Abstract
Purpose:
We examined gene expression, germline variant, and somatic mutation features associated with pathologic response to neoadjuvant durvalumab plus chemotherapy in basal-like triple negative breast cancer (bTNBC).
Experimental Design:
Germline and somatic whole exome DNA and RNA sequencing, PD-L1 immunohistochemistry, and stromal tumor infiltrating lymphocytes scoring were performed on 57 patients. We validated our results using 162 patients from the GeparNuevo randomized trial.
Results:
Gene set enrichment analysis showed that pathways involved in immunity (adaptive, humoral, innate), JAK-STAT signaling, cancer drivers, cell cycle, apoptosis and DNA repair were enriched in cases with pathologic complete response (pCR), whereas epithelial-mesenchymal transition, extracellular matrix, and TGFβ pathways were enriched in cases with residual disease (RD). Immune-rich bTNBC with RD was enriched in CCL-3,−4,−5,−8,−23, CXCL-1,−3,−6,−10, and interleukins-1,−23,−27,−34 and had higher expression of macrophage markers compared to immune-rich cancers with pCR that were enriched in IFNγ, interleukins-2,−12,−21, chemokines CXCL-9,−13, CXCR5, and activated T and B cell markers (GZMB, CD79A). In the validation cohort, an immune-rich 5-gene signature showed higher expression in pCR cases in the durvalumab arm (p=0.040) but not in placebo arm (p=0.923) or in immune-poor cancers. Independent of immune markers, tumor mutation burden was higher, and PI3K, DNA damage repair, MAPK, and WNT/β-Catenin signaling pathways were enriched in germline and somatic mutations in cases with pCR.
Conclusion:
The TGFβ pathway is associated with immune-poor phenotype and RD in bTNBC. Among immune-rich bTNBC RD, macrophage/neutrophil chemoattractants dominate the cytokine milieu, and IFNγ and activated B cells and T cells dominate immune-rich cancers with pCR.
Introduction
Multiple randomized trials demonstrated increased pathological complete response (pCR, ypT0is/ypN0) rates when an anti-PD-1 (pembrolizumab) or anti-PD-L1 (atezolizumab, durvalumab) antibody is included with standard of care neoadjuvant chemotherapy in triple negative breast cancer (TNBC) (1–5). Patients who achieve pCR have excellent long-term survival, and two of these randomized trials also reported statistically significant improvement in recurrence free survival with immunotherapy (2,6). The pCR rates after combined neoadjuvant immunotherapy + chemotherapy range between 44–65% depending on the type of chemotherapy regimen indicating that many patients continue to have residual disease (RD) after therapy.
Several studies examined molecular predictors of pCR in neoadjuvant immunotherapy + chemotherapy trials and demonstrated that cancers with higher levels of tumor infiltrating lymphocytes (TILs) and greater expression of PD-L1 protein on immune cells have higher pCR rates compared to cancers with lesser immune infiltration (7). The presence of TILs and PD-L1 expression are associated with the expression of a broad range of immune gene expression signatures that also predict pCR (8,9). Tumor mutation burden (TMB) recently emerged as an independent predictor for pCR (10). However, all of these markers are predictive of pCR with or without immunotherapy (11–13), no validated molecular markers exist that could identify patients who selectively benefit from inclusion of immunotherapy with their neoadjuvant chemotherapy. We recently reported that MHC class II protein expression on tumor cells may identify cancers that are selectively benefitting from neoadjuvant immunotherapy, but this observation will require independent validation (14).
The goal of the current study was to comprehensively characterize molecular features of basal-like TNBC (bTNBC) that achieved pCR after neoadjuvant anti-PD-L1 therapy plus chemotherapy compared to cases with RD in the overall study population and in the immune-rich subset. We focused on the basal-like subset of TNBC (bTNBC) to minimize molecular heterogeneity in our relatively small sample set and because of the clinical differences between bTNBC and other less frequent TNBC molecular subtypes (15,16). We performed whole exome and whole transcriptome RNA sequencing along with histologic assessment of pre-treatment needle biopsies collected during a single arm Phase II clinical trial to identify candidate markers of response (7). We assessed the association between our candidate response markers on the chemotherapy alone and chemotherapy plus durvalumab arms of the GeparNuevo randomized trial.
Materials & Methods
Patient population and biospecimens
Pretreatment core needle biopsies for research were obtained from patients with stage I-III TNBC who enrolled in a single arm neoadjuvant clinical trial (NCT02489448) and received durvalumab concurrent with weekly nab-paclitaxel x 12 followed by durvalumab plus dose dense doxorubicin/cyclophosphamide x 4 treatments. Primary efficacy results were previously published (7). Sixty female patients were enrolled in the trial, 2 patients were not evaluable for pathologic response and one patient withdrew consent, therefore the biomarker population includes 57 patients (pCR n=26, RD n=31) Supplementary Figure S1. All patients provided written informed consent for research on their donated tissues, including germline DNA sequencing. Ethical approval was obtained from the Yale Human Investigations Committee (Yale University, HIC# 1409014537). The validation data included targeted mRNA sequencing results of 2,559 transcripts generated from pretreatment biopsies of 162 patients enrolled in the GeparNuevo trial (NCT02685059) who received durvalumab or placebo every plus nab-paclitaxel x 12 weeks, followed by durvalumab or placebo plus epirubicin/cyclophosphamide x 4 treatments (2). The GeparNuevo protocol was approved by the respective ethics committee, institutional review board, and national competent authority. All studies were conducted in accordance with the Declaration of Helsinki.
Isolation of RNA and DNA
For the Yale cohort, RNA and DNA were extracted from one biopsy collected in RNAlater™ (Qiagen, Germantown, MD, USA) and stored at −80C. After homogenization with the TissueLyser II bead-milling system (Qiagen), DNA was isolated using the AllPrep DNA/RNA/miRNA universal kit, and the flow-through RNA was extracted with RNeasy Plus Kit (Qiagen) following the manufacturer’s instructions. The quality and concentration of isolated DNA and RNA were tested on the Agilent 2100 Bioanalyzer system. DNA and RNA sequencing were performed at the Yale Center for Genome Analysis.
RNA sequencing and data processing
Paired-end sequencing of 100bp fragments of total RNA for a targeted depth of 50 million reads was performed using the Illumina NovaSeq platform. Quality was assessed using FastqQC v 0.11.5 (17), adapter sequences were trimmed with Trimmomatic v0.36 (RRID:SCR_011848) (18). Sequencing reads were aligned to human genome, hg38, with STAR v2.5.3a (RRID:SCR_004463) (19) using two-pass mode and default parameters; alignment quality and strandedness was checked using RSeQC v2.6.4 (RRID:SCR_005275) (20). Gene expression was quantified using RSEM v1.3.0 (RRID:SCR_013027) (21) and ENSEMBL release 91 (RRID:SCR_002344) was used to annotate reads to human genes. One specimen was excluded due to poor RNA quality. Molecular subtyping was performed using the AIMS, SCMOD2 and PAM50 methods. All methods identified the same 6 cases as non-basal-like and outlier analysis using principal component analysis (PCA) and uniform manifold approximation and projection (UMAP) of transcriptomic data confirmed these cases as distinct from the remaining samples. (Supplementary Figure S1; Supplementary Figure S2). All six non-basal cases had RD. In order to work with a molecularly homogeneous bTNBC set we excluded these 6 cases from further analysis resulting in 50 bTNBC cases (n=25 pCR, n=25 RD).
Gene expression differences between pCR and RD samples were determined using “DESeq2” R package (RRID:SCR_000154) (22). To adjust for variable tissue composition from case to case, we added a previously published stromal score that quantifies tumor stromal content calculated using the ESTIMATE R package (23). Differentially expressed genes were defined as log-fold change >1, the Benjamini-Hochberg method was used to adjust for multiple comparisons and adjusted p<0.05 was considered significant. Gene set enrichment analysis was implemented using the fgsea R package (24). To quantify biological and immune processes, the NanoString Hallmarks of Cancer and Biological Pathways and Processes gene sets and a collection of previously published immune gene signatures (5,8) were used (Supplementary Data File Tables S1–S2). Immune signature-based classification into immune high versus immune low status was performed using the median values of the Tumor Inflammation Signature (TIS), GeparSixto, NHI 5-gene, STAT1 gene signatures and expression of IFNγ single gene, respectively. Gene signature expressions were compared between pCR and RD groups using the Mann-Whitney test. Multivariate association between gene signatures expressions and pCR were assessed using logistic regression adjusted for age (continuous variable), tumor size (T1 vs ≥ T2), nodal status (N0 vs N1-N3), and stromal score (from ESTIMATE algorithm). Benjamini-Hochberg corrected P< 0.05 was considered significant.
For the GeparNuevo validation cohort, formalin-fixed and paraffin-embedded (FFPE) tissues were processed using an HTG EdgeSeq instrument (HTG Molecular Inc, Tucson, AZ, USA) with the Oncology Biomarker Panel according to the manufacturer’s instructions as previously described (2,10). Fisher’s exact test and Pearson’s chi-square were used to evaluate categorical variables (pCR status; sTILs (High; Low)) (10). Univariate and multivariate logistic regression models with adjustments for clinical covariates (as previously described) were used for assessment of predictive value of genes for pCR (10).
Whole Exome Sequencing
Genomic DNA (1 μg) from tumor biopsies and matched peripheral blood buffy coats of 57 patients were sheared to a mean fragment length of 140 bp and exomes were captured using the NimbleGen SeqCap EZ v2 kit. The resulting library was sequenced on an Illumina HiSeq 4000 instrument in paired-end 75-cycles mode to achieve an average target sequencing depth of 232x for tumor samples and 207x for matched normal samples. Reads were filtered by Illumina CASAVA 1.8.2 software, trimmed at the 30 end using FASTX v0.0.13 (RRID:SCR_005534), and aligned to the human reference genome (GRCh38) by Burrows-Wheeler Aligner v0.7.15a (RRID:SCR_010910) (25). PCR duplicates were removed with MarkDuplicates (Picard v 2.17.11, http://broadinstitute.github.io/picard/, RRID:SCR_006525) algorithm. Indelrealigner and RealignerTargetCreator kits of GATK (v3.4) (26) were used to align indel regions. Mutect (v.1.1.4) (RRID:SCR_000559) (27) was used to identify somatic single nucleotide variants (SNV). We used IndelGenotyper (36.3336) of GATK (v3.4) (RRID:SCR_001876) for somatic indel calling. We applied the HaplotypeCaller algorithm of GATK (26) to call high quality germline variants with default parameters. To control for the false positive rate of germline variants calling, we filtered low-quality variants with the following criteria: DP < 4, QD < 2.0, FS > 60.0, MQ < 35.0, MQRankSum < −12.5 and ReadPosRankSum < −8.0. Tumor mutation burden (TMB) was calculated as the total number of exonic somatic mutation divided by total length of exome capture probes (34MB).
The functional impact of germline missense variants was predicted using MetaSVM (28) and annotation from the ClinVar database (RRID:SCR_006169) (29). We considered a missense variant as high functional impact if it was classified as deleterious by MetaSVM or Pathogenic/Likely-Pathogenic in ClinVar. Loss-of-function (LoF) variants including frameshift indels, stop gain, and stop loss variants were also considered as high functional impact, as well as variants annotated as high-confidence loss of function in gnomAD (30). We used 723 cancer census genes from the Catalogue Of Somatic Mutations In Cancer database (COSMIC, release v94, 28th May 2021) (RRID:SCR_002260) (31) to generate the oncoplots for both germline and somatic mutations. Associations between pCR and gene or pathway level germline variants and somatic mutations were assessed using logistic regression. For the gene-level analysis, only genes affected in at least 5 out of 57 cases were considered for the association test.
To assess mutations at the pathway level, we collected 107 canonical biological pathways from the NanoString Hallmarks, NanoString Metabolic, and MSigDB Pathway databases (Supplementary Data File Table S2) and considered a pathway mutated if it had ≥1 member gene with mutation. To assess significance of pathway level mutation, we first calculated odds ratios of the response category (pCR or RD) versus the gene or pathway status (mutated versus wildtype) using logistic regression, next we randomly permuted the pCR or RD labels for 1000 iterations and the odds ratio for each gene or pathway was recalculated. The proportion of random permutations showing an odds ratio greater than the odds ratio of the unperturbed data was defined as the p-value.
PD-L1 Immunohistochemistry and Stromal TILs Assessment
Stromal TILs (sTILs) were assessed on FFPE hematoxylin and eosin stained 4 μm sections. The slides were digitally scanned and independently scored by two pathologists. The sTILs score was calculated as the area occupied by mononuclear inflammatory cells over the total intratumoral stromal area (32). Immune high cancers were defined as sTILs ≥ 30%(33). PD-L1 protein expression was assessed with chromogenic immunohistochemistry (IHC) using the VENTANA PD-L1 (SP263) Assay following the manufacturer’s instructions. PD-L1 positivity was defined as ≥1% tumor and/or immune cells staining positive (32,34). Mann-Whitney test was used for sTILs scores and the Fisher’s exact test was use for PD-L1 IHC positivity to determine if there were significant (p<0.05) differences between pCR and RD.
Data and materials availability
All data associated with this study are presented in this paper or Supplemental Materials. The whole exome and transcriptomic data from the Yale clinical trial (NCT02489448) are deposited in National Center for Biotechnology Information (NCBI) database of Genotypes and Phenotypes (dbGaP) under bioproject #PRJNA558949. To access the GeparNuevo (NCT02685059) dataset please refer to https://gbg.de/en/research/trafo.php.
Results
Differentially expressed genes and enriched pathways between bTNBC with pCR and RD
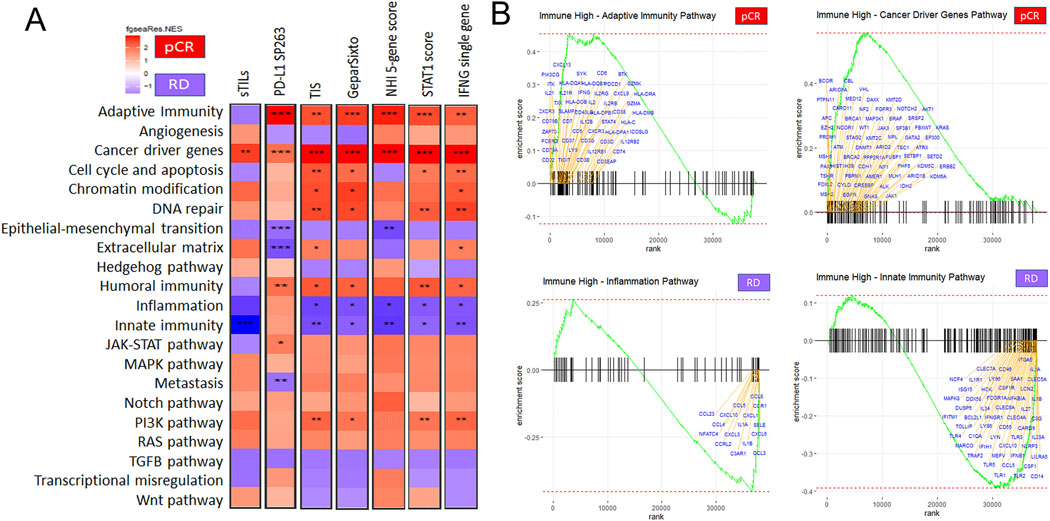
One hundred and forty-three and 66 genes were significantly overexpressed in cancers that achieved pCR and RD, respectively (Figure 1A; Supplementary Data File Table S3). Gene set enrichment analysis showed that adaptive immunity (p<0.001), cancer driver genes (p<0.01), cell cycle & apoptosis (p<0.05), DNA repair (p<0.05), humoral immunity (p<0.001), innate immunity (p<0.001), and JAK-STAT pathways (p<0.001) were enriched in patients with pCR (Figure 1B; Supplementary Data File Table S4). Epithelial-mesenchymal transition (p<0.05), extracellular matrix (p<0.01), and TGFβ (p<0.05) pathways were enriched in patients with RD (Figure 1B; Supplementary Data File Table S4). The leading-edge genes from the enriched pathways showed that genes that regulate T cell and B cell activities drove the pathway enrichment in pCR, and genes that impacted macrophages, fibroblasts, and cancer cell response to cytotoxic therapy (i.e., decreased DNA repair machinery) drove pathway enrichment in RD (Figure 1C, Supplementary Data File Table S4). These findings confirm that high levels of immune gene expression are characteristics of highly chemotherapy sensitive cancers. However, a subset of immune-rich bTNBC fail to achieve pCR and what drives this difference remains unknown.
Figure 1. Differentially expressed genes and pathways between pCR and RD in basal-like TNBC treated with neoadjuvant durvalumab and standard of care chemotherapy.
(A) Volcano plot of differentially expressed genes. Statistically significant genes are in red, top 50 significant genes annotated in blue. (B) Pathway enrichment results. (C) Enrichment score plots of the leading-edge genes from significantly enriched pathways. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Differentially expressed genes and pathways between immune-rich TNBC with pCR and RD
We also performed differential gene and pathway expression analysis between cases with pCR versus RD restricted to immune-rich cancers only. To assess how the results might depend on the definition of immune richness, we applied two histology based methods including sTILs ≥ 30% and PD-L1 IHC positivity, and 5 immune gene signatures dichotomized at the median (Tumor Inflammation Signature > 0.1249, GeparSixto gene signature > 0.26, NHI 5-gene score > 0.0014, STAT1 score > 0.07695, and IFNγ gene expression ≥ 0.20) to define immune-rich status (Supplementary Data File Tables S1–S2 and S5) (5,8). The ranking of differentially expressed genes varied substantially depending on how immune-rich status was defined (Figure 2 A–G; Supplementary Data File Tables S6–S13). However, gene set enrichment analysis revealed highly consistent differences at pathway level between cancers with RD versus pCR, regardless of how immune-rich status was defined (Figure 3A; Supplementary Data File Tables S14–S20). The pathways that were significantly enriched in RD despite high immune infiltration included inflammation (p<0.05) and innate immunity (p<0.05), the TGFβ pathway and epithelial mesenchymal transition were also consistently enriched but failed to reach statistical significance. In cancers with pCR, adaptive immunity (p<0.05) and cancer driver gene pathways (p<0.01) were significantly enriched, several other pathways including, DNA repair, cell cycle, apoptosis, chromatin modifications, PIK3A, and RAS were also consistently enriched but failed to reach statistical significance. Examination of the leading-edge genes from the significantly enriched pathways revealed a substantially different cytokine/chemokine milieu with RD compared to pCR (Figure 3B; Supplementary Data File Tables S14–S20). In cancers with RD, the dominant chemokines were CCL-3,−4,−5,−8,−23, CXCL-1,−3,−6,−10, and cytokines were interleukins (IL)-1A/B,−23A,−27,−34. The chemokines CCL-3,−5 and CXCL-1, −6 are major chemoattractants for tumor associated macrophages and neutrophils that can exerting pro-tumorigenic effects (35). IL-34 promotes macrophage differentiation and IL-1 is the classical macrophage derived proinflammatory cytokine. Indeed, the leading-edge genes of the innate immunity pathway enriched in RD included all the hallmarks of a strong macrophage presence; high expression of CSF1, CSF1R, CD14, scavenger receptor MARCO, and Toll like receptors (TLR)-1,−2,−3,−4,−5,−6.
Figure 2. Differentially expressed genes between pCR and RD in basal-like immune-rich TNBC.
(A-G) Volcano plots of differentially expressed genes in immune high cancers defined by sTILs ≥ 30 %, PD-L1 positive, TIS > 0.1249, GS > 0.26, NHI 5-gene score > 0.0014, STAT1 score > 0.07695, or IFNG ≥ 0.20, respectively. Statistically significant genes are in red, top 50 significant genes annotated in blue. sTILs = stromal tumor infiltrating lymphocytes. TIS = Tumor inflammation signature. GS = GeparSixto signature. IFNG = interferon gamma single gene.
Figure 3. Pathway enrichment differences between pCR and RD in basal-like immune-rich TNBC.
(A) Heatmaps of pathway enrichment results for 21 Cancer Hallmarks Pathways in each of the different ways immune-high status was defined (as on Figure 2). Pathways enriched in pCR are red and those enriched in RD are blue. (B) Enrichment score plots of the leading-edge genes from the pathways that were significantly and consistently enriched in cancers with pCR or RD. IFNG single gene expression was used to define the immune-rich cancers for this analysis. sTILs = stromal tumor infiltrating lymphocytes. TIS = Tumor inflammation signature. GS = GeparSixto signature. IFNG = interferon gamma. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
In immune-rich cancers with pCR, the dominant cytokines were IFNγ, IL-2,−12A/B,−21, and chemokines CXCL-9 and CXCL-13 and its receptor CXCR5. IFNγ and IL-2 are the quintessential immune growth factors that play critical roles in activating and sustaining T cell response. IL-12 induces T helper cell differentiation and increases the cytotoxic activity of T cells; it also inhibits tumor associated macrophages and myeloid-derived suppressor cells. IL-21 regulates differentiation of B cells into plasma cells and increases cytotoxicity of T cells. CXCL-9 is a chemoattractant for activated T-cells, and CXCL-13 is a chemoattractant for B cells. Consistent with this highly immune activating cytokine milieu, the leading-edge genes also included many T cell (CD3, CD5, CD6, CD7, CD40LG) and B cell (MS4A1, CD19, CD38, CD22, CD37, CD79A) markers, human leukocyte antigen class II (HLA-D) molecules that present antigens to T cells, and granzymes that mediate cytotoxicity.
When we compared immune-poor cancers with pCR versus RD we identified different sets of differentially expressed genes, with < 40% overlap with the genes associated with response in immune-rich cancers, suggesting that different processes are involved in determining response or resistance depending on the immune microenvironment. Cancers with pCR were enriched in the adaptive (p<0.01), humoral (p<0.05), and innate immunity pathways (p<0.05) despite belonging to the overall immune-poor subset. Patients with RD were enriched in the angiogenesis (p<0.01), extracellular matrix (p<0.05), and RAS pathways (p<0.05) (Supplementary Data File Tables S21–S34; Supplementary Figure S4).
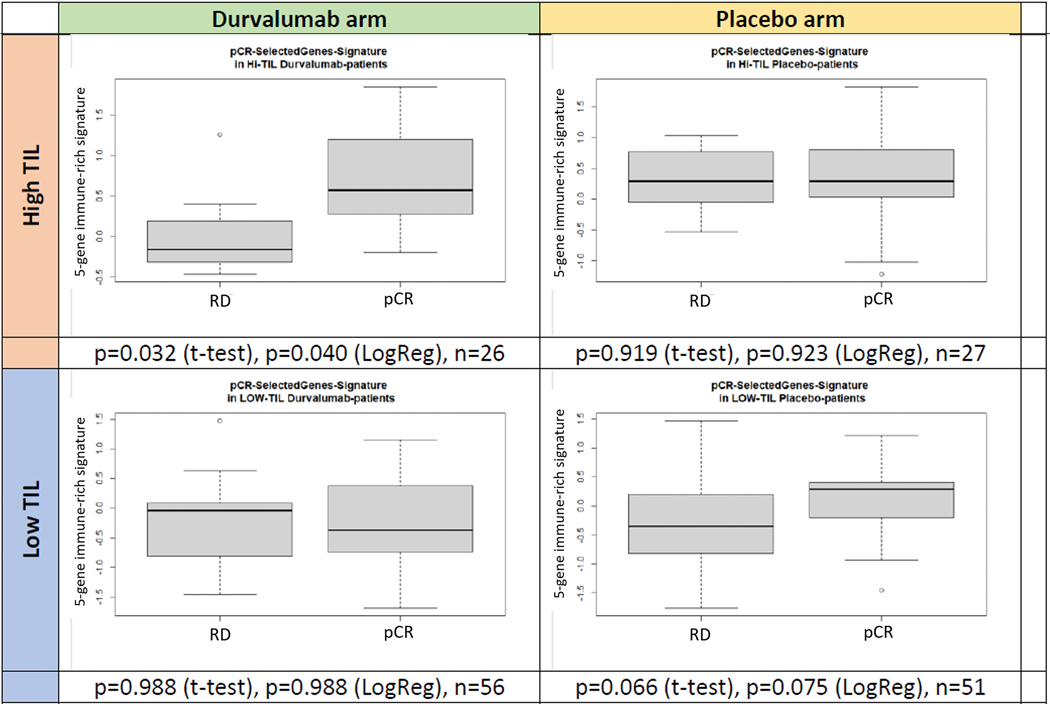
The GeparNuevo randomized trial used an essentially identical durvalumab plus chemotherapy arm as our study and therefore represents an ideal validation cohort that also provides an opportunity to test the immunotherapy-specific predictive role of our response associated genes. We tested if the leading-edge genes that distinguished immune-rich cancers with pCR from those with RD in the Yale cohort were also differentially expressed between pCR and RD in immune-rich cancers from the GeparNuevo trial. Only 36 of the leading-edge genes associated with pCR in immune-rich bTNBC had expression data available from the GeparNuevo samples. Supplementary data File Table S35 lists the gene level validation results. Most importantly, we observed that from our gene list IFNG and IL21 were significantly positively associated with pCR in the chemotherapy alone arm and CXCL9, CXCL13, CD79A, and cytotoxins GZMA and GZMB were positively associated with pCR only in the durvalumab arm. Chemokines CXCL1 and CXCL3 were positively associated with RD in chemotherapy alone arm whereas CSF1, Toll-like receptor TLR3, CCL5, CXCL10, and CCL4 were associated with RD in durvalumab arm only. An immune-rich pCR signature created from the mean value of the scaled expression of IFNG, IL2, IL21, CD79A, and GZMB that individually showed a weak association with pCR (P<0.2) in GeparNuevo, showed significantly higher expression in cases with pCR in the durvalumab arm (p=0.040) but not in the placebo arm (p=0.923) or in immune-poor cancers irrespective of treatment (Figure 4).
Figure 4. Expression of a 5-gene index of leading-edge genes associated with pCR in immune-rich bTNBC discovered in the Yale cohort and tested in the two arms of the GeparNuevo trial.
The five genes include IFNG, IL2, IL21, CD79A, and GZMB. Expression levels are shown in the durvalumab+chemotherapy and chemotherapy alone (i.e placebo) arms, each stratified by tumor infiltrating lymphocyte (TIL).
Germline and somatic mutation landscape associated with response
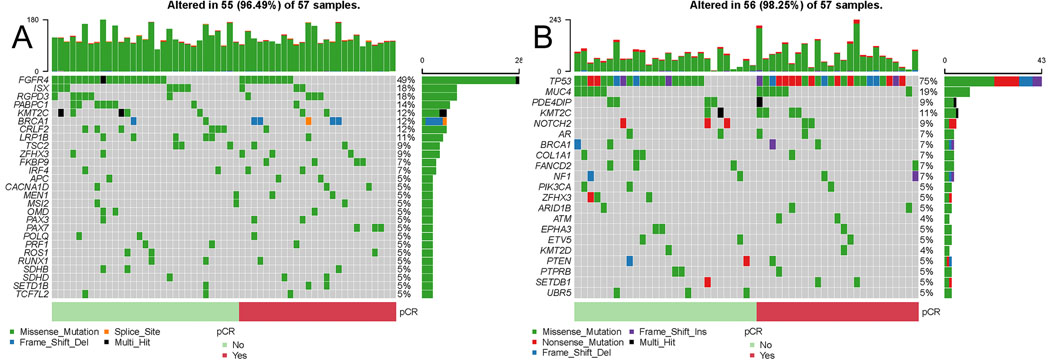
We identified 206 protein coding genes with high functional impact germline variants affecting at least 5 (out of 57) patients. Among genes affected by germline variants, there were no significant differences in variant frequency by pathologic response after adjusting for multiple comparison (Figure 5A, Supplementary Data File Tables S36). Eight patients had germline BRCA1/2 mutation (5 pCR, 3 RD, p=0.2). Somatic mutations affected 3422 distinct genes, among these 1342 were mutated in only one cancer (Figure 5B, Supplementary Data File Tables S37). The most frequently mutated gene was TP53 (24 pCR, 22 RD). There was no statistically significant difference in somatic mutation frequencies by pathologic response for any gene after adjustment for multiple comparison. Next, we assessed associations between response and pathway level mutations separately for somatic mutations and high functional impact germline variants. Four pathways were significantly enriched in high functional impact germline variants including PI3K, DNA damage repair, MAPK, and WNT/β-Catenin signaling pathways (p<0.05) (Table 1; Supplementary Data File Table S38). These pathways were more frequently affected in cases with pCR. Somatic mutations were enriched in 22 pathways (p<0.05) including the same 4 pathways identified in the germline analysis (Table 1; Supplementary Data File Table S39). Higher TMB was significantly associated with pCR and was independent of immune gene signature expression (Table 2; Supplementary Data File Tables S40).
Figure 5. Pathways affected by germline variants or somatic mutations in bTNBC with pCR and RD.
(A, B) Oncoplots of germline and somatic mutations in COSMIC genes ordered by pathologic response. (C) Significantly Germline and somatic variants pathway associations. Pathways with permutation P-values <0.05 are shown. (D) Multivariate analysis of association between pathologic response and tumor mutation burden (TMB) and the GeparSixto and tumor inflammation (TIS) gene signatures.
Table 1.
Germline and somatic variants pathway associations. Pathways with permutation P-values <0.05 are shown.
| Pathway | Source | Mutation | pCR (n=26) | RD (n=31) | Mutation Rate in pCR | Mutation Rate in RD | Permutation P value | OR | OR Lower 95% CI |
|---|---|---|---|---|---|---|---|---|---|
| PI3K | Nanostring metabolic |
Germline | 16 | 9 | 0.6154 | 0.2903 | 0.0082 | 3.9111 | 2.2232 |
| DNA Damage Repair | Nanostring metabolic |
Germline | 10 | 5 | 0.3846 | 0.1613 | 0.0307 | 3.2500 | 1.7254 |
| MAPK | Nanostring metabolic |
Germline | 8 | 4 | 0.3077 | 0.1290 | 0.0435 | 3.0000 | 1.5141 |
| WNT_BETA _CATENIN _SIGNALING |
MSigDB Hallmark |
Germline | 16 | 13 | 0.6154 | 0.4194 | 0.0493 | 2.2154 | 1.2870 |
| INTERFERON_ALPHA _RESPONSE |
MSigDB Hallmark |
Somatic | 12 | 5 | 0.4615 | 0.1613 | 0.0048 | 4.4571 | 2.3808 |
| MAPK | Nanostring metabolic |
Somatic | 25 | 23 | 0.9615 | 0.7419 | 0.0076 | 8.6957 | 2.8966 |
| Myc | Nanostring metabolic |
Somatic | 24 | 22 | 0.9231 | 0.7097 | 0.0089 | 4.9091 | 2.1290 |
| DNA Damage Repair | Nanostring metabolic |
Somatic | 25 | 23 | 0.9615 | 0.7419 | 0.0131 | 8.6957 | 2.8966 |
| Transcriptional Regulation | Nanostring metabolic |
Somatic | 25 | 23 | 0.9615 | 0.7419 | 0.0175 | 8.6957 | 2.8966 |
| Wnt_pathway | Nanostring hallmarks |
Somatic | 24 | 23 | 0.9231 | 0.7419 | 0.0184 | 4.1739 | 1.7971 |
| Cell Cycle | Nanostring metabolic | Somatic | 24 | 23 | 0.9231 | 0.7419 | 0.0184 | 4.1739 | 1.7971 |
| Transcriptional_misregulation | Nanostring hallmarks |
Somatic | 24 | 23 | 0.9231 | 0.7419 | 0.0217 | 4.1739 | 1.7971 |
| WNT_BETA _CATENIN _SIGNALING |
MSigDB Hallmark |
Somatic | 24 | 23 | 0.9231 | 0.7419 | 0.0217 | 4.1739 | 1.7971 |
| MYC_ TARGETS_V2 |
MSigDB Hallmark |
Somatic | 7 | 3 | 0.2692 | 0.0968 | 0.0250 | 3.4386 | 1.6221 |
| DNA_REPAIR | MSigDB Hallmark |
Somatic | 25 | 25 | 0.9615 | 0.8065 | 0.0268 | 6.0000 | 1.9645 |
| HYPOXIA | MSigDB Hallmark |
Somatic | 16 | 12 | 0.6154 | 0.3871 | 0.0287 | 2.5333 | 1.4670 |
| INTERFERON_GAMMA _RESPONSE |
MSigDB Hallmark |
Somatic | 15 | 10 | 0.5769 | 0.3226 | 0.0313 | 2.8636 | 1.6481 |
| UV _RESPONSE _UP |
MSigDB Hallmark |
Somatic | 12 | 8 | 0.4615 | 0.2581 | 0.0320 | 2.4643 | 1.3956 |
| APICAL _SURFACE |
MSigDB Hallmark |
Somatic | 8 | 4 | 0.3077 | 0.1290 | 0.0333 | 3.0000 | 1.5141 |
| Cell_cycle_and_apoptosis | Nanostring hallmarks |
Somatic | 25 | 24 | 0.9615 | 0.7742 | 0.0344 | 7.2917 | 2.4113 |
| Cytokine & Chemokine Signaling | Nanostring metabolic |
Somatic | 25 | 24 | 0.9615 | 0.7742 | 0.0344 | 7.2917 | 2.4113 |
| E2F _TARGETS |
MSigDB Hallmark |
Somatic | 25 | 24 | 0.9615 | 0.7742 | 0.0344 | 7.2917 | 2.4113 |
| PI3K | Nanostring metabolic |
Somatic | 25 | 24 | 0.9615 | 0.7742 | 0.0360 | 7.2917 | 2.4113 |
Table 2.
Multivariate analysis of TMB and inflammatory gene signatures (GeparSixto; TIS).
| Modela | Multivariate |
|---|---|
| pCR~TMB | OR 1.62 (1.09 – 2.61) |
| P=0.0279 | |
| pCR~GeparSixto | OR 2.86 (1.37 – 6.80) |
| P=0.0091 | |
| pCR~TIS | OR 3.06 (1.44 – 7.56) |
| P=0.0073 | |
| pCR~TMB+GeparSixto | OR 1.83 (1.16 – 3.29) |
| P=0.0213 | |
| pCR~TMB+TIS | OR 1.81 (1.15 – 3.28) |
| P=0.0249 |
Model+Age+Tsize+Nstatus
Discussion
Low levels of TILs and low expression of a broad range of immune genes were associated with lack of pCR after chemotherapy plus durvalumab therapy. The lower immune infiltration was accompanied by higher expression of TGFβ and mesenchymal features of the cancer. TGFβ is an important negative regulator of cellular immunity and has been implicated in immune evasion and resistance to PD-L1 blockade in multiple cancer models (36–38). These observations suggest that targeting TGFβ could remove a barrier to immune infiltration and create a more immune competent tumor microenvironment in immune-cold cancers.
While immune-rich cancers more frequently achieve pCR, a substantial minority continues to have viable residual cancer at surgery. We, therefore, examined transcriptional differences between immune-rich bTNBC that had pCR versus those with RD. Immune-rich cancers that achieved pCR were characterized by activated T cells and B cells, high expression of immunoglobulins, granzymes, granulysin, and HLA class II antigens. The dominant cytokines in the tumor microenvironment were IFNγ, IL-2,−12,−21, CXCL-9,−13 and CXCR5. These are classic chemoattractants and activators of T cells, B cells, and mediators of adaptive immunity (39). In contrast, immune-rich bTNBCs with RD were enriched in genes associated with myeloid/macrophage activity including monocyte chemoattractants CCL5 and CXCL10, interleukins that these cells secrete (IL-1,−23,−27,−34), and classic toll-like receptors (TLR-1,−2,−3,−4,−5,−6) that provide pathogen recognition and subsequent activation of innate immunity. RD samples were also enriched in genes inhibiting the complement system including CD46 and CD55. An impaired complement system can hinder both antibody-dependent and -independent cell death and diminish macrophage-mediated phagocytosis. These results suggest that some immune-rich cancers have a possibly dysfunctional innate, rather than adaptive immune response to the cancer that makes these cancers less responsive to cytotoxic and PD-L1 directed therapies. Altering the cytokine environment by targeting IL-1 (40) and the macrophage monocyte lineage might alter the balance between a dysfunctional innate and more effective adaptive immune response in otherwise immune rich TNBC.
In a single arm anti-PDL1 plus chemotherapy trial it is not possible to determine which response marker, if any, is selectively predictive of benefit from the combination versus individual components. Multiple studies have demonstrated that higher immune infiltration that can be captured by a large number of different immune gene signatures due to their highly correlated co-expression is associated with higher pCR rate to chemotherapy with or without immune checkpoint inhibitors. Our results indicated that there are significant differences in the cytokine and immune milieu of immune rich cancers that achieved pCR with durvalumab plus chemotherapy versus those that did not. We therefore tested if the leading-edge genes that distinguished immune-rich cancers with pCR from those with RD in the Yale cohort were also overrepresented in cancers with pCR in immune-rich cancers from the GeparNuevo trial, and if any of the genes could predict benefit selectively from durvalumab. We could only perform partial validation due to many missing genes in the GeparNuevo data, but reassuringly several genes showed a similar trend as seen in the Yale cohort and several cytokines showed a differential predictive role by treatment arm. We created a 5-gene signature from genes individually weekly associated with pCR that showed significantly higher expression in immune rich TNBC with pCR in the durvalumab arm (p=0.040) but not in the placebo arm (p=0.923) or in immune-poor cancers irrespective of treatment.
Our study has limitations, as we could only partially validate our observations in the similar GeparNuevo trial due to missing information on many candidate genes. The GeparNuevo data was generated on a different RNAseq platform and represent targeted sequencing with a different dynamic range than whole transcriptome RNAseq. These differences decrease the power and accuracy of our validation attempt. We also recognize that the candidate genes were identified in the Yale cohort but the 5-gene durvalumab plus chemotherapy predictive signature itself was selected from the GeparNuevo data, and therefore further validation on independent data will be required. Due to co-linear expression of many immune genes, it is entirely possible that other genes could also provide the same, or even better, response discriminating function. However, the combined analysis of these two trials suggests that there are immunological differences between immune-rich TNBC that achieve pCR and those that do not. These differences can inspire new therapeutic strategies and may hold the key for developing new biomarkers for treatment selection.
We also examined associations between pathologic response and germline variants in coding genes and somatic mutations. We found no statistically significant differences in somatic mutation or germline variant frequencies by pathologic response for any gene. However, TMB was significantly higher in cancers with pCR. When mutations were mapped to biological pathways, we found that cancers with pCR had significantly more germline variants and somatic mutations in the PI3K, DNA damage repair, MAPK, and WNT/β-Catenin pathways. There were no statistically significantly more frequently mutated pathways in cases with RD. The more frequent mutations in cancer relevant signaling pathways and DNA repair genes might lead to a more immunogenic cancer, however we found no positive correlation between TMB and immune gene expression, similar to an earlier study (41). It is more likely that mutations in these cancer relevant pathways directly lead to increased chemotherapy sensitivity due to DNA damage repair deficiency (42–44).
In conclusion, genes in the TGFβ pathway are associated with immune-attenuated phenotype and lack of pCR. Among immune-rich cancers that fail to achieve pCR, macrophage/neutrophil and innate immunity related chemoattractants dominate the cytokine milieu, whereas in cancers with pCR, IFNγ and activated B and T cells and adoptive immunity related markers dominate the tumor microenvironment. Inhibitors of complement cascade blockers, TGFβ inhibitors and modulators of tumor associated macrophages may improve immunotherapy efficacy in basal-like TNBC.
Supplementary Material
Translational Relevance Statement.
We found that high tumor mutation burden and immune-rich microenvironment are independently associated with pathologic complete response (pCR) to anti-PD-L1 therapy plus chemotherapy in basal-like triple negative breast cancer (bTNBC), whereas lack of pCR and immune-poor phenotype are associated with higher expression of TGFβ pathway and epithelial/mesenchymal markers. Immune-rich bTNBC with residual disease are characterized by higher expression of CCL-3,−4,−5,−8,−23, CXCL-1,−3,−6,−10, interleukins-1,−23,−27,−34 and more abundant in macrophage markers. Immune-rich bTNBC with pCR are characterized by activated T and B cell markers and expression of IFNγ, interleukins-2,−12,−21, CD79A, and GZMB. No mutation in single genes was associated with response, but cancers with pCR had significantly more germline variants and somatic mutations in the PI3K, DNA damage repair, MAPK, and WNT/β-Catenin pathways that could affect chemotherapy sensitivity.
Acknowledgments
This work was supported by a National Cancer Institute grant R01CA219647 to L. Pusztai, a Susan Komen Foundation Leadership Award (SAC160076) to L. Pusztai, and investigator awards from the Breast Cancer Research Foundation (AWDR11559) to L. Pusztai and D. Rimm, and research grant M82 from H.W. & J. Hector-Foundation (Mannheim, Germany) to T. Karn.
Conflict of Interest Statement:
M.M., T.Q., T.P., V.Y., Y.B., E.R, X.L., V.G., D.W., E.I., M.R., and J.F. have no competing interests. A.S. has received honoraria and consulting fees from Astra Zeneca. L.P. has received consulting fees and honoraria from Pfizer, Astra Zeneca, Merck, Novartis, Genentech, Eisai, Pieris, Immunomedics, Seagen, Almac and Biotheranostics. D.R. has served as an advisor for Astra Zeneca, Amgen, Cell Signaling Technology, Cepheid, Danaher, Daiichi Sankyo, Genoptix/Novartis, GSK, Konica Minolta, Merck, NanoString, PAIGE.AI, Roche, Sanofi, and Ventana. Amgen, Cepheid, NavigateBP, NextCure, and Konica Minolta fund research in DR’s lab. K.B. serves on the Scientific Advisory Board of CDI Labs. B.S. reports non-financial support from HTG Molecular Diagnostics and personal fees from Novartis, outside the submitted work and has a patent EP18209672 pending. S.L. reports grants and other from Abbvie, Amgen, Celgene, Novartis Roche, AstraZeneca, Pfizer, Daiichi-Sankyo, grants from Immunomedics, personal fees from Chugai, Seattle Genetics, PriME/ Medscape, Lilly, Samsung, BMS, Puma, MSD, Pierre Fabre, Merck outside the submitted work, and has patents EP14153692.0 and EP18209672 pending. C.D. received personal fees from Novartis, Roche, MSD Oncology, Daiichi Sankyo, grants from Myriad Genetics, Sividon Diagnostics / Myriad, outside the submitted work, and has patents EP18209672, EP20150702464 pending. T.K. has a patent EP18209672 pending.
References
- 1.Nanda R, Liu MC, Yau C, Shatsky R, Pusztai L, Wallace A, et al. Effect of Pembrolizumab Plus Neoadjuvant Chemotherapy on Pathologic Complete Response in Women With Early-Stage Breast Cancer: An Analysis of the Ongoing Phase 2 Adaptively Randomized I-SPY2 Trial. JAMA Oncol 2020;6(5):676–84 doi 10.1001/jamaoncol.2019.6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loibl S, Untch M, Burchardi N, Huober J, Sinn BV, Blohmer JU, et al. A randomised phase II study investigating durvalumab in addition to an anthracycline taxane-based neoadjuvant therapy in early triple negative breast cancer - clinical results and biomarker analysis of GeparNuevo study. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO 2019. doi 10.1093/annonc/mdz158. [DOI] [PubMed] [Google Scholar]
- 3.Schmid P, Cortes J, Pusztai L, McArthur H, Kummel S, Bergh J, et al. Pembrolizumab for Early Triple-Negative Breast Cancer. The New England journal of medicine 2020;382(9):810–21 doi 10.1056/NEJMoa1910549. [DOI] [PubMed] [Google Scholar]
- 4.Mittendorf EA, Zhang H, Barrios CH, Saji S, Jung KH, Hegg R, et al. Neoadjuvant atezolizumab in combination with sequential nab-paclitaxel and anthracycline-based chemotherapy versus placebo and chemotherapy in patients with early-stage triple-negative breast cancer (IMpassion031): a randomised, double-blind, phase 3 trial. Lancet 2020;396(10257):1090–100 doi 10.1016/S0140-6736(20)31953-X. [DOI] [PubMed] [Google Scholar]
- 5.Pusztai L, Yau C, Wolf DM, Han HS, Du L, Wallace AM, et al. Durvalumab with olaparib and paclitaxel for high-risk HER2-negative stage II/III breast cancer: Results from the adaptively randomized I-SPY2 trial. Cancer Cell 2021;39(7):989–98 e5 doi 10.1016/j.ccell.2021.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmid P, Cortes J, Dent R, Pusztai L, McArthur H, Kümmel S, et al. VP7–2021: KEYNOTE-522: Phase III study of neoadjuvant pembrolizumab + chemotherapy vs. placebo + chemotherapy, followed by adjuvant pembrolizumab vs. placebo for early-stage TNBC. Annals of Oncology 2021;32(9):1198–200 doi 10.1016/j.annonc.2021.06.014. [DOI] [Google Scholar]
- 7.Foldi J, Silber A, Reisenbichler E, Singh K, Fischbach N, Persico J, et al. Neoadjuvant durvalumab plus weekly nab-paclitaxel and dose-dense doxorubicin/cyclophosphamide in triple-negative breast cancer. NPJ Breast Cancer 2021;7(1):9 doi 10.1038/s41523-021-00219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Warren S, Pelekanou V, Wali V, Cesano A, Liu M, et al. Immune profiling of pre- and post-treatment breast cancer tissues from the SWOG S0800 neoadjuvant trial. J Immunother Cancer 2019;7(1):88 doi 10.1186/s40425-019-0563-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinn BV, Loibl S, Hanusch CA, Zahm DM, Sinn HP, Untch M, et al. Immune-related Gene Expression Predicts Response to Neoadjuvant Chemotherapy but not Additional Benefit from PD-L1 Inhibition in Women with Early Triple-negative Breast Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2021;27(9):2584–91 doi 10.1158/1078-0432.Ccr-20-3113. [DOI] [PubMed] [Google Scholar]
- 10.Karn T, Denkert C, Weber KE, Holtrich U, Hanusch C, Sinn BV, et al. Tumor mutational burden and immune infiltration as independent predictors of response to neoadjuvant immune checkpoint inhibition in early TNBC in GeparNuevo. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO 2020;31(9):1216–22 doi 10.1016/j.annonc.2020.05.015. [DOI] [PubMed] [Google Scholar]
- 11.Wimberly H, Brown JR, Schalper K, Haack H, Silver MR, Nixon C, et al. PD-L1 Expression Correlates with Tumor-Infiltrating Lymphocytes and Response to Neoadjuvant Chemotherapy in Breast Cancer. Cancer immunology research 2015;3(4):326–32 doi 10.1158/2326-6066.CIR-14-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pelekanou V, Barlow WE, Nahleh ZA, Wasserman B, Lo YC, von Wahlde MK, et al. Tumor-Infiltrating Lymphocytes and PD-L1 Expression in Pre- and Posttreatment Breast Cancers in the SWOG S0800 Phase II Neoadjuvant Chemotherapy Trial. Mol Cancer Ther 2018;17(6):1324–31 doi 10.1158/1535-7163.MCT-17-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmed FS, Gaule P, McGuire J, Patel K, Blenman K, Pusztai L, et al. PD-L1 Protein Expression on Both Tumor Cells and Macrophages are Associated with Response to Neoadjuvant Durvalumab with Chemotherapy in Triple-negative Breast Cancer. Clinical Cancer Research 2020;26(20):5456–61 doi 10.1158/1078-0432.Ccr-20-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez-Ericsson PI, Wulfkhule JD, Gallagher RI, Sun X, Axelrod ML, Sheng Q, et al. Tumor-specific major histocompatibility-II expression predicts benefit to anti-PD-1/L1 therapy in patients with HER2-negative primary breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2021. doi 10.1158/1078-0432.CCR-21-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertucci F, Finetti P, Viens P, Birnbaum D. Difference in therapeutic response between basal and nonbasal triple-negative breast cancers. The oncologist 2013;18(9):1060–1 doi 10.1634/theoncologist.2013-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masuda H, Baggerly KA, Wang Y, Zhang Y, Gonzalez-Angulo AM, Meric-Bernstam F, et al. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clinical cancer research : an official journal of the American Association for Cancer Research 2013;19(19):5533–40 doi 10.1158/1078-0432.Ccr-13-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Institute B. FastQC: A Quality Control tool for High Throughput Sequence Data. http://wwwbioinformaticsbabrahamacuk/projects/fastqc/ 2020.
- 18.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30(15):2114–20 doi 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1):15–21 doi 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics 2012;28(16):2184–5 doi 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 21.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011;12:323 doi 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 2014;15(12):550 doi 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612 doi 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sergushichev AA. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv 2016:060012 doi 10.1101/060012. [DOI] [Google Scholar]
- 25.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25(14):1754–60 doi 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20(9):1297–303 doi 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31(3):213–9 doi 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong C, Wei P, Jian X, Gibbs R, Boerwinkle E, Wang K, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet 2015;24(8):2125–37 doi 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 2018;46(D1):D1062–D7 doi 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536(7616):285–91 doi 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Research 2018;47(D1):D941–D7 doi 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Annals of oncology : official journal of the European Society for Medical Oncology 2015;26(2):259–71 doi 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park JH, Jonas SF, Bataillon G, Criscitiello C, Salgado R, Loi S, et al. Prognostic value of tumor-infiltrating lymphocytes in patients with early-stage triple-negative breast cancers (TNBC) who did not receive adjuvant chemotherapy. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO 2019;30(12):1941–9 doi 10.1093/annonc/mdz395. [DOI] [PubMed] [Google Scholar]
- 34.Reisenbichler ES, Han G, Bellizzi A, Bossuyt V, Brock J, Cole K, et al. Prospective multi-institutional evaluation of pathologist assessment of PD-L1 assays for patient selection in triple negative breast cancer. Mod Pathol 2020;33(9):1746–52 doi 10.1038/s41379-020-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poeta VM, Massara M, Capucetti A, Bonecchi R. Chemokines and Chemokine Receptors: New Targets for Cancer Immunotherapy. Frontiers in immunology 2019;10 doi ARTN 37910.3389/fimmu.2019.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larson C, Oronsky B, Carter CA, Oronsky A, Knox SJ, Sher D, et al. TGF-beta: a master immune regulator. Expert Opin Ther Targets 2020;24(5):427–38 doi 10.1080/14728222.2020.1744568. [DOI] [PubMed] [Google Scholar]
- 37.Tauriello DVF, Palomo-Ponce S, Stork D, Berenguer-Llergo A, Badia-Ramentol J, Iglesias M, et al. TGF beta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018;554(7693):538-+ doi 10.1038/nature25492. [DOI] [PubMed] [Google Scholar]
- 38.Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang YL, et al. TGF beta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554(7693):544-+ doi 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohli K, Pillarisetty VG, Kim TS. Key chemokines direct migration of immune cells in solid tumors. Cancer gene therapy 2021. doi 10.1038/s41417-021-00303-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voronov E, Apte RN. Targeting the Tumor Microenvironment by Intervention in Interleukin-1 Biology. Current pharmaceutical design 2017;23(32):4893–905 doi 10.2174/1381612823666170613080919. [DOI] [PubMed] [Google Scholar]
- 41.Safonov A, Jiang T, Bianchini G, Gyorffy B, Karn T, Hatzis C, et al. Immune Gene Expression Is Associated with Genomic Aberrations in Breast Cancer. Cancer research 2017;77(12):3317–24 doi 10.1158/0008-5472.CAN-16-3478. [DOI] [PubMed] [Google Scholar]
- 42.Jiang T, Shi W, Wali VB, Pongor LS, Li C, Lau R, et al. Predictors of Chemosensitivity in Triple Negative Breast Cancer: An Integrated Genomic Analysis. PLoS medicine 2016;13(12):e1002193 doi 10.1371/journal.pmed.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD, et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep 2018;23(1):239–54 e6 doi 10.1016/j.celrep.2018.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma P, Barlow WE, Godwin AK, Parkes EE, Knight LA, Walker SM, et al. Validation of the DNA Damage Immune Response Signature in Patients With Triple-Negative Breast Cancer From the SWOG 9313c Trial. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2019;37(36):3484–92 doi 10.1200/jco.19.00693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.