Summary
Understanding the evolutionary history of the highly diverse ray-finned fishes has been challenging, and the development of more universal primers for phylogenetic analyses may help overcoming these challenges. We developed FishPIE, a nested PCR primer set of 82 phylogenetically informative exon markers, and tested it on 203 species from 31 orders of Actinopterygii. We combined orthologous sequences of the FishPIE markers obtained from published genomes and transcriptomes and constructed the phylogeny of 710 species belonging to 190 families and 60 orders. The resulting phylogenies had topologies comparable to previous phylogenomic studies. We demonstrated that the FishPIE markers could address phylogenetic questions across broad taxonomic levels. By incorporating the newly sequenced taxa, we were able to shed new light on the phylogeny of the highly diverse Cypriniformes. Thus, FishPIE holds great promise for generating genetic data for broad taxonomic groups and accelerating our understanding of the fish tree of life.
Subject areas: Evolutionary biology, Evolutionary processes, Phylogenetics
Graphical abstract

Highlights
-
•
FishPIE is a nested PCR primer set of 82 markers for fish phylogenetic analysis
-
•
The markers can be broadly applied to all orders of ray-finned fishes
-
•
Their phylogenetic performance is comparable to that of genomic analyses
Evolutionary biology; Evolutionary processes; Phylogenetics
Introduction
Elucidating the phylogenetic relationships among the major lineages of ray-finned fishes (class Actinopterygii) is fundamental to explaining the tremendous diversity of this ancestral group of vertebrates. With over 34,000 species recorded (Fricke et al., 2021), the group dominates the Vertebrata and has evolved extraordinary morphological and ecological diversity. Many approaches have been used for phylogenetic analyses of the group. Numerous studies based on Sanger sequencing of a few to dozens of genetic markers (e.g., Dettaï and Lecointre, 2004, 2005; Chen et al., 2007; Betancur-R et al., 2013, 2017; Near et al., 2013; Smith et al., 2016) and recent studies based on genome-scale approaches (e.g., Alfaro et al., 2018; Hughes et al., 2018) have provided valuable information and successfully resolved many phylogenetic questions in fishes. However, some relationships, such as those within Cypriniformes and Ovalentaria, remain recalcitrant despite using hundreds to thousands of markers, as different studies yielded disparate topologies (c.f. Alfaro et al., 2018; Hughes et al., 2018). To resolve these difficult phylogenetic questions and to further improve our understanding of actinopterygian phylogeny, it may be important to sample more exhaustively instead of simply sequencing even more markers. Genomic approaches often present challenges to taxonomically extensive investigation. Genome sequencing is expensive, and genome assembly demands extensive computational resources. The transcriptome sequencing approach (e.g. Hughes et al., 2018) is less expensive and less demanding for computational resources, but it is not suitable for ethanol-preserved or dried samples. The sequence capture approach has fewer requirements on sample quality and is applicable to even highly degraded DNA. However, the sequence capture efficiency is strongly influenced by the probe to target DNA sequence divergence (Bragg et al., 2016; Paijmans et al., 2016). Considering that the Actinopterygii is a highly diverse and ancient group, designing universal capture probes and efficiently capturing sequences across the divergent lineages can be challenging. Multiple probe sets suitable for different lineages may be needed for studies with broad taxonomic coverage (e.g. Hughes et al., 2021), significantly increasing the budget for probe synthesis. More importantly, many studies do not require hundreds of markers (Rokas et al., 2003; Spinks et al., 2009). A solution to these problems is to recuperate the traditional PCR-based approach, which can be conducted in almost every molecular lab. The major shortcomings of this method are that performing PCR for many loci and samples is labor-intensive (due to often unpredictable PCR success rates), universal PCR primers are insufficient, and standard Sanger sequencing is costly. However, these weaknesses can be largely overcome by (1) leveraging the many publicly available fish genomes and transcriptomes to design universal PCR primers with high PCR success rates, (2) carefully selecting phylogenetically informative markers, and (3) sequencing the PCR products by Sanger sequencing or by next-generation sequencing only when many makers are necessary for resolving specific questions.
Here, we report a set of carefully selected informative nuclear protein-coding loci (NPCL), called FishPIE (stands for Fish Phylogenetically Informative Exons), to bridge the gap between the traditional markers sequencing and the more advanced high-throughput methods for fish phylogenetic analyses. From 131 fish genomes, we identified orthologous NPCL, which were further screened based on (1) sequence length, (2) PCR primer specificity and PCR amplification rate across 203 species from 67 families in 31 orders of ray-finned fishes, and (3) phylogenetic signal. Combined with orthologs identified from the genomes and transcriptomes of other fish species, our phylogenetic analyses yielded comparable resolutions to phylogenomic studies using over a thousand markers. Therefore, FishPIE would be an excellent alternative approach for phylogenetic projects for various fish clades.
Results and discussion
Summary of FishPIE
A total of 2093 orthologs were identified from the genomes of 131 species (Table S1), of which 102 loci were selected for primer design, and 82 of them exhibited a high PCR success rate (>75%) in 203 species (Tables S2 and S3). Among the 203 samples tested, 124 were preserved in 95% ethanol, 72 were frozen tissues, and seven were dried fin clips, whereas the average number of successfully amplified markers were 64.8, 54.3, and 69.4, respectively. We managed to obtain about 50% of the markers from specimens that had been frozen or ethanol-preserved for over 5 years and fin clips that have been dried for six months. This demonstrates that our method is applicable to preserved specimens and old specimens. The average PCR product sizes ranged from 544 to 1143 bp after trimming primer sequences and ambiguous regions (Table S3). We obtained orthologous sequences of FishPIE from RNA-seq data (268 species) and genome sequencing data (151 species) from public databases (Table S2). The trimmed alignments of the combined dataset totaled 58,386 bp, with GC content averaged at 47.66% (Table S4). The markers were functionally diverse; 80 of the markers could be annotated to 74 gene families in the PANTHER database (Table S3). Seven of the markers have been previously identified and often used for fish phylogenetic analyses, namely ENC1 (Li et al., 2007), PLAGL2 (Li et al., 2007), PTCHD4 (Broughton et al., 2013), RAG1 (Sullivan et al., 2006), RAG2 (Sullivan et al., 2006), RNF213 (Li et al., 2009), and VCPIP1 (Betancur-R et al., 2013). By examining the gene trees, we did not find issues with paralogs owing to whole-genome duplication in fish.
Broad taxonomic application
The FishPIE primers could be applied to diverse taxonomic groups. PCR success rates were higher in species from the “crown” cohorts Otomorpha (81.1%, N = 104) and Euteleosteomorpha (71.4%, N = 85) than those from Elopomorpha (46.1%, N = 5), Osteoglossomorpha (47.9%, N = 7), and outgroups (47.0%) (Figures S1 and S2). The low success rate in Elopomorpha and Osteoglossomopha may be due to severe DNA degradation in some of the samples, particularly those sourced from distant-water fishery and not well preserved on the cruise. Nonetheless, the former two cohorts cover over 96% of fish species (Fricke et al., 2021). All FishPIE could be retrieved from the genome or transcriptome data obtained from NCBI SRA or TSA. We observed no marked difference in retrieval rate among cohorts (53.4% to 69.3%), except that slightly fewer markers could be retrieved from the outgroups (45.1%) (Figure S1 and Table S2). Therefore, FishPIE can be used in conjunction with other sequencing approaches to increase taxon coverage.
Evolutionary rates
Evolutionary rate (using genetic distance as proxy) is one of the important attributes to consider when selecting markers for phylogenetic analyses. We estimate the substitution rate of each marker based on a chronogram calibrated using 14 calibration points. Figure 1 shows the box plot of substitution rates of the FishPIE markers, whereas detailed information is shown in Table S4. The mean substitution rates widely ranged from 0.578 × 10−4 to 3.241 × 10−4 substitution per lineage per million years, with an average of 1.471 × 10−4. The rates are comparable to those of the seven previously identified markers, ranging from 0.591 × 10−4 to 2.394 × 10−4. FishPIE markers can thus provide substantial information for resolving phylogenies at different evolutionary timescales, and researchers can select a subset of markers with suitable substitution rates for their questions.
Figure 1.
Boxplots of the substitution rates the 82 FishPIE markers, showing the minimum, Q1, Q2, Q3 and maximum, from left to right. See also Figures S1 and S2 and Table S1. Reference genomes used for initial ortholog identification, related to Figure 1, Table S2. Marker occupancy matrix showing taxonomic classification, source and preservation method (if applicable) of sample analyzed in this study, related to Figure 1, Table S3. Details of FishPIE markers including primer information, annealing temperatures, sizes of PCR products, and gene annotation, related to Figure 1, Table S4. FishPIE marker details showing alignment length, GC content, parsimony informative sites, the proportion of parsimony informative sites, and mean substitution rates in the full dataset (combining sequences generated by this study and those obtained from publicly available transcriptomic and genomic data). Related to Figure 1
Informativeness for phylogenetic reconstruction
To examine if enough information had been collected to resolve the target phylogeny, we constructed multiple phylogenetic trees by concatenating FishPIE markers one by one in ascending order of tree distances (normalized RF index), which are calculated between the respective gene trees and the tree constructed by concatenating all markers. The plots showed that the decrease in tree distance leveled off with 40 markers (Figure 2). The number of markers corroborates with Capella-Gutierrez et al. in suggesting that not all relationships require a genome-scale dataset to resolve (Capella-Gutierrez et al., 2014). As our dataset shows high phylogenetic accuracy in the different taxonomic levels analyzed (see below), the convergence of tree topology at about 40 markers implies that the 82 FishPIE markers should be more than adequate for resolving a considerable portion of fish phylogeny.
Figure 2.
Plot of tree distances (normalized RF index) of ML trees constructed by concatenating FishPIE markers one by one against a reference tree, showing a plateau at 40 markers. See also Figure S3
To further demonstrate that the phylogenetic informativeness of FishPIE is comparable to that of genome-scale analysis, we compared the ML tree constructed based on the nucleotide sequences of 82 FishPIE markers and 1,429 NPCL (identified by Hughes et al., 2018 as well as our analyses) obtained from 479 publicly available genomes and transcriptomes. The resulting trees were similar, with a normalized RF distance of 0.296. Most of the conflicts pertained to the relationships of closely-related species, particularly those of rapid radiation (e.g. Cichlidae, see Figure S3).
Overall phylogenetic performance
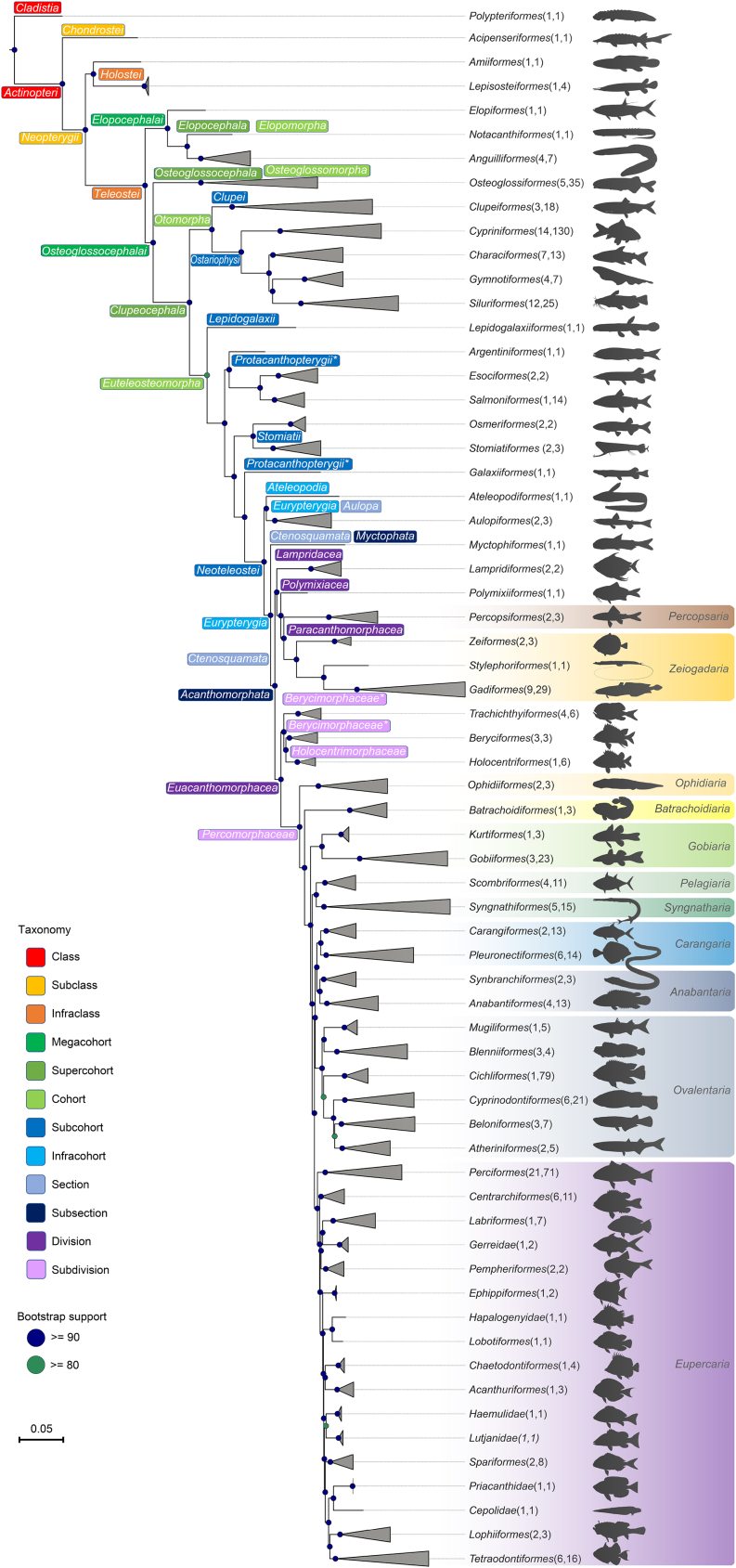
The concatenated gene trees and species trees were generally well supported (Figures S4–S8). The mean ultrafast bootstrap (BP) supports of the nucleotide and peptide maximum likelihood (ML) trees were 96.26% and 97.00%, respectively. The mean posterior probability (PP) of both Bayesian inference (BI) trees was 0.96, and the mean PP of the ASTRAL tree was 0.87. The tree topologies were highly similar, except for the ASTRAL tree, with some conflicting nodes (to be discussed below). The tree distances between the ASTRAL tree and the others ranged from 0.221 to 0.229, whereas the tree distances ranged from 0.047 to 0.105, among others (Table S5). Here, we presented an order-level backbone tree based on ML analysis of peptide data (Figure 3), as it had the shortest tree distances from others at this level (data not shown), whereas the other trees were shown in Figures S9–S13.
Figure 3.
Order-level backbone tree of ray-fin fish based on ML analysis of peptide sequences
Numbers in brackets following order names indicate the number of families and number of species analyzed. See also Figures S4–S18 and Tables S5 and S6.
Our tree topologies were largely coherent with those of genomic-scale studies at the order level (Alfaro et al., 2018; Hughes et al., 2018), except at a few recalcitrant nodes. One of those concerns the relationships within Otophysa. Our peptide BI and ML trees depicted a sister relationship between Gymnotiformes and Siluriformes, the same as the peptide ML tree based on 1,105 exon markers (Hughes et al., 2018), but our nucleotide BI, ML, and ASTRAL trees showed that Characiformes and Siluriformes were sisters, more consistent with Betancur-R et al. based on one mitochondrial and 20 nuclear loci (Betancur -R et al., 2013). Our phylogenies consistently showed that Galaxiiformes was sister to Neoteleostei, which was not recovered in two previous large-scale studies (Betancur -R et al., 2017; Hughes et al., 2018) though the gene genealogy interrogation analysis of the former study did recover such alternative topology. The relationships among the basal orders of Acanthomorpha were congruent and well supported in all our phylogenetic trees except the coalescence tree but were slightly different from previous large-scale studies (Betancur-R et al., 2017; Hughes et al., 2018) regarding the sister group of Acanthopterygii and of Percomorpha. Besides, all our phylogenies showed that Gobaria was more distantly related within percomorphs than Syngnatharia + Pelagaria, which is consistent with the phylogeny of Alfaro et al., whereas it was the opposite in Hughes et al. and Betancur-R et al. (Betancur-R et al., 2017; Alfaro et al., 2018; Hughes et al., 2018). Discordance was also observed in the relationship of the ovalentarian orders, where each tree depicted a unique pattern, though with weak statistical supports. These relationships were also poorly resolved in previous phylogenomic studies using over 1100 markers (Alfaro et al., 2018; Hughes et al., 2018). These discrepancies among studies and poorly resolved relationships may be due to differential and/or inadequate taxon coverage. Thus, future research may require more extensive taxon sampling instead of merely adding markers in addressing these phylogenetic uncertainties.
To further quantify the phylogenetic performance of FishPIE, we calculated for each gene tree the phylogenetic accuracy index defined as the average of ML BP supports for the widely accepted monophyly of 16 genera, families, and orders (see Table S6). If the monophyly of the clade was not recovered in the gene tree, the BP support would be considered a negative value. All these clades were well supported in the concatenated gene tree and ASTRAL species tree except for Euteleosteomorpha (Figures S4–S8). The phylogenetic performance of FishPIE was satisfactory at all three levels. The average accuracy index was highest at the genus level (74.71), followed by the family level (69.51), whereas the order level had the lowest score (55.68) (Table S6). Among the markers, ash1l, prdm11, and LOC100333177 showed full support for all the clades available for analysis. Another 19 markers, including the commonly used RAG2 marker, had an overall average accuracy index of over 90. Therefore, these markers can be useful for broad-spectrum phylogenetic analyses. Some markers may be suitable for elucidating certain taxonomic levels only. For instance, stox2a′s accuracy index was 100 at the order level but was less than 70 in the other two levels. Similarly, trip11, pwwp2b, and socs6 might be more suitable for phylogenetic analyses at or above the family level, whereas ppl, mycbp2, and elfn1 might be more informative for examining relationships at the family level or below.
Implications for phylogeny of cypriniformes
To examine FishPIE’s resolution power more closely at the family to genus levels, we strategically increased the sampling density of Cypriniformes, particularly for the suborders Cobitoidei and Cyprinitoidei. As the phylogenetic relationships in these taxa have been contentious (Tang et al., 2006; Šlechtová et al., 2007; Mayden et al., 2009; Tao et al., 2013, 2019; Stout et al., 2016), our analyses provide new implications for such discussions. The Cypriniformes section of each tree is shown in Figures S14–S18, with suborder, subfamily, and tribe classification annotated. The taxonomic classification herein follows Tan and Armbruster (2018).
In Cobitoidei (loach fishes), all nodes received full support in the nucleotide BI tree, whereas only 1–4 out of 20 nodes did not receive full support in the other trees. This suggests that FishPIE is phylogenetically informative at various taxonomic levels in this ancient lineage (Rabosky et al., 2018). In agreement with previous studies (e.g., Tang et al., 2006; Šlechtová et al., 2007; Stout et al., 2016), our phylogenetic trees consistently showed that Catostomidae and Gyrinochelidae occupied the basal positions in this suborder, followed by Botiidae. We also found that several cobitids nested within Nemacheilidae with strong supports in all trees, and the two families together were sister to Gastromyzontidae. Tang et al. also revealed a sister relationship between Nemacheilidae and Cobitidae based on mitochondrial DNA analyses, though with weak support (Tang et al., 2006). In contrast, the relationships depicted by 219 anchor hybrid enrichment markers suggested that Gastromyzontidae and Nemacheilidae were more closely related, followed by Cobitidae (Stout et al., 2016). Nonetheless, the taxon sampling of this study was not as comprehensive as the current study. Morphologically, Nemacheilidae shares some osteological similarities with Balitoridae (Sawada, 1982), but many of the characters used were later considered to be homoplasies (Nalbant and Bianco, 1998). Hence, the relationships among these “advanced” cobtitoid families require further elucidation.
In Cyprinoidei (carp-like fishes), 96.97% to 100% of the nodes received strong supports (>90% BP or >0.9 PP) in the BI and ML trees, though only 63.63% of the nodes were strongly supported in the ASTRAL tree. Most of the nodes with moderate supports pertained to relationships among species or closely related genera in Xenocyprididae. Thus, the phylogenetic resolution of FishPIE is also high in this diverse suborder, but the resolution for recent and rapid radiation may be less optimal. Here, all families were found to be monophyletic with strong supports in all trees. The relationships among cyprinoid families were generally consistent with results from previous studies except in a few nodes. In all our trees, Paedocyprinidae was the most distantly related, followed by Danionidae, whereas Cyprinidae was sister to a clade formed by the remaining families. In a UCE study that included these families, the positions of Danionidae and Cyprinidae were swapped, also with strong statistical support (Stout et al., 2016). Our phylogenetic trees showed a sister relationship between Acheilognathidae + Tincidae and Gobionidae with moderate (85%–96% BP in ML trees) to full supports (in BI trees), and together they were sister to Leuciscidae, though the relationships were poorly resolved in the ASTRAL tree. Such relationships (without Tinicidae) were also depicted in Stout et al. (2016), but placements of these families varied across studies (Mayden et al., 2009; Tang et al., 2013; Tao et al., 2013, 2019). For instance, Tao et al. (2013), using four nuclear gene segments, suggested that Gobionidae was more closely related to Leuciscidae. Although we deem results from phylogenomic analyses to be more vigorous, more detailed investigations, particularly regarding the placement of the enigmatic Tincidae, are necessary to clarify these relationships.
At the subfamily level, there is strong support for the genus Anabarilius, previously classified as Xenocyprididae incertae sedis (Tan and Armbruster, 2018), to be nested within Xenocypridinae in all analyses, suggesting it is a member of this subfamily. Our phylogenetic analyses also indicated the paraphyly of Schizothoracinae with strong supports, with Schizopygopsinae nested inside all trees. A reticulate relationship between these two subfamilies was also revealed in some gene trees (Wen et al., 2020), which may be attributable to the polyploidization in and the introgression between these families. All other cyprinid subfamilies were strongly supported to be monophyletic in our phylogenetic trees.
At the generic level, Schizopygopsis and Gymnocypris of Schizopygopsinae, Schizothorax and Schizopyge of Schizothoracinae, Cirrhinus of Labeoninae, and Culter of Xenocypridinae were not monophyletic with strong supports in all trees. Although the reasons for the non-monophyly of the schizopygopsinid and schizothoracinid genera might be attributable to polyploidization and introgression as proposed by Wen et al. The sampling of the latter two genera was limited in the current study, and further investigation is required to rectify their classification (Wen et al., 2020).
Taken together, given adequate taxon sampling, FishPIE exhibits satisfactory resolution at the family and genus levels, which are the levels that remain largely unexplored in many fish lineages. The FishPIE markers can thus contribute to building the fish tree of life in the future.
Application notes
The selection of makers is perhaps the most critical decision for any molecular phylogenetic investigation. The FishPIE markers present advantages in several aspects. Although the total number of characters of FishPIE is less than those of phylogenomic studies that generally analyzed over 10,000 sites, FishPIE demonstrated comparable phylogenetic resolution at different taxonomic levels and yielded tree topology highly similar to that based on thousands of markers. FishPIE demonstrated a high retrieval rate from previous NGS datasets, making it easy to capitalize on existing data. With PCR as a routine protocol in molecular laboratories, the FishPIE markers offer a smooth transition from traditional protocol to the high-throughput approach. Besides, the initial investment for conducting a phylogenetic analysis using FishPIE would primarily be the cost of primers, which was less than USD1000 for applying all markers on >200 samples. It would be at least three times higher for other phylogenomic toolkits for fish, such as UCEs, AHE, and exon capture. Besides, for users who opt for NGS, our protocol for pooling PCR products for library construction and pooling libraries for sequencing can markedly lower the sequencing cost (see Star Methods), and we found that a reliable assembly of all 82 FishPIE markers would require only 100MB Illumina raw reads per specimen. Hence, FishPIE has a good cost-performance ratio. This study proposed new phylogenetic hypotheses for several fish lineages and questioned the monophyly of some taxonomic groups. More exhaustive taxon sampling is needed to address these recalcitrant fish phylogenetic questions. We hope that by contributing this novel marker set, our understanding of fish phylogeny can progress speedily.
Limitations of the study
In this work, we tested FishPIE marker mostly using Illumina sequencing, and the performance of our primer set in Sanger sequencing has not been comprehensively tested in all specimens. The effects of different parameters of phylogenetic inference were not explored in this study. The divergence times and evolutionary rates are estimates, and new fossil records may change our calibration points and thus these estimates, but we do not anticipate significant change in the relative rates of our markers.
STAR★Method
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical commercial assays | ||
| Universal Genomic DNA Kit | Cowin Biosciences | Cat#CW2298M |
| PCR mix | Guangzhou Dongsheng Biotech | Cat#P2011 |
| DM2000 DNA marker | Cowin Biosciences | Cat#CW0632M |
| VAHTS DNA Clean Beads (60mL) | Vazyme | Cat#N411-02 |
| Next dsDNA Fragmentase | NEB | Cat#M0348L |
| VAHTSTM Turbo End Repair/dA-Tailing module for Illumina | Vazyme | Cat#N201-02 |
| T4 DNA Ligase (Rapid) | Vazyme | Cat#N103-01 |
| 2x Pfu MasterMix (with dye) | Cowin Biosciences | Cat#CW0686 |
| KOD FX | TOYOBO | Cat#KFX-101 |
| Deposited data | ||
| BioProject | This paper | PRJNA789926 |
| Scripts, sequences and newick trees | This paper | https://figshare.com/s/f2b2cd2557aebc4e5f40 |
| Software and algorithms | ||
| BLASTN | Altschul et al. (1990) | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| MAFFT ver 7.455 | Katoh and Standley (2013) | https://mafft.cbrc.jp/alignment/software/ |
| primer3 ver 2.4.0 | Koressaar and Remm (2007) | https://primer3.ut.ee/ |
| SOAPdenovo ver 2.04 | Luo et al. (2012) | https://github.com/aquaskyline/SOAPdenovo2.git |
| Trinity ver 2.8.5 | Grabherr et al. (2011) | https://github.com/trinityrnaseq/trinityrnaseq/ |
| IQtree2 ver 2.0.5 | Minh et al. (2020) | https://github.com/iqtree/iqtree2 |
| trimAL ver 1.4.rev15 | Capella-Gutiérrez et al. (2009) | https://vicfero.github.io/trimal/ |
| ExaBayes ver 1.5.1 | Aberer et al. (2014) | https://cme.h-its.org/exelixis/web/software/exabayes/manual/index.html |
| ASTRAL-III ver. 5.6.3. | Zhang et al. (2018) | https://github.com/smirarab/ASTRAL |
| PAML | Yang (2007) | http://evomics.org/resources/software/molecular-evolution-software/paml/ |
| R package phangorn | Schliep (2010) | https://cran.r-project.org/web/packages/phangorn/ |
| OrthoFinder ver. 2.5.4 | Emms and Kelly (2019) | https://github.com/davidemms/OrthoFinder/releases |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead author, Xidong Mu (muxd@prfri.ac.cn).
Materials availability
This study did not generate new reagents.
Method details
Ortholog identification
The genomes of 131 fish species (Table S1) were first fragmented into sequences of 10 kb by iterating over a 5 kb sliding window. The sequences from each species were then aligned against the zebrafish genome (GCF_000002035.6) using BLASTN (Altschul et al., 1990) with a maximum e-value of 1e−10 and a minimum alignment length of 1000 bp. One-to-one orthologs were detected by reciprocal best hit using BLASTN with a maximum e-value of 1e−10. We then retained orthologs that (1) were within an annotated protein-coding region in the zebrafish genome, (2) with exon length >1000 bp, and (3) can find a match in 80% of the fish species tested. 2093 genes satisfied these criteria.
Primer design
The sequences of each orthologs were aligned by MAFFT ver 7.455 (Katoh and Standley, 2013) with a maximum number of iterative refinement of 1000 and under the localpair mode. Conserved regions with the least the number of gaps and most conserved sites calculated within 50 bp windows with sliding steps of 25 bp were identified using a customized script. Using the nested PCR strategy that exhibited high efficiency of obtaining target PCR amplicons (Shen et al., 2013), we deigned two pairs of primers from these orthologs alignments manually, facilitated by primer3 ver 2.4.0 (Koressaar and Remm, 2007) within the conserved regions. We selected 20–40 bp long primers whose 3′ ends of the primers were conserved and do not contain ambiguous sites, and that produce products that are >800bp with 40%–70% GC content. The primers were then mapped onto the 131 fish genomes to assess specificity using BLASTN with a maximum e-value of 10 and default parameters. Primers that matched with non-target region(s) with >95% similarity, and those that did not align with the target region with >90% for more than 80% of the primer length, and those that exhibited mismatch in the final three bases in the 3′end were considered unspecific. We ranked the nested PCR primer sets of each ortholog according to the number of species to which they were specific, and then selected 102 loci of the top-ranking primer sets as the FishPIE testing primer set.
Primer testing
We tested the primers on 203 fish species from 31 orders (Table S2) for each of the 102 loci. Fresh, frozen, ethanol-preserved, and, formalin-preserved samples, and dried fin clips (Table S2) were obtained from the National Freshwater Genetic Resource Center, which is a national depository of biological samples and life specimens of aquatic organisms. The genomic DNA was extracted using Universal Genomic DNA Kit (Cowin Biosciences, China). PCRs were conducted using protocol detailed in Method S1, with the annealing temperature listed in Table S3. PCR success was assessed by gel electrophoresis on 1% agarose gel.
Equi-quantity PCR products of the same species were then pooled, fragmented and treated with Taq DNA polymerase for end repairing, 5′ phosphorylation and dA-tailing, followed by T-A ligation adding adaptors to both ends (see Method S2 for details). The products were sequenced in Illumina Hiseq 4000 platform, using pair-end 150-bp sequencing method. The resulting reads were assembled using SOAPdenovo ver 2.04 (Luo et al., 2012) with default settings and multiple k-mers. We tested k-mer size every 4 bp from 33 to 127bp. We then matched the resulting sequences against the reference genome of the most closely related species (Table S1) using BLASTN with maximum e-value of 1e−10. For each of the marker, we selected the assembled contig that was the (1) longest and (2) the most similar to the reference sequences.
Testing NPCL markers for phylogenetic utility
To examine the phylogenetic utility of the selected loci in resolving phylogenetic relationships at different taxonomic levels, we increased the taxonomic coverage of the dataset by obtaining orthologous sequences of these loci from genomic and transcriptomic data from NCBI SRA (Table S2). Genomic sequencing reads were assembled using SOAPdenovo2 with default settings and multiple k-mers (we tested k-mer size every 4 bp from 33 to 127bp), while transcriptomes were assembled using Trinity ver 2.8.5 (Grabherr et al., 2011) using default parameters. The resulting sequences were mapped against the reference genome of the most closely related species (see Table S2) using BLASTN with maximum e-value of 1e−10. The reciprocal best hit of the selected loci were deemed valid sequences. We filtered potential paralogous sequences by manually examining each gene tree. The gene trees were constructed by aligning the sequences of each selected locus using MAFFT, followed by phylogenetic analysis using IQtree2 ver 2.0.5 (Minh et al., 2020) with edge-linked proportional partition model, 1000 ultrafast bootstrap replicates, and 1000 replicates for SH approximate likelihood ratio test.
We examined the phylogenetic resolution of all orthologous markers in concert. The selected orthologous sequences were aligned again using MAFFT with a maximum number of iterative refinement of 1000 and under the localpair mode, and alignments were trimmed using trimAL ver 1.4.rev15 (Capella-Gutiérrez et al., 2009) using the “gappyout” setting. Alignments were then translated to peptide sequences. For both nucleotide and peptide datasets, the best substitution model for each marker was selected using IQtree2, and was then used for species tree reconstruction using the same software with gene-partition (same set of branch lengths but different evolutionary rates) and 1000 ultrafast bootstrapping. We conducted Bayesian inference using ExaBayes ver 1.5.1 (Aberer et al., 2014) using four chains and ASDSF <5.00%, ran for at least 10000 generations. Coalescent species tree was constructed using nucleotide gene trees inferred using IQtree2 as inputs in ASTRAL-III ver. 5.6.3. (Zhang et al., 2018) with 1000 bootstrap replicates.
Substitution rates of FishPIE
To examine substitution rates, time calibrated chronogram was constructed based on codons which included fourfold degenerate sites (4DTv) SNPs, in a Bayesian framework using MCMCTree of the PAML package (Yang, 2007) using 14 calibration points obtained from the TimeTree web resource (www.timetree.org, see Table S7 for calibration points) (Kumar et al., 2017). The nucleotide ML tree was used as the input tree. HKY85 + G model was used as the substitution model, global clock was selected as the clock model. The Markov chains were run for 102,000 generations, sampled every 10 generations, with first 2,000 generations discarded as burnin. Convergence plot of two MCMC chains were used to check for convergence of MCMC runs.
Raw pairwise distance between each taxon was calculated by the dist.dna function of the R package ape for each marker, and substitution rates were calculated as substitution per site per million years.
Phylogenetic informativeness of FishPIE marker set
To test if the number of markers in the FishPIE set were adequate for yielding credible phylogeny, we examined the convergence of tree topologies as the number of markers increased. As this analysis was time-consuming, we scaled down the dataset by selecting only individuals with >50% of the markers sequenced and at most two representatives from a genus. We accessed the concordance of phylogenetic signal among the FishPIE markers by calculating tree distance (normalized RF index) between each nucleotide gene tree and a reference species tree using the RF.dist function of the R package phangorn (Schliep, 2010). The gene trees were computed using IQtree2 with default settings, while the reference species tree was reconstructed using the same approach using a concatenated dataset. We then constructed multiple phylogenetic trees by concatenating FishPIE markers one by one in an ascending order of tree distances based on the respective gene trees. Tree distances between individual reconstruction and the reference species tree were calculated as aforementioned.
To further test the phylogenetic informativeness of FishPIE, we compared the phylogeny of 479 fish species constructed using FishPIE markers and using genome-scale analyses. We first selected from each of the 29 orders a genome with the most comprehensive annotation (see Table S8). We then used OrthoFinder ver. 2.5.4 (Emms and Kelly, 2019) to identify 324 single-copy orthologs of protein sequences using default parameters. These orthologs were combined with the 1105 exon markers obtained from Hughes et al. to form a seed marker set (Hughes et al., 2018). We assembled the genomes and transcriptomes of 479 species using SOAPdenovo2 and Trinity, respectively. Default parameters were used for these assemblies. We then identified orthologs of the seed marker set from these assemblies by reciprocal best hit analysis using BLASTN with a maximum e-value of 1e−10. Orthologs were aligned using MAFFT with a maximum number of iterative refinement of 1000 and under the localpair mode. The alignments were then concatenated, and we used IQtree2 to select the best substitution model of each marker based on BIC, and then construct the maximum likelihood phylogeny with 1000 ultrafast bootstrap and 1000 replicates for SH test.
Using the same method, we constructed the maximum likelihood phylogeny of the same 479 species based on FishPIE marker sequences.
Annotation of FishPIE markers
Representative sequences of each ortholog were searched against Uniport and nr database using BLASTP with an E-value cutoff of 1e−10. Genes were tentatively identified according to the best hits against known sequences. KEGG, Gene Ontology, PANTHER and KOG databases were used to infer gene functions.
Acknowledgments
This study was supported by the National Key Research and Development Program of China (2018YFD0900705), Guangdong Basic and Applied Basic Research Foundation (2020A1515011359), Guangzhou Scientific Planning Programs (201904010409), Guangdong Provincial Special Fund for Modern Agriculture Industry Technology Innovation Team (2021KJ150, 2021KJ134), Guangdong Scientific Planning Programs (2019B030316029), China-ASEAN Maritime Cooperation Fund (CAMC-2018F), and National Freshwater Genetic Resource Center (FGRC18537). The authors thank SCGene Co. Ltd. for technical support.
Author contributions
Conceptualization, XD Mu and KY Ma; Resources, YX Yang, JH Sun, Y Liu, C Liu, DE GU, M Fang, W Li, C Zhang, F Liu, and XJ Wang; Methodology: XD Mu, YX Yang, JH Sun, Y Liu, M Xu, and JH Chen; Investigation, XD Mu, YX Yang, JH Sun, Y Liu, M Xu, and JH Chen; Formal Analysis, XD Mu, CW Shao, KH Chu, JH Chen, and KY Ma; Writing—Original Draft, KY Ma and XD Mu wrote the manuscript; Writing—Review & Editing, all authors; Funding Acquisition, XD Mu.
Declaration of interests
The authors declare that they have no conflict of interest.
Published: September 16, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105025.
Contributor Information
Xidong Mu, Email: muxd@prfri.ac.cn.
Ka Yan Ma, Email: majx26@mail.sysu.edu.cn.
Supplemental information
Data and code availability
The sequencing data have been deposited in the GenBank: PRJNA789926.
Sequence alignments are available in FigShare repository (https://figshare.com/s/f2b2cd2557aebc4e5f40).
All original code has been deposited at Github and FigShare repository, and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Aberer A.J., Kobert K., Stamatakis A. ExaBayes: massively parallel Bayesian tree inference for the whole-genome era. Mol. Biol. Evol. 2014;31:2553–2556. doi: 10.1093/molbev/msu236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro M.E., Faircloth B.C., Harrington R.C., Sorenson L., Friedman M., Thacker C.E., Oliveros C.H., Černý D., Near T.J. Explosive diversification of marine fishes at the Cretaceous–Palaeogene boundary. Nat. Ecol. Evol. 2018;2:688–696. doi: 10.1038/s41559-018-0494-6. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Betancur-R R., Broughton R.E., Wiley E.O., Carpenter K., López J.A., Li C., Holcroft N.I., Arcila D., Sanciangco M., Cureton Ii J.C., et al. The tree of life and a new classification of bony fishes. PLoS Curr. 2013;5:1–45. doi: 10.1371/currents.tol.53ba26640df0ccaee75bb165c8c26288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur-R R., Wiley E.O., Arratia G., Acero A., Bailly N., Miya M., Lecointre G., Ortí G. Phylogenetic classification of bony fishes. BMC Evol. Biol. 2017;17:162. doi: 10.1186/s12862-017-0958-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg J.G., Potter S., Bi K., Moritz C. Exon capture phylogenomics: efficacy across scales of divergence. Mol. Ecol. Resour. 2016;16:1059–1068. doi: 10.1111/1755-0998.12449. [DOI] [PubMed] [Google Scholar]
- Broughton R.E., Betancur-R R., Li C., Arratia G., Ortí G. Multi-locus phylogenetic analysis reveals the pattern and tempo of bony fish evolution. PLoS Curr. 2013;5 doi: 10.1371/currents.tol.2ca8041495ffafd0c92756e75247483e. ecurrents.tol.2ca8041495ffafd0c92756e75247483e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S., Kauff F., Gabaldón T. A phylogenomics approach for selecting robust sets of phylogenetic markers. Nucleic Acids Res. 2014;42:e54. doi: 10.1093/nar/gku071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.J., Ruiz-Carus R., Ortí G. Relationships among four genera of mojarras (Teleostei: perciformes: Gerreidae) from the western Atlantic and their tentative placement among percomorph fishes. J. Fish. Biol. 2007;70:202–218. doi: 10.1111/j.1095-8649.2007.01395.x. [DOI] [Google Scholar]
- Dettaï A., Lecointre G. In search of nothothenioid (Teleostei) relatives. Antarct. Sci. 2004;16:71–85. doi: 10.1017/S0954102004. [DOI] [Google Scholar]
- Dettaï A., Lecointre G. Further support for the clades obtained by multiple molecular phylogenies in the acanthomorph bush. C. R. Biol. 2005;328:674–689. doi: 10.1016/j.crvi.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Emms D.M., Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke R., Eschmyer W.N., van der Lann R. 2021. Eschmeyer’s Catalog of Fishes: Genera, Species, References. Eschmeyer’s Catalog of Fishes: Genera, Species, References.http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp Available from. [Google Scholar]
- Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes L.C., Ortí G., Huang Y., Sun Y., Baldwin C.C., Thompson A.W., Arcila D., Betancur-R R., Li C., Becker L., et al. Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data. Proc. Natl. Acad. Sci. USA. 2018;115:6249–6254. doi: 10.1073/pnas.1719358115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes L.C., Ortí G., Saad H., Li C., White W.T., Baldwin C.C., Crandall K.A., Arcila D., Betancur-R R. Exon probe sets and bioinformatics pipelines for all levels of fish phylogenomics. Mol. Ecol. Resour. 2021;21:816–833. doi: 10.1111/1755-0998.13287. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koressaar T., Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Suleski M., Hedges S.B. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017;34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- Li B., Dettaï A., Cruaud C., Couloux A., Desoutter-Meniger M., Lecointre G. RNF213, a new nuclear marker for acanthomorph phylogeny. Mol. Phylogenet. Evol. 2009;50:345–363. doi: 10.1016/j.ympev.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Li C., Ortí G., Zhang G., Lu G. A practical approach to phylogenomics: the phylogeny of ray-finned fish (Actinopterygii) as a case study. BMC Evol. Biol. 2007;7:44. doi: 10.1186/1471-2148-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y., et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayden R.L., Chen W.J., Bart H.L., Doosey M.H., Simons A.M., Tang K.L., Wood R.M., Agnew M.K., Yang L., Hirt M.V., et al. Reconstructing the phylogenetic relationships of the earth's most diverse clade of freshwater fishes - order Cypriniformes (Actinopterygii: Ostariophysi): a case study using multiple nuclear loci and the mitochondrial genome. Mol. Phylogenet. Evol. 2009;51:500–514. doi: 10.1016/j.ympev.2008.12.015. [DOI] [PubMed] [Google Scholar]
- Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., Lanfear R. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbant T.T., Bianco P.G. The loaches of Iran and adjacent regions with description of six new species (Cobitoidea) Ital. J. Zool. 1998;65:109–123. doi: 10.1080/11250009809386803. [DOI] [Google Scholar]
- Near T.J., Dornburg A., Eytan R.I., Keck B.P., Smith W.L., Kuhn K.L., Moore J.A., Price S.A., Burbrink F.T., Friedman M., Wainwright P.C. Phylogeny and tempo of diversification in the superradiation of spiny-rayed fishes. Proc. Natl. Acad. Sci. USA. 2013;110:12738–12743. doi: 10.1073/pnas.1304661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paijmans J.L.A., Fickel J., Courtiol A., Hofreiter M., Förster D.W. Impact of enrichment conditions on cross-species capture of fresh and degraded DNA. Mol. Ecol. Resour. 2016;16:42–55. doi: 10.1111/1755-0998.12420. [DOI] [PubMed] [Google Scholar]
- Rabosky D.L., Chang J., Title P.O., Cowman P.F., Sallan L., Friedman M., Kaschner K., Garilao C., Near T.J., Coll M., Alfaro M.E. An inverse latitudinal gradient in speciation rate for marine fishes. Nature. 2018;559:392–395. doi: 10.1038/s41586-018-0273-1. [DOI] [PubMed] [Google Scholar]
- Rokas A., Williams B.L., King N., Carroll S.B. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- Sawada Y. Phylogeny and zoogeography of the superfamily Cobitoidea (Cyprinoidei, Cypriniformes) Mem. Fac. Fish. Sci. Hokkaido Univ. 1982;28:65–223. doi: 10.1002/ece3.5553. [DOI] [Google Scholar]
- Schliep K.P. phangorn: phylogenetic analysis in R. Bioinformatics. 2010;27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X.X., Liang D., Feng Y.J., Chen M.Y., Zhang P. A versatile and highly efficient toolkit including 102 nuclear markers for vertebrate phylogenomics, tested by resolving the higher level relationships of the Caudata. Mol. Biol. Evol. 2013;30:2235–2248. doi: 10.1093/molbev/mst122. [DOI] [PubMed] [Google Scholar]
- Slechtová V., Bohlen J., Tan H.H. Families of Cobitoidea (Teleostei; Cypriniformes) as revealed from nuclear genetic data and the position of the mysterious genera Barbucca, psilorhynchus, Serpenticobitis and Vaillantella. Mol. Phylogenet. Evol. 2007;44:1358–1365. doi: 10.1016/j.ympev.2007.02.019. [DOI] [PubMed] [Google Scholar]
- Smith W.L., Stern J.H., Girard M.G., Davis M.P. Evolution of venomous cartilaginous and ray-finned fishes. Integr. Comp. Biol. 2016;56:950–961. doi: 10.1093/icb/icw070. [DOI] [PubMed] [Google Scholar]
- Spinks P.Q., Thomson R.C., Lovely G.A., Shaffer H.B. Assessing what is needed to resolve a molecular phylogeny: simulations and empirical data from emydid turtles. BMC Evol. Biol. 2009;9:56. doi: 10.1186/1471-2148-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout C.C., Tan M., Lemmon A.R., Lemmon E.M., Armbruster J.W. Resolving Cypriniformes relationships using an anchored enrichment approach. BMC Evol. Biol. 2016;16:244–313. doi: 10.1186/s12862-016-0819-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J.P., Lundberg J.G., Hardman M. A phylogenetic analysis of the major groups of catfishes (Teleostei: Siluriformes) using rag1 and rag2 nuclear gene sequences. Mol. Phylogenet. Evol. 2006;41:636–662. doi: 10.1016/j.ympev.2006.05.044. [DOI] [PubMed] [Google Scholar]
- Tan M., Armbruster J.W. Phylogenetic classification of extant genera of fishes of the order Cypriniformes (Teleostei: Ostariophysi) Zootaxa. 2018;4476:6–39. doi: 10.11646/zootaxa.4476.1.4. [DOI] [PubMed] [Google Scholar]
- Tang K.L., Agnew M.K., Hirt M.V., Lumbantobing D.N., Sado T., Teoh V.H., Yang L., Bart H.L., Harris P.M., He S., et al. Limits and phylogenetic relationships of East Asian fishes in the subfamily Oxygastrinae (Teleostei: Cypriniformes: Cyprinidae) Zootaxa. 2013;3681:101–135. doi: 10.11646/zootaxa.3681.2.1. [DOI] [PubMed] [Google Scholar]
- Tang Q., Liu H., Mayden R., Xiong B. Comparison of evolutionary rates in the mitochondrial DNA cytochrome b gene and control region and their implications for phylogeny of the Cobitoidea (Teleostei: Cypriniformes) Mol. Phylogenet. Evol. 2006;39:347–357. doi: 10.1016/j.ympev.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Tao W., Mayden R.L., He S. Remarkable phylogenetic resolution of the most complex clade of Cyprinidae (Teleostei: Cypriniformes): a proof of concept of homology assessment and partitioning sequence data integrated with mixed model Bayesian analyses. Mol. Phylogenet. Evol. 2013;66:603–616. doi: 10.1016/j.ympev.2012.09.024. [DOI] [PubMed] [Google Scholar]
- Tao W., Yang L., Mayden R.L., He S. Phylogenetic relationships of Cypriniformes and plasticity of pharyngeal teeth in the adaptive radiation of cyprinids. Sci. China Life Sci. 2019;62:553–565. doi: 10.1007/s11427-019-9480-3. [DOI] [PubMed] [Google Scholar]
- Wen Y., Chai J., Ma W., Murphy R.W., He S., Chen Z., Zhang Y., Lu X. Polyploidization, hybridization, and maternal and paternal lineages in Cyprinids (Teleostei: Cypriniformes) Research Square. 2020 doi: 10.21203/rs.3.rs-119099/v1. [DOI] [Google Scholar]
- Yang Z. Paml 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhang C., Rabiee M., Sayyari E., Mirarab S. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinf. 2018;19:153. doi: 10.1186/s12859-018-2129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data have been deposited in the GenBank: PRJNA789926.
Sequence alignments are available in FigShare repository (https://figshare.com/s/f2b2cd2557aebc4e5f40).
All original code has been deposited at Github and FigShare repository, and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.