Abstract
Progress towards understanding catalytically “dead” protein kinases—or pseudokinases—in biology and disease has hastened over the past decade. An especially lively area for structural biology, pseudokinases appear strikingly similar to their kinase relatives, despite lacking key catalytic residues. Distinct active- and inactive-like conformation states, crucial for regulating bona fide protein kinases, are conserved in pseudokinases and appear essential for function. Here, we discuss recent structural data on conformational transitions and nucleotide binding by pseudokinases, from which some common principles emerge. For pseudokinases and bona fide kinases alike, a conformational toggle appears to control their ability to interact with signaling effectors. We also discuss how biasing this conformational toggle may provide opportunities to target pseudokinases pharmacologically in disease.
Keywords: cell signaling, pseudokinase, kinase, protein conformation, allostery, conformational disruptor
Pseudokinases: New Challenges in Cell Signaling
Enzymatic protein phosphorylation is one of the most pervasive post-translational modifications in eukaryotes [1, 2], Protein kinases (see Glossary) catalyze transfer of the γ-phosphate group from adenosine-5’-triphosphate (ATP) to Ser, Thr, or Tyr side chains of protein substrates, regulating many biological processes. As with all enzymes, precise orientation of substrates (ATP and substrate protein), cofactors (e.g. Mg2+), and key conserved residues is essential for catalytic function [3]. It was therefore a surprise when a significant proportion (~10%) of protein kinases in the deduced human kinome were found to lack one or more of the conserved catalytic residues [4]. This observation suggested the existence of a class of pseudokinases incapable of catalyzing phosphotransfer. The fact that these pseudokinases are conserved across species [5] argued further that they may have evolved new non-catalytic functions [6, 7]. The past two decades have seen significant progress both in understanding pseudokinases themselves [8] and in appreciating non-catalytic functions of their bona fide kinase relatives [9]. The field has been illuminated particularly by a growing set of crystal structures of pseudokinase and kinase domains caught in different signaling ‘states’. We summarize some common principles emerging from these studies, which reflect the versatility and resourcefulness of the kinase fold.
What Makes a Pseudokinase?
Several pseudokinases were identified through experiments demonstrating lack of catalytic activity. Most, however, were recognized based on loss of one or more highly conserved motifs (Figure 1 A) crucial for substrate orientation and/or catalysis [3]. The Gly-loop (GxGxxG) in protein kinase domains engages ATP phosphates with its backbone amides, and lies above the ATP-binding site between the largely β-sheet N-lobe and α-helical C-lobe (Figure 1B). In active kinases, the invariant β3 lysine (K) salt bridges with the αC glutamate (E) in helix αC to position the β3 lysine for interaction with ATP (Figure 1B). The aspartate side-chain in the HRD (His-Arg-Asp) motif at the beginning of the catalytic loop serves as a catalytic base, abstracting a proton from the substrate’s hydroxyl group to promote nucleophilic attack of the ATP γ-phosphate. At the beginning of the activation loop, the aspartate of the DFG (Asp-Phe-Gly) motif positions key divalent cations (Mg2+) to interact with ATP (Figure 1B).
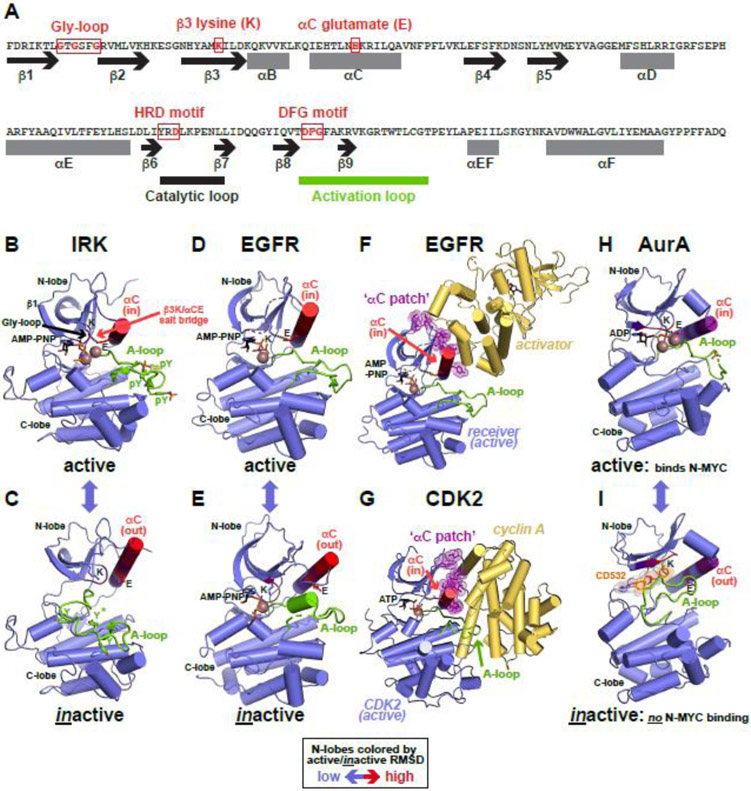
Figure 1. Kinases and conformational toggling.
(A) Key residues and sequence motifs in a protein kinase domain. An abbreviated primary sequence of the bona fide kinase protein kinase A (PKA) is shown, with secondary structural elements represented as black arrows (β strands) or grey rectangles (α helices), and marked. The key motifs are indicated and boxed in red. The activation loop is marked as a thick green line and catalytic loop as thick black line.
(B,C) Conformational transition between active (B) and inactive (C) conformations of IRK (PDB IDs: 1IR3 [101] and 1IRK [102]). Kinase domains are colored slate blue, and the N-lobe colored according to the backbone position root mean square deviation (RMSD) of alpha carbon positions between active and inactive conformation states – red representing the greatest changes. Activation loops are green, with YxxxYY motif tyrosine side-chains shown, phosphorylated in (B). The β3 lysine (K) and αC glutamate (E) are marked, with the interaction shown by a dotted line in (B). Bound AMP-PNP and Mg2+ (brown) are shown in (B), but are absent in (C). The αC position is assigned ‘in’ (active) or ‘out’ (inactive).
(D,E) Transition between the active (D) and inactive (E) EGFR kinase domain conformations (PDB IDs: 3VJO [103] and 2GS7 [104]), labeled as in (B) and (C). Both structures have AMP-PNP bound. The Mg2+ ion in (D) is positioned based on its location in (E).
(F) Activation of the EGFR kinase domain via the purple ‘αC patch’. The (active) receiver molecule within the asymmetric dimer is the same as in (D), and the activator is yellow. Interfacial residues in the receiver N-lobe are shown (purple), and constitute the ‘αC patch’. From PDB ID 3VJO [103].
(G) Activation of CDK2 by cyclin A, represented as for EGFR in (F), with the αC patch highlighted (PDB ID: 1FIN [44]).
(H,I) Conformational toggling of the Aurora A (AurA) kinase domain from the active ATP-bound conformation (H) to an inactive conformation (I) when bound to the conformational disruptor CD532. Labels are as in (B). PDB IDs are 1MQ4 [105] and 4J8M [76] respectively. AurA binds N-MYC in the active conformation (H), but not the disrupted conformation (I).
Are All Pseudokinases Really Phosphotransfer-Inactive?
One of the earliest potential pseudokinase families investigated biochemically and structurally was the WNK family, which regulate ion homeostasis and blood pressure [10]. The name ‘With-No-lysine(K)’ reflects replacement of the β3 lysine with cysteine, suggesting ATP-binding defects. Surprisingly, however, WNK1 has significant in vitro kinase activity [11], albeit much slower (kcat ~ 1 min−1 [12]) than activated protein kinase A (kcat ~1,000 min−1 [13]) and more in line with receptor tyrosine kinases prior to activation. A lysine in strand β2 (in the Gly-loop) compensates for loss of the β3 lysine to allow ATP binding [14]. WNKs are now considered bona fide protein kinases, and this early discovery suggested that other pseudokinases might also retain activity through non-canonical structural solutions [15].
Indeed, CASK, a calcium/calmodulin (CaM)-activated serine-threonine kinase, was reported to function as a Mg2+-independent kinase [16] through uniquely placed basic side-chains. Other examples followed, including the EGFR family member ErbB3/HER3 [17], JAK2 [18], KSR1/2 [19], and TRIB2 [20]. In each case, reported activity was very weak compared with canonical kinases, noting the caveat that ideal substrates and activating conditions may not have been employed. Importantly, at least for ErbB3, later studies showed that small molecule inhibition of the kinase does not impact function [21]. It is now clear that many pseudokinases completely lack the ability to catalyze phosphotransfer, although other related enzymatic activities are now emerging for a subset [22-24] as described in Box 1.
Box 1. Pseudokinases with Unexpected Catalytic Activities.
Beyond protein phosphorylation, several fascinating examples of novel chemistries have recently been discovered in divergent pseudokinases (Figure I), further highlighting the versatility of the kinase fold. It will be interesting to follow additional examples as more enigmatic enzymes with the kinase fold are studied.
Sugar substrates:
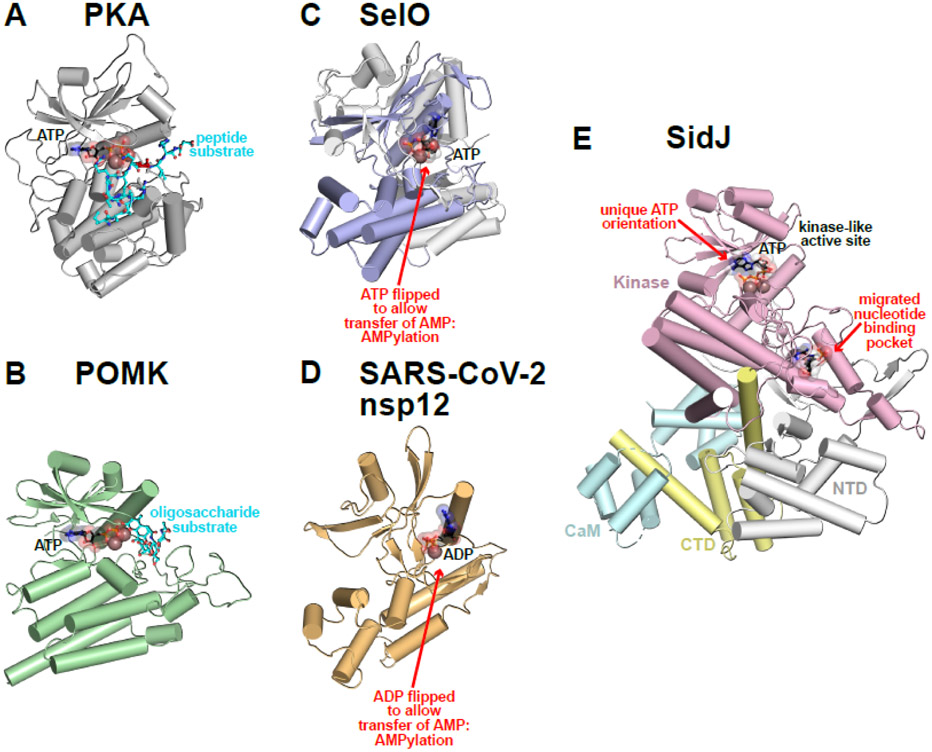
Two members of the human kinome are now known to phosphorylate glycan, rather than peptide, substrates. FAM20B was shown to direct 2-O-phosphorylation of xylose within glycosaminoglycans to promote chain elongation, which appears to be essential for normal skeletal development [85-87]. A second example is protein O-mannose kinase (POMK), which transfers phosphate to a mannose unit on glycan-modified α-dystroglycan (Figure IB) – a process that is impaired in some forms of muscular dystrophy [88, 89].
Protein AMPylation:
A highly conserved selenocysteine-containing protein SelO was recently discovered to co-opt the kinase fold to transfer AMP onto protein side-chain hydroxyls [24, 90], a reaction previously thought to be the preserve of FIC domain-containing proteins [91]. Structural studies of SelO [24] revealed that the key is ATP binding to the enzyme in a ‘flipped’ orientation (Figure IC), allowing it to transfer AMP rather than the terminal phosphate group to glutaredoxin in a process that protects yeast from oxidative stress. Remarkably, recent work on the SARS-CoV-2 nsp12 protein has shown that its pseudokinase-like N-terminal nidovirus RdRp-associated nucleotidyltransferase, or NiRAN, domain resembles SelO structurally (Figure ID) [92]. The enzymatic activity of the NiRAN domain is required for viral propagation, and has been shown to transfer nucleotide monophosphates to protein substrates in a divalent cation-dependent way [93]. Very recent studies have shown that NiRAN transfers RNA to protein substrates as an intermediate step in the process of capping the 5’ end of the SARS-CoV-2 RNA genome [94].
Protein Glutamylation:
Bacterial effector pseudokinases can also play unique roles, as exemplified by the Legionella pneumophila effector SidJ. SidJ has a divergent pseudokinase domain that, when activated by host-cell calmodulin, catalyzes transfer of free glutamate onto glutamic acid side chains of its co-effectors in the SidE family, regulating SidE ubiquitination activity for precise Legionella replication [22, 95, 96]. Key to this activity are both a flipped mode of ATP binding and a second migrated nucleotide binding site (Figure IE). SidE is thought first to become adenylated by the kinase-like active site, and then to associate (via AMP) with the migrated nucleotide binding site, where glutamylation ensues [23].
Moonlighting metabolic enzymes:
A handful of metabolic kinases have also been associated with phosphorylation of proteins in addition to the small molecule metabolites they are typically associated with [97]. Perhaps the most fiercely debated example is the pyruvate kinase isoform M2, reported to phosphorylate several protein substrates including histone H3 (thus promoting cell proliferation). Although contaminating adenine nucleotides within commercial preparations of radiolabeled phosphoenolpyruvate have been argued to be a potential experimental confounder [98], PK-M2 and other ‘moonlighting’ metabolic kinases remain a highly controversial possibility. Additional examples continue to be described [99, 100], in some cases with altered nucleotide selectivity [100].
Which Came First: The Kinase or the Pseudokinase?
An early question in field was whether kinases or pseudokinases evolved first [25]. Pseudokinases occur in all eight major phylogenetic groups of protein kinases [4], and in atypical groups. Some groups, like the receptor guanylyl cyclase (RGC) family [26], contain only pseudokinases. Although the mean fraction of the kinome represented by pseudokinases is ~10% across the tree of life [5], they are enriched in certain species – reaching 50% in the protist Giardia lamblia, for example. Certain pseudokinase families are expanded in particular domains of life: IRAK-like and non-RD [27] pseudokinases in plants, tyrosine kinase-like (TKL) pseudokinases in fungi, and PknB-like pseudokinases in bacteria. Kwon et al. [5] suggested an analogy between expansion of these families and tyrosine kinase expansion in metazoans, providing alternatives modes of directing signaling specificity. Although pseudokinases are now generally considered to have arisen through duplication of genes encoding active kinases, the possibility that some active kinases instead evolved from ancestral pseudokinases is intriguing.
Dead Kinases in Disease
Most human pseudokinases are well conserved, and several are dysregulated in disease [8]. For example, a V617F mutation in the JAK2 pseudokinase domain (JH2) causes myeloproliferative neoplasms [28]. Mutations in ErbB3’s pseudokinase domain are linked to gastrointestinal cancers [29, 30]. Moreover, altered expression of Tribbles family pseudokinases (TRIB1/2/3) tips the balance between oncogenic and tumor suppressive roles of this pseudokinase, which controls degradation of the C/EBPα transcription factor [31].
The important biological roles of these ‘dead’ enzymes are presumably achieved through non-catalytic molecular mechanisms. Several appear to function as allosteric regulators. Some, notably ErbB3 and the JAK JH2 domain, directly regulate related active kinases [32]: ErbB3 allosterically activates an adjacent EGFR kinase in ErbB3/EGFR receptor dimers [30], and in JAKs, the JH2 pseudokinase domain regulates the adjacent catalytically-competent JH1 kinase domain in the same protein [33, 34]. In another example discussed below, the STRADα pseudokinase allosterically activates the tumor suppressor kinase LKB1/STK11 [35]. Other pseudokinases may function as ‘switchable adaptors’ in signaling complex assembly, or as dominant-negative-like signaling inhibitors as discussed elsewhere [8, 32]. It is important to note that these non-catalytic functions are not solely the preserve of pseudokinases – they are also now known to contribute to functions of bona fide protein kinases [8, 9]. The existence of pseudokinases thus reflects the dual fortes of the kinase fold: the ability to act both as a molecular switch [36] and as a regulated catalyst of phosphotransfer reactions (or other reactions outlined in Box 1).
Conformational Switching is Crucial for Kinase and Pseudokinase Function
Structures of roughly half of all human protein kinase domains have now been determined [37]. Conserved features led Louise Johnson and colleagues [38] to comment early on, after the first line in Anna Karenina, that “all active kinases are alike; each inactive kinase is inactive after its own fashion”. The large structural database underlines this insight and shows how each protein kinase family is inactive in its own way, making the opening sentence of Tolstoy’s novel even more apt.
Active/canonical protein kinases
Canonical protein kinases have been known for some time to undergo a ‘conformational toggle’ between active and inactive conformations [39, 40], illustrated in Figure 1B, C for the insulin receptor tyrosine kinase domain (IRK) [41]. Active kinases are all conformationally very similar (compare Figure 1B and D, for example), but are subject to a variety of family-specific distortions in their inactive forms. Changes are largely restricted to the activation loop and the αC region of the N-lobe (compare Figure 1C and E). Conformational changes in the N-lobes in Figures 1 and 2 are illustrated by coloring according to root mean square deviation (RMSD) of alpha carbon positions between active and inactive states (red represents the largest change).
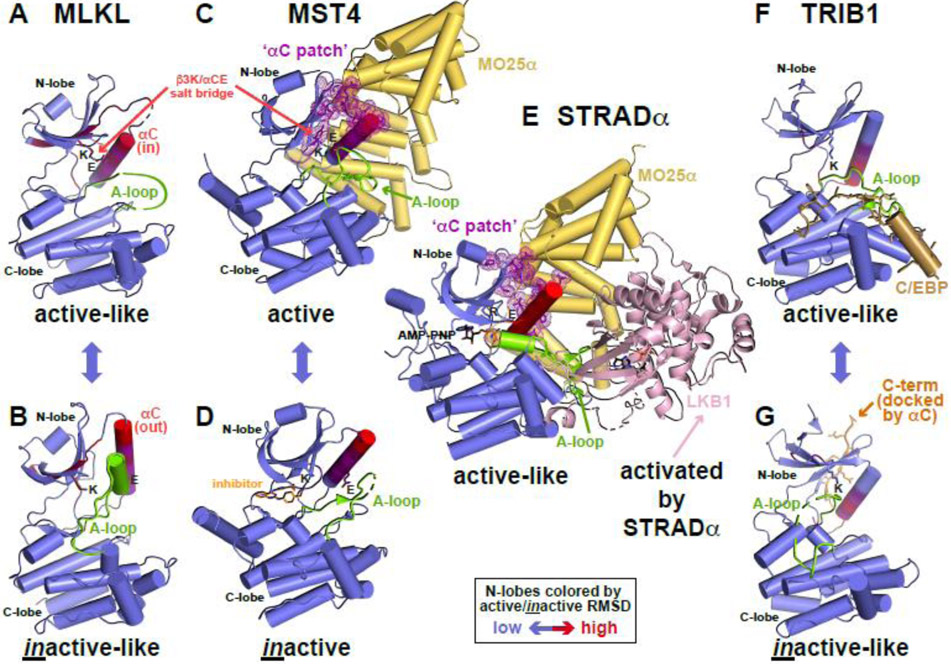
Figure 2. Conformational toggling of pseudokinases.
(A,B) Conformational transition between active-like (A) and inactive-like (B) conformations of MLKL (PDB IDs: 7JW7 [47] and 7JXU [47]), labeled and colored as in Figure 1, with N-lobe colored according to the RMSD between states in an N-lobe overlay.
(C) Allosteric activation of the bona fide protein kinase MST4 by MO25α (PDB ID: 4FZA [48]), resembling EGFR and CDK2 activation in Figure 1. MO25α (yellow) binds to residues in the MST4 N-lobe that constitute the αC patch (C), which in turn coincides with the most variable region in the MST4 N-lobe between active and inactive conformations.
(D) MST4 kinase domain in an inactive conformation (PDB ID: 3GGF [106]), bound to a quinazoline inhibitor [106], which is shown in orange.
(E) Conformational modulation of the pseudokinase STRADα by MO25α to activate LKB1. MO25α binds the αC patch of STRADα to stabilize it in the active-like ATP-bound conformation. The two molecules form a heterotrimer with the bona fide kinase LKB1/STK11 (pink) with STRADα recognizing LKB1 like a pseudosubstrate and allosterically activating it. PDB ID: 2WTK [35].
(F,G) Conformational transition between active-like (F) and inactive-like (G) TRIB1 (PDB IDs: 5CEM [52] and 6DC0 [51]), as shown for MLKL in (A). In the active-like state (F), TRIB1 binds the degron of C/EBP family transcription factors (shown in sand color) like a pseudosubstrate, and promotes release of the C-terminal fragment of TRIB1 (orange in G) that docks adjacent to αC – near the αC patch – in the inactive-like conformation.
For the IRK active-to-inactive transition (Figure 1B, C):
The activation loop is distorted, inserting one of its tyrosines (starred in Figure 1C) into the active site, preventing access of protein substrate.
The ATP-binding site is blocked by the A-loop conformation, with the DFG motif in the so-called ‘out’ [42] conformation.
Helix αC is rotated away from the ATP-binding site to the ‘out’ position, breaking the β3 K/αC E salt-bridge.
These distortions are all reversed when the activation loop tyrosines become auto-phosphorylated (Figure 1B) and IRK is stabilized in its ATP-bound ‘active’ conformation.
Moving to a different tyrosine kinase family, the active conformation is once again very similar for the kinase domain of epidermal growth factor receptor (EGFR, Figure 1D), but the inactive form is quite different (Figure 1E):
The A-loop is distorted in a very different way, acquiring a short 310 helix that interacts with and displaces helix αC (Figure 1E).
Helix αC is thus rotated away (out) from the ATP-binding site (which is retained), and the key β3 Lys/αC Glu salt-bridge is broken.
The DFG motif remains in the ‘in’ conformation in inactive EGFR, which also retains ATP-binding ability. Notably, EGFR activation does not involve activation loop phosphorylation, instead relying on allosteric changes induced within asymmetric kinase domain dimers. In these dimers, the C-lobe of one kinase domain in Figure 1F (‘activator’) docks onto a site in the ‘receiver’ N-lobe represented as the purple ‘αC patch’. These interactions promote an allosteric transition in the receiver to the familiar active conformation [43].
This allosteric regulation mode closely resembles activation of cyclin-dependent kinases (CDKs). Cyclins bind a similar ‘αC patch’ site on the CDK N-lobe (Figure 1G) [44], allosterically promoting transition of helix αC from the ‘out’ to ‘in’ position, among other changes. As pointed out by Jura et al. [43], the αC patch is a key interaction site on the N-lobe of many different kinases, to which allosteric activators and inhibitors bind to control kinase conformation. Importantly, the αC patch coincides with the region in the N-lobe of kinase domains that differs most when toggling between active and inactive states (hence the red color in Figures 1 and 2). This toggling can alternatively be driven ‘externally’ (e.g. by binding of a cyclin or dimerization partner) or ‘internally’, through mutation or by binding of small molecules that selectively stabilize one state. In such ‘internally’ controlled cases, interactions mediated by the αC patch might be regulated [9]. Aurora A provides one intriguing example, switched from an active conformation that binds N-MYC (Figure 1H) to an inactive conformation (Figure 1I) that does not when it binds conformation disrupting ATP-competitive inhibitors such as CD532 [45].
Pseudokinases
Although few pseudokinases are well understood, conformational toggling seems to be a key feature. The mixed lineage kinase domain-like (MLKL) pseudokinase, a terminal effector of necroptosis, provides one example. Recent studies of human MLKL using conformation-specific monobodies argue that A-loop phosphorylation by the upstream activator RIPK3 [46] toggles the pseudokinase domain of MLKL from the inactive-like conformation (Figure 2B) into an active-like conformation (Figure 2A). In the inactive state, the MLKL ‘executioner domain’ is kept at bay by intramolecular interactions with the pseudokinase domain [47]. The conformational toggle ‘releases’ the executioner domain to kill the cell by permeabilizing the plasma membrane from the inside – aided by MLKL oligomerization. Here, pseudokinase domain conformational toggling regulates interaction with another domain within the same molecule.
Conformational toggling of the pseudokinase domain is also functionally crucial for the Ste20-family adaptor protein-α (STRADα). STRADα has several close bona fide kinase relatives, including MST4. These are allosterically regulated in a similar manner to EGFR and CDK (Figure 1) by binding of a helical scaffolding protein called MO25 (for mouse protein-25) to the N-lobe αC patch of MST4, resulting in its allosteric activation (Figure 2C, D) [48, 49]. MO25 binds the same αC patch on STRADα’s pseudokinase domain [50], stabilizing its active-like conformation and enhancing its ability to bind ATP. These changes allow STRADα to engage the LKB1 tumor suppressor kinase (pink in Figure 2E) as a pseudosubstrate and to cooperate with MO25α in activating LKB1 [35]. Here, an allosteric mechanism used to activate bona fide kinases (MST3, MST4 and others) is employed to ‘activate’ the STRADα pseudokinase (Figure 2E).
Studies of the Tribbles pseudokinase TRIB1 also suggest that its regulation involves an inactive-to-active conformational toggle. A degron within C/EBP transcription factors binds the TRIB1 pseudokinase like a pseudosubstrate (colored sand in Figure 2F), displacing and ordering the pseudokinase A-loop (compare with Figure 2G). The resulting allosteric changes resemble an inactive-to-active kinase domain transition, with key alterations in αC close to the αC patch [51]. Without bound substrate (Figure 2G), a C-terminal extension of TRIB1 (orange in Figure 2G) docks alongside αC [52]. Binding of the C/EBP degron prevents this C-term docking, displacing it so that it can bind/recruit the COP1 E3 ubiquitin ligase. These allosteric changes regulate TRIB1’s ability to mediate COP1 binding to C/EBP transcription factors to promote their ubiquitinylation and degradation in adipocyte and myeloid cell development.
Similar lessons are also being learned from the large number of pseudokinases in plants where kinomes can be expanded up to 5-fold and pseudokinases are particularly prevalent [5]. So-called non-RD kinases [27], which lack the HRD motif arginine, are particularly expanded in plants. They are associated with innate immune functions, like their IRAK and RIPK kinase relatives in humans. Some non-RD bona fide kinases have been shown to promote antibacterial immunity in plants independent of phosphotransfer activity [53]. The findings suggest that a conformational toggle controls signaling. The pathogen recognition receptor (PRR) BIR2, has an intracellular pseudokinase domain thought to be phosphorylated by an associated receptor (BAK1), leading to a conformational toggle that alters its interaction properties [54] to drive signaling.
Pseudokinase signaling by G-protein-like conformational switching?
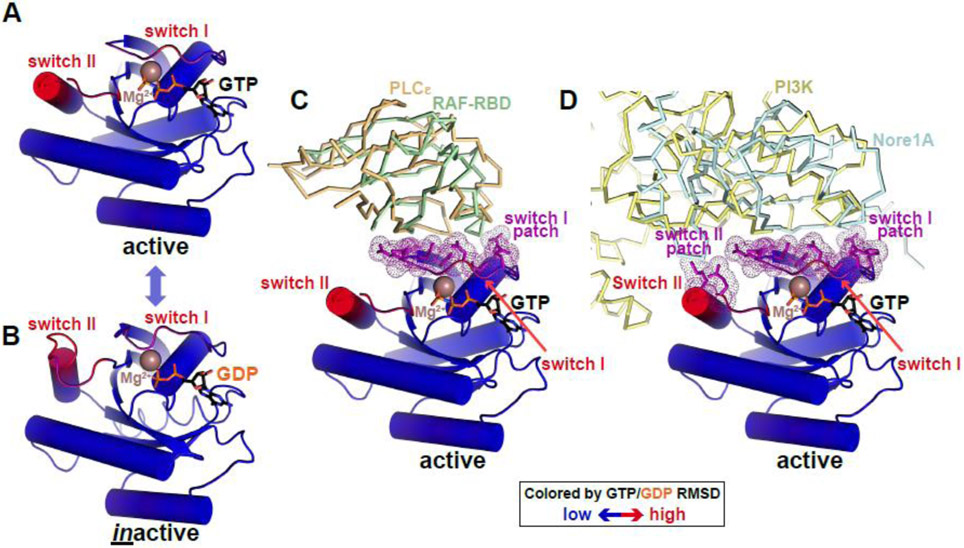
Pseudokinase/kinase conformational toggling can be controlled by nucleotide or inhibitor binding (e.g. Aurora A), binding of other proteins (e.g. EGFR, STRADα, and TRIB1), or phosphorylation (e.g. IRK and MLKL) or other modifications. Signaling through conformational toggling is not a new concept and is perhaps best known for guanine nucleotide-binding (G) proteins such as RAS. Like protein kinases, G proteins adopt a characteristic nucleotide-binding fold with conserved motifs spatially arranged to catalyze GTP hydrolysis. Like kinases, G proteins toggle between ‘active’ and ‘inactive’ states – bound to GTP and GDP respectively [55]. Two so-called ‘switch’ regions (I and II) display the majority of the resulting conformational changes in RAS (Figure 3A, B, colored by RMSD), although the changes are substantially smaller than in kinases and pseudokinases. Switch I might be considered analogous to the kinase domain A-loop, and switch II to αC. These regions also correspond to the common surface bound by RAS effectors such as RAF (Figure 3C), phosphoinositide-3-kinase-gamma (Figure 3D), and other proteins that mediate RAS signaling. The ‘switch I’ and ‘switch II’ patches responsible for these interactions (purple in Figure 3) might be considered analogous to the αC patch in pseudokinases and kinase discussed above.
Figure 3. Conformational switching in RAS: an analogy for pseudokinases?
(A,B) RAS as a prototypical G protein conformational switch. RAS is represented in cartoon form, colored according to backbone position RMSD when GTP- and GDP-bound forms are overlaid (blue is no difference, red is maximum difference: ~5Å). Regions undergoing the greatest conformational changes were originally named switch I and switch II [107] – as labelled. Bound nucleotides are shown in black sticks and marked, and Mg2+ ions are brown spheres. Active RAS is from PDB ID 4G0N [108] and inactive RAS from PDB ID 4Q21 [107].
(C) Active GTP-bound RAS binds effector proteins phospholipase Cε and RAF (through its RAS-binding domain or RBD) via switch I. Interfacial residues in RAS are colored purple, and correspond with switch I. PLCε is shown as orange ribbons (PBD ID: 2C5L [109]) and RAF-RBD as green ribbons (PDB ID: 4G0N [108]).
(D) Active GTP-bound RAS uses both switch I and switch II to bind phosphatidylinositol-3-kinase (PI3K) and Nore1A, with interfacial residues (purple) coinciding with the two switch regions just as the kinase αC patch coincides with regions of maximum change in the kinase N-lobe. PI3K is shown in pale yellow ribbons (PDB ID: 1HE8 [110]) and Nore1A is shown as pale blue ribbons (PDB ID: 3DDC [111]).
Just as stabilizing the active state of RAS (when bound to GTP) promotes its interaction with RAF effectors via switch I and switch II, we suggest that stabilizing the active-like conformation of a pseudokinase (or kinase) can promote association with pseudokinase effectors. The pseudokinase transition may be driven by small molecule binding, phosphorylation, or other changes. Conformational toggling may enhance binding of some effectors but inhibit interaction with others (as with MLKL and the executioner domain [47]). Along similar lines, one pseudokinase domain (from IRAK3) was recently observed crystallographically to form symmetric ‘head-to-head’ dimers only in the (likely ATP-bound) active-like conformation [56], engaging the αC patch of each pseudokinase domain. In such cases, interactions with effectors might be regulated by controlling the inactive-to-active toggle as in RAS. Indeed, it is interesting in this context of this analogy that several pseudokinases can bind GTP [57].
Nucleotide-Binding Properties of Pseudokinase Domains
The possibility that pseudokinases signal through conformational toggling suggests opportunities for pharmacological manipulation. The ability to target a given pseudokinase will likely depend on its nucleotide binding ability, which varies across the family. Pseudokinases were previously characterized based on their ATP- and divalent cation-binding properties [57]. It is also informative to group pseudokinases based on how many (and which) of the motifs shown in Figure 1A they lack [5]. Doing so with an eye to nucleotide-binding properties shows how kinases and pseudokinases are remarkably resilient with respect to ATP binding. ATP binding is retained in the absence of kinase activity by many pseudokinases. Recent structural studies have revealed several interesting compensatory mechanisms for retaining ATP binding in the face of degraded motifs.
Altered ATP-binding modes – making do without canonical motifs
A few pseudokinases have defects in all four ATP-binding motifs: the Gly-loop, β3 lysine, HRD motif, and DFG motif. We exclude the αC glutamate here, since its position changes with activation state and it serves largely to buttress the β3 lysine. Unsurprisingly, no pseudokinase that lacks all four motifs binds nucleotides [32, 57]. Well characterized examples include the MviN from M. tuberculosis [57] and ROP8 from T. gondii [58].
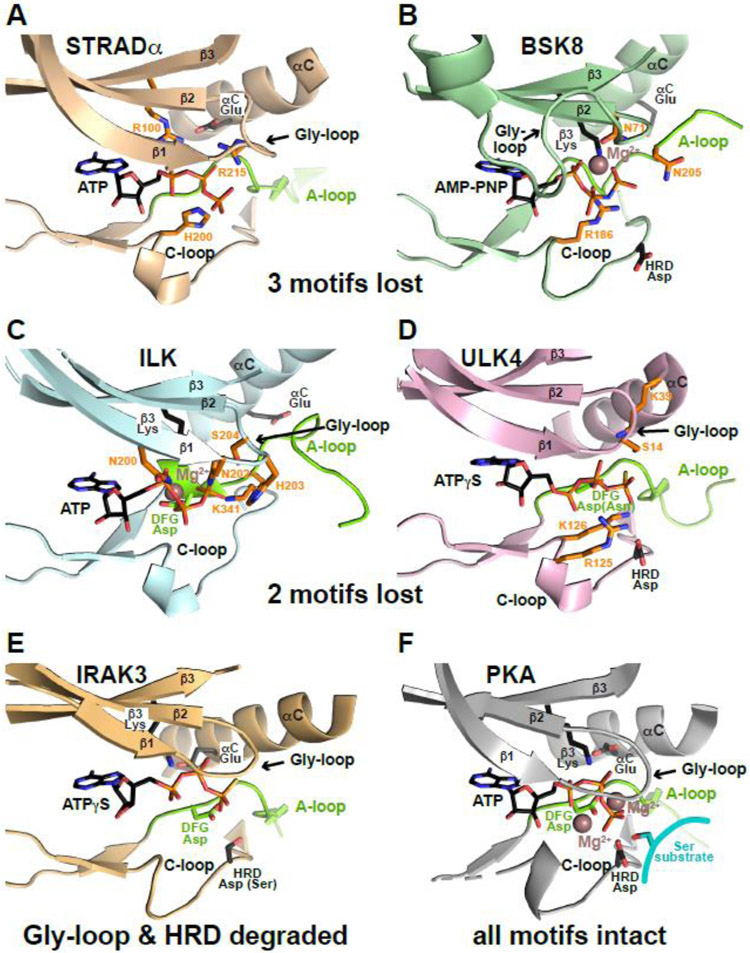
Loss of 3 motifs
Several pseudokinases with three degraded motifs also fail to bind nucleotides, as might be expected, including GCN2 and VRK3 [57]. However, a similar number retain ATP binding. including STRADα [50], MLKL [59], BSK8 [60], and EphB6 [61]. ATP binding has been visualized structurally for STRADα and BSK8. In STRADα (Figure 4A), only the Gly-loop remains intact. STRADα binds ATP without divalent cations, and the ATP phosphates interact directly with an arginine (R100, replacing the β3 lysine) and with several other ‘compensating’ basic residues including R215 (which replaces the DFG glycine) and H200 in the catalytic loop [50]. The plant (non-RD) pseudokinase BSK8 takes a different approach [60] (Figure 4B). BSK8 retains only the β3 lysine. It binds ATP in a Mg2+-dependent manner, chelating a Mg2+ ion with its much-altered Gly-loop (plus the β3 lysine). An arginine in the catalytic loop (R186) and an asparagine in the A-loop (N205) also directly engage ATP phosphates. Another plant pseudokinase, the tomato atypical receptor kinase 1 (TARK1) shows similar characteristics, standing out with BSK8 as the only two well-characterized pseudokinases that retain just one key motif, but nonetheless retain cation-dependent ATP binding [57].
Figure 4. Resilience of pseudokinase ATP binding.
ATP binding sites are shown for a series of pseudokinase domains that retain ATP binding (A-E) and for PKA (F) for comparison. Bound ATP, AMP-PNP or ATPγS is shown. The part of the activation loop shown is colored green (as is the DFG Asp when present), and preserved ATP-interacting side-chains are shown in black sticks. ‘Compensatory’ side-chains that make up for lost ATP-interacting residues by contacting the nucleotide as described in the text are colored orange. Mg2+ is shown as brown sphere in (B, C, F), but is absent in (A, D, E), which show cation-independent ATP binding.
MLKL resembles BSK8 in retaining only the β3 lysine, but its ATP binding is cation-independent [57]. Although binding has yet to be visualized structurally, human MLKL has several basic side-chains positioned to play roles similar to compensating residues in STRADα – notably a lysine (K331) that replaces the HRD aspartate and an arginine two residues away (R333) [62]. Models of EphB6, in which the DFG motif is replaced by the sequence RLG, also suggest a similar mechanism for Mg2+-independent ATP binding [61, 63].
Loss of 2 motifs
Although some pseudokinases with two missing motifs fail to bind nucleotides, such as PEAK1 (which has Gly-loop and DFG motif defects [64]), others have found new ‘tricks’ to bind ATP. For example, the ILK pseudokinase has no HRD aspartate and has a defective Gly-loop. It makes up for these deficits through an unusual set of interactions between its much-altered Gly-loop and the ATP phosphates (Figure 4C). Although ILK’s ATP-binding is Mg2+-dependent, the β3 lysine also directly engages both the β and γ phosphates of ATP, as do unique basic and other residues in the ILK catalytic and A-loops (e.g. K341) [65]. ULK4 has lost the other two motifs with β3 lysine and DFG motif defects. A lysine at the N-terminal cap of αC compensates for the lost β3 lysine in Mg2+- independent binding by this pseudokinase (Figure 4D) [66, 67], reminiscent of the compensating β2 lysine in WNK1 [14]. In addition, two ULK4-specific basic residues in the catalytic loop (R125 and K126) interact directly with ATP. Unlike the situation with WNKs, however, these compensatory changes in ULK4 fail to resurrect kinase activity.
Mg2+-independent ATP binding (albeit weak) has been reported for TRIB2 [20], which lacks the Gly-loop and DFG motif and may use a combination of compensatory mechanisms seen in the examples above. IRAK3 has also lost two motifs (with a degraded Gly-loop and HRD motif) and binds ATP relatively weakly (although strongly enough to be occupied in cells) – with a dissociation constant of several hundred micromolar [56]. A recent crystal structure (PDB: 6ZIW) showed that the IRAK3 ATP-binding site is largely unaltered (compare Figures 4E and F), with no compensating interactions, contrasting with the other cases described above and likely explaining the reduced affinity.
Loss of just 1 motif
Most pseudokinases that have only one motif degraded retain robust nucleotide binding. How they do it varies according to which motif is degraded. Losing just the HRD motif typically allows retention of ATP binding that resembles canonical binding, but with minor distortions in the binding site, as seen structurally for ErbB3 [17, 68], JAKs [69], and ROP5B from T. gondii [70]. KSR2 also retains a similar mode of ATP binding despite having the β3 lysine replaced with an arginine [19]. As reported for CASK [16], losing just the DFG motif aspartate (and Mg2+ binding) can also be overcome with basic residues located to substitute for the cation’s role in ATP binding.
Self-sabotage of ATP binding
These examples illustrate the resilience of many pseudokinases which retain ATP binding by compensating for loss of key motifs through a variety of mechanisms. Other mechanisms are also seen – including binding ATP in an inverted orientation as seen for the intriguing secreted Fam20A pseudokinase [71, 72] that allosterically activates its paralog Fam20C to phosphorylate extracellular matrix proteins.
An interesting contrasting subset of pseudokinases fail to bind ATP despite retaining most (or even all) of the motifs in Figure 1A. Interestingly, most of these are from the subfamily of receptor tyrosine kinases (RTKs) that bind WNTs – namely ROR1, ROR2, RYK, and PTK7 [57, 63]. ROR1 and ROR2 both retain all motifs listed in Figure 1A but have a slightly altered Gly-loop and the DFG phenylalanine is replaced by leucine. RYK and PTK7 both also have slightly altered Gly-loops, and their DFG motifs are replaced with the sequences DNA and ALG respectively. The plant non-RD pseudokinase BIR2 also falls into this category, failing to bind ATP despite having defects only in the Gly-loop and the HRD motif.
A common feature of these examples, also shared by the VRK3, MviN, Pragmin, and other pseudokinases, is that the potential ATP-binding site is occluded by hydrophobic side-chains. In BIR2, this arises because the altered β1/β2 loop/Gly-loop directly occludes the ATP-binding site. The WNT-binding RTK pseudokinases have an additional feature. They all resemble inactive IRK (Figure 1C), with which they share sequence similarity, in having a YxxxYY motif in their A-loops [63]. As discussed above, a key characteristic of IRK in its inactive conformation is that the distorted activation loop occludes the ATP-binding site [41]. The ROR1, ROR2, RYK, and PTK7 pseudokinase domains all faithfully retain this characteristic [63, 73], and further project a series of bulky side-chains from around the vestigial ATP-binding site to occlude it (compare Figure 5A with Figure 5B, C). IRK and its YxxxYY-motif containing relatives are unusual among bona fide kinase domains in occluding their own ATP binding site in the inactive state [41] in this way. The homology to this group of RTKs presumably explains the failure of ROR1, ROR2, RYK, and PTK7 pseudokinases to bind ATP. By contrast, ErbB3’s pseudokinase domain adopts a structure very similar to that of the inactive EGFR kinase domain [17, 68], which (like ErbB3) retains ATP binding.
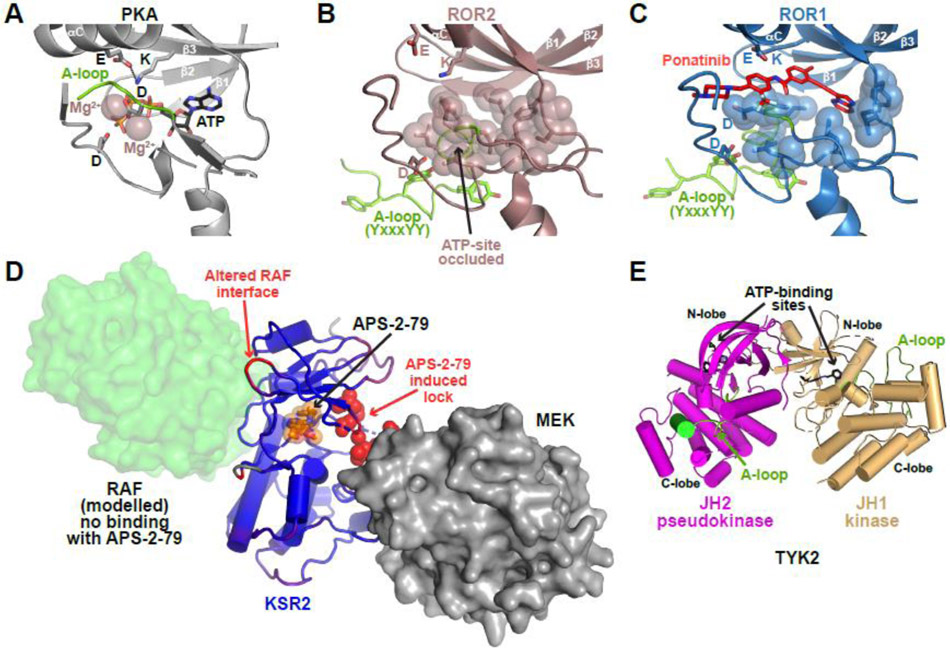
Figure 5. Modulating pseudokinases with small molecules.
(A-C) Comparison of the PKA ATP-binding site (A) with those of the ROR2 (B) and ROR1 (C), shown in the same orientation. Hydrophobic side-chains occluding the ROR2 and ROR1 ATP-binding sites are shown as sticks and transparent spheres. The activation loop section containing the YxxxYY motif that also contributes to active site occlusion is green. Ponatinib (red) binds ROR1 (but not ROR2) [63] in a cavity above the occluded (vestigial) ATP-binding site (C: PDB ID: 6TU9 [63]). Similar cavities exist in ROR2 (B: PDB ID: 3ZZW), RYK, and PTK7, that may also be targetable. ATP-binding motifs marked in Figure 1A are shown in each binding site (β3 lysine, αC glutamate, HRD and DFG aspartates).
(D) Conformational disruption of KSR2 by APS-2-79 (orange sticks/spheres). The KSR2 pseudokinase domain is blue, with APS-2-79-induced conformational changes colored red according to RMSD changes between drug- and ATP-bound states (comparing PDB IDs: 5KKR [77] and 2Y4I [19]). KSR2 forms an inhibitory complex with MEK (grey surface representation) that is stabilized by the APS-2-79-induced ‘lock’ in KSR2 (residues I809-Q814: colored red), ordered by compound binding [77], APS-2-79-induced alterations on the opposite face of KSR2 appear to inhibit its association with RAF (transparent green surface, modeled based on the B-RAF dimer in PDB ID: 1UWH [112]).
(E) N-lobe-mediated contacts between the JH2 pseudokinase domain of TYK2 (magenta) and the JH1 (bona fide) kinase domain in the same TYK2 molecule (sand), from PDB ID: 4OLI [82]. The activation loop is colored green and each ATP-binding site is occupied by an ATP-competitive TYK2 inhibitor that binds both domains [82]. Selective binding of deucravacitinib/BMS-986165 to the JH2 domain [80] stabilizes this configuration and indirectly inhibits JH1 activation as described in the text.
Importantly, the autoinhibited-like conformations of ROR1, ROR2, RYK, and PTK7 provide an opportunity for trans-phosphorylation of these pseudokinases on A-loop YxxxYY motif tyrosines (Figure 5B,C) to induce conformational changes that could resemble IRK’s inactive-to-active-like conformational switch [63]. PhosphoSitePlus [74] reports phosphorylation of these tyrosines in at least ROR2 and PTK7. Analysis of the conformational dynamics of these pseudokinase domains [63] argued that they could share IRK’s ability to undergo a phosphorylation-driven inactive-to-active-like conformational transition. We speculate that such a transition would restructure their αC patch and activation loop to promote the ability of the pseudokinase domains to bind downstream signaling effectors as a key part of their function.
Pharmacological targeting of pseudokinases with conformational disruptors and stabilizers
If pseudokinases signal through conformational toggling, compounds that selectively stabilize the active-like versus inactive-like state (or vice versa) could be valuable therapeutics. This idea is becoming increasingly important in the field of bona fide protein kinase inhibition, where ATP-competitive inhibitors with different ‘state selectivities’ differentially affect phosphotransfer-independent kinase functions [9]. Aurora A kinase provides one illustration. Beyond its phosphotransfer-dependent functions, Aurora A binds to and prevents proteolytic degradation of the transcription factor N-MYC. When the MYCN gene is amplified in neuroblastoma, this N-MYC stabilization has oncogenic consequences [75]. ATP-competitive Aurora A inhibitors that bind/stabilize Aurora A’s active conformation block phosphotransfer activity, but do not prevent it from stabilizing N-MYC [45] – and have no influence on neuroblastoma cell growth. Only compounds that disrupt or distort Aurora A’s active conformation prevent N-MYC binding [45, 76] and inhibit N-MYC-driven growth of neuroblastoma cells. One such ‘conformational disruptor’ (CD532) stabilizes an αC-out/DFG-in inactive Aurora A conformation [76] (Figure 1I). This work showed that even a relatively small deviation from the active conformation could prevent Aurora A from stabilizing overexpressed N-MYC, illustrating how inhibitor-driven conformational switching can inhibit a kinase domain’s phosphotransfer-independent functions.
The ability to control non-catalytic functions of bona fide kinases in this conformation-specific way suggests that similar small molecular pharmacological modulation of pseudokinases may be possible [32]. Dhawan et al. [77] developed small molecules (e.g. APS-2-79) that bind the ATP-binding site of KSR2’s pseudokinase domain and suppress ERK signaling, while also enhancing the efficacy of MEK inhibitors. These inhibitory compounds stabilize a unique ‘inactive’ KSR2 conformation that fails to heterodimerize with (and thus activate) RAF (Figure 5D). The unique KSR2 conformation also binds more strongly to MEK, and sequesters it away from RAF phosphorylation to antagonize signaling [77]. The conformational changes induced by APS-2-79 are quite small, but, as with Aurora A, are sufficient to bias conformation and/or dynamics to modulate key interactions. The same group has also developed compounds that bind the STRADα pseudokinase domain and are predicted to stabilize an inactive-like conformation that will fail to activate LKB1 [78].
Interestingly, work with ROR1 has shown that similar modes of targeting may also be possible for pseudokinases that do not bind ATP. Sheetz et al. [63] screened for kinase inhibitor-like molecules that bind ROR1/ROR2/RYK/PTK7 pseudokinases and identified the Abl inhibitor ponatinib as a robust ROR1 binder. A crystal structure showed how ponatinib binds the vestigial ROR1 ATP-binding site (Figure 5C), and hydrogen-deuterium exchange mass spectrometry studies revealed that ponatinib binding induces conformational changes in ROR1 that resemble a partial inactive-to-active transition that might modulate function [63]. The ROR2, RYK, and PTK7 pseudokinases all retain the narrow cavity above the occluded ATP-binding site to which ponatinib binds (Figure 5C), suggesting that they could also be targeted in this way. Along similar lines, several kinase inhibitor-like molecules were identified that bind to and stabilize the TRIB1 pseudokinase domain [51], which also does not bind ATP (with 2 motifs degraded). Stabilization was sensitive to C/EBPα degron binding in one case, suggesting conformational selectivity. Kinase inhibitor stabilizers and destabilizers of TRIB2 have also been identified [79].
The first pseudokinase-specific molecule to enter clinical development was deucravacitinib or BMS-986165 [80], which binds selectively to the ATP-binding site of the TYK2 JH2 pseudokinase domain. Deucravacitinib is currently undergoing clinical trials for psoriasis (currently Phase III) as well as other inflammatory and autoimmune diseases such as Crohn’s disease, ulcerative colitis, and systemic lupus erythematosus (currently Phase II) [32, 81]. In JAK family proteins, the JH2 pseudokinase domain interacts with the adjacent JH1 (bona fide) kinase domain to its C-terminus and inhibits it [82, 83], in the configuration shown in Figure 5E with N-lobes adjacent. This provides an interesting contrast with the activating interactions seen for ErbB3/EGFR and STRADα/LKB1 pseudokinase/kinase pairs (Figures 1F and 2E) discussed above. A recent structure of intact JAK1 [84] suggests that, by stabilizing the JH2/JH1 interaction, deucravacitinib inhibits (JH2-mediated) TYK2 dimerization and ‘release’ of the active JH1 kinase domain.
Concluding Remarks
Domains with the kinase fold have typically been designated pseudokinases based on the absence of conserved residues known to be crucial for the activity of canonical kinases like PKA. Loss of these residues can have at least three consequences: retention of kinase activity through compensating changes (as in WNK), alternative catalytic activity (see Box 1), or catalytic inactivity with alternative functions. All three scenarios are played out in the human kinome, and we have attempted here to identify some common principles in how the kinase fold is used for non-catalytic functions. An emerging theme is that pseudokinases and bona fide kinases alike show different propensities to interact with effectors depending on whether they adopt their active-like or inactive-like conformations. Toggling between these states controls catalytic activity of bona fide kinases in ways that are increasingly well understood. It is now appreciated that the same alterations can control non-catalytic activity of both bona fide kinases, as recently reviewed [9], and pseudokinases. Further analysis of structure and function will be required to establish whether this use of conformational toggling by the kinase fold is common to all pseudokinases – or whether this versatile fold has been endowed with other unexpected tricks in some areas of biology.
Figure I. Old Domain, New Tricks.
(A) PKA represents a prototypical protein kinase (PDB ID: 1ATP [113]). The kinase domain is shown as a grey cartoon. Bound ATP and peptide substrates are shown as black and cyan sticks respectively, and Mg2+ ions are shown as brown spheres.
(B) POMK transfers the γ-phosphate from ATP to the mannose unit of glycan-modified α-dystroglycan. The kinase domain is shown as a pale green cartoon. A trisaccharide substrate, colored cyan, traverses the canonical peptide binding site to place the mannose O6 hydroxyl in a phosphoacceptor position next to ATP (present as ADP in the crystal) and the catalytic base aspartate (PDB ID: 5GZ9 [114]).
(C) SelO transfers AMP from ATP onto Ser/Thr/Tyr residues of redox regulatory proteins. The ATP analog AMP-PNP is present in a ‘flipped’ orientation compared to the canonical binding mode in active protein kinases – also seen in the secreted Fam20A pseudokinase [72]. The core kinase domain is colored pale blue, and the SelO-specific sequences are light gray (PDB ID: 6EAC [24]).
(D) The NiRAN domain from nsp12 in the SARS-CoV-2 replication-transcription complex is shown (PDB ID: 6XEZ [92]). The NiRAN domain resembles SelO structurally and in its ‘flipped’ mode of nucleotide binding, with Mg2+-ADP bound in this structure.
(E) The Legionella effector protein SidJ polyglutamylates SidE family ubiquitin ligases during viral amplification. The core pseudokinase domain is shown as a pale pink cartoon, and the SidJ N- and C-terminal domains (NTD and CTD respectively) are colored light yellow (NTD) and gray (CTD). The bound yeast calmodulin (CaM), which activates SidJ, is colored light cyan (PDB ID: 7MIR [23]).
Outstanding Questions.
Do all (or most) pseudokinases undergo conformational toggling?
How is conformational toggling of pseudokinases controlled in normal signaling? By phosphorylation? By other interacting proteins? By dimerization?
Do pseudokinase domains take an active or passive role in defining their own conformation states during signaling?
If conformational toggling is a common mechanism for signaling by pseudokinase domains, what are the ‘effector’ proteins?
What other mechanisms do pseudokinase domains employ to regulate signaling?
Do some pseudokinases function as passive platforms for complex assembly, as has been suggested?
Is nucleotide hydrolysis an important factor for function of ATP-binding pseudokinases, or does nucleotide binding play only a stabilization role for these domains? Or, could these pseudokinases also resemble G proteins in this sense?
Do some pseudokinases bind other nucleotides or small molecule metabolites in the cell?
For pseudokinases that are cell surface receptors, how does the intracellular domain transmit signals?
Can all pseudokinases dysregulated in disease be targeted with small molecule conformational disruptors?
HIGHLIGHTS.
Approximately 10% of proteins (58) in the human kinome are pseudokinases, which display the protein kinase fold but lack key conserved residues or have been observed experimentally to lack protein kinase activity.
Several pseudokinases are implicated in human diseases, making them potential drug targets.
A few domains first thought to qualify as pseudokinases retain kinase activity through compensatory changes or have alternative catalytic activities.
Many pseudokinases have compensatory mutations that allow retained ATP binding, despite being inactive kinases.
Like their bona fide kinase relatives, several pseudokinases toggle conformationally between active-like and inactive-like ‘states’, controlling their ability to interact with signaling effectors and suggesting analogies to regulation by small G proteins.
Insights into conformation-dependent signaling by pseudokinases sets the stage for their pharmacological targeting with conformational disruptors, with one pseudokinase-binding small molecule (targeting TYK2) in clinical development.
Acknowledgments
The authors are grateful for funding from NIGMS through R35 GM122845 (M.A.L.) and NSF Graduate Research Fellowship DGE1122492 (J.B.S.). We thank members of the Lemmon and Ferguson laboratories for comments on the manuscript.
GLOSSARY
- αC helix:
key conserved helix within the N-lobe of protein kinase domains that adopts different positions in the active and inactive states.
- αC glutamate:
key conserved glutamate in αC of nearly all protein kinases that forms a crucial salt-bridge with the β3 lysine in the active conformation.
- Activation loop:
a loop that follows strand β8 in the protein kinase fold and regulates kinase activity by controlling access to the substrate binding sites. Generally considered to be the region between the DFG and APE motifs.
- Allostery:
molecular communication between two or more distinct sites in a protein, where modification or binding of ligand to one site alters the conformation of the other site(s).
- β3 lysine:
key conserved lysine residue within the N-lobe of active protein kinases that forms a crucial salt-bridge with the αC glutamate and contacts ATP in the active conformation.
- Conformational disruptor:
a small molecule that binds to a pseudokinase domain and distorts conformation away from the pro-signaling state.
- Conformational toggle:
switch between two conformations with different catalytic of interaction capabilities.
- DFG motif:
a conserved Asp-Phe-Gly motif found in nearly all active kinases in which the crucial aspartate residue contributes to Mg2+ coordination and ATP binding.
- Effector:
a downstream binding partner that propagates signaling. RAS effectors, for example, bind the active GTP-bound form of RAS.
- Gly-loop (GxGxxG):
key conserved motif within the N-lobe of active kinases that is crucial for ATP binding and orientation.
- HRD motif:
a conserved His-Arg-Asp motif found in active kinases, in which the crucial aspartate functions as a catalytic base in the phosphotransferase reaction.
- Human kinome:
the collection of eukaryotic protein kinases within the human genome, first described by Manning et al. [4]. Excluding atypical protein kinases and including pseudokinases, there are roughly 550 members of the human kinome.
- Motif:
short recurring sequence of amino acid residues within a protein, usually with functional significance.
- Non-RD kinase:
kinases that lack the arginine in the HRD motif, concentrated among kinases involved in regulation of innate immunity.
- Phosphotransfer:
reaction catalyzed by protein kinases.
- Protein kinase:
eukaryotic protein kinases catalyze the transfer of phosphate from ATP to Ser, Thr, or Tyr side chains on protein substrates. All kinase domains possess the quintessential bilobal fold illustrated in Figure 1.
- Pseudokinase:
a protein containing a kinase domain that lacks enzymatic activity. Roughly 60 pseudokinases are found within the human kinome.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no conflicts of interest with the content of this article.
REFERENCES
- 1.Burnett G and Kennedy EP (1954) The enzymatic phosphorylation of proteins. J. Biol. Chem 211, 969–980 [PubMed] [Google Scholar]
- 2.Pawson T and Scott JD (2005) Protein phosphorylation in signaling--50 years and counting. Trends Biochem. Sci 30, 286–290 [DOI] [PubMed] [Google Scholar]
- 3.Adams JA (2001) Kinetic and catalytic mechanisms of protein kinases. Chem. Rev 101, 2271–2290 [DOI] [PubMed] [Google Scholar]
- 4.Manning G, et al. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 5.Kwon A, et al. (2019) Tracing the origin and evolution of pseudokinases across the tree of life. Sci. Signal 12, eaav3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reiterer V, et al. (2014) Day of the dead: pseudokinases and pseudophosphatases in physiology and disease. Trends Cell Biol. 24, 489–505 [DOI] [PubMed] [Google Scholar]
- 7.Zeqiraj E and van Aalten DM (2010) Pseudokinases-remnants of evolution or key allosteric regulators? Curr. Opin. Struct. Biol 20, 772–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mace PD and Murphy JM (2021) There's more to death than life: non-catalytic functions in kinase and pseudokinase signaling. J. Biol. Chem 296, 100705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kung JE and Jura N (2016) Structural basis for the non-catalytic functions of protein kinases. Structure 24, 7–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson FH, et al. (2001) Human hypertension caused by mutations in WNK kinases. Science 293, 1107–1112 [DOI] [PubMed] [Google Scholar]
- 11.Xu B, et al. (2000) WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J. Biol. Chem 275, 16795–16801 [DOI] [PubMed] [Google Scholar]
- 12.Sengupta S, et al. (2012) Interactions with WNK (with no lysine) family members regulate oxidative stress response 1 and ion co-transporter activity. J. Biol. Chem 287, 37868–37879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bastidas AC, et al. (2012) Role of N-terminal myristylation in the structure and regulation of cAMP-dependent protein kinase. J. Mol. Biol 422, 215–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Min X, et al. (2004) Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension. Structure 12, 1303–1311 [DOI] [PubMed] [Google Scholar]
- 15.Dar AC (2013) A pickup in pseudokinase activity. Biochem. Soc. Trans 41, 987–994 [DOI] [PubMed] [Google Scholar]
- 16.Mukherjee K, et al. (2008) CASK functions as a Mg2+-independent neurexin kinase. Cell 133, 328–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi F, et al. (2010) ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc. Natl. Acad. Sci. U. S. A 107, 7692–7697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ungureanu D, et al. (2011) The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nat. Struct. Mol. Biol 18, 971–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brennan DF, et al. (2011) A Raf-induced allosteric transition of KSR stimulates phosphorylation of MEK. Nature 472, 366–369 [DOI] [PubMed] [Google Scholar]
- 20.Bailey FP, et al. (2015) The Tribbles 2 (TRB2) pseudokinase binds to ATP and autophosphorylates in a metal-independent manner. Biochem. J 467, 47–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novotny CJ, et al. (2010) Overcoming resistance to HER2 inhibitors through state-specific kinase binding. Nat. Chem. Biol 12, 923–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Black MH, et al. (2019) Bacterial pseudokinase catalyzes protein polyglutamylation to inhibit the SidE-family ubiquitin ligases. Science 364, 787–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osinski A, et al. (2021) Structural and mechanistic basis for protein glutamylation by the kinase fold. Mol. Cell 81, 4527–4539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sreelatha A, et al. (2018) Protein AMPylation by an evolutionarily conserved pseudokinase. Cell 175, 809–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boudeau J, et al. (2006) Emerging roles of pseudokinases. Trends Cell Biol. 16, 443–452 [DOI] [PubMed] [Google Scholar]
- 26.Edmund AB, et al. (2019) The pseudokinase domains of guanylyl cyclase-A and -B allosterically increase the affinity of their catalytic domains for substrate. Sci. Signal 12, eaau5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dardick C and Ronald P (2006) Plant and animal pathogen recognition receptors signal through non-RD kinases. PLoS Pathog. 2, e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones AV, et al. (2005) Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood 106, 2162–2168 [DOI] [PubMed] [Google Scholar]
- 29.Jaiswal BS, et al. (2013) Oncogenic ERBB3 mutations in human cancers. Cancer Cell 23, 603–617 [DOI] [PubMed] [Google Scholar]
- 30.Littlefield P, et al. (2014) Structural analysis of the EGFR/HER3 heterodimer reveals the molecular basis for activating HER3 mutations. Sci. Signal 7, ra114. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 31.Eyers PA, et al. (2017) Tribbles in the 21st Century: The evolving roles of Tribbles pseudokinases in biology and disease. Trends Cell Biol. 27, 284–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kung JE and Jura N (2019) Prospects for pharmacological targeting of pseudokinases. Nat. Rev. Drug Discov 18, 501–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammaren HM, et al. (2015) ATP binding to the pseudokinase domain of JAK2 is critical for pathogenic activation. Proc. Natl. Acad. Sci. U. S. A 112, 4642–4647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toms AV, et al. (2013) Structure of a pseudokinase-domain switch that controls oncogenic activation of Jak kinases. Nat. Struct. Mol. Biol 20, 1221–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeqiraj E, et al. (2009) Structure of the LKB1-STRAD-MO25 complex reveals an allosteric mechanism of kinase activation. Science 326, 1707–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor SS, et al. (2021) From structure to the dynamic regulation of a molecular switch: A journey over 3 decades. J. Biol. Chem 296, 100746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Modi V and Dunbrack RL (2022) Kincore: a web resource for structural classification of protein kinases and their inhibitors. Nucl. Acids Res 50, D654–D664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noble ME, et al. (2004) Protein kinase inhibitors: insights into drug design from structure. Science 303, 1800–1805 [DOI] [PubMed] [Google Scholar]
- 39.Huse M and Kuriyan J (2002) The conformational plasticity of protein kinases. Cell 109, 275–282 [DOI] [PubMed] [Google Scholar]
- 40.Nolen B, et al. (2004) Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell 15, 661–675 [DOI] [PubMed] [Google Scholar]
- 41.Hubbard SR (2013) The insulin receptor: both a prototypical and atypical receptor tyrosine kinase. Cold Spring Harb. Perspect. Biol 5, a008946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levinson NM, et al. (2006) A Src-like inactive conformation in the abl tyrosine kinase domain. PLoS Biol. 4, e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jura N, et al. (2011) Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol. Cell 42, 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeffrey PD, et al. (1995) Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature 376, 313–320 [DOI] [PubMed] [Google Scholar]
- 45.Richards MW, et al. (2016) Structural basis of N-Myc binding by Aurora-A and its destabilization by kinase inhibitors. Proc. Natl. Acad. Sci. U. S. A 113, 13726–13731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petrie EJ, et al. (2018) Conformational switching of the pseudokinase domain promotes human MLKL tetramerization and cell death by necroptosis. Nat. Commun 9, 2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garnish SE, et al. (2021) Conformational interconversion of MLKL and disengagement from RIPK3 precede cell death by necroptosis. Nat. Commun 12, 2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi Z, et al. (2013) Structure of the MST4 in complex with MO25 provides insights into its activation mechanism. Structure 21, 449–461 [DOI] [PubMed] [Google Scholar]
- 49.Hao Q, et al. (2014) Structural insights into regulatory mechanisms of MO25-mediated kinase activation. J. Struct. Biol 186, 224–233 [DOI] [PubMed] [Google Scholar]
- 50.Zeqiraj E, et al. (2009) ATP and MO25alpha regulate the conformational state of the STRADalpha pseudokinase and activation of the LKB1 tumour suppressor. PLoS Biol. 7, e1000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jamieson SA, et al. (2018) Substrate binding allosterically relieves autoinhibition of the pseudokinase TRIB1. Sci. Signal 11, eaau0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murphy JM, et al. (2015) Molecular mechanism of CCAAT-enhancer binding protein recruitment by the TRIB1 pseudokinase. Structure 23, 2111–2121 [DOI] [PubMed] [Google Scholar]
- 53.Bender KW, et al. (2021) Activation loop phosphorylaton of a non-RD receptor kinase initiates plant innate immune signaling. Proc. Natl. Acad. Sci. U. S. A 118, e2108242118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Halter T, et al. (2014) The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr. Biol 24, 134–143 [DOI] [PubMed] [Google Scholar]
- 55.Gasper R and Wittinghofer F (2019) The Ras switch in structural and historical perspective. Biol. Chem 401, 143–163 [DOI] [PubMed] [Google Scholar]
- 56.Lange SM, et al. (2021) Dimeric structure of the pseudokinase IRAK3 suggests an allosteric mechanism for negative regulation. Structure 29, 238–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murphy JM, et al. (2014) A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem. J 457, 323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qiu W, et al. (2009) Novel structural and regulatory features of rhoptry secretory kinases in Toxoplasma gondii. EMBO J. 28, 969–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murphy JM, et al. (2013) The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity 39, 443–453 [DOI] [PubMed] [Google Scholar]
- 60.Grütter C, et al. (2013) Structural characterization of the RLCK family member BSK8: a pseudokinase with an unprecedented architecture. J. Mol. Biol 425, 4455–4467 [DOI] [PubMed] [Google Scholar]
- 61.Liang LY, et al. (2021) The intracellular domains of the EphB6 and EphA10 receptor tyrosine pseudokinases function as dynamic signalling hubs. Biochem. J 478, 3351–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murphy JM, et al. (2014) Insights into the evolution of divergent nucleotide-binding mechanisms among pseudokinases revealed by crystal structures of human and mouse MLKL. Biochem J 457, 369–377 [DOI] [PubMed] [Google Scholar]
- 63.Sheetz JB, et al. (2020) Structural insights into pseudokinase domains of receptor tyrosine kinases. Mol. Cell 79, 390–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ha BH and Boggon TJ (2018) The crystal structure of pseudokinase PEAK1 (Sugen kinase 269) reveals an unusual catalytic cleft and a novel mode of kinase fold dimerization. J. Biol. Chem 293, 1642–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fukuda K, et al. (2009) The pseudoactive site of ILK is essential for its binding to alpha-Parvin and localization to focal adhesions. Mol. Cell 36, 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khamrui S, et al. (2020) High-resolution structure and inhibition of the schizophrenia-linked pseudokinase ULK4. J. Am. Chem. Soc 142, 33–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Preuss F, et al. (2020) Nucleotide binding, evolutionary insights, and interaction partners of the pseudokinase Unc-51-like kinase 4. Structure 28, 1184–1196 [DOI] [PubMed] [Google Scholar]
- 68.Jura N, et al. (2009) Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc. Natl. Acad. Sci. U. S. A 106, 21608–21613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bandaranayake RM, et al. (2012) Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F. Nat. Struct. Mol. Biol 19, 754–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reese ML and Boothroyd JC (2011) A conserved non-canonical motif in the pseudoactive site of the ROP5 pseudokinase domain mediates its effect on Toxoplasma virulence. J. Biol. Chem 286, 29366–29375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cui J, et al. (2015) A secretory kinase complex regulates extracellular protein phosphorylation. Elife 4, e06120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cui J, et al. (2017) Structure of Fam20A reveals a pseudokinase featuring a unique disulfide pattern and inverted ATP-binding. Elife 6, e23990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Artim SC, et al. (2012) Assessing the range of kinase autoinhibition mechanisms in the insulin receptor family. Biochem. J 448, 213–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hornbeck PV, et al. (2015) PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucl. Acids Res. 43(Database issue), D512–D520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Otto T, et al. (2009) Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell 15, 67–78 [DOI] [PubMed] [Google Scholar]
- 76.Gustafson WC, et al. (2014) Drugging MYCN through an allosteric transition in Aurora kinase A. Cancer Cell 26, 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dhawan NS, et al. (2016) Small molecule stabilization of the KSR inactive state antagonizes oncogenic Ras signalling. Nature 537, 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith RHB, et al. (2021) Type II binders targeting the "GLR-out" conformation of the pseudokinase STRADalpha. Biochemistry 60, 289–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Foulkes DM, et al. (2018) Covalent inhibitors of EGFR family protein kinases induce degradation of human Tribbles 2 (TRIB2) pseudokinase in cancer cells. Sci. Signal 11, eaat7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wrobleski ST, et al. (2019) Highly selective inhibition of tyrosine kinase 2 (TYK2) for the treatment of autoimmune diseases: Discovery of the allosteric inhibitor BMS-986165. J. Med. Chem 62, 8973–8995 [DOI] [PubMed] [Google Scholar]
- 81.Papp K, et al. (2018) Phase 2 trial of selective tyrosine kinase 2 inhibition in psoriasis. N. Engl. J. Med 379, 1313–1321 [DOI] [PubMed] [Google Scholar]
- 82.Lupardus PJ, et al. (2014) Structure of the pseudokinase-kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition. Proc. Natl. Acad. Sci. U. S. A 111, 8025–8030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shan Y, et al. (2014) Molecular basis for pseudokinase-dependent autoinhibition of JAK2 tyrosine kinase. Nat. Struct. Mol. Biol 21, 579–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Glassman CR, et al. (2022) Structure of a Janus kinase cytokine receptor complex reveals the basis for dimeric activation. Science, eabn8933: 89 10.1126/science.abn8933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koike T, et al. (2009) FAM20B is a kinase that phosphorylates xylose in the glycosaminoglycan-protein linkage region. Biochem. J 421, 157–162 [DOI] [PubMed] [Google Scholar]
- 86.Wen J, et al. (2014) Xylose phosphorylation functions as a molecular switch to regulate proteoglycan biosynthesis. Proc. Natl. Acad. Sci. U. S. A 111, 15723–11578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Worby CA, et al. (2021) The ABCs of the atypical Fam20 secretory pathway kinases. J. Biol. Chem 296, 100267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yoshida-Moriguchi T, et al. (2013) SGK196 is a glycosylation-specific O-mannose kinase required for dystroglycan function. Science 341, 896–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhu Q, et al. (2016) Structure of protein O-mannose kinase reveals a unique active site architecture. Elife 5, e22238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sheetz JB and Lemmon MA (2018) Flipping ATP to AMPlify kinase functions. Cell 175, 641–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Casey AK and Orth K (2018) Enzymes Involved in AMPylation and deAMPylation. Chem. Rev 118, 1199–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen J, et al. (2020) Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell 182, 1560–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Slanina H, et al. (2021) Coronavirus replication-transcription complex: Vital and selective NMPylation of a conserved site in nsp9 by the NiRAN-RdRp subunit. Proc. Natl. Acad. Sci. U. S. A 118, e2022310118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Park GJ, et al. (2022) The mechanism of RNA capping by SARS-CoV-2. bioRxiv, 10.1101/2022.1102.1107.479471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bhogaraju S, et al. (2019) Inhibition of bacterial ubiquitin ligases by SidJ-calmodulin catalysed glutamylation. Nature 572, 382–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sulpizio AG, et al. (2019) Glutamylation of bacterial ubiquitin ligases by a legionella pseudokinase. Trends Microbiol. 27, 967–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pan C, et al. (2021) Moonlighting functions of metabolic enzymes and metabolites in cancer. Mol. Cell 81, 3760–3774 [DOI] [PubMed] [Google Scholar]
- 98.Hosios AM, et al. (2015) Lack of evidence for PKM2 protein kinase activity. Mol. Cell 59, 850–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu R, et al. (2021) Choline kinase alpha 2 acts as a protein kinase to promote lipolysis of lipid droplets. Mol. Cell 81, 2722–2735 [DOI] [PubMed] [Google Scholar]
- 100.Xu D, et al. (2020) The gluconeogenic enzyme PCK1 phosphorylates INSIG1/2 for lipogenesis. Nature 580, 530–535 [DOI] [PubMed] [Google Scholar]
- 101.Hubbard SR (1997) Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 16, 5572–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hubbard SR, et al. (1994) Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature 372, 746–754 [DOI] [PubMed] [Google Scholar]
- 103.Yoshikawa S, et al. (2013) Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor. Oncogene 32, 27–38 [DOI] [PubMed] [Google Scholar]
- 104.Zhang X, et al. (2006) An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125, 1137–1149 [DOI] [PubMed] [Google Scholar]
- 105.Nowakowski J, et al. (2002) Structures of the cancer-related Aurora-A, FAK, and EphA2 protein kinases from nanovolume crystallography. Structure 10, 1659–1667 [DOI] [PubMed] [Google Scholar]
- 106.Record CJ, et al. (2010) Structural comparison of human mammalian ste20-like kinases. PLoS One 5, e11905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Milburn MV, et al. (1990) Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science 247, 939–945 [DOI] [PubMed] [Google Scholar]
- 108.Fetics SK, et al. (2015) Allosteric effects of the oncogenic RasQ61L mutant on Raf-RBD. Structure 23, 505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bunney TD, et al. (2006) Structural and mechanistic insights into ras association domains of phospholipase C epsilon. Mol. Cell 21, 495–507 [DOI] [PubMed] [Google Scholar]
- 110.Pacold ME, et al. (2000) Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell 103, 931–943 [DOI] [PubMed] [Google Scholar]
- 111.Stieglitz B, et al. (2008) Novel type of Ras effector interaction established between tumour suppressor NORE1A and Ras switch II. EMBO J. 27, 1995–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wan PT, et al. (2004) Cancer Genome Project. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 116, 855–867 [DOI] [PubMed] [Google Scholar]
- 113.Zheng J, et al. (1993) 2.2 A refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor. Acta Crystallogr. D. Biol. Crystallogr 49, 362–365 [DOI] [PubMed] [Google Scholar]
- 114.Nagae M, et al. (2017) 3D structural analysis of protein O-mannosyl kinase, POMK, a causative gene product of dystroglycanopathy. Genes Cells 22, 348–359 [DOI] [PubMed] [Google Scholar]