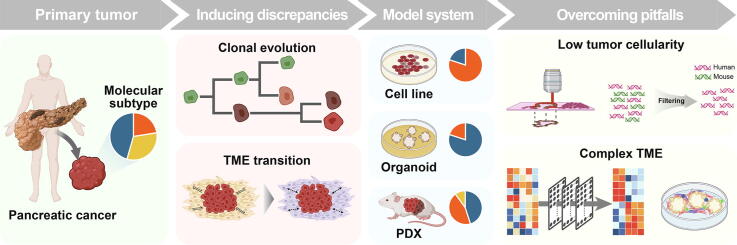
Graphical abstract
Keywords: Pancreatic ductal adenocarcinoma, Cancer genomics, Model system, Clonal evolution, Tumor microenvironment, Bioinformatics
Abstract
Pancreatic ductal adenocarcinoma (PDAC) is characterized by aggressive tumor behavior and poor prognosis. Recent next-generation sequencing (NGS)-based genomic studies have provided novel treatment modes for pancreatic cancer via the identification of cancer driver variants and molecular subtypes in PDAC. Genome-wide approaches have been extended to model systems such as patient-derived xenografts (PDXs), organoids, and cell lines for pre-clinical purposes. However, the genomic characteristics vary in the model systems, which is mainly attributed to the clonal evolution of cancer cells during their construction and culture. Moreover, fundamental limitations such as low tumor cellularity and the complex tumor microenvironment of PDAC hinder the confirmation of genomic features in the primary tumor and model systems. The occurrence of these phenomena and their associated complexities may lead to false insights into the understanding of mechanisms and dynamics in tumor tissues of patients. In this review, we describe various model systems and discuss differences in the results based on genomics and transcriptomics between primary tumors and model systems. Finally, we introduce practical strategies to improve the accuracy of genomic analysis of primary tissues and model systems.
1. Introduction
Pancreatic cancer is a notoriously devastating disease with a 5-year survival rate of about 10 % [1]. Unfortunately, the incidence and mortality of pancreatic ductal adenocarcinomas (PDAC), the most common pancreatic cancer accounting for 90 % of pancreatic cancer, are increasing worldwide [2], [3]. In 2020, this disease was the seventh leading cause (4.7 %) of all cancer deaths in both men and women and had the 14th highest incidence of all cancers.
To characterize this fatal cancer, state-of-the-art genomic approaches have been extensively applied to human and animal tissues and in vitro cultured cells [4]. Nevertheless, the intra-tumoral complexity of PDAC limits the advances in treatment efficacy, and targeted therapy is still under development [5]. Additionally, resolving the inter-tumoral and inter-systematic variability remains a challenge to the precise understanding of genomic and transcriptomic profiles.
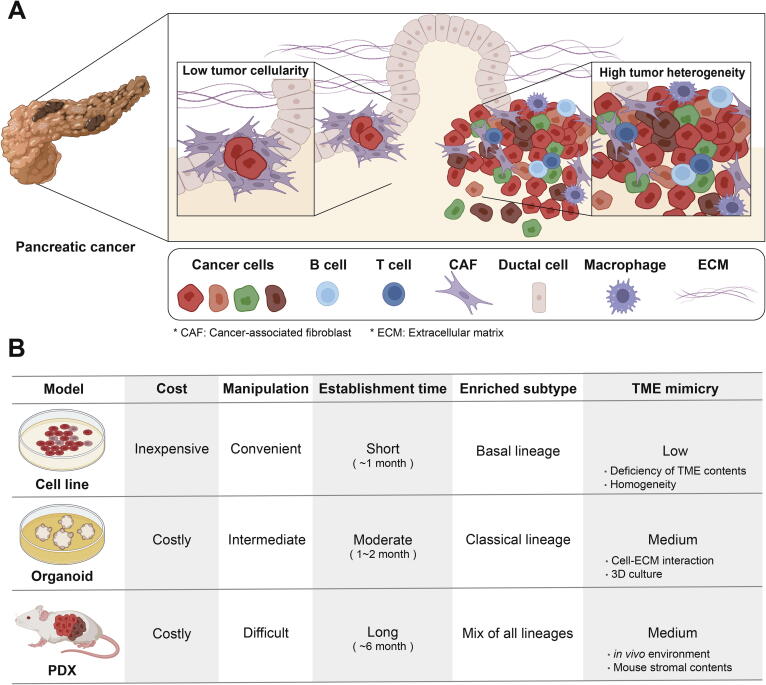
PDAC is one of the cancers with the lowest tumor cellularity, which is the proportion of tumor cells in a tissue [6]. Large-scale genomic studies have shown that the median cellularity of PDACs was only 26 % compared to that of all cancers (81.1 %) [7]. This low tumor cellularity results in the heterogeneity and complexity of the tumor microenvironment (TME) consisting of various non-cancer cells (Fig. 1A) [5], [8]. For example, the diverse cells include cancer-associated fibroblasts (CAFs), macrophages, stellate cells, lymphocytes, and a high proportion of normal pancreatic acinar cells [5], [8], [9], [10]. The presence of this complex composition hinders the accurate genomic characterization of cancer cells in PDAC as non-cancer cell contaminants. Furthermore, the cellularity issue in PDAC studies is aggravated when tumor tissues are exposed to conditions of selection and expansion of cells within the TME, therefore, clonal drift diversifies the genomic features of PDACs [11], [12].
Fig. 1.
Characteristics of PDAC tissue and patient-derived model. (A) PDAC exhibits low tumor cellularity surrounded by plenty of non-cancerous normal cell components. Also, PDAC is characterized by high tumor heterogeneity due to complex TME and diverse cancer cell clones with genetic alterations. (B) Representative PDAC patient-derived model systems are cell line, PDX, and organoid. Cell line is the most basic and easiest to handle. PDX has the advantage of being able to perform various in vivo tests, and organoid is a model that supplements disadvantages of 2D culture in cell lines. Model systems have biased proportions of PDAC molecular subtypes.
To explore the complexity of PDAC and develop the relevant therapeutics, various model systems such as cell lines, patient-derived xenografts, and organoids, have been constructed. Extensive genomic studies on these model systems showed that distinct genomic characteristics between primary tumor tissue and model system were a cautionary consideration in the interpretation of the experimental results [13], [14]. This discrepancy is mainly due to the phenomenon of activated clonal evolution relative to the primary tissue, occurring in model systems [4], [15]. The differential selection of a variety of tumor cell clone types from primary tumor tissues for the construction and culturing of the model system leads to differences in mutation frequency, copy number variation (CNV), and gene expression between the primary tumor tissue and model system [15], [16]. Therefore, understanding the clonal transition from a primary tumor tissue to a model system is essential for acquiring accurate insights into the biology of PDAC and its therapeutic opportunities.
In this review, we summarize the current status and statistics of patient-derived model systems for PDAC. Genomic profile differences between primary tumor tissue and model system are introduced in two perspectives: (1) landscape of structural variations (SVs), DNA mutations, and CNVs; and (2) characterization of molecular subtypes. Finally, we discuss the approaches to avoid the pitfalls of analyzing the profiles in PDAC. Therefore, we show that caution should be exercised when interpreting genomic data from the model systems of PDAC.
2. Patient-derived model systems of PDAC
2.1. Cell line
Cell lines are the most classical and widely used model among patient-derived model systems. The advantage of cell lines is that they are inexpensive and convenient for maintenance and handling (Fig. 1B) [10]. In addition, since the cell populations are much more homogeneous than primary tumor tissues, cell lines ensure reproducibility by making it convenient to conduct repeated experiments [17]. However, the cell line system, unfortunately, does not reflect the environment of the primary tumor, meaning that cell lines often fail to reproduce the experimental results obtained in tumor tissues [18], [19]. Patient-derived cell lines (PDCLs) might include the TME of the corresponding primary tumor in early passages but rapidly lose the complexity upon repeated subculturing [20]. Thus, the use of cell lines is more appropriate for investigations into the biology of cancer cells rather than the cancer environment.
PANC-1, MIA PaCa-2, AsPC-1, and Capan-1 are examples of pancreatic cancer cell lines bearing mutations at KRAS G12 which is frequently mutated in the primary tumor, but the cell lines have distinct KRAS genotypes [21]. PANC-1 and AsPC-1 have the KRAS G12D mutation whose allelic status is heterozygous and homozygous, respectively [22]. Furthermore, MIA PaCa-2 and Capan-1 have homozygous KRAS G12C and KRAS G12V mutations, respectively. As a result, the cell lines of pancreatic cancer exhibit different phenotypes such as cell adhesion, growth rate, tumorigenicity, and even drug resistance [21]. Therefore, careful choice of cell lines considering the pair of genotype and phenotype is necessary for the purpose and context of experiments.
The landscape of genotype and phenotype (e.g., drug sensitivity) have been comprehensively examined in the cancer cell line consortiums; for instance, the Cancer Cell Line Encyclopedia (CCLE), established in 2012 [23]. In 2019, this database was expanded to include the genomic data (e.g., RNA-seq and whole exome sequencing (WES)) of the cell lines in addition to the existing drug screening information [24]. As a result, the availability of genome-wide information for cell lines has offered an opportunity to reinforce insights into the primary tumors. As of March 7, 2022, CCLE includes 58 pancreatic cancer samples and 55 PDAC samples. The PDAC cell lines account for a relatively smaller proportion than lung cancer (275 samples) and breast cancer (83 samples) lines in the database. Considering that the survival rates of PDAC are steady and the incidence is increasing gradually, the construction of PDAC cell lines is warranted for further research. Beyond KRAS G12 mutation, the cell lines with diverse genotypes should be available for expanding the choice to examine the heterogenous cancer cell types. Currently, genome-wide genotyping followed by characterization of phenotypes derived from complex genotypes has become a common practice for cancer research using cell lines.
To comprehensively search pancreatic cancer genotypes, the catalogue of somatic mutations in cancer (COSMIC) revealed that the proportion of the samples with cancer mutations in 32 pancreatic cancer cell lines is as follows: KRAS (94 %), TP53 (91 %), SMAD4 (34 %), and CDKN2A (25 %) [25]. A database for primary tumors, the international cancer genome consortium (ICGC), described that KRAS mutations were found in 92.16 %, 89.51 %, and 66.67 % of cohorts in Canadian (PACA-CA), Australian (PACA-AU), and The Cancer Genome Atlas (PAAD-US), respectively [26]. These results show that COSMIC cell line data have a similar or higher proportion of samples with KRAS mutation compared to KRAS mutation rates in primary tumors. In contrast, TP53 mutations account for a lower proportion of the ICGC data (PACA-CA: 78.93 %, PACA-AU: 65.98 %, and PAAD-US: 54.24 %) than the COSMIC cell lines data. The frequent observation of TP53 mutations in the cell lines suggests that cell lines have higher tumor cellularity than primary tumor tissue through the rapid growth of the clones bearing TP53 mutations.
2.2. Organoid
Organoids are advantageous as models due to their three-dimensional (3D) cell culture (Fig. 1B) [10], [17]. Three-dimensional cultures can be prevented from attaching to the culture dish using a matrix such as collagen or Matrigel [4], allowing them to mimic in vivo conditions, mainly the extracellular matrix (ECM). In addition, it is possible to establish organoid cultures with relatively fewer cells. However, organoids cannot fully account for the action of environment-related stromal cells including immune cells, and the subtypes of cancer organoids could be altered during culture [27], [28]. To overcome these limitations, attempts to coculture organoids with CAFs or use media that includes immune cells while culturing organoids have been increasing [4]. Since the construction of organoids has not been standardized, the protocols for quality control are still actively under development [29].
The characteristics of PDAC organoids are known to change dynamically according to genomic studies and drug screening [30], [31], [32], [33], [34], [35]. Seino et al. analyzed 39 PDAC patient-derived organoids through WES and microarray [31]. Generally, the proportion of frequently mutated genes such as KRAS, TP53, SMAD4, and CDKN2A in organoids is similar to that seen in patient tumors. However, the detailed genomic profiles such as variant allele frequency, structural variation, and CNV showed discrepancies between patient tumors and organoids [14], [32], [34]. Another example is a study conducted in 2021 on single-cell RNA-seq for 24 organoids derived from PDAC patients [36]. Investigators observed a lack of cells with basal-like characteristics in the PDAC organoid, indicating that the in vitro condition made the PDAC organoid acquire classical characteristics due to selective pressure. These results support that the characteristics of tumor cells are not static during the process of organoid construction from the primary tumor.
To take advantage of organoids, several biobanks have been established by academia and industry [37]. The Human Cancer Models Initiative (HCMI) is a representative database containing genomic data and patient information regarding various cancer-derived models, including organoids (https://ocg.cancer.gov/programs/HCMI). Pancreatic cancer organoid accounts for 28 samples of 127 3D organoids, making it the second most represented in this database after colon cancer, among 18 cancer types. Taking the incidence into account, the proportion of pancreatic cancer organoids is relatively similar to or slightly higher than that of other cancer types, indicating that a number of PDAC organoids were constructed despite the lower incidence than other cancers. Tiriac et al. established PDAC organoid using the endoscopic ultrasound-guided fine needle biopsy sampling method with a success rate of 87 % (33/38) [38]. Methods using a core needle are known to increase the accuracy of PDAC diagnosis by conserving the tissue structure. Therefore, reducing the normal cell content using these methods could be a solution to increase the success rate for the establishment of PDAC organoids.
2.3. Patient-derived xenograft
Patient-derived xenografts (PDXs) are representative of in vivo model systems (Fig. 1B) [17]. This model is established by transplanting human primary tumor tissue into immunocompromised mice [4]. A majority of PDAC PDXs (1833/1965 = 93 %) are constructed through subcutaneous implantation, and establishing orthotopic PDXs are relatively infrequent compared to subcutaneous PDXs although they have been used for tissue-specific research by imitating pancreatic TME [14], [15], [34], [39], [40], [41], [42], [43], [44], [45]. PDX is expected to have a TME similar to that of primary tumor tissue, and thus, it can accurately predict drug response by facilitating in vivo experimentation [39].
However, while PDX is constructed from the primary tumor and cultured across the passage, the human stroma is lost and is replaced with mouse stroma [40]. Tumor-stroma interaction in PDACs is known as a desmoplastic reaction, which is a universal characteristic of PDACs [46]. This interaction occurs between complex cell components and ECM, normal epithelial cells, stromal fibroblasts, and tumor cells. It can determine the genomic variation, drug resistance, tumor growth, and invasion. In summary, the replacement of human stroma with mouse stroma triggers different tumor-stroma interactions from the primary tumor and could provide a mouse-specific tissue environment in PDX, which can induce strong clonal evolution of the tumor [47].
Regarding clonal selection, the occurrence of evolution in a mouse-specific manner during the progression from the primary tumor to PDX is controversial. Through CNV analysis as an evolutionary genomic signature, Ben-David et al. analyzed CNVs of 1,110 PDX samples and identified the augmented changes in CNVs during engraftment and passaging PDX [15], distinctively from primary tumors. For instance, the patient-specific recurrent CNVs disappeared during PDX progression. These results suggest that clonal evolution occurred through mouse-specific selective pressure. Since alterations in the genomic characteristics of PDX determined drug response, these results suggested that PDX might not adequately explain the primary tumor.
In contrast, another study showed that clonal evolution does not occur in PDX and is consistent with the CNV profile of the primary tumor [41]. In this study, CNV profiling of 509 PDXs matched with patients revealed the strong conservation of CNVs during the engraftment process from patient tumors to late-passage PDXs. These results reveal that mouse-specific genomic alterations and clonal evolution did not severely occur during PDX construction.
This general debate on PDX is likely to be more serious with PDAC PDX. The stroma proportion of PDAC is higher than that of other cancer types because PDAC has low tumor cellularity and a heterogeneous environment. Our group performed an integrative genomics analysis of 36 PDAC PDXs and matched tissue samples (unpublished data). We confirmed that the CNV and variant allele frequency of cancer-associated genes are discordant between the primary tumor and PDX. Therefore, when PDAC PDX is used as a model system, the issue of clonal evolution should be seriously considered.
An increase in the importance of PDX in preclinical testing has led to the establishment of large-scale databases. Patient-Derived Model Repository (PDMR), Jackson Laboratory, and the EurOPDX Consortium are the representative databases of PDX [42], [43], [44]. EurOPDX and Jackson Laboratory hold 34 and 15 pancreatic cancer PDXs, respectively. In contrast, 466 PDXs derived from 47 patients with pancreatic cancer are available in PDMR. PDMR has abundant PDX samples because various passages are included. However, the number of patients is small in the PDMR database because PDX is difficult to engraft. The establishment success rate of PDAC PDXs is approximately 62 %, which is higher than that of breast cancer (13–27 %) [17], [40]. This shows that PDAC PDX has a high engraftment rate among cancer types. In 2017, a study demonstrated that engraftment rates and tumor growth rates of PDX have a correlation with the prognosis of patients [39]. In this study, PDAC PDXs derived from patients with poor prognosis have high engraftment rates and rapid xenograft growth rates. These results suggest that aggressive and metastatic tumors are stably engrafted while constructing PDXs. In other words, primary tissues with high normal cell contents may have low PDX engraftment rates because those subtypes are correlated with favorable prognoses. Therefore, although PDAC is known as an aggressive cancer type, the establishment of PDAC PDX can be difficult because primary tumor tissues often have low tumor cellularity.
3. Clonal evolution by genetic alterations in model systems
Clonal evolution is driven by genetic variations such as somatic mutations, chromosomal rearrangement, CNV, and epigenetic modification [48]. Since these variations accelerate tumor cell proliferation, specific clones of tumor cells are selected by genetic selection pressure. When PDAC is generated from normal cells, tumor progression occurs through the three pancreatic intraepithelial neoplasia (PanIN) stages [49]. In the PanIN-1 stage, KRAS mutation is introduced in over 99 % of the samples [50]. CDKN2A mutation occurs during early PanIN-2 stage. In PanIN-3, mutations leading to inactivation of TP53 and SMAD4 are accumulated and subsequently cause the generation of invasive cancer. These genetic alterations that accumulate as tumors are initiated and progress, driving clonal evolution.
Clonal evolution occurs during passaging and constructing model systems as well as tumor progression and metastasis in the patient [51]. PDX was not accurately reflected in the heterogeneity of metastatic tumors due to clonal evolution analysis from primary tumors [45]. Mutations of APC, TP53, and TCF7L2 genes were commonly identified in primary tumor and PDX, but mutations of ROBO1, SMAD3, and KMT2C were not represented in PDX. In addition, the KRAS Q22K mutation found in the primary tumor did not appear in PDX. Therefore, it is important to map the genetic characteristics different from primary tumors in the model system for elucidating the evolution of PDAC.
Since clonal evolution occurs by the accumulation of genetic alterations during tumor progression, analysis of genetic alterations such as SV, mutation, and CNV enables us to track clonal evolution (Table 1). First, SV discrepancy between primary tumor and model system indicates genetic alteration resulting from clonal evolution. One study observed inconsistent SV between primary tumor tissue and model system in PDAC using whole genome sequencing [14]. SVs within most chromosomes were mismatched between primary tumor and PDX pairs, and only 40 % of the total samples showed a high SV concordance score. Additionally, they confirmed that PDX had at least twice as many insertions and deletions as the primary tumor. These results indicate that variants could be accumulated by deficiency of the DNA repair pathway in PDX. The SV discrepancy between the primary tumor and the model system indicates that there is structural heterogeneity, which can be explained as the result of clonal evolution by selective pressure.
Table 1.
Discrepancies in genetic alteration between primary tumor and model system. Clonal evolution occurs due to accumulation of genetic alterations while model system is constructed from primary tumor. Clonal evolution leads to discrepancies in genetic alteration profiles between primary tumor and model system. This table summarizes the differences in genetic alterations between the two groups.
| Genetic alteration | Primary tumor | Model system | Reference |
|---|---|---|---|
| SV | |||
| SV events concordance |
|
|
[14], [52] |
| Insertion and deletion |
|
|
[14], [52] |
| Mutation | |||
| Significantly mutated genes |
|
|
[34], [50], [53], [54] |
| KRAS mutation genotype |
|
|
[35], [54] |
| CNV | |||
| Loci and concordance |
|
|
[14], [54], [55] |
| Recurrence |
|
|
[15] |
| Copy number of CDKN2A | CNV mean log2 ratio was approximately −1.5 for CDKN2A and CDKN2B. |
|
[56] |
Second, clonal evolution also can be traced by identifying the accumulation of the mutation. Romero-Calvo et al. conducted targeted capture DNA sequencing while constructing primary tumors, PDXs, organoids, PDX-derived organoids, and cell lines [34]. KRAS, TP53, and CDKN2A mutations in the primary tumor were maintained in the model system. However, the model system revealed a high variant allele frequency (VAF) compared to the primary tumors; 57.69 and 12.44 are the medians of PDX and primary tumors, respectively. In another study, KRAS mutant allele frequency (MAF) increased by passages of PDAC organoids. The first passage had G12V, G12D, and G12R mutations in KRAS with MAF 33 %, 9 %, and 1 %, respectively [35]. In passage 3, the MAF of G12R was steeply increased to 51 %, whereas G12V and G12D variants disappeared. In passage 4, the pattern of passage 3 was maintained. This result supports the expansion of clones with particular variant alleles through clonal evolution during culture and establishment in the model system.
Finally, copy number amplification or deletion provided evidence of clonal evolution. When CNV was estimated for primary tumor and PDX pairs, most pairs had similar ploidy, but a few pairs showed that PDX ploidy was twice higher than that of primary tumors [14]. This indicated that primary tumor and PDX pairs had CNV concordance across the entire genome, but not in local chromosome regions. These results were also reproduced in organoids. Another study described that the clonal evolution of subclones in primary tumors plays an important role in developing the CNV environment of PDX [15]. They investigated whether the recurrent CNV identified in the patient tumor tissue was maintained by selection pressure during PDX construction and passage. Interestingly, repeatedly occurring CNV in the TCGA data tended to be lost during PDX passages. This suggested that the acquisition and maintenance of patient-specific CNV may not exist in the model environment due to clonal evolution. These clonal dynamics should be considered for experimental design and interpretation, and caution is warranted in using the model system for further preclinical steps such as drug response tests.
4. Inconsistencies in molecular subtypes between primary tissue and model system
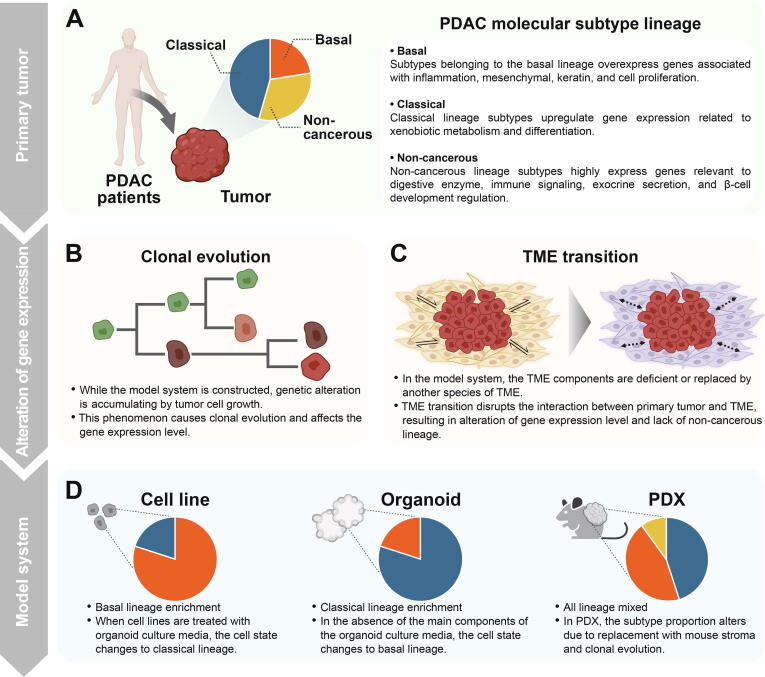
Gene expression profiles have been primarily used to classify molecular subtypes of PDAC primary tumor tissue. Based on the expression profiles, representative molecular subtypes for PDAC primary tumors were defined in Collison et al., Moffitt et al., and Bailey et al. [57], [58], [59]. The molecular subtypes of PDAC are divided into three major lineages (Fig. 2A). First, basal lineage includes quasimesenchymal (QM-PDA), basal-like, and squamous. These subtypes typically have a poor prognosis compared to the subtypes of other lineages. The transcriptional signatures of the basal lineage subtypes are characterized by over-expression of inflammation, mesenchymal, keratin, and cell proliferation genes. Second, classical lineage contains the classical and pancreatic progenitor. Classical lineage subtypes highly express GATA6 and genes associated with transcription factor network, xenobiotic metabolism, and differentiation [36], [58], [59]. Finally, non-cancer cell-related lineage consists of aberrantly differentiated endocrine exocrine (ADEX), immunogenic, and exocrine-like. These subtypes have been argued to be subtypes caused by normal cell contamination [59], [60]. Tumors in the subtypes upregulated genes relevant to tumor cell-derived digestive enzymes, immune signaling, exocrine secretion, and β-cell development regulation [57], [59]. Exceptionally, Chan-Seng-Yue et al. and Topham et al. defined the hybrid and discordant subtypes for samples that do not belong to these three representative lineages [61], [62]. In the model systems, clonal evolution and TME reshaping lead to inconsistent subtypes compared to primary tumor tissues (Fig. 2B, C). Therefore, the model system-specific gene expression profiling followed by subtyping is essential for using the model system as a preclinical tool to correctly reflect patients’ subtypes.
Fig. 2.
Transition of molecular subtypes during model system establishment. (A) PDAC molecular subtypes based on gene expression are largely divided into three lineages: basal, classical, and non-cancerous. (B-C) Clonal evolution and TME transition occur during model system construction. These phenomena alter gene expression levels of both cancer and stromal cells, leading to differences in molecular characteristics of cancer cells compared to primary tissues. Also, model systems do not perfectly mimic patient’s TME. (D) Unique properties of culture conditions and clonal evolution in each model system yield skewed proportions of three PDAC subtype lineages.
Cancerous subtypes of PDAC make up a high proportion of the model system. The relatively poor proliferating capability of normal cells results in more cancer cells in model systems. Accordingly, TME transitions in PDCL reduce the proportion of normal cells involved in digestive enzyme secretion. As the subtype associated with endocrine and exocrine activities was characterized by high normal cell content, this subtype was deficient in PDCL [53]. To explain the difference, attempts have been made to define the specific subtype for the model system [32], [63]. Most of the subtypes identified in the model system were similar to the basal and classical lineages defined in PDAC primary tumors, meaning that the construction of the model system does not modify the characteristics of the cancer cell itself but alters the cellular composition from the primary tumor (Fig. 2D). Taken together, the model system is lack of subtypes such as ADEX and immunogenic due to insufficient TME components, thereby basal and classical subtypes are abundant because of clonal evolution or expansion.
The model system consists of mostly subtypes known as basal and classical lineages, but each model has different proportions for these two subtypes. Previous studies have confirmed that the basal type was predominant in cell lines. In a study by Moffitt et al., all 17 PDAC cell lines were found to belong to the basal-like subtype [58]. Consistently, independent research for subtyping of PDAC cell lines showed that the basal lineage subtype accounted for 63 % (23/36) of the total samples [63]. In contrast, 80 % (35/44) of PDAC organoids were classified as a classical lineage subtype [32].
In order to examine the dynamic alteration of subtypes in the model systems, single-cell RNA-seq (scRNA-seq) technology has been actively utilized for dissecting heterogeneous cellular states. scRNA-seq data showed that the cell state of the organoids shifted from the classical to basal when PDAC organoids were cultured without major components of the organoid medium such as mEGF and Wnt3A [64]. Furthermore, the basal score of PDAC organoids increased when the cell line media was treated to the organoid instead of the organoid media, whereas growing the cell line in organoid media reduced the basal score. Autocrine TGFB and paracrine IFNG were the key shifting factors from a classical state into a basal or intermediate cell state that was a precursor to the basal cell state, indicating that secretory TME components can determine the subtype state of tumor cells. Notably, ADEX and immunogenic subtypes are not well-maintained in the ex vivo model system due to the TME component deficiency, and subtype transition occurs depending on culture media and condition.
A drug sensitivity study using scRNA-seq revealed that precise subtyping of the model system is crucial for the proper use and interpretation of the system [36]. When PDAC organoids were treated with drugs such as 5-fluorouracil and gemcitabine, the classical subtype organoids showed sensitive drug response and had highly differentiated cell states. In contrast, basal-like organoids had proliferating gene signatures and exhibited poor drug response, suggesting the subtype-specific drug response. Since the classical subtype constituted a majority of PDAC organoids, drug tests using organoids could lead to biased results and provide inaccurate insights. Moreover, a few primary tumor subtypes (i.e., ADEX, immunogenic subtype) are rarely present in the model systems, and even the molecular subtypes shift due to culture conditions. Therefore, inconsistency of molecular subtypes with the primary tumor should be considered when using the model system.
5. Strategies to overcome pitfalls of genomics analysis of PDAC model systems
The heterogeneity of PDAC is augmented by the high proportion of normal cells and low tumor cellularity in the PDAC tissue [65]. These characteristics reduce the detection sensitivity of genetic variants, interrupting interpretations of PDAC progression. In addition, these issues hinder the classification of subtypes and drug responses, preventing the development of personalized medicine. The model system has emerged to disentangle these issues, but the limitation of the model system is that it cannot perfectly mimic the TME of the patient tumor tissue. Since TME is altering dynamically during the construction of model systems, tracing the TME transition is crucial to correctly interpreting PDAC. Therefore, it is necessary to understand the TME and tumor heterogeneity of the model system in combination with the primary tumor.
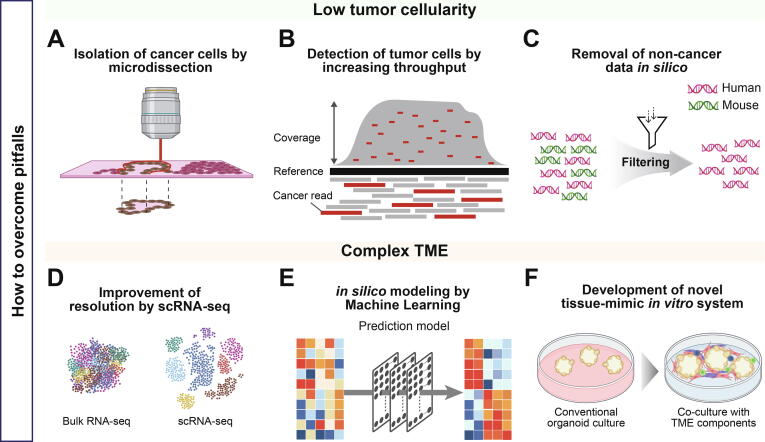
To overcome the obstacles posed by low tumor cellularity in tissue, previous studies have used microdissection as an attempt to analyze the high tumor cellularity state [58], [61]. Recently, an elaborate and convenient technique known as laser microdissection (LMD) has been used to focus on the region of interest [66] (Fig. 3A). For instance, a study by Immervoll et al., showed 67 % mutation to be detected in the 12th codon of KRAS after DNA extraction from whole tissue [67]. In contrast, the frequency of this mutation was 91 % in samples that underwent LMD. LMD facilitates accurate analysis of tumors specifically and decreases normal cell contamination.
Fig. 3.
Advanced genomic analysis strategies for PDAC model systems. Low tumor cellularity and complex TME of PDAC are obstacles to performing precise genomic analysis. (A) Issues of low tumor cellularity issue can be physically resolved by resecting tumor cells through microdissection. (B) Acquisition of more genomic data by increasing sequencing depth, improving chances of detecting genomic information of cancer cells. (C) Filtering out genomic data of other species helps to focus on human-derived cells in cases of low cellularity caused by including non-human genomic data such as PDX. (D) scRNA-seq provides highest resolution to unravel complex TME at a single-cell level. (E) Classification accuracy can be improved by subtype prediction model using computational approaches such as artificial intelligence. (F) Addition of stromal components on 3D cultures is an experimental strategy to mimic complex TME of tissues.
Another approach to overcome the low tumor cellularity issue is to increase the sequencing depth (Fig. 3B). By increasing the sequencing depth, the elevated chance of detecting nucleic acids derived from cancer cells improves genomic interpretation for low abundance cancer cells [68]. Furthermore, the classification of gene expression-based subtypes could be incorrect in samples with low tumor cellularity due to the inclusion of normal tissue-associated gene expression, suggesting that measurement of low abundant tumor RNAs is critical for the correct classification of PDAC tumor tissues. Previous studies have already adopted strategies to increase the sequencing depth of the tumor compared to that of the normal control or to perform targeted sequencing of specific genes [52], [54], [69], [70]. A study by the TCGA network described deep coverage sequencing (mean ∼ 30,000X) being conducted for the hotspot codon of KRAS to improve the detection of somatic mutations in samples with low cellularity [54]. In addition, they performed targeted sequencing (∼644X) for PDAC-specific mutated genes. As the model systems have high tumor cellularity compared to the primary tumors, excessive sequencing depth is not necessary for the samples in the model systems. Therefore, optimal sequencing depth should be produced mainly based on the tumor cellularity of the primary tumor in order to compare genomic characteristics between primary tumors and model systems in pairs.
We previously discussed that human stroma is replaced by mouse stroma, resulting in an interaction between the mouse stroma and the transplanted tumor in PDX. Mouse stroma is one of the causes of genetic discrepancies between PDX and primary tumors. In addition, even though tissues from xenograft models are cautiously resected for sequencing, cross-contamination may occur because human or mouse DNA or RNA are mixed [71]. Therefore, in order to prevent errors in interpretation, genomic data derived from xenografts and human tissues should be separated (Fig. 3C). One previous study inferred that unfiltered mouse-derived reads caused more false-positive variant calls [72]. Filtered samples exhibited a proper correlation between the predicted allele frequency and the real allele frequency. Thus, analysis of the xenograft should be followed by filtering genomic data from non-human sources.
The development of scRNA-seq technologies has resolved the cellular complexity of the model system at single-cell resolution (Fig. 3D). These methods can help to accurately understand the transcriptomic discrepancy and TME between the tumor and the model system. In a scRNA-seq study for PDAC organoid, the proliferating cells either re-entered the cell cycle stage or entered the differentiation phase and acquired unique characteristics of pancreas tissue [36]. This suggested that the TME dynamics during organoid establishment can be delineated at a single cell level. Subsequently, drug tests showed that the cellular ratio between the classical and basal cell types determined the drug sensitivity in an organoid. Drug-sensitive organoids were made of cells with high expression of differentiation genes. In contrast, the cells in drug-resistant organoids upregulated the expression of genes related to proliferation and cell cycling. This demonstrated that the drug response in the model system could be inferred by tracking TME dynamics at the single cell level. Ultimately, identifying the TME dynamics of organoids using scRNA-seq can improve the reliability when organoids are used as a preclinical model.
Recently, prediction modeling methods are developed using machine learning and deep learning. Based on the prediction of the improved computational model, it is possible to accurately understand the model system by resolving the difference in classification with the primary tumor (Fig. 3E). In a previous study, the authors predicted the molecular subtype of pancreatic cancer cell line based on gene expression through prediction model establishment [73]. They applied the nearest template prediction method to predict the subtype of the cell lines. This prediction model revealed a 96 % classification accuracy for the PAAD TCGA tumor test set. In addition, they predicted the subtype of the PAAD cell line using the prediction model. Ten of the cell lines used in this study overlapped the Moffitt et al. study [58]. These ten cell lines were known as the basal-like subtype in Moffitt et al. study. However, this study predicted that eight cell lines were basal and two cell lines were classical. Interestingly, these two cell lines had a relatively high classical score in the Moffit et al. study [58], suggesting that accurate subtype prediction could be achieved by identifying samples with ambiguous subtypes based on cross-classification between primary tumor and model system.
In addition to the strategies to enhance understanding of current systems, a recently advanced new system has simulated more closely patient tumor tissues (Fig. 3F). The novel model system, termed ‘assembloid’, has emerged to improve organoids by adding stromal components such as CAF, endothelial cell, immune cell, and muscle layer [74]. The TME-enhanced assembloid was able to complement the limitations of current organoids in which subtypes were shifted depending on the culture conditions. The bladder tumor assembloid prohibited shifting to the basal subtype and maintained the parental tumor subtype during in vitro culture. Therefore, PDAC assembloid construction is expected to prevent shifting to the dominant classical subtype in the organoid.
6. Summary and outlook
In recent years, various model systems for PDAC have emerged to provide experimental platforms for studying and treating the devastating disease. Since the model system represents the patient’s tumor tissue and is used as a preclinical tool, an accurate understanding of the model system is necessary for the development of cancer therapies. In this review, we have described the differences between the primary tumor and the model system. First, the heterogeneity and cellularity of PDAC amplify the differences in genetic alteration such as SV, mutation, and CNV. The genetic discrepancies are accumulating during tumor progression and subclone selection due to clonal evolution. Second, the molecular subtypes are imperfectly matched between the primary tumor and the model system. The PDAC model system does not perfectly mimic the TME of the patient’s tumor tissue. As a result, the model systems preferentially include cancerous subtypes, and the culture conditions convert the subtypes during culture. The unstable and inconsistent subtypes of the model systems could lead to biased results of drug response tests on PDAC due to subtype-specific drug sensitivities. Therefore, these genomic and transcriptomic differences should be carefully considered and interpreted when the model system is used as a preclinical tool.
This review has described challenges and strategies to overcome the issues associated with the genomic analysis of PDAC tumor tissue and model systems. To alleviate the low tumor cellularity issue in PDAC analysis, we recommend microdissection and an increase in NGS depth. Additionally, we have discussed the computational and experimental approaches to accurately understand the model system along with the patient’s tumor tissue. Discriminating mouse data from human data in silico can reduce cross-contamination in the PDX model, and scRNA-seq can increase the resolution to investigate clonal evolution and subtype transition. Finally, the development of a novel model system such as an assembloid will be required to mimic patient tumor tissue as closely as possible.
Although large-scale genomic studies have been conducted on PDAC, model system studies are still insufficient and the interpretation of the results is controversial. Since there is still no targeted therapy for PDAC, the development of an accurate drug testing system is an urgent need to identify effective drug candidates. Finally, taking clonal evolution and TME transition into account, unbiased genomic analysis of the model system will enable us to move a step forward in conquering deadly cancer.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Science and ICT (NRF-2019R1C1C1008181) and the Ministry of Education (NRF-2021R1A6A1A10044950). We appreciate the helpful advice from Dr. Jae Yun Moon and clinical comments by Won-Gun Yun.
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Sarantis P., Koustas E., Papadimitropoulou A., Papavassiliou A.G., Karamouzis M.V. Pancreatic ductal adenocarcinoma: treatment hurdles, tumor microenvironment and immunotherapy. World J Gastrointest Oncol. 2020;12:173–181. doi: 10.4251/wjgo.v12.i2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 4.Garcia P.L., Miller A.L., Yoon K.J. Patient-derived xenograft models of pancreatic cancer: overview and comparison with other types of models. Cancers (Basel) 2020;12(5):1327. doi: 10.3390/cancers12051327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Juiz N.A., Iovanna J., Dusetti N. Pancreatic cancer heterogeneity can be explained beyond the genome. Front Oncol. 2019;9:246. doi: 10.3389/fonc.2019.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semaan A., Bernard V., Lee J.J., Wong J.W., Huang J., Swartzlander D.B., et al. Defining the comprehensive genomic landscapes of pancreatic ductal adenocarcinoma using real-world endoscopic aspiration samples. Clin Cancer Res. 2021;27:1082–1093. doi: 10.1158/1078-0432.CCR-20-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aran D., Sirota M., Butte A.J. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. doi: 10.1038/ncomms9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karamitopoulou E. Tumour microenvironment of pancreatic cancer: immune landscape is dictated by molecular and histopathological features. Br J Cancer. 2019;121:5–14. doi: 10.1038/s41416-019-0479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleeff J., Korc M., Apte M., La Vecchia C., Johnson C.D., Biankin A.V., et al. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022. doi: 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- 10.Carter E.P., Roozitalab R., Gibson S.V., Grose R.P. Tumour microenvironment 3D-modelling: simplicity to complexity and back again. Trends Cancer. 2021;7:1033–1046. doi: 10.1016/j.trecan.2021.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Pelosi E., Castelli G., Testa U. Pancreatic cancer: molecular characterization, clonal evolution and cancer stem cells. Biomedicines. 2017;5:65. doi: 10.3390/biomedicines5040065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cros J., Raffenne J., Couvelard A., Poté N. Tumor heterogeneity in pancreatic adenocarcinoma. Pathobiology. 2018;85:64–71. doi: 10.1159/000477773. [DOI] [PubMed] [Google Scholar]
- 13.Xie T., Musteanu M., Lopez-Casas P.P., Shields D.J., Olson P., Rejto P.A., et al. Whole exome sequencing of rapid autopsy tumors and xenograft models reveals possible driver mutations underlying tumor progression. PLoS ONE. 2015;10:e0142631. doi: 10.1371/journal.pone.0142631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gendoo D.M.A., Denroche R.E., Zhang A., Radulovich N., Jang G.H., Lemire M., et al. Whole genomes define concordance of matched primary, xenograft, and organoid models of pancreas cancer. PLOS Comput Biol. 2019;15:e1006596. doi: 10.1371/journal.pcbi.1006596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ben-David U., Ha G., Tseng Y.Y., Greenwald N.F., Oh C., Shih J., et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat Genet. 2017;49:1567–1575. doi: 10.1038/ng.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peille A.L., Vuaroqueaux V., Wong S.S., Ting J., Klingner K., Zeitouni B., et al. Evaluation of molecular subtypes and clonal selection during establishment of patient-derived tumor xenografts from gastric adenocarcinoma. Commun Biol. 2020;3:367. doi: 10.1038/s42003-020-1077-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behrens D., Walther W., Fichtner I. Pancreatic cancer models for translational research. Pharmacol Ther. 2017;173:146–158. doi: 10.1016/j.pharmthera.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Gillet J.P., Varma S., Gottesman M.M. The clinical relevance of cancer cell lines. J Natl Cancer Inst. 2013;105:452–458. doi: 10.1093/jnci/djt007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Namekawa T., Ikeda K., Horie-Inoue K., Inoue S. Application of prostate cancer models for preclinical study: advantages and limitations of cell lines, patient-derived xenografts, and three-dimensional culture of patient-derived cells. Cells. 2019;8:74. doi: 10.3390/cells8010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fredebohm J., Boettcher M., Eisen C., Gaida M.M., Heller A., Keleg S., et al. Establishment and characterization of a highly tumourigenic and cancer stem cell enriched pancreatic cancer cell line as a well defined model system. PLoS ONE. 2012;7:e48503. doi: 10.1371/journal.pone.0048503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deer E.L., González-Hernández J., Coursen J.D., Shea J.E., Ngatia J., Scaife C.L., et al. Phenotype and genotype of pancreatic cancer cell lines. Pancreas. 2010;39:425–435. doi: 10.1097/MPA.0b013e3181c15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konishi H., Karakas B., Abukhdeir A.M., Lauring J., Gustin J.P., Garay J.P., et al. Knock-in of mutant K-ras in nontumorigenic human epithelial cells as a new model for studying K-ras mediated transformation. Cancer Res. 2007;67:8460–8467. doi: 10.1158/0008-5472.CAN-07-0108. [DOI] [PubMed] [Google Scholar]
- 23.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghandi M., Huang F.W., Jané-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R., et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569:503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.International Cancer Genome Consortium, Hudson T.J., Anderson W., Artez A., Barker A.D., Bell C., et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jabs J, Zickgraf FM, Park J, Wagner S, Jiang X, Jechow K, et al. Screening drug effects in patient-derived cancer cells links organoid responses to genome alterations. Mol Syst Biol. 2017;13:955. 10.15252/msb.20177697. [DOI] [PMC free article] [PubMed]
- 28.Xu H., Lyu X., Yi M., Zhao W., Song Y., Wu K. Organoid technology and applications in cancer research. J Hematol Oncol. 2018;11:116. doi: 10.1186/s13045-018-0662-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J., Koo B.K., Knoblich J.A. Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol. 2020;21:571–584. doi: 10.1038/s41580-020-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang L., Holtzinger A., Jagan I., BeGora M., Lohse I., Ngai N., et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat Med. 2015;21:1364–1371. doi: 10.1038/nm.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seino T., Kawasaki S., Shimokawa M., Tamagawa H., Toshimitsu K., Fujii M., et al. Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell. 2018;22:454–467.e6. doi: 10.1016/j.stem.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Tiriac H., Belleau P., Engle D.D., Plenker D., Deschênes A., Somerville T.D.D., et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 2018;8:1112–1129. doi: 10.1158/2159-8290.CD-18-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Driehuis E., van Hoeck A., Moore K., Kolders S., Francies H.E., Gulersonmez M.C., et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc Natl Acad Sci U S A. 2019;116(52):26580–26590. doi: 10.1073/pnas.1911273116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romero-Calvo I., Weber C.R., Ray M., Brown M., Kirby K., Nandi R.K., et al. Human organoids share structural and genetic features with primary pancreatic adenocarcinoma tumors. Mol Cancer Res. 2019;17:70–83. doi: 10.1158/1541-7786.MCR-18-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seppälä T.T., Zimmerman J.W., Sereni E., Plenker D., Suri R., Rozich N., et al. Patient-derived organoid Pharmacotyping is a clinically tractable strategy for precision medicine in pancreatic cancer. Ann Surg. 2020;272:427–435. doi: 10.1097/SLA.0000000000004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krieger T.G., Le Blanc S., Jabs J., Ten F.W., Ishaque N., Jechow K., et al. Single-cell analysis of patient-derived PDAC organoids reveals cell state heterogeneity and a conserved developmental hierarchy. Nat Commun. 2021;12:5826. doi: 10.1038/s41467-021-26059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao J., Yang M., Atteh L., Liu P., Mao Y., Meng W., et al. A pancreas tumor derived organoid study: from drug screen to precision medicine. Cancer Cell Int. 2021;21:398. doi: 10.1186/s12935-021-02044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tiriac H., Bucobo J.C., Tzimas D., Grewel S., Lacomb J.F., Rowehl L.M., et al. Successful creation of pancreatic cancer organoids by means of EUS-guided fine-needle biopsy sampling for personalized cancer treatment. Gastrointest Endosc. 2018;87:1474–1480. doi: 10.1016/j.gie.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pergolini I., Morales-Oyarvide V., Mino-Kenudson M., Honselmann K.C., Rosenbaum M.W., Nahar S., et al. Tumor engraftment in patient-derived xenografts of pancreatic ductal adenocarcinoma is associated with adverse clinicopathological features and poor survival. PLoS ONE. 2017;12:e0182855. doi: 10.1371/journal.pone.0182855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hidalgo M., Amant F., Biankin A.V., Budinská E., Byrne A.T., Caldas C., et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014;4:998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woo X.Y., Giordano J., Srivastava A., Zhao Z.M., Lloyd M.W., de Bruijn R., et al. Conservation of copy number profiles during engraftment and passaging of patient-derived cancer xenografts. Nat Genet. 2021;53:86–99. doi: 10.1038/s41588-020-00750-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.The NCI. Patient-Derived Models Repository (PDMR), https://pdmr.cancer.gov/; 2022 [Internet]. Frederick National Laboratory for Cancer Research. last modified Sep 1, 2021, accessed March 12, 2022.
- 43.Krupke D.M., Begley D.A., Sundberg J.P., Richardson J.E., Neuhauser S.B., Bult C.J. The mouse tumor biology database: a comprehensive resource for mouse models of human cancer. Cancer Res. 2017;77:e67–e70. doi: 10.1158/0008-5472.CAN-17-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dudová Z., Conte N., Mason J., Stuchlík D., Peša R., Halmagyi C., et al. The EurOPDX Data Portal: an open platform for patient-derived cancer xenograft data sharing and visualization. BMC Genomics. 2022;23:156. doi: 10.1186/s12864-022-08367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dang H.X., Krasnick B.A., White B.S., Grossman J.G., Strand M.S., Zhang J., et al. The clonal evolution of metastatic colorectal cancer. Sci Adv. 2020;6:eaay9691. doi: 10.1126/sciadv.aay9691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mahadevan D., Von Hoff D.D. Tumor-stroma interactions in pancreatic ductal adenocarcinoma. Mol Cancer Ther. 2007;6:1186–1197. doi: 10.1158/1535-7163.MCT-06-0686. [DOI] [PubMed] [Google Scholar]
- 47.Shi J., Li Y., Jia R., Fan X. The fidelity of cancer cells in PDX models: characteristics, mechanism and clinical significance. Int J Cancer. 2020;146:2078–2088. doi: 10.1002/ijc.32662. [DOI] [PubMed] [Google Scholar]
- 48.Connor A.A., Gallinger S. Pancreatic cancer evolution and heterogeneity: integrating omics and clinical data. Nat Rev Cancer. 2022;22:131–142. doi: 10.1038/s41568-021-00418-1. [DOI] [PubMed] [Google Scholar]
- 49.Yachida S., Iacobuzio-Donahue C.A. Evolution and dynamics of pancreatic cancer progression. Oncogene. 2013;32:5253–5260. doi: 10.1038/onc.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanda M., Matthaei H., Wu J., Hong S.M., Yu J., Borges M., et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology. 2012;142:730–733.e9. doi: 10.1053/j.gastro.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu Y., Yang G., Huang H., Fu Z., Cao Z., Zheng L., et al. Preclinical models of pancreatic ductal adenocarcinoma: challenges and opportunities in the era of precision medicine. J Exp Clin Cancer Res. 2021;40:8. doi: 10.1186/s13046-020-01787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waddell N., Pajic M., Patch A.M., Chang D.K., Kassahn K.S., Bailey P., et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Knudsen E.S., Balaji U., Mannakee B., Vail P., Eslinger C., Moxom C., et al. Pancreatic cancer cell lines as patient-derived avatars: genetic characterisation and functional utility. Gut. 2018;67:508–520. doi: 10.1136/gutjnl-2016-313133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raphael B.J., Hruban R.H., Aguirre A.J., Moffitt R.A., Yeh J.J., Stewart C., et al. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32(2):185–203.e13. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalinina T., Güngör C., Thieltges S., Möller-Krull M., Penas E.M., Wicklein D., et al. Establishment and characterization of a new human pancreatic adenocarcinoma cell line with high metastatic potential to the lung. BMC Cancer. 2010;10:295. doi: 10.1186/1471-2407-10-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seppälä T.T., Zimmerman J.W., Suri R., Zlomke H., Ivey G.D., Szabolcs A., et al. Precision medicine in pancreatic cancer: patient derived organoid pharmacotyping is a predictive biomarker of clinical treatment response. Clin Cancer Res. 2022 doi: 10.1158/1078-0432.CCR-21-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collisson E.A., Sadanandam A., Olson P., Gibb W.J., Truitt M., Gu S., et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moffitt R.A., Marayati R., Flate E.L., Volmar K.E., Loeza S.G., Hoadley K.A., et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bailey P., Chang D.K., Nones K., Johns A.L., Patch A.M., Gingras M.C., et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 60.Collisson E.A., Bailey P., Chang D.K., Biankin A.V. Molecular subtypes of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2019;16:207–220. doi: 10.1038/s41575-019-0109-y. [DOI] [PubMed] [Google Scholar]
- 61.Chan-Seng-Yue M., Kim J.C., Wilson G.W., Ng K., Figueroa E.F., O’Kane G.M., et al. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat Genet. 2020;52:231–240. doi: 10.1038/s41588-019-0566-9. [DOI] [PubMed] [Google Scholar]
- 62.Topham J.T., Karasinska J.M., Lee M.K.C., Csizmok V., Williamson L.M., Jang G.H., et al. Subtype-discordant pancreatic ductal adenocarcinoma tumors show intermediate clinical and molecular characteristics. Clin Cancer Res. 2021;27:150–157. doi: 10.1158/1078-0432.CCR-20-2831. [DOI] [PubMed] [Google Scholar]
- 63.Song L., Qi S., Hu W., Fang Z., Yu D., Liu T., et al. Integrative analysis reveals clinically relevant molecular fingerprints in pancreatic cancer. Mol Ther Nucleic Acids. 2021;26:11–21. doi: 10.1016/j.omtn.2021.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raghavan S., Winter P.S., Navia A.W., Williams H.L., DenAdel A., Lowder K.E., et al. Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer. Cell. 2021;184:6119–6137.e26. doi: 10.1016/j.cell.2021.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heid I., Steiger K., Trajkovic-Arsic M., Settles M., Eßwein M.R., Erkan M., et al. Co-clinical assessment of tumor cellularity in pancreatic cancer. Clin Cancer Res. 2017;23:1461–1470. doi: 10.1158/1078-0432.CCR-15-2432. [DOI] [PubMed] [Google Scholar]
- 66.Funel N., Giovannetti E., Pollina L.E., del Chiaro M., Mosca F., Boggi U., et al. Critical role of laser microdissection for genetic, epigenetic and proteomic analyses in pancreatic cancer. Expert Rev Mol Diagn. 2011;11:695–701. doi: 10.1586/erm.11.62. [DOI] [PubMed] [Google Scholar]
- 67.Immervoll H., Hoem D., Kugarajh K., Steine S.J., Molven A. Molecular analysis of the EGFR-RAS-RAF pathway in pancreatic ductal adenocarcinomas: lack of mutations in the BRAF and EGFR genes. Virchows Arch. 2006;448:788–796. doi: 10.1007/s00428-006-0191-8. [DOI] [PubMed] [Google Scholar]
- 68.Shin H.T., Choi Y.L., Yun J.W., Kim N.K.D., Kim S.Y., Jeon H.J., et al. Prevalence and detection of low-allele-fraction variants in clinical cancer samples. Nat Commun. 2017;8:1377. doi: 10.1038/s41467-017-01470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Witkiewicz A.K., McMillan E.A., Balaji U., Baek G., Lin W.C., Mansour J., et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aguirre A.J., Nowak J.A., Camarda N.D., Moffitt R.A., Ghazani A.A., Hazar-Rethinam M., et al. Real-time genomic characterization of advanced pancreatic cancer to enable precision medicine. Cancer Discov. 2018;8:1096–1111. doi: 10.1158/2159-8290.CD-18-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Conway T., Wazny J., Bromage A., Tymms M., Sooraj D., Williams E.D., et al. Xenome–a tool for classifying reads from xenograft samples. Bioinformatics. 2012;28 doi: 10.1093/bioinformatics/bts236. i172–i178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Woo X.Y., Srivastava A., Graber J.H., Yadav V., Sarsani V.K., Simons A., et al. Genomic data analysis workflows for tumors from patient-derived xenografts (PDXs): challenges and guidelines. BMC Med Genomics. 2019;12:92. doi: 10.1186/s12920-019-0551-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu K., Chen B., Aran D., Charalel J., Yau C., Wolf D.M., et al. Comprehensive transcriptomic analysis of cell lines as models of primary tumors across 22 tumor types. Nat Commun. 2019;10:3574. doi: 10.1038/s41467-019-11415-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim E., Choi S., Kang B., Kong J., Kim Y., Yoon W.H., et al. Creation of bladder assembloids mimicking tissue regeneration and cancer. Nature. 2020;588:664–669. doi: 10.1038/s41586-020-3034-x. [DOI] [PubMed] [Google Scholar]