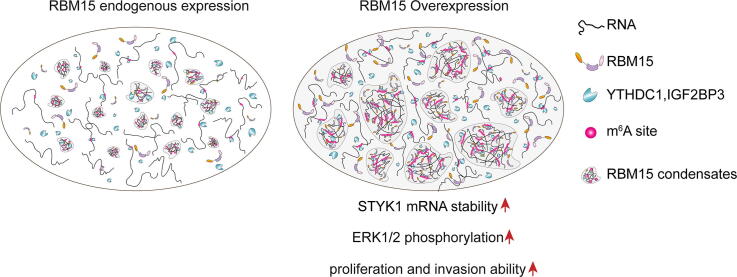
Graphical abstract
Keywords: RBM15, N6-methyladenine modification, Phase separation, STYK1
Abstract
RBM15 expression is recurrently upregulated in several types of malignant tissues, and its high expression level is typically associated with poor prognosis. However, whether and how RBM15 is involved in the tumor progression remains unclear. In this study, we found that overexpressing RBM15 in NIH3T3 cells was able to enhance proliferation rate in vitro and induced subcutaneous tumor formation in vivo. Moreover, we imaged the subcellular localization of RBM15 with our home-built structured illumination super-resolution microscopy, and revealed that RBM15 formed substantial condensates dispersed in the nucleus, undergoing dynamic fusion and fission activities. These condensates were partially colocalized with m6A-modified transcripts in the nucleus. In addition, we confirmed that RBM15 formed “liquid-like” droplets in a protein/salt concentration-dependent manner in vitro, and the addition of RNA further enhanced its phase-separation propensity. To identify downstream targets of RBM15, we performed meRIP-seq and RNA-seq, revealing that RBM15 preferentially bound to and promoted the m6A modification on the mRNA of Serine/threonine/tyrosine kinase 1 (STYK1), thereby enhancing its stability. The upregulated STYK1 expression caused MAPK hyperactivation, thereby leading to oncogenic transformation of NIH3T3 cells.
1. Introduction
RNA-binding motif protein 15 (RBM15) was originally identified in infants with acute megakaryocytic leukemia (AMKL). The infants carried the (1;22) (p13;q13) translocation, which resulted in the fusion of the RBM15 and MKL1 genes [1], [2]. Several subsequent studies reported cases of infantile AMKL with RBM15-MKL1 gene fusion [3], [4], [5], [6], [7], [8], [9], [10], [11]. RBM15 is a ubiquitously expressed gene abundant in the hematopoietic system, which comports with its hematopoietic function [12]. The conserved functional domains of RBM15 include an RNA recognition motif (RRM) N-terminal and a C-terminal Spen paralog and ortholog C-terminal (SPOC). The RRM domain is critical for the binding of RNA molecules, whereas the SPOC domain has been identified in multiple cancer- and apoptosis-related proteins, but its function remains unclear [13]. RBM15 participates in several different aspects of RNA metabolism. For example, RBM15 facilitates the export of mRNA from the nucleus to the cytoplasm [14]. In addition, RBM15 regulates alternative splicing of transcripts [15]. RBM15-knockout mice exhibits embryonic lethality on approximately embryonic day 9.5, suggesting that RMB15 is essential for mice embryonic development [16].
The N6-methyladenine (m6A) modification is a ubiquitous, abundant, and reversible epigenetic modification in RNA molecules. The dynamics and functions of m6A depend on m6A methyltransferases (“writers”), demethylases (“erasers”), and recognition proteins (“readers”). Specifically, the m6A methyltransferase complex consists of a catalytic subunit methyltransferase-like 3 (METTL3) and a regulatory subunit, methyltransferase-like 14 (METTL14), as well as other regulatory subunits, which catalyze the addition of a methyl group at the N6 position of adenosine (A) to form m6A [17]. A recent study identified RBM15 as a regulatory subunit of the methyltransferase complex that was necessary to recruit METTL3 onto its bound transcripts [18], [19], [20]. YTH family members YTH domain-containing (YTHDC1/2) [21], YTH structure N6-methyladenine RNA-binding protein 1/2/3 (YTHDF1/2/3) [22], [23], heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1) [24], and insulin-like growth factor 2 mRNA-binding protein 1–3 (IGF2BP1/2/3) [25] recognize m6A and participate in downstream biological processes. The downstream biological processes mediated by m6A include mRNA transport and degradation [23], [26], alternative splicing [27], [28], and transcription regulation [29]. Notably, one recent study suggested that IGF2BP1/2/3 stabilize RNA transcripts by recognizing the m6A modification, ensuring effective protein production from m6A-marked transcripts [25]. Although previous works have extensively studied the contributions of m6A “writers” and “erasers” to tumor progression [30], [31], [32], [33], the role of m6A regulatory subunits on carcinogenesis remained unknown.
In this study, we found that RBM15 over-expression significantly promoted the proliferation of NIH3T3 cells both in vitro and in vivo. Mechanistically, RBM15 formed “liquid-like” condensates in the nucleus and in vitro. These condensates were partially colocalized with m6A transcripts and promoted the deposition of the m6A modification onto multiple transcripts, such as STYK1 mRNA, which in turn enhanced their stability. The upregulation of STYK1 in turn activated the downstream MAPK signaling pathway. In brief, this work revealed that upregulated RBM15 was able to enhance cell proliferation by depositing m6A modification onto related transcripts.
2. Materials and methods
2.1. Cell culture
HeLa (CCL2), 293 T (crl-3216), and NIH3T3 cells (crl-1658) were purchased from ATCC. All cells were cultured in DMEM (Thermo Fisher, Cat. No.: 11965–092) supplemented with 10 % fetal bovine serum (FBS) and kept in a humid incubator at 37 °C and supplemented with 5 % CO2.
2.2. Plasmid construction
The RBM15 coding sequence was amplified from HeLa cDNA and ligated to a mEmerald-C1 empty vector to generate mEmerald-RBM15 expression vector. The coding sequences of mEmerald-RBM15 and mEmerald were subcloned into a pCDH-CMV-mcs-EF1a-puro vector (System Biosciences Cat. No.: CD510B-1) to generate the lentiviral transfer plasmids pCDH-EGFP and pCDH-EGFP-RBM15. For construction of different RBM15 truncated mutations (ΔN, ΔRRM, and ΔSPOC), Gibson Assembly® Master Mix was used. The primers for the RBM15 truncated mutations were designed on NEBuilder (https://nebuilder.neb.com/). The RBM15 coding sequence was cloned into a pRSFDuet vector (Merk, 71341) to obtain the bacterial expression plasmid. The sequences of all above vectors were verified by Sanger sequencing.
2.3. Generation of stable cell lines
2 μg of the transfer plasmids (pCDH-EGFP or pCDH-EGFP-RBM15), 1 μg of the envelope plasmid pMD2.G (Addgene, Cat. No.: 12259), and 1.5 μg of the packaging plasmid psPAX2 (Addgene, Cat. No.: 12260) were cotransfected into 293 T cells. Forty-eight hours post-transfection, the lentiviral particles in the supernatant were collected and used to infect NIH3T3 cells. Stably transduced NIH3T3 cells (i.e. RBM15 over-expressing or empty vector over-expressing cells) were selected by fluorescence-activated cell sorting (FACS) and selection antibiotics.
2.4. Live-cell imaging
For live-cell imaging, 35-mm glass-bottom dishes (NEST, Cat. No.: 801001) were coated with 200 μL of 0.05 mg/mL collagen (Thermo Fisher, Cat. No.: A1048301) for 1 h in the incubator. 1 × 105 cells were plated on 35-mm glass-bottom dishes 24 h before imaging. The live-cell 3D images were captured using the home-built structured illumination microscopy (SIM), or lattice light-sheet microscopy (LLSM). The detail of SIM and LLSM was described in our previous studies [34], [35], [36].
2.5. Transwell assay
Transwell assay was performed according to the protocol described in our previous report [36]. In brief, NIH3T3 cells were resuspended in serum-free DMEM to obtain a final density of 1x105 cells/mL. 200 μL of the cell suspension were seeded in the upper insert of a BioCoat Matrigel invasion chamber (Corning, Cat. No.: 354480). The inserts were placed on 24-well plates containing 520 µL of DMEM supplemented with 10 % FBS. The cells were fixed with 3.7 % formaldehyde, permeabilized with methanol, and stained with 0.1 % crystal violet 12 h post-seeding. The cells on the upper side of the filter were swapped with cotton. The cells on the lower side of the filter were counted by confocal microscopy with a 20 × objective.
2.6. Proliferation curve
200 μL of cell suspension (104 cells/mL) was dispensed in a 96-well plate and cultured for an appropriate length of time in the incubator. The cell numbers of different groups were counted by TC20 Automated Cell Counter (Bio-Rad).
2.7. RNA isolation and real-time PCR
Total RNA was isolated by TRIzol (Invitrogen, Cat. No.: 15596026). The procedures for total RNA extraction were performed according to the manufacturer’s instructions. First-strand cDNA synthesis was performed with a PrimeScriptTM RT reagent kit (Takara, RR037B) according to the manufacturer’s instruction. RT-qPCR was used to analyze the abundance of transcripts in the cDNA using AceQ Universal SYBR qPCR Master Mix (Vazyme, Cat. No.: Q712). The reactions were carried out on a Bio-Rad CFX96 Touch Real-Time PCR detection system. Relative expression values were calculated from the 2-ΔΔCT method. The express level was normalized to the reference gene GAPDH.
2.8. Immunoblot and immunofluorescence
For immunoblotting, cells were lysed with RIPA buffer (Thermo Scientific, Cat. No.: 89901) or Pierce IP lysis buffer (Thermo Scientific, Cat. No.: 87788) supplemented with protease cocktail inhibitors (TargetMol, Cat. No.: C0001), and the protein concentration in the supernatant was determined by BCA assay (Thermo Scientific, Cat. No.: 23225). 20 μg cell lysates were separated in a precast NuPAGETM Bis-Tris gel (Thermo Fisher, Cat. No.: NP0321PK2) and transferred onto a PVDF membrane. The PVDF membrane was incubated with primary antibodies for 12 h at 4 °C and was washed 3 times in PBS + 0.1 % Triton X-100 (PBST). The corresponding horseradish peroxidase (HRP)-linked secondary antibodies were diluted 5000-fold in PBST and incubated with the PVDF membrane. Pierce ECL Western blotting substrate was used to detect the chemofluorescence signal. Chemoluminescence was captured using a Tanon-5200 chemiluminescent imaging system (Tanon Science & Technology).
For immunofluorescence, cells were seeded on 35-mm glass-bottom dishes and fixed in 4 % formaldehyde for 10 min. The cells were then rinsed three times with PBS for 5 min each time and blocked with blocking buffer (5 % normal donkey serum (Sigma, Cat. No.: D9663) in PBST for 30 min at room temperature. The cells were incubated with primary antibodies diluted in blocking buffer at 4 °C overnight. The samples were rinsed three times with PBST for 5 min each time, incubated with Alexa 488/560 fluorochrome-conjugated secondary antibodies diluted in blocking buffer for 1 h at room temperature in a dark room, and then rinsed three times with PBST for 5 min each time.
2.9. meRIP and RNA-seq
The procedures for the meRIP and RNA-seq were described in detail in our previous paper [36]. meRIP-seq and RNA-seq assays were performed by Shanghai Jiayin Biotechnology Co., ltd.
2.10. TMT labeling of peptides and mass spectrometry
100 μg of desalted peptides were resuspended in 100 μL of 100 mM TEAB (pH = 8.0). Each 0.8-mg vial of TMT reagent was reconstituted in 41 μL of acetonitrile. The resolved TMT reagents were added to the corresponding peptide samples and incubated for 1 h at 25 °C. The labeling reaction was quenched with 8 μL of 5 % hydroxylamine for 15 min at 25 °C. The TMT-labeled peptides were then vacuum dried and desalted using C18 ZipTip columns. The desalted peptides were resuspended in 5 μL of 2 % MeCH/0.1 % FA and analyzed by online nanoflow liquid chromatography-tandem mass spectrometry (LC-MS/MS) using an Orbitrap Exploris™ 480 mass spectrometer (Thermo Scientific) coupled online to a Proxeon Easy-nLC 1200 system (Thermo Scientific). The collected data were analyzed using the SEQUEST HT search engine of Thermo Proteome Discoverer software (2.4.1.15) and searched against the UniProt database containing human reference proteome sequences.
2.11. RBM15 protein purification
pRSFDuet-RBM15 was transformed into chemically competent BL21 (DE3) E. coli cells (Transgene Cat. No.: CD601-02). To express recombinant proteins, the transformed BL21 cells were grown to an OD600 = 0.8 and then induced with 0.5 mM isopropyl-β-d-1-thiogalactoside (IPTG) for 16 h at 16 °C. One liter of bacteria was collected and lysed with an EmulsiFlex-C5 cell homogenizer (Avestin, Ottawa, Canada) in 100 mL of ice-cold lysis buffer (50 mM Tris, pH 7.5; 500 mM NaCl; 10 % (v/v) glycerol; 20 mM imidazole, freshly supplemented with protease inhibitor cocktail; and 1 mM PMSF). The cell lysates were applied to nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen, Germany) at a flow rate of 1 mL/min. The nickel column was washed with 3 volumes of lysis buffer and 3 volumes of 1 × SUMO Protease Buffer (50 mM Tris pH 8.0, 500 mM NaCl, 1 mM DTT). To remove SUMO tag on beads, 50 μL of Ulp1 (2 mg/mL) was added to the column and incubated for 12 h at 4℃. The purified RBM15 protein was eluted and further concentrated to 100 μM using an Amicon Ultra15 filter (Millipore, UFC903024).
2.12. In vitro phase separation
To set up the in vitro phase separation assays, 100 μM RBM15 was diluted into a series of salt and protein concentrations in a glass-bottom 96-well plate (Corning, Cat. No.: 13539050). The plates were then incubated for the desired time, and images were captured by an Olympus FV3000 confocal microscope with a 60 × oil objective. To test how RNA affected the phase separation of RBM15, 10 μM purified RBM15 was incubated with total or predigested RNA for two hours at 150 mM NaCl.
2.13. Bioinformatics analyses of clinical samples
A Kaplan-Meier curve was plotted (https://gepia2.cancer-pku.cn/#survival) based on the RBM15 expression level in patients. The Kaplan-Meier plot parameters were as follows: Methods, overall survival; Cutoff, Median, High 50 %, Low 50 %; Hazards ratio: Yes; 95 % Confidence Interval dot line: No; Axis Unit: Month; Datasets Selection: LGG, KICH, LAUD, ACC, PAAD, and THCA. The quantitative criteria to consider RBM15 abundance were medians in different types of cancers. In another word, if the RBM15 abundance in a patient was bigger than median of specific tumor, it was recognized as high and vice versa.
RBM15 expression abundance was retrieved from an online server Gepia 2.0. The RBM15 expression profiles in Fig. 1B were plotted using the following parameters: Differential methods: ANOVA; log2 fold of change cutoff: 1; q-value: 0.01; log scale: No; Match Normal Data: Match TCGA normal and GTEx data; Database selection: All tumor types. By using of the ANOVA statistical analysis, if RBM15 was identified as upregulated expression compared to paired normal tissues in specific tumor type, then this tumor was labeled in red; in contrast, if RBM15 was identified as downregulated expression in specific tumor, it was labeled in green. However, downregulated expression of RBM15 was not identified in all tumor types. If RBM15 expression remained relatively stable in specific tumor type, the tumor was labeled in black.
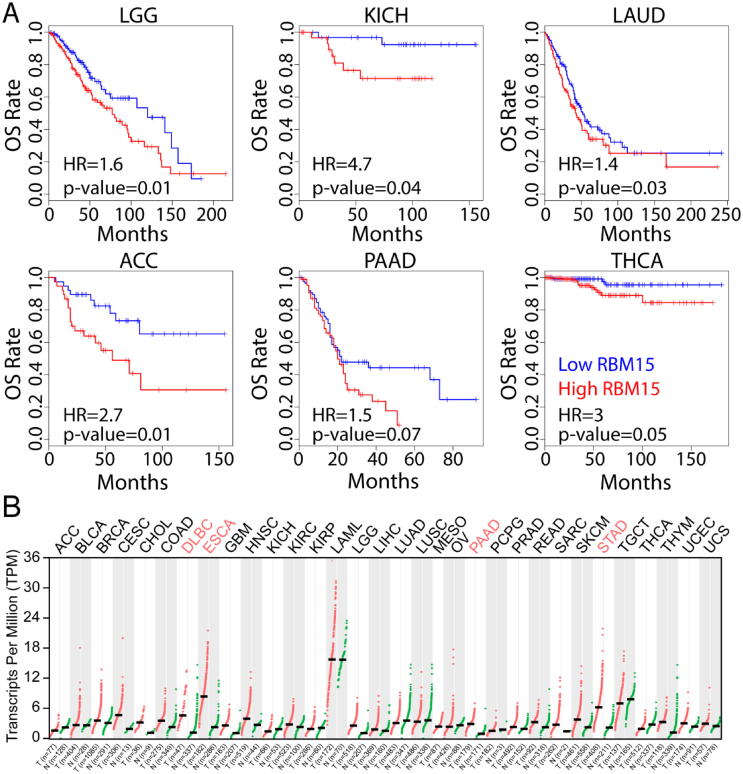
Fig. 1.
The abundance and mutational signatures of RBM15 in clinical samples. (A) High abundance of RBM15 is associated with short overall survival (OS) in different types of cancers. (B) The expression level of RBM15 in the indicated types of cancers. All data were from The Cancer Genome Atlas (TCGA). ACC, Adrenocortical carcinoma; BLCA, Bladder Urothelial Carcinoma; BRCA, Breast invasive carcinoma; CESC, Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, Cholangio carcinoma; COAD, Colon adenocarcinoma; DLBC, Lymphoid Neoplasm Diffuse Large B-cell Lymphoma; ESCA, Esophageal carcinoma; GBM, Glioblastoma multiforme; HNSC, Head and Neck squamous cell carcinoma; KICH, Kidney Chromophobe; KIRC Kidney renal clear cell carcinoma; KIRP, Kidney renal papillary cell carcinoma; LAML, Acute Myeloid Leukemia; LGG, Brain Lower Grade Glioma; LIHC, Liver hepatocellular carcinoma; LUAD, Lung adenocarcinoma; LUSC Lung squamous cell carcinoma; MESO, Mesothelioma; OV, Ovarian serous cystadenocarcinoma; PAAD, Pancreatic adenocarcinoma; PCPG, Pheochromocytoma and Paraganglioma; PRAD, Prostate adenocarcinoma; READ, Rectum adenocarcinoma; SARC, Sarcoma; SKCM, Skin Cutaneous Melanoma; STAD, Stomach adenocarcinoma; TGCT, Testicular Germ Cell Tumors; THCA, Thyroid carcinoma; THYM, Thymoma; UCEC, Uterine Corpus Endometrial Carcinoma; UCS, Uterine Carcinosarcoma; UVM, Uveal Melanoma.
2.14. Animal experiments setup
Five-week old male NOD-SCID mice were purchased from Beijing Vital River Laboratory Animal Technology. Mouse experiments were approved by the Animal Care and Use Committee of Institute of Biophysics, Chinese Academy of Sciences (Approval No. SYXK2021132). The mice were subcutaneously injected with 200 μL suspension of 2 × 106 cells/mL in DMED medium. The mice were sacrificed 4 weeks after xenograft and the tumors were dissected for Hematoxylin and Eosin staining and tumor size measurement.
2.15. FRAP data acquisition and analysis
RBM15 condensates in HeLa cells were photobleached using a 60 × oil immersion objective mounted on an Olympus FV1200 inverted confocal microscopy to measure their dynamic properties. The droplets (n = 30) were bleached to measure the exchange rate between the condensates and the surrounding environment. A frame was collected before bleaching, and then forty-eight frames were captured at 200 ms/frame after bleaching. The interval between images was 1 s. The collected data were imported in to Image J to obtain the mean intensities over time from the following three areas: the bleached spots, regions outside of the cell, and the whole nucleus. The mean intensities were uploaded to the web-based tool easyFRAP (https://easyfrap.vmnet.upatras.gr/) to calculate the recovery time and the mobile fraction. The FRAP curves were fitted with an exponential equation.
2.16. siRNA procedures
2 × 105 cells were plated in a 6-well plate 12 h prior to transfection. Cells were transfected with 10 nM siRNA duplexes against STYK1 using Lipofectamine RNAiMax (Invitrogen, 13778150). The siRNA sequences for STYK1 were designed and synthesized by Ribobio (Guangzhou, China). The siRNA sequences were as follows: si-STYK1-1: 5′- agaaaggcuucuccugaga-dTdT-3′; si-STYK1-2: 5′- caaggaaaggcuuugucuc-dTdT-3′. The knockdown efficacies were validated by RT-qPCR 72 h post transfection. The level of phospho-ERK was detected 72 h post transfection.
3. Results
3.1. RBM15 promotes the proliferation and invasion of cells
To study the correlation between RBM15 abundance and clinical prognosis, we assessed the overall survival (OS) associated with high RBM15 abundance in patients, and compared it to patients with low RBM15 abundance based on the GEPIA2.0 database [37]. We found that the OS of patients with high RBM15 abundance was significantly shorter than that of patients with low RBM15 abundance in six different cancer types (Fig. 1A). In addition, we compared the expression level of RMB15 in cancer tissues with paired normal tissues, and found that the expression level of RBM15 was significantly higher in the four types of cancer tissues than that in the paired normal tissues, and there is no downregulated expression of RBM15 among all of the 32 types of cancers (Fig. 1B). By using the cBioPortal [38], we further revealed that RBM15 gene was frequently amplified or mutated in multiple cancer samples (Fig. 1C). Thus, we speculated the high abundance of RBM15 might associate with the progression of certain types of cancers.
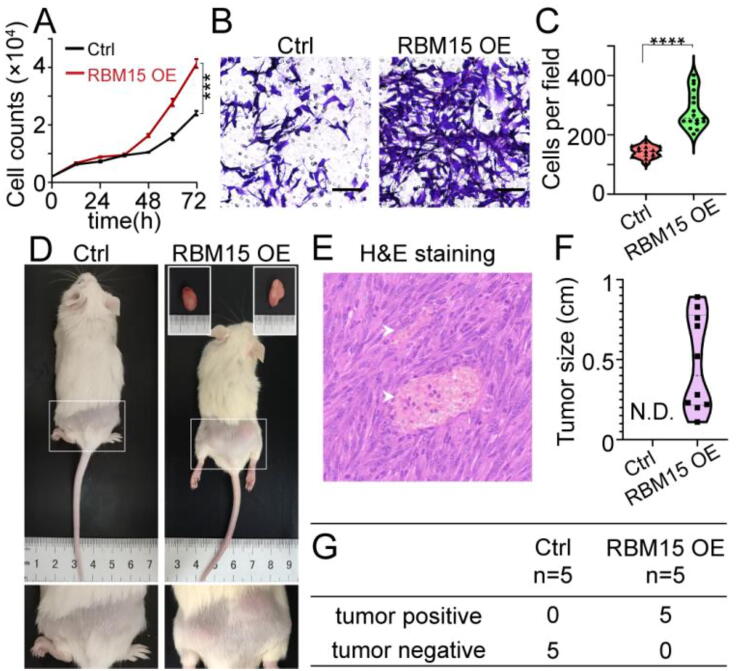
We chose the NIH3T3 murine embryonic fibroblast cell line to test whether RBM15 promoted the proliferation and invasion, as it had been widely used to identify and study novel oncogenes in>26,000 publications [39], [40]. We created the NIH3T3 cell lines stably over-expressing RBM15 (RMB15 OE) or an empty vector (control). The proliferation and Transwell assay revealed that RBM15 over-expression significantly enhanced the proliferating and migrating ability of NIH3T3 cells in vitro (Fig. 2A-C). Furthermore, subcutaneous injection of the NIH3T3 cells into NOD-SCID mice revealed that RBM15 over-expression NIH3T3 cells could grow into tumors (Fig. 2D & 2F-G). Moreover, we observed hyalinized vessels in the H&E-stained slides dissected from the RBM15 OE group (Fig. 2E). These results demonstrated that RBM15 could transform NIH3T3 cells, suggesting that RBM15 potentially played the oncogenic role during the progression of certain cancers.
Fig. 2.
RBM15 promotes the proliferation and invasion of NIH3T3 cells both in vitro and in vivo. (A, B) Proliferation (A) or Transwell assay (B) of NIH3T3 cells stably expressing RBM15 or empty vector (Ctrl). (C) Quantification of the number of migrated cells in (B). (D) Representative images showing subcutaneous tumor xenografts of NOD-SCID mice. Tumors from the RBM15 group were dissected as indicated and shown in the insets of upper right panel. Arrowheads indicated the hyalinized vessels. (E) Hematoxylin and eosin (H&E) stained tumor slides shown in (D). (F, G) Statistical analysis of the tumor diameter (F) and incidence (G) in the mice shown in (D). Scale bar, 100 μm for the image shown in (E). ****, P < 0.001. N.D., not detected.
3.2. RBM15 forms “liquid-like” condensates both in vivo and in vitro
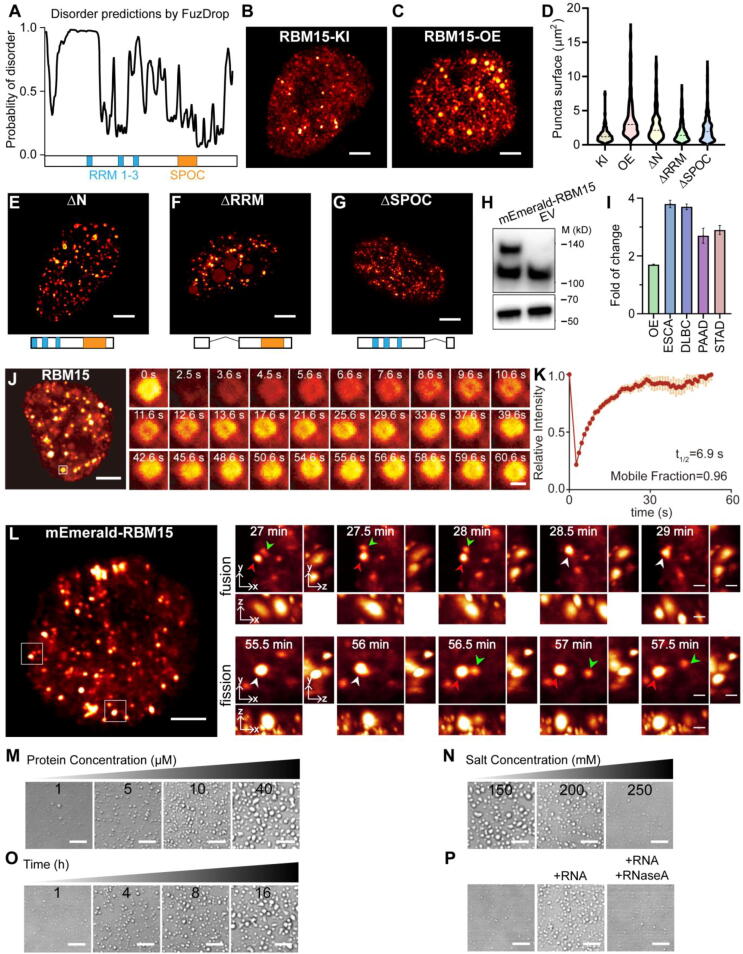
To gain insights into how escalated RBM15 resulted in oncogenic effect, we analyzed the protein domain structure and disordered regions of its amino acid composition. We found that RBM15 was composed of very rich intrinsically disordered sequences (IDRs) as predicted with FuzDrop[41], rather than well-defined structured domains (Fig. 3A), suggesting that RBM15 had a high potential to undergo phase separation. To check if RBM15 formed phase separated condensates under endogenous expression level, we created a knock-in cell line of mEmerald-RBM15 using CRISPR method, and then imaged endogenous RBM15 with super-resolution microscopy. The 3D-SIM live cell image showed that RBM15 was able to form small but abundant condensates scattered over the nucleoplasm (Fig. 3B and Video S1). Moreover, over-expressing RBM15 could significantly enlarge the size of these condensates (Fig. 3C, 3D and Video S2), presumably due to the enhanced phase separation under higher RBM15 concentration. We further tested the contribution of different sub-domains to the phase separation propensity of RBM15 by over-expressing its truncated mutants of RBM15-ΔN, RBM15-ΔRRM, and RBM15-ΔSPOC, respectively. We observed that no matter which domain we deleted, the truncated RBM15 proteins still tended to condensate into small droplets dispersed in nucleoplasm (Fig. 3E-G), although the deletion of RRM domain led to mislocalization of partial RBM15 into the nucleolus. In addition, the condensates of RBM15 with partial IDR deletion (i.e. ΔN, ΔRRM, and ΔSPOC) were a bit smaller compared to the ones of full-length RBM15 (Fig. 3D-3G).
Fig. 3.
Characterization of RBM15 condensates. (A) Protein domain structure and disordered regions prediction of RBM15 in silico with FuzDrop (https://fuzdrop.bio.unipd.it/). (B, C) Representative 3D-SIM images showing the distribution of RBM15 condensates inside nucleus of the NIH3T3 cells with knock-in mEmerald-RBM15 (i.e. RBM15-KI) (B) and over-expressing mEmerald-RBM15 (i.e. RBM15-OE) (C). (D) Comparison of the RBM15 condensates size shown in (B), (C), and (E-G). (E-G) Representative images of RBM15 after deletion of the N-terminal (E, ΔN), RRM domain (F, ΔRRM), and SPOC domain (G, ΔSPOC). (H) Immunoblotting of RBM15 in cells transfected with mEmerald-RBM15 or empty vector of mEmerald using an anti-RBM15 primary antibody. The lower band represents endogenous RBM15. (I) The fold-change of RBM15 transcript in the RBM15 over-expression NIH3T3 cells vs control cells (OE column), as well as RBM15 transcript in different types of tumor samples vs paired normal tissues (ESCA, DLBC, PAAD, and STAD). (J, K) Fluorescence recovery after photobleaching (FRAP) analysis of the RBM15 condensates in live cells. (L) Time-lapse LLSM images showing the fusion and fission activities of RBM15 condensates in live cells. Arrow heads indicate the fusion and fission. (M) The condensation of purified RBM15 protein at different concentrations for 1 h. (N) The condensation of 40 μM purified RBM15 incubated at different salt concentrations for 2 h. (O) 1 μM RBM15 was incubated from 1 to 16 h at 150 mM NaCl concentration. (P) 1 μM RBM15 was incubated with 10 ng/µL intact or degraded RNA for 2 h at 150 mM NaCl concentration. Scale bar, 8 μm in (M−P); 5 μm in (B, C, E-G, J, L); 1 μm in the inset images of (J and L).
Next, we quantified the escalation of RBM15 protein and transcript in RBM15 over-expression stable cell line and in clinical samples. As shown in Fig. 3H and 3I, RBM15 over-expression led to ∼ 1.6-fold and ∼ 2-fold increases in the abundance of protein (Fig. 3H) and transcript (Fig. 3I, OE column), respectively. In fact, the escalation of RBM15 transcript in RBM15 over-expression stable cell line was still slightly lower than that assessed from clinical cases (Fig. 3I), indicating that the phase separation of RBM15 protein in clinical samples might be more prominent than that in RBM15 over-expression stable cell line.
We further tested if RBM15 condensates possessed “liquid-like” property. As shown in Fig. 3J and 3 K, FRAP assay revealed that RBM15 condensates in the nucleus were rapidly exchanging their content with the surrounding nucleoplasm. Moreover, using the time-lapse 3D live-cell LLSM imaging, we observed that RBM15 condensates underwent constant fusion and fission activities (Fig. 3L and Video S3). These results illustrated that RBM15 condensates formed in live cells possesses typical characteristics of “liquid-like” droplets.
To further characterize the phase separation property of RBM15, we purified the full-length RBM15 protein (Fig. 3M) and quantified its phase separation propensity in vitro. As expected, RBM15 formed “liquid-like” droplets in a protein concentration-dependent manner (Fig. 3N). Consistent with other studies of phase separation in vitro [42], [43], the phase separation propensity of RBM15 was repressed as the salt concentration increased (Fig. 3O), but RBM15 still retained the ability to phase separate even at the high salt concentration up to 300 mM. Moreover, at physiological salt concentration of 150 mM, prolonging the incubation time enhanced the phase separation of RBM15 (Fig. 3P). Because RBM15 is an RNA-binding protein, we tested the influence of RNA molecules to the phase separation of RBM15. Of note, we found that mixing RNA molecules with RBM15 proteins could significantly enhanced its phase separation, whereas adding RNase A would impair this enhancement (Fig. 3Q). These results confirmed that the phase separation propensity of RBM15 protein was robust and strong, and revealed the positive regulation of RNA molecules to the phase separation of RBM15.
3.3. Location and composition of RBM15 condensates
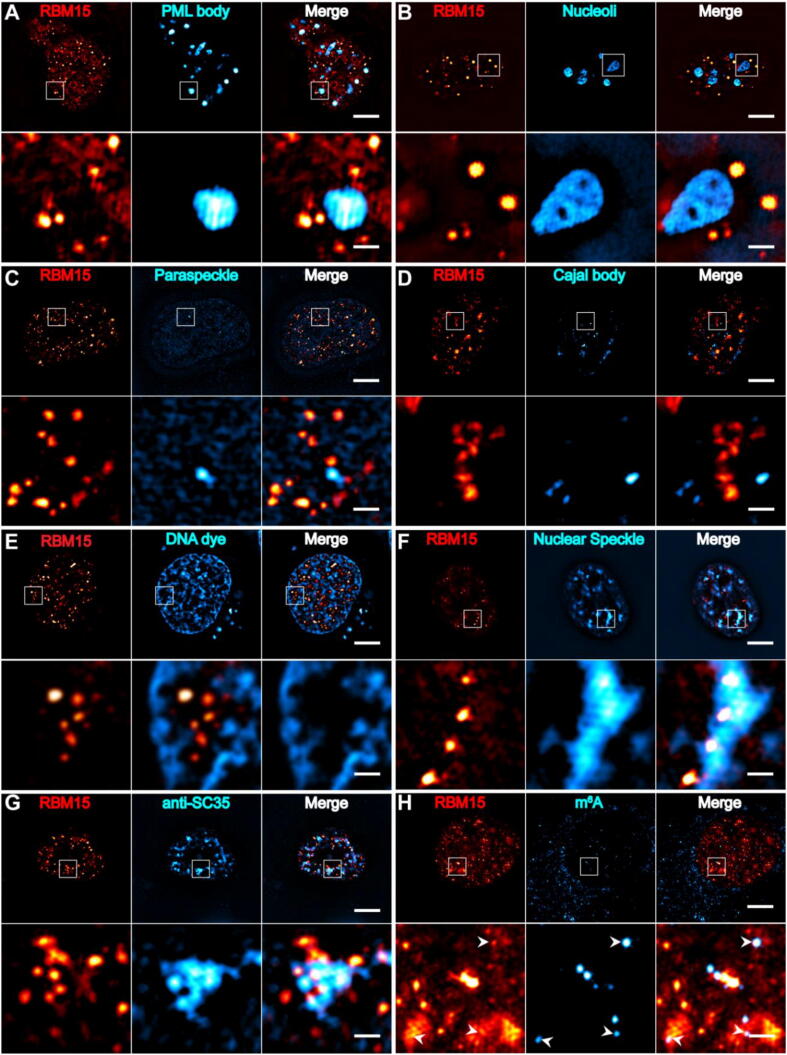
It is known that there are numbers of membrane-less subnuclear bodies including nucleoli, Cajal bodies, nuclear speckles (NSs), paraspeckles, promyelocytic leukaemia (PML) bodies, etc. It was straightforward to ask whether the RBM15 condensates were part of or different from these subnuclear bodies. To this end, we transfected the NIH3T3 cells with B23-mCherry, PML-mCherry, p54-mCherry, collin-mCherry, and SRRM1-mCherry, respectively, or stained the chromatin with CyTRAK OrangeTM dye, and then the cells were immunostained with RBM15 antibody. These specimens allowed us to examine the colocalization of RBM15 condensates with the PML bodies, nucleoli, paraspeckles, Cajal bodies, chromatin, and NSs, respectively. The two-color 3D-SIM images showed that the RBM15 condensates did no overlap with the PML bodies, nucleoli, paraspeckles, Cajal bodies or chromatin at all (Fig. 4A-E), but RBM15 condensates were prone to contact or partially overlap with NSs (Fig. 4F). To rule out the possibility that the overlap between RBM15 condensates and NSs was due to overexpressing the NSs’ marker of SRRM1-mCherry, we immunostained the cells with the antibodies against RBM15 and SC35 (an essential component of NSs) simultaneously. As shown in Fig. 4G, the RBM15 condensates indeed contacted with or partially embedded into the NSs.
Fig. 4.
The colocalization of RBM15 condensates with different nuclear structures. (A-F) Cells were labeled with mEmerald-RBM15 as well as PML bodies (A, PML-mCherry), nucleoli (B, B23-mCherry), paraspeckles (C, p54-mCherry), Cajal bodies (D, Collin-mCherry), chromatin (E, DNA dye) and nuclear speckles (F, SRRM1-mCherry). (G, H) Representative immunofluorescence images of NIH3T3 cells labeled with anti-SC-35(G) or anti-m6A (H). White arrowheads in (H) indicated that RBM15 condensates were colocalize with m6A. Scale bar, 5 μm; 1 μm in the cropped images.
Previous studies reported that RBM15 was able to promote the methyltransferase complex to deposit m6A modification on RNA transcripts [18], and the NSs were found to be in association with the actively transcribed genes [44]. Thus, an intriguing question was that if the RBM15 condensates enhanced the m6A modification of the transcripts of highly expressing cancer-related genes that subsequently altered the downstream pathways. To this end, we first checked the distribution relation between m6A modified RNA transcripts and RBM15 condensates by immunostaining the cells with m6A and RBM15 antibodies simultaneously. Fig. 4H showed that RBM15 condensates partially colocalized with the puncta rich of m6A modified transcripts in the nucleus, indicating that RBM15 was involved in depositing m6A modification onto RNA transcripts in the “liquid-like” condensation microenvironment.
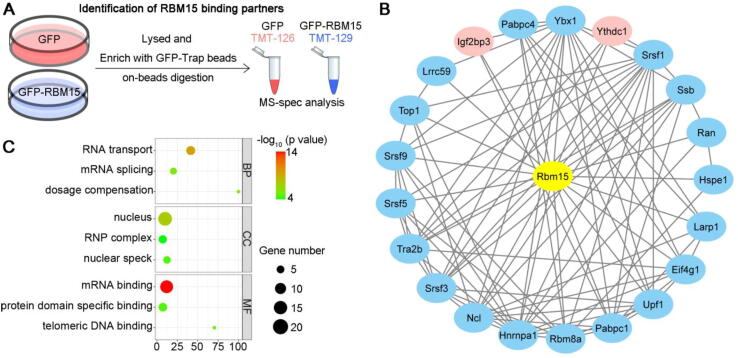
Subsequently, we employed the relative proteomic quantitation of tandem mass tags (TMT) multiplex labeling to dissect the composition of RBM15 condensates. We first used GFP-Trap beads to enrich GFP or GFP-RBM15 in lysates of 293 T cells over-expressing GFP or GFP-RBM15, respectively. Then, we digested the enriched proteins on beads using trypsin and labeled the digested peptides with TMT reagents (Fig. 5A). Although total 613 proteins were detected in the raw data of TMT mass spectrometry, only 38 out of which were identified as high-confidence binding partners of RBM15 after setting the relatively stringent threshold of TMT-129/TMT-126 > 2.0 and p-value < 0.05 (Supplementary Table 1). 21 out of the 38 interaction partners of RBM15 could be grouped as a focal and functionally related network according to the STRING database (Fig. 5B). Interestingly, YTHDC1 and IGF2BP3, that were known as the “reader” proteins of m6A modification, were among the most enriched interaction partners of RBM15 (Fig. 5C, red nodes). Moreover, Gene Ontology analysis suggested that these binding partners were highly related to the pathways of RNA transport and splicing (Fig. 5C), in which the m6A modification had been proven to play important role. Taken together, the super-resolution imaging and proteomic data implied that RBM15 condensates were a new type of subnuclear body, which might facilitate the deposition of m6A modification onto the related RNA transcripts.
Fig. 5.
Characterization of RBM15 condensates. (A) Sample preparation schematics for tandem mass tag (TMT)-labeled proteomics. Cells stably expressing GFP or GFP-RBM15 were lysed, immunoprecipitated by GFP-Trap, and digested on beads. The digested peptides were desalted, subjected to TMT-labeling, and mass spectroscopy spectra (MS-spec). (B) The interaction data from the RBM15 immunoprecipitation-MS-spec and STRING database were imported into Cytoscape for visualization. Red nodes, key components in the m6A pathway. (C) Gene Ontology analysis of the interaction partners of RBM15. BP, biological process; CC, cellular components; and MF, molecular function. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. RBM15-STYK1 m6A-MAPK signaling cascade results in oncogenic transformation of RBM15 overexpressing cells
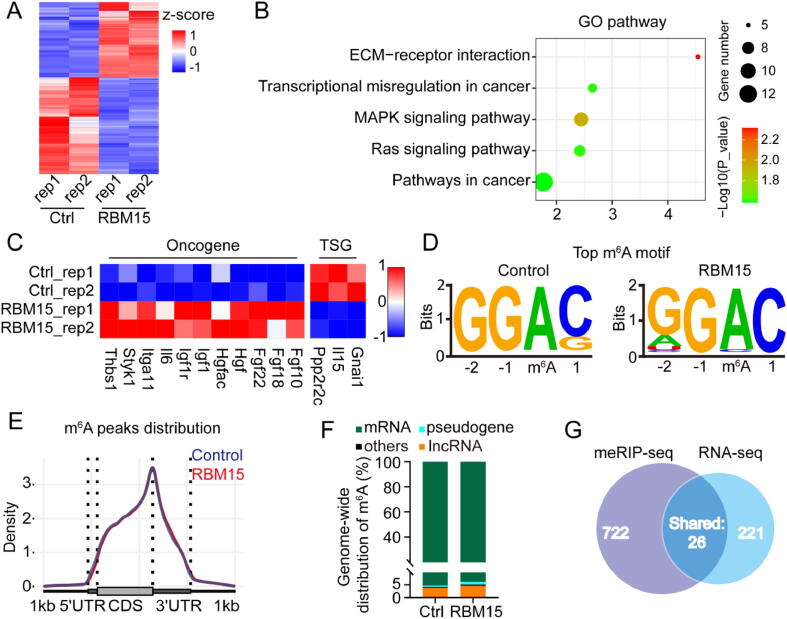
Based on the results of above experiments, we speculated that the high-level RBM15 might cause abnormal m6A modification of specific transcripts related to carcinogenesis, thus manifesting its oncogenic effect observed in Fig. 1, Fig. 2. To determine which transcripts were modulated by RBM15, we performed RNA-seq and methylated RNA immunoprecipitation sequencing (meRIP-seq) for the NIH3T3 cells over-expressing RBM15 (RBM15 OE) or empty vector (control), respectively. The RNA-seq data revealed that RBM15 over-expression led to 564 differentially expressed genes whose expression was changed by > 2-fold (Fig. 6A, Supplementary Table 2). Gene Ontology analysis indicated that the upregulated genes were mainly involved in cancer-related pathways, such as the mitogen-activated protein kinase (MAPK) signaling and extracellular matrix (ECM)-receptor interaction (Fig. 6B). For instance, the expression of eleven representative oncogenes in the MAPK pathways was significantly enhanced, whereas the expression of three typical tumor suppressor genes (TSGs) was repressed, respectively (Fig. 6C).
Fig. 6.
Characterization of transcriptomic gene expression and m6A modification changes caused by RBM15. (A) Heatmap showing differentially expressed genes between the NIH3T3 cells of over-expressing RBM15 (RBM15 OE) and empty vector (control cells, Ctrl) as determined from RNA-seq data. (B) Gene Ontology analysis of the differentially expressed genes is shown in (A). (C) Heatmap showing representative oncogenes or TSGs in (A). (D) The most common consensus m6A motif in the meRIP-seq data as determined with Homer software. (E) The probability distribution of m6A modification sites along transcripts. Transcripts are grouped into 5′UTR, CDS, and 3′UTR regions. (F) The proportion of different types of transcripts computed from meRIP-seq data. (G) Venn diagram depicting the number of the upregulated differential genes as determined by RNA-seq, and the number of genes whose transcripts were m6A hyper-methylated as determined by meRIP-seq, as well as overlapping genes identified in both assay.
On the other hand, the meRIP-seq data from both RBM15 OE and control cells revealed that (1) “GGAC” was the most frequent m6A consensus motif (Fig. 6D); (2) the m6A modification was enriched in the 3’-UTR (Fig. 6E); and (3) the major type of transcript with m6A modification was mRNA (Fig. 6F). These results were consistent with the common features of m6A modification reported in previous studies [22], [45], suggesting that RBM15 over-expression did not alter the characteristics of m6A modification. We then used the MeTDiff software [46] to determine the differentially methylated m6A sites, and found that RBM15 OE resulted in 748 hyper-methylated sites, presumably due to the enhanced recruitment of m6A methyltransferase by RBM15. According to the interaction partners of RBM15 identified in Fig. 5B, these hyper-methylated m6A sites in RBM15 OE cells were most likely recognized and bound by the m6A “reader” proteins of IGF2BP3 or YTHDC1. Recent studies reported that both IGF2BP3 and YTHDC1 were able to stabilize their bounded transcripts via recognition of m6A modification [25], [47]. These results inspired us to perform a joint analyzation of the hyper-methylated sites via meRIP-seq and the upregulated genes via RNA-seq. As shown in Fig. 6G, we identified 26 upregulated genes from RNA-seq data, which possessed hyper-methylated m6A sites on their transcripts.
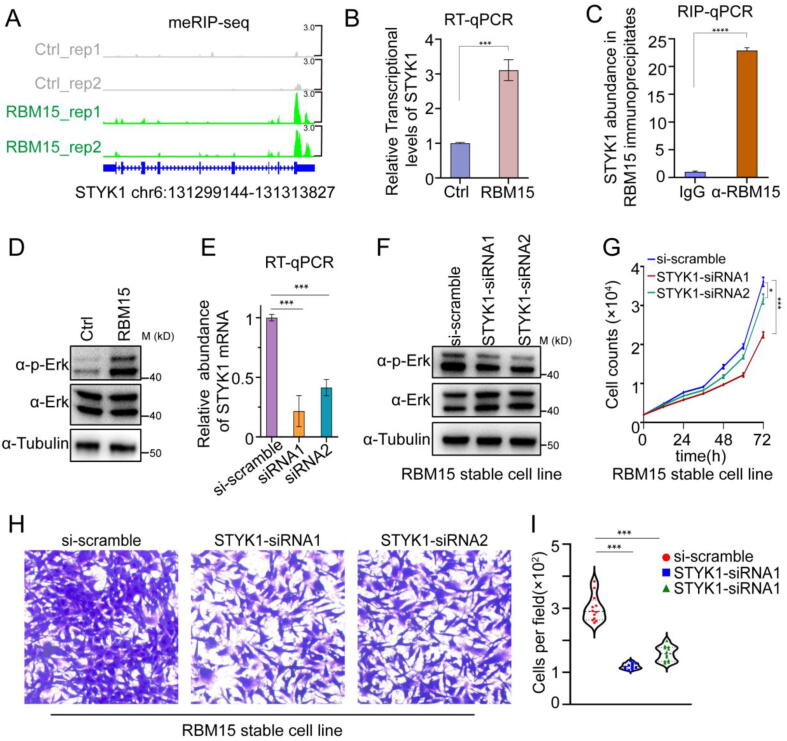
Among the 26 genes, we found that the mRNA of Serine/Threonine/Tyrosine Kinase 1 (STYK1), a tyrosine receptor kinase, was subject to the most significant increases in its m6A modification (Fig. 7A) and total amount (Fig. 7B). Moreover, the RIP-qPCR results showed that the abundance of STYK1 in RBM15 antibody immunoprecipitation was significantly higher than that in normal IgG control, implying that RBM15 directly bound to the STYK1 transcript (Fig. 7C). Previous studies revealed that STYK1 could activate MAPK signal transduction pathway to induce malignant transformation of a variety of cell lines, e.g., NIH3T3 [48], [49], [50]. Therefore, we examined the phosphorylated extracellular signal-regulated kinase 1/2 (p-ERK1/2) status, one of the most important signaling molecules in MAPK pathway, in both RBM15 OE and control cells. As expected, RBM15 OE cells presented much higher p-ERK1/2 level than control cells (Fig. 7D). To validate if the STYK1 was responsible for the escalated p-ERK1/2 and the malignant transformation, we depleted the expression of STYK1 in RBM15 OE cells by siRNA duplexes (Fig. 7E). As a result, we found that knockdown of STYK1 in RBM15 OE cells not only decreased the p-ERK1/2 level (Fig. 7F), but also significantly suppressed the invasion and proliferation abilities (Fig. 7G-I). Taken together, these results revealed that RBM15 OE caused the m6A-hypermethylated STYK1 that promoted the phosphorylation of ERK1/2, transforming the noncarcinogenic NIH3T3 cell line into tumor-forming cells.
Fig. 7.
The role of RBM15-STYK1 m6A-MAPK axis during tumor progression. (A) IGV snapshots of m6A peaks in the STYK1 locus. (B) RT–qPCR analysis of STYK1 abundance in the RBM15 OE and control cells (Ctrl). (C) RIP-qPCR quantification of RBM15-bound STYK1 using either RBM15 antibody (α-RBM15) or rabbit normal IgG (IgG). (D) Immunoblot images showing the difference of p-Erk and total Erk in RBM15 OE and control cells (Ctrl). (E) Validation of the efficiency of STYK1 knockdown in RBM15 OE cells. (F) Immunoblot images showing the difference of p-Erk and total Erk in RBM15 OE cells treated with 10 nM scramble siRNA (si-scramble) or siRNA duplexes against STYK1 (STYK1-siRNA1/2). (G, H) The proliferation curve (G) or Transwell assay (H) of the cell samples presented in (E). (I) Quantification of the number of migrated cells in (H).*, P < 0.05; ***, P < 0.001, ****, P < 0.0001.
4. Discussion
Emerging evidences had revealed that abnormal m6A modifications are highly correlated with tumor progression [30]. However, these studies mainly focused on modulating the methyltransferase complex, such as “writers” (METTL3/METTL14) [31], [33], [51] or “erasers” (FTO/ALKBH5) [32], [52], [53], [54] of the m6A modification, which inevitably altered transcriptome-wide m6A status. As a result, the conclusions of these studies were sometimes contradictory and difficult to interpret. In contrast, we revealed that RBM15, the key regulatory subunit of the m6A methyltransferase complex, preferentially bound to a subset of transcripts containing uridine (U)-enriched consensus motif, and promoted the deposition of m6A modifications onto these transcripts, many of which are closely related to megakaryocyte maturation and carcinogenesis [55], [56], [57]. Therefore, targeting RBM15 might be a more specific and effective way to modulate m6A modifications.
Although the high expression level of RBM15 had been recurrently observed in several different types of cancers [58], [59], [60], [61], whether the abundant RBM15 altered the m6A status of related transcripts, and how they influenced the cancer progression remains unclear. In this work, we first revealed that endogenous RBM15 formed “liquid-like” phase-separated condensates dispersed in the nucleus, which were different from other known membrane-less subnuclear bodies, and underwent dynamic fusion and fission activities. Moreover, the condensation propensity of RBM15 could be significantly enhanced by increasing its concentration or by adding the RNA molecules. We further found that the m6A “reader” proteins of both IGF2BP3 and YTHDC1 were highly enriched in the RBM15 condensates, which were known to enhance the stability of their bounded transcripts [25], [47]. These results inspired us to focus on these genes whose transcripts were differentially upregulated and m6A hyper-methylated. This guided us to select out the candidate STYK1 and confirmed that the m6A modification on STYK1 mRNA was significantly boosted by RBM15 OE, contributing to the elevation of its total mRNA level. Subsequently, the high abundance of STYK1 enhanced the phosphorylation of ERK1/2 to activate MAPK pathways, which was critical for the oncogenic transformation of NIH3T3 cells.
5. Conclusion
In conclusion, the RBM15-STYK1 m6A-MAPK signaling axis we dissected here might explain, at least in part, how high abundance RBM15 facilitated the cancer progression from the perspective of phase separation.
Authors contribution
D.L. and X.W. conceived and supervised this project. A.J. prepared samples and performed experiments. A.J., S.Z., and X.W. analyzed the data and composed the figures and videos under the supervision of D.L. D.L. and X.W. wrote the manuscript with the input from all the authors. All authors discussed the results and commented on the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by grants from the Ministry of Science and Technology (MOST; 2021YFA1300303), the National Natural Science Foundation of China (NSFC; 31827802, 32125024), Chinese Academy of Sciences (CAS; ZDBS-LY-SM004, XDA16021401).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.08.068.
Contributor Information
Xinyu Wang, Email: wangxinyu@ibp.ac.cn.
Dong Li, Email: lidong@ibp.ac.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Mercher T., et al. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc Natl Acad Sci USA. 2001;98(10):5776–5779. doi: 10.1073/pnas.101001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma Z., et al. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat Genet. 2001;28(3):220–221. doi: 10.1038/90054. [DOI] [PubMed] [Google Scholar]
- 3.Langenberg-Ververgaert K., et al. TERT promotor variant associated with poor clinical outcome in a patient with novel RBM15-MKL1 fusion-positive pediatric acute megakaryoblastic leukemia. Pediatr Blood Cancer. 2020 doi: 10.1002/pbc.28542. [DOI] [PubMed] [Google Scholar]
- 4.Saito Y., et al. Acute megakaryoblastic leukaemia with t(1;22)(p13.3;q13.1)/RBM15-MKL1 in an adult patient following a non-mediastinal germ cell tumour. Br J Haematol. 2020 doi: 10.1111/bjh.16900. [DOI] [PubMed] [Google Scholar]
- 5.Wiseman D.H., Bonney D.K., Wynn R.F. Hemophagocytosis by leukemic megakaryoblasts in acute myeloid leukemia (megakaryoblastic) with t(1;22)(p13;q13);RBM15-MKL1. J Pediatr Hematol Oncol. 2012;34(7):576–580. doi: 10.1097/MPH.0b013e318245a027. [DOI] [PubMed] [Google Scholar]
- 6.Torres L., et al. Acute megakaryoblastic leukemia with a four-way variant translocation originating the RBM15-MKL1 fusion gene. Pediatr Blood Cancer. 2011;56(5):846–849. doi: 10.1002/pbc.22765. [DOI] [PubMed] [Google Scholar]
- 7.Hsiao H.H., et al. RBM15-MKL1 (OTT-MAL) fusion transcript in an adult acute myeloid leukemia patient. Am J Hematol. 2005;79(1):43–45. doi: 10.1002/ajh.20298. [DOI] [PubMed] [Google Scholar]
- 8.Mercher T., et al. Recurrence of OTT-MAL fusion in t(1;22) of infant AML-M7. Genes Chromosom Cancer. 2002;33(1):22–28. doi: 10.1002/gcc.1208. [DOI] [PubMed] [Google Scholar]
- 9.de Rooij J.D.E., et al. Pediatric non-Down syndrome acute megakaryoblastic leukemia is characterized by distinct genomic subsets with varying outcomes. Nat Genet. 2017;49(3):451–456. doi: 10.1038/ng.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hara Y., et al. Prognostic impact of specific molecular profiles in pediatric acute megakaryoblastic leukemia in non-Down syndrome. Genes Chromosom Cancer. 2017;56(5):394–404. doi: 10.1002/gcc.22444. [DOI] [PubMed] [Google Scholar]
- 11.de Rooij J.D.E., et al. Recurrent abnormalities can be used for risk group stratification in pediatric AMKL: a retrospective intergroup study. Blood. 2016;127(26):3424–3430. doi: 10.1182/blood-2016-01-695551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raffel G.D., et al. Ott1(Rbm15) has pleiotropic roles in hematopoietic development. PNAS. 2007;104(14):6001–6006. doi: 10.1073/pnas.0609041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez-Pulido L., et al. SPOC: A widely distributed domain associated with cancer, apoptosis and transcription. BMC Bioinf. 2004;5 doi: 10.1186/1471-2105-5-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zolotukhin A.S., et al. Nuclear export factor RBM15 facilitates the access of DBP5 to mRNA. Nucleic Acids Res. 2009;37(21):7151–7162. doi: 10.1093/nar/gkp782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L., et al. Cross-talk between PRMT1-mediated methylation and ubiquitylation on RBM15 controls RNA splicing. Elife. 2015;4 doi: 10.7554/eLife.07938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao N., et al. Hematopoietic stem cells lacking Ott1 display aspects associated with aging and are unable to maintain quiescence during proliferative stress. Blood. 2012;119(21):4898–4907. doi: 10.1182/blood-2012-01-403089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J., et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patil D.P., et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537(7620):369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knuckles P., et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32(5–6):415–429. doi: 10.1101/gad.309146.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie Y.B., et al. RBM15 Modulates the Function of Chromatin Remodeling Factor BAF155 Through RNA Methylation in Developing Cortex. Mol Neurobiol. 2019;56(11):7305–7320. doi: 10.1007/s12035-019-1595-1. [DOI] [PubMed] [Google Scholar]
- 21.Xu C., et al. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol. 2014;10(11):927–929. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- 22.Dominissini D., et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 23.Wang X., et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alarcon C.R., et al. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell. 2015;162(6):1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang H., et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roundtree I.A., et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife. 2017;6 doi: 10.7554/eLife.31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haussmann I.U., et al. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540(7632):301–304. doi: 10.1038/nature20577. [DOI] [PubMed] [Google Scholar]
- 28.Xiao W., et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016;61(4):507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Liu J., et al. N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science. 2020;367(6477):580–586. doi: 10.1126/science.aay6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lan Q., et al. The Critical Role of RNA m(6)A Methylation in Cancer. Cancer Res. 2019;79(7):1285–1292. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 31.Lin S., et al. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell. 2016;62(3):335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su R., et al. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell. 2020;38(1):79–96 e11. doi: 10.1016/j.ccell.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng W., et al. Multiple Functions and Mechanisms Underlying the Role of METTL3 in Human Cancers. Front Oncol. 2019;9:1403. doi: 10.3389/fonc.2019.01403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo Y., et al. Visualizing Intracellular Organelle and Cytoskeletal Interactions at Nanoscale Resolution on Millisecond Timescales. Cell. 2018;175(5):1430–1442 e17. doi: 10.1016/j.cell.2018.09.057. [DOI] [PubMed] [Google Scholar]
- 35.Xu G., et al. Viral tegument proteins restrict cGAS-DNA phase separation to mediate immune evasion. Mol Cell. 2021;81(13):2823–2837 e9. doi: 10.1016/j.molcel.2021.05.002. [DOI] [PubMed] [Google Scholar]
- 36.Wang X.Y., et al. N-6-methyladenosine modification of MALAT1 promotes metastasis via reshaping nuclear speckles. Dev Cell. 2021;56(5):p. 702-+. doi: 10.1016/j.devcel.2021.01.015. [DOI] [PubMed] [Google Scholar]
- 37.Tang Z., et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cerami E., et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discovery. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soda M., et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–U3. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 40.NIH3T3 Transformation Assay, in Encyclopedia of Cancer, S. M, Editor. 2008, Springer: Berlin, Heidelberg.
- 41.Hatos A., et al. FuzDrop on AlphaFold: visualizing the sequence-dependent propensity of liquid-liquid phase separation and aggregation of proteins. Nucleic Acids Res. 2022;50(W1):W337–W344. doi: 10.1093/nar/gkac386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patel A., et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162(5):1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 43.Hyman A.A., Weber C.A., Julicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 44.Kim J., et al. Gene expression amplification by nuclear speckle association. J Cell Biol. 2020;219(1) doi: 10.1083/jcb.201904046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyer K.D., et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui X., et al. MeTDiff: A Novel Differential RNA Methylation Analysis for MeRIP-Seq Data. IEEE/ACM Trans Comput Biol Bioinform. 2018;15(2):526–534. doi: 10.1109/TCBB.2015.2403355. [DOI] [PubMed] [Google Scholar]
- 47.Cheng Y.M., et al. N-6-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer Cell. 2021;39(7):p. 958-+. doi: 10.1016/j.ccell.2021.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi W., Fu Y., Wang Y. Downregulation of GLUT3 impairs STYK1/NOK-mediated metabolic reprogramming and proliferation in NIH-3T3 cells. Oncol Lett. 2021;22(1):527. doi: 10.3892/ol.2021.12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J., et al. NOK/STYK1 interacts with GSK-3beta and mediates Ser9 phosphorylation through activated Akt. FEBS Lett. 2012;586(21):3787–3792. doi: 10.1016/j.febslet.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 50.Wang Z., et al. STYK1 promotes epithelial-mesenchymal transition and tumor metastasis in human hepatocellular carcinoma through MEK/ERK and PI3K/AKT signaling. Sci Rep. 2016;6:33205. doi: 10.1038/srep33205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen H., et al. RNA N(6)-Methyladenosine Methyltransferase METTL3 Facilitates Colorectal Cancer by Activating the m(6)A-GLUT1-mTORC1 Axis and Is a Therapeutic Target. Gastroenterology. 2021;160(4):1284–1300 e16. doi: 10.1053/j.gastro.2020.11.013. [DOI] [PubMed] [Google Scholar]
- 52.Huang Y., et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell. 2019;35(4):677–691 e10. doi: 10.1016/j.ccell.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jeschke J., et al. Downregulation of the FTO m(6)A RNA demethylase promotes EMT-mediated progression of epithelial tumors and sensitivity to Wnt inhibitors. Nat Cancer. 2021;2(6):611–628. doi: 10.1038/s43018-021-00223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen C., et al. RNA Demethylase ALKBH5 Selectively Promotes Tumorigenesis and Cancer Stem Cell Self-Renewal in Acute Myeloid Leukemia. Cell Stem Cell. 2020;27(1):64–80 e9. doi: 10.1016/j.stem.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao N., et al. Ott1 (Rbm15) regulates thrombopoietin response in hematopoietic stem cells through alternative splicing of c-Mpl. Blood. 2015;125(6):941–948. doi: 10.1182/blood-2014-08-593392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niu C., et al. c-Myc is a target of RNA-binding motif protein 15 in the regulation of adult hematopoietic stem cell and megakaryocyte development. Blood. 2009;114(10):2087–2096. doi: 10.1182/blood-2009-01-197921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jin S.L., et al. PRMT1-RBM15 axis regulates megakaryocytic differentiation of human umbilical cord blood CD34(+) cells. Exp Ther Medicine. 2018;15(3):2563–2568. doi: 10.3892/etm.2018.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Z., Mei Y., Hou M.X. Knockdown RBM15 Inhibits Colorectal Cancer Cell Proliferation and Metastasis Via N6-Methyladenosine (m6A) Modification of MyD88 mRNA. Cancer Biother Radiopharm. 2021 doi: 10.1089/cbr.2021.0226. [DOI] [PubMed] [Google Scholar]
- 59.Huang H., et al. Analysis and identification of m(6)A RNA methylation regulators in metastatic osteosarcoma. Mol Ther Nucleic Acids. 2022;27:577–592. doi: 10.1016/j.omtn.2021.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ahn J.H., et al. Phase separation drives aberrant chromatin looping and cancer development. Nature. 2021 doi: 10.1038/s41586-021-03662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao Z.Y., et al. N6-Methyladenosine Methylation Regulator RBM15 is a Potential Prognostic Biomarker and Promotes Cell Proliferation in Pancreatic Adenocarcinoma. Front Mol Biosci. 2022;9 doi: 10.3389/fmolb.2022.842833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.