Abstract
Multiple sclerosis (MS) is a chronic autoimmune disease in the central nervous system (CNS) marked by inflammation, demyelination, and axonal loss. Currently available MS medication is limited, thereby calling for a strategy to accelerate new drug discovery. One of the strategies to discover new drugs is to utilize old drugs for new indications, an approach known as drug repurposing. Herein, we first identified 421 MS-associated SNPs from the Genome-Wide Association Study (GWAS) catalog (p-value < 5 × 10−8), and a total of 427 risk genes associated with MS using HaploReg version 4.1 under the criterion r2 > 0.8. MS risk genes were then prioritized using bioinformatics analysis to identify biological MS risk genes. The prioritization was performed based on six defined categories of functional annotations, namely missense mutation, cis-expression quantitative trait locus (cis-eQTL), molecular pathway analysis, protein-protein interaction (PPI), genes overlap with knockout mouse phenotype, and primary immunodeficiency (PID). A total of 144 biological MS risk genes were found and mapped into 194 genes within an expanded PPI network. According to the DrugBank and the Therapeutic Target Database, 27 genes within the list targeted by 68 new candidate drugs were identified. Importantly, the power of our approach is confirmed with the identification of a known approved drug (dimethyl fumarate) for MS. Based on additional data from ClinicalTrials.gov, eight drugs targeting eight distinct genes are prioritized with clinical evidence for MS disease treatment. Notably, CD80 and CD86 pathways are promising targets for MS drug repurposing. Using in silico drug repurposing, we identified belatacept as a promising MS drug candidate. Overall, this study emphasized the integration of functional genomic variants and bioinformatic-based approach that reveal important biological insights for MS and drive drug repurposing efforts for the treatment of this devastating disease.
Keywords: Autoimmune disease, Bioinformatics, Drug repurposing, Genomic variants, Multiple sclerosis
Highlights
-
•
Utilizing genomic data from GWAS catalog to provide novel biological insight for drug repurposing for multiple sclerosis.
-
•
The leveraging of genomic information contribute to drug discovery for multiple sclerosis.
-
•
Our findings suggest the plausibility of genomic variant-driven drug repurposing for multiple sclerosis.
1. Introduction
Multiple sclerosis (MS) is a chronic autoimmune disease in the central nervous system (CNS) marked by inflammation, demyelination, and axonal loss since the onset of the disease. The onset of MS usually occurs between 20 and 40 years of age and more predominantly in women [1]. MS also causes a series of other heterogeneous symptoms due to varying involvements of the motor, sensor, visual, and autonomous systems. It is characterized by optic neuritis (optic nerve inflammation), Uhthoff's phenomenon (temporary fluctuation and worsened MS symptoms with increased body temperature), and Lhermitte's sign (abnormal electrical-shock-like sensation over the spinal cord or body parts during neck flexion) [2], and tends to develop in genetically susceptible individuals who are exposed to a diversity of triggering environmental factors (e.g., Epstein-Barr virus, tobacco use, and vitamin D deficiency) [3]. The genes involved in MS have long been sought after. A number of approaches to this problem have been applied with varying degrees of success. The candidate gene approach has been in use over several decades, where potentially MS-associated genes are selected based on autoimmune MS prognosis, involving class I and II immune-response-gene-controlling human leukocyte antigen (HLA) [4].
Treatments for MS have been divided into three categories: 1) acute relapse management; 2) disease-modifying therapies; and 3) symptomatic treatments [2]. One MS treatment available and approved is dimethyl fumarate (Tecfidera) [2,5]. So far, these medications can help people with MS that have fewer and less severe relapses. However, the problem is still arising from those medications, including resistance and toxicity [6]. Under such circumstances, drug repurposing emerges as one of the solutions to identify new candidate drugs for MS disease. In addition, further investigations such as clinical validation and in vivo experiments are needed to accelerate new discoveries for the treatment of MS disease, which aim to maximize the likelihood of success during pre-clinical development and validation [7].
The concept of drug repurposing is to find new indications for existing drugs that are already available on the market [8]. The drug repurposing approach has several advantages compared to the traditional ones, such as time and cost-effectiveness [9], safety profile (drugs have previously passed clinical trials), dosage, and the toxicity of existing drugs has been vetted [10]. Genome-wide association study (GWAS) can potentially be leveraged for precision drug repurposing by applying functional annotation [11]. Several studies were applied to the risk variants from GWAS, and were able to prioritize the biological risk genes based on the functional annotations to drive drug repurposing for various diseases, including chronic hepatitis B [12], atopic dermatitis [13], asthma [14], colorectal cancer [15] and rheumatoid arthritis [16]. Of interest to this study, GWAS has revolutionized MS genetic analyses, including the MS variants. These variants have consistently implicated genes associated with immunological processes, mostly lie in regulatory rather than coding areas, and are often associated with other autoimmune diseases [17]. This study aimed to implement bioinformatics strategies to identify biological MS candidate genes through an integrated gene network. Six functional annotations (missense mutation, cis-expression quantitative trait locus (cis-eQTL), molecular pathway analysis, protein-protein interaction (PPI), overlap knockout mouse phenotype, and primary immunodeficiency (PID)) were used to find biological MS risk genes. Finally, we overlapped the biological MS risk genes with the drug databases and prioritized the candidate drugs to be repurposed for MS disease.
2. Methods
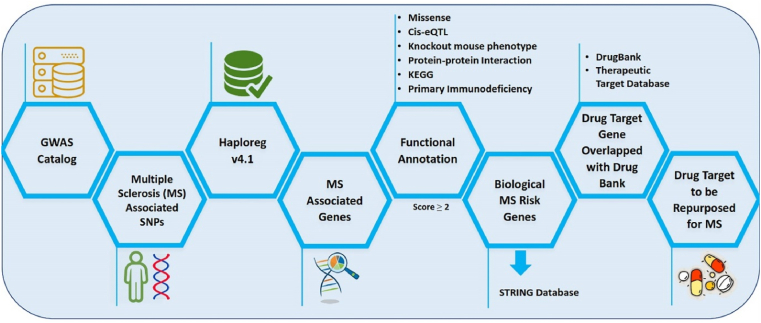
A detailed description of the study design of drug repurposing utilizing the genomic information for MS is provided in Fig. 1. MS-associated single nucleotide polymorphisms (SNPs) were obtained from the GWAS catalog under the criterion p-value < 5 x 10−8 and expanded using HaploReg (v4.1) based on the criterion of r2 ≥ 0.8 in Asian (ASN) populations. The data was retrieved from the 1000 Genome Project Phase I data [18,19]. Genes matching MS-associated SNPs are denoted as “MS-associated genes”. Then, genomic data were prioritized based on six functional annotation criteria. Every functional annotation is assigned a score of 1, and genes with a score ≥2 are defined as “biological MS risk genes”. Biological MS risk genes were used in advanced analysis using the STRING database to extend the list of candidate genes as drug-target genes. This study mapped an approved expanded list of drug-target genes in the DrugBank and the Therapeutic Target Database. The drug-target genes were checked with ClinicalTrials.gov to determine the clinical status.
Fig. 1.
Scheme of drug repurposing using genomic database for multiple sclerosis (MS).
2.1. Functional annotations of MS risk genes
Functional annotation describes a gene's biological identity by compiling the relevant biological information for a particular gene. Herein, six categories of functional annotations were used to build an assessment system representing the candidate genes most likely to be MS targets. The first category of annotation was missense or nonsense mutation according to HaploReg v4.1, which contains functional consequence annotations of the SNP database (db). HaploReg v4.1 also connected genetic variants to cis-expression quantitative trait loci (cis-eQTLs) [16]. If a gene has an MS risk SNP with cis-eQTL effect throughout the blood, the gene was assigned one point. Then, to gain an understanding of the relationship between a mutant gene and the phenotype, WebGestalt 2019 was used for functional enrichment analysis. The data source was the Mammalian Phenotype (MP) Ontology, which contains information on the mouse and other mammalian phenotypes [15]. The genes from human Ensembl ID were converted into mouse Ensembl ID using BioMart. Clusters of genes with FDR <0.05 in the enrichment analysis were considered significant. Specifically, the gene ontology (GO) biological process categories were analyzed for this stage. The result significance was set at FDR <0.05. Enrichment analysis was performed on molecular pathways using the Kyoto Encyclopedia of Genes and Genomes (KEGG). Genes enriched on the KEGG pathway (FDR <0.05) were assigned a score of 1. Primary immunodeficiency (PID) was the last annotation criterion. It refers to inborn immunity diseases that are genetic disorders associated with increased severity [13,15]. Data enrichment analysis was performed using the hypergeometric test; p < 0.05 was used in this stage as the significance criterion [13]. It is important to note that each functional annotation is assigned a score of 1, and genes with a score ≥2 are defined as “biological MS risk genes”. Biological MS risk genes were used in advanced analysis from the STRING database.
2.2. STRING database
The use of the STRING database (http://string-db.org) aimed to integrate functional interactions related to protein expressions by inputting and regulating data associated with the predicted protein-protein interactions [20,21]. The majority of protein networks in various diseases can be the targets of the diseases [22]. The biological MS risk genes were expanded using the STRING database to gain more candidate drug targets. This step emphasized that the genomic information of MS has given insight into the biological risk genes for MS.
2.3. Validation and drug discovery
The drug target gene's candidate was overlapped with drug databases such as the DrugBank (https://www.drugbank.ca/) and Therapeutic Target Database (http://db.idrblab.net/ttd/) to find the candidate drug to be repurposed for MS disease. DrugBank and Therapeutic Target Database are databases widely used to identify the drug target precisely. It also contributes to driving drug repurposing for various diseases [23]. Drug-target genes were used to analyze the database based on several criteria, such as drugs with pharmacological activity, effectiveness in humans, and approved annotations in clinical trials or drug experiments [24]. Furthermore, the identified drugs were reviewed with ClinicalTrials.gov (https://clinicaltrials.gov) to identify clinical examinations for MS or other diseases. The data visualization of drugs under clinical investigation for MS were built by using R (Chord diagram) with the circlize package (RStudio 4.0.3 program).
3. Results
The susceptibility of various MS genomic variants was retrieved from the genomic database. A variety of genomic databases can be used including GWAS databases. GWAS not only provides information on the susceptibility of diseases but also provides information on the biological insight of diseases. In this study, 420 MS-associated SNPs were obtained from the GWAS database (Table S1). The expansion was then performed using HaploReg v4.1 under the criterion r2 > 0.8, resulting in 427 MS-associated risk genes (Table S2).
3.1. Functional annotations of MS risk genes
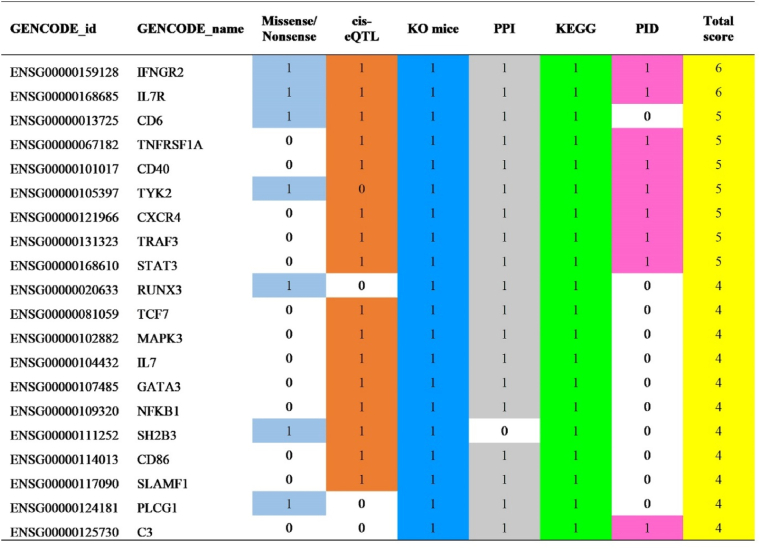
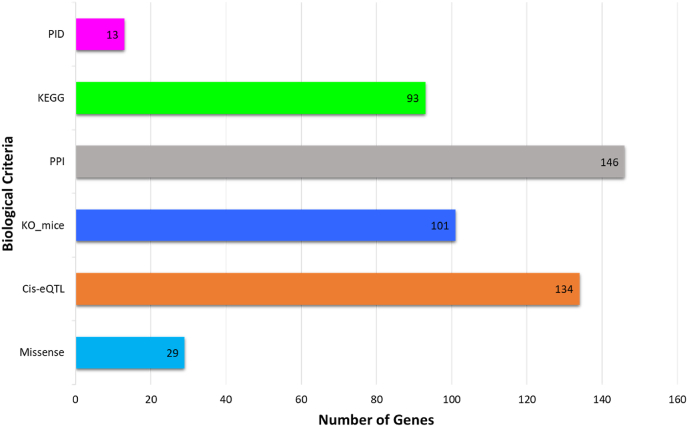
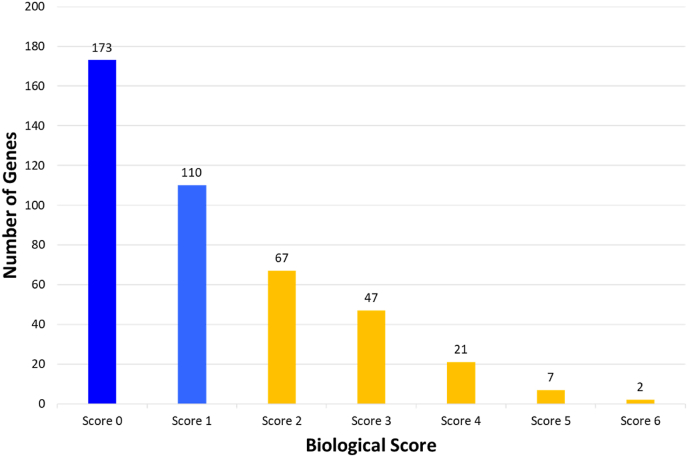
Six categories of biological functional annotations were applied to prioritize biological MS risk genes. One point was assigned to each category of functional annotation. The assessment of each of the 427 candidate genes under the six criteria was as follows: (1) genes with missense mutation MS risk variants (n = 29); (2) cis-eQTL genes (n = 134); (3) genes in knockout mouse phenotypes (n = 101); (4) engaged genes in terms of GO for evaluating PPI (n = 146); (5) genes overlapped with the KEGG pathways (n = 93); and (6) number of genes overlapped with the PID (n = 13) (Fig. 2 and Fig. 3) (Table S2). Biological scoring was conducted after data collection. There were 173 genes with a score of 0, 110 genes with a score of 1, 67 genes with a score of 2, 47 genes with a score of 3, 7 genes with a score of 5, and 2 genes with a score of 6. A total of 144 genes had a score >2 (Fig. 4). We found that interferon-gamma receptor 2 (IFNGR2) and interleukin 7 receptor (IL7R) were the top two biological MS risk genes, each with a score of 6.
Fig. 2.
Biological annotations prioritized for multiple sclerosis (MS) genes with score ≥2.
Fig. 3.
Histogram of the number of genes (y-axis) meeting each of the six biological criteria (x-axis) for drug prioritization..
Fig. 4.
Histogram of the number of genes (y-axis) meeting each of the six biological criteria (x-axis) for drug prioritization. It is shown that there were 173 genes with score 0, 110 genes with score 1, and 144 (67 + 47+21 + 7+2) genes with total score ≥2, denoted as “biological MS risk genes”.
3.2. Expansion of biological MS risk genes list
The STRING database was utilized to expand the investigation of biological MS risk genes. The expansion is based on the rationale that, the more biological MS risk genes we find, the more candidate drug targets for MS drug repurposing can be identified. We successfully obtained 194 genes on the list (Table S3). These genes were included on the list of new candidate drug genes for further analysis.
3.3. MS drug targets finding
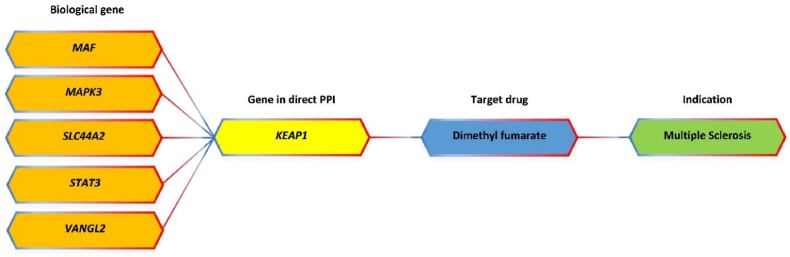
Finally, the drug target genes were prioritized based on the network analysis and the drug databases. Herein, we obtained 2904 gene pairs from the PPI network (Table S4) and 27 genes targeted by 68 new candidate drugs based on DrugBank and Therapeutic Target Database (Tabel S5). We found one drug, dimethyl fumarate that has been clinically approved for MS treatment (Fig. 5). This drug is an effective medicinal option, administered twice a day in MS medication [25,26]. This study emphasized that the biological functional annotation we applied can be validated through a known drug used in the clinic for MS disease.
Fig. 5.
Relationship between biological MS risk genes and available drugs for MS.
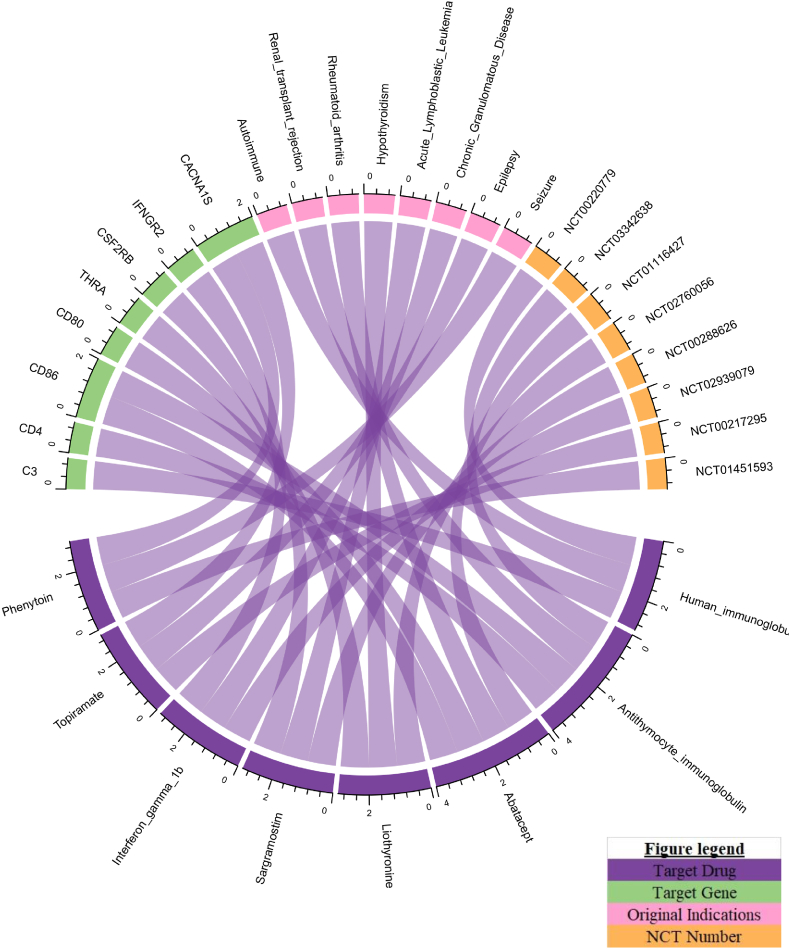
This study also found eight drug-target genes bound to 8 drugs approved for other diseases and under clinical investigation for MS, including human immunoglobulin G, antithymocyte immunoglobulin (rabbit), liothyronine, abatacept, topiramate, and phenytoin (Table S6). These drugs can potentially repurpose MS medication (Fig. 6). An example of a drug repurposed for MS is abatacept, which is approved for rheumatoid arthritis, targeting CD80 and CD86 gene pathways. This drug is currently under clinical investigation for MS in a phase II trial (NCT01116427) and has a considerable potential to be used for MS [27]. Thus, we would like to emphasize that integrating genomic variants and gene networking can potentially guide the drug repurposing for MS disease.
Fig. 6.
Relationship between biological MS genes, and drugs approved for other indications and under clinical investigation for MS.
4. Discussion
The focus of our present work is to narrow down candidate drugs for a debilitating disease, multiple sclerosis, through leveraging large bioinformatics datasets together with human genetics data. More specifically, we use genomic data and genetic mapping to guide drug repurposing for MS. In particular, this study focused on using new candidate drugs for MS by prioritizing candidate genes derived and identified from the GWAS database. Six categories of functional annotations were used to build an assessment system, in order to prioritize the MS risk genes as leads for new candidate drugs. We hypothesized that the broad strategy of genetic variant prioritization, using the functional annotations described in this study, would enable us to translate the risk genes to meaningful, actionable insights on MS. According to our analyses, we ensure the sensitivity of our study results by setting the threshold of a biological score ≥2 to screen a much higher number of genes as biological MS risk genes, and thereby as candidates for MS drug targets.
In this research, 27 drug-target gene products were found to bind to 68 drugs. In addition, 8 drug-target gene products were found to bind to 8 drugs, of which some were under clinical testing for MS, namely human immunoglobulin G (NCT00220779), anti-thymocyte immunoglobulin (rabbit) (NCT03342638), liothyronine (NCT02760056), abatacept (NCT01116427), topiramate (NCT00217295), and phenytoin (NCT01451593). From the data collected, one available drug approved for MS is dimethyl fumarate (Fig. 5). Dimethyl fumarate, also known as BG-12, was licensed as first-line therapy and oral therapy for MS in 2013. It is also known as neuroprotective and immunomodulatory [28,29]. The mechanism of action of dimethyl fumarate is to react with cysteine residues from KEAP1 (Kelch-like ECH-associated protein 1), which causes KEAP1 to be dissociated from the nuclear factor (erythroid-derived 2)-like 2 (Nrf2) pathway toward Nrf2 nuclear translocation. Nrf2 then binds antioxidant response element (ARE) and drives antioxidant target gene expression toward neuronal protection, reduces astrocyte activation, and prolongs cell life [30].
We identified eight promising targets overlapping with drugs that could potentially be repurposed to treat MS. These include C3, CD4, CD86, THRA, CSF2RB, CD80, IFNGR2, and CACNA1S. Among them, we highly proposed CD80 and CD86 as potential targets for MS, since these targets are closely related to IL7R as biological MS risk genes with high functional annotation scores (Fig. 6). The CD80/CD86 pathway is essential for controlling T cell activation and preserving immunological tolerance to self-antigens. The findings of functional and genome-wide investigations demonstrate that genes encode molecules that fit in. This pathway may increase the likelihood of developing autoimmune illnesses and may be viewed as a potential MS candidate gene [[31], [32], [33]]. In addition, we identified CD80- and CD86- targeting drugs, including anti-thymocyte immunoglobulin (rabbit), abatacept, and belatacept. Among these drugs, in fact, anti-thymocyte immunoglobulin (rabbit) (NCT03342638) and abatacept (NCT01116427) are currently under clinical investigation for MS. Therefore, from this perspective, targeting CD80 and CD86 might become novel therapeutic options for MS therapy. Further clinical evidence generation would be needed to validate these targets.
Combining results from GWAS and bioinformatic, gene-level annotation of human genetic variants is a powerful approach to identify candidate new drugs for MS. However, it is important to consider that this approach is not without limitations, such as not all the identified target genes can either be targeted and/or demonstrate the pharmacological activity with the desired profile for use in the clinic. The genes identified in this manner would therefore miss the theoretical druggability for the particular disease. Further investigation is thus required to determine the effect of the candidate drugs on the clinic. Hence, we suggest that the use of current findings with subsequent functional studies to ascertain the role of drug target genes discovered in this study. Preclinical and clinical validation is necessary and important to ensure whether our drug candidates produced the desired interaction (intended from the study), any undesired side effects, or suboptimal effects.
5. Conclusions
Our study utilizes MS functional genomic variants to open up additional avenues for the repurposing of existing drugs from other therapeutic classes and disease areas. Herein, we identify CD80 and CD86 as potential targets for MS treatment. The involvement of these genes with MS might be highly significant, thereby requiring further examination. Through targeting CD80 and CD86, belatacept could be a promising therapy option for MS therapy. However, additional studies from animal models and clinical trials are needed to confirm the biological mechanisms of the drug for new diseases. Overall, we combined the drug repurposing approach with integrated bioinformatics methodology to identify drugs with new indications for MS. Finally, this study emphasizes the vast potential of utilizing functional genomic variants as a basis to drive drug repurposing for MS disease.
Author's contribution
A.R.A., L.M.I., and W.A. conceived and designed the study. A.R.A. and W.A. performed the computational analysis. A.R.A. wrote the manuscript. A.R.A., L.M.I., W.A., D.A.P., A.B. and R.C. revised the manuscript. L.M.I and W.A. supervised and coordinated this study. All authors have read and approved the manuscript and made significant contributions to this study.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bbrep.2022.101337.
Contributor Information
Lalu Muhammad Irham, Email: lalu.irham@pharm.uad.ac.id.
Wirawan Adikusuma, Email: adikusuma28@gmail.com.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Data availability
We used the databases in the current study which were retrieved from GWAS (https://www.ebi.ac.uk/gwas/); ClinicalTrials.gov (https://clinicaltrials.gov/); Drug bank (https://www.drugbank.ca/); HaploReg Version 4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php); STRING database (http://string-db.org/); Therapeutic Targets Database (http://db.idrblab.net/ttd/).
References
- 1.Vidal-Jordana A., Montalban X. Multiple sclerosis: epidemiologic, clinical, and therapeutic aspects, neuroimaging clin. N. Am. 2017;27:195–204. doi: 10.1016/j.nic.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Doshi A., Chataway J. Multiple sclerosis, a treatable disease. Clin. Med. 2016;16:s53–s59. doi: 10.7861/clinmedicine.16-6-s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Handel A.E., Giovannoni G., Ebers G.C., Ramagopalan S.V. Environmental factors and their timing in adult-onset multiple sclerosis. Nat. Rev. Neurol. 2010;6:156–166. doi: 10.1038/nrneurol.2010.1. [DOI] [PubMed] [Google Scholar]
- 4.Howard J., Trevick S., Younger D.S. Epidemiology of multiple sclerosis, neurol. Clin. 2016;34:919–939. doi: 10.1016/j.ncl.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 5.Axisa P.P., Hafler D.A. Multiple sclerosis: genetics, biomarkers, treatments. Curr. Opin. Neurol. 2016;29:345–353. doi: 10.1097/WCO.0000000000000319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Editorial: challenges in developing new multiple sclerosis therapies. Racke M.K., editor. Ther. Adv. Neurol. Disord. 2008;1:63–65. doi: 10.1177/1756285608095831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vesterinen H.M., Connick P., Irvine C.M.J., Sena E.S., Egan K.J., Carmichael G.G., Tariq A., Pavitt S., Chataway J., Macleod M.R., Chandran S. Drug repurposing: a systematic approach to evaluate candidate oral neuroprotective interventions for secondary progressive multiple sclerosis, PLoS One. 2015;10 doi: 10.1371/journal.pone.0117705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta S.C., Sung B., Prasad S., Webb L.J., Aggarwal B.B. Cancer drug discovery by repurposing: teaching new tricks to old dogs. Trends Pharmacol. Sci. 2013;34:508–517. doi: 10.1016/j.tips.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Zhao M., Yang C.C. Drug repositioning to accelerate drug development using social media data: computational study on Parkinson disease. J. Med. Internet Res. 2018;20 doi: 10.2196/jmir.9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashburn T.T., Thor K.B. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 11.Reay W.R., Cairns M.J. Advancing the use of genome-wide association studies for drug repurposing. Nat. Rev. Genet. 2021;22:658–671. doi: 10.1038/s41576-021-00387-z. [DOI] [PubMed] [Google Scholar]
- 12.Irham L.M., Adikusuma W., Perwitasari D.A., Dania H., Maliza R., Faridah I.N., Santri I.N., Phiri Y.V.A., Chong R. The use of genomic variants to drive drug repurposing for chronic hepatitis B., Biochem. Biophys. Reports. 2022;31:101307. doi: 10.1016/j.bbrep.2022.101307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adikusuma W., Irham L.M., Chou W.H., Wong H.S.C., Mugiyanto E., Ting J., Perwitasari D.A., Chang W.P., Chang W.C. Drug repurposing for atopic dermatitis by integration of gene networking and genomic information, front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.724277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adikusuma W., Chou W.H., Lin M.R., Ting J., Irham L.M., Perwitasari D.A., Chang W.P., Chang W.C. Identification of druggable genes for asthma by integrated genomic network analysis. Biomedicines. 2022;10 doi: 10.3390/biomedicines10010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irham L.M., Wong H.S.C., Chou W.H., Adikusuma W., Mugiyanto E., Huang W.C., Chang W.C. Integration of genetic variants and gene network for drug repurposing in colorectal cancer. Pharmacol. Res. 2020;161 doi: 10.1016/j.phrs.2020.105203. [DOI] [PubMed] [Google Scholar]
- 16.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., Graham R.R., Manoharan A., Ortmann W., Bhangale T., Denny J.C., Carroll R.J., Eyler A.E., Greenberg J.D., Kremer J.M., Pappas D.A., Jiang L., Yin J., Ye L., Su D.F., Yang J., Xie G., Keystone E., Westra H.J., Esko T., Metspalu A., Zhou X., Gupta N., Mirel D., Stahl E.A., Diogo D., Cui J., Liao K., Guo M.H., Myouzen K., Kawaguchi T., Coenen M.J.H., Van Riel P.L.C.M., Van De Laar M.A.F.J., Guchelaar H.J., Huizinga T.W.J., Dieudé P., Mariette X., Bridges S.L., Zhernakova A., Toes R.E.M., Tak P.P., Miceli-Richard C., Bang S.Y., Lee H.S., Martin J., Gonzalez-Gay M.A., Rodriguez-Rodriguez L., Rantapää-Dahlqvist S., Ärlestig L., Choi H.K., Kamatani Y., Galan P., Lathrop M., Eyre S., Bowes J., Barton A., De Vries N., Moreland L.W., Criswell L.A., Karlson E.W., Taniguchi A., Yamada R., Kubo M., Liu J.S., Bae S.C., Worthington J., Padyukov L., Klareskog L., Gregersen P.K., Raychaudhuri S., Stranger B.E., De Jager P.L., Franke L., Visscher P.M., Brown M.A., Yamanaka H., Mimori T., Takahashi A., Xu H., Behrens T.W., Siminovitch K.A., Momohara S., Matsuda F., Yamamoto K., Plenge R.M. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawcer S., Franklin R.J.M., Ban M. Multiple sclerosis genetics. Lancet Neurol. 2014;13:700–709. doi: 10.1016/S1474-4422(14)70041-9. [DOI] [PubMed] [Google Scholar]
- 18.Devuyst O. The 1000 genomes project: welcome to a new world, Perit. Dial. Bar Int. 2015;35:676–677. doi: 10.3747/pdi.2015.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward L.D., Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44:D877–D881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., Jensen L.J., Von Mering C. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M., Chen H., Yin P., Song J., Jiang F., Tang Z., Fan X., Xu C., Wang Y., Xue Y., Han B., Wang H., Li G., Zhong D. Identification and clinical validation of key extracellular proteins as the potential biomarkers in relapsing-remitting multiple sclerosis, front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.753929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruffner H., Bauer A., Bouwmeester T. Human protein-protein interaction networks and the value for drug discovery, Drug Discov. Today Off. 2007;12:709–716. doi: 10.1016/j.drudis.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 23.Masoudi-Sobhanzadeh Y., Omidi Y., Amanlou M., Masoudi-Nejad A. Drug databases and their contributions to drug repurposing. Genomics. 2020;112:1087–1095. doi: 10.1016/j.ygeno.2019.06.021. [DOI] [PubMed] [Google Scholar]
- 24.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., Assempour N., Iynkkaran I., Liu Y., MacIejewski A., Gale N., Wilson A., Chin L., Cummings R., Le Di, Pon A., Knox C., Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papadopoulou A., D'Souza M., Kappos L., Yaldizli O. Dimethyl fumarate for multiple sclerosis. Expet Opin. Invest. Drugs. 2010;19:1603–1612. doi: 10.1517/13543784.2010.534778. [DOI] [PubMed] [Google Scholar]
- 26.Blair H.A. Dimethyl fumarate: a review in relapsing-remitting MS, Drugs. 2019;79:1965–1976. doi: 10.1007/s40265-019-01229-3. [DOI] [PubMed] [Google Scholar]
- 27.Khoury S.J., Rochon J., Ding L., Byron M., Ryker K., Tosta P., Gao W., Freedman M.S., Arnold D.L., Sayre P.H., Smilek D.E. ACCLAIM: a randomized trial of abatacept (CTLA4-Ig) for relapsing-remitting multiple sclerosis, Mult. Scler. 2017;23:686–695. doi: 10.1177/1352458516662727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gross C.C., Schulte-Mecklenbeck A., Klinsing S., Posevitz-Fejfár A., Wiendl H., Klotz L. Dimethyl fumarate treatment alters circulating T helper cell subsets in multiple sclerosis. Neurol. Neuroimmunol. NeuroInflammation. 2016;3 doi: 10.1212/NXI.0000000000000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linker R.A., Lee D.H., Ryan S., Van Dam A.M., Conrad R., Bista P., Zeng W., Hronowsky X., Buko A., Chollate S., Ellrichmann G., Brück W., Dawson K., Goelz S., Wiese S., Scannevin R.H., Lukashev M., Gold R. Fumaric acid esters exert neuroprotective effects in neuroinflammation via activation of the Nrf2 antioxidant pathway. Brain. 2011;134:678–692. doi: 10.1093/brain/awq386. [DOI] [PubMed] [Google Scholar]
- 30.Montes Diaz G., Hupperts R., Fraussen J., Somers V. Dimethyl fumarate treatment in multiple sclerosis: recent advances in clinical and immunological studies. Autoimmun. Rev. 2018;17:1240–1250. doi: 10.1016/j.autrev.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 31.Menezes S.M., Decanine D., Brassat D., Khouri R., Schnitman S.V., Kruschewsky R., López G., Alvarez C., Talledo M., Gotuzzo E., Vandamme A.M., Galvão-Castro B., Liblau R., Weyenbergh J.V. CD80+ and CD86+ B cells as biomarkers and possible therapeutic targets in HTLV-1 associated myelopathy/tropical spastic paraparesis and multiple sclerosis. J. Neuroinflammation. 2014;11 doi: 10.1186/1742-2094-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boylan M.T., Crockard A.D., McDonnell G.V., Armstrong M.A., Hawkins S.A. CD80 (B7-1) and CD86 (B7-2) expression in multiple sclerosis patients: clinical subtype specific variation in peripheral monocytes and B cells and lack of modulation by high dose methylprednisolone. J. Neurol. Sci. 1999;167:79–89. doi: 10.1016/S0022-510X(99)00132-X. [DOI] [PubMed] [Google Scholar]
- 33.Wagner M., Sobczyński M., Karabon L., Bilińska M., Pokryszko-Dragan A., Pawlak-Adamska E., Cyrul M., Kuśnierczyk P., Jasek M. Polymorphisms in CD28, CTLA-4, CD80 and CD86 genes may influence the risk of multiple sclerosis and its age of onset. J. Neuroimmunol. 2015;288:79–86. doi: 10.1016/j.jneuroim.2015.09.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We used the databases in the current study which were retrieved from GWAS (https://www.ebi.ac.uk/gwas/); ClinicalTrials.gov (https://clinicaltrials.gov/); Drug bank (https://www.drugbank.ca/); HaploReg Version 4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php); STRING database (http://string-db.org/); Therapeutic Targets Database (http://db.idrblab.net/ttd/).