Abstract
Background
Non-alcoholic fatty liver disease (NAFLD) is a spectrum of disease ranging from simple hepatic steatosis (NAFL) to non-alcoholic steatohepatitis (NASH) which may progress to cirrhosis and liver cancer. NAFLD is rapidly becoming a global health challenge, and there is a need for improved diagnostic- and prognostic tools and for effective pharmacotherapies to treat NASH. The molecular mechanisms of NAFLD development and progression remain incompletely understood, though ample evidence supports a role of microRNAs (miRNAs) – small non-coding RNAs regulating gene expression – in the progression of metabolic liver disease.
Scope of review
In this review, we summarise the currently available liver miRNA profiling studies in people with various stages of NAFLD. We further describe the mechanistic role of three of the most extensively studied miRNA species, miR-34a, miR-122 and miR-21, and highlight selected findings on novel NAFLD-linked miRNAs. We also examine the literature on exosomal microRNAs (exomiRs) as inter-hepatocellular or -organ messengers in NAFLD. Furthermore, we address the status for utilizing circulating NAFLD-associated miRNAs as minimally invasive tools for disease diagnosis, staging and prognosis as well as their potential use as NASH pharmacotherapeutic targets. Finally, we reflect on future directions for research in the miRNA field.
Major conclusions
NAFLD is associated with changes in hepatic miRNA expression patterns at early, intermediate and late stages, and specific miRNA species appear to be involved in steatosis development and NAFL progression to NASH and cirrhosis. These miRNAs act either within or between hepatocytes and other liver cell types such as hepatic stellate cells and Kupffer cells or as circulating inter-organ messengers carrying signals between the liver and extra-hepatic metabolic tissues, including the adipose tissues and the cardiovascular system. Among circulating miRNAs linked to NAFLD, miR-34a, miR-122 and miR-192 are the best candidates as biomarkers for NAFLD diagnosis and staging. To date, no miRNA-targeting pharmacotherapy has been approved for the treatment of NASH, and no such therapy is currently under clinical development. Further research should be conducted to translate the contribution of miRNAs in NAFLD into innovative therapeutic strategies.
Keywords: microRNA, Non-alcoholic fatty liver disease, Non-alcoholic steatohepatitis, Liver fibrosis, Cirrhosis
Abbreviations
- AMPK
AMP-activated protein kinase
- ASO
Antisense oligonucleotide
- ATM
Adipose tissue macrophages
- CVD
Cardiovascular disease
- DIO
Diet-induced obese
- ECM
Extracellular matrix
- EMT
Epithelial−mesenchymal transition
- ER
Endoplasmic reticulum
- EV
Extracellular vesicle
- exomiR
Exosomal miRNA
- FXR
Farnesoid X receptor
- HCC
Hepatocellular carcinoma
- HFD
High-fat diet
- HMGCR
3-Hydroxy-3-methyl-glutaryl-coenzyme A reductase
- HNF
Hepatocyte nuclear factor
- HSC
Hepatic stellate cells
- KO
Knockout
- LXR
Liver X receptor
- MCD
Methionine-choline deficient
- MiRNA
MicroRNA
- NAFLD
Non-alcoholic fatty liver disease
- NASH
Non-alcoholic steatohepatitis
- NF-κB
Nuclear factor-κB
- PPAR
Peroxisome proliferator-activated receptor
- ROS
Reactive oxygen species
- RT-qPCR
Reverse transcription quantitative polymerase chain reaction
- SHP
Small heterodimer partner
- SIRT
Sirtuin
- TGF
Transforming growth factor
- TNF
Tumour necrosis factor
- UPR
Unfolded protein response
- UTR
Untranslated region
1. Introduction
Non-alcoholic fatty liver disease (NAFLD) is a continuum of worsening clinical-histological liver phenotypes characterized by hepatic lipid accumulation, also termed steatosis or non-alcoholic fatty liver (NAFL). In a subset of patients with simple hepatic steatosis, NAFLD may progress to non-alcoholic steatohepatitis (NASH), as hepatocellular lipid accumulation becomes associated with hepatocyte injury and death, lobular inflammatory cell infiltration and varying degrees of fibrosis [1]. NASH may progress to cirrhosis (extensive fibrosis), causing scarring and stiffening of the liver, and to hepatocellular carcinoma (HCC) with a consequent need for liver transplantation to prevent liver failure and death [2].
NAFLD has become a major cause of chronic liver disease. The global prevalence of NAFLD is estimated to approximately 25% and has increased in parallel with the obesity and type 2 diabetes epidemic [2,3]. NAFLD is highly associated with metabolic risk factors like obesity, insulin resistance, type 2 diabetes mellitus, dyslipidemia and hypertension, and with extra-hepatic complications like cardiovascular disease (CVD) [1,2]. While the relationship between obesity and NAFLD is established, some cases of obesity, where a high body mass index is not associated with insulin-resistance, suggest that the relationship between metabolic risk factors and NAFLD is complex. Such “metabolically healthy” obesity is associated with modest hepatic lipid accumulation [4]. Conversely, NAFLD may also develop in lean, normal-weight people, with or without metabolically unhealthy body fat distribution [2,5]. “Classical” NAFLD associated with insulin-resistant obesity as well as “lean NAFLD” constitute a highly heterogenous spectrum of liver disease in terms of etiologies and risk of progression to NASH, cirrhosis and HCC [2,5]. Interestingly, the major risk factors thought to be involved in NAFLD that have a strong hepatic genetic component (variability in a range of genes of hepatic lipid metabolism, including PNPLA3 and TM6SF2) are associated with a moderate to strong risk to develop NASH and fibrosis, but only show a weak association with insulin resistance and type 2 diabetes [3]. Curiously, these risks even appear to be linked to a lower risk of CVD [3]. Thus, there is a clear need for an improved understanding of the molecular links between NAFLD etiologies and progression and cardio-metabolic co-morbidities.
NAFL and NASH are usually asymptomatic and go unnoticed for many years until cirrhosis or HCC ensue, or when the condition is incidentally detected during an abdominal scan for other diseases [1]. Liver biopsy remains the gold standard technique for NASH diagnosis and fibrosis staging but has several limitations and carries a risk to the patient due to its invasive nature [6]. Improved minimally invasive tools for NAFLD/NASH diagnosis and staging as well as methods for identification of at-risk cases for disease progression are highly needed. Moreover, effective pharmacotherapies to arrest or even reverse progression of NASH to cirrhosis and HCC are still lacking [1,7,8]. Timely identification of at-risk NASH cases as well as development of effective treatments rely on an in-depth understanding of the molecular mechanisms underlying the progression of NAFLD. One class of small non-coding RNAs that function to regulate gene expression is microRNAs (miRNAs), and ample evidence supports a role of miRNAs in the pathophysiological mechanisms behind the evolution of NAFLD. In this review, we summarize the current knowledge on the contribution of miRNAs to NAFL development and progression to fibrotic NASH, with specific focus on three of the most extensively studied NAFLD-linked miRNA species. We also review the literature on the mechanistic role of exosomal miRNAs (exomiRs) in NAFLD pathogenesis. We finally address the status of the clinical utility of circulating miRNAs as biomarkers of disease stage and targets of NASH pharmacotherapies.
2. Pathophysiological mechanisms of progressive NAFLD
Numerous intra- and extrahepatic mechanisms, including adipose tissue dysfunction and altered gut microbiota, act on a background of varying genetic pre-disposition to drive NAFLD development [1,3,[8], [9], [10], [11], [12]]. Hepatocellular lipid accumulation forms the basis of NAFL and arises when the liver’s capacity for lipid- and carbohydrate handling is exceeded. Accumulated toxic lipid species induce lipotoxicity, which is likely involved in the development of insulin resistance and cellular stress, injury and ultimately death by interfering with multiple cellular pathways [1,8,9]. Toxic lipid species may induce endoplasmic reticulum (ER) stress and trigger the unfolded protein response (UPR), an adaptive cell-protective stress response functioning to re-establish cellular homeostasis. In NAFLD, chronic UPR activation might aggravate lipid accumulation and insulin resistance, and fuel the progression to NASH by inducing inflammation, oxidative stress and hepatocellular death [11]. NASH is also associated with mitochondrial dysfunction and reduced hepatocyte capacity to oxidize excess lipids. This aggravates lipotoxicity and leads to elevated production of reactive oxygen species (ROS), resulting in oxidative stress, inflammation and fibrosis [8,9]. During NASH development, liver-resident macrophages, also termed Kupffer cells, are activated and secrete cytokines and chemokines [12]. This causes alterations in the immune landscape of the liver, including infiltration by immune cells such as monocytes and neutrophils and contributes to the liver’s chronic inflammatory state [13]. Liver resident macrophages have also been shown to contribute to insulin resistance independently of their inflammatory status via production of non-inflammatory factors which directly regulate hepatocyte insulin signalling and hence liver metabolism [14]. Factors from liver resident macrophages, as well as from stressed hepatocytes, contribute to fibrosis by activating hepatic stellate cells (HSCs) to transform into myofibroblasts and deposit excessive amounts of extracellular matrix (ECM) proteins [10].
Profound gene expression reprogramming, including altered miRNA expression, is parallel to the pathophysiological mechanisms driving NAFLD. Since the initial report on changes in hepatic miRNA expression patterns in human NASH [15], miRNAs have been in the spotlight of research into the molecular mechanisms by which gene expression is reprogrammed in NAFLD and may contribute to the development and progression of this increasingly prevalent disease.
3. microRNAs in NAFLD
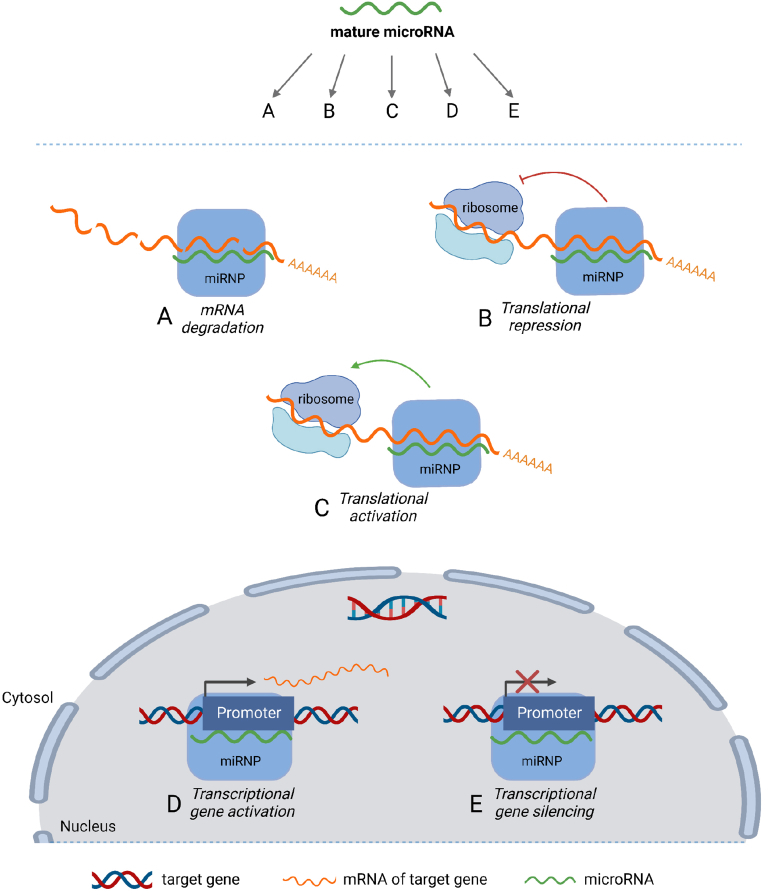
miRNAs were discovered as post-transcriptional repressors of target gene expression [16]. Indeed, the established view was that miRNAs act to degrade target mRNAs and/or represses their translation (Figure 1). However, since miR-369 was shown to act as either an activator or a repressor of Tumor Necrosis Factor α (TNFα) translation depending on cell cycle state [17], many studies have emerged supporting a role of miRNAs as activators of target expression via interaction with either the 3′ or the 5′ untranslated region (UTR) of target transcripts [[18], [19], [20], [21], [22], [23], [24], [25], [26], [27]]. Moreover, some miRNAs reside in the nucleus and control target expression by transcriptional gene activation or silencing by post-transcriptional regulation of the non-coding transcriptome or by regulation of alternative mRNA splicing [28]. Apart from functioning as cell-endogenous regulators of gene expression, miRNAs are also found outside cells, circulating in blood in the form of exosomes (exomiRs) and other types of extracellular vesicles (EV) [29].
Figure 1.
Mechanisms of miRNA-mediated gene expression regulation. A mature ∼22 base pair miRNA is loaded into the microRNA ribonucleotide complex (miRNP) after enzymatic processing in one of the miRNA maturation pathways. This miRNA may then, via the miRNP, affect gene expression through several different mechanisms depending on the miRNA-mRNA pair in question and the cellular context. Binding of the miRNP to target mRNA sequences can lead to mRNA degradation (A) or repression of mRNA translation into protein (B) – the two canonical mechanisms of miRNA action. Binding of some miRNA species to certain targets have been shown to increase translation (C) rather than repress it. Furthermore, some miRNA species have been shown to bind to gene promoter sequences in the nucleus, acting as either activators (D) or silencers (E) of transcription.
3.1. Hepatic miRNA expression profiling in human NAFLD
While studies examining rodent experimental models of NAFL or NASH are numerous, relatively few studies have been published assessing hepatic miRNome-wide expression changes in human NAFLD. A summary of microRNA expression in NAFLD, NASH, and cirrhosis is provided in Table 1. A locked nucleic acid (LNA)-based microarray was used to compare hepatic miRNA expression profiles between a group of obese people without hepatic steatosis (n = 15) and a group showing a high degree of intrahepatic lipid accumulation (n = 15) [30]. Among the 42 differentially expressed miRNAs in the steatotic vs. the non-steatotic group, the eight miRNA species found most significantly regulated had not previously been linked to NAFLD (upregulated hsa-miR-3663-5p, miRPlus-I137∗, miR-576-5p, miR-892a, miR-3924, miR-106b∗, miR-103a-2∗ and miR-1282) [30]. A reverse transcription quantitative polymerase chain reaction (RT-qPCR)-based array was used to profile miRNAs in livers of obese women undergoing gastric bypass surgery, comparing a group without hepatic steatosis (n = 19) and a group with NAFLD (n = 17) [31]. Five miRNAs were reported to be differentially expressed between the two groups (upregulated miR-146b-5p and downregulated miR-139-5p, miR-30b-5p, miR-122-5p and miR-422a). Interestingly, treatment of cultured primary human hepatocytes with palmitate alone or in combination with high glucose changed expression levels of all five miRNA species [31]. Other studies have examined hepatic miRNA profiles of more advanced NAFLD stages. Microarrays were used to assess miRNA expression in liver biopsies from people with metabolic syndrome, comparing a group with normal liver histology (n = 23) with a group with NASH (n = 23). Among the 46 differentially expressed miRNA species in the NASH group, miR-224, miR-34a, miR-200a, miR-146b and miR-222 were upregulated and miR-617, miR-641, miR-198, miR-765 and miR-601 were downregulated [15]. miRNA sequencing was used to compare a group of people without steatosis and fibrosis (n = 10) with a group with severe NAFLD fibrosis or cirrhosis (stage F3–F4 in the NASH Clinical Research Network histological scoring system [32]) (n = 15) [33]. Forty-three miRNA species were found to be differentially expressed (log2 fold change cut-off = 1), and nine were subsequently validated by RT-qPCR (upregulated miR-150, miR-224, miR-183, miR-31 and miR-182 and downregulated miR-590, miR-219a, miR-17 and miR-378i) [33]. To the best of our knowledge, only a single study has been published assessing global miRNA expression patterns for the entire NAFLD spectrum, spanning from the non-steatotic through NAFL and NASH to the cirrhotic state [34]. This analysis was performed on samples from obese men and women undergoing gastric bypass surgery. However, the examined cohorts were small (n = 3–12 in sequencing screen, n = 4–5 for qPCR validation of selected miRNAs, though excluding the cirrhosis group), and only a few miRNAs were found to be regulated between consecutive NAFLD stages: miR-375 was downregulated in cirrhosis vs. NASH according to the differential expression analysis (not confirmed by qPCR due to lack of RNA from cirrhosis group), and a trend analysis identified three miRNAs to gradually increase (miR-301a-3p and miR-34a-5p) or decrease (miR-375) with NAFLD progression towards the cirrhotic state [34].
Table 1.
Overview of human microRNAs with altered expression in NAFLD, NASH and cirrhosis.
| microRNA | Regulation in NAFLD | Regulation in NASH | Regulation in fibrosis/cirrhosis | References |
|---|---|---|---|---|
| miR-103-2∗ | Upregulated | – | – | Soronen et al., 2016 |
| miR-106b∗ | Upregulated | – | – | Soronen et al., 2016 |
| miR-122 | Upregulated in simple steatosis, else downregulated | Downregulated | – | Cheung et al., 2008 |
| Latorre et al., 2017 | ||||
| Miyaaki et al., 2014 | ||||
| Pirola et al., 2015 | ||||
| miR-1282 | Upregulated | – | – | Soronen et al., 2016 |
| miR-139-5p | Downregulated | – | – | Latorre et al., 2017 |
| miR-146b | – | Upregulated | – | Cheung et al., 2008 |
| miR-146b-5p | Upregulated | – | – | Latorre et al., 2017 |
| miR-150 | – | – | Upregulated | Leti et al., 2015 |
| miR-17 | – | – | Downregulated | Leti et al., 2015 |
| miR-182 | – | – | Upregulated | Leti et al., 2015 |
| miR-183 | – | – | Upregulated | Leti et al., 2015 |
| mIR-198 | – | Downregulated | – | Cheung et al., 2008 |
| miR-200a | – | Upregulated | – | Cheung et al., 2008 |
| mIR-21 | – | Upregulated | Upregulated | Cheung et al., 2008 |
| Dattaroy et al., 2015 | ||||
| Loyer et al., 2016 | ||||
| Rodrigues et al., 2017 | ||||
| Zhang et al., 2013 | ||||
| miR-219a | – | – | Downregulated | Leti et al., 2015 |
| miR-222 | – | Upregulated | – | Cheung et al., 2008 |
| miR-224 | – | -Upregulated | Upregulated | Cheung et al., 2008 |
| Leti et al., 2015 | ||||
| miR-26a | Downregulated | – | – | Xu et al., 2020 |
| miR-301a-3p | Upregulateda | Upregulateda | Upregulateda | Guo et al., 2016 |
| miR-30b-5p | Downregulated | – | – | Latorre et al., 2017 |
| miR-31 | – | – | Upregulated | Leti et al., 2015 |
| miR-34a | Upregulated | Upregulated | – | Castro et al., 2013 |
| Cheung et al., 2008 | ||||
| Liu et al., 2018 | ||||
| miR-34a-5p | Upregulateda | Upregulateda | Upregulated | Guo et al., 2016 |
| miR-3663-5p | Upregulated | – | – | Soronen et al., 2016 |
| miR-375 | Downregulateda | Downregulateda | Downregulatedb | Guo et al., 2016 |
| miR-378 | Upregulated | Upregulated | – | Zhang, Duan et al., 2019 |
| Zhang, Hu et al., 2019 | ||||
| miR-378i | – | – | Downregulated | Leti et al., 2015 |
| miR-3924 | Upregulated | – | – | Soronen et al., 2016 |
| miR-422a | Downregulated | – | – | Latorre et al., 2017 |
| miR-576-5p | Upregulated | – | – | Soronen et al., 2016 |
| miR-590 | – | – | Downregulated | Leti et al., 2015 |
| miR-601 | – | Downregulated | – | Cheung et al., 2008 |
| miR-617 | – | Downregulated | – | Cheung et al., 2008 |
| miR-641 | – | Downregulated | – | Cheung et al., 2008 |
| miR-765 | – | Downregulated | – | Cheung et al., 2008 |
| miR-892a | Upregulated | – | – | Soronen et al., 2016 |
| miRPlus-I137∗ | Upregulated | – | – | Soronen et al., 2016 |
Found by trend analysis to gradually increase or decrease with disease progression from the non-steatotic state through NAFL and NASH to cirrhosis.
Found to be significantly downregulated in cirrhosis vs. NASH by the differential expression analysis and found to gradually decrease from early NAFLD towards the cirrhotic state by the trend analysis.
3.2. Mechanistic insights into the role of miRNAs in progressive NAFLD
Little agreement exists between the findings of published human studies, both the global hepatic miRNA expression studies reviewed above and studies assessing one or a few selected miRNAs (included in the meta-analysis [35]). Nevertheless, a few miRNA species were reported to be differentially expressed in human progressive NAFLD by several studies. These include miR-34a, miR-122 and miR-21, which are also among the most extensively studied miRNAs in the molecular pathways governing NAFLD development and progression.
3.2.1. miR-34a
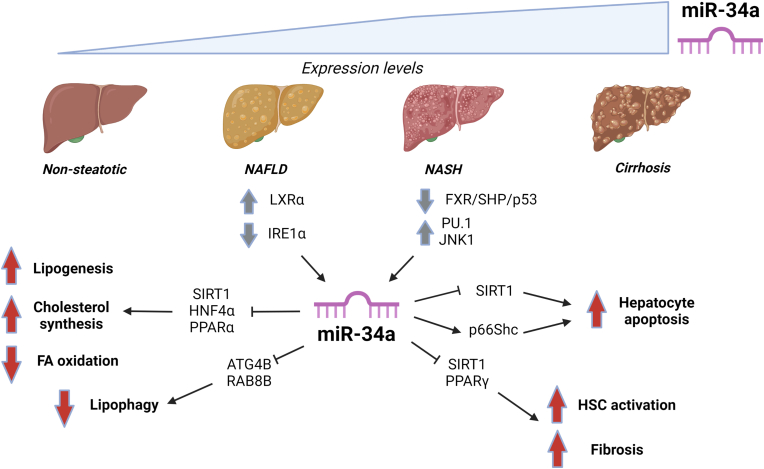
Hepatic miR-34a expression is elevated in people with NAFLD and appears to increase with disease progression (Figure 2) [[34], [35], [36]]. Liver miR-34a expression is controlled by a signalling pathway involving the bile acid receptor Farnesoid X Receptor (FXR), Small Heterodimer Partner (SHP) and p53 [37]. Repression of this pathway may contribute to miR-34a upregulation in NAFLD, since FXR expression is decreased in livers of people with NASH [38].
Figure 2.
miR-34a in NAFLD/NASH. Expression of miR-34a was shown to increase with NAFLD progression and to be controlled through pathways involving FXR, PU.1, JNK1, LXRα and IRE1α. Altered expression/activities of these upstream regulators in NAFLD might contribute to the elevated levels of miR-34a with disease progression. Several miR-34a targets were identified in NAFLD, including SIRT1, HNF4α, PPARα, ATG4B and RAB8B in hepatic lipid metabolic pathways (repressing fatty acid (FA) oxidation and inducing lipogenesis and cholesterol synthesis), p66Shc to stimulate hepatocyte apoptosis, and SIRT1 and PPARγ to stimulate hepatic stellate cell (HSC) activation and fibrosis. Grey arrows: Regulation of upstream factors found to control miR-34a expression in NAFLD/NASH. Red arrows: Increased or decreased activities in pathways/processes as a result of miR-34a actions, leading to unfavourable effects on liver metabolic function and progression of NAFLD.
The role of miR-34a in NAFLD development and progression to NASH was examined in mice either overexpressing or deficient in hepatocyte miR-34a and in a mouse model of NASH induced by a diet high in fat, cholesterol and fructose [39]. miR-34a was shown to regulate progressive NAFLD by acting on several metabolic pathways, including activation of lipid absorption, lipogenesis, inflammation and apoptosis as well as inhibition of fatty acid oxidation [39].
The first hepatic target of miR-34a to be identified was the NAD+-dependent deacetylase Sirtuin 1 (SIRT1) [37], a metabolic regulator controlling many cellular processes deregulated in NAFLD [40]. miR-34a represses SIRT1 function both directly by decreasing its protein levels via 3’ UTR interaction [36,37,41] and indirectly by downregulating the NAD+-synthesizing enzyme Nicotinamide Phosphoribosyltransferase (NAMPT), resulting in decreased substrate availability and activity of SIRT1 [42].
3.2.1.1. miR-34a in hepatic lipid metabolism and steatosis
Several studies have implicated miR-34a in the control of hepatic lipid metabolic pathways by targeting key transcription factors. In vivo antagonism of miR-34a in obese mice normalized SIRT1 activity, causing deacetylation of several regulators of hepatic lipid metabolism and downstream upregulation of β-oxidation genes and downregulation of lipogenesis genes (Figure 2). This effect was paralleled by reduced degrees of liver steatosis [42]. SIRT1 repression by miR-34a has also been implicated in cholesterol metabolism and its derangement in NAFLD. miR-34a overexpression and silencing experiments in a human liver cell line suggested that miR-34a contributes to increased hepatic cholesterol synthesis observed in human NAFLD by maintaining 3-Hydroxy-3-Methyl-Glutaryl-Coenzyme A Reductase (HMGCR) – the rate-limiting enzyme of cholesterol synthesis – in its dephosphorylated active form via SIRT1 inhibition and downstream AMP-activated Protein Kinase (AMPK) inactivation [43]. Blocking miR-34a in fatty acid-treated cultured liver cells in vitro and high-fat diet (HFD)-fed mice in vivo increased expression of Peroxisome Proliferator-Activated Receptor α (PPARα) and its downstream targets of fatty acid β-oxidation and transport as well as increased phosphorylation (activation) of AMPK, another inducer of fatty acid oxidation. This effect was paralleled by a reduction in hepatocellular steatosis [44]. Similarly, hepatocyte deletion of miR-34a attenuated the development of steatohepatitis in mice fed a diet high in fat, cholesterol and fructose, whereas overexpression aggravated NAFLD development [39].
Loss-of-function experiments in mouse models of obesity and diabetes, as well as assessment of human NASH liver biopsies, provided evidence that miR-34a regulates hepatic lipid and lipoprotein metabolism by directly targeting Hepatocyte Nuclear Factor 4α (HNF4α) [45]. miR34a was also found to inhibit lipophagy, a type of autophagy which degrades intracellular lipid droplets in lysosomes [46], in a pathway controlled by the nuclear hormone receptor Liver X Receptor α (LXRα). LXRα is a major regulator of lipogenesis and cholesterol homeostasis upregulated in NAFLD and was found to inhibit autophagy of accumulated lipid droplets both in hepatocytes in vitro and in mouse livers in vivo. miR-34a became transactivated by LXRα upon its pharmacological activation and directly repressed two factors important for the autophagic process, Autophagy Related 4B Cysteine Peptidase (ATG4B) and the small GTPase RAB8B, thereby inhibiting lipophagy. These findings were supported by gene expression analysis on liver biopsies from people with NAFLD [46]. miR-34a expression is also controlled by Inositol-Requiring Enzyme 1α (IRE1α), an enzyme inducing the ER stress-activated UPR. IRE1α is critical for maintaining lipid homeostasis in the healthy liver by enzymatic degradation of the precursors of several miRNAs, including miR-34a. However, upon HFD-feeding in mice and in human hepatosteatosis, IRE1α is enzymatically inactivated by S-nitrosylation, resulting in upregulation of the targeted miRNAs and worsening of steatosis via de-repression of downstream miRNA targets [47].
3.2.1.2. miR-34a in progressive NAFLD
miR-34a was initially shown to contribute to hepatocyte death in progressive NAFLD through the pro-apoptotic miR-34a/SIRT1/p53 axis [36]. Later studies showed that c-Jun N-terminal Kinase 1 (JNK1) was an upstream activator of the miR-34a/SIRT1/p53 axis induced by certain pro-apoptotic bile acids found elevated in human NASH livers [48]. In diet-induced obese (DIO) rats, miR-34a augments liver cell apoptosis by increasing expression of p66Shc, a mitochondrial adaptor protein upregulated in human NASH livers which conveys oxidative stress signals into apoptosis [41].
A few studies have assessed the contribution of miR-34a up-regulation to HSC activation and liver fibrosis. Using a chemically induced rat model of liver fibrosis combined with in vitro experiments in human hepatocytes and co-cultured HSCs, it was demonstrated that the miR-34a/SIRT1/p53 signalling pathway is activated specifically in hepatocytes and contributed to hepatocyte apoptosis and subsequent HSC activation [49]. The function of the miR-34a/SIRT1/p53 axis in rat liver fibrosis induced by fructose, a known risk factor of human liver fibrosis, was assessed [50]. Activation of this axis resulted in hepatocyte epithelial to mesenchymal transition (EMT) via activation of Transforming Growth Factor (TGF)-β1/SMAD signalling [50]. In vitro studies suggest that hepatocytes can undergo EMT, i.e., transdifferentiation, to myofibroblasts in liver fibrosis in response to TGF-β stimulation, thereby contributing to the pool of ECM-depositing cells. However, it is controversial whether hepatocytes undergo full conversion to fibrogenic myofibroblasts in vivo (reviewed in [51]). PU box-binding protein 1 (PU.1) was shown to be one of the most upregulated transcription factors in livers of obese mouse models and elevated in human NASH compared to NAFL livers. Pharmacological inhibition of PU.1 improved the metabolic dysfunction in the DIO mice and alleviated steatosis, inflammation and fibrosis in mice fed a NASH-promoting diet [52]. Regulation of miRNA expression may contribute to PU.1-mediated metabolic changes in NASH, since PU.1 was shown to be an upstream HSC activator in mice by stimulating transcription of miR-34a, leading to SIRT1 suppression [53] (Figure 2). Decreased expression of hepatic SIRT1 has been observed both in mouse models of fibrosis and in human cirrhosis and it was suggested to be involved in HSC activation and fibrosis through a pathway involving the transcriptional repressor Enhancer of Zeste Homolog 2 (EZH2) and the anti-fibrogenic nuclear receptor PPARγ [54], which was also demonstrated to be a direct miR-34a target in cultured human HSCs [55].
3.2.2. miR-122
3.2.2.1. miR-122 expression changes in progressive NAFLD
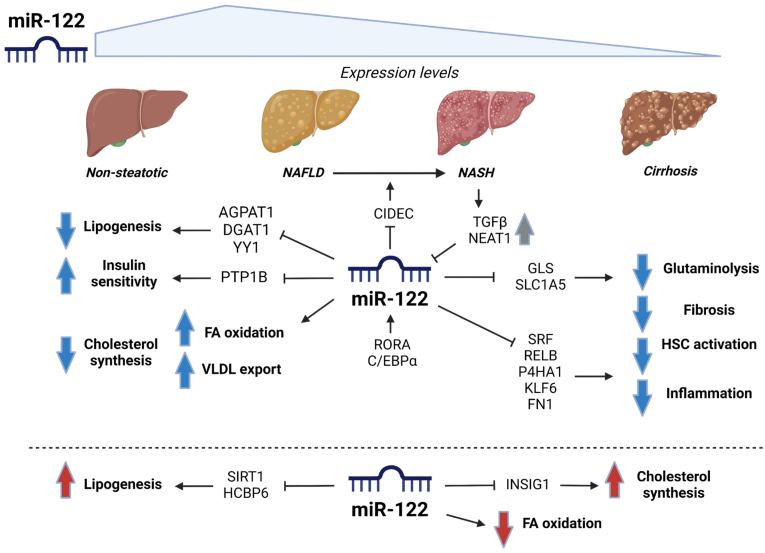
miR-122 is the most abundant hepatic miRNA species [56] and plays a fundamental role in many aspects of liver physiology [57]. Human expression studies suggest that hepatic miR-122 levels increase during early NAFLD development and then gradually decrease with progression to NASH and with advancement of fibrosis (Figure 3). Increased miR-122 levels were reported in simple steatosis vs. the healthy liver [58], and in severe vs. mild steatosis [59], whereas levels of miR-122 were reduced in livers of people with obesity and NAFLD compared to non-steatotic controls [31], in NASH compared to “simple” steatosis [58] or healthy controls [15] and in severe fibrosis compared to mild fibrosis [59].
Figure 3.
miR-122 in NAFLD/NASH. Expression levels of miR-122 were found to increase during early NAFLD development but decline with progression towards NASH and cirrhosis. miR-122 expression is controlled by pathways involving the transcription factors RORA and C/EBPα and the pro-fibrogenic factors TGF-β and NEAT1. Data regarding the role of miR-122 in early NAFLD are conflicting: miR-122 was shown to target both positive (AGPAT1 and DGAT1) and negative (SIRT1, HCBP6, YY1 and Insig1) regulators/effectors of lipogenesis and to affect hepatic lipid metabolic pathways (cholesterol synthesis, VLDL export and fatty acid (FA) oxidation) in both a pro- (lower part of figure) and anti-steatotic (upper left part of figure) direction. miR-122 was found to contribute to maintaining insulin sensitivity by targeting PTP1B, a negative regulator of insulin signalling. Loss of miR-122 has been implicated in fibrosis development by de-repression of the targets FN1 (a fibrosis constituent) and SRF and KLF6 (pro-fibrogenic transcription factors), P4HA1 (a collagen maturation enzyme) and GLS/SLC1A5 (effectors in the glutaminolytic pathway, a process progressively deregulated with NASH fibrosis). miR-122 was shown to protect against hepatic inflammation by targeting RELB, a member of the NF-κB family of transcription factors. Finally, miR-122 was found to target CIDEC, a factor associated with NAFLD to NASH progression. Grey arrows: Regulation of upstream factors found to control miR-122 expression in NAFLD/NASH. Blue/red arrows: Increased or decreased activities in pathways/processes as a result of miR-122 actions, leading to favourable (blue) or unfavourable (red) effects on liver metabolic function. Note that the downregulation of miR-122 levels with NAFLD progression leads to opposite effects of those illustrated.
3.2.2.2. Is miR-122 a pro- or an anti-steatotic miRNA?
Conflicting results have been reported regarding the consequences of experimental miR-122 ablation and the role of miR-122 as an inducer or repressor of steatosis. Liver-specific- and germline Mir122a knockout (KO) mice developed hepatic steatosis, which progressed to steatohepatitis and fibrosis and ultimately to development of HCC-like, malignant tumours with age [60,61]. Re-establishment of miR-122 expression reduced disease symptoms and tumorigenesis [61]. The steatotic Mir122a−/− livers were characterized by increased synthesis and decreased secretion of triglyceride in very-low-density lipoprotein (VLDL) as well as by alterations in several genes involved in fatty acid and triglyceride synthesis and VLDL export [60,61]. These results suggest that miR-122 plays a protective role against NAFLD development. Studies assessing the effects of transient miR-122 inhibition either suggested a pro- [62,63] or an anti-steatotic [64] function of miR-122 in the liver: miR-122 antisense oligonucleotide (ASO) treatment in lean mice activated hepatic AMPK and β-oxidation and reduced fatty acid and cholesterol synthesis. Moreover, miR-122 ASO reduced liver steatosis and expression of lipogenic genes in DIO mice and reduced plasma cholesterol levels in both groups of mice [62]. In line with these findings, antagomiR-122 injection in mice was shown to reduce levels of plasma cholesterol as well as expression of cholesterol biosynthesis genes and HMGCR activity in the liver [63]. AntagomiR-122 injection in mice increased liver triglyceride content, paralleled by a decreased fatty acid oxidation [64]. Conversely, pharmacological activation of hepatic miR-122 expression and secretion in two DIO mouse models with a novel agonist of the RAR Related Orphan Receptor A (RORA, a nuclear receptor shown to stimulate miR-122 expression in a free fatty acid-dependent manner [64]) resulted in reduced hepatic steatosis [65]. Likewise, studies of miR-122 inhibition or overexpression in vitro, omitting potential extrahepatic factors, also yielded conflicting results, either reporting a pro- [66] or an anti-steatotic [15,67] function of miR-122 in hepatocytes.
3.2.2.3. Targets of miR-122 in hepatic lipid metabolism
Several direct miR-122 targets have been identified in lipid metabolic pathways and the upstream signalling cascades controlling their activity. Among these are the triglyceride biosynthesis enzymes 1-Acylglycerol-3-Phosphate O-Acyltransferase 1 (AGPAT1) [64,68] and Diacylglycerol O-Acyltransferase 1 (DGAT1) [64] (Figure 3). Cell Death Inducing DFFA Like Effector C (CIDEC), a protein associated with lipid droplets and recently implicated in the progression of NAFL to NASH [69], was also found to be a direct miR-122 target [60] (Figure 3). Downregulation of miR-122 expression in livers of DIO mice in vivo and TNFα-treated hepatocytes in vitro induced insulin resistance via de-repression of a direct miR-122 target Protein Tyrosine Phosphatase 1B (PTP1B), one of the negative regulators of insulin signalling [70]. Other upstream controllers of lipid metabolism identified as direct miR-122 targets are SIRT1 (upstream negative regulator of lipogenic genes via the Liver Kinase B1 (LKB1)/AMPK axis) [66] and Yin Yang 1 (YY1, transcriptional repressor of the FXR/SHP axis which inhibits lipogenesis) [67] (Figure 3). In addition, miR-122 represses expression of Hepatitis C virus Core-Binding Protein 6 (HCBP6), which acts upstream of Sterol Regulatory Element-Binding Protein (SREBP)-1 and Fatty Acid Synthase (FASN) to control lipid synthesis [71]. Sirtuin 6 (SIRT6) is a key miR-122 target in the liver, where the two factors inhibited each other’s expression in a negative feedback loop to fine-tune fatty acid β-oxidation [72]. miR-122 may also stimulate cholesterol synthesis by a non-canonical mechanism where both its mature and precursor forms repress synthesis of the inhibitor Insulin-induced gene 1 (Insig1) protein by modulating polyadenylation site usage in its transcript [73].
3.2.2.4. miR-122 in NASH pathogenesis
Many studies have causally implicated miR-122 expression changes in the mechanisms governing NAFL to NASH progression. miR-122 downregulation contributes to NASH-related fibrosis by affecting pathways of HSC activation [[74], [75], [76]]. The long non-coding RNA Nuclear Paraspeckle Assembly Transcript 1 (NEAT1) is upregulated in HSCs from livers of mice with chemically induced liver fibrosis. NEAT1 contributes to HSC activation via repression of miR-122 expression and hence de-repression of its target Krüppel-Like Factor 6 (KLF6), a pro-fibrotic transcription factor involved in HSC activation via stimulation of TGF signalling (Figure 3). Importantly, analysis of gene expression in human cirrhosis liver biopsies showed the same pattern of NEAT1/KLF6 upregulation and mir-122 downregulation, suggesting that the NEAT1/miR-122/KLF6 axis also contributes to fibrogenesis in human NASH [75]. miR-122 expression was also found to be suppressed by TGF-β signalling in HSCs and fibrogenic fibroblasts [76]. Moreover, miR-122 had an anti-fibrotic function in these cells by repressing expression of fibrosis-related genes Fibronectin 1 (FN1, direct miR-122 target), alpha Smooth Muscle Actin (α-SMA) and Type 1 Collagen α1 (COL1A1) (both indirect miR-122 targets via repression of the key fibrogenic transcription factor Serum Response Factor, SRF). Transcription factor CCAAT/Enhancer-Binding Protein α (C/EBPα) was shown to control miR-122 expression in murine HSCs, and miR-122 negatively controlled collagen production via direct repression of subunit alpha-1 of Prolyl 4-Hydroxylase (P4HA1), a key collagen maturation enzyme [74].
Hepatic glutamine metabolism is progressively de-regulated in NASH with fibrosis severity, and it has been suggested that this de-regulation, with increased glutaminolysis as a key characteristic, is a causal mechanism of fibrogenesis [77]. Progressive loss of miR-122 may be one of the mechanisms behind these changes, as loss of miR-122 expression in mice enhances glutaminolysis, whereas miR-122 overexpression in cultured hepatocytes blocks this pathway. Two important glutaminolytic enzymes, GLS and SLC1A5, were also upregulated in HSCs of fibrotic mouse livers and in human NASH livers [77] and directly targeted by miR-122 [78] (Figure 3). However, while myofibroblasts primarily account for the increased glutamine breakdown in fibrotic livers [77], studies of the role of miR-122 in glutamine metabolism was only made on whole livers in vivo and hepatocytes in vitro [78]. Therefore, it remains to be determined whether miR-122 affects the fibrogenic phenotype of HSCs/myofibroblasts via regulation of glutamine metabolism in these cells.
Two recent studies provided evidence of miR-122 as a regulator of NASH-associated liver inflammation [60,61]. miR-122 KO mice had an elevated production of pro-inflammatory cytokines and chemokines in hepatocytes, and RELB, a member of the Nuclear Factor-κB (NF-κB) transcription factor family, was shown to be a direct miR-122 target [68]. NF-κB signalling is involved in a range of processes related to NASH progression, including hepatocyte death, inflammatory responses in Kupffer cells and HSCs and activation and survival of HSCs [79]. Studies in humans and in mouse models have suggested that cholesterol acts as a lipotoxic molecule in NASH pathogenesis [80]. miR-122 may be involved, as cholesterol loading of liver cells in vitro stimulated release of exosomes and increased the exosomal level of miR-122 relative to that of other miRNAs. Treatment of in vitro-differentiated macrophages with the secreted exosomes promoted an M1 pro-inflammatory phenotype of the cells, an effect which was blunted if exosomes were derived from anti-miR-122 treated hepatocytes [81].
3.2.3. miR-21
3.2.3.1. Does miR-21 promote hepatic steatosis?
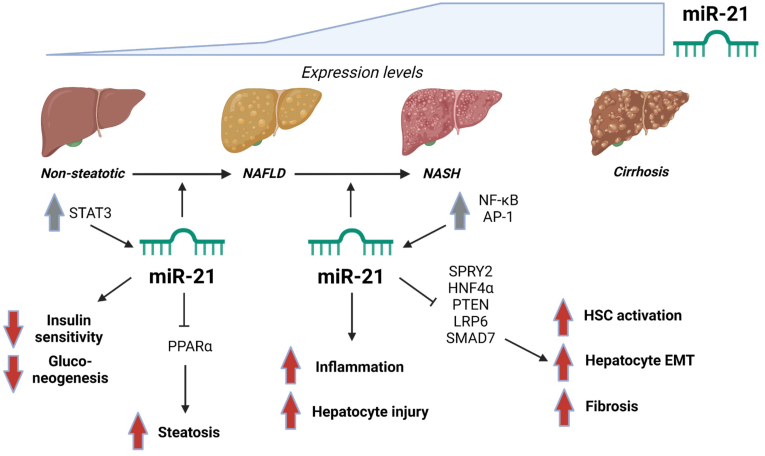
Some studies suggest that miR-21 contributes to changes in liver energy metabolism associated with early NAFLD [82,83] (Figure 4). A mechanistic insight into hepatocyte miR-21 upregulation during steatosis development was provided by sodium oleate treatment of cultured hepatocytes. Stimulation induced vesicular steatosis and a cell-autonomous inflammatory response leading to phosphorylation, activation and differential binding of the transcription factor Signal Transducer and Activator of Transcription 3 (STAT3) to promoters of several NAFLD-regulated genes, including miR-21 and miR-122. Conversely, STAT3 inhibition restored miRNA expression changes and strongly reduced sodium oleate-induced steatosis [82]. Using germline and hepatocyte-specific conditional miR-21 KO mice, it was shown that hepatocyte miR-21 is kept inactive under normal physiological conditions [83]. Upon metabolic stress by HFD feeding, miR-21 becomes activated and promotes development of hepatic steatosis and insulin resistance and impedes gluconeogenesis, likely by affecting expression of key upstream regulators and downstream enzymes of lipid and glucose metabolic pathways [83]. In contrast to these reports, other studies suggest that miR-21 is important for major pathogenic processes related to later NAFLD stages rather than for steatosis development [84,85] (Figure 4). MiR-21 ablation by pharmacological inhibition or KO in different mouse models of NASH-associated liver damage did not affect the degree of hepatic steatosis but rather reduced NASH features such as hepatocyte injury, inflammation and fibrosis [84]. In partial agreement, miR-21 KO methionine-choline deficient (MCD) diet-fed mice displayed markedly reduced levels of steatosis compared to control mice on the same diet [85]. Thus miR-21 might play a major role in progression rather than initiation of NAFLD. However, despite the MCD diet being frequently used to recapitulate human NASH in mice, the metabolic profile of MCD diet-fed mice is very different from that of people with NASH [1,85]. Therefore, miR-21 function was also assessed in fast-food diet-fed mice, a model more closely mimicking metabolic and histopathological features of human NASH [1]. miR-21 KO in this model led to a slight reduction in hepatic steatosis, which was more pronounced when mice were treated with obeticholic acid [85] – one of several investigational FXR agonists under clinical development for the treatment of NASH [1,8]. miR-21 was found to affect hepatic lipid metabolism through PPARα inhibition [84,85], and to directly target PPARα in human liver cells [86]. By inhibiting expression of PPARα and likely a range of other targets, hepatocyte miR-21 may assist other miRNAs, including miR-34a, in promoting steatogenesis during early human NAFLD development.
Figure 4.
miR-21 in NAFLD/NASH. Expression levels of miR-21 were found to increase with NAFLD progression. miR-21 expression is controlled through pathways involving STAT3, NF-κB and AP-1. Some studies demonstrate a role of miR-21 in early NAFLD pathogenesis, where PPARα is a direct miR-21 target, while other studies suggest that miR-21 is involved in later NAFLD stages only. miR-21 was shown to decrease hepatic insulin sensitivity and gluconeogenesis and to promote steatosis. miR-21 has also been implicated in NAFLD progression to NASH by inducing fibrosis via hepatic stellate cell (HSC) activation and epithelial–mesenchymal transition (EMT) of hepatocytes by repression of SPRY2, HNF4α, PTEN, LRP6 and SMAD7. Some studies point to a central role of miR-21 in aggravating hepatocyte injury as well as liver inflammation and fibrosis, while other studies suggest that miR-21 might be dispensable for these processes. Grey arrows: Regulation of upstream factors found to control miR-21 expression in NAFLD/NASH. Red arrows: Increased or decreased activities in pathways/processes as a result of miR-21 actions, leading to unfavourable effects on liver metabolic function and progression of NAFLD.
3.2.3.2. miR-21 in NASH pathogenesis
Human liver expression profiling studies suggest a function of miR-21 in NASH pathogenesis. While hepatic miR-21 expression was not significantly changed in early stages of human NAFLD [35,84], miR-21 levels were upregulated in human NASH [15,35,84,85,87] and cirrhosis [88] compared to healthy controls and/or people with simple steatosis (Figure 4). Moreover, examination of experimental mouse NASH and fibrosis models or liver biopsies from people with NASH have shown that miR-21 is upregulated primarily in non-parenchymal liver cells, including HSCs [88,89], inflammatory cells and biliary cells [84].
miR-21 contributes to fibrosis in the liver by activating HSCs via several molecular pathways. In human cirrhosis livers and rats with chemically induced liver cirrhosis, miR-21 induced HSC activation and EMT of hepatocytes to myofibroblasts by activating Extracellular Signal Regulated Kinase (ERK)-signalling via repression of two target genes Sprouty2 (SPRY2, inhibitor of receptor tyrosine kinase signalling) and HNF4α [90]. miR-21 mediates activation of HSCs in vitro induced by the pro-fibrotic agent Platelet-Derived Growth Factor (PDGF)-BB, possibly by repressing Phosphatase and Tensin Homolog (PTEN) expression and thereby activating Phosphoinositide 3 (PI3) kinase/AKT signalling, a known pro-fibrotic signalling pathway [91]. miR-21 is also implicated in Wnt/β-catenin signalling, another pathway involved in liver fibrosis [92], as miR-21 inhibited expression of a co-receptor for the classical Wnt/β-catenin signalling pathway, Low-density Lipoprotein-Related Receptor 6 (LRP6), in livers of MCD-fed mice [93]. It was suggested that miR-21 stimulates fibrosis by activating TGF-β signalling, possibly via SMAD7 repression [88], and that miR-21 maintains its upregulation in activated HSCs via a feedback loop involving the Programmed Cell Death Protein 4 (PDCD4) and the transcription factor Activation protein-1 (AP-1), which induces miR-21 transcription [88] (Figure 4). A more recent study showed that miR-21 expression in NASH is induced by a NADPH Oxidase 2 (NOX-2)- and NF-κB-dependent mechanism and promotes HSC activation and fibrogenesis via activation of the TGF-β pathway [87] (Figure 4). In contrast to the reports of hepatic miR-21 as an important pro-fibrotic factor in NASH pathogenesis, another study demonstrated that miR-21 was dispensable for HSC activation and liver fibrosis [89]. Despite miR-21 being the most upregulated miRNA in HSCs isolated from the livers of several mouse models of liver fibrosis, neither miR-21 KO nor acute miR-21 inhibition in these mice affected the degree of liver fibrosis or HSC activation. Moreover, experiments involving HSC-specific deletion of Dicer, one of the enzymes responsible for miRNA maturation, suggested that miRNAs are dispensable for HSC activation and liver fibrosis [89].
3.3. Novel miRNAs linked to NAFLD pathogenesis
Several recent studies disclose a mechanistic role in progressive NAFLD of novel hepatic miRNA species with relevance for human fatty liver disease. A negative feed-back loop was identified between miR-26a and the PKR-like ER Kinase (PERK)-orchestrated ER stress response pathway [94]. PERK inhibits the downstream target eukaryotic Initiation Factor 2α (eIF2α) to halt translation and hence alleviate ER workload and restore cellular homeostasis. MiR-26a expression was downregulated in liver biopsies from people with NAFLD, whereas markers of ER stress were upregulated. Extensive miR-26a loss- and gain-of-function experiments in mice suggested that in the healthy liver, miR-26a expression is induced by ER stress, and miR-26a functions to alleviate this condition by targeting eIF2α. However, during prolonged metabolic stress such as in NAFLD, miR-26a expression is repressed, possibly by a post-transcriptional mechanism, thereby worsening ER stress and fueling its adverse effects on liver metabolism [94]. miR-378 has been implicated in the pathogenic mechanisms of both early and later NAFLD stages [[95], [96], [97], [98]]. Liver miR-378 expression is upregulated in DIO mice as well as in human NAFLD/NASH [96,97]. LXRα was further shown to uncouple miR-378 expression from that of its host gene PPAR gamma coactivator 1-β (PGC1β) - a positive regulator of mitochondrial function and fatty acid oxidation - by activating miR-378 transcription but repressing PGC1β transcription [96]. Moreover, previous studies in mice provided evidence that miR-378 also directly targets the transcription factor and mitochondrial regulator Nuclear Respiratory Factor 1 (NRF1) and thereby inhibits fatty acid oxidation [98]. Accordingly, upregulated miR-378 in the liver may aid its own transcriptional activator LXRα in promoting development of hepatic steatosis by repressing key controllers and effectors of hepatic lipid metabolic pathways [95,96,98]. Another direct target of hepatic miR-378 is PRKAG2, the gene encoding AMPK subunit γ2 [97]. MiR-378 has also been implicated in NASH pathogenesis and shown to promote hepatic inflammation, necrosis and apoptosis by activating NF-κB/TNFα inflammatory signaling through repression of AMPK and the downstream p65 NF-κB inhibitor SIRT1 [97]. miR-144 expression was recently shown to increase specifically in liver macrophages and hepatocytes from obese and insulin-resistant mice and humans [99,100] and to impair the antioxidant response to hepatic lipid accumulation by targeting a master mediator of this process, Nuclear factor erythroid 2-related factor (NRF2). miR-144 targeting of NRF2 function was found to occur both directly by downregulation of NRF2 protein levels [99] and indirectly by repression of Immuno-responsive gene 1 (IRG1) and a resulting shift in levels of tricarboxylic acid cycle metabolites and downstream inhibition of NRF2 activity [100]. Interestingly, the transcription factor GATA-binding protein 4 (GATA4) was found to drive the increased transcription of miR-144 in livers of obese insulin-resistant individuals compared to lean controls, likely induced by ERK signaling [99].
3.4. miRNAs mediating hepatic cell–cell crosstalk in NAFLD
An inverse directionality of changes in hepatic and circulating levels has been observed with NAFLD progression for many miRNA species, including miR-122, miR-192 and miR-375 [35,58]. These miRNAs may be among the exomiRs mechanistically involved in NAFLD pathogenesis both inside and outside the liver. miRNAs released from stressed hepatocytes might promote NAFLD progression by acting on specific molecular pathways in non-parenchymal liver cells. EVs released upon palmitate treatment of cultured hepatocytes were shown to be internalised into HSCs and induced a phenotypical switch from the quiescent to the activated myofibroblastic state. This was found to be through EV-derived miR-128 directly targeting and downregulating PPARγ, a key factor in the maintenance of the quiescent HSC state [101]. Exosomes secreted from palmitate-treated hepatocytes were also found to contain a different panel of miRNAs than those isolated from vehicle-treated cells, including a five-to ten-fold increase in the levels of miR-34a, miR-122 and miR-192. Treatment of cultured HSCs with these exosomes or with a miR-192 mimic enhanced the expression of several fibrosis markers, suggesting that miR-192-containing exosomes from hepatocytes activate HSCs in response to lipotoxicity [102]. Recently, serum miR-192 levels were found to correlate positively with markers of hepatocellular injury, steatosis and ballooning degeneration as well as lobular inflammation in human NAFLD. Exosomes isolated from people with NASH, high-fat high-cholesterol fed rats (model of progressive NAFLD [1]) or palmitate-treated cultured human hepatoma cells carry markers of hepatocyte origin and contain higher levels of miR-192 than those isolated from control groups. Moreover, exomiR-192 promotes expression of inflammatory cytokines and M1 (pro-inflammatory) activation of macrophages in culture by repressing Rapamycin-insensitive companion of mTOR (RICTOR) and hence activating downstream Forkhead Box O1 (FOXO1) signalling [103]. miRNAs also appear to mediate inflammatory cell-to-hepatocyte crosstalk in NAFLD. miR-223, an anti-inflammatory, granulocyte-specific miRNA elevated in liver biopsies from people with NASH and HFD-fed mice, has a hepatoprotective role by limiting NAFLD to NASH/HCC progression [104,105]. Interestingly, miR-223 is transferred to hepatocytes via EVs released from neutrophils infiltrating the NASH-affected liver and is internalized selectively by a Low-density lipoprotein receptor (LDLR)- and Apolipoprotein E (APOE)-dependent mechanism [106]. Within the hepatocytes, miR-223 acted to inhibit inflammatory and fibrogenic gene expression and thus ameliorated NASH [106].
3.5. exomiRs may mediate organ crosstalk in NAFLD
Metabolic homeostasis is achieved by a complex interplay between organs, and derangements in this inter-organ communication is a key factor contributing to metabolic disease [107]. The adipose tissue compartment might be an extra-hepatic component of the pathogenesis of progressive NAFLD. This is not merely due to its function as a main site for lipid storage, but also due to the release of soluble factors affecting hepatic energy metabolism. The composition of factors released from the adipose tissues may change during prolonged metabolic stress associated with conditions like obesity and NAFLD, which may have deleterious effects on liver metabolism [8,108]. MiRNAs could be among these adipose tissue-derived factors, as expression levels of several miRNAs, including miR-122, miR-192, miR-27a and miR-375, were changed in exosomes from DIO vs. lean mice [109]. The liver and epididymal white adipose tissues were potential sources of these exomiRs, since miRNA expression changes in these tissues reflected changes in exosomal miRNA content. Interestingly, treatment of lean mice with exosomes isolated from obese mice or exosomes from lean mice transfected with mimics of the four miRNAs adversely affected whole-body metabolism, with induction of hepatic steatosis by the latter approach [109]. EV-mediated adipose tissue-to-liver communication appears to be bi-directional, as lipid overload in HFD-fed mice induces the release of hepatocyte-derived miRNA-containing EVs which, in turn, regulate adipogenesis and lipogenesis in the adipose tissues [110]. ExomiRs released from adipose tissues in mice, and possibly also in humans, can affect gene expression in the liver: a transgenic human exomiR, hsa-miR-302f, released from adipocytes of donor mice repressed the luminescent signal from recipient mouse hepatocytes engineered to express a 3′UTR reporter containing a recognition site specific for hsa-miR-302f [111]. Adipose tissue-derived macrophages (ATM) from obese mice secrete exosomes with an altered miRNA content, such as elevated miR-155 level, compared to lean mice. Administration of ATM-derived exosomes from obese mice to lean recipient mice caused glucose intolerance and insulin resistance, and conversely, obese mice demonstrated improved metabolic health when receiving ATM-derived exosomes isolated from lean mice. ExomiR-155 was taken up in vitro by both adipocytes, myocytes and hepatocytes and impaired cellular insulin actions when overexpressed [112]. The authors concluded that secretion of exomiRs from ATMs could be a novel mechanism for adipose tissue inflammation-induced insulin resistance in organs such as the liver and skeletal muscle [112].
ExomiR-122 is another circulating miRNA which may be involved in the bi-directional crosstalk between the liver and extra-hepatic tissues in progressive NAFLD [58,64,109,113,114]. While hepatic miR-122 levels are decreased, circulating miR-122 levels are elevated in human NAFLD and appear to increase with disease progression [35]. miR-122 was discovered close to the plasma membrane of lipid-laden hepatocytes in liver biopsies from people with NAFLD, suggesting that this miRNA was ready for export into the circulation [58]. More recently, free fatty acids were shown to induce active miR-122 secretion from the liver [64]. MiR-122 secreted from cultured hepatocytes repressed a miR-122-sensitive luciferase reporter and impaired lipid-droplet formation in cultured mouse myoblasts. Results further suggested that liver-secreted miR-122 can affect energy metabolism in peripheral tissues such as skeletal muscle [64]. Liver-derived exomiR-122 may promote development of metabolic cardiomyopathy by negatively affecting myocardial mitochondrial function via repression of ADP-ribosylation factor-like 2 (ARL2) [114]. A recent review proposed a model to explain the apparent paradox of increasing circulating but decreasing intrahepatic levels of miR-122 with NAFLD progression [113]: In NAFLD, other tissues such as the adipose tissues may start to produce and secrete miR-122, thereby aiding the failing liver in keeping up circulating miR-122 levels. With progressive adipose tissue dysfunction, this compensation will eventually fail, and a systemic lack of miR-122 will aggravate liver fibrosis and favour HCC development [113]. Hence, circulating miRNAs may both contribute to and delay NAFLD progression based on cellular origin and specific metabolic conditions.
4. Clinical utility of miRNAs in NAFLD
4.1. Circulating miRNAs as NAFLD biomarkers
Extracellular miRNAs have proven useful as biomarkers of specific human diseases due to the distinct signatures of miRNAs originating from different cell types under specific physiological and pathophysiological conditions [29]. Indeed, miRNA profiling in blood samples has been examined thoroughly for its potential to improve NAFLD/NASH diagnostics. Several recent meta-analyses have been performed to evaluate the status in this field. A systematic literature review of publications from 2008 to early 2018 was performed to make a meta-analysis of the numerous published studies that evaluate circulating miRNA expression profiles in various human NAFLD stages. The results showed that elevated miR-122, miR-34a and mir-192 appeared to be useful in distinguishing NASH from NAFL. Sufficient data were only available to evaluate the diagnostic accuracy for miR-122 and miR-34a, which was only moderate in terms of distinguishing NAFLD from the healthy control state and NASH from NAFL, respectively. Too few studies were available to examine which miRNAs could distinguish fibrosis stages in NASH [35]. A more recent systematic literature review and meta-analysis evaluated the use of serum miRNA profiles to diagnose total NAFLD, NAFL and NASH and assessed the diagnostic efficacy of the three miRNAs with the largest sample size, miR-34a, miR-122 and miR-99a, as serological biomarkers for total NAFLD. They found that overall, serum miRNAs showed better diagnostic efficacy for NASH than for NAFLD and had a high accuracy for discriminating NASH from NAFL. While both miR-34a, miR-122 and miR-99a were moderately performing in terms of diagnosing total NAFLD, miR-34a showed the lowest heterogeneity and the most stable efficacy. However, miR-34a was not evaluated separately for its diagnostic efficacy for NASH [115].
Decreased plasma content of miR-122 is a risk factor for mortality in people with NAFLD [116]. The differential relationship between circulating miR-34a, miR-122, miR-192 and miR-200a and histological features and pathogenic factors of NAFLD was recently assessed. MiR-34a, miR-122 and miR-192 were shown to independently associate with hepatic steatosis as well as with pathogenic factors such as insulin resistance and known NAFLD-predisposing polymorphisms. MiR-200a was only additionally associated with a single pre-disposing genetic variant [117]. All four miRNAs showed a strong, increasing association with progressive fibrosis. The predictive value of miR-34a alone or all four miRNAs combined was compared to that of FIB-4 (a validated, though only moderately performing score for liver fibrosis [118]). While miR-34a and the four miRNAs combined predicted stage 4 with lower and stage 3+ with similar sensitivity and specificity as FIB-4, the miRNAs outperformed FIB-4 in predicting early stages of fibrosis. However, the diagnostic accuracy was still only moderate [117]. Thus, although miR-34a, miR-122 and miR-192 are among the best miRNA candidates for minimally invasive biomarkers of NAFLD progression, their combined diagnostic performance is insufficient in terms of accuracy and discriminative value regarding different types of liver disease or specific NAFLD stages. This was also the conclusion drawn by a recent review on miRNAs as biomarkers of human NAFLD [119].
Development and implementation of techniques to selectively isolate circulating EVs of hepatic origin may aid identification of a panel of miRNA biomarkers for accurate NAFLD diagnosis and staging. Indeed, a method for selective isolation of hepatocyte-derived EVs from human plasma was recently developed [120]. As a first approach to utilise this method for NAFLD diagnostics, it was used for hepatocyte-derived EV isolation from plasma samples from healthy people and people with NAFL or NASH, followed by miRNA isolation [121]. Levels of three selected miRNA species, miR-122, miR-192 and miR-128, were significantly increased in hepatocyte-derived EVs from plasma of people with NASH compared to controls. Moreover, they trended positively with disease severity and showed improved performance in terms of distinguishing NAFLD from controls compared to the same miRNAs isolated from total plasma cell-free RNA or global plasma-derived EVs [121]. However, the utilised method isolates EVs of hepatocyte origin only [120]. Moreover, only three selected miRNA species were assessed in a small cohort of people (n = 6–14) for which very limited clinical data were available. Development of techniques to selectively capture EVs from other cell and tissue types involved in NAFLD pathogenesis, combined with miRNome-wide screening for the best candidate marker species, may identify a miRNA panel with high diagnostic efficacy, thereby improving NAFLD diagnostics. Using a diagnostic algorithm, NIS4™, which combines quantification of miRNAs with that of other circulating factors associated with NAFLD, improves the diagnostic and prognostic value of circulating miRNAs in NAFLD [122]. NIS4 quantifies a panel of four factors in blood (miR-34a, the two fibrosis markers alpha-2 macroglubulin (A2MG) and Chitinase-3-like protein 1 (YKL-40) and the long-term glycaemic control marker Haemoglobin A1c). In a large (N > 700 patients) pooled validation cohort, the diagnostic performance of NIS4 in terms of identifying NASH with a high risk of long-term severe liver outcomes among individuals with metabolic risk factors was compared to that of six non-invasive tests already used in the clinic. NIS4 significantly outperformed five of these tests and accurately identified at-risk cases independently of age, sex and body mass index (BMI), making it a promising candidate for replacing liver biopsy for at-risk patient identification across the broad spectrum of NASH and fibrosis [122]. Another possible utilization of circulating miRNAs in the clinic would be to combine quantification of plasma-derived miRNAs with the use of standard non-invasive diagnostic methods (reviewed in [6]) to refine NAFLD/NASH diagnosis and fibrosis staging.
4.2. miRNAs as targets of NASH pharmacotherapies
miRNAs are attractive pharmacotherapeutic targets due to their capacity to simultaneously control multiple downstream pathways in a cell- and tissue-specific manner. Several pharmaceutical companies are developing miRNA-targeting agents to treat specific human diseases. This is accomplished with chemically stabilised oligonucleotides, either mimicking one or more specific miRNA species suppressed by the progression of disease or in the form of AntagomiRs to impede target interactions by miRNAs overexpressed and/or mechanistically involved in disease pathogenesis [123]. Various strategies for delivering such therapeutics to specific cell types exist [124]. Conjugation with the Asialoglycoprotein receptor ligand N-Acetylgalactosamine (GalNAc) is a widely used approach to specifically target oligonucleotides to hepatocytes [125]. Other delivery systems have been used in non-clinical experiments in vitro and in vivo to specifically target oligonucleotides to HSCs or Kupffer cells [126] and could, in the future, be used for therapeutic oligonucleotides mimicking or silencing miRNA species involved in NASH and/or fibrosis to treat more advanced NAFLD stages. To date, the only miRNA-targeting agent that has made it to clinical development for the treatment of NASH in type 2 diabetes/pre-diabetes is AZD4076 (RG-125) [127]. RG-125 is an GalNAc-conjugated oligonucleotide that represses mir-103/miR-107, two homologous miRNA species linked to insulin resistance and NAFLD [128]. However, RG-125 development was halted in phase II in 2017 after serious adverse events were reported for one of the other miRNA-targeting agents under clinical development by the company alliance behind RG-125 [129].
5. Future perspectives for understanding and utilizing miRNAs in NAFLD treatment
A growing body of literature endorses the concept of miRNAs as contributors in human NAFLD pathogenesis. MiRNAs are not only key to unravelling the mechanisms behind the evolution of the disease but also appealing as biomarkers for NAFLD diagnosis and staging or as targets of pharmacotherapies to arrest or reverse disease progression. However, numerous challenges in the field remain to be overcome, a few of which will be discussed in the following sections.
A limited number of global miRNA profiling studies in human NAFLD have been reported, characterizing hepatic miRNA expression changes in early or later NAFLD stages [15,30,31,33]. To the best of our knowledge, only a single study has sought to cover the broad spectrum from NAFL through NASH to cirrhosis [34]. Distinct sets of miRNA species were found differentially expressed in these studies. For instance, the three miRNA species in focus in this review due to their identified mechanistic role in progressive NAFLD, miR-34a, miR-122 and miR-21, were each only found by a few of the studies. These divergent findings may be due to differences in NAFLD stages examined, the relatively small cohort sizes, as well as to the use of different miRNA detection platforms, in most cases limited to pre-selected miRNA species. Importantly, inter-individual clinical variability likely contributes to differences in findings between the studies, given the fact that NAFLD is a highly heterogenous condition in terms of pre-disposing genetic factors and metabolic risk factors such as metabolically unhealthy obesity, insulin resistance and subclinical inflammation [[2], [3], [4], [5]]. It is possible that distinct, yet possibly partially overlapping, sets of microRNA species are involved in NAFLD pathogenesis in groups of people with specific genetic backgrounds and metabolic phenotypes. Unbiased profiling studies are needed to identify the miRNA species that are consistently differentially expressed and hence most likely to contribute to the pathophysiological changes in early, intermediate, and late human NAFLD stages. These should compare metabolically well-characterized individuals (in larger cohorts to increase statistical power) across the spectrum of NAFLD and use appropriate statistical analyses to adjust for potentially confounding clinical variability among individuals. For this, platforms such as RNA sequencing that allow sufficiently sensitive detection of any expressed miRNA species must be employed. Moreover, standard methods for normalization of miRNA expression data obtained using different techniques remain to be established [130]. Post-screening functional analysis of differentially expressed miRNA species should include liver cells of human origin, preferably non-immortalised cells, given the fact that many NAFLD-linked miRNAs also function as oncomiRs or tumour suppressors and are associated with HCC [131,132]. Similar methodological adjustments would aid identification of extracellular miRNAs important for interorgan crosstalk in NAFLD.
Animal loss- and gain-of function studies combined with experiments assessing miRNA function in vitro have shed light onto how single deregulated miRNA species contribute to the pathogenesis of progressive NAFLD and identified specific downstream targets involved. However, miRNAs function as part of highly complex gene-regulatory networks where a single miRNA species has the potential to target hundreds of gene transcripts, each of which may contain recognition sites for a multitude of different miRNAs [[133], [134], [135]]. Moreover, miRNAs often function in nodes where several miRNA species, belonging to the same or different miRNA families, cooperatively target common downstream genes and pathways [135,136]. Among miRNA targets are also transcription factors, often controlling expression of other miRNA species or even that of their own targeting miRNA, forming functional feedback loops [72,88,94,98]. Accordingly, gradual changes in expression of a multitude of genes, among these clusters of miRNAs as well as their upstream regulators and downstream targets, are what drives the development of complex diseases such as progressive NAFLD. The progression of NAFLD varies widely between individuals [2], and the pattern of gene expression changes taking place during its course expectedly depends on various factors such as genetic background, nutritional factors and coexistence of various metabolic comorbidities [2,8]. Likewise, animal models used to assess miRNA contributions to NAFLD pathogenesis differ in genetic background and diet as well as method for NAFLD induction and consequently differ in the degree of resemblance to human NAFLD/NASH [1]. This notion may explain the discrepancies between experimental animal studies assessing the contribution of specific miRNA species to NAFLD pathogenic processes, for instance the studies of miR-21 or miR-122 reviewed above, and it calls for future research on carefully controlled experimental models to replicate specific human NAFLD stages. The use of rodent models to study the functional role of specific miRNA species in human NAFLD must also consider the degree of conservation of those miRNA species between the employed model organism and humans. Importantly, what matters in this respect is the degree to which a potential miRNA−target interaction is conserved in the cellular context under investigation. Moreover, functional redundancy of a specific miRNA−target interaction may exist in the experimental model organism, meaning that additional miRNA species may need to be depleted before any effects of the miRNA inhibition can be observed.
Most studies addressing the involvement of miRNAs in NAFLD pathogenesis have focused on the downstream effects of miRNA actions rather than the upstream regulatory mechanisms responsible for gain or loss of miRNA expression during disease progression. An important goal for future studies will be to expand our knowledge of the intricate gene regulatory networks and the upstream mechanisms of their de-regulation in NAFLD by identifying key transcription factors driving global expression changes of miRNAs and downstream protein targets at clearly defined early, intermediate and late NAFLD stages. Investigation of a hitherto unexplored ground of the cis-regulatory DNA elements, i.e., enhancers, transcriptionally controlling miRNA expression changes in human NAFLD will be at the forefront of this research.
miRNAs appear to contribute to NAFLD pathogenesis not only by acting as endogenous regulators of gene expression within hepatocytes, hepatic stellate cells or Kupffer cells, but also by mediating hepatic intercellular crosstalk after release [[101], [102], [103]]. Future research should determine which circulating miRNAs are merely biomarkers of specific organ dysfunction in NAFLD and which miRNA species are mechanistically involved in intercellular and inter-organ crosstalk driving disease progression. While miRNA-biomarkers of NAFLD diagnosis may prove clinically useful in staging and identification of at-risk patients for disease progression, miRNAs found to be mechanistically involved in NAFLD – or their upstream regulators, for most miRNA species yet to be identified – may serve as targets of NASH pharmacotherapies. Since CVD remains the most common cause of death in people with NAFLD, even in NASH [2], developing a therapy which not only targets molecular pathways in the liver but also those of the cardiovascular system would be of great benefit. Identifying miRNAs mediating crosstalk between these two organ systems as well as unravelling the mechanisms of their expressional control and their downstream actions may be an important step on the path towards reaching this goal.
Author contributions
Idea and concept: MYH & RB. Literature search: MYH, MD & RB. Manuscript writing: MYH, MD & RB. Manuscript editing: MYH, MD, JTT & RB.
Acknowledgements
MD was awarded a postdoc stipend from the Danish Diabetes Academy (NNF17SA0031406). JTT is funded by the Independent Research Fund Denmark (DFF 0134-00217B). Support for this work was provided by the Novo Nordisk Foundation Center for Basic Metabolic Research (https://cbmr.ku.dk), which is an independent research center at the University of Copenhagen, and partially funded by an unrestricted donation from the Novo Nordisk Foundation (NNF18CC0034900). Figures were created with BioRender.com.
Conflict of Interest
Authors declare no conflict of interest.
Data availability
No data was used for the research described in the article.
References
- 1.Friedman S.L., Neuschwander-Tetri B.A., Rinella M., Sanyal A.J. Mechanisms of NAFLD development and therapeutic strategies. Nature Medicine. 2018;24(7):908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Younossi Z.M. Non-alcoholic fatty liver disease - a global public health perspective. Journal of Hepatology. 2019;70(3):531–544. doi: 10.1016/j.jhep.2018.10.033. [DOI] [PubMed] [Google Scholar]
- 3.Stefan N., Cusi K. A global view of the interplay between non-alcoholic fatty liver disease and diabetes. The Lancet Diabetes & Endocrinology. 2022;10(4):284–296. doi: 10.1016/S2213-8587(22)00003-1. [DOI] [PubMed] [Google Scholar]
- 4.Stefan N. Causes, consequences, and treatment of metabolically unhealthy fat distribution. The Lancet Diabetes & Endocrinology. 2020;8(7):616–627. doi: 10.1016/S2213-8587(20)30110-8. [DOI] [PubMed] [Google Scholar]
- 5.Albhaisi S., Chowdhury A., Sanyal A.J. Non-alcoholic fatty liver disease in lean individuals. JHEP Reports. 2019;1(4):329–341. doi: 10.1016/J.JHEPR.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinou E., Pericleous M., Stefanova I., Kaur V., Angelidi A.M. Diagnostic modalities of non-alcoholic fatty liver disease: from biochemical biomarkers to multi-omics non-invasive approaches. Diagnostics. 2022;12(2):407. doi: 10.3390/DIAGNOSTICS12020407. Page 407 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotter T.G., Rinella M. Nonalcoholic fatty liver disease 2020: the state of the disease. Gastroenterology. 2020;158(7):1851–1864. doi: 10.1053/j.gastro.2020.01.052. [DOI] [PubMed] [Google Scholar]
- 8.Pafili K., Roden M. Non-alcoholic fatty liver disease (NAFLD) from pathogenesis to treatment concepts in humans. Molecular Metabolism. 2020 doi: 10.1016/j.molmet.2020.101122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buzzetti E., Pinzani M., Tsochatzis E.A. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD) Metabolism. 2016;65(8):1038–1048. doi: 10.1016/j.metabol.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 10.Higashi T., Friedman S.L., Hoshida Y. Hepatic stellate cells as key target in liver fibrosis. Advanced Drug Delivery Reviews. 2017;121:27–42. doi: 10.1016/j.addr.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lebeaupin C., Vallée D., Hazari Y., Hetz C., Chevet E., Bailly-Maitre B. Endoplasmic reticulum stress signalling and the pathogenesis of non-alcoholic fatty liver disease. Journal of Hepatology. 2018;69(4):927–947. doi: 10.1016/j.jhep.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Tacke F. Targeting hepatic macrophages to treat liver diseases. Journal of Hepatology. 2017;66(6):1300–1312. doi: 10.1016/j.jhep.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 13.Huby T., Gautier E.L. Immune cell-mediated features of non-alcoholic steatohepatitis. Nature Reviews Immunology. 2021 doi: 10.1038/S41577-021-00639-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morgantini C., Jager J., Li X., Levi L., Azzimato V., Sulen A., et al. Liver macrophages regulate systemic metabolism through non-inflammatory factors. Nature Metabolism. 2019;1(4):445–459. doi: 10.1038/s42255-019-0044-9. 4 1. [DOI] [PubMed] [Google Scholar]
- 15.Cheung O., Puri P., Eicken C., Contos M.J., Mirshahi F., Maher J.W., et al. Nonalcoholic steatohepatitis is associated with altered hepatic microRNA expression. Hepatology. 2008;48(6):1810–1820. doi: 10.1002/hep.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 17.Vasudevan S., Tong Y., Steitz J.A. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 18.Bluhm B., Ehlen H.W.A., Holzer T., Georgieva V.S., Heilig J., Pitzler L., et al. miR-322 stabilizes MEK1 expression to inhibit RAF/MEK/ERK pathway activation in cartilage. Development. 2017;144(19):3562–3577. doi: 10.1242/dev.148429. [DOI] [PubMed] [Google Scholar]
- 19.Cordes K.R., Sheehy N.T., White M.P., Berry E.C., Morton S.U., Muth A.N., et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460(7256):705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hochreuter M.Y., Altıntaş A., Garde C., Emanuelli B., Kahn C.R., Zierath J.R., et al. Identification of two microRNA nodes as potential cooperative modulators of liver metabolism. Hepatology Research. 2019;49(12):1451–1465. doi: 10.1111/hepr.13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin C.-C., Liu L.-Z., Addison J.B., Wonderlin W.F., Ivanov A.V., Ruppert J.M. A KLF4-miRNA-206 autoregulatory feedback loop can promote or inhibit protein translation depending upon cell context. Molecular and Cellular Biology. 2011;31(12):2513–2527. doi: 10.1128/MCB.01189-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu H., Buchan R.J., Cook S.A. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovascular Research. 2010;86(3):410–420. doi: 10.1093/cvr/cvq010. [DOI] [PubMed] [Google Scholar]
- 23.Mortensen R.D., Serra M., Steitz J.A., Vasudevan S. Posttranscriptional activation of gene expression in Xenopus laevis oocytes by microRNA-protein complexes (microRNPs) Proceedings of the National Academy of Sciences of the United States of America. 2011;108(20):8281–8286. doi: 10.1073/pnas.1105401108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy A.J., Guyre P.M., Pioli P.A. Estradiol suppresses NF-kappa B activation through coordinated regulation of let-7a and miR-125b in primary human macrophages. Journal of Immunology. 2010;184(9):5029–5037. doi: 10.4049/jimmunol.0903463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu X., Zhang L., Wen G., Zhao H., Luong L.A., Chen Q., et al. Upregulated sirtuin 1 by miRNA-34a is required for smooth muscle cell differentiation from pluripotent stem cells. Cell Death & Differentiation. 2015;22(7):1170–1180. doi: 10.1038/cdd.2014.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X., Zuo X., Yang B., Li Z., Xue Y., Zhou Y., et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158(3):607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ørom U.A., Nielsen F.C., Lund A.H. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Molecular Cell. 2008;30(4):460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Catalanotto C., Cogoni C., Zardo G. MicroRNA in control of gene expression: an overview of nuclear functions. International Journal of Molecular Sciences. 2016;17(10):1712. doi: 10.3390/ijms17101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J., Li S., Li L., Li M., Guo C., Yao J., et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics, Proteomics & Bioinformatics. 2015;13(1):17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soronen J., Yki-Järvinen H., Zhou Y., Sädevirta S., Sarin A.-P., Leivonen M., et al. Novel hepatic microRNAs upregulated in human nonalcoholic fatty liver disease. Physiological Reports. 2016;4(1):1–14. doi: 10.14814/phy2.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Latorre J., Moreno-Navarrete J., Mercader J., Sabater M., Rovira Ò., Gironès J., et al. Decreased lipid metabolism but increased FA biosynthesis are coupled with changes in liver microRNAs in obese subjects with NAFLD. International Journal of Obesity. 2017;4121(4):620–630. doi: 10.1038/ijo.2017.21. [DOI] [PubMed] [Google Scholar]
- 32.Kleiner D.E., Brunt E.M., Van Natta M., Behling C., Contos M.J., Cummings O.W., et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41(6):1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 33.Leti F., Malenica I., Doshi M., Courtright A., Van Keuren-Jensen K., Legendre C., et al. High-throughput sequencing reveals altered expression of hepatic microRNAs in nonalcoholic fatty liver disease-related fibrosis. Translational Research. 2015;166(3):304–314. doi: 10.1016/j.trsl.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo Y., Xiong Y., Sheng Q., Zhao S., Wattacheril J., Flynn C.R. A micro-RNA expression signature for human NAFLD progression. Journal of Gastroenterology. 2016;51(10):1022–1030. doi: 10.1007/s00535-016-1178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu C.H., Ampuero J., Gil-Gómez A., Montero-Vallejo R., Rojas Á., Muñoz-Hernández R., et al. miRNAs in patients with non-alcoholic fatty liver disease: a systematic review and meta-analysis. Journal of Hepatology. 2018;69(6):1335–1348. doi: 10.1016/j.jhep.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 36.Castro R.E., Ferreira D.M.S., Afonso M.B., Borralho P.M., MacHado M.V., Cortez-Pinto H., et al. MiR-34a/SIRT1/p53 is suppressed by ursodeoxycholic acid in the rat liver and activated by disease severity in human non-alcoholic fatty liver disease. Journal of Hepatology. 2013;58(1):119–125. doi: 10.1016/j.jhep.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Lee J., Padhye A., Sharma A., Song G., Miao J., Mo Y.-Y., et al. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. Journal of Biological Chemistry. 2010;285(17):12604–12611. doi: 10.1074/jbc.M109.094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiao Y., Lu Y., Li X.Y. Farnesoid X receptor: a master regulator of hepatic triglyceride and glucose homeostasis. Acta Pharmacologica Sinica. 2015;36(1):44–50. doi: 10.1038/aps.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Y., Zhu Y., Hu S., Pan X., Bawa F.C., Wang H.H., et al. Hepatocyte miR-34a is a key regulator in the development and progression of non-alcoholic fatty liver disease. Molecular Metabolism. 2021;51 doi: 10.1016/J.MOLMET.2021.101244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding R.-B., Bao J., Deng C.-X. Emerging roles of SIRT1 in fatty liver diseases. International Journal of Biological Sciences. 2017;13(7):852–867. doi: 10.7150/ijbs.19370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shan W., Gao L., Zeng W., Hu Y., Wang G., Li M., et al. Activation of the SIRT1/p66shc antiapoptosis pathway via carnosic acid-induced inhibition of miR-34a protects rats against nonalcoholic fatty liver disease. Cell Death & Disease. 2015;6 doi: 10.1038/cddis.2015.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi S.-E., Fu T., Seok S., Kim D.-H., Yu E., Lee K.-W., et al. Elevated microRNA-34a in obesity reduces NAD+ levels and SIRT1 activity by directly targeting NAMPT. Aging Cell. 2013;12(6):1062–1072. doi: 10.1111/acel.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Min H.-K., Kapoor A., Fuchs M., Mirshahi F., Zhou H., Maher J., et al. Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metabolism. 2012;15(5):665–674. doi: 10.1016/j.cmet.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding J., Li M., Wan X., Jin X., Chen S., Yu C., et al. Effect of miR-34a in regulating steatosis by targeting PPARalpha expression in nonalcoholic fatty liver disease. Scientific Reports. 2015;5 doi: 10.1038/srep13729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y., Zalzala M., Xu J., Li Y., Yin L., Zhang Y. A metabolic stress-inducible miR-34a-HNF4alpha pathway regulates lipid and lipoprotein metabolism. Nature Communications. 2015;6 doi: 10.1038/ncomms8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim Y.S., Nam H.J., Han C.Y., Joo M.S., Jang K., Jun D.W., et al. LXRα activation inhibits autophagy and lipophagy in hepatocytes by dysregulating ATG4B and Rab-8B, reducing mitochondrial fuel oxidation. Hepatology: Hep. 2020 doi: 10.1002/hep.31423. [DOI] [PubMed] [Google Scholar]
- 47.Wang J.M., Qiu Y., Yang Z., Kim H., Qian Q., Sun Q., et al. IRE1alpha prevents hepatic steatosis by processing and promoting the degradation of select microRNAs. Science Signaling. 2018;11(530) doi: 10.1126/scisignal.aao4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferreira D.M.S., Afonso M.B., Rodrigues P.M., Simao A.L., Pereira D.M., Borralho P.M., et al. c-Jun N-terminal kinase 1/c-Jun activation of the p53/microRNA 34a/sirtuin 1 pathway contributes to apoptosis induced by deoxycholic acid in rat liver. Molecular and Cellular Biology. 2014;34(6):1100–1120. doi: 10.1128/mcb.00420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian X.-F., Ji F.-J., Zang H.-L., Cao H. Activation of the miR-34a/SIRT1/p53 signaling pathway contributes to the progress of liver fibrosis via inducing apoptosis in hepatocytes but not in HSCs. PLoS One. 2016;11(7) doi: 10.1371/journal.pone.0158657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song L., Chen T., Zhao X., Xu Q., Jiao R., Li J., et al. Pterostilbene prevents hepatocyte epithelial-mesenchymal transition in fructose-induced liver fibrosis through suppressing miR-34a/Sirt1/p53 and TGF-β1/Smads signalling. British Journal of Pharmacology. 2019;176(11):1619–1634. doi: 10.1111/bph.14573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Munker S., Wu Y.-L., Ding H.-G., Liebe R., Weng H.-L. Can a fibrotic liver afford epithelial-mesenchymal transition? World Journal of Gastroenterology. 2017;23(26):4661–4668. doi: 10.3748/wjg.v23.i26.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Q., Yu J., Wang L., Tang Y., Zhou Q., Ji S., et al. Inhibition of PU.1 ameliorates metabolic dysfunction and non-alcoholic steatohepatitis. Journal of Hepatology. 2020;73(2):361–370. doi: 10.1016/j.jhep.2020.02.025. [DOI] [PubMed] [Google Scholar]
- 53.Liu Q., Yongming Z., Songzhu Y., Wu Y., Jiantao W., Weiwei Y., et al. PU.1-deficient mice are resistant to thioacetamide-induced hepatic fibrosis: PU.1 finely regulates Sirt1 expression via transcriptional promotion of miR-34a and miR-29c in hepatic stellate cells. Bioscience Reports. 2017;37(6) doi: 10.1042/BSR20170926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li M., Hong W., Hao C., Li L., Wu D., Shen A., et al. SIRT1 antagonizes liver fibrosis by blocking hepatic stellate cell activation in mice. Federation of American Societies for Experimental Biology Journal. 2018;32(1):500–511. doi: 10.1096/fj.201700612R. [DOI] [PubMed] [Google Scholar]
- 55.Li X., Chen Y., Wu S., He J., Lou L., Ye W., et al. MicroRNA-34a and microRNA-34c promote the activation of human hepatic stellate cells by targeting peroxisome proliferator-activated receptor γ. Molecular Medicine Reports. 2015;11(2):1017–1024. doi: 10.3892/mmr.2014.2846. [DOI] [PubMed] [Google Scholar]
- 56.Hou J., Lin L., Zhou W., Wang Z., Ding G., Dong Q., et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19(2):232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 57.Hu J., Xu Y., Hao J., Wang S., Li C., Meng S. MiR-122 in hepatic function and liver diseases. Protein Cell. 2012;3(5):364–371. doi: 10.1007/s13238-012-2036-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pirola C.J., Gianotti T.F., Castaño G.O., Mallardi P., Martino J.S., Ledesma M.M.G.L., et al. Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut. 2015;64(5):800–812. doi: 10.1136/gutjnl-2014-306996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miyaaki H., Ichikawa T., Kamo Y., Taura N., Honda T., Shibata H., et al. Significance of serum and hepatic microRNA-122 levels in patients with non-alcoholic fatty liver disease. Liver International. 2014;34(7):e302–e307. doi: 10.1111/liv.12429. [DOI] [PubMed] [Google Scholar]
- 60.Hsu S.H., Wang B., Kota J., Yu J., Costinean S., Kutay H., et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. Journal of Clinical Investigation. 2012;122(8):2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsai W.C., Hsu S. Da., Hsu C.S., Lai T.C., Chen S.J., Shen R., et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. Journal of Clinical Investigation. 2012;122(8):2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Esau C., Davis S., Murray S.F., Yu X.X., Pandey S.K., Pear M., et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metabolism. 2006;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 63.Krützfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., et al. Silencing of microRNAs in vivo with “antagomirs. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 64.Chai C., Rivkin M., Berkovits L., Simerzin A., Zorde-Khvalevsky E., Rosenberg N., et al. Metabolic circuit involving free fatty acids, microRNA 122, and triglyceride synthesis in liver and muscle tissues. Gastroenterology. 2017;153(5):1404–1415. doi: 10.1053/j.gastro.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 65.Chai C., Cox B., Yaish D., Gross D., Rosenberg N., Amblard F., et al. Agonist of RORA attenuates nonalcoholic fatty liver progression in mice via up-regulation of microRNA 122. Gastroenterology. 2020;159(3):999–1014e9. doi: 10.1053/j.gastro.2020.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Long J.K., Dai W., Zheng Y.W., Zhao S.P. MiR-122 promotes hepatic lipogenesis via inhibiting the LKB1/AMPK pathway by targeting Sirt1 in non-alcoholic fatty liver disease. Molecular Medicine. 2019;25(1):26. doi: 10.1186/s10020-019-0085-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu G., Rui C., Chen J., Sho E., Zhan S., Yuan X., et al. MicroRNA-122 inhibits lipid droplet formation and hepatic triglyceride accumulation via Yin Yang 1. Cellular Physiology and Biochemistry. 2017;44(4):1651–1664. doi: 10.1159/000485765. [DOI] [PubMed] [Google Scholar]
- 68.Hsu K.H., Wei C.W., Su Y.R., Chou T., Lin Y.L., Yang F.C., et al. Upregulation of RelB in the miR-122 knockout mice contributes to increased levels of proinflammatory chemokines/cytokines in the liver and macrophages. Immunology Letters. 2020;226:22–30. doi: 10.1016/j.imlet.2020.06.015. [DOI] [PubMed] [Google Scholar]
- 69.Sans A., Bonnafous S., Rousseau D., Patouraux S., Canivet C.M., Leclere P.S., et al. The differential expression of Cide family members is sssociated with Nafld progression from steatosis to steatohepatitis. Scientific Reports. 2019;9(1):7501. doi: 10.1038/s41598-019-43928-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Y.M., Seo S.Y., Kim T.H., Kim S.G. Decrease of microRNA-122 causes hepatic insulin resistance by inducing protein tyrosine phosphatase 1B, which is reversed by licorice flavonoid. Hepatology. 2012;56(6):2209–2220. doi: 10.1002/hep.25912. [DOI] [PubMed] [Google Scholar]
- 71.Gao L.-L., Li M., Wang Q., Liu S.-A., Zhang J.-Q., Cheng J. HCBP6 modulates triglyceride homeostasis in hepatocytes via the SREBP1c/FASN Pathway. Journal of Cellular Biochemistry. 2015;116(10):2375–2384. doi: 10.1002/jcb.25188. [DOI] [PubMed] [Google Scholar]
- 72.Elhanati S., Ben-Hamo R., Kanfi Y., Varvak A., Glazz R., Lerrer B., et al. Reciprocal regulation between SIRT6 and miR-122 controls liver metabolism and predicts hepatocarcinoma prognosis. Cell Reports. 2016;14(2):234–242. doi: 10.1016/j.celrep.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 73.Norman K.L., Chen T.C., Zeiner G., Sarnow P. Precursor microRNA-122 inhibits synthesis of Insig1 isoform mRNA by modulating polyadenylation site usage. RNA. 2017;23(12):1886–1893. doi: 10.1261/rna.063099.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li J., Ghazwani M., Zhang Y., Lu J., Li J., Fan J., et al. MiR-122 regulates collagen production via targeting hepatic stellate cells and suppressing P4HA1 expression. Journal of Hepatology. 2013;58(3):522–528. doi: 10.1016/j.jhep.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu F., Jiang Z., Chen B., Dong P., Zheng J. NEAT1 accelerates the progression of liver fibrosis via regulation of microRNA-122 and Kruppel-like factor 6. Journal of Molecular Medicine. 2017;95(11):1191–1202. doi: 10.1007/s00109-017-1586-5. [DOI] [PubMed] [Google Scholar]
- 76.Zeng C., Wang Y.L., Xie C., Sang Y., Li T.J., Zhang M., et al. Identification of a novel TGF-β-miR-122-fibronectin 1/serum response factor signaling cascade and its implication in hepatic fibrogenesis. Oncotarget. 2015;6(14):12224–12233. doi: 10.18632/oncotarget.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Du K., Chitneni S.K., Suzuki A., Wang Y., Henao R., Hyun J., et al. Increased glutaminolysis marks active scarring in nonalcoholic steatohepatitis progression. CMGH. 2020;10(1):1–21. doi: 10.1016/j.jcmgh.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sengupta D., Cassel T., Teng K. yu., Aljuhani M., Chowdhary V.K., Hu P., et al. Regulation of hepatic glutamine metabolism by miR-122. Molecular Metabolism. 2020;34:174–186. doi: 10.1016/j.molmet.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luedde |., Schwabe T.F. NF-κB in the liver – linking injury, fibrosis and hepatocellular carcinoma. Nature Reviews Gastroenterology & Hepatology. 2011;8(2):108–118. doi: 10.1038/nrgastro.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ioannou G.N. The role of cholesterol in the pathogenesis of NASH. Trends in Endocrinology and Metabolism. 2016;27(2):84–95. doi: 10.1016/j.tem.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 81.Zhao Z., Zhong L., Li P., He K., Qiu C., Zhao L., et al. Cholesterol impairs hepatocyte lysosomal function causing M1 polarization of macrophages via exosomal miR-122-5p. Experimental Cell Research. 2020;387(1) doi: 10.1016/j.yexcr.2019.111738. [DOI] [PubMed] [Google Scholar]
- 82.Belloni L., Di Cocco S., Guerrieri F., Nunn A.D.G., Piconese S., Salerno D., et al. Targeting a phospho-STAT3-miRNAs pathway improves vesicular hepatic steatosis in an in vitro and in vivo model. Scientific Reports. 2018;8(1) doi: 10.1038/s41598-018-31835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Calo N., Ramadori P., Sobolewski C., Romero Y., Maeder C., Fournier M., et al. Stress-activated miR-21/miR-21∗ in hepatocytes promotes lipid and glucose metabolic disorders associated with high-fat diet consumption. Gut. 2016;65(11):1871–1881. doi: 10.1136/gutjnl-2015-310822. [DOI] [PubMed] [Google Scholar]
- 84.Loyer X., Paradis V., Hénique C., Vion A.C., Colnot N., Guerin C.L., et al. Liver microRNA-21 is overexpressed in non-alcoholic steatohepatitis and contributes to the disease in experimental models by inhibiting PPARα expression. Gut. 2016;65:1882–1894. doi: 10.1136/gutjnl-2014-308883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rodrigues P.M., Afonso M.B., Simão A.L., Carvalho C.C., Trindade A., Duarte A., et al. miR-21 ablation and obeticholic acid ameliorate nonalcoholic steatohepatitis in mice. Cell Death & Disease. 2017;8(4) doi: 10.1038/cddis.2017.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kida K., Nakajima M., Mohri T., Oda Y., Takagi S., Fukami T., et al. PPARalpha is regulated by miR-21 and miR-27b in human liver. Pharmaceutical Research. 2011;28(10):2467–2476. doi: 10.1007/s11095-011-0473-y. [DOI] [PubMed] [Google Scholar]
- 87.Dattaroy D., Pourhoseini S., Das S., Alhasson F., Seth R.K., Nagarkatti M., et al. Micro-RNA 21 inhibition of SMAD7 enhances fibrogenesis via leptin-mediated NADPH oxidase in experimental and human nonalcoholic steatohepatitis. American Journal of Physiology - Gastrointestinal and Liver Physiology. 2015;308(4):298–312. doi: 10.1152/ajpgi.00346.2014.-Hepatic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Z., Zha Y., Hu W., Huang Z., Gao Z., Zang Y., et al. The autoregulatory feedback loop of MicroRNA-21/programmed cell death protein 4/Activation protein-1 (MiR-21/PDCD4/AP-1) as a driving force for hepatic fibrosis development. Journal of Biological Chemistry. 2013;288(52):37082–37093. doi: 10.1074/jbc.M113.517953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Caviglia J.M., Yan J., Jang M.K., Gwak G.Y., Affo S., Yu L., et al. MicroRNA-21 and Dicer are dispensable for hepatic stellate cell activation and the development of liver fibrosis. Hepatology. 2018;67(6):2414–2429. doi: 10.1002/hep.29627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao J., Tang N., Wu K., Dai W., Ye C., Shi J., et al. MiR-21 simultaneously regulates ERK1 signaling in HSC activation and hepatocyte EMT in hepatic fibrosis. PLoS One. 2014;9(10) doi: 10.1371/journal.pone.0108005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wei J., Feng L., Li Z., Xu G., Fan X. MicroRNA-21 activates hepatic stellate cells via PTEN/Akt signaling. Biomedicine & Pharmacotherapy. 2013;67(5):387–392. doi: 10.1016/j.biopha.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 92.Jarman E.J., Boulter L. Targeting the Wnt signaling pathway: the challenge of reducing scarring without affecting repair. Expert Opinion on Investigational Drugs. 2020;29(2):179–190. doi: 10.1080/13543784.2020.1718105. [DOI] [PubMed] [Google Scholar]
- 93.Wang X.M., Wang X.Y., Huang Y.M., Chen X., Lü M.H., Shi L., et al. Role and mechanisms of action of microRNA-21 as regards the regulation of the WNT/β-catenin signaling pathway in the pathogenesis of non-alcoholic fatty liver disease. International Journal of Molecular Medicine. 2019;44(6):2201–2212. doi: 10.3892/ijmm.2019.4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xu H., Tian Y., Tang D., Zou S., Liu G., Song J., et al. An endoplasmic reticulum stress-microRNA-26a feedback circuit in NAFLD. Hepatology: Hep. 2020;31428 doi: 10.1002/hep.31428. [DOI] [PubMed] [Google Scholar]
- 95.Jeon T. Il., Park J.W., Ahn J., Jung C.H., Ha T.Y. Fisetin protects against hepatosteatosis in mice by inhibiting miR-378. Molecular Nutrition & Food Research. 2013;57(11):1931–1937. doi: 10.1002/mnfr.201300071. [DOI] [PubMed] [Google Scholar]
- 96.Zhang T., Duan J., Zhang L., Li Z., Steer C.J., Yan G., et al. LXRalpha promotes hepatosteatosis in part through activation of microRNA-378 transcription and inhibition of Ppargc1beta expression. Hepatology. 2019;69(4):1488–1503. doi: 10.1002/hep.30301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang T., Hu J., Wang X., Zhao X., Li Z., Niu J., et al. MicroRNA-378 promotes hepatic inflammation and fibrosis via modulation of the NF-κB-TNFα pathway. Journal of Hepatology. 2019;70(1):87–96. doi: 10.1016/j.jhep.2018.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang T., Zhao X., Steer C.J., Yan G., Song G. A negative feedback loop between microRNA-378 and Nrf1 promotes the development of hepatosteatosis in mice treated with a high fat diet. Metabolism, Clinical and Experimental. 2018;85:183–191. doi: 10.1016/j.metabol.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Azzimato V., Jager J., Chen P., Morgantini C., Levi L., Barreby E., et al. Liver macrophages inhibit the endogenous antioxidant response in obesity-associated insulin resistance. Science Translational Medicine. 2020;12(532) doi: 10.1126/SCITRANSLMED.AAW9709. [DOI] [PubMed] [Google Scholar]
- 100.Azzimato V., Chen P., Barreby E., Morgantini C., Levi L., Vankova A., et al. Hepatic miR-144 drives fumarase activity preventing NRF2 activation during obesity. Gastroenterology. 2021;161(6):1982–1997. doi: 10.1053/J.GASTRO.2021.08.030. e11. [DOI] [PubMed] [Google Scholar]
- 101.Povero D., Panera N., Eguchi A., Johnson C.D., Papouchado B.G., de Araujo Horcel L., et al. Lipid-induced hepatocyte-derived extracellular vesicles regulate hepatic stellate cell via microRNAs targeting PPAR-gamma. CMGH. 2015;1(6):646–663. doi: 10.1016/j.jcmgh.2015.07.007. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee Y.S., Kim S.Y., Ko E., Lee J.H., Yi H.S., Yoo Y.J., et al. Exosomes derived from palmitic acid-treated hepatocytes induce fibrotic activation of hepatic stellate cells. Scientific Reports. 2017;7(1):3710. doi: 10.1038/s41598-017-03389-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu X., Pan Q., Cao H., Xin F., Zhao Z., Yang R., et al. Lipotoxic hepatocyte-derived exosomal microRNA 192-5p activates macrophages through Rictor/Akt/Forkhead Box Transcription Factor O1 signaling in nonalcoholic fatty liver disease. Hepatology. 2020;72(2):454–469. doi: 10.1002/hep.31050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.He Y., Hwang S., Cai Y., Kim S.J., Xu M., Yang D., et al. MicroRNA-223 ameliorates nonalcoholic steatohepatitis and cancer by targeting multiple inflammatory and oncogenic genes in hepatocytes. Hepatology. 2019;70(4):1150–1167. doi: 10.1002/HEP.30645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Calvente C.J., Tameda M., Johnson C.D., Del Pilar H., Lin Y.C., Adronikou N., et al. Neutrophils contribute to spontaneous resolution of liver inflammation and fibrosis via microRNA-223. The Journal of Clinical Investigation. 2019;129(10):4091–4109. doi: 10.1172/JCI122258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.He Y., Rodrigues R.M., Wang X., Seo W., Ma J., Hwang S., et al. Neutrophil-to-hepatocyte communication via LDLR-dependent miR-223-enriched extracellular vesicle transfer ameliorates nonalcoholic steatohepatitis. The Journal of Clinical Investigation. 2021;131(3) doi: 10.1172/JCI141513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Priest C., Tontonoz P. Inter-organ cross-talk in metabolic syndrome. Nature Metabolism. 2019;1(12):1177–1188. doi: 10.1038/s42255-019-0145-5. [DOI] [PubMed] [Google Scholar]
- 108.Azzu V., Vacca M., Virtue S., Allison M., Vidal-Puig A. Adipose tissue-liver cross talk in the control of whole-body metabolism: implications in nonalcoholic fatty liver disease. Gastroenterology. 2020;158(7):1899–1912. doi: 10.1053/j.gastro.2019.12.054. [DOI] [PubMed] [Google Scholar]
- 109.Castaño C., Kalko S., Novials A., Párrizas M. Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proceedings of the National Academy of Sciences of the United States of America. 2018;115(48):12158–12163. doi: 10.1073/pnas.1808855115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhao Y., Zhao M.-F., Jiang S., Wu J., Liu J., Yuan X.-W., et al. Liver governs adipose remodelling via extracellular vesicles in response to lipid overload. Nature Communications. 2020;11(1) doi: 10.1038/S41467-020-14450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Thomou T., Mori M.A., Dreyfuss J.M., Konishi M., Sakaguchi M., Wolfrum C., et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542(7642):450–455. doi: 10.1038/nature21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ying W., Riopel M., Bandyopadhyay G., Dong Y., Birmingham A., Seo J.B., et al. Adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell. 2017;171(2):372–384. doi: 10.1016/j.cell.2017.08.035. e12. [DOI] [PubMed] [Google Scholar]
- 113.Baranova A., Maltseva D., Tonevitsky A. Adipose may actively delay progression of NAFLD by releasing tumor-suppressing, anti-fibrotic miR-122 into circulation. Obesity Reviews. 2019;20(1):108–118. doi: 10.1111/obr.12765. [DOI] [PubMed] [Google Scholar]
- 114.Wang Y., Jin P., Liu J., Xie X. Exosomal microRNA-122 mediates obesity-related cardiomyopathy through suppressing mitochondrial ADP-ribosylation factor-like 2. Clinical Science. 2019;133(17):1871–1881. doi: 10.1042/CS20190558. [DOI] [PubMed] [Google Scholar]
- 115.Xin S., Zhan Q., Chen X., Xu J., Yu Y. Efficacy of serum miRNA test as a non-invasive method to diagnose nonalcoholic steatohepatitis: a systematic review and meta-analysis. BMC Gastroenterology. 2020;20(1) doi: 10.1186/S12876-020-01334-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Akuta N., Kawamura Y., Arase Y., Saitoh S., Fujiyama S., Sezaki H., et al. Circulating MicroRNA-122 and fibrosis stage predict mortality of Japanese patients with histopathologically confirmed NAFLD. Hepatology Communications. 2020;4(1):66. doi: 10.1002/HEP4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ezaz G., Trivedi H.D., Connelly M.A., Filozof C., Howard K., L.Parrish M., et al. Differential associations of circulating microRNAs with pathogenic factors in NAFLD. Hepatology Communications. 2020;4(5):670–680. doi: 10.1002/hep4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Albhaisi S., Sanyal A.J. Applying non-invasive fibrosis measurements in NAFLD/NASH: progress to date. Pharmaceutical Medicine. 2019;33(6):451–463. doi: 10.1007/s40290-019-00305-z. [DOI] [PubMed] [Google Scholar]
- 119.Newman L.A., Sorich M.J., Rowland A. Role of extracellular vesicles in the pathophysiology, diagnosis and tracking of non-alcoholic fatty liver disease. Journal of Clinical Medicine. 2020;9(7):2032. doi: 10.3390/jcm9072032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rodrigues A.D., van Dyk M., Sorich M.J., Fahmy A., Useckaite Z., Newman L.A., et al. Exploring the use of serum-derived small extracellular vesicles as liquid biopsy to study the induction of hepatic cytochromes P450 and organic anion transporting polypeptides. Clinical Pharmacology and Therapeutics (St. Louis) 2021;110(1):248–258. doi: 10.1002/CPT.2244. [DOI] [PubMed] [Google Scholar]
- 121.Newman L.A., Useckaite Z., Johnson J., Sorich M.J., Hopkins A.M., Rowland A. Selective isolation of liver-derived extracellular vesicles redefines performance of miRNA biomarkers for non-alcoholic fatty liver disease. Biomedicines. 2022;10(1) doi: 10.3390/BIOMEDICINES10010195/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Harrison S.A., Ratziu V., Boursier J., Francque S., Bedossa P., Majd Z., et al. A blood-based biomarker panel (NIS4) for non-invasive diagnosis of non-alcoholic steatohepatitis and liver fibrosis: a prospective derivation and global validation study. The Lancet Gastroenterology and Hepatology. 2020;1253(11):1–16. doi: 10.1016/S2468-1253(20)30252-1. [DOI] [PubMed] [Google Scholar]
- 123.Bajan S., Hutvagner G. RNA-based therapeutics: from antisense oligonucleotides to miRNAs. Cells. 2020;9(1):137. doi: 10.3390/cells9010137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Roberts T.C., Langer R., Wood M.J.A. Advances in oligonucleotide drug delivery. Nature Reviews Drug Discovery. 2020;19(10):673–694. doi: 10.1038/s41573-020-0075-7. 19:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Debacker A.J., Voutila J., Catley M., Blakey D., Habib N. Delivery of oligonucleotides to the liver with GalNAc: from research to registered therapeutic drug. Molecular Therapy. 2020;28(8):1759–1771. doi: 10.1016/J.YMTHE.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kumar V., Xin X., Ma J., Tan C., Osna N., Mahato R.I. Therapeutic targets, novel drugs, and delivery systems for diabetes associated NAFLD and liver fibrosis. Advanced Drug Delivery Reviews. 2021;176 doi: 10.1016/J.ADDR.2021.113888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Regulus Therapeutics Inc RG-125(AZD4076), a microRNA therapeutic targeting microRNA-103/107 being developed for the treatment of NASH in patients with type 2 diabetes/pre-diabetes. 2015. http://ir.regulusrx.com/news-releases/news-release-details/rg-125azd4076-microrna-therapeutic-targeting-microrna-103107 enters phase I clinical development [press release]
- 128.Trajkovski M., Hausser J., Soutschek J., Bhat B., Akin A., Zavolan M., et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474(7353):649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 129.Regulus Therapeutics Inc Regulus announces pipeline updates and advancements. 2017. http://ir.regulusrx.com/news-releases/news-release-details/regulus-announces-pipeline-updates-and-advancements [press release]
- 130.Madadi S., Schwarzenbach H., Lorenzen J., Soleimani M. MicroRNA expression studies: challenge of selecting reliable reference controls for data normalization. Cellular and Molecular Life Sciences. 2019;76(18):3497–3514. doi: 10.1007/s00018-019-03136-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rajesh Y., Sarkar D. Molecular mechanisms regulating obesity-associated hepatocellular carcinoma. Cancers. 2020;12(5):1290. doi: 10.3390/cancers12051290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang C., Wang P., Li Y., Huang C., Ni W., Chen Y., et al. Role of microRNAs in the development of hepatocellular carcinoma in nonalcoholic fatty liver disease. The Anatomical Record. 2019;302(2):193–200. doi: 10.1002/ar.23954. [DOI] [PubMed] [Google Scholar]
- 133.Friedman R.C., Farh K.K.-H., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Research. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hon L.S., Zhang Z. The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biology. 2007;8(8):R166. doi: 10.1186/gb-2007-8-8-r166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xu J., Shao T., Ding N., Li Y., Li X. miRNA-miRNA crosstalk: from genomics to phenomics. Briefings in Bioinformatics. 2017;18(6):1002–1011. doi: 10.1093/bib/bbw073. [DOI] [PubMed] [Google Scholar]
- 136.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.