Abstract
The mechanistic target of rapamycin complex 1 (mTORC1) controls growth by regulating anabolic and catabolic processes in response to environmental cues, including nutrients1,2. Amino acids signal to mTORC1 through the Rag GTPases, which are regulated by several protein complexes, including GATOR1 and GATOR2. GATOR2, which has five components (WDR24, MIOS, WDR59, SEH1L, SEC13), is required for amino acids to activate mTORC1 and interacts with the leucine and arginine sensors Sestrin2 and CASTOR1, respectively3–5. Despite this central role in nutrient sensing, GATOR2 remains mysterious as its subunit stoichiometry, biochemical function, and structure are unknown. Here, we used electron cryomicroscopy to determine the three-dimensional structure of the human GATOR2 complex. We find that GATOR2 adopts a large (1.1 MDa), two-fold symmetric, cage-like architecture, supported by an octagonal scaffold and decorated with eight pairs of WD40 β-propellers. The scaffold contains two WDR24, four MIOS, and two WDR59 subunits circularized via two distinct types of junctions involving non-catalytic RING domains and α-solenoids. Integration of SEH1L and SEC13 into the scaffold through β-propeller blade donation stabilizes the GATOR2 complex and reveals an evolutionary relationship to the nuclear pore and membrane coating complexes6. The scaffold orients the WD40 β-propeller dimers, which mediate interactions with Sestrin2, CASTOR1, and GATOR1. Our work reveals the structure of an essential component of the nutrient sensing machinery and provides a foundation for understanding GATOR2 function within the mTORC1 pathway.
Introduction
The mechanistic target of rapamycin (mTORC1) pathway is a central regulator of cell growth that responds to the availability of amino acids. A critical step in mTORC1 activation is the nutrient-dependent translocation of mTORC1 to the lysosomal surface7. The heterodimeric Rag GTPases (RagA or B bound to RagC or D) recruit mTORC1 to the lysosome, and a number of proteins that directly modulate the Rags collectively convey cytosolic and lysosomal amino acid availability to mTORC11,2,8,9. Among these are the antagonistic GATOR1 and GATOR2 complexes, which directly interact with each other and, together with the KICSTOR complex, mediate cytosolic amino acid sensing3,10,11. The GATOR1 complex inhibits mTORC1 signaling by stimulating GTP hydrolysis by RagA/B3,12. In contrast, GATOR2 promotes mTORC1 activation when amino acids are abundant and is the receptor for the leucine sensors, Sestrin1/2, and arginine sensor, CASTOR14,5. During nutrient deprivation, these amino acid sensors inhibit mTORC1 via their associations with GATOR2. The counterbalance between the two GATOR complexes enables mTORC1 to dynamically respond to fluctuating levels of amino acids in the cell. Despite its central position in the nutrient sensing pathway, the structural and biochemical basis by which GATOR2 regulates mTORC1 remains unclear.
Results
Structural Determination of Human GATOR2
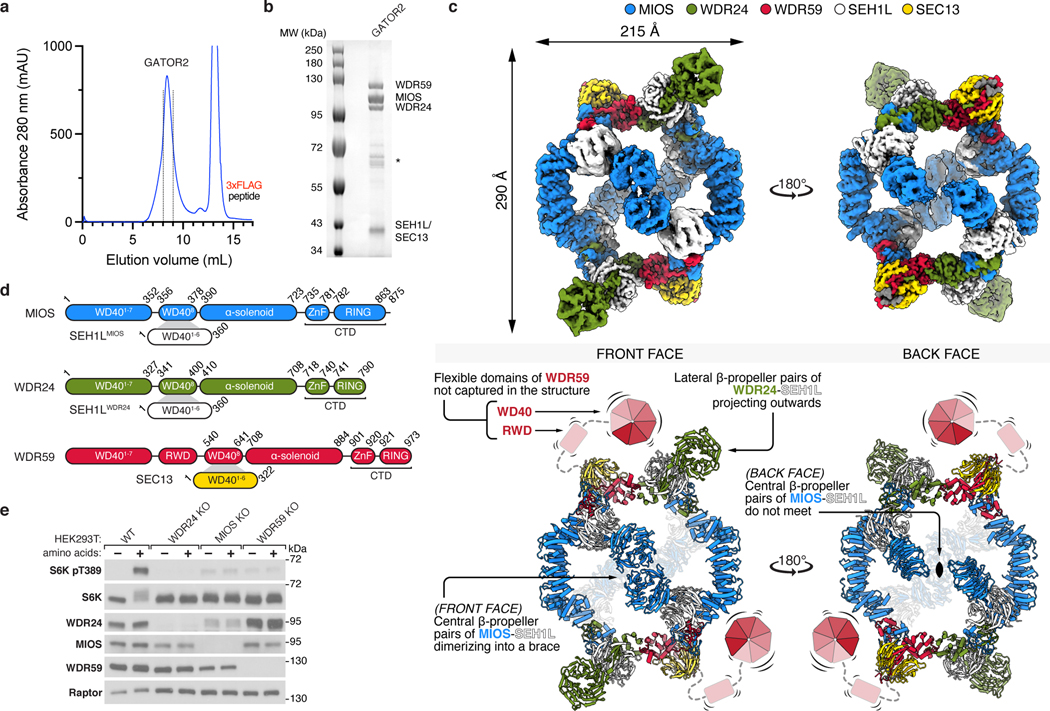
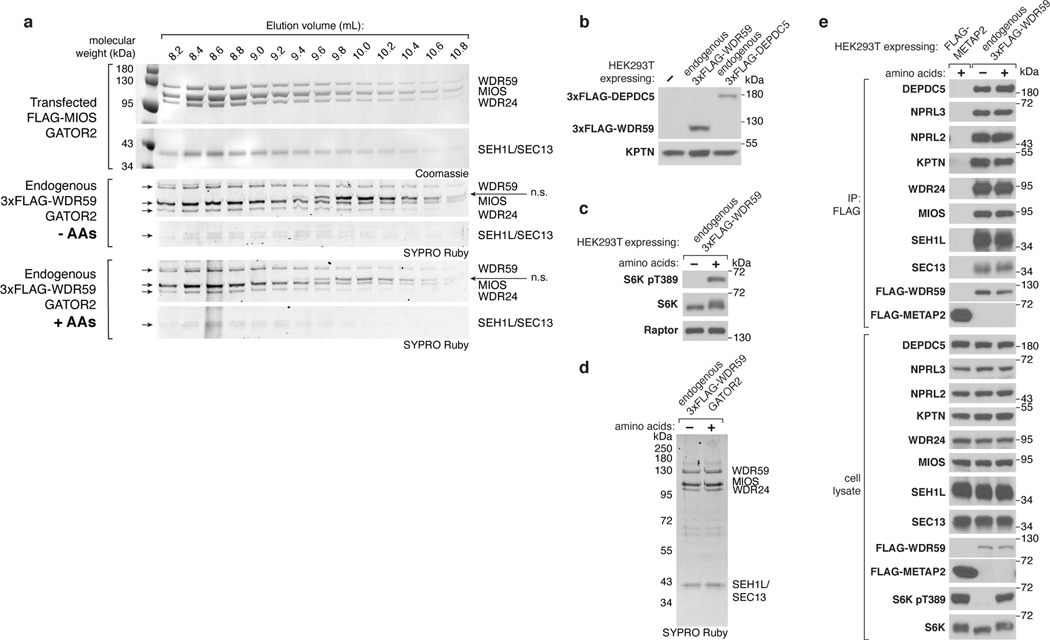
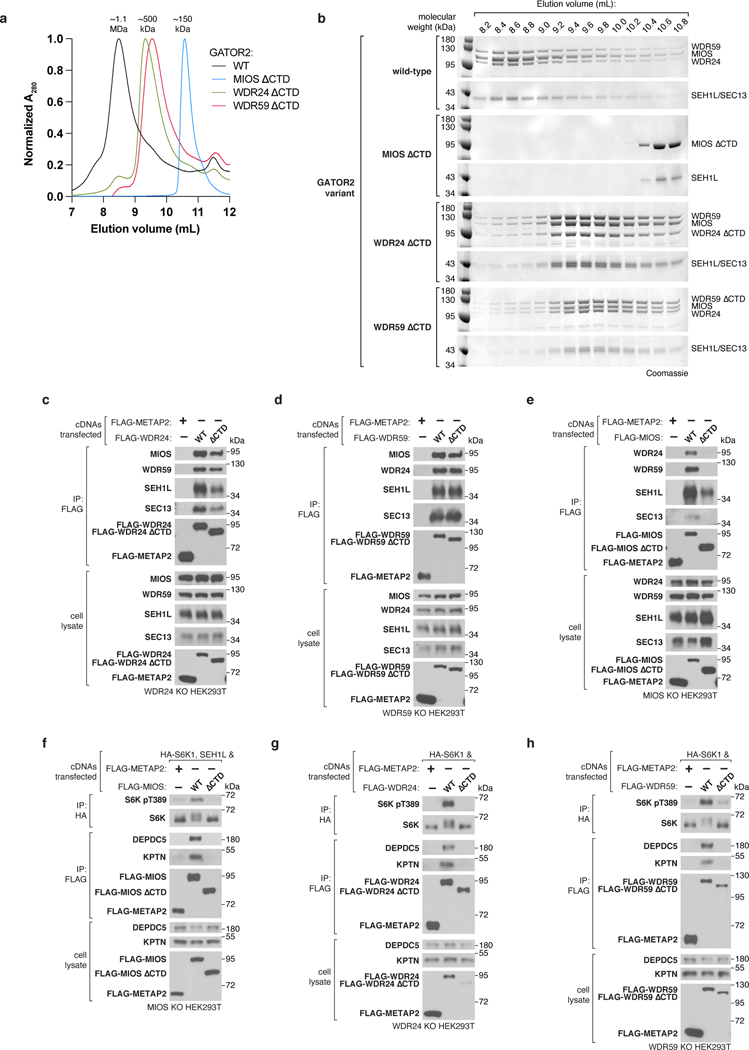
To determine the structure of GATOR2, we first transiently co-expressed all five components in suspension-adapted HEK293T cells and purified the complex by affinity chromatography followed by size-exclusion chromatography (SEC) (Fig. 1a,b). Transiently-expressed GATOR2 and GATOR2 isolated from endogenously-tagged cell lines were comparable in size, composition, and subunit stoichiometry, all of which were unaffected by amino acid availability (Extended Data Fig. 1a–e).
Figure 1: Structure of human GATOR2.
(a) Size-exclusion chromatography profile of human GATOR2 used for structural analyses. mAU, milli-absorbance unit.
(b) Coomassie Blue stained SDS-PAGE analysis of purified GATOR2. Asterisk indicates the CCT chaperonin complex that co-purifies with GATOR2.
(c) Cryo-EM structure of the human GATOR2 complex. Two views of the experimental map (top) are shown next to the corresponding views of the molecular model (bottom). The C2 symmetry axis is perpendicular to the viewing plane, and marked with a filled ellipse.
(d) Domain organization of GATOR2 components. Numbers denote the boundaries of the indicated domains. Gray trapezoids indicate β-blade donation by WDR24, MIOS, or WDR59 to complete β-propellers of SEH1L or SEC13.
(e) Loss of core GATOR2 components renders mTORC1 signaling insensitive to amino acid availability. HEK293T cell clones of the indicated genotype were starved for amino acids for 60 min and restimulated with amino acids for 15 min prior to harvest. Cell lysates were analyzed by immunoblotting for the levels and phosphorylation states of the indicated proteins.
Data in (e) are representative of two independent experiments. For gel source data, see Supplementary Figure 1.
We performed single particle cryo-EM analysis of the GATOR2 complex and determined its structure at an overall resolution of 3.7 Å (Supplementary Information). While the octagonal scaffold was resolved to ~3 Å, the auxiliary WD40 β-propellers showed a high degree of flexibility, and ranged in resolution between 4 and 6 Å. With local masking and variability clustering, we built a composite map of the entire complex, and used that map for de novo building of the model (Supplementary Information). We were able to trace and build >60% of WDR24, MIOS, and WDR59, including their C-terminal RING domains. SEH1L and SEC13 were re-built after docking in models of their S. cerevisiae orthologues. The N-terminal β-propeller and predicted RWD domain of WDR59 were not visualized in our cryo-EM map, likely due to high flexibility of these regions.
Architecture of GATOR2
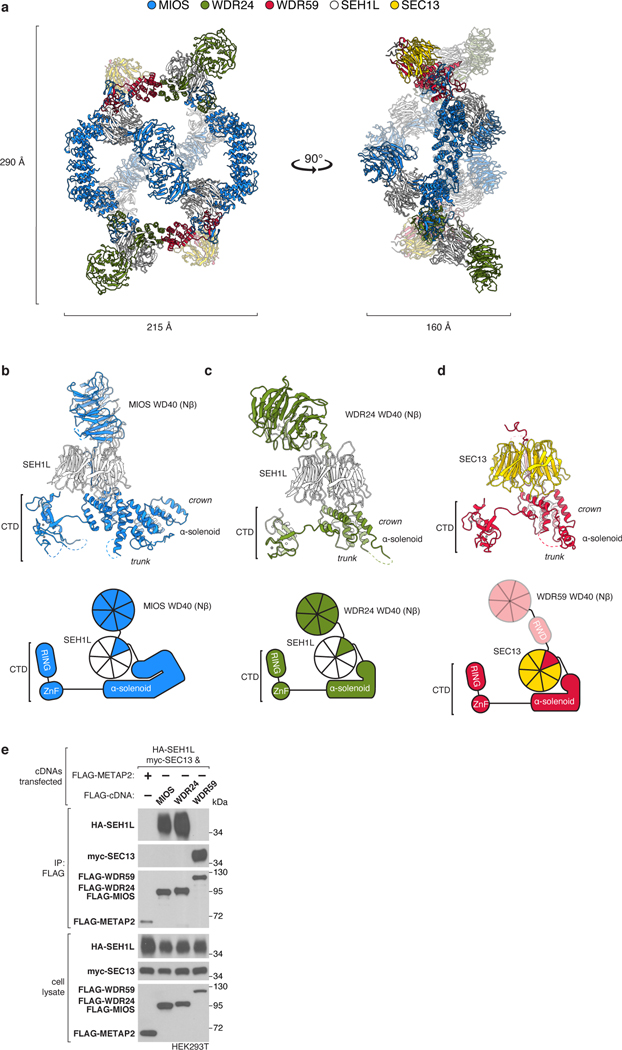
GATOR2 adopts a two-fold rotationally symmetric, cage-like architecture of approximately 290 × 215 × 160 Å built from two WDR24-SEH1L, four MIOS-SEH1L, and two WDR59-SEC13 heterodimers for a total molecular weight of 1.1 MDa (Fig. 1c, Extended Data Fig. 2a, Supplementary Video 1). These subunits assemble together to form an octagonal scaffold that is decorated with protruding pairs of WD40 β-propellers. The core GATOR2 components (WDR24, MIOS, WDR59) share a similar fold, reminiscent of the Ancestral Coatomer Element 1 (ACE1), wherein the N-terminal β-propeller of each subunit is followed by a three-stranded β-blade and then an α-helical solenoid (the “trunk”) that doubles back on itself (the “crown”)6. Each protein culminates in a C-terminal zinc-binding domain (CTD), composed of a zinc finger (ZnF) and a RING domain (Extended Data Fig. 2b–d). The six-bladed β-propeller proteins, SEH1L and SEC13, are incorporated into the GATOR2 scaffold through a β-blade donation by WDR24 or MIOS and WDR59, respectively (Fig. 1d, Extended Data Fig. 2e). Two types of subunit junctions mediate the assembly of the octagonal scaffold: (1) heterodimeric CTD-CTD interactions (MIOS-WDR24 and MIOS-WDR59), and (2) α-solenoid crown-crown interactions (MIOS-MIOS and WDR24-WDR59). The angled MIOS solenoidal domain and the CTD-CTD junctions form the scaffold vertices, allowing for its circularization. The surface of GATOR2 is highly charged with interspersed lipophilic patches (Extended Data Fig. 3a–c). Consistent with their key roles in GATOR2 assembly, WDR24, MIOS, and WDR59 are each required for mTORC1 to sense amino acid availability (Fig. 1e).
The 16 β-propellers of GATOR2 form eight propeller pairs that project either towards the middle of the octagon (central propeller pairs), or away from it (lateral propeller pairs) (Fig. 1c). The four central pairs are composed of the MIOS and SEH1L β-propeller dimers, such that each face of the scaffold is decorated with two of these pairs, positioned exactly in opposite corners of the octagon. The angles at which these β-propeller pairs emerge from the scaffold are different for the two scaffold faces. On one side, the two MIOS propellers meet in the middle, forming a “brace” that spans across the scaffold. On the other, they remain separated like poised boxing “gloves.” The lateral β-propellers of WDR24 and SEH1L form a dimer that projects away from the scaffold, on its braced face. Correspondingly, SEC13, and likely the unvisualized WDR59 β-propeller and RWD domain, protrude laterally from the scaffold on the gloved face.
CTD Dimers Link MIOS to WDR24 and WDR59
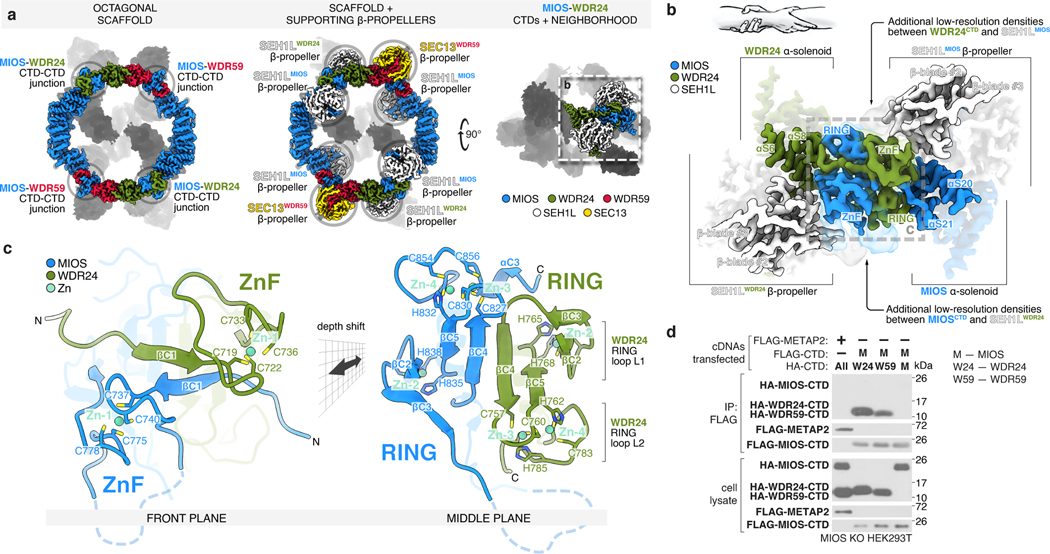
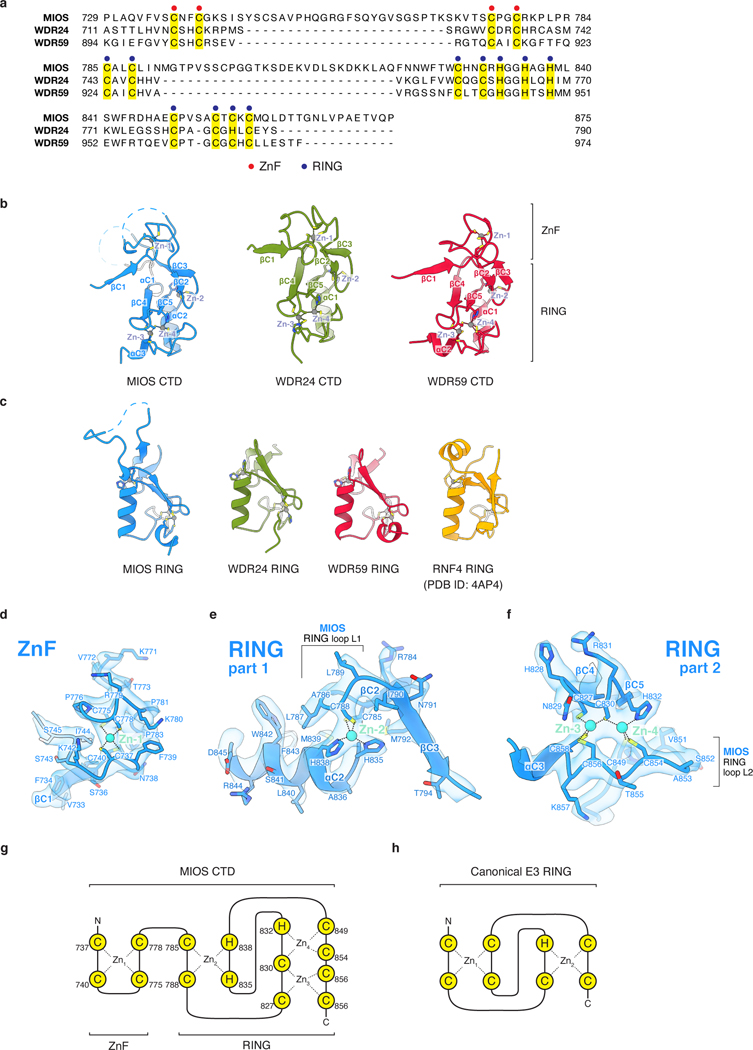
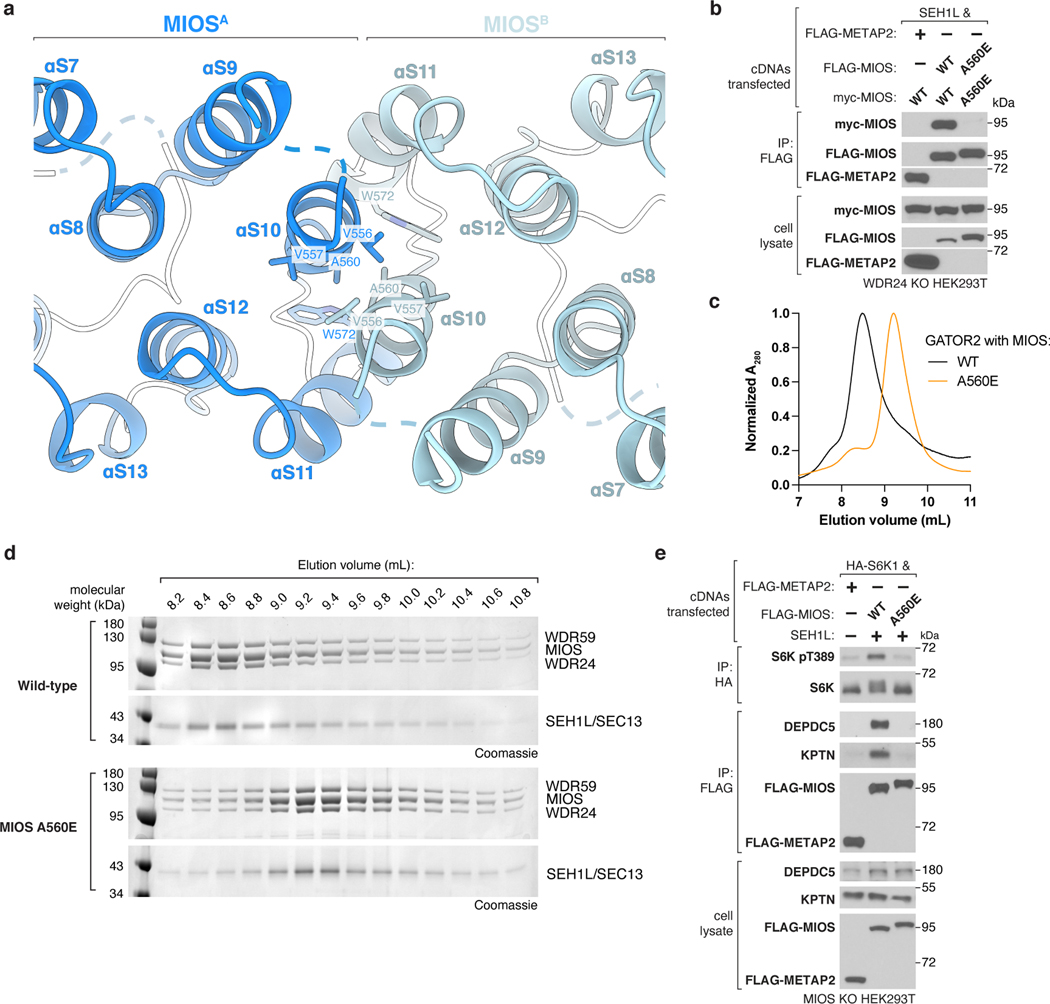
Each core subunit, WDR24, MIOS, and WDR59, contains a homologous, conserved CTD composed of a four-cysteine ZnF followed by a RING domain (Extended Data Fig. 4a–h). The eight GATOR2 RING domains assemble into four heterodimeric CTD-CTD junctions that hold the scaffold together: two between the C-termini of MIOS and WDR24 and two between those of MIOS and WDR59 (Fig. 2a, Extended Data Fig. 5a). A linker connects the α-solenoid of each core protein to its CTD, crossing over the dimerization partner CTD and forming a “forearm shake” between the two proteins (Fig. 2b). The resulting pair of cross-over linkers forms an anti-parallel β-sheet that supports two ZnF domains (one from each protomer in the junction) (Fig. 2c, Extended Data Fig. 4d). Each RING domain is sandwiched between the α-solenoid and the RING domain of its associated CTD partner. The paired RING domains interact directly via an anti-parallel four-stranded β-sheet, supported by hydrophobic interactions between bulky side chains at the subunit interface (Extended Data Fig. 5b, c). This mode of dimerization is distinct from that observed in dimeric RING E3 ligases13, and in fact constitutes a novel class of dimerization domains involving zinc fingers (confirmed by PDB and SCOP database searches with FATCAT)14.
Figure 2: MIOS interacts with WDR24 and WDR59 via dimerization of C-terminal Zn-binding domains (CTDs).
(a) The GATOR2 octagonal scaffold contains four CTD-CTD junctions, supported by interactions with adjacent SEH1L and SEC13 β-propellers.
(b) The CTDs of MIOS and WDR24 cross over one another and dimerize via a RING-RING interface, RING-solenoid interactions, and contacts between ZnFs and associated SEH1L propellers forming a “forearm shake,” as cartooned.
(c) Two views of the molecular model of the WDR24-MIOS CTD-CTD junction. Each CTD coordinates four zinc ions: one in the ZnF and three in the RING. Zinc ions are colored teal. For a detailed representation of the MIOS CTD, see Extended Data Figure 4.
(d) The MIOS CTD interacts with WDR24 and WDR59 CTDs. Anti-FLAG immunoprecipitates were prepared from MIOS-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed by immunoblotting for the indicated proteins. HA, hemagglutinin.
Data in (d) are representative of two independent experiments. For gel source data, see Supplementary Figure 1.
The RING domains exhibit the ‘cross-brace’ topology found in many RING E3 ubiquitin ligases (Extended Data Fig. 4c). But unlike such canonical RING domains, each GATOR2 RING coordinates three zinc ions using eleven side chains (Extended Data Fig. 4e–h). Like the GATOR2 RING domains, the RING domain of RAG1 also contains a binuclear zinc cluster15, but coordination of the additional zinc ion occurs at the canonical Zn1 site and requires side chains outside of the core RING. Thus, together, the ZnF and RING domains in each core component of GATOR2 form a C-terminal domain that folds through the coordination of four zinc ions (one in the ZnF and three in the RING).
Contacts between the zinc fingers and the adjacent SEH1L or SEC13 subunit provide further support to the CTD-CTD junctions: the WDR24 ZnF interacts with the MIOS-associated SEH1L (SEH1LMIOS) whereas the MIOS ZnF engages the SEH1L associated with WDR24 (SEH1LWDR24) (Fig. 2a,b). The two MIOS-WDR59 CTD-CTD junctions share a similar topology and make analogous contacts with neighboring subunits, in line with the domain architecture common to the three core proteins (Extended Data Fig. 5a). The extensive network of protein-protein contacts between CTDs, the adjacent α-solenoids, and the β-propellers of SEH1L and SEC13 suggest that these CTD-CTD junctions are likely stable interfaces.
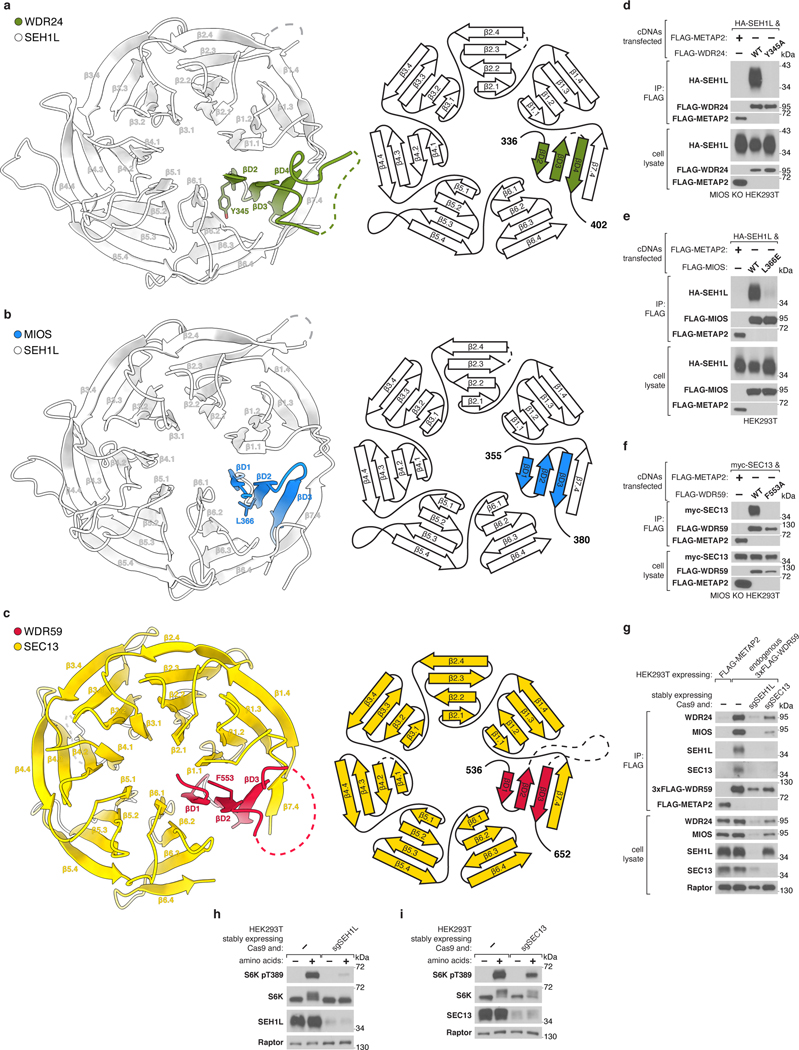
To understand the role of the GATOR2 CTDs in scaffold formation, we generated truncated variants of the core subunits expressing either (1) the CTD alone plus the preceding solenoidal α-helix (“CTD”) or (2) lacking the CTD (“∆CTD”). Consistent with our cryo-EM structure, the MIOS CTD was sufficient to bind corresponding CTDs of WDR24 and WDR59 but was unable to homodimerize (Fig. 2d). The isolated CTDs were sufficient to mediate MIOS-WDR24 and MIOS-WDR59 dimerization and, likewise, were required for these interactions (Extended Data Fig. 5d–i).
Deletion of the WDR24 or WDR59 CTDs resulted in the formation of complexes that contained all five components but were smaller than wild-type GATOR2, consistent with division of the scaffold (Extended Data Fig. 6a–d). Furthermore, GATOR2 lacking the MIOS CTD failed to incorporate either WDR24, WDR59 or SEC13 (Extended Data Fig. 6e). The CTDs of MIOS, WDR24, and WDR59 were all required for GATOR2 to interact with the GATOR1 and KICSTOR complexes and to complement the mTORC1 signaling defect of the respective knock-out cells (Extended Data Fig. 6f–h). Thus, the CTDs are critical structural elements required for GATOR2 assembly, its association with GATOR1 and KICSTOR, and amino acid signaling to mTORC1.
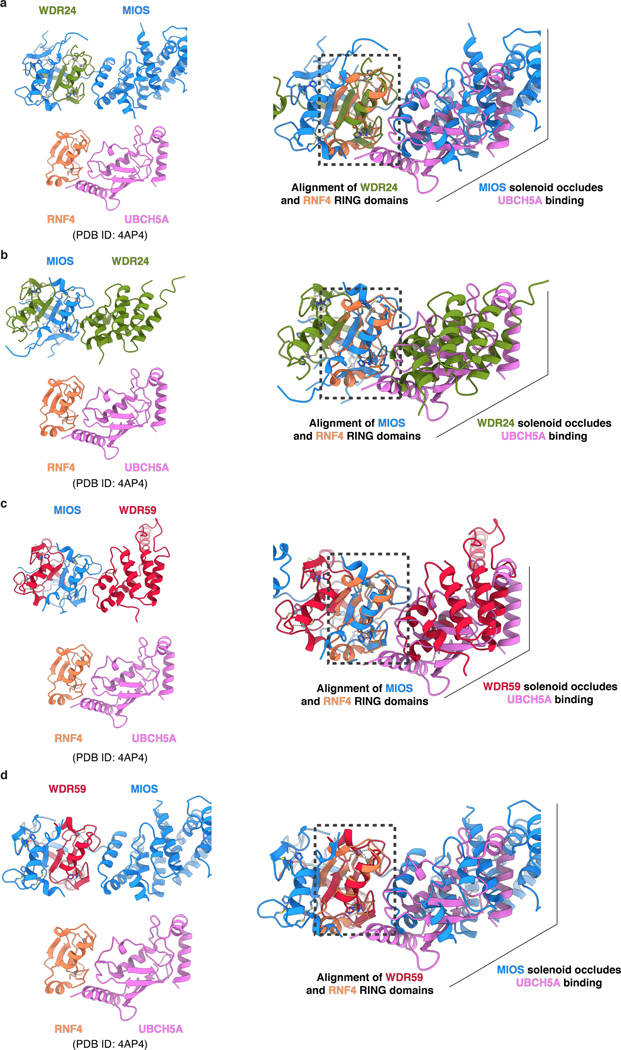
RING domains are typically found in E3 ubiquitin ligases, which catalyze the transfer of ubiquitin from an E2 enzyme to a substrate acceptor lysine13. Given the presence of multiple RING domains, it has been speculated that GATOR2 may contain E3 ligase activity16. RING domains bind ubiquitin-charged E2 using a well-defined, conserved interaction surface formed by the L1 and L2 zinc-coordinating loops and the central α-helix13. Structural alignment of each GATOR2 RING domain with the RING domain of the RNF4-UBCH5A co-structure17 revealed that the adjacent α-solenoid of a neighboring subunit in the GATOR2 scaffold completely occludes the putative E2-interacting surface of each GATOR2 RING (Extended Data Fig. 7a–d). As such, a RING-E2 interaction would result in a substantial structural clash, precluding any E2 from engaging GATOR2 without compromising the integrity of the complex.
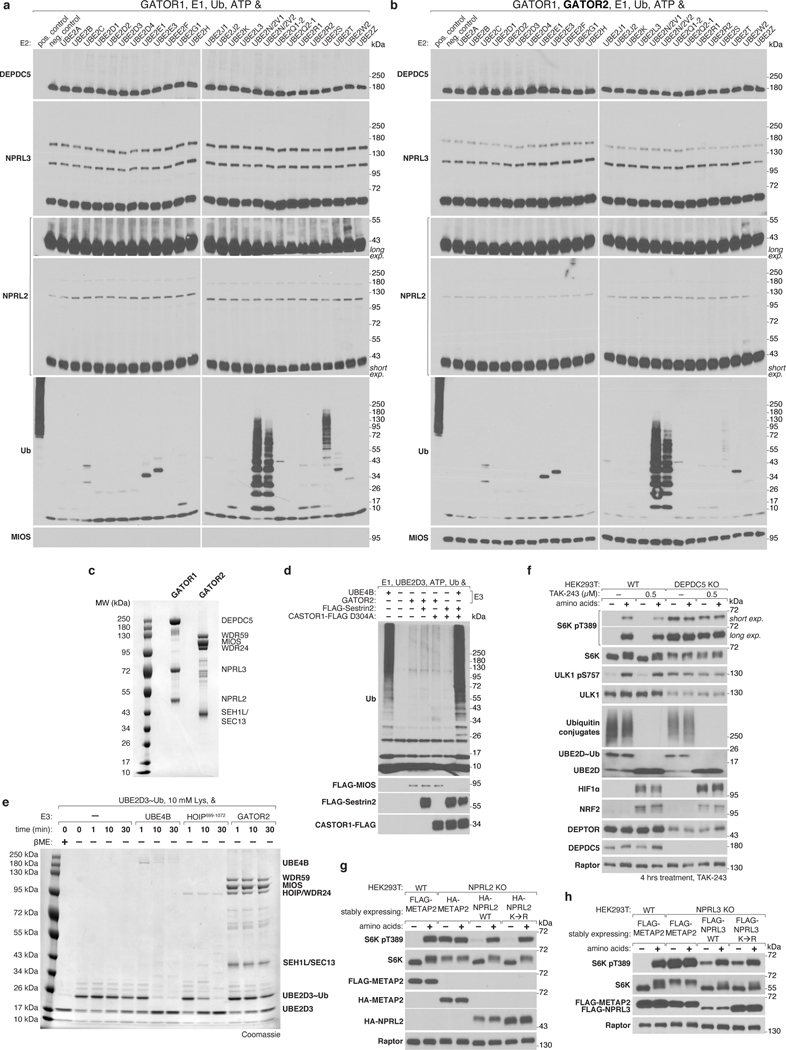
Despite the structural implausibility of GATOR2 having E3 ligase activity, we directly tested this by incubating GATOR2 with GATOR1, its most likely target, and a panel of E2 enzymes. We observed no GATOR2-dependent modification of GATOR1 and no meaningful GATOR2-dependent ubiquitin chain formation (Extended Data Fig. 8a–d). Likewise, GATOR2 did not promote ubiquitin discharge from an E2~Ub conjugate (Extended Data Fig. 8e). Treatment of cells with TAK-243, an inhibitor of the ubiquitination cascade that targets E1 enzymes, did not affect GATOR2-dependent activation of mTORC1 by amino acids despite disrupting E2-ubiquitin charging, ablating polyubiquitin chain accumulation, and stabilizing short-lived proteins such as HIF1α and NRF2 (Extended Data Fig. 8f). GATOR2 interacts with GATOR1 components NPRL2 and NPRL3, which contain its GAP activity12. We synthesized non-modifiable NPRL2 and NPRL3 constructs in which all the lysines were replaced with arginines and the native N-termini were obfuscated by epitope tags. Expression of these variants in knock-out cells restored amino acid sensitive mTORC1 signaling, indicating that lysine ubiquitination of these proteins is dispensable for amino acid sensing by mTORC1 (Extended Data Fig. 8g,h). Taken together, our structural and signaling data strongly argue against GATOR2-catalyzed ubiquitin transfer playing a role in mTORC1 activation.
α-Solenoids Circularize the Scaffold
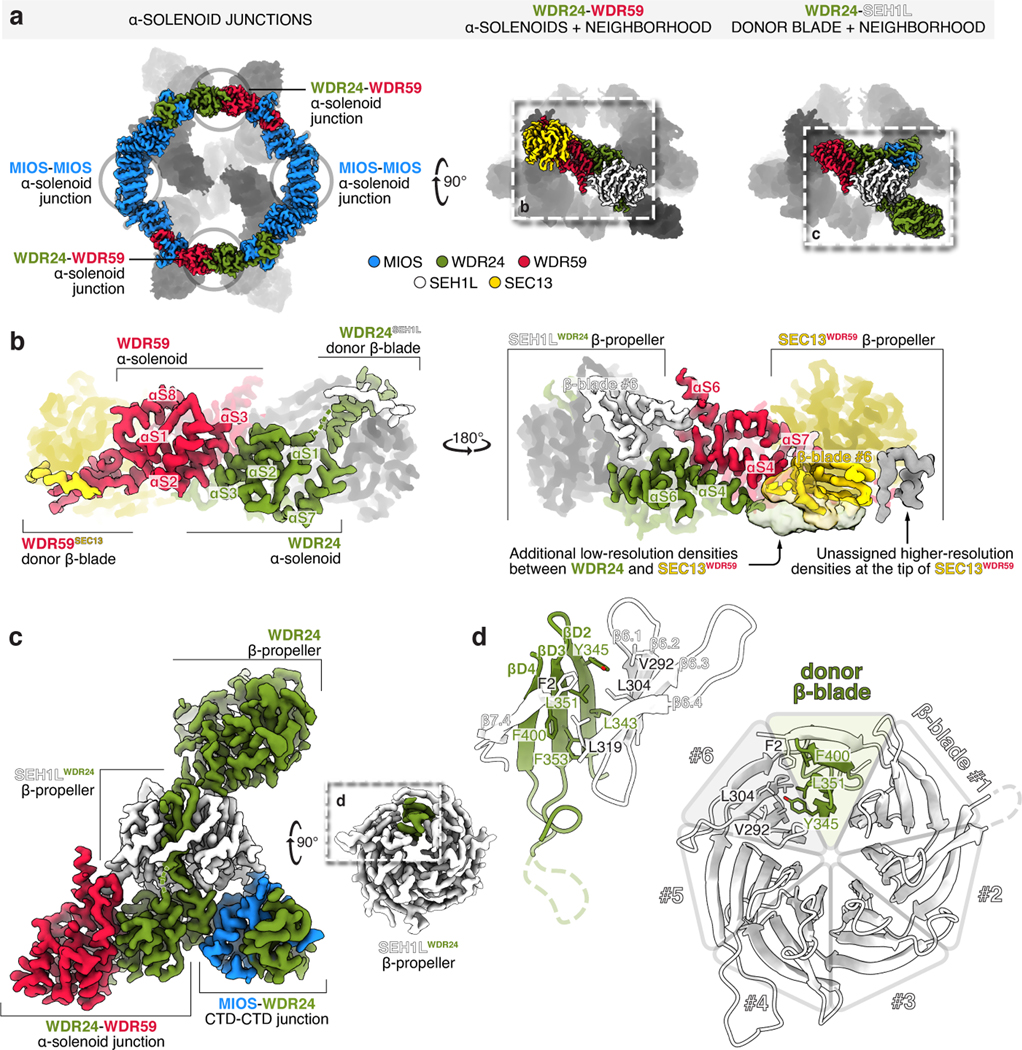
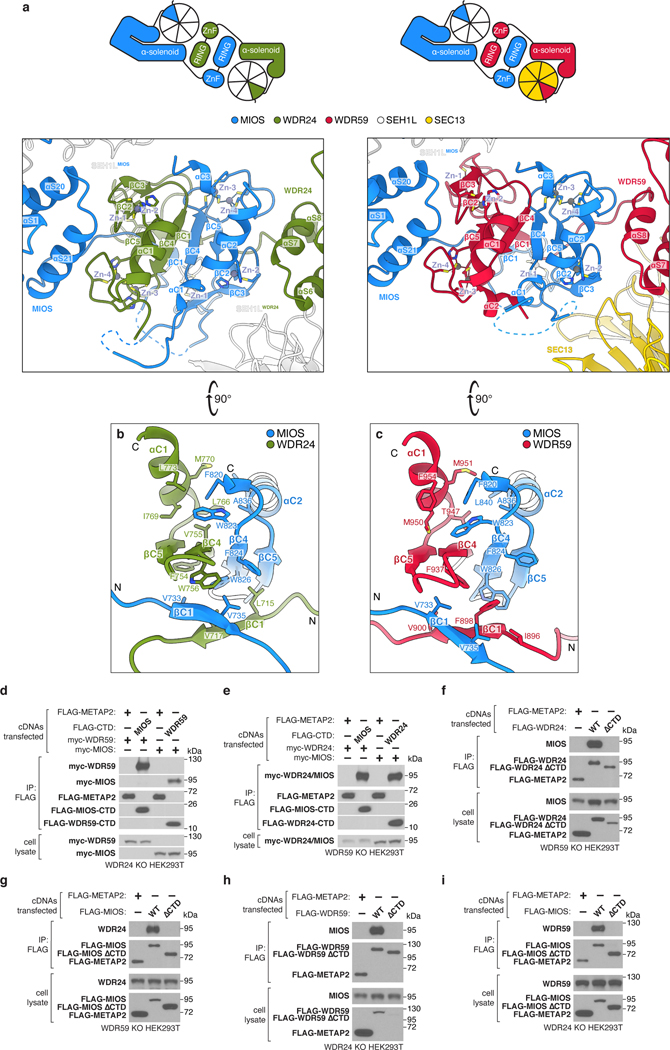
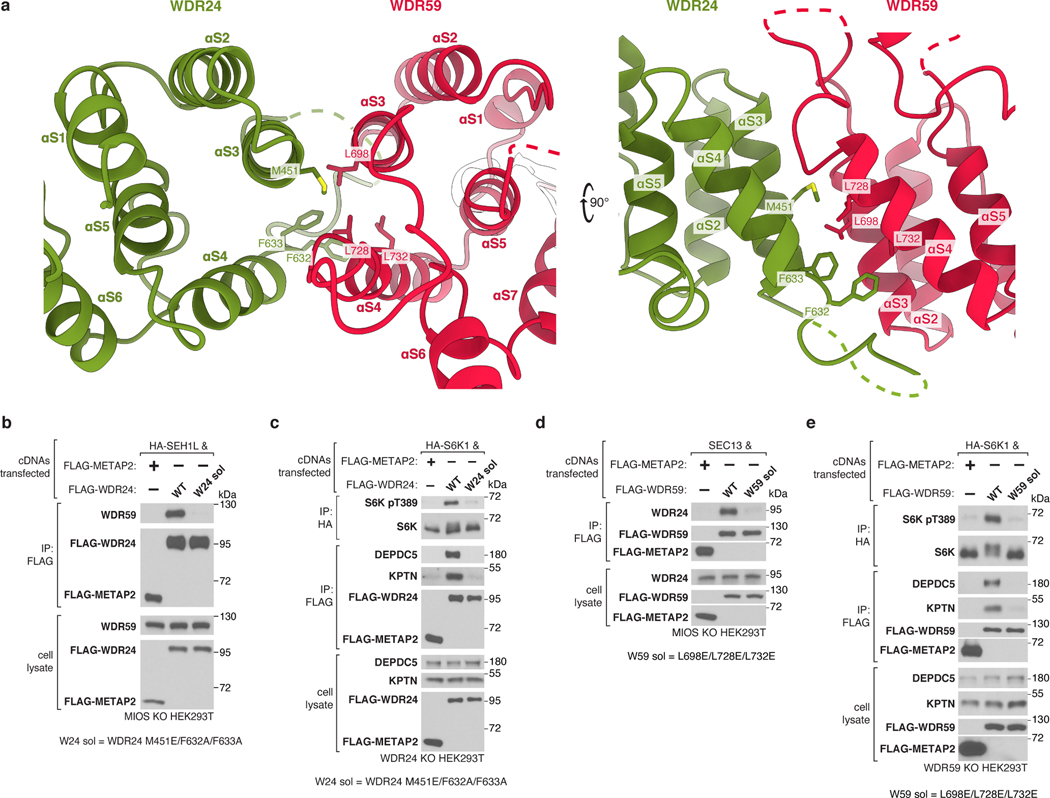
Completion of the GATOR2 scaffold requires two homomeric associations between the α-solenoid crown regions of MIOS subunits and two heteromeric interactions between those of WDR24 and WDR59 (Fig. 3a,b). Mutations at these α-solenoid interfaces on either WDR24 (M451E/F632A/F633A) or WDR59 (L698E/L728E/L732E) disrupted the WDR24-WDR59 interaction and GATOR2 containing these variants did not co-immunoprecipitate GATOR1 and KICSTOR or restore mTORC1 signaling in knock-out cells (Extended Data Fig. 9a–e).
Figure 3: α-solenoid junctions, supported by β-propellers of SEH1L and SEC13, complete the GATOR2 scaffold.
(a) Four α-solenoid junctions, stabilized by adjacent SEH1L and SEC13 β-propellers, complete the GATOR2 scaffold.
(b) Two views of the WDR24-WDR59 α-solenoid junction. The helical interface is supported by contacts between WDR59 and SEH1LWDR24 and between WDR24 and SEC13WDR59. For a detailed representation of the helical interface, see Extended Data Figure 9.
(c) WDR24 completes SEH1LWDR24 by β-blade donation. The incorporated SEH1L interacts with the WDR24 β-propeller, the WDR24 solenoidal domain, the WDR59 solenoidal domain, and the MIOS CTD.
(d) Detailed view of the β-blade donation from WDR24 to complete SEH1LWDR24. WDR24 contributes the three innermost strands of the seventh SEH1L propeller blade.
Likewise, substitution of a conserved Ala to Glu (A560E) in MIOS disrupted the homodimerization between MIOS subunits (Extended Data Fig. 10a,b). Purification of this GATOR2 variant revealed a subunit stoichiometry identical to that of the wild-type complex, yet it was smaller in size, indicating that this mutation partitioned GATOR2 into two equal halves (Extended Data Fig. 10c,d). GATOR2 containing MIOS A560E also failed to interact with GATOR1 and KICSTOR or restore mTORC1 activation in knock-out cells (Extended Data Fig. 10e). Collectively, these results underscore that the integrity of the GATOR2 scaffold is required for GATOR1 and KICSTOR binding and amino acid signaling to mTORC1.
Blade Donation Recruits SEH1L and SEC13
MIOS and WDR24 associate stoichiometrically with SEH1L while each copy of WDR59 interacts with SEC13 (Fig. 1c, Extended Data Fig. 2b–e). Both SEH1L and SEC13 form open six-bladed β-propellers. We found that each core GATOR2 component donates an extra β-blade to complete its associated SEH1L or SEC13 (Fig. 3c,d). Specifically, three β-strands contributed by MIOS/WDR24/WDR59 are belted inside the β-propeller by the N-terminal β-strand of either SEH1L or SEC13, which functions as the outermost strand in the seventh β-blade (Extended Data Fig. 11a–c). The donated β-blade interacts with the neighboring blades 1 and 6 of SEH1L or SEC13 via conserved, hydrophobic side chains, mutation of which destabilized the interaction (Extended Data Fig. 11d–f). This mode of a protein-protein interaction via blade donation is analogous to the recruitment of SEH1L and SEC13 to the nuclear pore complex via NUP85 and NUP145C or of SEC13 to COP-II coats by SEC31 and SEC16, suggesting an ancestral relationship between these complexes6,18–20.
In addition to buttressing the CTD-CTD junctions, SEH1L and SEC13 also contribute to the α-solenoid interfaces between WDR24 and WDR59. Loop β6.3-β6.4 of SEH1LWDR24 interacts with two WDR59 elements protruding from the WDR24-WDR59 α-solenoid junction: αS6 and loop αS3-αS4. On the opposite side of the WDR24-WDR59 α-solenoid junction, a large section of unmodeled density preceding αS4 of WDR24 interacts with the sixth and seventh blades of SEC13WDR59 (Fig. 3b). Thus, each SEH1LMIOS bridges the MIOS-WDR24 and MIOS-WDR59 CTD-CTD junctions while each SEH1LWDR24 and SEC13WDR59 rigidify the scaffold through contacts with all three core GATOR2 proteins (Fig. 3c). In line with this key structural role, partial depletion of either SEH1L or SEC13 destabilized the entire complex and impaired mTORC1 activation by amino acids (Extended Data Fig. 11g–i).
β-propellers engage sensors and GATOR1
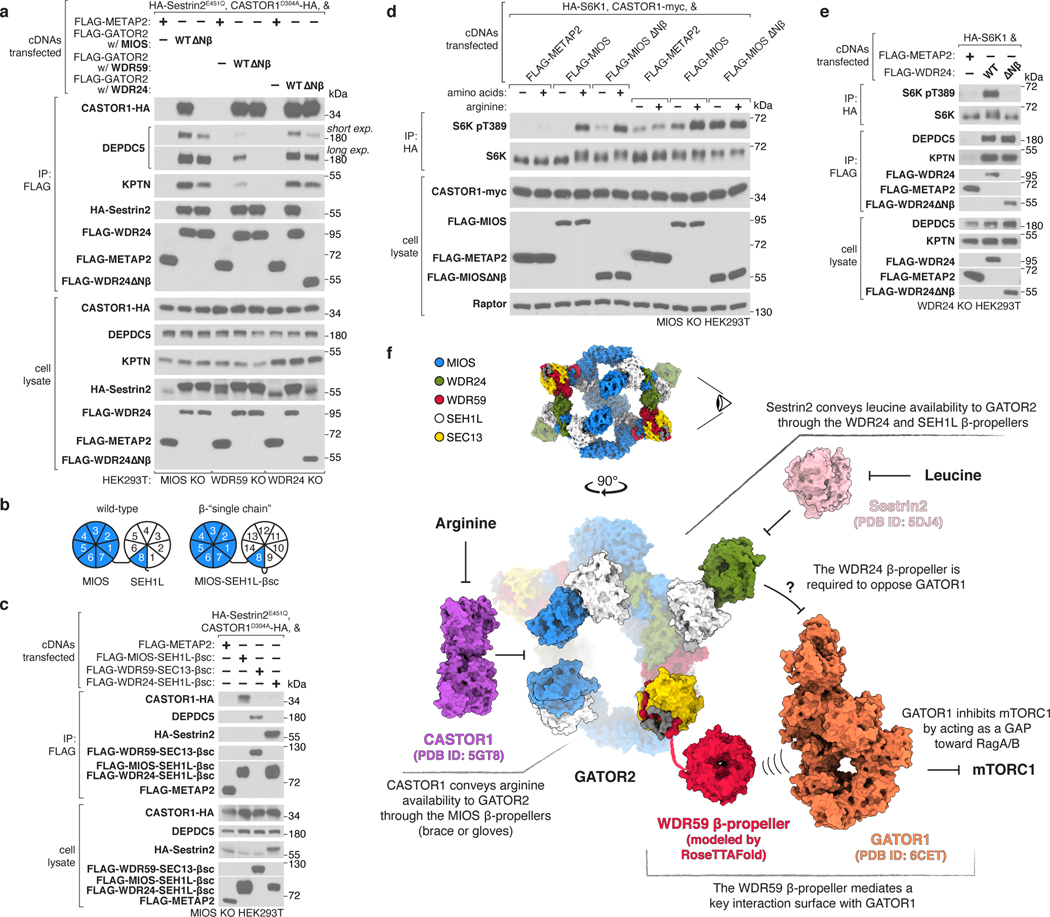
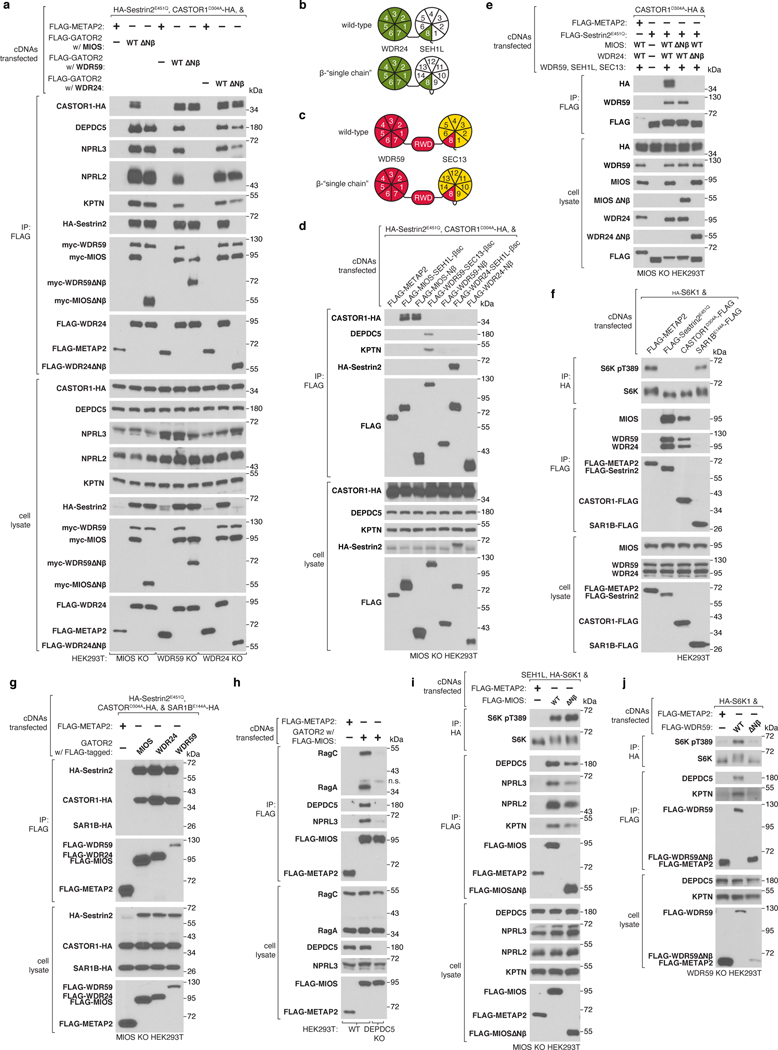
Given the spatial arrangement of the eight N-terminal β-propellers belonging to the core GATOR2 components, we hypothesized that they may contribute to the interactions between GATOR2 and other components of the amino acid sensing pathway upstream of mTORC1. Indeed, we found that the MIOS β-propeller is specifically required for GATOR2 to associate with CASTOR1, the WDR59 β-propeller necessary to bind GATOR1 and KICSTOR, and the WDR24 β-propeller essential for GATOR2 to co-immunoprecipitate Sestrin2 (Fig. 4a, Extended Data Fig. 12a).
Figure 4: The GATOR2 β-propellers receive and transmit amino acid availability.
(a) The β-propellers of MIOS, WDR59, and WDR24 are required for the interactions of GATOR2 with CASTOR1, GATOR1 and KICSTOR, as well as Sestrin2, respectively. Anti-FLAG immunoprecipitates were prepared from MIOS-deficient, WDR59-deficient or WDR24-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed by immunoblotting for the indicated proteins. CASTOR1 D304A and Sestrin2 E451Q mutants do not bind arginine or leucine, respectively, and bind GATOR2 irrespective of amino acid availability. HA, hemagglutinin.
(b) Schematic comparing MIOS Nβ and SEH1L β-propeller organization in wild-type and β-single chain (βsc) constructs.
(c) The GATOR2 single chain β-propeller pairs of MIOS-SEH1L, WDR59-SEC13, and WDR24-SEH1L are sufficient to interact with CASTOR1, GATOR1, and Sestrin2, respectively. Anti-FLAG immunoprecipitates were prepared from MIOS-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (a).
(d) MIOS Nβ is necessary for arginine deprivation, but not total amino acid deprivation, to inhibit mTORC1 signaling. MIOS-deficient HEK293T cells that transiently expressed the indicated cDNAs were starved of either all amino acids or arginine for 60 min and restimulated with amino acids or arginine for 15 min prior to harvest. Anti-HA immunoprecipitates were prepared and analyzed as in (a).
(e) WDR24 Nβ is required for activation of mTORC1 but not for the GATOR2-GATOR1 or GATOR2-KICSTOR interactions. Anti-FLAG and anti-HA immunoprecipitates were prepared from WDR24-deficient HEK293T cells that transiently expressed the indicated cDNAs and analyzed as in (a).
(f) Schematic of the integration of amino acid availability by GATOR2 β-propellers. The eye symbol indicates the viewing angle of GATOR2 in the bottom panel. The WDR59 Nβ was modeled using RoseTTAFold28. Models for CASTOR1 (PDB ID: 5GT8)29, Sestrin2 (PDB ID: 5DJ4)30, and GATOR1 (PDB ID: 6CET)12 were obtained from the PDB.
Data in (a), (c), (d), and (e) are representative of two independent experiments. For gel source data, see Supplementary Figure 1.
To demonstrate the sufficiency of these interaction surfaces, we generated “single chain” β-propeller dimers by fusing the sequences of β-propeller pairs into a single polypeptide chain, such that each construct contained a core GATOR2 N-terminal β-propeller, a donor β-blade and its corresponding SEH1L or SEC13 subunit (Fig. 4b, Extended Data Fig. 12b,c). We found that the MIOS-SEH1L single chain propeller dimer interacted with CASTOR1, the WDR59-SEC13 dimer with GATOR1 and KICSTOR, and the WDR24-SEH1L dimer with Sestrin2, thereby establishing these domains as distinct, modular surfaces that directly engage other components of the nutrient sensing machinery (Fig. 4c, Extended Data Fig. 12d). The MIOS β-propeller was sufficient to associate with CASTOR1, indicating that SEH1L is dispensable for this interaction but not for that between the WDR24 β-propeller and Sestrin2 (Extended Data Fig. 12d).
As Sestrin2 and CASTOR1 bind different GATOR2 components, we evaluated whether these nutrient sensors can simultaneously occupy a single GATOR2 particle. Indeed, GATOR2 was necessary to mediate the interaction between Sestrin2 and CASTOR1 (Extended Data Fig. 12e). We also examined the reported association between GATOR2 and SAR1B, which has been suggested to be an alternative leucine sensor for the mTORC1 pathway21. We did not detect an interaction in co-immunoprecipitation experiments and SAR1B overexpression did not inhibit mTORC1 signaling (Extended Data Fig. 12f,g). We were also unable to detect a stable, GATOR1-independent interaction between GATOR2 and the Rag GTPases (Extended Data Fig. 12h).
Because the β-propellers directly interact with the amino acid sensors, GATOR1, and KICSTOR, we hypothesized that they contribute to the regulation of mTORC1 by nutrients. Whereas the MIOS N-terminal β-propeller was not required for the inhibition of mTORC1 caused by total amino acid starvation, it was necessary for mTORC1 signaling to respond to arginine deprivation, establishing the CASTOR1-MIOS Nβ interaction as one path through which mTORC1 senses the absence of arginine (Fig. 4d, Extended Data Fig. 12i). Consistent with its role in coupling GATOR2 to GATOR1 and KICSTOR, expression of GATOR2 lacking the WDR59 β-propeller failed to rescue mTORC1 signaling in WDR59 knock-out cells (Extended Data Fig. 12j).
Surprisingly, while the β-propeller of WDR24 is necessary for mTORC1 activation, it is not required for the association between GATOR2 and GATOR1 or KICSTOR, indicating that its essential function is not to link the complexes together (Fig. 4e). The WDR24 β-propeller is also the Sestrin2-binding site, but despite extensive mutagenesis efforts, we were unable to generate a WDR24 variant that restores mTORC1 activity to WDR24 knock-out cells and also cannot bind Sestrin2. This finding suggests that the WDR24 β-propeller mediates an essential GATOR2 function and that Sestrin2 inhibits it, likely by directly interfering with the same surface of the propeller critical for the essential function. Thus, GATOR2 uses the MIOS and WDR24 β-propellers to receive inputs from the cytosolic amino acid sensors and the WDR24 and WDR59 propellers to transduce amino acid availability to mTORC1 (Fig. 4f).
Discussion
As a receptor for the cytosolic amino acid sensors Sestrin1/2 and CASTOR1, GATOR2 plays a pivotal role in the regulation of mTORC1 by nutrients. Despite this, its architecture and the identities of its functionally relevant domains have remained elusive. Here, we present the cryo-EM structure of human GATOR2 and reveal that this nutrient sensing hub adopts an open, cage-like architecture built on a continuous, octagonal scaffold. We find that the GATOR2 RING domains form portions of larger zinc-binding domains that dimerize to construct the scaffold, impairing any potential role of the RING domains in ubiquitin ligase activity. Instead, the GATOR2 β-propellers mediate distinct interactions with components of the amino acid sensing machinery, providing a conceptual framework for understanding how GATOR2 integrates and communicates upstream signals to mTORC1.
The combination of β-propeller and α-solenoid domains as building blocks of large multiprotein assemblies is common to components of membrane coating complexes, including COP-II vesicle coats, the nuclear pore complex, and the HOPS (Homotypic fusion and Protein Sorting) and CORVET (class C cORe Vacuole/Endosome Tethering) complexes, which has led to the suggestion that these proteins share a common protocoatomer ancestor22. These complexes form branched, lattice-like architectures that spread across membranes or encircle vesicles. Unlike the components of the nuclear pore complex and COP-II coats, the core GATOR2 proteins contain C-terminal zinc binding domains critical for complex formation. Indeed, RING dimerization may constrain the diversity and size of architectures into which the GATOR2 components can assemble. Notably, the HOPS complex components VPS18 and VPS41 both contain C-terminal RING domains that mediate complex assembly, suggesting RING dimerization may be a general mechanism that specifies the architecture of protocoatomer-derived complexes23. The evolutionary relationship between GATOR2 and membrane coating complexes also raises questions of how GATOR2 interacts with the lysosomal surface and whether it may regulate other aspects of lysosomal function. For instance, exposed basic or lipophilic surfaces may contribute to membrane interaction (Extended Data Fig. 3a–c). Intriguingly, one report suggests that WDR24 controls the lysosomal pH independently of its role in regulating mTORC1 activity24.
GATOR2 uses distinct surfaces to simultaneously engage Sestrin2 and CASTOR1, providing a structural basis for the parallel detection and integration of leucine and arginine availability by mTORC1. CASTOR1 forms a dimer in solution and requires two intact GATOR2-binding sites to convey arginine deficiency to mTORC1, suggesting it likely binds GATOR2 through a bipartite interaction25. The approximation of two MIOS β-propellers in either the “brace” or the “gloves” offers adjacent, equivalent surfaces capable of interacting with CASTOR1. Interestingly, we found that the MIOS β-propeller is not absolutely required for mTORC1 activation. Deletion of the MIOS β-propeller, however, partially destabilizes the interaction between GATOR2, GATOR1, and KICSTOR, indicating that these β-propellers may contribute to auxiliary interactions between the complexes. Functional coupling of GATOR2 to GATOR1 and KICSTOR is critical for proper nutrient sensing as disruption of these interactions compromises mTORC1 activation26. Simultaneous binding of CASTOR1 to two MIOS β-propellers may allosterically modulate GATOR2 in a manner that influences the interaction between GATOR complexes and renders GATOR2 a less effective opponent of GATOR1.
In contrast, the WDR24 β-propeller contributes to the Sestrin2 binding site and is uniquely dispensable for the stable association of GATOR2 with GATOR1 and KICSTOR but absolutely required for mTORC1 activation, suggesting that Sestrin2 likely directly interferes with a key function of GATOR2. WD40 β-propellers, like the WDR24 β-propeller, scaffold protein-protein interactions and have not been associated with enzymatic activity27. Consistently, the expression levels of GATOR1 and GATOR2 are approximately equal in the cell (Extended Data Fig. 1b), suggesting that GATOR2 does not function in a catalytic fashion on GATOR1. Accordingly, the WDR24 β-propeller may make multiple mutually exclusive interactions: to Sestrin1/2 in the absence of leucine or, in its presence, a productive interaction that opposes GATOR1. Among GAPs, GATOR1 is distinct in that it engages the Rag GTPases in at least two distinct manners: a functional and an ‘inhibitory’ mode12. GATOR2 may function to regulate the relative frequency of these two states, thereby transducing amino acid availability to the Rag GTPases via GATOR1. Further structural studies are needed to reveal the nature of the GATOR2-GATOR1 interaction and the manner in which the GATOR2 β-propellers contribute to nutrient sensing on the lysosomal surface.
Materials
Reagents were obtained from the following sources: antibodies against S6K pT389 (9234), S6K (2708), MIOS (13557), WDR59 (53385), NPRL2 (37344), RagA (4357), RagC (3360), ULK1 pS757 (6888), ULK1 (8054), ubiquitin (3933), HIF-1α (14179), DEPTOR (11816), Myc epitope tag (2278), Flag epitope tag (14793), HA epitope tag (3724), HRP-linked anti-mouse secondary antibody (7076) and HRP-linked anti-rabbit secondary antibody (7074) were from Cell Signaling Technology; antibody against ubiquitin-conjugated proteins (FK2) was from Millipore Sigma (04–263); antibody against UBE2D was from Santa Cruz Biotechnology (sc-166278); antibodies against SEH1L (ab218531) and DEPDC5 (ab185565, ab213181) were from Abcam; antibodies against SEC13 (15397–1-AP), WDR24 (20778–1-AP), KPTN (16094–1-AP) and NRF2 (16396–1-AP) were from Proteintech; Flag-M2 antibody used for preparing anti-Flag magnetic beads was from Millipore Sigma (F1804); antibody against NPRL3 was from Novus Biologicals (NBP1–88447); antibody against Raptor (09–217) was from EMD Millipore. InstantBlue Coomassie Protein Stain was from Abcam; Anti-Flag M2 affinity gels, ATP, N-ethylmaleimide, and amino acids were from Millipore Sigma; DMEM, FreeStyle 293 Expression Medium, inactivated fetal serum (IFS), SYPRO Ruby Protein Stain, Dynabeads M-270 Epoxy, and anti-HA magnetic beads were from Thermo Fisher Scientific; XtremeGene9, PhosSTOP, and Complete Protease Cocktail were from Roche; amino acid-free RPMI was from US Biologicals; PEI MAX 40K was from Polysciences; E2Select Ubiquitin Conjugation Kit, UBE1, UBE2D3, ubiquitin charged UBE2D3, HOIP catalytic domain, UBE4B, ubiquitin, and ubiquitin conjugation reaction buffer were obtained from R&D Systems; and TAK-243 was obtained from Chemietek.
Cell lines and tissue culture
Adherent HEK293T cells were cultured in DMEM (Thermo Fisher Scientific) with 10% IFS (Thermo Fisher Scientific) and 4.5 g/L glucose containing 2 mM GlutaMAX (Thermo Fisher Scientific), 100 IU/mL penicillin, and 100 μg/mL streptomycin. Adherent cell lines were maintained at 37°C and 5% CO2. Suspension-adapted HEK293T cell lines were grown in FreeStyle 293 (Thermo Fisher Scientific) supplemented with 1% IFS, 100 IU/mL penicillin, and 100 μg/mL streptomycin. Suspension cells were grown in a Multitron Pro shaker operating at 37°C, 8% CO2, 80% humidity, and 125 rpm. All cell lines were obtained from ATCC (American Type Culture Collection) and validated and tested for mycoplasma.
Transfections, cell lysis, and immunoprecipitation experiments
To harvest samples, cells were washed once with ice-cold PBS and then lysed with lysis buffer (1% Triton X-100, 40 mM HEPES pH 7.4, 10 mM β-glycerol phosphate, 10 mM pyrophosphate, 2.5 mM MgCl2) and 1 tablet of EDTA-free protease inhibitor (Roche) per 25 mL buffer. Cell lysates were clarified by centrifugation at 21,000 x g at 4°C for 10 min.
For anti-Flag immunoprecipitations, magnetic beads bound to antibody recognizing the Flag epitope tag were prepared in-house by coupling Dynabeads M-270 Epoxy (Thermo Fisher Scientific) to Flag M2 antibody (Millipore Sigma) as previously described31. For anti-HA immunoprecipitations, anti-HA coupled magnetic beads (Thermo Fisher Scientific) were used. Beads were washed three times prior to use with Triton X-100 lysis buffer, and were then incubated with the supernatant of each clarified lysate for 1 h at 4°C. Each immunoprecipitation used 10 μL of anti-Flag Dynabeads or 30 μL of anti-HA magnetic beads. Following immunoprecipitation, beads were washed one time with Triton X-100 lysis buffer and two times with Triton X-100 lysis buffer supplemented to contain 150–500 mM NaCl. Immunoprecipitated proteins were denatured by addition of SDS-PAGE sample buffer and boiling for 5 min at 95°C. Immunoprecipitated proteins were then resolved by 4–20% SDS-PAGE before analysis by immunoblotting.
For experiments requiring transfection of DNA into HEK293T cells, 2×106 cells were plated in 10-cm culture dishes, or 5×106 cells were plated in 15-cm culture dishes. 24 h later, cells were transfected with the appropriate pRK5-based cDNA expression plasmids using the polyethylenimine method, as previously described32. The total amount of plasmid DNA in each transfection was normalized to 5 μg with empty pRK5 (10 cm plates) or 20 μg of empty pRK5 (15 cm plates). 36 h following transfection, cells were lysed as described above.
For experiments which required amino acid starvation, cells were incubated in amino acid-free RPMI for 60 min, as previously described3. To restimulate cells following starvation, an amino acid mixture prepared from individual powders of amino acids (Millipore Sigma) was added to cell culture media for 15 min3. Starvations for individual amino acids were performed similarly.
For experiments which required inhibition of the ubiquitination cascade, cells were treated with 0.5 μM TAK-243 (UAE inhibitor; Chemietek), or DMSO as a control, for a total of 4 h (including amino acid deprivation) prior to harvest. Cells were washed once in ice-cold PBS and harvested in RIPA (1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris pH 8.0, 150 mM NaCl) supplemented with 10 mM N-ethylmaleimide, 1 tablet of EDTA-free protease inhibitor (Roche) per 25 mL buffer, and 1 tablet of PhosSTOP per 10 mL buffer. Samples were collected in sample buffer prepared without reducing agent to preserve the thioester linkage between UBE2D3 and ubiquitin.
Protein expression and purification
For large scale purification, 5 L of suspension-adapted HEK293T cells grown in FreeStyle 293 Expression Medium (Thermo Fisher Scientific) supplemented with 1% IFS were transiently transfected with cDNAs encoding N-terminally Flag-tagged MIOS (Addgene #184561) and tag-free WDR24 (Addgene #154213), WDR59 (Addgene #154214), SEH1L (Addgene #154216), and SEC13 (Addgene #154215) on the pRK5 vector. Cells were transfected at a density of 2×106/mL with 1 mg total cDNA and 3 mg PEI MAX 40k (Polysciences) per L culture. 48 h after transfection, cells were harvested, washed in ice-cold PBS, and then lysed in Triton Lysis Buffer (TLB; 1% Triton X-100, 50 mM HEPES pH 7.4, 100 mM NaCl, 50 mM arginine, 50 mM glutamic acid, 2.5 mM MgCl2) with EDTA-free protease inhibitor cocktail (Roche; 1 tablet per 25 mL buffer) and 1 mM ATP (100 mL of lysis buffer was used per L cell culture). The lysate was cleared by ultracentrifugation at 50,000 x g for 20 minutes. Pre-washed anti-Flag M2 affinity gel (Millipore Sigma) was added to the clarified lysate (2 mL slurry per L culture) and incubated for 2 h at 4°C on a nutator. The beads were washed once in TLB, twice in TLB supplemented with 400 mM NaCl, once in CHAPS buffer (CB; 0.1% CHAPS, 50 mM HEPES pH 7.4, 150 mM NaCl, 50 mM arginine, 50 mM glutamic acid, 2 mM MgCl2), once in CB supplemented with 20 mM MgCl2 and 5 mM ATP, and once again in CB. The GATOR2 complex was eluted from the beads by competitive elution in CB supplemented with 0.5 mg/ml FLAG peptide (sequence DYKDDDDK). Eluate was collected with bead separator columns, concentrated with a 100 kDa MWCO centrifugal filter (Millipore), and centrifuged at 21,000 x g for 10 min at 4°C. Further purification was performed by size-exclusion chromatography (SEC) using a TSKgel G4000SWxl column (Tosoh) pre-equilibrated in CB supplemented with 2 mM DTT. Elution fractions were resolved by SDS-PAGE (4–12% Bolt gels, Thermo Fisher Scientific) and stained with InstantBlue Coomassie Protein Stain (Abcam). Pure protein fractions were pooled, concentrated by centrifugal ultrafiltration, supplemented with 10% glycerol, and snap frozen in liquid nitrogen before storage at −80°C. For analysis by cryo-EM, GATOR2 samples were prepared immediately following SEC. Purified GATOR2 was largely free of contaminants except for minor amounts of the CCT chaperonin complex, which assists in folding WD40 β-propellers33.
For small scale purification of GATOR2 complexes used in analytical SEC experiments (Extended Data Fig. 6a,b and 10c,d), 500 mL of transfected cell culture gave yields sufficient for analysis. GATOR1 was expressed and purified as previously described and both Sestrin2 and CASTOR1 D304A mutant were expressed and purified using the same protocol12.
Purification and analysis of endogenous GATOR2
Suspension-adapted HEK293T cells that express endogenously-tagged WDR5910 were pelleted, rinsed in PBS, and then resuspended in RPMI lacking amino acids for 60 min. Cells were restimulated with an amino acid mixture for 15 minutes. The cell cultures were harvested and the GATOR2 complex was isolated as described above. Elution fractions from SEC were separated by 4–12% SDS-PAGE and analyzed by immunoblotting. The peak elution fractions were separated by 4–12% SDS-PAGE and the gel was stained with SYPRO Ruby to examine the composition and stoichiometry of the endogenous protein complex.
In vitro ubiquitination assays
In vitro ubiquitination assays were performed using the E2Select Ubiquitin Conjugation Kit (R&D Systems) following the manufacturer’s instructions. These reactions contained 500 nM GATOR1 and, where indicated, 250 nM GATOR2. In vitro ubiquitination assays containing Sestrin2 and CASTOR1 were performed with 100 nM UBE1 (R&D Systems), 1 μM UBE2D3 (R&D Systems), 1 mg/mL Ubiquitin (R&D Systems), 50 nM GATOR2 or UBE4B (R&D Systems), 2.5 μM Sestrin2, 2.5 μM CASTOR1 D304A, 2 mM ATP in Ubiquitin Reaction Conjugation Buffer (R&D Systems). All reactions were incubated at 37°C for 1 h before quenching with sample buffer. Per the manufacturer, “the positive control wells contain an E3 Ligase (Itch), substrate (S5a), and compatible E2.” Reactions were separated by 4–20% SDS-PAGE and analyzed by immunoblotting.
Ubiquitin discharge assays
Ubiquitin charged-UBE2D3 (R&D Systems; 6.25 μM final) was incubated with 10 mM free lysine and 100 nM of either UBE4B (R&D Systems), HOIP catalytic domain (residues 699–1072), or GATOR2 at 22°C. All reaction components were diluted in ubiquitin conjugation reaction buffer (R&D Systems). Reactions were quenched with sample buffer without reducing agent to preserve the E2~Ub thioester linkage. Reactions were separated by SDS-PAGE (4–12% Bolt gels, Thermo Fisher Scientific) and stained with InstantBlue Coomassie Protein Stain (Abcam).
Electron cryomicroscopy
Cryo-EM specimens were prepared with a Vitrobot Mark IV (Thermo Fisher Scientific). 3 μL of 3 mg/mL GATOR2 complex was applied to glow-discharged gold 300 square mesh Quantifoil R 1.2/1.3 holey carbon grids (Quantifoil). The grids were then blotted from both sides for 3 s at 95% humidity, before being plunge-frozen in liquid ethane.
The resulting GATOR2 grids were screened for favorable ice thickness and particle distribution on a Talon Arctica electron microscope (Thermo Fisher Scientific) at MIT.nano, operated at 200 kV. A preliminary dataset of 873 movies was collected with a Falcon III direct detection camera (Thermo Fisher Scientific) in linear mode. The total dose on the specimen was 54 e- per Å2 fractionated over 40 frames with a calibrated pixel size of 1.6 Å.
Following initial reconstructions of the GATOR2 complex (see below), two high-quality GATOR2 datasets were collected using two different Titan Krios electron microscopes (Thermo Fisher Scientific) with Gatan K3 direct detection cameras at the end of BioQuantum energy filters (slit width 20 eV), at: (1) the University of Massachusetts School of Medicine (UMass), and (2) MIT.nano. Specific microscope settings used for data collection are listed in Supplementary Information Table 1. Fully automated data collection was carried out using either SerialEM34 (UMass) or EPU (MIT.nano) operating software.
Image processing — preliminary GATOR2 reconstitution
Movies recorded with a Talos Arctica microscope (MIT.nano) were corrected for drift using Relion’s MotionCor2 implementation35,36, and contrast transfer function (CTF) parameters were determined using GCTF37. An initial subset of 500 particles was selected manually, and used inside SPHIRE-crYOLO to train a model for automated particle picking38.
A search box size of 192 pixels, corresponding to 307 Å, yielded a total of 97,265 high-confidence GATOR2 particles. After extraction (288 pixels / 461 Å), these particles were used in reference-free 2D classifications in Relion39. The resulting 2D classes allowed for the generation of an initial 3D reconstruction of GATOR2 de novo, which was then used as a reference for classifications in 3D. A final set of 25,942 particles was auto-refined in Relion, producing a low-resolution (9.2 Å) map of GATOR2, which then served as the starting point for the subsequent higher-resolution cryo-EM work (see below).
Image processing — global GATOR2 reconstitution
Data processing workflow of the final GATOR2 reconstitution is illustrated in Supplementary Fig. 2. Two large movie datasets recorded with Titan Krios microscopes (UMass: 4,103 and MIT.nano: 28,792) were corrected for drift using Relion’s MotionCor2 implementation, and contrast transfer function (CTF) parameters were determined using GCTF. For each motion-corrected micrograph dataset, we applied two particle search models in SPHIRE-crYOLO: (1) a general model pre-calculated by the software developers, and (2) a trained model, based on a manually-picked subset of 500 GATOR2 particles. The resulting particle sets (UMass: 472,167 and 441,452; MIT.nano: 3,131,961 and 3,192,293) were extracted and downscaled for further processing in Relion. An initial round of referenced 2D classification (based on the preliminary 9.2 Å map of GATOR2), followed by reference-free classifications in 2D, eliminated contaminating non-GATOR2 particles (including those of the CCT chaperonin) and those corresponding to partially disassembled GATOR2 complexes. The remaining particles were re-classified in 3D, and the best classes from different crYOLO sets (“general” and “trained”) were combined using a strict distance cut-off (200 Å) to discard duplicates. The resulting clean sets of particles (UMass: 210,387; MIT.nano: 567,597) were re-extracted at full size, and used for initial refinement and reconstruction that led to maps resolved at 5.8 Å (UMass dataset) and 4.6 Å (MIT.nano dataset). Iterative cycles of CTF and aberration refinements in Relion40 (per-particle defocus, per-micrograph astigmatism, beam-tilt, higher-order aberrations) improved the maps significantly. Further alignment of the GATOR2 map to C2 symmetry and correction of per-particle motion in Relion41 produced maps extending to a resolution of 4.50 Å (UMass) and 4.07 Å (MIT.nano). At this point, the two particle sets (UMass and MIT.nano) were merged after adjusting for differences in pixel size between them42. Following additional refinements of CTF parameters in Relion, the combined dataset of 784,651 particles reached 3.96 Å resolution. This particle set was further refined in cryoSPARC43 using iterative cycles of non-uniform refinement and CTF refinement — until convergence. The final map obtained from these particles was estimated at 3.66 Å resolution (with C2 symmetry applied), according to ‘gold standard’ Fourier shell correlation (FSC) of 0.143 (Supplementary Fig. 2,3c). Local resolution was estimated in cryoSPARC to extend from 2.5 to 6.5 Å (Supplementary Fig. 3c).
Image processing — building a composite map of GATOR2
The rather broad resolution span of the global GATOR2 map suggested a high degree of motion within the complex. To analyze the extent of that motion, we performed variability analysis in cryoSPARC44, which revealed that these motions were not reflective of specific conformational changes within the GATOR2 structure, but instead they were the result of individual subunit loss across various sections of the complex. We found that the most labile subunits of GATOR2 were those of SEH1L β-propellers engaged with MIOS. As such, the MIOS-SEH1L β-propeller pairs were the most poorly resolved structures in the global GATOR2 map (between 4.5 and 6.5 Å). Another poorly-resolved feature of the complex was the lateral propeller of WDR24, which is attached to the octagonal scaffold via SEH1L.
To address these occupancy issues, we performed C2 symmetry expansion, masking, and local refinements in cryoSPARC (without particle subtraction) — of 5 overlapping sections of GATOR2 (see Supplementary Fig. 2). The resulting local maps were then subjected to further variability analyses and clustering in cryoSPARC — to extract particle sets with the highest occupancy of the otherwise poorly resolved features. While it was possible to successfully recover the β-propeller dimers of WDR24-SEH1L and MIOS-SEH1L — on the “glove” side of GATOR2, the “brace” β-propellers of MIOS-SEH1L proved highly challenging. Because the brace β-propellers interact with one another right at the C2 symmetry axis, we decided to widen that mask to include the entire brace, spanning two pairs of MIOS-SEH1L β-propellers. In order to reconstruct a map that shows a fully-occupied brace — and because the new brace mask crosses over the C2 symmetry axis — we used a non-expanded set of particles for these specific calculations.
The resulting five local maps (4 from symmetry expanded particles, and 1 from non-expanded) and analyses of their motion variability allowed us to identify distinct sets of rigid regions within GATOR2 that move as semi-independent bodies (Supplementary Fig. 3c, 4a). With Phenix (phenix.combine_focused_maps)45 we combined these individual rigid sections into a high-occupancy composite map of GATOR2 (Supplementary Fig. 4b). The interpretation of the local maps was further improved after applying automated post-processing routines with DeepEMhancer46.
Model building and refinement
All model building tasks were performed using the C2-symmetric composite map of GATOR2. Coordinates from the following structures were docked into the cryo-EM reconstruction with UCSF Chimera47: P. pastoris Sec13 (4L9O)48, S. cerevisiae Sec13 (3JRP)49, S. cerevisiae Seh1 (3F3F)50, and rebuilt by hand using Coot51. All α-solenoid and CTD junctions were built de novo, while β-propellers of MIOS and WDR24 were rebuilt from models predicted by RoseTTAFold28. Specifically, for MIOS β-propeller building, we used the better-resolved glove cryo-EM map. A backbone-only version of this model was then directly rigid-body fit into the low resolution cryo-EM map of the brace MIOS β-propeller. Please note that only one protomer unit of GATOR2 was built manually, and after its refinement with phenix.real_space_refine52, the second protomer model was generated by copying the first promoter with C2 symmetry operators in Chimera. Model refinement was carried out with restraints applied to (1) secondary structure elements and (2) zinc-coordinating residues. Model quality was evaluated using MolProbity53 and EMRinger54. The final model contains 7,272 amino acids, and 32 zinc ions coordinated by Cys and His residues in CTD-CTD junctions (Supplementary Information Table 1). Figures of the map and the final model were rendered in UCSF ChimeraX55. Surface calculations were performed in ChimeraX, with atomic lipophilicity values from Ghose et al.56 Potential membrane-interacting residues were predicted using the DREAMM algorithm57.
Cloning
Truncation constructs were prepared by PCR amplification using Q5 High-Fidelity Polymerase (NEB) and re-cloned into pRK5. Truncations referred to in the manuscript were defined by the following boundaries:
WDR24 ∆CTD: 1–700
WDR24 CTD: 685–790
WDR24 ∆Nβ: 340–790
WDR24 Nβ: 1–339
MIOS ∆CTD: 1–725
MIOS CTD: 704–875
MIOS ∆Nβ: 351–875
MIOS Nβ: 1–355
WDR59 ∆CTD: 1–890
WDR59 CTD: 873–974
WDR59 ∆Nβ: 363–974
WDR59 Nβ: 1–363
To generate single chain β-propeller dimer constructs, fragments with the following boundaries were prepared by PCR amplification using Q5 High-Fidelity Polymerase (NEB) and assembled into pRK5 by Gibson Assembly (NEB).
WDR24-SEH1L-βsc = WDR24 1–404 in frame with SEH1L 1–360
MIOS-SEH1L-βsc = MIOS 1–380 in frame with SEH1L 1–360
WDR59-SEC13-βsc = WDR59 1–654 in frame with SEC13 1–321
To generate point mutants in cDNAs for structural validation, site-directed mutagenesis reactions were performed using QuikChange kits (Agilent), according to manufacturer’s instructions.
Human SAR1B and variants of NPRL2 (Addgene #184562) and NPRL3 (Addgene #184563) in which all lysines were substituted to arginine were synthesized as gBlock gene fragments (Integrated DNA Technologies).
Generation of cells with loss-of-function mutations GATOR2 components
Generation of HEK293T cells with reduced expression of GATOR2 components was performed as previously described10. The following oligonucleotide pairs were used to clone respective sgRNAs into the pX330 vector:
sgMIOS_F: caccgATCACATCAGTAAACATGAG
sgMIOS_R: aaacCTCATGTTTACTGATGTGATc
sgWDR24_F: caccgGCACAGCTCAGGTTAAGCGA
sgWDR24_R: aaacGCACAGCTCAGGTTAAGCGAc
sgWDR59_F: caccgCGGGGGAGATGGCGGCGCGA
sgWDR59_R: aaacTCGCGCCGCCATCTCCCCCGc
Generation of HEK293T cells that stably expressed the indicated sgRNAs was performed as previously described10. The following oligonucleotide pairs were used to clone respective sgRNAs into the pLenti viral vectors:
sgSEH1L_F: caccgCATTCCCCCATGATCGCCGT
sgSEH1L_R: aaacACGGCGATCATGGGGGAATGc
sgSEC13_F: caccgAGTCGTGTCCCGCATGCTCG
sgSEC13_R: aaacCGAGCATGCGGGACACGACTc
All cells were validated for decreased expression of relevant proteins by immunoblotting.
Generation of cells that stably expressed cDNAs
Cells that stably expressed cDNAs were generated as described previously10. The lentiviral expression plasmids used were: pLJM60-FLAG-METAP2, pLJM60-HA-METAP2, pLJM60-FLAG-NPRL3, pLJM60-FLAG-NPRL3 total K➜R, pLJM60 HA-NPRL2, and pLJM60-HA-NPRL2 total K➜R.
Extended Data
Extended Data Figure 1: GATOR2 integrity and composition are insensitive to amino acid availability.
(a) Amino acids do not regulate the size of the GATOR2 complex. Elution fractions from size-exclusion chromatography of transfected GATOR2 or endogenously-tagged GATOR2 were analyzed by SDS-PAGE. Gels were stained with Coomassie Blue or SYPRO Ruby, as indicated. n.s. indicates a non-specific co-purifying contaminant. Lane labels indicate elution volumes from TSKgel G4000SWxl column.
(b) Analyses of endogenously tagged-WDR59 and -DEPDC5 expression in cell lysates.
(c) Analysis of mTORC1 signaling in cell lysates from which endogenously-tagged GATOR2 was purified in (a).
(d) Amino acids do not regulate the subunit composition or stoichiometry of endogenous GATOR2. Peak elution fractions from size-exclusion chromatography of endogenously-tagged GATOR2 were analyzed by SDS-PAGE and the gel was stained with SYPRO Ruby.
(e) The subunit composition of GATOR2 and the interaction between GATOR2 and GATOR1 or KICSTOR are not regulated by amino acids. Anti-FLAG immunoprecipitates were prepared from HEK293T cells with the indicated genotype and were analyzed by immunoblotting for the indicated proteins.
Data in (a)-(e) are representative of two independent experiments. For gel source data, see Supplementary Figure 1.
Extended Data Figure 2: Modular Architecture of GATOR2.
(a) Two orthogonal views of the molecular model of GATOR2.
(b), (c), (d) Molecular models (top) and corresponding domain schematics (bottom) of three distinct building blocks of GATOR2: MIOS-SEH1L (b), WDR24-SEH1L (c), and WDR59-SEC13 (d). Domains not resolved in our cryo-EM structure are drawn with increased transparency for reference.
(e) Each core GATOR2 subunit (MIOS, WDR24, WDR59) associates with either SEH1L or SEC13. Anti-FLAG immunoprecipitates were prepared from HEK293T cells that transiently expressed the indicated cDNAs and were analyzed by immunoblotting for the indicated proteins. HA, hemagglutinin.
Data in (e) are representative of two independent experiments. For gel source data, see Supplementary Figure 1.
Extended Data Figure 3: Surface properties of GATOR2.
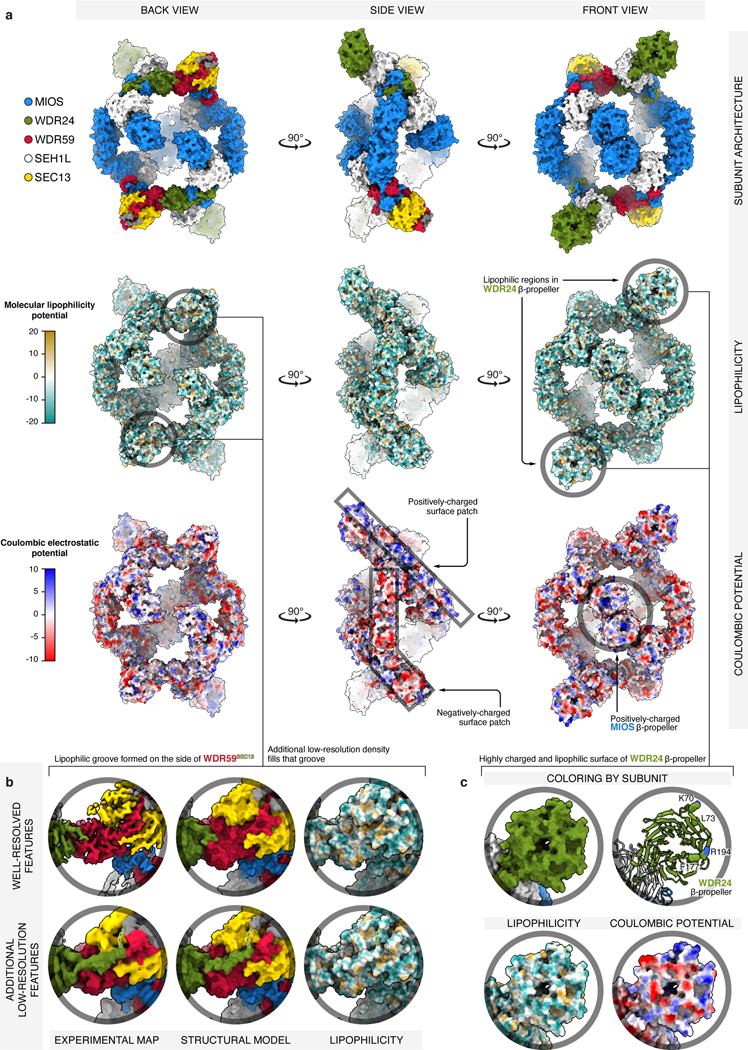
a) Three orthogonal views of GATOR2 are shown in surface representation and colored by: subunit identity (top), molecular lipophilicity potential (middle), coulombic electrostatic potential (bottom). Surface calculations were performed in ChimeraX55, with atomic lipophilicity values from Ghose et al.56 The most lipophilic surfaces are drawn as orange and the least lipophilic as cyan. Negatively charged surfaces are drawn as red, and positively as blue. GATOR2 is highly charged at its surface, with the only exposed lipophilic patches of significant size found near the donor β-blade of WDR59, and the β-propeller of WDR24 (see panels (b) and (c) for details). Furthermore, there are two major surface patches where either negative or positive charge dominates. The mostly-positive patch stretches from the side of the β-propeller of WDR24, through the WDR24-MIOS CTD, to MIOS glove β-propeller. The mostly-negative patch starts with the MIOS α-solenoid, passes through the MIOS-WDR59 CTD, and ends at the tip of SEC13. The center of MIOS β-propellers appears highly positively charged.
(b) Detailed view of the lipophilic patch near the donor β-blade of WDR59. While it might appear that the surface of the WDR59 β-blade forms a lipophilic groove, that groove is in fact occupied by protein density extending from the α-solenoid of WDR24. This WDR24 density likely continues to the top of the SEC13WDR59 β-propeller (green dashed line), where we found two better-resolved stretches of amino-acid sequence (colored in gray). Yet, for these particular sections of GATOR2, we were unable to build a high-confidence structural model or to unambiguously deduce the amino-acid sequence.
(c) Detailed view of the highly lipophilic and charged surface of the WDR24 β-propeller. The WDR24 β-propeller contains strong patches of negatively and positively charged surface. There are additional sporadic sections of highly lipophilic surface. Residues predicted by the DREAMM algorithm as membrane-penetrating are drawn as spheres57.
Extended Data Figure 4: GATOR2 CTDs contain RING domains.
(a) Sequence alignment of the homologous C-terminal regions of MIOS, WDR24, and WDR59. Zinc-coordinating residues are highlighted in yellow. Zinc-coordinating residues in the ZnF are indicated by red dots. Zinc-coordinating residues in the RING domain are indicated with blue dots.
(b) Side-by-side comparison of the structurally-aligned MIOS, WDR24, and WDR59 CTDs.
(c) Structural comparison of the MIOS, WDR24, and WDR59 RING domains with the E3 ligase RNF4 RING domain (PDB ID: 4AP4)17.
(d), (e), and (f) Detailed views of the experimental map and the corresponding molecular model surrounding the zinc coordination sites within the MIOS CTD ZnF (d) and RING domain, (e) and (f).
(g) Schematic of zinc ion coordination in the MIOS CTD. The MIOS CTD coordinates a total of four zinc ions with 15 amino-acid side chains — similarly to the CTDs in WDR24 and WDR59.
(h) Cartoon representation of the zinc ion coordination in a canonical RING domain associated with E3 ubiquitin ligase activity. The RING domain coordinates two zinc ions with eight side chains.
Extended Data Figure 5: CTD-CTD junctions link core GATOR2 proteins.
(a) Structural comparison (top) and schematic representations (bottom) of the MIOS-WDR24 and MIOS-WDR59 CTD-CTD junctions. Auxiliary interactions between CTDs and SEH1L or SEC13 contribute to the interface. This mode of interaction is distinct from that reported for dimeric RING E3 ligases58–61.
(b) View of the interface formed between the βC4 strands of the MIOS and WDR24 CTDs. Additional hydrophobic side chains from the MIOS and WDR24 βC1 strands, the MIOS αC2 helix and the WDR24 αC1 helix contribute to the interaction.
(c) View of the interface formed between the βC4 strands of the MIOS and WDR59 CTDs. Additional hydrophobic side chains from the MIOS and WDR59 βC1 strands, the MIOS αC2 helix and the WDR59 αC1 helix contribute to the interaction.
(d) The CTDs of MIOS and WDR59 are sufficient to mediate the MIOS-WDR59 interaction. Anti-FLAG immunoprecipitates were prepared from WDR24-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed by immunoblotting for the indicated proteins.
(e) The CTDs of MIOS and WDR24 are sufficient to mediate the MIOS-WDR24 interaction. Anti-FLAG immunoprecipitates were prepared from WDR59-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (d).
(f) The WDR24 CTD is necessary for the MIOS-WDR24 interaction. Anti-FLAG immunoprecipitates were prepared from WDR59-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (d).
(g) The MIOS CTD is necessary for the MIOS-WDR24 interaction. Anti-FLAG immunoprecipitates were prepared from WDR59-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (d).
(h) The WDR59 CTD is necessary for the MIOS-WDR59 interaction. Anti-FLAG immunoprecipitates were prepared from WDR24-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (d).
(i) The MIOS CTD is necessary for the MIOS-WDR59 interaction. Anti-FLAG immunoprecipitates were prepared from WDR24-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (d).
Data in (d)-(i) are representative of two independent experiments. For gel source data, see Supplementary Figure 1.
Extended Data Figure 6: GATOR2 CTDs are required for complex assembly and function.
(a) Size-exclusion chromatography profiles for wild-type GATOR2 (WT, black) or GATOR2 without individual CTDs: WDR24 CTD (WDR24 ∆CTD, green), WDR59 CTD (WDR59 ∆CTD, red), or MIOS CTD (MIOS ∆CTD, blue). The y-axis indicates normalized absorbance at 280 nm. Estimated molecular weights are indicated above each GATOR2 peak.
(b) Elution fractions from size-exclusion chromatography of wild-type GATOR2 or GATOR2 without the MIOS CTD (MIOS ∆CTD), the WDR24 CTD (WDR24 ∆CTD), and the WDR59 CTD (WDR59 ∆CTD), corresponding to panel (a). The gel image of wild-type GATOR2 elution fractions was reproduced from Extended Data Fig. 1a. Lane labels indicate elution volume from TSKgel G4000SWxl column.
(c), (d) The CTDs of WDR24 and WDR59 appear to be dispensable for the assembly of GATOR2 when analyzed by immunoprecipitation and immunoblotting. Anti-FLAG immunoprecipitates were prepared from WDR24-deficient (c) or WDR59-deficient (d) HEK293T cells that transiently expressed the indicated cDNAs and were analyzed by immunoblotting for the indicated proteins.
(e) The MIOS CTD is required for MIOS to immunoprecipitate WDR24, WDR59 and SEC13. Anti-FLAG immunoprecipitates were prepared from MIOS-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (c).
(f), (g), and (h) The CTDs of MIOS (f), WDR24 (g), and WDR59 (h) are necessary for mTORC1 activation and for the GATOR2-GATOR1 and GATOR2-KICSTOR interactions. Anti-FLAG and anti-HA immunoprecipitates were prepared from MIOS-deficient (f), WDR24-deficient (g), or WDR59-deficient (h) HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (c). HA, hemagglutinin.
Data in (a)-(h) are representative of two independent experiments. For gel source data, see Supplementary Figure 1.
Extended Data Figure 7: GATOR2 RING domains are incompatible with E2 binding.
Structural alignment between GATOR2 CTD-CTD junctions and the RNF4-UBCH5A complex (PDB ID: 4AP4)17. Individual CTD pairs are drawn (left) and superimposed with RNF4-UBCH5A (right) as follows: (a) WDR24-MIOS, (b) MIOS-WDR24, (c) MIOS-WDR59, and (d) WDR59-MIOS.
Extended Data Figure 8: GATOR2 complex does not exhibit E3 ubiquitin ligase activity.
(a), (b) In vitro ubiquitination assay does not demonstrate GATOR2-dependent ubiquitin chain formation or modification of GATOR1. GATOR1 (500 nM) was incubated with a panel of E2 enzymes in the absence (a) or presence of GATOR2 (250 nM; b). Reactions were incubated at 37°C for 1 h and analyzed by immunoblotting for the indicated proteins.
(c) Coomassie-stained SDS-PAGE analysis of GATOR1 and GATOR2 complexes assayed in (a) and (b).
(d) Ubiquitin chains formed in the presence of GATOR2 and UBE2D3 are not affected by GATOR2 inhibitors Sestrin2 or CASTOR1. UBE4B was used as a positive control for E3-dependent ubiquitin chain forming activity. Reactions were analyzed as in (a).
(e) GATOR2 does not promote ubiquitin discharge from E2~Ub conjugates. Ubiquitin charged-UBE2D3 (UBE2D3~Ub) was incubated in the presence of 10 mM lysine and UBE4B, HOIP catalytic domain, or GATOR2 (100 nM each) at 22°C for the indicated amounts of time. Samples were harvested in SDS-PAGE buffer without reducing agent, except where otherwise indicated, to preserve the E2~Ub thioester linkage.
(f) Pharmacological inhibition of the ubiquitination cascade does not affect activation of mTORC1 by amino acids. HEK293T cells were treated with 0.5 μM TAK-243 for a total of 4 h. Cells were starved of amino acids for 60 min and restimulated with amino acids for 15 min prior to harvest. Samples were harvested in SDS-PAGE sample buffer without reducing agent to preserve the E2~Ub thioester linkage and analyzed by immunoblotting.
(g) NPRL2 lysine residues are not required for activation and regulation of mTORC1 by amino acids. NPRL2-deficient cells stably expressing the indicated constructs were starved of amino acids for 60 min and restimulated with amino acids for 15 min prior to harvest and were analyzed by immunoblotting for the indicated proteins. HA, hemagglutinin.
(h) NPRL3 lysine residues are not required for activation and regulation of mTORC1 by amino acids. NPRL3-deficient cells stably expressing the indicated constructs were starved of amino acids for 60 min and restimulated with amino acids for 15 min prior to harvest and were analyzed as in (g).
Data in (a), (b), (d)-(h) are representative of two independent experiments. For gel source data, see Supplementary Figure 1.
Extended Data Figure 9: WDR24-WDR59 α-solenoid junction is required for GATOR2 assembly and function.
(a) Two views of the WDR24-WDR59 α-solenoid junction.
(b) Mutations in the WDR24 α-solenoid domain disrupt the WDR24-WDR59 interaction. Anti-FLAG immunoprecipitates were prepared from MIOS-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed by immunoblotting for the indicated proteins. HA, hemagglutinin.
(c) The WDR24-WDR59 interaction is required for activation of mTORC1 and the GATOR2-GATOR1 and GATOR2-KICSTOR interactions. Anti-FLAG and anti-HA immunoprecipitates were prepared from WDR24-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (b).
(d) Mutation of the WDR59 solenoidal domain disrupts the WDR24-WDR59 interaction. Anti-FLAG immunoprecipitates were prepared from MIOS-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (b).
(e) The WDR24-WDR59 interaction is required for activation of mTORC1 and the GATOR2-GATOR1 and GATOR2-KICSTOR interactions. Anti-FLAG and anti-HA immunoprecipitates were prepared from WDR24-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (b).
Data in (b)-(e) are representative of two independent experiments. For gel source data, see Supplementary Figure 1.
Extended Data Figure 10: MIOS-MIOS α-solenoid homodimerization is required for GATOR2 assembly and function.
(a) Structure of the MIOS-MIOS α-solenoid junction. The two interacting MIOS protomers are colored as either darker (MIOSA) or lighter blue (MIOSB).
(b) Mutagenesis of the interface between the two MIOS α-solenoids (MIOS A560E) disrupts MIOS homodimerization. Anti-FLAG immunoprecipitates were prepared from WDR24-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed by immunoblotting for the indicated proteins.
(c) Size-exclusion chromatography traces comparing the elution profile of GATOR2 containing wild-type MIOS with GATOR2 containing MIOS A560E. The trace for wild-type GATOR2 was reproduced from Extended Data Fig. 6a.
(d) SDS-PAGE analysis of elution fractions from size-exclusion chromatography in (c). The gel image of wild-type GATOR2 elution fractions was reproduced from Extended Data Fig. 1a. Lane labels indicate elution volume from TSKgel G4000SWxl column.
(e) Integrity of the MIOS α-solenoid junction is necessary for mTORC1 activation and the GATOR2-GATOR1 and GATOR2-KICSTOR interactions. Anti-FLAG and anti-HA immunoprecipitates were prepared from MIOS-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (b). HA, hemagglutinin.
Data in (b)-(e) are representative of two independent experiments. For gel source data, see Supplementary Figure 1.
Extended Data Figure 11: Core GATOR2 proteins incorporate SEH1L and SEC13 by blade donation.
(a) Structure (left) and schematic (right) of the SEH1LWDR24 β-propeller completed by the donated WDR24 β-blade.
(b) Structure (left) and schematic (right) of the SEH1LMIOS β-propeller completed by the donated MIOS β-blade.
(c) Structure (left) and schematic (right) of the SEC13WDR59 β-propeller completed by the donated WDR59 β-blade.
(d) Mutation of the donated WDR24 β-blade disrupts association of WDR24 with SEH1L. Anti-FLAG immunoprecipitates were prepared from MIOS-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed by immunoblotting for the indicated proteins. HA, hemagglutinin.
(e) Mutation of the donated MIOS β-blade disrupts association of MIOS with SEH1L. Anti-FLAG immunoprecipitates were prepared from HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (d).
(f) Mutation of the donated WDR59 β-blade disrupts association of WDR59 with SEC13. Anti-FLAG immunoprecipitates were prepared from MIOS-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (d).
(g) Depletion of SEH1L or SEC13 destabilizes GATOR2 proteins and disrupts complex formation. SEH1L or SEC13 were depleted by CRISPR/Cas9 in a HEK293T clone expressing endogenously tagged 3xFLAG-WDR59. Anti-FLAG immunoprecipitates were prepared and analyzed as in (d).
(h), (i) Depletion of SEH1L (h) or SEC13 (i) renders mTORC1 signaling resistant to amino acid availability. HEK293T cells of the indicated genotype were starved of amino acids for 60 min and restimulated with amino acids for 15 min prior to harvest and analysis by immunoblotting.
Data in (d)-(i) are representative of two independent experiments. For gel source data, see Supplementary Figure 1.
Extended Data Figure 12: Functional analysis of the core GATOR2 β-propellers.
(a) The β-propellers of MIOS, WDR59, and WDR24 are required for interactions between GATOR2 and CASTOR1, GATOR1 and KICSTOR, and Sestrin2, respectively. Anti-FLAG immunoprecipitates were prepared from MIOS-deficient, WDR59-deficient or WDR24-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed by immunoblotting for the indicated proteins. CASTOR1 D304A and Sestrin2 E451Q mutants do not bind arginine or leucine, respectively, and bind GATOR2 irrespective of amino acid availability. HA, hemagglutinin.
(b) Schematic comparing WDR24 Nβ and SEH1L β-propeller organization in wild-type and β-single chain (βsc) constructs.
(c) Schematic comparing WDR59 Nβ and SEC13 β-propeller organization in wild-type and β-single chain (βsc) constructs. The fused construct contains the intervening RWD domain.
(d) The GATOR2 single chain β-propeller pairs of MIOS-SEH1L, WDR59-SEC13, and WDR24-SEH1L are sufficient to interact with CASTOR1, GATOR1 and KICSTOR, and Sestrin2, respectively. Anti-FLAG immunoprecipitates were prepared from MIOS-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (a).
(e) GATOR2 mediates the interaction between Sestrin2 and CASTOR1, indicating that they can co-occupy the same GATOR2 particle. Anti-FLAG immunoprecipitates were prepared from MIOS-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (a).
(f) Sestrin2 and CASTOR1, but not SAR1B, interact with GATOR2 and suppress mTORC1 signaling. Anti-FLAG and anti-HA immunoprecipitates were prepared from HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (a).
(g) GATOR2 interacts with Sestrin2 and CASTOR1, but not SAR1B. Anti-FLAG immunoprecipitates were prepared from HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (a).
(h) GATOR2 does not immunoprecipitate the Rag GTPases independent of GATOR1. Anti-FLAG immunoprecipitates were prepared from wild-type of DEPDC5-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (a).
(i) MIOS Nβ is not absolutely required for activation of mTORC1 or the GATOR2-GATOR1 and GATOR2-KICSTOR interactions. Anti-FLAG and anti-HA immunoprecipitates were prepared from MIOS-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (a).
(j) WDR59 Nβ is necessary for activation of mTORC1 and the GATOR2-GATOR1 and GATOR2-KICSTOR interactions. Anti-FLAG and anti-HA immunoprecipitates were prepared from WDR59-deficient HEK293T cells that transiently expressed the indicated cDNAs and were analyzed as in (a).
Data in (a), (d)-(j) are representative of two independent experiments. For gel source data, see Supplementary Figure 1.
Supplementary Material
Acknowledgements
We thank all members of the Sabatini Laboratory for helpful insights and K. Linde-Garelli for assistance with protein purifications. We thank M. Baek, I. Anishchenko, and D. Baker for structural predictions, and the high-performance computing team at Whitehead Institute (C. Andrew, P. McCabe, R. Taylor, P. Macfarlane, and W. Pena) for the installation and maintenance of data-processing servers. We thank R. Levinson for the forearm shake illustration. This work was supported by grants to D.M.S. from the NIH (R01 CA103866, R01 CA129105, and R01 AI47389), the Department of Defense (W81XWH-21–1-0260, and TS200035), the Lustgarten Foundation, and Leo Foundation, and fellowship support from the NIH to M.L.V. (T32 GM007753 and F30 CA228229), K.B.R. (K99 CA255926), L.C. (F31 CA180271), R.A.S (F31 GM121093) and fellowships to M.L.V and X.G. from the Koch Institute for Integrative Cancer Research at MIT. K.B.R. was also supported by the Tuberous Sclerosis Association, and Charles A. King Trust.
Footnotes
Competing Interests
D.M.S. is a shareholder of Navitor Pharmaceuticals, which is targeting for therapeutic benefit the amino-acid-sensing pathway upstream of mTORC1.
Additional Information
Supplementary Information is available for this paper.
Materials are available from the Whitehead Institute (sabadmin@wi.mit.edu) upon request. Reprints and permissions information is available at www.nature.com/reprints.
Data availability
The data that support the findings of this study are available from the corresponding authors and the Whitehead Institute (sabadmin@wi.mit.edu) upon request. Cryo-EM maps were deposited in the Electron Microscopy Data Bank under accession number EMD-26519. The atomic model of GATOR2 was deposited in the Protein Data Bank under accession number 7UHY. Plasmids generated in this study are available from Addgene.
References:
- 1.Liu GY & Sabatini DM mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Bio 21, 183–203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Condon KJ & Sabatini DM Nutrient regulation of mTORC1 at a glance. J Cell Sci 132, jcs222570 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bar-Peled L. et al. A Tumor Suppressor Complex with GAP Activity for the Rag GTPases That Signal Amino Acid Sufficiency to mTORC1. Science 340, 1100–1106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolfson RL et al. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 351, 43–48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chantranupong L. et al. The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell 165, 153–164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brohawn SG, Leksa NC, Spear ED, Rajashankar KR & Schwartz TU Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science 322, 1369–1373 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sancak Y. et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogala KB et al. Structural basis for the docking of mTORC1 on the lysosomal surface. Science 366, 468–475 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anandapadamanaban M. et al. Architecture of human Rag GTPase heterodimers and their complex with mTORC1. Science 366, 203–210 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfson RL et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature 543, 438–442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng M, Yin N. & Li MO SZT2 dictates GATOR control of mTORC1 signalling. Nature 543, 433–437 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen K. et al. Architecture of the human GATOR1 and GATOR1–Rag GTPases complexes. Nature 556, 64–69 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metzger MB, Pruneda JN, Klevit RE & Weissman AM RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. BBA - Molecular Cell Research 1843, 47–60 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Jaroszewski L, Iyer M, Sedova M. & Godzik A. FATCAT 2.0: towards a better understanding of the structural diversity of proteins. Nucleic Acids Res 48, gkaa443- (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bellon SF, Rodgers KK, Schatz DG, Coleman JE & Steitz TA Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster. Nat Struct Biol 4, 586–591 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Dokudovskaya S. et al. A Conserved Coatomer-related Complex Containing Sec13 and Seh1 Dynamically Associates With the Vacuole in Saccharomyces cerevisiae. Molecular & Cellular Proteomics 10, M110.006478–17 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plechanovová A, Jaffray EG, Tatham MH, Naismith JH & Hay RT Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489, 115–120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsia K-C, Stavropoulos P, Blobel G. & Hoelz A. Architecture of a Coat for the Nuclear Pore Membrane. Cell 131, 1313–1326 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whittle JRR & Schwartz TU Structure of the Sec13–Sec16 edge element, a template for assembly of the COPII vesicle coat. J Cell Biology 190, 347–361 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fath S, Mancias JD, Bi X. & Goldberg J. Structure and Organization of Coat Proteins in the COPII Cage. Cell 129, 1325–1336 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Chen J. et al. SAR1B senses leucine levels to regulate mTORC1 signalling. Nature 596, 281–284 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Rout MP & Field MC The Evolution of Organellar Coat Complexes and Organization of the Eukaryotic Cell. Annual Review of Biochemistry 86, 637–657 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Hunter MR, Scourfield EJ, Emmott E. & Graham SC VPS18 recruits VPS41 to the human HOPS complex via a RING–RING interaction. Biochemical Journal 474, 3615–3626 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai W, Wei Y, Jarnik M, Reich J. & Lilly MA The GATOR2 Component Wdr24 Regulates TORC1 Activity and Lysosome Function. PLoS Genetics 12, e1006036–28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saxton RA, Chantranupong L, Knockenhauer KE, Schwartz TU & Sabatini DM Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. 536, 229–233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu X. et al. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 358, 813–818 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jain BP & Pandey S. WD40 Repeat Proteins: Signalling Scaffold with Diverse Functions. Protein J 37, 391–406 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Baek M. et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science eabj8754 (2021) doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y, Wang C, Xiao Q. & Guo L. Crystal structures of arginine sensor CASTOR1 in arginine-bound and ligand free states. Biochem Bioph Res Co 508, 387–391 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Saxton RA et al. Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science 351, 53–58 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cristea IM & Chait BT Conjugation of Magnetic Beads for Immunopurification of Protein Complexes. Cold Spring Harb Protoc 2011, pdb.prot5610 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boussif O. et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc National Acad Sci 92, 7297–7301 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuéllar J. et al. Structural and functional analysis of the role of the chaperonin CCT in mTOR complex assembly. Nat Commun 10, 2865 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mastronarde DN SerialEM: A Program for Automated Tilt Series Acquisition on Tecnai Microscopes Using Prediction of Specimen Position. Microsc Microanal 9, 1182–1183 (2003). [Google Scholar]
- 35.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zivanov J, Nakane T. & Scheres SHW A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. Iucrj 6, 5–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wagner T. et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun Biology 2, 218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zivanov J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zivanov J, Nakane T. & Scheres SHW Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. Iucrj 7, 253–267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheres SH Beam-induced motion correction for sub-megadalton cryo-EM particles. Elife 3, e03665 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilkinson ME, Kumar A. & Casañal A. Methods for merging data sets in electron cryo‐microscopy. Acta Crystallogr Sect D 75, 782–791 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Punjani A. & Fleet DJ 3D variability analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J Struct Biol 213, 107702 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Liebschner D. et al. Macromolecular structure determination using X‐rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr Sect D 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanchez-Garcia R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun Biology 4, 874 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pettersen EF et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Bharucha N. et al. Sec16 influences transitional ER sites by regulating rather than organizing COPII. Mol Biol Cell 24, 3406–3419 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brohawn SG & Schwartz TU Molecular architecture of the Nup84–Nup145C–Sec13 edge element in the nuclear pore complex lattice. Nat Struct Mol Biol 16, 1173–1177 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Debler EW et al. A Fence-like Coat for the Nuclear Pore Membrane. Mol Cell 32, 815–826 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Emsley P, Lohkamp B, Scott WG & Cowtan K. Features and development of Coot. Acta Crystallogr Sect D Biological Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Afonine PV et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr Sect D Struct Biology 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D Biological Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barad BA et al. EMRinger: side chain–directed model and map validation for 3D cryo-electron microscopy. Nat Methods 12, 943–946 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goddard TD et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghose AK, Viswanadhan VN & Wendoloski JJ Prediction of Hydrophobic (Lipophilic) Properties of Small Organic Molecules Using Fragmental Methods: An Analysis of ALOGP and CLOGP Methods. J Phys Chem 102, 3762–3772 (1998). [Google Scholar]
- 57.Chatzigoulas A. & Cournia Z. Predicting protein-membrane interfaces of peripheral membrane proteins using ensemble machine learning. Biorxiv 2021.06.28.450157 (2021) doi: 10.1101/2021.06.28.450157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Linke K. et al. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ 15, 841–848 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Liew CW, Sun H, Hunter T. & Day CL RING domain dimerization is essential for RNF4 function. Biochem J 431, 23–29 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mace PD et al. Structures of the cIAP2 RING Domain Reveal Conformational Changes Associated with Ubiquitin-conjugating Enzyme (E2) Recruitment*. J Biol Chem 283, 31633–31640 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Brzovic PS, Rajagopal P, Hoyt DW, King M-C & Klevit RE Structure of a BRCA1–BARD1 heterodimeric RING–RING complex. Nat Struct Biol 8, 833–837 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors and the Whitehead Institute (sabadmin@wi.mit.edu) upon request. Cryo-EM maps were deposited in the Electron Microscopy Data Bank under accession number EMD-26519. The atomic model of GATOR2 was deposited in the Protein Data Bank under accession number 7UHY. Plasmids generated in this study are available from Addgene.