Abstract
Combinatorial selections are powerful strategies for identifying biopolymers with specific biological, biomedical, or chemical characteristics. Unfortunately, most available software tools for high-throughput sequencing analysis have high entrance barriers for many users because they require extensive programming expertise. FASTAptameR 2.0 is an R-based reimplementation of FASTAptamer designed to minimize this barrier while maintaining the ability to answer complex sequence-level and population-level questions. This open-source toolkit features a user-friendly web tool, interactive graphics, up to 100 times faster clustering, an expanded module set, and an extensive user guide. FASTAptameR 2.0 accepts diverse input polymer types and can be applied to any sequence-encoded selection.
Keywords: MT: Bioinformatics, Next-generation sequencing, combinatorial selection, aptamer, SELEX, sequence analysis, ribozyme, phage display, synthetic biology, directed evolution
Graphical abstract
Combinatorial selections strategies are tools to design sequences with desirable properties. However, existing tools that analyze these data require significant computational expertise. The corresponding author and colleagues designed the webtool FASTAptameR 2.0 to minimize this technical barrier and allow researchers to still answer nuanced questions about their sequence populations.
Introduction
Combinatorial selections are powerful strategies for identifying biopolymers with specific characteristics such as target specificity or affinity, catalytic properties, or biological function. The strength and adaptability of this approach were recognized with the 2018 Nobel Prize in Chemistry for Francis Arnold, George Smith, and Gregory Winter.1 While these biopolymers are generally composed of nucleotides or amino acids, the molecular alphabets can be extended or modified to include non-canonical amino acids2 and chemically modified nucleotides such as AEGIS,3 Hachimoji,4 and others.5 Selection strategies for nucleic acids have been applied to aptamers,6,7 (deoxy)ribozymes,8, 9, 10 synthetic genetic polymers (XNAs),11,12 and other combinatorial chemistries. Selection strategies for peptides and proteins can be accomplished by selecting for bioactivity in cells or whole organisms13 or by displaying on phage particles,14 ribosomes,15 mRNA,16 whole bacteria,17 and other platforms. The genes that encode the evolving proteins can be translated from nucleic acid libraries according to the standard genetic code or to natural or artificial genetic codes.18 DNA sequence libraries have even been used as barcodes to track lipid nanoparticle formulations19, 20, 21 and combinatorial chemical synthesis.22,23 In short, any platform that links polymer sequence (genotype) with a selectable or screenable property (phenotype) can be adapted to combinatorial selections.
Under optimal circumstances, the evolutionary dynamics of populations undergoing selection reflect the relative fitness of each species, with high-fitness sequences typically enriching during selection and low-fitness sequences depleting. Thus, common analytic tasks of any combinatorial selection include counting the number of occurrences for each sequence,24,25 calculating enrichment of sequences between two or more rounds,25, 26, 27 filtering sequences based on the number of reads present in one or multiple rounds,28 clustering related sequences,25,29, 30, 31 and in some cases analyzing predicted structure motifs.29,32, 33, 34, 35, 36 High-throughput sequencing (HTS) provides large volumes of data for these analyses and can yield high-resolution insights. Many specialized bioinformatics toolkits have been developed to enable this analytical workflow,37 and several of these tools include graphical user interfaces to visualize HTS data during the analysis.36,38,39 However, some of these toolkits require significant computational resources or coding expertise that together constitute barriers to entry for the average molecular biologist. Our lab previously developed and released the FASTAptamer toolkit25 to address the primary, sequence-level needs in the field, such as those outlined above. FASTAptamer is an open-source toolkit consisting of five Perl scripts that can be used to count, normalize, and rank reads in a FASTQ or FASTA file, compare populations for sequence distribution, cluster related sequences, calculate fold-enrichment, and search for sequence motifs.25 Since its publication, the FASTAptamer toolkit has been used and cited extensively for diverse types of molecular and biological selections on populations of functional nucleic acids and protein/peptides (see Supplementary Information), thereby demonstrating its ability to address many of the first-level bioinformatics needs of the field.
Although FASTAptamer can analyze sequences from many types of biomolecules, its original application was targeted at aptamers, which are structured nucleic acids capable of binding to a molecular target, usually with high specificity and affinity. Aptamers are generated through an iterative, in vitro selection process termed SELEX (Systematic Evolution of Ligands by EXponential enrichment).6,7 After a determined number of selection rounds, the sequences of the enriched aptamers have traditionally been obtained by cloning the aptamer libraries into a plasmid and sequencing each clone one at a time. With HTS, millions of sequence reads from multiple rounds of selection can be determined, and this information can be used to identify aptamer candidates for further characterization.40, 41, 42, 43, 44, 45, 46, 47 HTS investigation of in vitro selection pools has revealed the distribution and relative frequencies of individual sequences and groups of related sequences as the populations evolve through the course of the experiment.48,49 Such data can inform on the success of the selection,45,50 aptamer-target interactions,51, 52, 53, 54 the mutation and fitness landscape,29,44,55 structure-function relations, biological constraints, and more.
While the initial release of FASTAptamer is generally user-friendly, it also has some limitations. First, as the FASTAptamer modules are Perl scripts, they must be run using a command line, which creates a modest barrier for practitioners of combinatorial selections who are unfamiliar or uncomfortable with a command line interface. Second, depending on the parameters used, the clustering module is time-consuming and computationally intensive.31 Third, while the output data from FASTAptamer can be downloaded for offline visualization, it does not allow for visualization of results within the platform, which can constrain data exploration.
To address these limitations, we describe here the development of FASTAptameR 2.0, an R-based reimplementation of FASTAptamer. This program improves upon the original version while keeping the features that made FASTAptamer an accessible, easy-to-use toolkit for the analysis of HTS datasets. Like FASTAptamer, FASTAptameR 2.0 does not need external dependencies (especially when used through the web tool) and is easy to install and launch. FASTAptameR 2.0 is portable across multiple platforms, open source, and comes with a detailed user guide that includes screenshots of the user interface and sample output tables and graphs for each module (see Supplementary Information). Further, the generalizable outputs can be used as downstream inputs to this program or any other bioinformatics program that supports FASTA files. It has a user-friendly interface that can be accessed online at https://fastaptamer2.missouri.edu/ or in a downloadable form as a Docker image from Docker Hub (https://hub.docker.com/repository/docker/skylerkramer/fastaptamer2), and the code can be accessed from GitHub at https://github.com/SkylerKramer/FASTAptameR-2.0.56 Additional improvements in FASTAptameR 2.0 include a faster clustering algorithm with speeds nearly 100X faster than FASTAptamer in some cases (e.g., for larger, more complex libraries) and an expanded set of interconnected modules (shown in Figure 1) that can be used to interactively analyze and visualize HTS data from new perspectives with custom, user-defined pipelines. Collectively, these improvements make exploration of HTS data from combinatorial selections significantly more accessible.
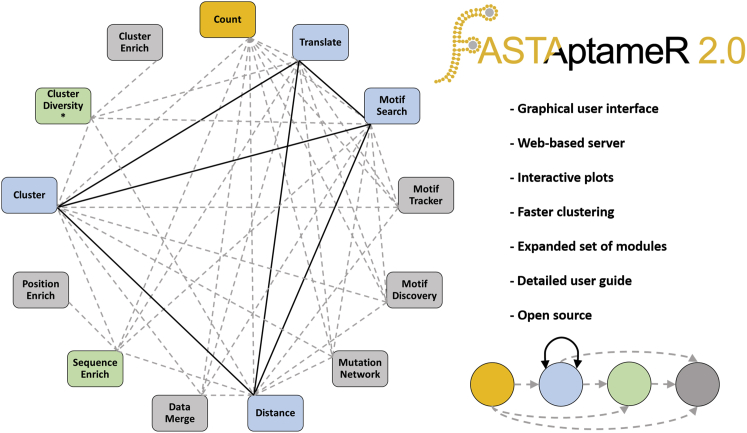
Figure 1.
General overview, module connectivity, and major new features of FASTAptameR 2.0
The gold node (the Count module) is the required first step of every user-customized workflow. Blue nodes (such as the Distance module) are either intermediate or final steps of workflows. Gray nodes (such as the Motif Tracker module) are final steps of workflows, and green nodes (such as the Sequence Enrich module) exclusively feed into gray nodes. Solid black edges are bidirectional, whereas dashed gray edges are unidirectional. The asterisk with the Cluster Diversity module is to indicate that the population must be clustered at some step before its use. Most module outputs are downloadable as FASTA, CSV, or both.
Results
Count module
The first step in analyzing sequence data from combinatorial selections is nearly always to determine the read count (abundance) of each unique sequence. This information can indicate whether the population is relatively diverse with little convergence or has converged on one or a few dominant sequences. Either of these scenarios is immediately evident when analyzing the population with the Count module, which, as in FASTAptamer, is the entry point into FASTAptameR 2.0.
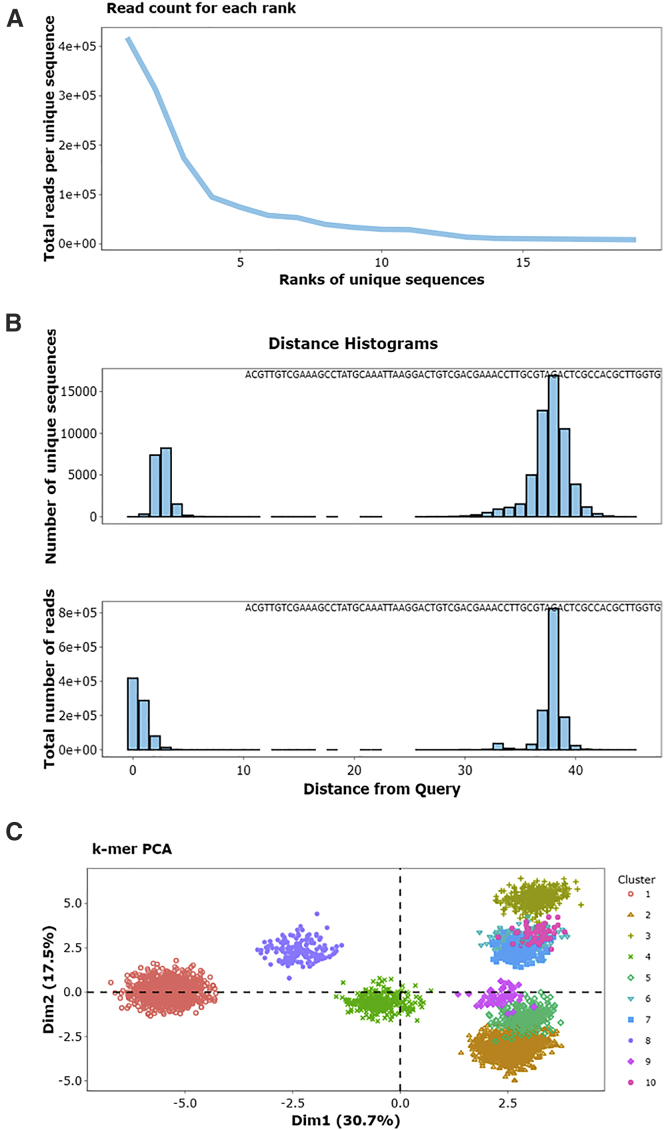
This module serves two main purposes. First, it condenses the original file size by returning a FASTA file with a single entry for each unique sequence. Second, it provides summary statistics for each unique sequence in the input population as three key metrics: abundance, rank by abundance, and reads per million (RPM), which is the read count divided by the population size in millions. It then incorporates these statistics into the sequence identifier for each entry. For example, a sequence with an identifier of “>4-94978-43966.9” is the fourth most abundant sequence in its population, has 94,978 identical reads, and is present at a frequency of 43,966.9 RPM. The distribution of those statistical values across a given population provides the first insights into the degree to which that population has converged, which can be seen by visualizing the relationship between rank and abundance (Figure 2A, generated with the 70HRT14 population from the original FASTAptamer publication51,57, 58, 59; see Materials and Methods). A slowly decreasing function suggests that the population has not converged onto a small set of sequences, whereas a steeply decreasing line suggests convergence.
Figure 2.
Example plots in a Cluster module workflow, using the 70HRT14 population as an example
(A) This line plot of the relationship between sequence rank and abundance (from the Count module) suggests that the population is dominated by a few sequences (convergence) due to its relatively steep slope and the magnitude of the y axis. (B) These histograms of LEDs suggest that many sequences in this population are similar to its most abundant sequence and that the region of sequence space surrounding the most abundant sequence is well-sampled, which can indicate biochemical significance. For the top plot, each unique sequence is equally weighted, whereas each unique sequence in the bottom plot is weighted by abundance. (C) A 3-mer matrix was generated from clustered sequences and visualized as a PCA plot where colors correspond to clusters.
A new feature of FASTAptameR 2.0 is the ability to identify overlaps among two or more populations. To this end, multiple populations from the Count module or from several other, downstream modules can be merged and visualized together in the Data Merge module. Supported merge types return the set of all sequences from every population (union), the set of all sequences that are shared between every population (intersection), or the set of all sequences from the first population with information from the other population(s) appended to it (left join).
Distance module
The Distance module is a new feature of FASTAptameR 2.0 that computes the Levenshtein edit distance (LED) between a query sequence and every other sequence in the population. An in-house precursor of this module was previously used in an in vivo selection.60 Distance analysis can be especially useful when monitoring the accumulation of point mutants, evaluating the effectiveness of a mutagenesis protocol, or monitoring diversity near the beginning of a selection from sequences that densely sample local sequence space (e.g., via mutagenic PCR or doped resynthesis). The distribution of these LEDs can then be visualized as a histogram of distances (Figure 2B) to provide additional perspectives on overall sequence relatedness within the population. For output libraries, an isolated cluster will be seen close to zero distance when the population consists predominantly of sequences that are closely related to the query (such as when the query is part of a cluster of sequences that have come to dominate the population) or after the accumulation of new mutations during the course of the selection (drift or divergent evolution from the founder sequences). A second peak will appear at large distances from the query when the remaining sequences are evolutionarily unrelated to the query, such as when many different founding members of a random sequence population are independently selected (Figure 2B). To illustrate, this analysis was applied to the 70HRT14 population, using the most abundant sequence as the query. Plotting this distribution by equally weighting each unique sequence (top plot of Figure 2B) reveals that many of these sequences are similar to the query, that the region of sequence space immediately surrounding this query is well-sampled (which can indicate its biochemical significance), that most species are within three mutations relative to the query, and that nearly all sequences related to the query are within an LED of 7. This visualization provides guidance in setting the maximum LED value to use in the Cluster module (see below). In contrast, plotting the distribution after weighting the data by sequence abundance (bottom plot of Figure 2B) shows that variants within one or two mutations of the query are far more abundant than those with higher-order mutations.
Mutation network module
Fitness landscapes and evolutionary histories can sometimes be revealed by looking at mutational intermediates and how they rise and fall during selection. The Mutation Network module is a new feature of FASTAptameR 2.0 that uses Dijkstra’s shortest path algorithm to discover the shortest evolutionary path between two query sequences in a population. The maximum number of mutations per evolutionary step can be defined by the user, thereby allowing for highly constrained, incremental steps (e.g., no more than one mutation per step) or for larger, more saltatory steps. If all intermediates for a given path are present, the module then returns a data table for the intermediates along that path. This functionality allows researchers to better understand evolutionary trajectories in the fitness landscape created in the experiment.
Cluster algorithm and validation
The Cluster module groups closely related sequences into “clusters,” thereby setting the stage for computing local fitness landscapes and further simplifying downstream analysis. The clustering algorithms for FASTAptamer and FASTAptameR 2.0 are both iterative, greedy processes that start by considering the most abundant, unclustered sequence as a cluster seed during each iteration. Given that the Count module sorts the population by abundance, the first sequence in that output becomes the cluster seed for the first iteration. All unique, unclustered sequences within a predetermined LED are added to this cluster. After considering all unique sequences in the population, the most abundant sequence that remains unclustered becomes the seed of the second cluster, and all unclustered sequences within the LED are added to that cluster. These steps are iterated until a predetermined stop condition is met (see below).
Clustering can be computationally intense and slow, a problem that has been observed for FASTAptamer and other platforms.31 FASTAptameR 2.0 significantly reduces the clustering runtime by changing the underlying data structure for the computations. The original implementation stores clustered sequences in arrays (a static data structure), whereas the FASTAptameR 2.0 implementation uses linked lists (a dynamic data structure). The list structure more efficiently handles memory requirements, which grow with the size and complexity of the population. FASTAptameR 2.0 offers additional means of reducing clustering time, such as by allowing the user to filter out sequences with abundance less than a user-defined threshold or to set a maximum number of clusters for the module to generate.
The FASTAptameR 2.0 clustering algorithm was benchmarked against FASTAptamer by comparing runtimes on an Ubuntu subsystem (v18.04) on a desktop computer with 16 GB RAM and an Intel I7 processor. All 72,921 unique sequences from the 70HRT14 population were used to generate the top 30 clusters with a maximum LED of seven. While the original implementation finished in 35 min 27 s, the FASTAptameR 2.0 implementation finished in 24.6 s, roughly 86 times faster. When clustering times were compared for a number of other scenarios, FASTAptameR 2.0 was always significantly faster than FASTAptamer, and the magnitude of this difference grew with the size and complexity of the population being analyzed. Therefore, the FASTAptameR 2.0 clustering algorithm is strictly better than the algorithm used in the original FASTAptamer.
Outputs from the Cluster module can be visualized with the Cluster Diversity module, which is another new addition to FASTAptameR 2.0. This module uses all unique sequences within each of the user-defined clusters to create a k-mer matrix. This matrix is subsequently visualized as a two-dimensional PCA plot (Figure 2C). The k-mer plot for the top 10 clusters of the 70HRT14 population shows most clusters in well-defined regions with separation from most or all of the other clusters. The separations among the clusters reflect their origins from independent, unrelated founder sequences present in the initial population, while the spread of each cluster reflects the sampling of local sequence space around each founder sequence, resulting from the accumulation of functional point mutations through neutral drift and purifying selection.
Sample plots from a cluster-specific workflow are shown in Figure 2. The panels show evidence of a convergent population (Figure 2A), quantify the distance from a query sequence to the rest of the population and suggest LED values required for clustering (Figure 2B), and provide a visualization of the separation between and diversity within clusters (Figure 2C).
Motif discovery
Combinatorial selections often converge upon one or more sequence or structural motifs that are present in many otherwise unrelated members of the population. The new Motif Discovery module is included in FASTAptameR 2.0 to provide a preliminary assessment of shared sequence motifs. This module uses an implementation of the Fast String-Based Clustering (FSBC) algorithm31 for de novo discovery for contiguous, over-enriched motifs in D/RNA sequences. There are many other excellent tools dedicated to de novo motif discovery, such as the MEME suite61 for sequence-based approaches and Infernal62 for RNA using both sequence-based and predicted structural similarities. FASTA-formatted output from any of the FASTAptameR 2.0 modules can be exported and analyzed by those other dedicated platforms.
Individual- and population-level tracking
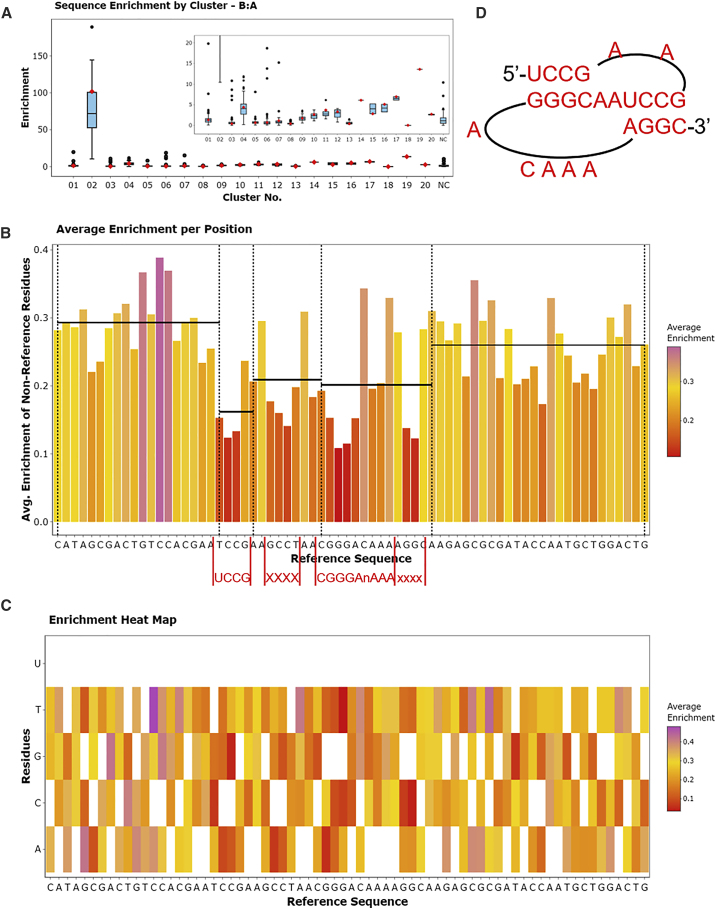
Another new feature of FASTAptameR 2.0 is the ability to track individual motifs, sequences, or clusters across multiple rounds of a selection experiment. The Motif Tracker module tracks query motifs or sequences, and the Enrich module tracks how every sequence changes between populations, while the Cluster Enrich module allow the user to monitor the collective behavior of the cluster as a whole, analogous to the collective evolutionary dynamics of a viral quasi-species. These three modules additionally calculate how families or species enrich, which can indicate how they performed in the selection experiment. An in-house precursor of Motif Tracker was previously used in a selection for 2′-modified RNA aptamers with affinity for HIV-1 RT.63 In the case when two clustered files are supplied to the Enrich module, the enrichment values of individual sequences can be grouped by clustering and visualized as a boxplot (Figure 3A). In the example shown in Figure 3A, Cluster 2 is enriched relative to the other clusters, suggesting that this set of sequences has a motif important for target binding (specifically the family 1 pseudoknot, or F1Pk, in this case). For each cluster in the boxplot, individual points that are well above or well below the median value represent species that are enriching or depleting relative to the cluster as a whole. Both the enriched and depleted species can be highly informative, as species that carry strongly advantageous variations may be emerging as future dominant species for that cluster, while species with strongly disadvantageous variations can illuminate critical portions of the biomolecule, as illustrated by the Position Enrich module.
Figure 3.
Cluster and position enrichment plots
(A) The cluster boxplot showing how clustered sequences in 70HRT14 enrich in 70HRT15. Cluster 2 of 70HRT14, for example, is highly enriched in 70HRT15 due to the presence of the F1Pk, which is implicated in target binding to HIV-1 RT. The 25th and 75th quartiles are respectively represented by the bottom and top of each box. The line in the middle of the box represents the median. Whiskers are at most 1.5 ∗ IQR (interquartile range), and any points beyond that are shown as outliers. The red marker indicates where the seed sequence of the cluster falls. (B) The x axis of the bar plot also shows the user-defined reference sequence, and the y axis shows the average enrichment of each non-reference residue at each position. The red text below this panel shows the portion of the query sequence that matches the linear F1Pk motif. Black horizontal lines show the left-inclusive average enrichment score of each user-defined region. The regions corresponding to the F1Pk motif have the lowest regional average of enrichment scores, indicating the importance of this motif for this selection experiment. (C) The x axis of the heatmap shows the user-defined reference sequence, and the y axis shows all possible residues at each position. Colors depict the average enrichment of each possible non-reference residue. (D) The experimentally determined secondary structure of the F1Pk motif.
Position enrichment
For a set of closely related sequences, mutations in some positions contribute directly to enrichment or depletion, while mutations at some other positions have little consequence. Uncovering these relationships can be enormously valuable for delineating the contributions of those positions to macromolecular functions. The Position Enrich module is a new feature of FASTAptameR 2.0 that calculates the average enrichment or depletion at each position for all sequences that do not match the corresponding user-defined reference residue at that position. This calculation is visualized as a bar plot (Figure 3B). Relatively short bars indicate functional conservation at that position, such that deviations from the reference residue identity contribute to depletion. Exceptionally tall bars may indicate positive selection for improved function relative to the reference sequence. As a result, highly conserved sections are immediately visible as regions with low bars because mutations in these positions contribute to depletion. The module calculates local averages across user-defined intervals and displays them as horizontal lines across those intervals, making the conserved and non-conserved regions especially evident from visual inspection (Figure 3B). Position Enrich further resolves enrichment and depletion patterns for each of the available substitute residues (e.g., three alternative nucleotides or 19 alternative amino acids when using standard alphabets) and displays the resulting patterns as a heatmap (Figure 3C). As in the Translate module, nonstandard nucleotides and amino acids can be analyzed with the Position Enrichment module.
To generate the plots in Figure 3, the 70HRT14 and 70HRT15 populations were counted and clustered following the workflow of Figure 2, although in this case the Count module was also used to omit any sequences that were not exactly 70 nucleotides long. The Enrich module created the boxplot from the full set of clustered sequences in both populations. The Enrich module then calculated enrichment scores for the first cluster from 70HRT15, which carries the F1Pk motif, and for the corresponding cluster from the preceding round of selection (second cluster from 70HRT14). The Position Enrich module used the output from the Sequence Enrich module as input, and the most abundant sequence in 70HRT15 was used as the reference sequence. The segments within the 70-nucleotide random region that contain the pseudoknot64 at the functional core of the aptamer are shown in Figure 3D.
Expanded sequence support
While populations of any sequence type (e.g., nucleotides or amino acids) could be fed into the original release of FASTAptamer, it did not allow for sequence translations. The Translate module of FASTAptameR 2.0 translates nucleotide sequences to amino acids according to either the standard genetic code or any of 15 alternative genetic codes such as those used by vertebrate mitochondria, mycoplasma, and other organisms. This module also supports complete customization of the genetic code used for translation to support nonstandard nucleotide input and/or nonstandard amino acid output, both of which are useful for applications in synthetic biology. Thus, FASTAptameR 2.0 explicitly supports all linear biopolymers of diverse biological origins.
Discussion
The integration of HTS with combinatorial selection experiments has created many opportunities and challenges for bioinformatics analyses. Though many tools exist to aid these analyses,24,25,28,30,31,34, 35, 36,38,39 usually they are not designed for users without a relatively strong computational background. As such, a typical practitioner of combinatorial selections may need to devote significant time and effort to tasks such as software installation and dependency handling before they can even learn to properly use the tool, constituting a serious barrier to data exploration. A notable exception is the REVERSE platform,65 which offers a user-friendly web service to analyze populations of RNA sequences from selection and evolution experiments. This tool is easy to use, supports preprocessing functionality, and offers helpful documentation, although it lacks the abilities to handle expanded/customized alphabets or to fully customize user workflows.
FASTAptameR 2.0 was designed with non-computational users in mind and according to best practices in the field of bioinformatics.66, 67, 68, 69 Like its predecessor FASTAptamer, FASTAptameR 2.0 is a powerful open-source toolkit to analyze combinatorial selection populations and is accompanied by an extensive user guide. The program simplifies data analysis by minimally requiring a web browser and internet access. For the web-based version, the UI can be accessed from any browser operating with any operating system. Alternatively, the user may choose to download the software and run it locally, which is compatible on any system with a functional Docker installation and does not require internet access. Further, the outputs are designed to be modular so that this platform can be easily integrated into existing workflows or used to develop custom ones. Module inputs and outputs are standard file types (e.g., FASTQ/A and CSV).
Modules in this platform can be used for a wide range of functions on subsets of individual populations or across many populations. FASTAptameR-Count, the starting point of the platform, counts and ranks unique sequences. FASTAptameR-Translate translates nucleotide sequences according to standard, nonstandard, or user-defined genetic codes. The trio of modules FASTAptameR-Motif_Search, FASTAptameR-Motif_Tracker, and FASTAptameR-Motif_Discovery serve the three functions of identifying occurrences of motifs, tracking motifs or sequences across multiple populations, and identifying over-enriched motifs, respectively. FASTAptameR-Distance computes the LED between all sequences and a query sequence. FASTAptameR-Mutation_Network identifies the shortest mutational path between two sequences in a population. FASTAptameR-Data_Merge merges sequences from multiple populations. FASTAptameR-Sequence_Enrich and FASTAptameR-Position_Enrich assess sequence trajectories across populations and provide insights into which residues contribute to the enrichment scores. The three linked modules of FASTAptameR-Cluster, FASTAptameR-Cluster_Diversity, and FASTAptameR-Cluster_Enrich cluster sequences, provide cluster-level metadata, and assess how they change across populations.
FASTAptameR 2.0 features substantial improvements relative to its predecessor. By increasing user accessibility, improving the original modules, and providing additional tools for data analysis, FASTAptameR 2.0 further lowers the technical barrier for analysis and exploration of HTS datasets and allows the user to gain more insights from their combinatorial selection experiments.
Materials and methods
Data description
Data from the original FASTAptamer publication57 were used to build and test FASTAptameR 2.0. In brief, these data are two populations of RNA aptamers selected to target HIV-1 reverse transcriptase after 14 and 15 rounds of a SELEX experiment (designated 70HRT14 and 70HRT15, respectively).51,58 These populations were trimmed via cutadapt70 and filtered for high-quality reads via the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). These FASTQ files are available at SkylerKramer/AptamerLibrary: Data for FASTAptameR 2.0 (Zenodo).59
Implementation
FASTAptameR 2.0 is written in the R programming language71 and made interactive with the Shiny package.72 The platform uses ggplot273 to build plots and plotly74 to make them interactive. The entire program (i.e., code, dependencies, and supporting files) is wrapped into a Docker image and deployed on a web server at the University of Missouri - Columbia. The web server has been tested in Google Chrome, FireFox, and Safari. Beta testers at five institutions confirmed platform independence and the absence of external dependencies.
Data availability statement
All code and supporting files are available at https://github.com/SkylerKramer/FASTAptameR-2.0.56 The Docker image is available at https://hub.docker.com/repository/docker/skylerkramer/fastaptamer2. Finally, the web-accessible version of FASTAptameR 2.0 is available at https://fastaptamer2.missouri.edu/. All data analyzed in this manuscript are available at https://github.com/SkylerKramer/AptamerLibrary.59 FASTAptameR 2.0 is distributed under a GNU General Public License version 3.0.
Acknowledgments
This research was supported by NASA Exobiology grant NNX17AE88G to D.H.B., US National Institutes of Health BD2K Training Grant T32HG009060 grant to S.T.K., US National Institute of Health R35-GM126985 grant to D.X., and University of Missouri Life Sciences Fellowship and Wayne L. Ryan Graduate Fellowship from the Ryan Foundation to P.R.G.
Feedback from beta testing was provided by Austin Prater, Marc Johnson, Carolyn Robinson, and Laurie Agosto from the University of Missouri - Columbia; Uli Müller from the University of California - San Diego; Andrej Lupták from the University of California - Irvine; and Elisa Biondi from the Foundation for Applied Molecular Evolution. The authors thank Rebecca Burke-Agüero for critical discussions and technical assistance early in the development of a faster clustering algorithm and Matt Stanley for network support during launch. The authors also thank members of the Burke and Xu research teams and the NASA Interdisciplinary Consortium for Astrobiology Research “Bringing RNA to Life” (NASA grant 80NSSC21K0596) for feedback, discussion, and advice throughout the project.
Author contributions
S.T.K. developed the front end and back end, prepared the tool for deployment, interacted with beta testers, and wrote the manuscript and user guide with input from P.R.G., K.K.A., D.X., and D.H.B. D.H.B. supervised the project, recruited and interacted with beta testers, conceived the project with P.R.G., and edited the manuscript. The authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.08.030.
Supplemental information
References
- 1.Gibney E., Van Noorden R., Ledford H., Castelvecchi D., Warren M. ’Test-tube’ evolution wins chemistry nobel prize. Nature. 2018;562:176. doi: 10.1038/d41586-018-06753-y. [DOI] [PubMed] [Google Scholar]
- 2.Strack R. Noncanonical amino acids on display. Nat. Methods. 2020;17:461. doi: 10.1038/s41592-020-0839-3. [DOI] [PubMed] [Google Scholar]
- 3.Yang Z., Chen F., Chamberlin S.G., Benner S.A. Expanded genetic alphabets in the polymerase chain reaction. Angew. Chem. Int. Ed. Engl. 2010;49:177–180. doi: 10.1002/anie.200905173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoshika S., Leal N.A., Kim M.-J., Kim M.-S., Karalkar N.B., Kim H.-J., Bates A.M., Watkins N.E., SantaLucia H.A., Meyer A.J., et al. Hachimoji DNA and RNA: a genetic system with eight building blocks. Science. 2019;363:884–887. doi: 10.1126/science.aat0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hwang G.T., Romesberg F.E. Unnatural substrate repertoire of a, b, and x family DNA polymerases. J. Am. Chem. Soc. 2008;130:14872–14882. doi: 10.1021/ja803833h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 7.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 8.Pitt J.N., Ferré-D'Amaré A.R. Rapid construction of empirical RNA fitness landscapes. Science. 2010;330:376–379. doi: 10.1126/science.1192001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pressman A.D., Liu Z., Janzen E., Blanco C., Müller U.F., Joyce G.F., Pascal R., Chen I.A. Mapping a systematic ribozyme fitness landscape reveals a frustrated evolutionary network for self-aminoacylating RNA. J. Am. Chem. Soc. 2019;141:6213–6223. doi: 10.1021/jacs.8b13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokobayashi Y. Applications of high-throughput sequencing to analyze and engineer ribozymes. Methods. 2019;161:41–45. doi: 10.1016/j.ymeth.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Burmeister P.E., Lewis S.D., Silva R.F., Preiss J.R., Horwitz L.R., Pendergrast P.S., et al. Direct in vitro selection of a 2’-O-methyl aptamer to VEGF. Chem. Biol. 2005;12:25–33. doi: 10.1016/j.chembiol.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 12.Taylor A.I., Holliger P. Directed evolution of artificial enzymes (XNAzymes) from diverse repertoires of synthetic genetic polymers. Nat. Protoc. 2015;10:1625–1642. doi: 10.1038/nprot.2015.104. [DOI] [PubMed] [Google Scholar]
- 13.Szardenings M., Törnroth S., Mutulis F., Muceniece R., Keinänen K., Kuusinen A., Wikberg J.E. Phage display selection on whole cells yields a peptide specific for melanocortin receptor 1. J. Biol. Chem. 1997;272:27943–27948. doi: 10.1074/jbc.272.44.27943. [DOI] [PubMed] [Google Scholar]
- 14.Dias-Neto E., Nunes D.N., Giordano R.J., Sun J., Botz G.H., Yang K., Setubal J.C., Pasqualini R., Arap W. Next-generation phage display: integrating and comparing available molecular tools to enable cost-effective high-throughput analysis. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villemagne D., Jackson R., Douthwaite J.A. Highly efficient ribosome display selection by use of purified components for in vitro translation. J. Immunol. Methods. 2006;313:140–148. doi: 10.1016/j.jim.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Cotten S.W., Zou J., Wang R., Huang B., Liu R. Ribosome Display and Related Technologies. Springer; New York: 2011. mRNA display-based selections using synthetic peptide and natural protein libraries; pp. 287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Granhøj J., Dimke H., Svenningsen P. A bacterial display system for effective selection of protein-biotin ligase BirA variants with novel peptide specificity. Sci. Rep. 2019;9:4118. doi: 10.1038/s41598-019-40984-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie J., Schultz P.G. Adding amino acids to the genetic repertoire. Curr. Opin. Chem. Biol. 2005;9:548–554. doi: 10.1016/j.cbpa.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 19.Dahlman J.E., Kauffman K.J., Xing Y., Shaw T.E., Mir F.F., Dlott C.C., Langer R., Anderson D.G., Wang E.T. Barcoded nanoparticles for high throughput in vivo discovery of targeted therapeutics. Proc. Natl. Acad. Sci. USA. 2017;114:2060–2065. doi: 10.1073/pnas.1620874114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sago C.D., Lokugamage M.P., Paunovska K., Vanover D.A., Monaco C.M., Shah N.N., Gamboa Castro M., Anderson S.E., Rudoltz T.G., Lando G.N., et al. High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing. Proc. Natl. Acad. Sci. USA. 2018;115:E9944–E9952. doi: 10.1073/pnas.1811276115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paunovska K., Sago C.D., Monaco C.M., Hudson W.H., Castro M.G., Rudoltz T.G., Kalathoor S., Vanover D.A., Santangelo P.J., Ahmed R., et al. A direct comparison of in vitro and in vivo nucleic acid delivery mediated by hundreds of nanoparticles reveals a weak correlation. Nano Lett. 2018;18:2148–2157. doi: 10.1021/acs.nanolett.8b00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brenner S., Lerner R.A. Encoded combinatorial chemistry. Proc. Natl. Acad. Sci. USA. 1992;89:5381–5383. doi: 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Favalli N., Bassi G., Scheuermann J., Neri D. DNA-encoded chemical libraries - achievements and remaining challenges. FEBS Lett. 2018;592:2168–2180. doi: 10.1002/1873-3468.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thiel W.H., Giangrande P.H. Analyzing HT-SELEX data with the galaxy project tools a web based bioinformatics platform for biomedical research. Methods. 2016;97:3–10. doi: 10.1016/j.ymeth.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alam K.K., Chang J.L., Burke D.H. FASTAptamer: a bioinformatic toolkit for high-throughput sequence analysis of combinatorial selections. Mol. Ther. Nucleic Acids. 2015;4 doi: 10.1038/mtna.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho M., Xiao Y., Nie J., Stewart R., Csordas A.T., Oh S.S., Thomson J.A., Soh H.T. Quantitative selection of DNA aptamers through microfluidic selection and high-throughput sequencing. Proc. Natl. Acad. Sci. USA. 2010;107:15373–15378. doi: 10.1073/pnas.1009331107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schütze T., Wilhelm B., Greiner N., Braun H., Peter F., Mörl M., Erdmann V.A., Lehrach H., Konthur Z., Menger M., et al. Probing the SELEX process with next-generation sequencing. PLoS One. 2011;6 doi: 10.1371/journal.pone.0029604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiel W.H. Galaxy workflows for web-based bioinformatics analysis of aptamer high-throughput sequencing data. Mol. Ther. Nucleic Acids. 2016;5:e345. doi: 10.1038/mtna.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen Quang N., Bouvier C., Henriques A., Lelandais B., Ducongé F. Time-lapse imaging of molecular evolution by high-throughput sequencing. Nucleic Acids Res. 2018;46:7480–7494. doi: 10.1093/nar/gky583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoinka J., Berezhnoy A., Sauna Z.E., Gilboa E., Przytycka T.M. Springer International Publishing; 2014. AptaCluster a Method to Cluster HT-SELEX Aptamer Pools and Lessons from its Application; pp. 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato S., Ono T., Minagawa H., Horii K., Shiratori I., Waga I., Ito K., Aoki T. FSBC: Fast string-based clustering for HT-SELEX data. BMC Bioinf. 2020;21:263. doi: 10.1186/s12859-020-03607-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoinka J., Zotenko E., Friedman A., Sauna Z.E., Przytycka T.M. Identification of sequence-structure RNA binding motifs for SELEX-derived aptamers. Bioinformatics. 2012;28:i215–i223. doi: 10.1093/bioinformatics/bts210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoinka J., Berezhnoy A., Dao P., Sauna Z.E., Gilboa E., Przytycka T.M. Large scale analysis of the mutational landscape in HT-SELEX improves aptamer discovery. Nucleic Acids Res. 2015;43:5699–5707. doi: 10.1093/nar/gkv308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dao P., Hoinka J., Takahashi M., Zhou J., Ho M., Wang Y., Costa F., Rossi J.J., Backofen R., Burnett J., Przytycka T.M. AptaTRACE elucidates RNA sequence-structure motifs from selection trends in HT-SELEX experiments. Cell Syst. 2016;3:62–70. doi: 10.1016/j.cels.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caroli J., Taccioli C., De La Fuente A., Serafini P., Bicciato S. APTANI: a computational tool to select aptamers through sequence-structure motif analysis of HT-SELEX data. Bioinformatics. 2016;32:161–164. doi: 10.1093/bioinformatics/btv545. btv545. [DOI] [PubMed] [Google Scholar]
- 36.Shieh K.R., Kratschmer C., Maier K.E., Greally J.M., Levy M., Golden A. AptCompare: optimized de novo motif discovery of RNA aptamers via HTS-SELEX. Bioinformatics. 2020;36:2905–2906. doi: 10.1093/bioinformatics/btaa054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen Quang N., Perret G., Ducongé F. Applications of high-throughput sequencing for in vitro selection and characterization of aptamers. Pharmaceuticals. 2016;9:76. doi: 10.3390/ph9040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoinka J., Dao P., Przytycka T.M. AptaGUI - a graphical user interface for the efficient analysis of HT-SELEX data. Mol. Ther. Nucleic Acids. 2015;4:e257. doi: 10.1038/mtna.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoinka J., Backofen R., Przytycka T.M. AptaSUITE: a full-featured bioinformatics framework for the comprehensive analysis of aptamers from HT-SELEX experiments. Mol. Ther. Nucleic Acids. 2018;11:515–517. doi: 10.1016/j.omtn.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gotrik M.R., Feagin T.A., Csordas A.T., Nakamoto M.A., Soh H.T. Advancements in aptamer discovery technologies. Acc. Chem. Res. 2016;49:1903–1910. doi: 10.1021/acs.accounts.6b00283. [DOI] [PubMed] [Google Scholar]
- 41.Berezhnoy A., Stewart C.A., Mcnamara J.O., 2nd, Thiel W., Giangrande P., Trinchieri G., Gilboa E., Gilboa E. Isolation and optimization of murine IL-10 receptor blocking oligonucleotide aptamers using high-throughput sequencing. Mol. Ther. 2012;20:1242–1250. doi: 10.1038/mt.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thiel W.H., Bair T., Peek A.S., Liu X., Dassie J., Stockdale K.R., Behlke M.A., Miller F.J., Giangrande P.H. Rapid identification of cell-specific, internalizing RNA aptamers with bioinformatics analyses of a cell-based aptamer selection. PLoS One. 2012;7 doi: 10.1371/journal.pone.0043836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valenzano S., De Girolamo A., DeRosa M.C., McKeague M., Schena R., Catucci L., Pascale M. Screening and identification of DNA aptamers to tyramine using in vitro selection and high-throughput sequencing. ACS Comb. Sci. 2016;18:302–313. doi: 10.1021/acscombsci.5b00163. [DOI] [PubMed] [Google Scholar]
- 44.Hamada M. In silico approaches to RNA aptamer design. Biochimie. 2018;145:8–14. doi: 10.1016/j.biochi.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi M., Wu X., Ho M., Chomchan P., Rossi J.J., Burnett J.C., Zhou J. High throughput sequencing analysis of RNA libraries reveals the influences of initial library and PCR methods on SELEX efficiency. Sci. Rep. 2016;6:33697. doi: 10.1038/srep33697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blind M., Blank M. Aptamer selection technology and recent advances. Mol. Ther. Nucleic Acids. 2015;4:e223. doi: 10.1038/mtna.2014.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Komarova N., Barkova D., Kuznetsov A. Implementation of high-throughput sequencing (HTS) in aptamer selection technology. Int. J. Mol. Sci. 2020;21:8774. doi: 10.3390/ijms21228774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jijakli K., Khraiwesh B., Fu W., Luo L., Alzahmi A., Koussa J., Chaiboonchoe A., Kirmizialtin S., Yen L., Salehi-Ashtiani K. The in vitro selection world. Methods. 2016;106:3–13. doi: 10.1016/j.ymeth.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 49.Kinghorn A., Fraser L., Liang S., Shiu S., Tanner J. Aptamer bioinformatics. Int. J. Mol. Sci. 2017;18:2516. doi: 10.3390/ijms18122516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zimmermann B., Gesell T., Chen D., Lorenz C., Schroeder R. Monitoring genomic sequences during SELEX using high-throughput sequencing: neutral SELEX. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ditzler M.A., Lange M.J., Bose D., Bottoms C.A., Virkler K.F., Sawyer A.W., Whatley A.S., Spollen W., Givan S.A., Burke D.H. High-throughput sequence analysis reveals structural diversity and improved potency among RNA inhibitors of HIV reverse transcriptase. Nucleic Acids Res. 2013;41:1873–1884. doi: 10.1093/nar/gks1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alam K.K., Chang J.L., Lange M.J., Nguyen P.D.M., Sawyer A.W., Burke D.H. Poly-target selection identifies broad-spectrum RNA aptamers. Mol. Ther. Nucleic Acids. 2018;13:605–619. doi: 10.1016/j.omtn.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dupont D.M., Larsen N., Jensen J.K., Andreasen P.A., Kjems J. Characterisation of aptamer-target interactions by branched selection and high-throughput sequencing of SELEX pools. Nucleic Acids Res. 2015;43:e139. doi: 10.1093/nar/gkv700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spiga F.M., Maietta P., Guiducci C. More DNA-aptamers for small drugs: a capture-SELEX coupled with surface plasmon resonance and high-throughput sequencing. ACS Comb. Sci. 2015;17:326–333. doi: 10.1021/acscombsci.5b00023. [DOI] [PubMed] [Google Scholar]
- 55.Levay A., Brenneman R., Hoinka J., Sant D., Cardone M., Trinchieri G., Przytycka T.M., Berezhnoy A. Identifying high-affinity aptamer ligands with defined cross-reactivity using high-throughput guided systematic evolution of ligands by exponential enrichment. Nucleic Acids Res. 2015;43:e82. doi: 10.1093/nar/gkv534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kramer S. Zenodo; 2022. SkylerKramer/FASTAptameR-2.0: FASTAptameR-2.0. [Google Scholar]
- 57.Burke D.H., Scates L., Andrews K., Gold L. Bent pseudoknots and novel RNA inhibitors of type 1 human immunodeficiency virus (HIV-1) reverse transcriptase. J. Mol. Biol. 1996;264:650–666. doi: 10.1006/jmbi.1996.0667. [DOI] [PubMed] [Google Scholar]
- 58.Whatley A.S., Ditzler M.A., Lange M.J., Biondi E., Sawyer A.W., Chang J.L., Franken J.D., Burke D.H. Potent inhibition of HIV-1 reverse transcriptase and replication by nonpseudoknot, “UCAA-motif” RNA aptamers. Mol. Ther. Nucleic Acids. 2013;2:e71. doi: 10.1038/mtna.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kramer S. Zenodo; 2022. SkylerKramer/AptamerLibrary: Data for FASTAptameR 2.0. [Google Scholar]
- 60.Salamango D.J., Alam K.K., Burke D.H., Johnson M.C. In vivo analysis of infectivity, fusogenicity, and incorporation of a mutagenic viral glycoprotein library reveals determinants for virus incorporation. J. Virol. 2016;90:6502–6514. doi: 10.1128/JVI.00804-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bailey T.L., Johnson J., Grant C.E., Noble W.S. The MEME suite. Nucleic Acids Res. 2015;43:W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nawrocki E.P., Eddy S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gruenke P.R., Alam K.K., Singh K., Burke D.H. 2’-fluoro-modified pyrimidines enhance affinity of RNA oligonucleotides to HIV-1 reverse transcriptase. RNA. 2020;26:1667–1679. doi: 10.1261/rna.077008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tuerk C., Macdougal S., Gold L. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc. Natl. Acad. Sci. USA. 1992;89:6988–6992. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weiss Z., DasGupta S. REVERSE: a user-friendly web server for analyzing next-generation sequencing data from in vitro selection/evolution experiments. bioRxiv. 2022 doi: 10.1093/nar/gkac508. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prlić A., Procter J.B. Ten simple rules for the open development of scientific software. PLoS Comput. Biol. 2012;8 doi: 10.1371/journal.pcbi.1002802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.List M., Ebert P., Albrecht F. Ten simple rules for developing usable software in computational biology. PLoS Comput. Biol. 2017;13 doi: 10.1371/journal.pcbi.1005265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sandve G.K., Nekrutenko A., Taylor J., Hovig E. Ten simple rules for reproducible computational research. PLoS Comput. Biol. 2013;9 doi: 10.1371/journal.pcbi.1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leprevost F.d.V., Barbosa V.C., Francisco E.L., Perez-Riverol Y., Carvalho P.C. On best practices in the development of bioinformatics software. Front. Genet. 2014;5:199. doi: 10.3389/fgene.2014.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. j. 2011;17:10. [Google Scholar]
- 71.R Core Team . R Foundation for Statistical Computing; 2019. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 72.Chang W., Cheng J., Allaire J., Sievert C., Schloerke B., Xie Y., Allen J., McPherson J., Dipert A., Borges B. 2021. Shiny: Web Application Framework for R. [Google Scholar]
- 73.Wickham H. Springer-Verlag; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 74.Sievert C. Chapman; Hall/CRC; 2020. Interactive Web-Based Data Visualization with R, Plotly, and Shiny. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All code and supporting files are available at https://github.com/SkylerKramer/FASTAptameR-2.0.56 The Docker image is available at https://hub.docker.com/repository/docker/skylerkramer/fastaptamer2. Finally, the web-accessible version of FASTAptameR 2.0 is available at https://fastaptamer2.missouri.edu/. All data analyzed in this manuscript are available at https://github.com/SkylerKramer/AptamerLibrary.59 FASTAptameR 2.0 is distributed under a GNU General Public License version 3.0.