysis may protect S. salivarius from being killed at low pH. To determine if urease is an essential determinant in protection of S. salivarius 57.I against lethal acidification in the presence of urea, the survival rates of the wild-type and the UreC-deficient strains in an acidic environment were compared in the presence of different concentrations of urea. The wild-type strain and its UreC-deficient derivative were grown to late exponential phase. Cells were washed once with 1% peptone and then resuspended in 1/10 of the culture volume in 1% peptone, pH 7.0, supplemented with 0, 10, 25, or 50 mM urea or in 1% peptone, pH 3.0, supplemented with the same concentrations of urea (Fig. 2). All cell suspensions were kept at room temperature with gentle rocking. An aliquot of the suspensions was removed at various time points and the viability of each sample was determined by plating serial dilutions on brain heart infusion agar. When wild-type cells were incubated at pH 7.0, no significant difference in the numbers of CFU was observed in the presence or absence of 10 mM urea over a 270-min period, with final pH values of 7.1 and 6.5, respectively. In the presence of 25 or 50 mM urea, the viability decreased approximately 10-fold over 270 min compared to that of cells with no urea added. Since the pH values of cell suspensions increased to 8.5 with 25 mM urea and 8.8 with 50 mM urea, the decline in viability was probably due to prolonged exposure to an excessively high pH (Fig. 2). When wild-type cells were suspended at pH 3.0, more than 95% of the population was killed after 90 min. The viability of the cells decreased further after 180 min to less than 0.01% of the population. An additional 90 min of incubation at pH 3.0 resulted in less than 0.001% survival, indicating that S. salivarius does not withstand prolonged exposure to pH 3.0. However, when urea was also provided in the system, survival was dramatically enhanced (Fig. 2). When 10 mM urea was introduced into the system, the viability was 100-fold higher than that of the no-urea control after 180 min at pH 3.0, despite the fact that the final pH value reached only 3.3 after 270 min. The enhancement of viability was more dramatic when 25 mM urea was added. With the addition of 25 mM urea, the viability was 1,000-fold greater than that of controls after 270 min at pH 3.0, and the pH value of the cell suspension reached 7.6. There was virtually no loss of viability with cells supplemented with 50 mM urea, and the final pH value was 8.3.
FIG. 2.
Effect of ureolysis on survival of the wild-type cells and the UreC-deficient mutant against lethal acidification. Cells were incubated at pH 7.0 (open symbols) or pH 3.0 (filled symbols), with no urea (□ and ■), 10 mM urea (▵ and ▴), 25 mM urea (◊ and ⧫), or 50 mM urea (○ and ●). The viability of each culture (in CFU) was determined. The graph shows the means and standard deviations of three independent experiments.
It is established that the cytoplasmic pH of streptococci is generally maintained at 0.5 to 1 pH unit higher than the extracellular environment (6) and that intracellular ureases are protected from inactivation by cell membrane barriers (11). Furthermore, in vitro analysis has shown that the purified urease enzyme retains about 50% of its activity at pH 4.0 (3). Therefore, it is not surprising that sufficient ammonia was produced through ureolysis at pH 3.0, even though in vitro analysis has indicated that the optimal pH for urease activity is about pH 7.0 (4). Also, the data support the hypothesis that the effects of ureolysis on viability were due in part to cytoplasmic alkalinization, since there was little change in the extracellular pH when cells were provided with as little as 10 mM urea. However, environmental alkalinization, as was seen with 25 and 50 mM urea, also would significantly enhance survival.
When the UreC-deficient mutant was incubated at pH 7.0, with or without any additional urea, the levels of viability paralleled those of the wild-type cells at pH 7.0 (Fig. 2), and the presence of urea did not influence the pH value of the cell suspensions (all three cultures had a final pH value of 6.5). At pH 3.0, the rate of killing of the UreC-deficient mutant was similar to that of the wild-type strain, and the presence of urea did not reverse the killing by low pH in this mutant. All four cell suspensions reached pH 3.4 by the end of 270 min. This result is consistent with the idea that ureolysis plays an important role in protecting S. salivarius against lethal acidification.
Urea provides a source of assimilable nitrogen to S. salivarius 57.I.
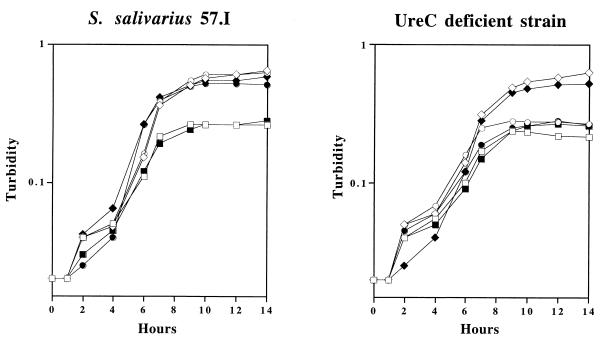
To investigate whether urea can serve as a nitrogen source through a urease-dependent pathway, overnight cultures of the wild-type strain and the UreC-deficient mutant were grown in the chemically defined medium FMC (13). The overnight cultures were concentrated by centrifugation, washed with equal volumes of FMC containing 0.005% Casamino Acids (FMC-CA), and then resuspended in the same medium in the original culture volume. The cultures were diluted 1:100 in fresh FMC-CA supplemented with 10 mM urea, 10 mM (NH4)2SO4, or no additional nitrogen source. Glucose (20 mM) or lactose (10 mM) was used as the carbohydrate source. All cultures were incubated at 37°C in a 5% CO2 environment, and the growth of each culture was monitored by measuring the optical density at 600 nm (Fig. 3). When wild-type cells and the UreC-deficient mutant were grown in the base medium (FMC-CA) supplemented with glucose or lactose, the cells apparently exhausted the nitrogen source within 8 h and achieved a final turbidity of only 0.25. When 10 mM (NH4)2SO4 or urea was provided in the medium, higher cell densities were achieved by wild-type organisms, with a final turbidity of 0.5 to 0.6, indicating that both (NH4)2SO4 and urea could serve as nitrogen sources for growth. When the UreC-deficient derivative was grown in the base medium supplemented with 10 mM (NH4)2SO4, growth was similar to that of the wild-type strain growing in the same medium. However, when urea was added to the base medium (FMC-CA), no further growth could be detected with the UreC-deficient mutant, indicating that urea served as a nitrogen source only through a urease-dependent pathway. The presence of urea in a saliva medium also enhanced the growth of S. salivarius 57.I but not the urease-deficient strain (data not shown).
FIG. 3.
Growth of wild-type cells and the UreC-deficient mutant in FMC. Cells were grown in defined medium supplemented with glucose (open symbols) or lactose (filled symbols) as the sole carbon source. □ and ■, no additional nitrogen source; ◊ and ⧫, 10 mM (NH4)2SO4; ○ and ●, 10 mM urea. The graph is representative of three independent experiments.
In conclusion, the results indicated that ureolysis can protect S. salivarius against extended exposure to acid and that S. salivarius utilizes urea as a nitrogen source exclusively through a urease-dependent pathway. Thus, the ureolytic phenotype may be an important ecological determinant and play significant roles in influencing the balance of oral microbial flora.
Acknowledgments
We thank Samir Bhagwat for critical reading of the manuscript.
This work was supported by PHS grant DE10362 from the National Institute for Dental and Craniofacial Research to R.A.B.
REFERENCES
- 1.Burne R A. Oral streptococci…products of their environment. J Dent Res. 1998;77:445–452. doi: 10.1177/00220345980770030301. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y M, Burne R A. Analysis of Streptococcus salivarius urease expression using continuous chemostat culture. FEMS Microbiol Lett. 1996;135:223–229. doi: 10.1111/j.1574-6968.1996.tb07993.x. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y M, Clancy K A, Burne R A. Streptococcus salivarius urease: genetic and biochemical characterization and expression in a dental plaque streptococcus. Infect Immun. 1996;64:585–592. doi: 10.1128/iai.64.2.585-592.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y M, Weaver C A, Mendelsohn D R, Burne R A. Transcriptional regulation of the Streptococcus salivarius 57.I urease operon. J Bacteriol. 1998;180:5769–5775. doi: 10.1128/jb.180.21.5769-5775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins C M, D'Orazio S E F. Bacterial ureases: structure, regulation of expression and role in pathogenesis. Mol Microbiol. 1993;9:907–913. doi: 10.1111/j.1365-2958.1993.tb01220.x. [DOI] [PubMed] [Google Scholar]
- 6.Dashper S G, Reynolds E C. pH regulation by Streptococcus mutans. J Dent Res. 1992;71:1159–1165. doi: 10.1177/00220345920710050601. [DOI] [PubMed] [Google Scholar]
- 7.Golub L M, Borden S M, Kleinberg I. Urea content of gingival crevicular fluid and its relationship to periodontal disease in humans. J Periodontal Res. 1971;6:243–251. doi: 10.1111/j.1600-0765.1971.tb00615.x. [DOI] [PubMed] [Google Scholar]
- 8.Kopstein J, Wrong O M. The origin and fate of salivary urea and ammonia in man. Clin Sci Mol Med. 1977;52:9–17. doi: 10.1042/cs0520009. [DOI] [PubMed] [Google Scholar]
- 9.Mobley H L T, Island M D, Hausinger R P. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mobley, H. L. T., R. M. Garner, and P. Bauerfield. Helicobacter pylori nickel-transport gene nixA: synthesis of catalytically active urease in Escherichia coli independent of growth conditions. Mol. Microbiol. 16:97–109. [DOI] [PubMed]
- 11.Sissons C H, Hancock E M. Urease activity in Streptococcus salivarius at low pH. Arch Oral Biol. 1993;38:507–516. doi: 10.1016/0003-9969(93)90187-q. [DOI] [PubMed] [Google Scholar]
- 12.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 13.Terleckyj B, Shockman G D. Amino acid requirements of Streptococcus mutans and other oral streptococci. Infect Immun. 1975;11:656–664. doi: 10.1128/iai.11.4.656-664.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weeks D L, Eskandari S, Scott D R, Sachs G. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science. 2000;287:482–485. doi: 10.1126/science.287.5452.482. [DOI] [PubMed] [Google Scholar]